Abstract
Objective
To identify genetic loci associated with features of nonalcoholic fatty liver disease. (NAFLD) histologic severity in a cohort of Hispanic boys.
Study design
234 eligible Hispanic boys age 2–17 years with clinical, laboratory and histological data enrolled in the Nonalcoholic Steatohepatitis Clinical Research Network (NASH CRN) were included in the analysis of 624,297 single nucleotide polymorphisms (SNPs). After elimination of 4 outliers and 22 boys with cryptic relatedness, association analyses were performed on 208 DNA samples with corresponding liver histology. Logistic regression analyses were carried out for qualitative traits and linear regression analyses were applied for quantitative traits.
Results
The median age and body mass index Z-score were 12.0 y (interquartile range, 11.0y to 14.0y) and 2.4 (interquartile range 2.1–2.6) respectively. The NAFLD activity score (NAS, scores 1–4 vs 5–8) was associated with SNP rs11166927 on chromosome 8 in the TRAPPC9 region (p = 8.7e-07). Fibrosis stage was associated with SNP rs6128907 on chromosome 20, near actin related protein 5 homolog (p=9.9e-07). In comparing our results in Hispanic boys to those of previously reported SNPs in adult NASH, two of 26 susceptibility loci were associated with NAS and two were associated with fibrosis stage.
Conclusions
In this discovery genome-wide association scan (GWAS), we found significant novel gene effects on histologic traits associated with NAS and fibrosis that are distinct from those previously recognized by adult NAFLD GWAS studies.
Keywords: Genome wide, Hispanic, fatty liver, fibrosis, pediatrics
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in children and adults in the developed world. The histologic expression of the disease ranges from steatosis to steatohepatitis, to marked fibrosis and cirrhosis. Multiple factors contribute to disease progression including environmental influences such as dietary intake, physical activity and genetic predisposition. Genome wide association research has shown that the heritability of complex traits and complex disorders, such as NAFLD, may be due to multiple genes of small effect size (1). The genetic susceptibility in NAFLD merits investigation given the implications for disease progression associated with unique pathogenic variants.
The basis for a genetic component in NAFLD is well founded. Familial clustering, epidemiologic data and twin studies demonstrate that inherited factors influence the likelihood and severity of pediatric NASH (2). The heritability of fat fraction as a continuous trait and the heritability of steatosis and fibrosis have been demonstrated using noninvasive means(3) (4). Furthermore, striking ethnic variability found in the prevalence of NAFLD provides further evidence for significant genetic factors impacting pathogenesis (5, 6).
Several large-scale genome-wide association studies (GWAS) have sought to identify common genetic variants associated with NAFLD susceptibility and disease progression. These studies have consistently reported strong associations between the non-synonymous amino acid substitution I148M in the patatin-like phospholipase domain containing 3 gene (PNPLA3), that encodes a triacylglycerol lipase expressed in adipocytes, with the risk of steatosis and steatohepatitis (7, 8). Additionally, single nucleotide polymorphisms (SNPs) near TM6SF2 and PPP1R3B have been associated with hepatic steatosis and variants within or near NCAN, GCKR and LYPLAL1, in addition to PNPLA3, emerged in a study relating these variants to severity of lobular inflammation and fibrosis (9). Smaller scale GWAS of similar sample size in adults provided additional evidence that genetic variants seen in adults with NAFLD were associated with key histologic features (10). The I148M variant of PNPLA3, the major genetic risk factor for NASH identified in adults, also is associated with liver enzyme elevations and degree of steatosis in obese children (11–14).
Epidemiologic data not only points to a genetic component of the disease but also highlights the influence of sex hormones. NAFLD is consistently more prevalent in boys than in girls, which suggests that sex hormones are associated with the predilection for pediatric NAFLD (15). Given the aforementioned effects from sex hormones and ethnicity, we aimed to minimize noise from heterogeneity in ethnicity and gender by focusing on Hispanic boys.
Methods
All Hispanic boys enrolled in the Nonalcoholic Steatohepatitis (NASH) Clinical Research Network (CRN) in the NAFLD Database I Study (n=234) were included in this discovery cohort. This multicenter, prospective, longitudinal cohort was established in 2002 by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and contains over 4,400 subjects. Clinical and histologic features of database participants have been described by Patton et al (16). These subjects met exclusion criteria for any other potential contributors to fatty liver disease. From the pediatric cohort, all Hispanic boys (age 2–17 y) with liver biopsies were included. Biopsy specimens were reviewed and scored centrally by the NASH CRN Pathology Committee. Specimens were scored according to the histology scoring system established by the NASH CRN (17). The Institutional Review Boards at each participating center approved the protocols in addition to the NASH CRN Steering Committee. All parents provided consent for their child’s participation and all children age >7 years provided assent.
Genotyping was performed at the Medical Genetics Institute at Cedars–Sinai Medical Center using Illumina OmniExpress chips technology (HumanCNV370-Quadv3 BeadChips; Illumina, San Diego, California(18, 19)). Genotypes were determined based on clustering of the raw intensity data for the two dyes using Illumina BeadStudio software. Two samples performed in duplicate yielded 100% concordance. Quality controls (QCs) were performed on the 234 samples and 624,297 single-nucleotide polymorphisms (SNPs) using PLINK(20). For SNPs on the 22 autosomal chromosomes, we applied the following filter criteria: genotype missing rate >0.02, minor allele frequency (MAF) <0.05, Hardy-Weinberg equilibrium (HWE) P value < 10−6, and heterozygosity >.53. We also performed quality control at an individual level to check for missing rate and cryptic relatedness (π̂). Cryptic relatedness refers to unknown more recent relatedness (as opposed to distant relatedness) and includes family relationships such as grandparent-grandchild and full sibling pairs. Population-based association studies assume independent (unrelated) individuals.
We observed no sample with missing rate > 0.02, but found 22 pairs of samples with π̂ ≥ 0.25. Principal component analysis (PCA) was then carried out using EIGENSTRAT (21) to examine potential population stratification among our study samples. 4 samples were identified as population outliers (spurious component analysis). We thereafter excluded 26 samples (22 cryptic relatedness and 4 PCA outliers) from further association analysis. To adjust for potential population stratification, we included the first 2 principal components as covariates in the model of association analysis. The final data set for the association analysis after QCs had 208 samples.
We evaluated the ability to detect an association between a SNP and NAS score by power calculation implemented in QUANTO version 1.2.4. Based on the mean and standard deviation of NAS (mean 4.27, SD 1.69) in a preliminary sample of patients enrolled into the NAFLD Database Study, we assessed the power using 208 independent individuals under an additive genetic model. For detectable effect size > 0.8, a sample size of 208 will have enough power (>0.83) to identify the association under additive model with minor allele frequency > 0.1.
Statistical Analyses
The GWAS was performed to identify genetic factors associated with specific features or combinations of liver histology. End points of interest in this study were NAFLD Activity Score (NAS), definite nonalcoholic steatohepatitis (NASH) as defined by prespecified histologic criteria, and fibrosis stage. NAS ranges in score from 1 to 8. In our association analysis, NAS was analyzed as a qualitative (binary) trait by comparing NAS ≤4 to NAS >4. This cutpoint was chosen based on clinical significance as NAS > 4 has been shown to correlate with the presence of NASH (22). Fibrosis stage was analyzed as a quantitative trait based on stage, which ranged from 0–4. The association between the end points of interest and each SNP was evaluated in the model with the first two PCs as covariates. For each SNP, association analyses were run both under an additive and a dominant genetic model using the PLINK software. Logistic regression analysis was carried out for NAS and definite NASH and linear regression analysis was run for fibrosis grade. We further examined SNPs with a p-value < 10−5 from the GWAS (referred below as “top SNPs”). We used SCAN (SNP and CNV Annotation Database http://www.scandb.org/newinterface/about.html) to annotate genes for the top SNPs.
We also examined previously identified genetic loci associated with NAFLD in adult populations to determine if they generalize to this group of Hispanic boys. We studied the 26 susceptibility loci that were reported in the previously published GWAS (10). Proxy SNPs found by SNAP (23) were used for SNPs not included in our current study. SNPs with a p-value < 0.05 in our analysis provided the degree of confirmation for the association with NAS and/or fibrosis grade.
With the association results obtained from additive and dominant genetic models, we generated Manhattan plots showing the −log10(p-value) along with the 22 autosomal chromosomes for NAS and fibrosis grade. We also generated regional plots for the region around the top association signals. The regional plot shows association p-values (−log10 scale) on the vertical axis along with recombination rates and all candidate genes in the region around the top SNPs (250 kilobases bilaterally) using LocusZoom (24). The plots also show the pairwise LD pattern with the most strongly associated SNP, which has the smallest P value in the region. SNPs in the plot are colored based on their r2 with the most strongly associated SNP.
We calculated quantile–quantile (QQ) plots for NAS and fibrosis using an additive model to assess for normality and these are shown (Figure 3; available at www.jpeds.com).
Results
A total of 234 overweight or obese Hispanic boys were included in the GWAS. Subjects with cryptic relatedness were eliminated (n=22) as were those with a spurious component analysis (n=4), leaving 208 subjects that met inclusion and exclusion criteria. Clinical and laboratory characteristics of the 208 subjects included are shown in Table I. The median age was 12 years (interquartile range [IQR], 11–14 years). Median BMI, waist-to-hip ratio, and serum aminotransferase values are elevated and consistent with that reported previously in pediatric NAFLD, along with elevated metabolic parameters of median HbA1c, glucose, and insulin. As regards diagnostic patterns, 22% of subjects had a definite NASH pattern (one group), 53 % with borderline NASH (zone 3 or 1), and 23% with NAFLD but no NASH (grouped together). The borderline zone 1 pattern is unique to children and may indicate an age-specific entity with a unique pathogenesis and natural history (25). The median NAS was 4 (IQR, 3–5). The majority of subjects had some degree of fibrosis, with 18% demonstrating advanced fibrosis.
Table 1.
Baseline Clinical Characteristics (N=208)
| Characteristic | Median (25th–75th%) | Actual # of subjects |
|---|---|---|
| Demographic Characteristics | ||
| Age (years) | 12.0 (11.0–14.0) | 208 |
| BMI (kg/m2) | 31.4 (27.8–35.5) | 207 |
| BMI Z-Score | 2.4 (2.1–2.6) | 208 |
| Waist/Hip Ratio | 1.00 (0.96–1.04) | 207 |
| Diabetes mellitus, n (%) | 4 (2) | 4 |
| Laboratory Measures | ||
| ALT (U/L) | 83 (61–138) | 208 |
| AST (U/L) | 51 (38–72) | 208 |
| Total cholesterol, mg/dL | 167 (142.5–187) | 207 |
| HDL cholesterol, mg/dL | 38 (32–43) | 207 |
| LDL cholesterol, mg/dL | 101 (83.3–118.8) | 206 |
| Triglycerides, mg/dL | 118 (82–157.5) | 207 |
| Serum glucose (mg/dl) | 86 (82–92) | 207 |
| Serum insulin(U/ml) | 26 (17–42) | 201 |
| HBA1c (%) | 5.3 (5.1–5.5) | 205 |
| Histologic characteristics | ||
| Steatosis, n (%) | NASH CRN Score | Number of Subjects (Percentage of total) |
| <5% | 0 | 8 (4) |
| 5%–33% | 1 | 52 (25) |
| 34%–66% | 2 | 60 (29) |
| >66% | 3 | 88 (42) |
| Lobular inflammation, n (%) | NASH CRN Score | |
| 0 | 0 | 0 (0) |
| <2 under 20x magnification | 1 | 113 (54) |
| 2–4 under 20x magnification | 2 | 81 (39) |
| >4 under 20_xmagnification | 3 | 14 (7) |
| Ballooning, n (%) | NASH CRN Score | |
| None | 0 | 123 (59) |
| Few | 1 | 54 (26) |
| Many | 2 | 31 (15) |
| NASH diagnosis, n (%) | ||
| No NASH | 48 (23) | |
| Borderline zone 3 pattern | 26 (13) | |
| Borderline zone 1 pattern | 84 (40) | |
| Yes, definite | 46 (22) | |
| Missing | 4 (2) |
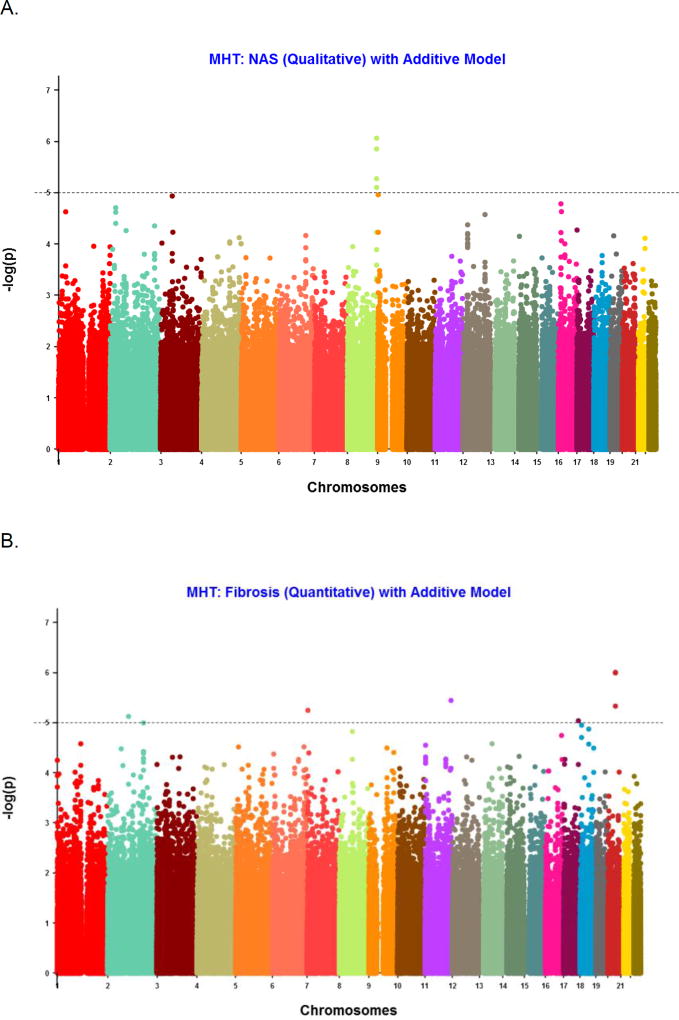
The results are depicted in the Manhattan plots (Figure 1 displaying the −log10(p-value)) for each variant on each of the 22 autosomal chromosomes for NAS and fibrosis stage. SNPs with p-value < 10−5 from the GWAS of NAS as a binary trait (NAS ≤4 to NAS >4) are shown (Table 3; available at www.jpeds.com). Under an additive genetic model, 4 SNPs had a p-value < 10−5, all located in the same region on chromosome 8 in the Trafficking Protein Particle Complex 9 (TRAPPC9) gene. The lowest p-value was observed for a variant rs11166927 (8.66 × 10−7 under an additive model (Figure 2A), and 6.06 × 10−7 with a dominant model). In addition, we found SNP rs1544175 on chromosome 1 in the Olfactory Receptor 2T4 (OR2T4) gene met the selection criterion with p-value (Pd = 9.61 × 10−6 ; Pa =1.14 × 10−4). However, given a median score of 4 (IQR 3–5), we undertook a secondary binary appropriation of NAS <3 versus ≥ 5, excluding the midrange to avoid misclassification. Two SNPs were associated with NASH (defined as a binary trait, definite vs borderline) on logistic regression: SNP rs6571631 on chromosome 14 near Rho guanine nucleotide exchange factor ARHGEF40 (p = 6.51 × 10−6) and rs6660749 on chromosome 1 near C1orf94 (8.21 ×10−6).
Figure 1.
Manhattan plots for NAS (A) and fibrosis (B). X-axis is chromosome number and y-axis is −log10(P value) for the variant association differing from the expected frequency. Each point represents a single SNP.
Figure 2.
(A) Regional plot for NAS. The −log10(P value) from association analysis is shown for all SNPs in the region around the top SNP rs11166927 for NAS. X-axis shows position of the SNPs along chromosome 8; y-axis in the left gives −log10(P value). P values were obtained by association analysis when including age and BMI Z-score as covariates and assuming an additive genetic model. Y-axis on the right shows recombination rate (cM/Mb). The r2 shows measure of the linkage disequilibrium between this SNP and target SNP. The genes TRAPPC9 and KCNK9 are located in the region as indicated at the bottom of the figure. (B) Regional plot for fibrosis. The −log10(P value) from association analysis is show for all SNPs in the region around the top SNP rs6128907 for fibrosis. X-axis shows position of the SNPs along chromosome 20; y-axis in the left gives −log10(P value). P values were obtained by association analysis when including age and BMI Z-score as covariates and assuming an additive genetic model. Y-axis on the right shows recombination rate (cM/Mb). The r2 shows measure of the linkage disequilibrium between this SNP and target SNP. The genes located in the region of ACTR5 are as indicated at the bottom of the figure.
Table 2 presents the SNPs associated with fibrosis stage. Using an additive model, seven SNPs associated with fibrosis grade, including rs11465670 on chromosome 2 in the interleukin 18 receptor accessory protein (IL18RAP) gene, rs688020 on chromosome 7 in sidekick homolog 1 (SDK1) gene, rs3935794 on chromosome 11 in v-ets erythroblastosis virus E26 oncogene homolog 1 (ETS1) gene, rs12942311 on chromosome 17 in RAB37 gene, and 3 SNPs (rs6128907, rs6124026, and rs6128918) on chromosome 20 in the ARP5 actin-related protein 5 homolog (ACTR5) gene (Figure 1B). When we tested the association under a dominant genetic model, association with SNPs rs3935794 and rs11465670 remained with p-value <10−5. In the latter analysis, we also found an association with rs17223990 on chromosome 2, rs231957 on chromosome 6 in the NADPH oxidase 3 (NOX3) gene, rs3935794 on chromosome 11 in the ETS1 gene, rs7202949 on chromosome 16, and rs454006 on chromosome 19 in the Protein Kinase C Gamma (PRKCG) gene.
Table 2.
Top SNPs from GWAS for Fibrosis in a Pediatric Cohort.
| SNP | Chr | Base pair | Gene | Minor | Major | MAF | P-value |
|---|---|---|---|---|---|---|---|
| rs6128907 | 20 | 37387862 | ACTR5 | C | T | 0.2067 | 9.918 × 10−7 |
| rs6124026 | 20 | 37399987 | ACTR5 | G | A | 0.1643 | 1.017 × 10−6 |
| rs3935794 | 11 | 128390677 | ETS1 | G | A | 0.0625 | 3.611 × 10−6 |
| rs6128918 | 20 | 37391432 | ACTR5 | A | G | 0.2067 | 4.685 × 10−6 |
| rs688020 | 7 | 4228553 | SDK1 | C | T | 0.2428 | 5.682 × 10−6 |
| rs11465670 | 2 | 103034440 | IL18RAP | C | T | 0.04327 | 7.515 × 10−6 |
| rs12942311 | 17 | 72710796 | RAB37 | C | T | 0.2212 | 9.148 × 10−6 |
To inspect associated regions in more detail, we generated regional plots that provided association analysis results along with recombination rates and all candidate genes in the region around the top SNPs (250 kilobases on each side) using the LocusZoom [21]. Most of the regional plots indicate a series of SNPs contributed to the association at each locus (Figure 2).
Generalization study for the previously published GWAS SNPs
Results in the current study were compared with results reported in our previous study of adult patients with NASH, as well as examining significant SNPs previously reported (7–14). We examined association results for 26 susceptibility loci in our Hispanic boys cohort (Table 4; available at www.jpeds.com). Two out of the 26 SNPs evaluated were found to be associated (p-value≤0.05) with NAS: rs2228603 located within the Neurocan (NCAN) gene on chromosome 19 which codes for the neurocan core protein and rs7324845, located within the intron of the lymphocyte cytosolic protein-1 (LCP1) gene on chromosome 13, encoding an actin-binding protein. Variants in NCAN had been previously associated with the existence of hepatic steatosis (9), inflammation and fibrosis (26) and variants in LCP1 were associated with NAFLD in adolescents (27).
Five out of the 26 SNPs were found to be associated with fibrosis grade when the additive model was used: rs2228603 (Pa = 0.013) encoding NCAN as aforementioned and rs2499604 (Pa = 0.0033) located on chromosome 1 and previously associated with elevated ALT values in NAFLD (10). Interestingly, variant rs5764034 encoding PNPLA3 and rs2074301 and rs2074303 encoding TM6SF2 were three of the variants associated with fibrosis, which have been previously reported in adult GWAS for NAFLD (9).
Discussion
NAFLD is largely defined by histology. We undertook this discovery GWAS to investigate potential genetic influences relating to histologic components of pediatric NAFLD in a discovery cohort of Hispanic boys with biopsy-proven NAFLD. Hispanic boys with NAFLD serve as an ideal discovery cohort given their higher propensity for NAFLD risk factors: obesity, diabetes and other aspects of the metabolic syndrome (28–31). The genetic homogeneity of this population, primarily Mexican-Americans, increases power to detect associations of interest that would require greater sample size in a more heterogeneous sample (32). Notably, alleles of patatin like phospholipase domain containing 3 (PNPLA3) with extensive prior associations with NAFLD were not associated with NAS in this study and fibrosis only at the p= 0.010 level. Likely, this is due to relative homogeneity within this cohort for the PNSPLA3 susceptibility variants.
This GWAS suggests novel associations using histologic parameters in Hispanic boys; however the threshold for these associations is variable and dependent on quality of results, potential for mechanistic association, sample size, and minor allele frequency (ours chosen to limit spurious results). Novel variants found in our study included significant variants associated with NAS and fibrosis. Specifically, SNPs within the C8orf17/ potassium channel, subfamily K, member 9 (TRAPPC9) gene locus on chromosome 8 were associated with NAS and SNPs within the ARP5 actin-related protein 5 homolog (ACTR5) gene locus on chromosome 20 were associated with fibrosis. The role of TRAPPC9 in NAFLD is unknown. Our association between 4 variants within chromosome 8 and the NAS in the intron regions of TRAPPC9 are novel. TRAPPC9 functions as an activator of NF-kappa-B through increased phosphorylation of the IKK complex(33). Mutations in TRAPPC9 have been associated with autosomal recessive mental retardation (34–36). No prior associations with NAFLD in adults or pediatric populations have been described. The role of ACTR5 is thought to be one of a chromatin remodeling complex involved in transcriptional regulation, DNA replication and probably DNA repair(37, 38). Its role in hepatic fibrosis also has not been reported.
Previous work has been done with regards to the genetic variants associated with histologic changes in children with NAFLD. In a study of 118 Italian children with histologically proven NAFLD, based on prior work that showed some hepatoprotective effects of the cannabinoid receptor in children with steatosis, a functional variant, the CB2 Q63R polymorphism, was associated with severity of inflammation (p=0.002) and the presence of NASH (p=0.02), though not associated with steatosis or fibrosis (39). Negatively associated with fibrosis (p = 0.012) was the rs13412852 TT genotype of LPIN1, independent of PNPLA3 genotype (previously mentioned as associated with hepatic steatosis) as well as clinical risk factors of age, waist circumference, hyperglycemia or ALT. This variant was also associated with a lower NAFLD activity score (NAS) severity (p= 0.026)(40). LPIN1 is involved in the metabolism of phospholipids and triacylglycerol; it is required for adipogenesis and lipid flux between adipose tissue and the liver. In the liver it also acts as an inducible transcriptional coactivator to regulate fatty acid metabolism. As opposed to the independent effects of LPIN1, in 2012, Santoro et al. reported that the rs1260326 variant in the glucokinase regulatory protein (GCKR) was associated with fat accumulation (detected by proton nuclear magnetic resonance, not histology) in the liver together with PNPLA3 to increase susceptibility in obese youths (41). All prior pediatric genetic studies highlight preselected candidate variants in pathways known to be involved in the development of NAFLD: lipid biosynthesis and metabolism or glucose metabolism.
Our study selected a group of individuals with similar ethnicity, BMI Z-score, sex and age range. This strategy serves to minimize haplotypes that would be present in a larger study of varied ethnic groups. Although this is a strength for a GWAS of this size, lack of a control group may have led to lack of association with previously reported SNPs related to NAFLD. A major limitation is the lack of validation for this discovery cohort. Other limitations include the lack of functional variants identified and an inability to determine genetic risk for Hispanics relative to non-Hispanics given the understandable lack of healthy controls. Additional clinical limitations include variation in time points between the time of biopsy and the collection of blood for DNA (range was approximately 6 months to 2 years, median 4.5 months) and the inability to control for lifestyle factors such as diet and physical activity. Generalizability to girls and adults is also limited.
This study highlights possible associations between variants and histologic findings within a well characterized disease state, but the functional implications of these variants is unknown, as is the mechanism by which these variants are implicated in the development or progression of NAFLD. Therefore, future directions should include validation cohorts of non-Hispanic children, adolescents and adults. Confirmation studies for the identified SNPs should be undertaken. By gaining a better understanding of the genetic causes of cellular changes as reflected histologically, we may better identify novel pathways of disease pathogenesis that are missed or minimized in adults, in part due to the unique pattern of borderline zone 1 histology seen in a subset of children with NAFLD. As children with NAFLD develop into adults accumulating additional environmental influences, recognizing underlying genetic propensity adds significantly to informed interventions beyond recommended lifestyle changes.
Supplementary Material
Acknowledgments
Supported by NIDDK (U01DK061734, U01DK061718, U01DK061728, U01DK061731, U01DK061732, U01DK061737, U01DK061738, U01DK061730, U01DK061713) and NICHD. Also supported by NIH CTSA awards (UL1 TR000040, UL1RR024989, UL1RR025761, M01RR00188, UL1RR024131, UL1RR025014, UL1RR031990, UL1RR025741, UL1RR029887, UL1RR24156, UL1RR025055, UL1RR031980). Genotyping supported in part by UL1 TR000124 and NIDDK DRC DK063491.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10(12):872–8. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 2.Dongiovanni P, Anstee QM, Valenti L. Genetic predisposition in NAFLD and NASH: impact on severity of liver disease and response to treatment. Curr Pharm Des. 2013;19(29):5219–38. doi: 10.2174/13816128113199990381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136(5):1585–92. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, et al. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology. 2015;149(7):1784–93. doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40(6):1387–95. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 6.Weston SR, Leyden W, Murphy R, Bass NM, Bell BP, Manos MM, et al. Racial and ethnic distribution of nonalcoholic fatty liver in persons with newly diagnosed chronic liver disease. Hepatology. 2005;41(2):372–9. doi: 10.1002/hep.20554. [DOI] [PubMed] [Google Scholar]
- 7.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naik A, Kosir R, Rozman D. Genomic aspects of NAFLD pathogenesis. Genomics. 2013;102(2):84–95. doi: 10.1016/j.ygeno.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology. 2010;139(5):1567–76. 76 e1–6. doi: 10.1053/j.gastro.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Romeo S, Sentinelli F, Cambuli VM, Incani M, Congiu T, Matta V, et al. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. J Hepatol. 2010;53(2):335–8. doi: 10.1016/j.jhep.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 12.Santoro N, Kursawe R, D'Adamo E, Dykas DJ, Zhang CK, Bale AE, et al. A common variant in the patatin-like phospholipase 3 gene (PNPLA3) is associated with fatty liver disease in obese children and adolescents. Hepatology. 2010;52(4):1281–90. doi: 10.1002/hep.23832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin YC, Chang PF, Hu FC, Yang WS, Chang MH, Ni YH. A common variant in the PNPLA3 gene is a risk factor for non-alcoholic fatty liver disease in obese Taiwanese children. J Pediatr. 2011;158(5):740–4. doi: 10.1016/j.jpeds.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Goran MI, Walker R, Le KA, Mahurkar S, Vikman S, Davis JN, et al. Effects of PNPLA3 on liver fat and metabolic profile in Hispanic children and adolescents. Diabetes. 2010;59(12):3127–30. doi: 10.2337/db10-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwimmer JB, McGreal N, Deutsch R, Finegold MJ, Lavine JE. Influence of gender, race, and ethnicity on suspected fatty liver in obese adolescents. Pediatrics. 2005;115(5):e561–5. doi: 10.1542/peds.2004-1832. [DOI] [PubMed] [Google Scholar]
- 16.Patton HM, Lavine JE, Van Natta ML, Schwimmer JB, Kleiner D, Molleston J, et al. Clinical correlates of histopathology in pediatric nonalcoholic steatohepatitis. Gastroenterology. 2008;135(6):1961–71 e2. doi: 10.1053/j.gastro.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–21. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 18.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37(5):549–54. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 19.Gunderson KL, Steemers FJ, Ren H, Ng P, Zhou L, Tsan C, et al. Whole-genome genotyping. Methods Enzymol. 2006;410:359–76. doi: 10.1016/S0076-6879(06)10017-8. [DOI] [PubMed] [Google Scholar]
- 20.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Brunt EM, Kleiner DE, Wilson LA, Belt P, Neuschwander-Tetri BA, Network NCR. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 2011;53(3):810–20. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwimmer JB, Behling C, Newbury R, Deutsch R, Nievergelt C, Schork NJ, et al. Histopathology of pediatric nonalcoholic fatty liver disease. Hepatology. 2005;42(3):641–9. doi: 10.1002/hep.20842. [DOI] [PubMed] [Google Scholar]
- 26.Gorden A, Yang R, Yerges-Armstrong LM, Ryan KA, Speliotes E, Borecki IB, et al. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75(1):34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams LA, White SW, Marsh JA, Lye SJ, Connor KL, Maganga R, et al. Association between liver-specific gene polymorphisms and their expression levels with nonalcoholic fatty liver disease. Hepatology. 2013;57(2):590–600. doi: 10.1002/hep.26184. [DOI] [PubMed] [Google Scholar]
- 28.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999–2010. JAMA. 2012;307(5):483–90. doi: 10.1001/jama.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwimmer JB, Deutsch R, Kahen T, Lavine JE, Stanley C, Behling C. Prevalence of fatty liver in children and adolescents. Pediatrics. 2006;118(4):1388–93. doi: 10.1542/peds.2006-1212. [DOI] [PubMed] [Google Scholar]
- 30.Beltran-Sanchez H, Harhay MO, Harhay MM, McElligott S. Prevalence and trends of metabolic syndrome in the adult U.S. population, 1999–2010. J Am Coll Cardiol. 2013;62(8):697–703. doi: 10.1016/j.jacc.2013.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity among adults: United States, 2011–2012. NCHS Data Brief. 2013;(131):1–8. [PubMed] [Google Scholar]
- 32.Manchia M, Cullis J, Turecki G, Rouleau GA, Uher R, Alda M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One. 2013;8(10):e76295. doi: 10.1371/journal.pone.0076295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu WH, Pendergast JS, Mo XM, Brambilla R, Bracchi-Ricard V, Li F, et al. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J Biol Chem. 2005;280(32):29233–41. doi: 10.1074/jbc.M501670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marangi G, Leuzzi V, Manti F, Lattante S, Orteschi D, Pecile V, et al. TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur J Hum Genet. 2013;21(2):229–32. doi: 10.1038/ejhg.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mir A, Kaufman L, Noor A, Motazacker MM, Jamil T, Azam M, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet. 2009;85(6):909–15. doi: 10.1016/j.ajhg.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet. 2009;85(6):897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kitayama K, Kamo M, Oma Y, Matsuda R, Uchida T, Ikura T, et al. The human actin-related protein hArp5: nucleo-cytoplasmic shuttling and involvement in DNA repair. Exp Cell Res. 2009;315(2):206–17. doi: 10.1016/j.yexcr.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 38.Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, et al. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci U S A. 2010;107(40):17274–9. doi: 10.1073/pnas.1008388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossi F, Bellini G, Alisi A, Alterio A, Maione S, Perrone L, et al. Cannabinoid receptor type 2 functional variant influences liver damage in children with non-alcoholic fatty liver disease. PLoS One. 2012;7(8):e42259. doi: 10.1371/journal.pone.0042259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valenti L, Motta BM, Alisi A, Sartorelli R, Buonaiuto G, Dongiovanni P, et al. LPIN1 rs13412852 polymorphism in pediatric nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr. 2012;54(5):588–93. doi: 10.1097/MPG.0b013e3182442a55. [DOI] [PubMed] [Google Scholar]
- 41.Santoro N, Zhang CK, Zhao H, Pakstis AJ, Kim G, Kursawe R, et al. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012;55(3):781–9. doi: 10.1002/hep.24806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.