Abstract
Background
Craniofacial anomalies involve defective pharyngeal arch development and neural crest function. Copy number variation at 1p35, containing histone deacetylase 1 (Hdac1), or 6q21-22, containing Hdac2, are implicated in patients with craniofacial defects, suggesting an important role in guiding neural crest development. However, the roles of Hdac1 and Hdac2 within neural crest cells remain unknown.
Results
The neural crest and its derivatives express both Hdac1 and Hdac2 during early murine development. Ablation of Hdac1 and Hdac2 within murine neural crest progenitor cells cause severe hemorrhage, atrophic pharyngeal arches, defective head morphogenesis, and complete embryonic lethality. Embryos lacking Hdac1 and Hdac2 in the neural crest exhibit decreased proliferation and increased apoptosis in both the neural tube and the first pharyngeal arch. Mechanistically, loss of Hdac1 and Hdac2 upregulates cyclin-dependent kinase inhibitors Cdkn1a, Cdkn1b, Cdkn1c, Cdkn2b, Cdkn2c, and Tp53 within the first pharyngeal arch.
Conclusions
Our results show that Hdac1 and Hdac2 function redundantly within the neural crest to regulate proliferation and the development of the pharyngeal arches via repression of cyclin-dependent kinase inhibitors.
Keywords: Histone Deacetylase, Neural Crest, Pharyngeal Arch, Craniofacial morphogenesis
Introduction
The neural crest (NC) is a migratory cell population unique to vertebrates, which delaminates from the dorsal neural tube early in development (Huang and Saint-Jeannet, 2004; Helms et al., 2005; Cordero et al., 2011; Munoz and Trainor, 2015). The NC migrates along the length of the developing embryo, colonizes a wide array of critical structures, and contributes to the development of almost every tissue and organ (Jiang et al., 2000). NC cells (NCCs) give rise to epithelial structures and can undergo epithelial-to-mesenchymal transition in order to form mesenchymal structures (Jiang et al., 2000). In mammals, the NC is divided into several regional subpopulations including the cranial-NC, cardiac-NC, and trunk-NC (Crane and Trainor, 2006; Bhatt et al., 2013). Trunk NCCs form the majority of the peripheral nervous system, skin melanocytes, and endocrine cells (Le Lievre and Le Douarin, 1975; Rothman et al., 1993). Cranial NCCs migrates from the fore-, mid-, and hindbrain to colonize anterior portions of developing embryos including the frontal prominence and cranial pharyngeal arches (PhAs) early in development (Saint-Jeannet and Moody, 2014; Twigg and Wilkie, 2015). These cells will go on to form the craniofacial skeleton, smooth muscle of the head and neck, and certain specialized organs/structures (Twigg and Wilkie, 2015). Similarly, the cardiac NC travels from the dorsal portion of the embryo to populate the caudal PhAs and pharyngeal mesoderm (Kirby and Hutson, 2010). Under guidance from cardiac progenitors, cardiac NCCs populate the developing outflow tract (aorta and pulmonary artery) in mammalian embryos enabling rotation and septation (Kirby et al., 1983; Vincent and Buckingham, 2010).
The PhAs are bilaterally paired structures numbered from 1 to 6, which contain a nerve, artery and mesenchymal component (Hall, 2015). The first PhA, often termed the mandibular or maxillary arch, gives rise to the numerous structures including maxillary artery, trigeminal nerve, muscles of mastication, and a mix of skeletal components including the mandible and maxilla (Benson et al., 1992). The second, third, fourth, and sixth arch give rise to other nerves, muscles, and bones in the craniofacial region along with critical components of the cardiac outflow tract and aortic arch (Kirby and Waldo, 1995).
Mirroring the complex roles and derivatives of the NC, numerous congenital anomalies and syndromes have been mapped to defects in NCC function (Firulli and Conway, 2004; Verzi et al., 2007; Conway and Kaartinen, 2011; Bonilla-Claudio et al., 2012). Cranial NC abnormalities have been identified in patients with thymic aplasia (Ohnemus et al., 2002), palate defects (van Limborgh et al., 1983), and craniofacial malformations (Conway and Kaartinen, 2011). Trunk NC defects have been implicated in pigment abnormalities and abnormal enteric innervation (Southard-Smith et al., 1998). Additionally, cardiac NC defects are found in patients with outflow tract septation defects (Chin et al., 2012), aortic arch anomalies (Theveniau-Ruissy et al., 2008), and cardiac conduction defects (Nakamura et al., 2006). These defects often present together and are grouped clinically into syndromes such as DiGeorge Syndrome (22q11.2) (McDonald-McGinn et al., 2015), Treacher-Collins Syndrome (Trainor, 2010), or CHARGE Syndrome (Siebert et al., 1985). Despite evolving understanding of the NC in mammalian development, epigenetic factors regulating its behavior remain largely unknown.
Studies of patients with craniofacial abnormalities of unknown origin identified ranges of chromosomal regions and histone-modifying genes that may be involved (Wilkie and Morriss-Kay, 2001). For instance, copy number variations at 1p35, a region including the class I histone deacetylase (HDAC) HDAC1, and 6q21-22, containing HDAC2 were found in patients with craniofacial abnormalities, suggesting that dose-dependent disruptions in epigenome homeostasis may influence NC behavior (Hudson et al., 2014; de Souza et al., 2015). HDACs, classified into five subfamilies based on their phylogenetic analysis and sequence homology, are a major family of histone modifying enzymes with roles in numerous biologic processes, including craniofacial development. A long-held assumption about HDACs is that they are ubiquitously expressed. However, recent studies demonstrate that HDACs are expressed in tissue and temporal specific manner during development. For instance, Hdac7 is endothelial specific and loss results in vascular defects (Chang et al., 2006). Hdac8, specifically expressed in fore and midbrain regions at E10 and E11.5, is critical for skull development (Haberland et al., 2009a; Murko et al., 2010). Similarly, murine embryos show restricted, distinct, and non-overlapping Hdac1 and Hdac2 mRNA expression areas at E11.5 and E13 (Murko et al., 2010). However, endogenous Hdac1 expression pattern during early murine development remains unknown. Hdac1 and Hdac2 are closely related, ~80% homologous, enzymatically active class I histone deacetylases (Gregoretti et al., 2004; Brunmeir et al., 2009). Global ablation of Hdac1 is embryonically lethal early in development due to proliferation arrest caused by upregulation of P21 (Cdkn1a) (Lagger et al., 2002). Hdac2-null mice show cardiac defects and variable levels of lethality at birth depending upon genetic background (Montgomery et al., 2007; Trivedi et al., 2007; Zimmermann et al., 2007; Guan et al., 2009; Hagelkruys et al., 2014). Hdac1 and Hdac2 function together in repressive complexes including Sin3a, NuRD, and Co-REST (Hassig et al., 1997; Laherty et al., 1997; Nagy et al., 1997; Zhang et al., 1999). Consequently, Hdac1 and Hdac2 are classically considered to have overlapping, redundant functions. Indeed, Hdac1 and Hdac2 redundantly regulate various cellular and developmental processes, including cardiac morphogenesis, adipogenesis, brain development, intestinal homeostasis, proliferation, and stem-cell self-renewal (Montgomery et al., 2007; Montgomery et al., 2009; Haberland et al., 2010; Jurkin et al., 2011; Jamaladdin et al., 2014; Zimberlin et al., 2015). In some cases, a single allele of Hdac1 or Hdac2 is enough to rescue the double knockout phenotype (Dovey et al., 2013; Winter et al., 2013; Hagelkruys et al., 2014). While the developmental roles of Hdac1 and Hdac2 are emerging, their function in NC progenitor cells remains unknown.
Using a novel conditional Hdac1Flox allele, a previously characterized conditional Hdac2Flox allele (Anokye-Danso et al., 2011), and Wnt1-Cre2 (Lewis et al., 2013), we genetically ablated Hdac1 and Hdac2 in NC progenitor cells. Loss of both Hdac1 and Hdac2 (Wnt1;1KO2KO) resulted in embryonic lethality around E11.5 secondary to vascular defects and atrophic PhAs. Furthermore, Wnt1;1KO2KO embryos show reduced proliferation of NC progenitor cells within the neural tube and first PhA. Mechanistically, ablation of Hdac1 and Hdac2 causes upregulation of cyclin-dependent kinase inhibitors Cdkn2b, Cdkn2c, Cdkn1b, Cdkn1c, Cdkn1a, and Tp53 within first PhA at E9.5. Interestingly, we observed that a single copy of Hdac1 in an Hdac2 knockout background (Wnt1;1Het2KO) or a single copy of Hdac2 in an Hdac1 knockout background (Wnt1;1KO2Het) was sufficient to rescue the craniofacial phenotype, first PhA phenotype, and embryonic lethality. These results reveal redundant and essential roles for Hdac1 and Hdac2 in the developing neural crest and its derivatives.
Results
The neural crest and its derivatives express both Hdac1 and Hdac2 during early murine development
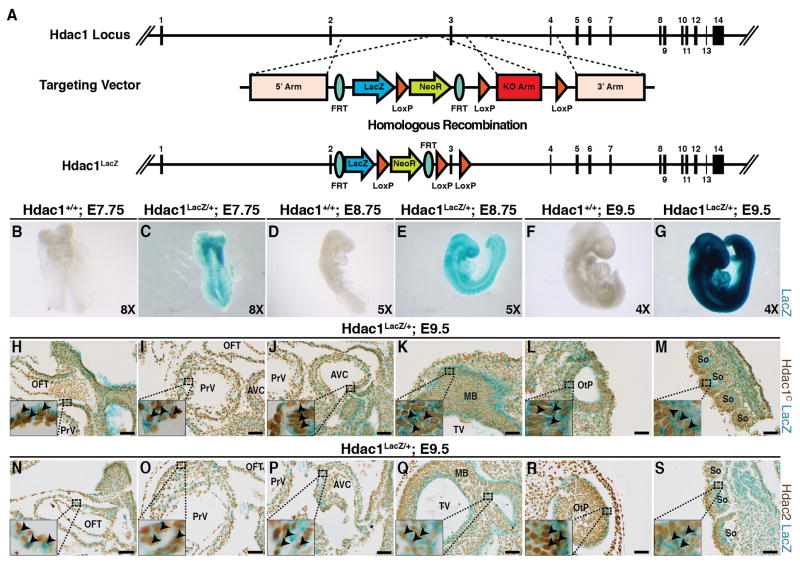
Hdac1 alleles were generated using a Knockout-first strategy (Skarnes et al., 2011) (Figs 1–2). This strategy generates an Hdac1 knock-in reporter allele (Tm1a, Hdac1LacZ, Fig 1A), Hdac1 reporter knockout allele (Tm1b, not shown), and Hdac1 conditional-ready allele (Tm1c, Hdac1Flox, Fig 2A) following exposure to site-specific recombinases Cre and Flp in different orders. In this study, the Tm1a knockout-first allele is utilized as a knock-in reporter allele, but in many cases exposure to Cre-recombinase (without exposure to Flp-recombinase) is necessary to generate a robust reporter allele (Skarnes et al., 2011). The flexibility of this allele is an advantage when compared to other published Hdac1 alleles; the reporter function enables tracking the spatiotemporal dynamics of in vivo Hdac1 during development (Fig 1A), while the conditional deletion of exon 3 completely abolishes Hdac1 expression in a tissue-specific manner (Fig 2).
Figure 1.
(A) Hdac1LacZ knock-in allele targeting map and generation scheme. (B–G) Whole-mount X-Gal staining for β-galactosidase (blue, LacZ) of Hdac1LacZ/+ and Hdac1+/+ at various developmental stages. B, ventral view of Hdac1+/+ at E7.75. C, ventral view of Hdac1LacZ/+ at E7.75. D, right lateral view of Hdac1+/+ embryo at E8.75. E, right lateral view of Hdac1LacZ/+ embryo at E8.75. F, right lateral view of Hdac1+/+ embryo at E9.5. G, right lateral view of Hdac1LacZ/+ embryo at E9.5.. (H–M) C-terminal Hdac1 (Hdac1) immunostaining (brown) on whole-mount X-Gal stained (LacZ, blue) E9.5 Hdac1Lacz/+ sagittal sections including the outflow tract (OFT, H), primitive ventricle (PrV, I), atrioventricular canal (AVC, J), midbrain and telencephalic vesicle (MB & TV, K), otic pit (OtP, L), and caudal somites (So, M) (N–S) Hdac2 immunostaining (brown) on whole-mount X-Gal stained (LacZ, blue) E9.5 Hdac1LacZ/+ sagittal sections including the OFT (N), PrV (O), AVC (P), MB & TV (Q), OtP (R), and So (S). Scale bars 50μm.
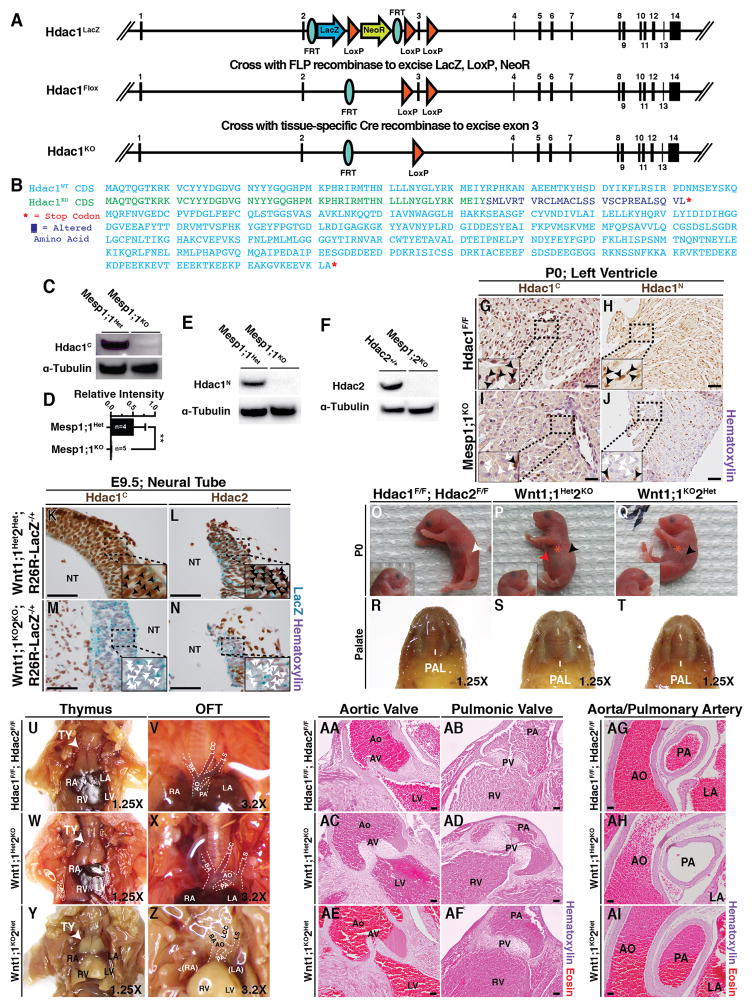
Figure 2.
(A) Hdac1Flox conditional allele targeting vector map and generation scheme. (B) Predicted aligned amino acid sequence for Hdac1WT and recombined Hdac1Flox (Hdac1KO) alleles consensus-determining sequences (CDS). (Blue, WT; Green, unchanged; Purple, frameshift mutation; Red, stop codon) (C–F) Western blot analysis of control (Mesp1;1Het, Hdac2+/+) and knockout (Mesp1;1KO, Mesp1;2KO) P0 heart lysate for Hdac1C (C), Hdac1N (E), and Hdac2 (F). Hdac1C was quantified and normalized to total input α-Tubulin and is presented as mean (SD) (**, p=0.007) (G–J) Hdac1C and Hdac1N immunostaining of Mesp1;1KO and Mesp1;1Het frontal heart sections at P0 (Black – present; White – absent). (K–N) Hdac1C (K,M) and Hdac2 (L,N) immunostaining of Wnt1;1Het2Het;R26R-LacZ−/+ and Wnt1;1KO2KO;R26R-LacZ−/+ neural tubes (NT) at E9.5 (Black, present; White, absent). (O–Q) Left lateral (main) and right lateral (inset) images of Hdac1F/F;Hdac2F/F (O), Wnt1;1Het2KO (P), and Wnt1;1KO2Het (Q) pups at birth (White, milk spot; Black, absent milk spot; Red, distended abdomen; Orange, neonatal distress). (R–T) Dissected/fixed P0 Hdac1F/F;Hdac2F/F (R), Wnt1;1Het2KO (S), and Wnt1;1KO2Het (T) palates (PAL). (U–Z) Dissected/fixed P0 Hdac1F/F;Hdac2F/F (U–V), Wnt1;1Het2KO (W–X), and Wnt1;1KO2Het (Y–Z) thymus (TY; U,W,Y) and outflow tract (OFT; V,X,Z). (AA–AF) Hematoxylin and Eosin P0 frontal sections of Hdac1F/F;Hdac2F/F (AA–AB), Wnt1;1Het2KO (AC–AD), and Wnt1;1KO2Het (AE–AF) aortic valves (AA,AC,AE) and pulmonic valves (AB,AD,AF). (AG–AI) Hematoxylin and Eosin staining of Hdac1F/F;Hdac2F/F (AG), Wnt1;1Het2KO (AH), and Wnt1;1KO2Het (AI) P0 hearts shows peri-arterial morphology in the aorta/pulmonary artery. Nuclear hematoxylin counterstain is present. Scale bars 100μM. AO, aorta; PA, pulmonary artery; PAL, palate; TY, thymus; BA, brachiocephalic artery; LCC, left common carotid artery; LS, left subclavian artery; RA, right atria; LA, left atria; RV, right ventricle; LV, left ventricle; PV, pulmonic valve; AV, aortic valve.
The insertion of a LacZ cassette between exons 2 and 3 (Fig 1A) results in the expression of β-galactosidase under the control of native Hdac1 promoter and enhancer elements which, in turn, causes blue staining following incubation with X-gal (Fig 1C, E, G) compared to controls (Fig 1B, D, F). X-Gal staining of Hdac1LacZ/+ and Hdac1+/+ embryos at E7.75 (Fig 1B–C), E8.75 (Fig 1D–E), and E9.5 (Fig 1F–G) shows broad LacZ staining (blue) in Hdac1LacZ/+ embryos compared to Hdac1+/+ controls. To further investigate the specificity of the Hdac1LacZ knock-in allele and the overlapping spatiotemporal expression pattern of Hdac1 and Hdac2 during development, we sectioned E9.5 Hdac1LacZ/+ embryos and performed immunostaining using a C-terminal Hdac1 antibody (Hdac1) (Fig 1H–M) and Hdac2 antibody (Fig 1N–S). Hdac1 staining (brown) and Hdac1LacZ staining (blue) overlapped in all examined developmental structures including the outflow tract (OFT, Fig 1H), primitive ventricle (PRV, Fig 1I), atrioventricular canal (AVC, Fig 1J), midbrain & telencephalic vesicle (MB & TV, Fig 1K), otic pit (OtP, Fig 1L), and caudal somites (So, Fig 1M). Similarly, immunostaining for Hdac2 (brown) and Hdac1LacZ staining (blue) overlapped in all examined developmental structures including the outflow tract (Fig 1N), primitive ventricle (Fig 1O), atrioventricular canal (Fig 1P), midbrain & telencephalic vesicle (Fig 1Q), otic pit (Fig 1R), and caudal somites (Fig 1S).
Hdac1 and Hdac2 function redundantly within the neural crest progenitor cells
While the genetic underpinnings of NC derived congenital defects are emerging, the role of Hdac1 and Hdac2 in this process is unknown. To investigate roles of Hdac1 and Hdac2 during craniofacial and cardiac outflow tract (OFT) development, we first validated the conditional Hdac1 and Hdac2 alleles used in this study (Fig 2A–J) before ablating Hdac1 and Hdac2 in the NC progenitor cells using Wnt1-Cre2 (Lewis et al., 2013) (Fig 2K–AI). Hdac1 conditional allele (Tm1c, Hdac1Flox) was generated from the Tm1a (Hdac1LacZ) allele by exposure to Flp-recombinase, with LoxP sites flanking exon 3 (Fig 2A). Exposure of Hdac1Flox to Cre-recombinase results in excision of exon 3, introducing a frameshift mutation and premature stop codon early in the Hdac1 deacetylase domain (Hdac1KO) (Fig 2B). To verify conditional allele performance, we used Mesp1-Cre (Saga et al., 1999), a marker of the cardiogenic mesoderm that contributes to the majority of the developing mammalian heart, to delete Hdac1 or Hdac2 (Fig 2C–J). To query whether excision of exon 3 may generate semi-functional Hdac1 protein fragments, we performed Hdac1 immunostaining and Western blot with C-terminal (Hdac1C) and N-terminal (Hdac1N) antibodies (Fig 2C–E, G–J). Western blot analysis of Mesp1-Cre;Hdac1F/+ (Mesp1;1Het) and Mesp1-Cre;Hdac1F/F (Mesp1;1KO) P0 heart lysate shows loss of Hdac1 protein levels assessed with both Hdac1N and Hdac1C antibodies (Fig 2C–E). Additionally, Mesp1-Cre efficiently mediated ablation of the already-established Hdac2 allele (Anokye-Danso et al., 2011) utilized in this study (Fig 2F). Hdac1 protein expression (black arrows) is lost (white arrows) in the left ventricle (within the Mesp1-Cre domain) by P0 following excision of exon 3 by Cre-recombinase using both Hdac1C and Hdac1N antibodies when compared to controls (Fig 2G–J). Next, we queried the efficacy of ablation by Wnt1-Cre2. To help understand the spatiotemporal dynamics of Wnt1-Cre2 expressing neural crest progenitor cells and their derivatives, a R26R-LacZ allele (Soriano, 1999) was included and X-gal staining was performed. The result of this staining is to mark any Wnt1-Cre2 expressing cells (and their descendants) as blue. Immunostaining analysis reveals loss of Hdac1 (Fig 2M) and Hdac2 (Fig 2N) in Wnt1;1KO2KO;R26R-LacZ−/+ neural tube compared to Wnt1;1Het2Het;R26R-LacZ−/+ neural tube (Fig 2K, L) at E9.5. Wnt1-Cre2; Hdac1F/F, Hdac2F/F mice (Wnt1;1KO2KO) displayed complete embryonic lethality while single-allele mice (Wnt1-Cre2; Hdac1F/F, Hdac2F/+, Wnt1;1KO2Het & Wnt1-Cre2; Hdac1F/+, Hdac2F/F, Wnt1;1Het2KO) were recovered in expected ratios at birth but were found to die by P21 (Table 1). The single-allele mice recovered at birth were found with varying perinatal defects ranging from cyanosis, distended abdomen, and neonatal lethality (Table 1, Fig 2O–Q). Wnt1;1KO2Het and Wnt1;1Het2KO mice were recovered without an observable milk spot compared to controls, indicating inability to feed (Fig 2O–Q). Additionally, some Wnt1;1Het2KO mice were recovered with a readily observable distended abdomen (Fig 2P) and dissection revealed trapped air in a distended stomach. Often, an absent milk spot indicates a craniofacial and/or palate defect; however, no craniofacial or palate defect was observed in the single-allele mice (Fig 2O–T). The NC also contributes to the thymus, the cardiac outflow tract vessels (aorta and pulmonary artery) and valves (aortic valve and pulmonary valve). Wnt1;1KO2Het and Wnt1;1Het2KO mice show normal thymic structure (Fig 2U, W, Y), outflow tract course and caliber (Fig 2V, X, Z), and valve morphology (Fig 2AA–AF). NC progenitor cells also make major contributions to arterial smooth muscle in the head, neck, and heart (Jiang et al., 2000; Crane and Trainor, 2006). Peri-arterial outflow tract morphology in Wnt1;1KO2Het and Wnt1;1Het2KO appeared similar to controls (Fig 2AG–AI).
Table 1.
Genotypes of samples at various developmental stages.
| Genotype | Observed (Expected)
|
||||
|---|---|---|---|---|---|
| P21 | P0 | E11.5 | E10.5 | E9.5 | |
| Hdac1F/+, Hdac2F/+ | 4 (5) | 13 (8) | 3 (4.625) | 7 (5.875) | 8 (8.75) |
| Hdac1F/+, Hdac2F/F | 7 (5) | 7 (8) | 3 (4.625) | 7 (5.875) | 8 (8.75) |
| Hdac1F/F, Hdac2F/+ | 10 (5) | 15 (8) | 1 (4.625) | 3 (5.875) | 5 (8.75) |
| Hdac1F/F, Hdac2F/F | 8 (5) | 10 (8) | 13 (4.625) | 5 (5.875) | 14 (8.75) |
| Wnt1-Cre2; Hdac1F/+, Hdac2F/+ | 11 (5) | 9 (8) | 6 (4.625) | 4 (5.875) | 10 (8.75) |
| Wnt1-Cre2; Hdac1F/+, Hdac2F/F | 0 (5) | 6a,b (8) | 4 (4.625) | 7 (5.875) | 13 (8.75) |
| Wnt1-Cre2; Hdac1F/F, Hdac2F/+ | 0 (5) | 4c,d (8) | 2 (4.625) | 5 (5.875) | 5 (8.75) |
| Wnt1-Cre2; Hdac1F/F, Hdac2F/F | 0 (5) | 0 (8) | 5e (4.625) | 9 (5.875) | 7 (8.75) |
|
| |||||
| Total (p value): | 40 (p<0.001) | 51 (p=0.004) | 37 (p=0.003) | 42 (p=0.71) | 70 (p=0.25) |
Wnt1-Cre2; Hdac1F/+; Hdac1F/+ mice were crossed with Hdac1F/F; Hdac2F/F mice and samples were collected at the designated time point.
2 pups found cyanotic with distended abdomen/pulmonary distress.
3 pups recovered dead.
2 pups recovered dead.
1 pup found cyanotic in pulmonary distress.
5 embryos found reabsorbing with hemorrhage.
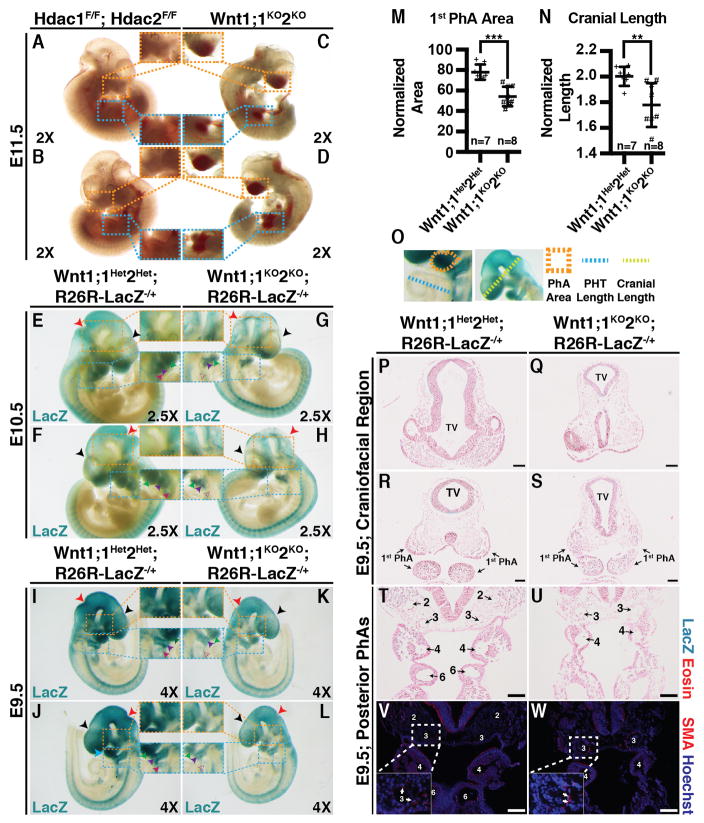
Wnt1;1KO2KO embryos were recovered in expected Mendelian ratios at E9.5, E10.5, and E11.5 (Table 1). However, Wnt1;1KO2KO pups were not recovered at P0, indicating that they suffer complete embryonic lethality (Table 1). The Wnt1;1KO2KO embryos recovered at E11.5 appeared grossly hemorrhagic with blood pooling in the cranial vesicles (orange) and primitive gut (blue), indicating recent fetal demise compared to Hdac1F/F, Hdac2F/F controls (Fig 3A–D) and suggesting defects in PhA-derived artery development. Wnt1;1KO2KO;R26R-LacZ−/+ embryos appeared smaller, with reduced LacZ staining intensity (orange) (Fig 3G, H, K, L) compared to Wnt1-Cre2;Hdac1F/+,Hdac2F/+;R26R-LacZ−/+ (Wnt1;1Het;2Het;R26R-LacZ−/+) controls at E10.5 (Fig 3E, F, I, J). Differential LacZ staining appears to be restricted to the developing head and PhAs, with LacZ staining intensity and distribution being similar in the developing torso and tail between Wnt1;1KO2KO;R26R-LacZ−/+ embryos (Fig 3G, H, K, L) and Wnt1;1Het2Het;R26R-LacZ−/+ controls (Fig 3E, F, I, J). Additionally, Wnt1;1KO2KO;R26R-LacZ−/+ embryos presented with abnormal craniofacial anatomy, an attenuated posterior nuchal angle (red), reduced frontal swelling (black), and focally decreased LacZ staining intensity (orange) (Fig 3G, H, K, L) when compared to Wnt1;1Het2Het;R26R-LacZ−/+ controls (Fig 3E, F, I, J). Wnt1;1KO2KO;R26R-LacZ−/+ first (green) and second (purple) PhAs appeared atrophic when compared with controls seen by a decrease in size, definition, and LacZ staining intensity (blue, inset) (Fig 3G, H, K, L). A significant reduction of LacZ staining at the location of the third PhA (magenta) was seen in Wnt1;1KO2KO;R26R-LacZ−/+ embryos (Fig 3G, H, K, L) compared to controls (Fig 3E, F, I, J). Quantification of the first PhA area (normalized to primitive heart tube length) and cranial length (normalized to primitive heart tube length) at E9.5 showed significant reduction (***, p=0.0001 & **, p=0.0076) in the normalized area and normalized length in Wnt1;1KO2KO;R26R-LacZ−/+ embryos compared to Wnt1;1Het2Het;R26R-LacZ−/+ controls (Fig 3M–O). Additionally, frontal sections of E9.5 Wnt1;1KO2KO;R26R-LacZ−/+ and Wnt1;1Het2Het;R26R-LacZ−/+ embryos revealed abnormal telencephalic vesicle (TV) morphology in experimental embryos (Fig. 3Q, S) compared to controls (Fig 3P, R), confirming the gross observations of craniofacial defects.
Figure 3.
(A–D) Wnt1;1KO2KO (C–D) and Hdac1F/F;Hdac2F/F (A–B) embryos at E11.5 (Orange – cranial blood; Blue – gut blood). (E–H) LacZ-stained Wnt1;1KO2KO;R26R-LacZ−/+ (G–H) and Wnt1;1Het2Het;R26R-LacZ−/+ (E–F) embryos at E10.5 (Red –nuchal angle; Blue, first pharyngeal arch; Orange, differential LacZ staining). (I–L) LacZ-stained Wnt1;1KO2KO;R26R-LacZ−/+ (K–L) and Wnt1;1Het2Het;R26R-LacZ−/+ (I–J) embryos at E9.5 (Red, posterior nuchal angle; Black, frontal swelling; Blue, first pharyngeal arch; Orange, differential LacZ staining). (M–O) First pharyngeal arch area (normalized to PHT length; M) and cranial length (normalized to PHT length; N) at E9.5 in Wnt1;1Het2Het;R26R-LacZ−/+ and Wnt1;1KO2KO;R26R-LacZ−/+. Presented as mean (SD) (***, p=0.0001; **, p=0.0076)(Schematic diagram, O). (P–S) Eosin and LacZ stained E9.5 Frontal sections of Wnt1;1Het2Het;R26R-LacZ−/+ (P,R) and Wnt1;1KO2KO;R26R-LacZ−/+ (Q,S) craniofacial morphology at two levels. (T–W) Eosin/LacZ stained (T–U) or α-smooth muscle actin (V–W, SMA) immunofluorescence of Wnt1;1Het2Het;R26R-LacZ−/+ (T,V) and Wnt1;1KO2KO;R26R-LacZ−/+ (U,W) E9.5 posterior pharyngeal arch arteries (2/3/4/6, Second/Third/Fourth/Sixth arch arteries; White arrows, SMA). Scale bars 100μM. PhA, pharyngeal arch; PHT, primitive heart tube; NT, neural tube; SMA, α-smooth muscle actin.
Eosin-stained frontal sections of the PhAs and PhA arteries (PhAAs) revealed that the sixth PhA and PhAA are missing in Wnt1;1KO2KO;R26R-LacZ−/+ embryos (Fig 3U, W) compared to controls (Fig 3T, V). The second PhA was visualized in a different section of Wnt1;1KO2KO;R26R-LacZ−/+ embryos (not shown). All other arches were found to be present, albeit smaller in double knockouts (Fig 3U, W) than controls (Fig 3T, V). Peri-arterial α-smooth muscle staining appears similar in both Wnt1;1KO2KO;R26R-LacZ−/+ and Wnt1;1Het2Het;R26R-LacZ−/+ embryos at E9.5 (Fig 3V–W).
Hdac1 and Hdac2 regulate proliferation in the neural tube and first pharyngeal arch by repressing cyclin-dependent kinase inhibitors
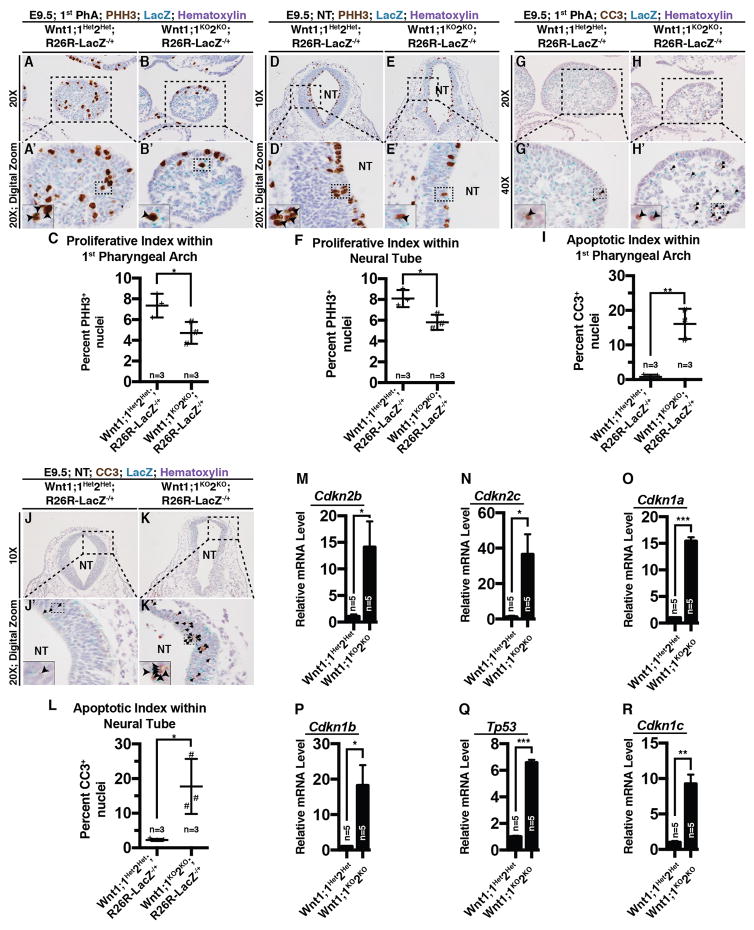
To investigate the cellular and molecular basis of defective neural crest and PhA development in Wnt1;1KO2KO embryos at E9.5, we performed phospho-histone H3 (PHH3, a marker of cellular proliferation) immunostaining on the first pharyngeal arch and the neural tube lacking both Hdac1 and Hdac2 (Fig 4A–B, D–E). Wnt1;1KO2KO;R26R-LacZ−/+ embryos exhibited reduced number of PHH3+ nuclei within the first PhA (Fig 4B, B′, C) and the neural tube (Fig 4E, E′, F) compared to controls (Fig 4A, A′, C, D, D′, F). Quantification of the number of PHH3-positive nuclei per one hundred total nuclei (proliferative index) is significantly reduced in both the first PhA (*, p=0.022; Fig 4C) and the neural tube (*, p=0.043; Fig 4F) of Wnt1;1KO2KO;R26R-LacZ−/+ embryos at E9.5.
Figure 4.
(A–B) Phospho-histone H3 (PHH3) immunostaining within the first pharyngeal arch in Wnt1;1Het2Het;R26R-LacZ−/+ (Ai, Ai′) and Wnt1;1KO2KO;R26R-LacZ−/+ (B, B′) at E9.5 (Black, PHH3+). (C) Quantification of PHH3 positive nuclei per 100 nuclei (proliferative index; presented as mean (SD); *, p=0.022). (D–E) PHH3 immunostaining within the neural tube (NT) in Wnt1;1Het2Het;R26R-LacZ−/+ (D, D′) and Wnt1;1KO2KO;R26R-LacZ−/+ (E, E′) embryos at E9.5 (Black, PHH3+). (F) Quantification of PHH3 positive nuclei per 100 nuclei (proliferative index; presented as mean(SD); *, p=0.043). (G–H) Cleaved caspase 3 (CC3) immunostaining within the first pharyngeal arch in Wnt1;1Het2Het;R26R-LacZ−/+ (G, G′) and Wnt1;1KO2KO;R26R-LacZ−/+ (H, H′) at E9.5 (Black, CC3+). (I) Quantification of CC3 positive nuclei per 100 nuclei (apoptotic index; presented as mean (SD); **, p=0.0038). (J–K) CC3 immunostaining within the NT in Wnt1;1Het2Het;R26R-LacZ−/+ (J, J′) and Wnt1;1KO2KO;R26R-LacZ−/+ (K, K′) at E9.5 (Black, CC3+). (L) Quantification of CC3 positive nuclei per 100 nuclei (apoptotic index; presented as mean (SD); *, p=0.028). (M–R) Relative mRNA levels of in E9.5 Wnt1;1Het2Het and Wnt1;1KO2KO microdissected first pharyngeal arches of Cdkn2b (M, *, p=0.042), Cdkn2c (N, *, p=0.032), Cdkn1a (O, ***, p=0.0008), Cdkn1b (P, *, p=0.035), Tp53 (Q, ***, p=0.0001), and Cdkn1c (R, **, p=0.0073). NT, neural tube.
Furthermore, Wnt1;1KO2KO;R26R-LacZ−/+ embryos show increased number of cleaved caspase 3 (CC3, a marker of apoptosis)-positive nuclei within the first PhA (Fig 4H, H′, I) and the neural tube (Fig 4K, K′, L) compared to controls (Fig 4G, G′, I, J, J′, L). Quantification of the number of CC3-positive nuclei per one hundred total nuclei (apoptotic index) is significantly increased in both the first PhA (**, p=0.0038; Fig 4I) and the neural tube (*, p=0.028; Fig 4L).
Given that there is a shift away from proliferation towards apoptosis in Wnt1;1KO2KO neural-crest enriched areas at E9.5, we wanted to characterize the molecular changes underlying this shift. Hence, we performed qPCR on an array of cyclin-dependent kinases, using the neural-crest derived first PhAs dissected from Wnt1;1KO2KO and Wnt1;1Het2Het embryos at E9.5. Our analysis revealed significant upregulation of Cdkn2b (Fig 4M, *, p=0.043), Cdkn2c (Fig 4N, *, p=0.032), Cdkn1a (Fig 4O, ***, p=0.0008), Cdkn1b (Fig 4P, *, p=0.035), Tp53 (Fig 4Q, ***, p=0.0001), and Cdkn1c (Fig 4R, **, p=0.0073) within Wnt1;1KO2KO;R26R-LacZ−/+ first PhAs compared to controls.
Discussion
Disruption of the pharyngeal arches and their corresponding arteries are a common source of congenital defects and syndromes with a multitude of etiologies and clinical courses (Siebert et al., 1985; Wilkie and Morriss-Kay, 2001; Trainor, 2010; McDonald-McGinn et al., 2015). While much of the cellular, molecular, and genetic basis of these defects is understood, the role of epigenetic regulators in development and congenital disease is just starting to be elucidated (Butler et al., 2012). HDACs are a group of enzymes that regulate the epigenome (Haberland et al., 2009b). HDACs are divided into five classes with class I HDACs (HDAC1, 2, 3, and 8) being structurally similar and having the most intrinsically active enzymatic domains (Brunmeir et al., 2009; Reichert et al., 2012). Hdac1 and Hdac2 have already been implicated in congenital craniofacial defects seen in humans (Hudson et al., 2014; de Souza et al., 2015; Matsumoto et al., 2015), yet no causal link has been drawn. Here we show that Hdac1 and Hdac2 regulate neural crest progenitor cell pluripotency, pharyngeal arch development, and craniofacial morphogenesis during earliest stages of development, which strengthens the association between these critical enzymes and human congenital defects. In contrast, Hdac3 and Hdac8, other class I HDACs, regulate neural crest-derived skull morphogenesis during late-gestation (Haberland et al., 2009a; Singh et al., 2011), suggesting a unique role of Hdac1 and Hdac2 within neural crest progenitor cells.
The cephalic neural crest has an established role in brain development, in addition to its critical role in craniofacial morphogenesis (Le Douarin et al., 2007). Interestingly, Hdac1 and Hdac2 have also been found to regulate brain development and function (Guan et al., 2009; Montgomery et al., 2009; Hagelkruys et al., 2014; Volmar and Wahlestedt, 2015). Specifically, Hdac1 and Hdac2 were found to regulate cerebellar foliation and cortical neuron organization in GFAP-Cre+ neuronal progenitor cells during development with loss of Hdac1 and Hdac2 resulting in postnatal lethality (Montgomery et al., 2009). While PhA defects in Wnt1;1KO2KO embryos suggest disrupted craniofacial development, defects in the formation of neuronal structures, such as the telencephalic vesicle, suggest that brain development may also be affected, which is an exciting area for future investigation.
Proliferation and self-renewal are essential to any progenitor cell population and when genetic or epigenetic insult disrupts these processes, progenitor cells have a choice: differentiate into an end-linage or undergo apoptosis. In this case, we observed that neural crest progenitor cells lacking Hdac1 and Hdac2 experienced arrested proliferation and apoptosis, indicating a depletion of this critical population. Working through what seems to be emerging as a common mechanism across multiple progenitor cell populations (Lagger et al., 2002; Brunmeir et al., 2009; Reichert et al., 2012; Hagelkruys et al., 2014; Moser et al., 2014), Hdac1 and Hdac2 are responsible for repressing cyclin-dependent kinase inhibitors in neural crest progenitor cells. Increased expression of cyclin-dependent kinase inhibitors places a brake on the cell cycle while decrease expression induces proliferation (Okamoto et al., 1995; Waldman et al., 1995; Chiarugi and Ruggiero, 1996; Shin et al., 2000; Swellam et al., 2004; Chu et al., 2008). Numerous reports have shown that Tp53 mediates cell cycle arrest and apoptosis through a variety of pathways (Polyak et al., 1997; Fridman and Lowe, 2003; Amaral et al., 2010; Mollereau and Ma, 2014; Chen, 2016). The significant upregulation of Tp53 in Wnt1;1KO2KO embryos, increased apoptosis, and resultant depletion of NC progenitor cells align with these reports. There is also evidence that inhibition of HDACs can result in increased p53 activity, resulting in elevated p21 (leading to cell cycle arrest) and Bax translocation and activation of apoptotic pathways (Roy et al., 2005). These results integrate the significant upregulation of Tp53 observed in Wnt1;1KO2KO embryos, decreased proliferation, and increased apoptosis. Hdac1 and Hdac2 function in NC progenitor cells to suppress Tp53 and other cyclin dependent kinase inhibitors, maintain a proliferative progenitor cell phenotype, and moderate NC progenitor contribution to craniofacial and PhA development.
A long-held assumption about HDACs, including Hdac1, is that they are ubiquitously expressed. However, recent studies demonstrate that HDACs are expressed in tissue and temporal specific manner during development. Hdac7, a class II HDAC, is endothelial specific and loss results in vascular defects (Chang et al., 2006). Hdac8, specifically expressed in fore and midbrain regions at E10 and E11.5, is critical for skull development (Haberland et al., 2009a). Similarly, murine embryos show restricted, distinct, and non-overlapping Hdac1 mRNA expression areas between E10 and E13 (Murko et al., 2010). For instance, Hdac1 mRNA is expressed in telencephalic vesicles but not in the spinal cord of E10 murine embryos. Similarly, rhombic fossa expresses Hdac1 mRNA at E10, but not at E13, suggesting spatiotemporal regulation of Hdac1 expression during murine development. Hence, the expression pattern of Hdac1, at both the gene-level and protein-level, is crucial to understanding how this highly conserved class I HDAC functions during mammalian development. To date, there are no models or reports that use a genetic approach to evaluate the endogenous Hdac1 expression under the control of native Hdac1 promoter and enhancer elements at the earliest stages of neural crest development. Using the Hdac1LacZ knock-in allele (Fig 1A), we show that Hdac1 is broadly expressed in all embryonic lineages at E7.5, E8.5, and E9.5 (Fig 1B–D). Additionally, overlapping Hdac1LacZ and Hdac1 protein expression confirmed the specificity of the Hdac1LacZ knock-in allele. This allele will enable dissection of the spatiotemporally specific expression patterning of Hdac1 in development and disease. HDAC1 expression variance is important in a number of human diseases, including several cancers (Kawai et al., 2003; Halkidou et al., 2004; Willis-Martinez et al., 2010). With this allele, the spatiotemporal expression dynamics of Hdac1 can be explored in mouse models of these diseases. Additionally, whether the expression pattern of Hdac1 changes later in development or beyond the neonatal period remains unknown and a topic for future investigation using the Hdac1LacZ knock-in allele.
Wnt1;1KO2KO embryos exhibited areas of extravascular pooling of blood in the cranial vesicles and primitive gut suggesting defective formation of NC derived PhAs and PhAAs. At E9.5 and E10.5, these embryos displayed reduced LacZ staining intensity in developing craniofacial structures. Additionally, there was decreased LacZ staining intensity within the first and second PhAs and absent LacZ staining within the third PhA at both E9.5 and E10.5. At E9.5, the first PhA was significantly smaller in the double knockouts than its counterpart in controls. Despite the reduced intensity in the cranial and pharyngeal regions, there was no observed change in the craniocaudal migration of neural crest cells, seen by comparable LacZ staining patterns in the primitive gut and developing tail region. These results suggest that loss of Hdac1 and Hdac2 in the neural crest reduced the proliferation or increase apoptosis of neural crest cells after they migrate along the craniocaudal axis. Further morphologic evaluation identified that the developing network of cranial vesicles is abnormal in Wnt1;1KO2KO samples when compared to controls (Fig 3D). The telencephalic vesicle is misshapen, narrow, and exhibits a disconnect between its cranial and caudal components, reinforcing the observation of disrupted craniofacial development in these samples. Additionally, we were unable to locate a sixth pharyngeal arch or PhAA in Wnt1;1KO2KO embryos (Fig 3E). The remaining PhAs and their corresponding arteries were visualized in double knockout embryos and, despite being smaller than comparable structures in controls, have similar early pockets of α-smooth muscle actin staining suggesting that at least initial specification of these structures proceeded normally. Whether the sixth arch never forms, is so small it is unrecognizable, or forms and then regresses cannot be determined yet the result is same – absence of this critical structure. The loss of the sixth arch combined with the other frank craniofacial and pharyngeal abnormalities (mainly the atrophic first and second PhAAs) would lead to defective formation of the maxillary artery and stapedial artery and is a likely cause of the pooling blood observed at E11.5. Our findings demonstrate that Hdac1 and Hdac2 function within the neural crest to support craniofacial and PhA development necessary to sustain mammalian embryonic development past E11.5.
Reinforcing the idea that epigenetic homeostasis is critical to developing cell populations, a single allele of either Hdac1 or Hdac2 was able to rescue Wnt1;1KO2KO embryos to birth, but was not sufficient to sustain them much further. There are several other reports of either partial or complete rescue of an Hdac1 and Hdac2 double knockout phenotype with the reintroduction of a single allele of either Hdac1 or Hdac2. In both the epidermis and in T-cells a single copy of Hdac1 was sufficient to rescue (fully or partially) the double knockout phenotype (Dovey et al., 2013; Winter et al., 2013) while Hdac2 was not. In contrast, a single copy of Hdac2 was sufficient to rescue the Hdac1/2 double knockout phenotype in the brain (Hagelkruys et al., 2014) yet Hdac1 was unable to rescue. In addition, Hdac1 and Hdac2 redundantly regulate cardiac morphogenesis, neuron development, oligodendrocyte differentiation, adipogenesis, and skeletal muscle homeostasis (Montgomery et al., 2007; Li and Richardson, 2009; Montgomery et al., 2009; Haberland et al., 2010; Moresi et al., 2012). These results suggest that Hdac1 and Hdac2 have a complex array of co-dependent, semi-independent, and truly independent roles during development (Kelly and Cowley, 2013). In this case, it appears that Hdac1 and Hdac2 are both able to partially rescue the double knockout phenotype. Single allele pups were found with a mix of abdominal distension, cyanosis, pulmonary distress (demonstrated by increased work of breathing and intercostal retractions), or perinatal lethality (Table 1, Fig 2O–Q). Interestingly, all examined neural-crest derived structures appeared normal including the palate (Fig 2R–T), cardiac outflow tract and thymus (Fig 2U–Z), aortic valve (Fig 2AA,AC,AE), pulmonic valve (Fig 2AB,AD,AF), and peri-arterial morphology in the outflow tract (Fig 2AG–AI). Hence, the dose-sensitive nature of Hdac1 and Hdac2 coupled with their potent ability to modulate progenitor cell behavior aligns them at a crossroads from which they may play a causal or contributory role in a variety of human congenital defects, including those arising from defective neural crest function and pharyngeal arch disruption. Work understanding the genetic basis of congenital defects in humans is only just now maturing and in the coming years we can expect to see a rapid expansion of our understanding of such developmental defects.
Experimental Procedures
Mice
Wnt1-Cre2 (Jackson Laboratory, 022137)(Lewis et al., 2013), Mesp1-Cre (Saga et al., 1999), R26R-LacZ (Soriano, 1999), and Hdac2Flox (Anokye-Danso et al., 2011) mice have been previously described. Frozen embryos of Hdac1tm1a(EUCOMM)Wtsi (strain EM:04097) were obtained from the European Mouse Mutant Archive (EMMA). The University of Massachusetts Medical School Transgenic Animal Facility regenerated cryopreserved embryos. The knockout-first allele, Hdac1tm1a(EUCOMM)Wtsi, mice were bred with wild type mice and are annotated Hdac1LacZ/+. Hdac1tm1a(EUCOMM)Wtsi (Hdac1LacZ) mice were bed with ACTB-FLPe (Jackson Laboratory, 003800) to generate Hdac1Flox mice. Hdac1Flox mice were bred with Hdac2Flox, R26R-LacZ, and Wnt1-Cre2 mice to generate mice for analysis. All animal protocols were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC).
Histology
Collected tissues were fixed in 2–4% paraformaldehyde in PBS at 4ºC overnight, ethanol dehydrated, embedded in paraffin, and section at 8μm thickness via microtome.
Antibodies and Reagents
The following antibodies were utilized in this study: Hdac1C (Abcam), Hdac1N (Sigma), Hdac2 (Invitrogen), α-tubulin (Sigma), PHH3 (Cell Signaling), SMA (Abcam), CC3 (Cell Signaling). Eosin Y and Harris modified hematoxylin were purchased from Fisher while X-gal was purchased from 5 Prime. Vectashield mounting medium, Vectastain Elite ABC reagent kit, and DAB Peroxidase Substrate kit were purchased from Vector Labs
Hematoxylin and Eosin Staining
Staining was performed through deparaffinizing sections in xylenes, rehydrating through an ethanol gradient, 30sec stain with 30% Harris modified hematoxylin (in diH2O), followed by 30sec counterstain with eosin Y. Slides were then rinsed and dehydrated through an ethanol gradient, cleared with xylenes, and mounted using Vectashield permanent mounting medium.
LacZ Staining
Tissue samples were dissected in cold PBS and quickly fixed in 2% paraformaldehyde in PBS for 30min at 4ºC. They were then washed in PBS at room temperature and stained over night in LacZ staining solution (5mM potassium ferricyanide, 5mM potassium ferrocyanide, 2mM MgCl2, 0.01% deoxycholic acid, 0.04% Nonidet P 40, 0.1% X-Gal, in 1X PBS) in the dark at 37ºC until blue (LacZ) staining developed. Following this, samples were washed in 1X PBS and fixed overnight in 4% paraformaldehyde in PBS at 4ºC.
Immunohistochemistry
Selected sections were deparaffinized in xylenes, pretreated using the Aptum Antigen Retriever 2100 and Aptum R-Buffer A or Sodium Citrate Buffer (pH 6.0). Immunohistochemistry was conducted with the Vectastain Elite ABC kit and DAB Peroxidase Substrate kit following manufacturer guidelines. Sections were incubated with primary antibody (Hdac1 1:100; Hdac2 1:100; PHH3 1:100, SMA 1:100, CC3 1:100, P21 1:100) overnight at 4ºC. If performed, counterstaining consisted of 30 sec incubation with 30% Harris modified hematoxylin (in diH2O) after 3,3′-diaminobenzidine development. All slides were ethanol-dehydrated, cleared with xylenes, and mounted using Vectashield permanent mounting medium.
Western Blotting
Tissue lysates were prepared in lysis buffer (20mM Tris-HCl pH 7.5, 15mM NaCl, 1mM Na2EDTA, 1mM EGTA, 1% Triton X-100, 1μg/mL leupeptin, 2.5mM sodium pyrophosphate, 1mM Na3VO4, and 1mM beta-glycerolphosphate; 1mM phenylmethylsulfonyl fluoride was added before use). Protein samples were resolved on a 4–12% SDS-Page acrylamide gel before transfer to a PVDF membrane. Primary antibodies against Hdac1C (1:1000), Hdac1N (1:1000), and Hdac2 (1:1000) were used. Primary antibodies were visualized with HRP-conjugated secondary antibodies. Blots were probed with α-tubulin (1:1000 or 1:2500) for loading control.
Morphometric Analysis
Quantitative morphologic analysis was performed using ImageJ software to measure and record anatomical features of interest. These measurements were normalized to the primitive heart tube length to control for any variability in embryo size and are presented as mean (SD).
Real Time quantitative PCR
Total RNA was extracted from microdissected first pharyngeal arches using the Qiagen Plus Micro RNeasy Kit according to the manufacturer guidelines. cDNA was reverse transcribed using iScript Reverse Transcription Supermix (BioRad) according to the manufacturer guidelines. Transcript expression was measured by qRT-PCR using SYBR Green PCR Master Mix (ThermoFisher). Signals were normalized to Gapdh controls and represented as relative expression ratios of experimental samples relative to Wnt1;1Het2Het controls using the ΔΔCt method. Primer sequences available upon request.
Statistical Analysis
Statistical significance between groups was determined using an unpaired two-tailed Student’s t-tests or a χ2 test. The level of significance (α) was set as 0.05 for all analyses.
Acknowledgments
Funding – This work was supported by National Heart, Lung, and Blood institute grant R01 HL118100 and institutional startup funds (to C.M.T.).
We gratefully acknowledge The University of Massachusetts Transgenic Core Facility for regeneration of cryopreserved embryos. This work was supported by National Heart, Lung, and Blood Institute grant R01 HL118100 and institutional startup funds (to C.M.T.). The authors declare that they have no conflicts of interest with the contents of this article.
References
- Amaral JD, Xavier JM, Steer CJ, Rodrigues CM. The role of p53 in apoptosis. Discov Med. 2010;9:145–152. [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MT, Dalen K, Mancuso AA, Kerr HH, Cacciarelli AA, Mafee MF. Congenital anomalies of the branchial apparatus: embryology and pathologic anatomy. Radiographics. 1992;12:943–960. doi: 10.1148/radiographics.12.5.1529135. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Diaz R, Trainor PA. Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;139:709–719. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- Butler JS, Koutelou E, Schibler AC, Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;4:163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Chen J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med. 2016;6:a026104. doi: 10.1101/cshperspect.a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarugi V, Ruggiero M. Role of three cancer “master genes” p53, bcl2 and c-myc on the apoptotic process. Tumori. 1996;82:205–209. [PubMed] [Google Scholar]
- Chin AJ, Saint-Jeannet JP, Lo CW. How insights from cardiovascular developmental biology have impacted the care of infants and children with congenital heart disease. Mech Dev. 2012;129:75–97. doi: 10.1016/j.mod.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- Conway SJ, Kaartinen V. TGFbeta superfamily signaling in the neural crest lineage. Cell Adh Migr. 2011;5:232–236. doi: 10.4161/cam.5.3.15498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero DR, Brugmann S, Chu Y, Bajpai R, Jame M, Helms JA. Cranial neural crest cells on the move: their roles in craniofacial development. Am J Med Genet A. 2011;155A:270–279. doi: 10.1002/ajmg.a.33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane JF, Trainor PA. Neural crest stem and progenitor cells. Annu Rev Cell Dev Biol. 2006;22:267–286. doi: 10.1146/annurev.cellbio.22.010305.103814. [DOI] [PubMed] [Google Scholar]
- de Souza KR, Mergener R, Huber J, Campos Pellanda L, Riegel M. Cytogenomic Evaluation of Subjects with Syndromic and Nonsyndromic Conotruncal Heart Defects. Biomed Res Int. 2015;2015:401941. doi: 10.1155/2015/401941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey OM, Foster CT, Conte N, Edwards SA, Edwards JM, Singh R, Vassiliou G, Bradley A, Cowley SM. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood. 2013;121:1335–1344. doi: 10.1182/blood-2012-07-441949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB, Conway SJ. Combinatorial transcriptional interaction within the cardiac neural crest: a pair of HANDs in heart formation. Birth Defects Res C Embryo Today. 2004;72:151–161. doi: 10.1002/bdrc.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene. 2003;22:9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. Redundant control of adipogenesis by histone deacetylases 1 and 2. J Biol Chem. 2010;285:14663–14670. doi: 10.1074/jbc.M109.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009a;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009b;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelkruys A, Lagger S, Krahmer J, Leopoldi A, Artaker M, Pusch O, Zezula J, Weissmann S, Xie Y, Schofer C, Schlederer M, Brosch G, Matthias P, Selfridge J, Lassmann H, Knoblich JA, Seiser C. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Development. 2014;141:604–616. doi: 10.1242/dev.100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- Hall JE. Guyton and Hall textbook of medical physiology. Elsevier Health Sciences; 2015. [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Helms JA, Cordero D, Tapadia MD. New insights into craniofacial morphogenesis. Development. 2005;132:851–861. doi: 10.1242/dev.01705. [DOI] [PubMed] [Google Scholar]
- Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev Biol. 2004;275:1–11. doi: 10.1016/j.ydbio.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Hudson C, Schwanke C, Johnson JP, Elias AF, Phillips S, Schwalbe T, Tunby M, Xu D. Confirmation of 6q21-6q22.1 deletion in acro-cardio-facial syndrome and further delineation of this contiguous gene deletion syndrome. Am J Med Genet A. 2014;164A:2109–2113. doi: 10.1002/ajmg.a.36548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaladdin S, Kelly RD, O’Regan L, Dovey OM, Hodson GE, Millard CJ, Portolano N, Fry AM, Schwabe JW, Cowley SM. Histone deacetylase (HDAC) 1 and 2 are essential for accurate cell division and the pluripotency of embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:9840–9845. doi: 10.1073/pnas.1321330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Jurkin J, Zupkovitz G, Lagger S, Grausenburger R, Hagelkruys A, Kenner L, Seiser C. Distinct and redundant functions of histone deacetylases HDAC1 and HDAC2 in proliferation and tumorigenesis. Cell Cycle. 2011;10:406–412. doi: 10.4161/cc.10.3.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Li H, Avraham S, Jiang S, Avraham HK. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int J Cancer. 2003;107:353–358. doi: 10.1002/ijc.11403. [DOI] [PubMed] [Google Scholar]
- Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans. 2013;41:741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Gale TF, Stewart DE. Neural crest cells contribute to normal aorticopulmonary septation. Science. 1983;220:1059–1061. doi: 10.1126/science.6844926. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Hutson MR. Factors controlling cardiac neural crest cell migration. Cell Adh Migr. 2010;4:609–621. doi: 10.4161/cam.4.4.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Brito JM, Creuzet S. Role of the neural crest in face and brain development. Brain Res Rev. 2007;55:237–247. doi: 10.1016/j.brainresrev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- Lewis AE, Vasudevan HN, O’Neill AK, Soriano P, Bush JO. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol. 2013;379:229–234. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Richardson WD. Genetics meets epigenetics: HDACs and Wnt signaling in myelin development and regeneration. Nat Neurosci. 2009;12:815–817. doi: 10.1038/nn0709-815. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Nozaki Y, Minami T, Jimbo EF, Shiraishi H, Yamagata T. 6q21-22 deletion syndrome with interrupted aortic arch. Hum Genome Var. 2015;2:15015. doi: 10.1038/hgv.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, Zackai EH, Emanuel BS, Vermeesch JR, Morrow BE, Scambler PJ, Bassett AS. 22q11.2 deletion syndrome. Nat Rev Dis Primers. 2015;1:15071. doi: 10.1038/nrdp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollereau B, Ma D. The p53 control of apoptosis and proliferation: lessons from Drosophila. Apoptosis. 2014;19:1421–1429. doi: 10.1007/s10495-014-1035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci U S A. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V, Carrer M, Grueter CE, Rifki OF, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc Natl Acad Sci U S A. 2012;109:1649–1654. doi: 10.1073/pnas.1121159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MA, Hagelkruys A, Seiser C. Transcription and beyond: the role of mammalian class I lysine deacetylases. Chromosoma. 2014;123:67–78. doi: 10.1007/s00412-013-0441-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz WA, Trainor PA. Neural crest cell evolution: how and when did a neural crest cell become a neural crest cell. Curr Top Dev Biol. 2015;111:3–26. doi: 10.1016/bs.ctdb.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Murko C, Lagger S, Steiner M, Seiser C, Schoefer C, Pusch O. Expression of class I histone deacetylases during chick and mouse development. Int J Dev Biol. 2010;54:1527–1537. doi: 10.1387/ijdb.092971cm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Colbert MC, Robbins J. Neural crest cells retain multipotential characteristics in the developing valves and label the cardiac conduction system. Circ Res. 2006;98:1547–1554. doi: 10.1161/01.RES.0000227505.19472.69. [DOI] [PubMed] [Google Scholar]
- Ohnemus S, Kanzler B, Jerome-Majewska LA, Papaioannou VE, Boehm T, Mallo M. Aortic arch and pharyngeal phenotype in the absence of BMP-dependent neural crest in the mouse. Mech Dev. 2002;119:127–135. doi: 10.1016/s0925-4773(02)00345-3. [DOI] [PubMed] [Google Scholar]
- Okamoto A, Hussain SP, Hagiwara K, Spillare EA, Rusin MR, Demetrick DJ, Serrano M, Hannon GJ, Shiseki M, Zariwala M, et al. Mutations in the p16INK4/MTS1/CDKN2, p15INK4B/MTS2, and p18 genes in primary and metastatic lung cancer. Cancer Res. 1995;55:1448–1451. [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- Reichert N, Choukrallah MA, Matthias P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell Mol Life Sci. 2012;69:2173–2187. doi: 10.1007/s00018-012-0921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman TP, Le Douarin NM, Fontaine-Perus JC, Gershon MD. Colonization of the bowel by neural crest-derived cells re-migrating from foregut backtransplanted to vagal or sacral regions of host embryos. Dev Dyn. 1993;196:217–233. doi: 10.1002/aja.1001960308. [DOI] [PubMed] [Google Scholar]
- Roy S, Packman K, Jeffrey R, Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005;12:482–491. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Saint-Jeannet JP, Moody SA. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014;389:13–27. doi: 10.1016/j.ydbio.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JY, Kim HS, Lee KS, Kim J, Park JB, Won MH, Chae SW, Choi YH, Choi KC, Park YE, Lee JY. Mutation and expression of the p27KIP1 and p57KIP2 genes in human gastric cancer. Exp Mol Med. 2000;32:79–83. doi: 10.1038/emm.2000.14. [DOI] [PubMed] [Google Scholar]
- Siebert JR, Graham JM, Jr, MacDonald C. Pathologic features of the CHARGE association: support for involvement of the neural crest. Teratology. 1985;31:331–336. doi: 10.1002/tera.1420310303. [DOI] [PubMed] [Google Scholar]
- Singh N, Trivedi CM, Lu M, Mullican SE, Lazar MA, Epstein JA. Histone deacetylase 3 regulates smooth muscle differentiation in neural crest cells and development of the cardiac outflow tract. Circ Res. 2011;109:1240–1249. doi: 10.1161/CIRCRESAHA.111.255067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Swellam M, El-Aal AA, AbuGabel KM. Deletions of p15 and p16 in schistosomal bladder cancer correlate with transforming growth factor-alpha expression. Clin Biochem. 2004;37:1098–1104. doi: 10.1016/j.clinbiochem.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Theveniau-Ruissy M, Dandonneau M, Mesbah K, Ghez O, Mattei MG, Miquerol L, Kelly RG. The del22q11.2 candidate gene Tbx1 controls regional outflow tract identity and coronary artery patterning. Circ Res. 2008;103:142–148. doi: 10.1161/CIRCRESAHA.108.172189. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, Floss T, Goettlicher M, Noppinger PR, Wurst W, Ferrari VA, Abrams CS, Gruber PJ, Epstein JA. Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- Twigg SR, Wilkie AO. New insights into craniofacial malformations. Hum Mol Genet. 2015;24:R50–59. doi: 10.1093/hmg/ddv228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Limborgh J, Lieuw Kie Song SH, Been W. Cleft lip and palate due to deficiency of mesencephalic neural crest cells. Cleft Palate J. 1983;20:251–259. [PubMed] [Google Scholar]
- Verzi MP, Agarwal P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Dev Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- Volmar C-H, Wahlestedt C. Histone deacetylases (HDACs) and brain function. Neuroepigenetics. 2015;1:20–27. [Google Scholar]
- Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45:279–285. doi: 10.1016/j.exger.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter M, Moser MA, Meunier D, Fischer C, Machat G, Mattes K, Lichtenberger BM, Brunmeir R, Weissmann S, Murko C, Humer C, Meischel T, Brosch G, Matthias P, Sibilia M, Seiser C. Divergent roles of HDAC1 and HDAC2 in the regulation of epidermal development and tumorigenesis. EMBO J. 2013;32:3176–3191. doi: 10.1038/emboj.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimberlin CD, Lancini C, Sno R, Rosekrans SL, McLean CM, Vlaming H, van den Brink GR, Bots M, Medema JP, Dannenberg JH. HDAC1 and HDAC2 collectively regulate intestinal stem cell homeostasis. FASEB J. 2015;29:2070–2080. doi: 10.1096/fj.14-257931. [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Kiefer F, Prudenziati M, Spiller C, Hansen J, Floss T, Wurst W, Minucci S, Gottlicher M. Reduced body size and decreased intestinal tumor rates in HDAC2-mutant mice. Cancer Res. 2007;67:9047–9054. doi: 10.1158/0008-5472.CAN-07-0312. [DOI] [PubMed] [Google Scholar]