Abstract
NLRX1 is unique among nucleotide-binding domain and leucine-rich repeat (NLR) proteins in its mitochondrial localization and capacity to negatively regulate MAVS- and STING-dependent antiviral innate immunity. However, some studies suggest a positive regulatory role for NLRX1 in inducing antiviral responses. We show that NLRX1 exerts opposing regulatory effects on virus activation of the transcription factors IRF1 and IRF3, potentially explaining these contradictory results. Whereas NLRX1 suppresses MAVS-mediated IRF3 activation, NLRX1 conversely facilitates virus-induced increases in IRF1 expression, thereby enhancing control of virus infection. NLRX1 has a minimal effect on NF-κB-mediated IRF1 transcription, and regulates IRF1 abundance post-transcriptionally by preventing translational shutdown mediated by the dsRNA-activated protein kinase PKR, thereby allowing virus-induced increases in IRF1 protein abundance.
The nucleotide-binding domain and leucine-rich repeat (NLR) family of proteins is recognized for its roles in inflammasome-mediated responses to both pathogen- and damage-associated molecular patterns (PAMPs and DAMPs)1. However, several NLR proteins, including NLRX1, NLRC3, NLRC5 and NLRP12, act as negative regulators of innate immunity with the capacity to check type I interferon (IFN) responses or NF-κB-induced pro-inflammatory cytokines2–5. NLRX1 is distinguished from other NLR members by its localization to mitochondria, where it interacts with the adaptor protein MAVS through its unique N-terminal X and nucleotide binding-oligomerization (NBD) domains, sequestering MAVS and suppressing virus-induced IFN responses mediated by the pathogen sensor RIG-I6. NLRX1 also negatively regulates lipopolysaccharide (LPS)-induced NF-κB activation, interacting with TRAF6 in unstimulated cells and being recruited to the NEMO/IKK signaling complex following LPS stimulation via its leucine-rich repeat domain3. Deletion or functionally knocking down NLRX1 results in heightened IFN responses to poly(I:C) or RNA viruses, as well as increased inflammatory responses3,6,7. Acting amazingly like a Swiss Army knife, NLRX1 also interacts with the adaptor protein STING through its NBD domain, thereby inhibiting IFN responses to DNA viruses mediated through the cGAS/cGAMP signaling pathway8. Abundant evidence thus supports a role for NLRX1 as a checkpoint inhibitor of early innate immune responses to both DNA and RNA viruses.
However, not all studies show NLRX1 exerts negative regulatory effects on innate immune responses to viruses. Sendai virus (SeV)-induced RIG-I- and MAVS-dependent phosphorylation of the transcription factor IRF3 and IFN-β and IP10 production were reported to be unchanged in Nlrx1−/− murine embryonic fibroblasts (MEFs), as were IL-6 and IFN-β responses to poly(I:C)9. Although inflammatory responses to influenza virus infection are increased in the lungs of Nlrx1−/− mice7, macrophage-mediated IFN responses were impaired secondary to enhanced apoptosis10. Overexpression of NLRX1 also enhanced, rather than inhibited, NF-κB signaling by amplifying production of reactive oxygen species (ROS) in response to several stimuli11. These findings are puzzling given that the preponderance of evidence favors a negative regulatory role for NLRX1. While they may reflect different experimental conditions, they have fueled controversy about the regulatory role of NLRX1.
Here we sought to understand how NLRX1 influences innate immune responses to virus infection of human hepatocytes. Hepatocytes are targeted for infection by several medically important viruses including hepatitis A virus (HAV) and hepatitis C virus (HCV), RNA viruses that cause inflammatory diseases of the liver12. We show that NLRX1 competes with dsRNA-activated protein kinase R (PKR) for binding to viral RNA, and that it promotes early innate immune antiviral responses by protecting NF-κB-driven increases in expression of the transcription factor IRF1 from PKR-mediated translational suppression. Hepatocytes deficient in NLRX1 expression have reduced accumulation of IRF1 but increased IRF3 dimer formation in response to virus infection, revealing opposing actions of NLRX1 on key signaling pathways. Our data unveil a novel and sophisticated regulation of early innate immune responses by NLRX1 that overall promotes immediate antiviral defense in hepatocytes.
RESULTS
NLRX1 is a positive immune regulator in hepatocytes
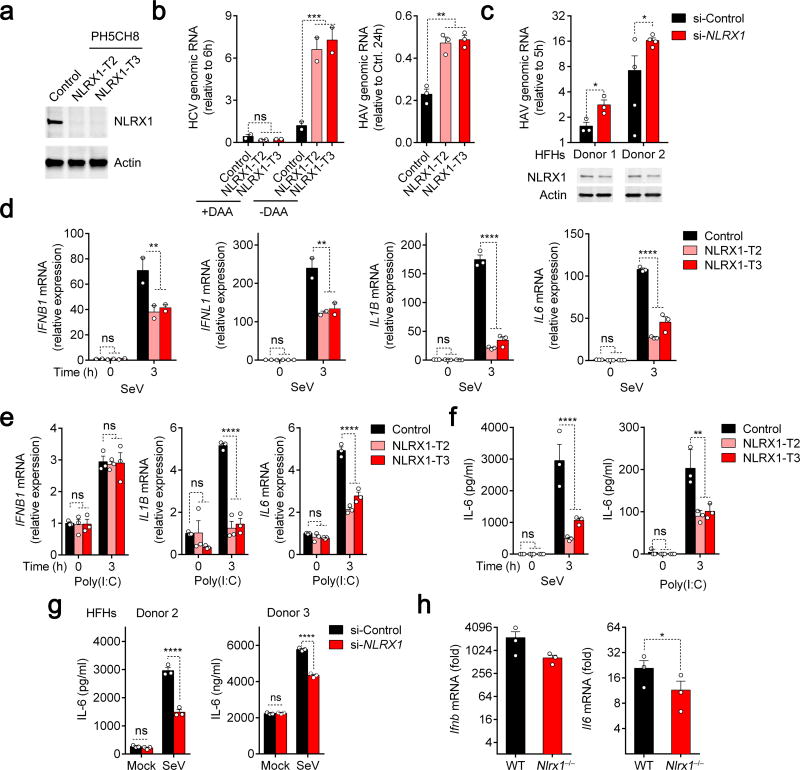
To determine how NLRX1 influences antiviral responses in hepatocytes, we used CRISPR/Cas9 gene editing to eliminate its expression in PH5CH8 cells, which are T-antigen transformed primary human hepatocytes with functional RIG-I and TLR3 signaling13,14. NLRX1 expression was detected in control cells transduced with a nontargeting (‘Scramble’) sgRNA, but not in either of two independent cell lines (NLRX1-T2 and NLRX1-T3) transduced with different NLRX1-specific sgRNAs (Fig. 1a, Supplementary Table 1). We initiated HCV replication in these cells by co-electroporating synthetic viral RNA and duplex miR-122, an essential HCV host factor lacking in PH5CH8 cells15. We demonstrated that subsequent increases in viral RNA were due to bona fide viral replication because they were blocked by a direct-acting antiviral (DAA) inhibitor (Fig. 1b, left). We also infected the cells directly with HAV16 (Fig. 1b, right). Replication of both viruses was enhanced in NLRX1-deficient cells. We similarly assessed HAV replication in primary human fetal hepatoblasts (HFHs). Partial RNAi-mediated depletion of NLRX1 boosted viral replication in cells from two donors (Fig. 1c), confirming an antiviral role for NLRX1 in human liver cells.
Figure 1.
NLRX1 is a positive regulator of innate immunity and an antiviral factor in hepatocytes. (a) Immunoblot confirmation of NLRX1 depletion in two independent PH5CH8-derived cell lines. (b) qRT-PCR of HCV and HAV genomic RNAs in NLRX1-deficient cells transfected with HCV JFH1-QL RNA (left, 24 h) or infected with HAV (right, 72 h). Data are from n=2 (left) or 3 (right) technical replicates and are representative of 3 independent experiments. (c) qRT-PCR of HAV genomic RNA in primary human fetal hepatoblasts (HFHs) with partial NLXR1 depletion at 72 h. n=3 and 4 technical replicates for donors 1 and 2, respectively. (d,e) qRT-PCR for IFNB1, IFNL1, IL1B and IL6 mRNA in NLRX1-deficient PH5CH8 cells infected with SeV (d) or stimulated with poly(I:C) added to medium for 3 h (e). Data are from 2 independent experiments with 4 technical replicates (d, IFNB1 and IFNL1), or are representative of 3 experiments, each with n=3 technical replicates (d, IL1B and IL6; e). (f,g) ELISA for IL-6 protein production in NLRX1-deficient PH5CH8 cells (f) and NLRX1-depleted HFHs (g). Data are representative of 3 experiments or donors, each with n=3 technical replicates. (h) qRT-PCR for Ifnb and Il6 mRNA in livers of Nlrx1−/− mice 3 h following hydrodynamic injection of HAV RNA (n=3 per group). For all qRT-PCR analyses, host gene expression was normalized to ACTB (human) or Actb (mouse). All data are shown as mean ± S.E.M. Unless otherwise indicated, comparisons were between control and NLRX1-depleted cells by two-way ANOVA (ns, not significant; *p < 0.05; ** p< 0.01; ***p < 0.001; ****p < 0.0001). Comparisons in (b right,c,g,h) were performed by t test (*p < 0.05).
Although innate immune responses restrict both HAV and HCV infection in PH5CH8 cells, it is difficult to document induction of antiviral cytokines in these cells following virus challenge. We thus employed a classic agonist of RIG-I signaling, SeV, to define the impact of NLRX1 deficiency on cytokine responses. We observed significant reductions in early (3 h) IFNB1, IFNL1, IL1B and IL6 mRNA responses in SeV-infected NLRX1-deficient cells (Fig. 1d). This effect was no longer evident at 8 h by which time these responses had substantially subsided (Supplementary Fig. 1a). NLRX1 deficiency also impaired IL1B and IL6, but not IFNB1 mRNA accumulation in response to the TLR3 agonist, poly(I:C), added to medium (Fig. 1e). NLRX1 deficiency consistently reduced the amount of IL-6 protein induced in response to SeV and poly(I:C) stimulation (Fig. 1f). Likewise, siRNA-mediated depletion of NLRX1 significantly impaired IL-6 protein increases induced by SeV infection of primary HFHs (Fig. 1g). Neither HAV nor HCV replicate in murine cells, but we observed an approximate 4-fold reduction in the early (3 h) intrahepatic Ifnb and Il6 mRNA responses to synthetic HAV RNA administered intravenously to Nlrx1−/− versus wild-type mice (Fig. 1h).
Supplementing PH5CH8 cells with an antioxidant (N-Acetyl-l-cysteine, NAC) did not abrogate the positive effect of NLRX1 deficiency on replication of an HCV reporter virus, nor did treating cells with inhibitors of ER stress (tauroursodeoxycholic acid, TUDCA) or autophagy (3-methyladenine, 3-MA) (Supplementary Fig. 1b). Thus, increased HCV replication in NLRX1-deficient cells is unlikely to be due to reduced ROS production11,17, or the influence of NLRX1 depletion on autophagy or the response to endoplasmic reticulum (ER) stress18,19.
To ascertain whether the proviral effect of NLRX1 deficiency results from a reduction in the early innate immune response, we designed a Transwell assay for soluble antiviral factors (Supplementary Fig. 1c, left). HCV replication was enhanced in indicator cells separated by a permeable membrane from NLRX1-deficient versus control PH5CH8 cells stimulated by a replication-competent HCV RNA (Supplementary Fig. 1c, right). Inhibition of JAK/STAT signaling with ruxolitinib (JAK1,2 inhibitor) and tofacitinib (JAK3 inhibitor) also substantially reduced the difference in HCV replication in NLRX1-T3 versus control cells (Supplementary Fig. 1d), consistent with viral replication being suppressed by cytokine responses involving JAK signaling. Depleting NLRX1 had no effect on HAV or HCV replication in RIG-I-deficient Huh-7.5 human hepatoma cells20 (Supplementary Fig. 1e), or in MAVS-deficient PH5CH8 cells (Supplementary Fig. 1f). Collectively, these results define NLRX1 as a positive regulator of soluble RIG-I- and MAVS-mediated antiviral responses in human hepatocytes.
NLRX1 differentially regulates IRF3 and IRF1 responses
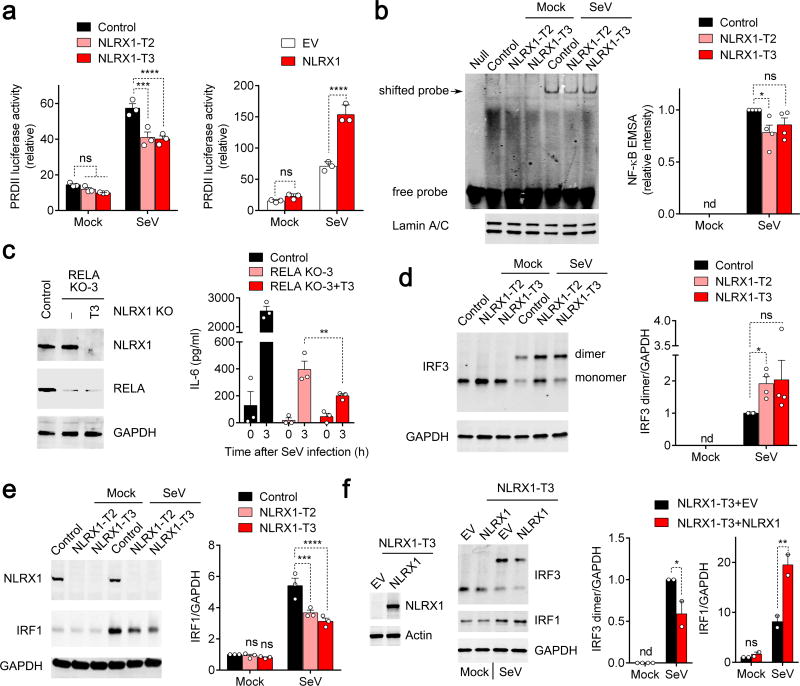
We carried out a series of dual luciferase promoter assays to determine if NLRX1 directly regulates cytokine gene transcription. NLRX1 deficiency had little effect on basal NF-κB (PRDII) promoter activity, but modestly reduced its activation by SeV (Fig. 2a, left). Consistent with this, NLRX1 overexpression enhanced PRDII activation triggered by SeV (Fig. 2a, right). In contrast, NLRX1 deficiency minimally affected activation of the IFNB1 promoter, and had no effect on an IRF3-responsive promoter (4*PRD(I/III)) (Supplementary Fig. 2a,b). We also examined the impact of NLRX1 deficiency on stability of IL6 mRNA, as IL-6 expression is regulated in part through 3′ untranslated RNA (3′UTR) sequences programmed for rapid mRNA turnover21. NLRX1 deficiency had no effect on IL6 mRNA decay in cells treated with actinomycin D (Supplementary Fig. 2c), nor did NLRX1 overexpression alter luciferase expression from mRNA transcripts containing the IL6 3′UTR (Supplementary Fig. 2d). In aggregate, these data suggest that NLRX1 has a positive but limited effect on SeV activation of NF-κB-responsive promoters, and no influence on IL6 message stability.
Figure 2.
Impact of NLRX1 deficiency on NF-κB signaling and IRF1 and IRF3 activation in SeV-infected PH5CH8 cells. (a) PRDII-Luc promoter activation in cells with NLRX1 depletion (left) or overexpression (right). Data are from n=3 technical replicates and are representative of 3 independent experiments. (b) NF-κB electrophoretic mobility shift assay (EMSA) with nuclear extracts from mock- and SeV-infected NLRX1-deficient PH5CH8 cells (left). Mean infrared fluoresence intensity measurements from 4 independent EMSA experiments (right). (c) Effects of NLRX1 depletion in the absence of RELA. Immunoblots for NLRX1 and RELA in NLRX1-RELA double-deficient (RELA KO+NLRX1-T3) PH5CH8 cells (left). ELISA quantitation of IL-6 protein abundance in SeV-infected (3 h) control, RELA-deficient and NLRX1-RELA double-deficient cells (right). Results are representative of 2 independent experiments, each with 3 technical repeats. (d,e) Immunoblots showing SeV-triggered IRF3 dimerization (d) and IRF1 expression in NLRX1-deficient cells (e). n=3–4 independent experiments. (f) SeV-induced IRF3 dimerization and increased IRF1 protein expression in NLRX1-reconstituted NLRX1-T3 cells. Data are representative of 2 independent experiments. All data are shown as mean ± S.E.M. Comparisons of control versus NLRX1-depleted cells in panels (a,e) and (b-d, f) were made by two-way ANOVA and t test, respectively. ns = nonsignificant, *p< 0.05; **p < 0.01; ***p< 0.001; ****p< 0.0001.
To more directly examine the influence of NLRX1 deficiency on NF-κB activation, we examined SeV-induced RELA signaling. NLRX1 deficiency minimally affected NFKBIA (IκBα) or RELA phosphorylation in SeV-infected PH5CH8 cells (Supplementary Fig. 2e). Electrophoretic mobility-shift assays (EMSA) with an NF-κB probe revealed a significant, but quantitatively small decrease in the intensity of SeV-induced band shifts in NLRX1-deficient versus control PH5CH8 cells (Fig. 2b). We conclude from these data that NLRX1 deficiency minimally suppresses SeV-induced NF-κB signaling in PH5CH8 cells, and that this subtle impairment of NF-κB signaling is unlikely to explain the marked reductions we observed in cytokine expression.
To further test this hypothesis, we assessed how depleting RELA influences IL-6 production upon SeV infection and the modulation of IL-6 expression by NLRX1 in PH5CH8 cells. As anticipated, RELA deficiency caused a large decrease in SeV-induced IL-6 protein expression (Fig. 2c). Importantly, however, eliminating NLRX1 expression resulted in a further reduction in IL-6 production in RELA-deficient cells (Fig. 2c). Thus, the negative impact of NLRX1 deficiency on the IL-6 response that we observed in RELA-replete cells is preserved in RELA-deficient cells. Taken collectively, these results indicate that NLRX1 regulates IL-6 production downstream and independently of NF-κB.
Although our data show IL-6 is strongly regulated by NF-κB in PH5CH8 cells (Fig. 2c), CRISPR/Cas9 depletion of either of two IRF family members, IRF1 and IRF3, suppressed early (3 h) IL1B and IL6 mRNA and IL-6 protein responses to SeV (Supplementary Fig. 2f–h). Because these transcription factors regulate the induction of IFNB1 and IFNL1 transcripts in human hepatoma cells22, we examined the impact of NLRX1 deficiency on IRF3 and IRF1 responses in PH5CH8 cells. IRF3 is constitutively expressed, and activated by phosphorylation leading to its dimerization and cytoplasmic-nuclear translocation23. NLRX1 deficiency significantly enhanced this response, leading to an increase in SeV-induced IRF3 dimer formation (Fig. 2d). In contrast, basal IRF1 protein expression is low, but is strongly induced by SeV infection. NLRX1 deficiency substantially reduced SeV-triggered increases in IRF1 abundance (Fig. 2e). Reconstituting NLRX1 expression in NLRX1-T3 cells with recombinant lentivirus reversed both of these changes (Fig. 2f). Collectively, these data reveal that NLRX1 differentially regulates SeV-induced IRF3 and IRF1 signaling in hepatocytes, suppressing the activation of IRF3 but enhancing increases in IRF1 expression.
NLRX1 promotion of IRF1 dominates in hepatocytes
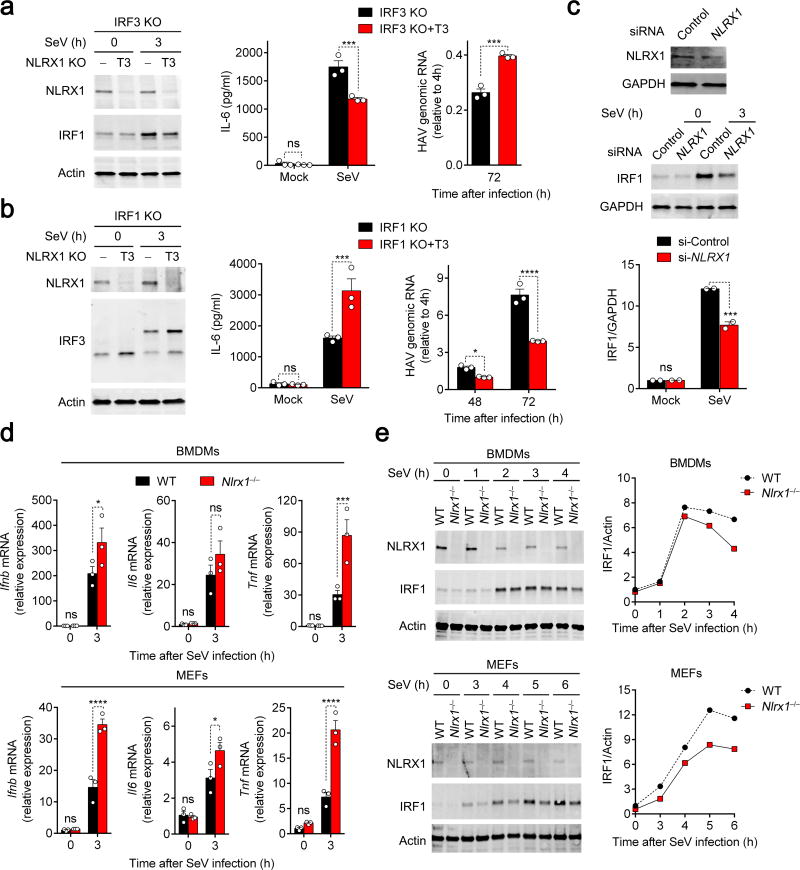
Because NLRX1 deficiency suppresses innate immune control of viral replication in hepatocytes, we reasoned that the positive regulation of IRF1 by NLRX1, rather than the negative regulation of IRF3, is likely to dominate cytokine responses in hepatocytes. To test this hypothesis, we depleted NLRX1 expression in IRF1- and IRF3-deficient PH5CH8 cells, and examined responses to SeV challenge in the resulting NLRX1-IRF3 and NLRX1-IRF1 double-deficient cells. In IRF3-deficient cells, IRF1 and IL-6 protein responses to SeV infection were suppressed and HAV replication was enhanced by depleting NLRX1 (Fig. 3a). Thus, the impact of NLRX1 depletion was similar in IRF3-deficient and wild-type PH5CH8 cells. In contrast, depleting NLRX1 in IRF1-deficient cells enhanced IRF3 dimer formation (as in IRF1-replete cells), but enhanced SeV-induced IL-6 production and also suppressed HAV replication (Fig. 3b). Thus, the effects of NLRX1 depletion on cytokine expression and viral replication were reversed in IRF1-deficient cells, suggesting that the NLRX1-IRF1 signaling axis is dominant in hepatocytes. Consistent with this conclusion, reduced IL-6 production in SeV-infected NLRX1-depleted HFHs (Fig. 1g) was accompanied by reductions in IRF1 protein accumulation (Fig. 3c).
Figure 3.
NLRX1/IRF1 signaling dominates the antiviral cytokine response in hepatocytes. (a,b) ELISA measurements of SeV-induced IL-6 production and qRT-PCR quantitation of HAV replication in IRF3-deficient and IRF3-NLRX1 double-deficient PH5CH8 cells (a), or IRF1-deficient and IRF1-NLRX1 double-deficient cells (b). Panels on the left show immunoblots for NLRX1 and IRF1 (a) or IRF3 dimerization at 3 h (b). See Supplementary Fig. 2f for immunoblots of IRF1-deficient and IRF3-deficient cells. Results are representative of 2–3 independent experiments, each with 3 technical repeats. (c) Immunoblots of NLRX1 and IRF1 in partial NLRX1-depleted HFHs infected with SeV for 3 h. Bottom panel shows quantification of immunoblots (infrared fluorescence intensities) from 2 independent experiments using cells from different donors. (d) qRT-PCR for Ifnb, Il6 and Tnf mRNA and (e) immunoblots of IRF1 in SeV-infected wild-type versus Nlrx1−/− BMDMs (top panels) and MEFs (bottom panels). Data are from n=3 technical replicates and are representative of 4 (BMDMs) and 2 (MEFs) independent experiments. All immunoblots are representative of 2–3 independent experiments or donors. Data are shown as mean ± S.E.M. ns = nonsignificant, *p< 0.05; **p < 0.01; ***p< 0.001; ****p< 0.0001 by t test.
Since most reports suggest NLRX1 suppresses, rather than enhances early innate immune responses in bone marrow-derived macrophages (BMDMs) and primary MEFs6–8, we sought to determine whether IRF1 signaling is negatively or positively regulated by NLRX1 in these cell types. Consistent with a suppressive effect on innate immunity, we found that loss of NLRX1 enhanced SeV-mediated Ifnb, Tnf, and Il6 mRNA responses in murine BMDMs and MEFs (Fig. 3d). Despite this, NLRX1 deficiency reduced SeV-triggered increases in IRF1 protein abundance in both cell types (Fig. 3e). Thus, the positive regulation of IRF1 signaling by NLRX1 is not specific to hepatocytes or human cells. The fact that cytokine responses were enhanced, while IRF1 induction was decreased in NLRX1-deleted BMDMs and MEFs, indicates that IRF1 does not play a dominant role in determining cytokine responses in these cells. Thus, the ultimate impact of NLRX1 on early antiviral responses is determined by whether IRF1 or IRF3 dominates in driving the response.
NLRX1 enhances IRF1 protein synthesis in infected cells
IRF1 is the founding member of the IRF family, and its expression is induced rapidly by viral infection24,25. However, the mechanisms mediating this response are poorly understood. Given its role in initiating IRF3-directed responses to SeV in hepatocytes20, we reasoned that RIG-I, and its adaptor, MAVS, may mediate the IRF1 response. Consistent with this notion, depleting MAVS significantly suppressed both IRF3 and IRF1 responses to SeV infection in PH5CH8 cells (Supplementary Fig. 3a). This loss of SeV-induced IRF3 dimerization and IRF1 protein expression was not reversed by additionally depleting NLRX1 in MAVS-deficient PH5CH8 cells (Supplementary Fig. 3a).
Consistent with previous studies showing that increased IRF1 transcription drives the IRF1 protein response to viral infection25, globally inhibiting transcription with actinomycin D completely abolished the IRF1 protein response to SeV (Supplementary Fig. 3b). To better understand how NLRX1 might regulate this IRF1 response, we assessed the roles of IRF3 and NF-κB. Importantly, IRF3 deletion had no impact on either IRF1 transcription or IRF1 protein expression (Supplementary Fig. 3c). In contrast, deleting NF-κB RELA significantly suppressed both IRF1 transcription and the increase in IRF1 protein expression in response to SeV in PH5CH8 cells (Supplementary Fig. 3d). siRNA-mediated silencing of NFKB1, which encodes NF-κB p50, modestly reduced the IRF1 protein response to SeV infection, and in combination with RELA deficiency severely reduced SeV-induced IRF1 transcript abundance (Supplementary Fig. 3e,f). Collectively, these data demonstrate that the immediate (3 h) increase in IRF1 protein expression in response to SeV infection is driven by MAVS and NF-κB signaling in PH5CH8 cells.
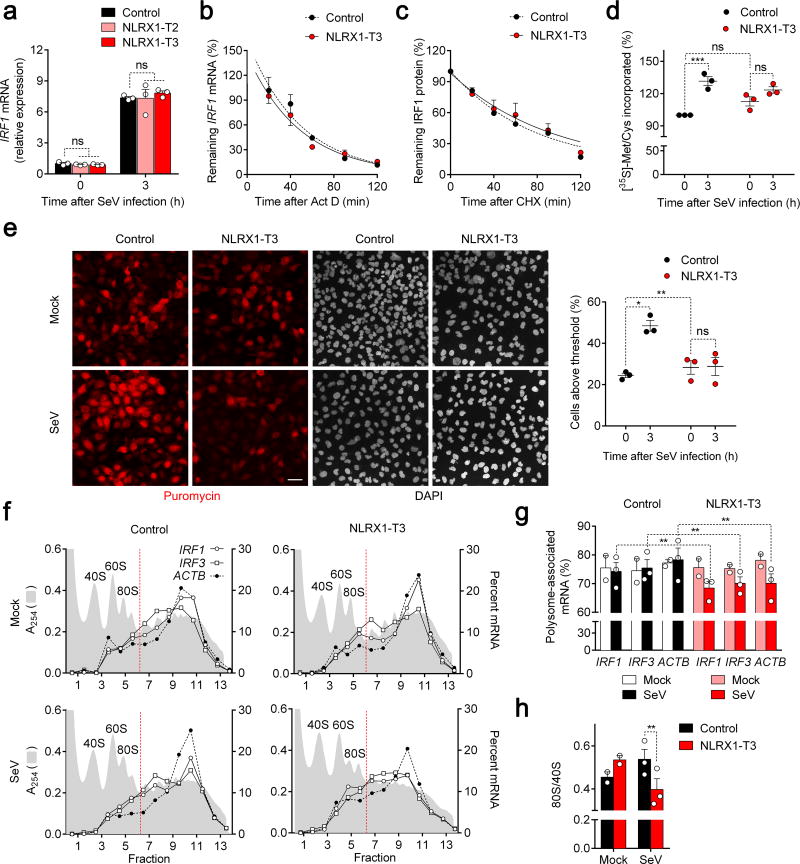
Importantly, although NLRX1 deficiency modestly enhanced SeV-induced IRF3 transcript abundance (Supplementary Fig. 4a), it had no effect on either IRF1 transcript abundance (Fig. 4a) or IRF1 mRNA stability (Fig. 4b). These data are in agreement with our earlier observations indicating only a modest impairment of NF-κB signaling in NLRX1-deficient cells, and collectively indicate that NLRX1 regulates IRF1 expression post-transcriptionally. To gain insight into whether this negative regulation of the IRF1 response was due to enhanced IRF1 degradation or reduced IRF1 protein synthesis, we globally inhibited protein synthesis in SeV-infected cells with cycloheximide (CHX) and monitored the subsequent decay of IRF1 protein. Despite a greater initial abundance of IRF1 in control PH5CH8 cells, the half-life (stability) of IRF1 protein was indistinguishable in NLRX1-deficient cells (Fig. 4c, Supplementary Fig. 4b). Taken collectively, these data suggest that the reduced IRF1 protein abundance in NLRX1-deficient cells results from impaired translation of IRF1 mRNA. Consistent with this, globally blocking protein synthesis with puromycin completely abolished the impact of NLRX1 deficiency on the IRF1 response to SeV infection (Supplementary Fig. 4c).
Figure 4.
NLRX1 facilitates immediate IRF1 antiviral responses by promoting global protein synthesis in SeV-infected cells. (a) qRT-PCR of SeV-induced IRF1 mRNA responses in control and NLRX1-deficient PH5CH8 cells. (n=3). (b) IRF1 mRNA stability in actinomycin D (Act D)-treated SeV-infected control vs. NLRX1-T3 PH5CH8 cells. (n=3). (c) IRF1 protein stability in cycloheximide (CHX)-treated cells following SeV infection assessed by quantitation of immunoblots. (n=6). See Supplementary Fig. 4b for representative immunoblots. (d) Nascent protein synthesis monitored by [35S]-Met/Cys incorporation in mock- versus SeV-infected control and NLRX1-T3 cells. [35S] incorporated into TCA-precipitated protein is shown relative to that in uninfected control cells (100%). Data are from n=6 technical replicates from three experiments. See Supplementary Fig. 4d for representative immunoblots. (e) Global protein synthesis imaged by confocal microscopy in mock- versus SeV-infected control and NLRX1-T3 cells. Puromycin was detected by staining with specific antibody, and nuclei counterstained with DAPI. Scale bar, 40 µm. The percentage of cells with puromycin labeling exceeding an arbitrary threshold in 3 experiments is shown on the right. An average of 176 cells were evaluated for each condition. (f) Representative polysome profiles of IRF1, IRF3 and ACTB mRNAs in mock- versus SeV-infected control and NLRX1-T3 cells. Absorbance at 254 nm (A254) is aligned with qRT-PCR quantitation of mRNA, plotted as percent of total mRNA in each fraction. (g) Proportion of IRF1, IRF3 and ACTB mRNAs associated with translationally-active polysomes (fractions 7–14) in SeV- (n=3) and mock-infected (n=2) control versus NLRX1-T3 cells. (h) 80S/40S ratios determined from the area under the curve (AUC) of 40S and 80S peaks of A254 traces in 2–3 experiments. Data are shown as mean ± S.E.M. ns = nonsignificant, *p< 0.05; **p < 0.01; ***p< 0.001 by two-way ANOVA (a,d,e,g) or t test (h).
To determine how NLRX1 might influence protein synthesis, we measured the incorporation of [35S]-Met/Cys into protein. Protein synthesis was qualitatively similar in NLRX1-T3 and control cells, both before and after SeV infection (Supplementary Fig. 4d). However, whereas SeV infection induced a significant, quantitative increase in protein synthesis in control cells, it had a much reduced, nonsignificant effect in NLRX1-T3 cells (Fig. 4d). The absence of any novel protein bands incorporating [35S]-Met/Cys (Supplementary Fig. 4d) indicated that this increase was due to enhanced cellular protein synthesis, not viral protein synthesis. As an independent measure of protein synthesis, we pulse-labeled NLRX1-T3 cells with a low concentration of puromycin, and used confocal microscopy to monitor its incorporation into nascent protein at a single-cell level (Fig 4e, left). SeV infection induced an increase in nascent protein synthesis in a large proportion of control cells, but not in NLRX1-T3 cells (Fig. 4e). Similar results in NLRX1-T2 cells, at both a whole cell culture and single-cell level of analysis, excluded the possibility that this reflects an off-target effect of NLRX1-T3 sgRNA (Supplementary Fig. 4e). siRNA-mediated depletion of NLRX1 expression in primary HFHs also substantially suppressed SeV-induced increases in nascent protein synthesis (Supplementary Fig. 4f).
Although NLRX1 exerts distinctly different regulatory effects on SeV-induced cytokine expression in MEFs versus human hepatocytes, depleting NLRX1expression reduced SeV-induced increases in IRF1 abundance in both cell types. Consistent with this finding, puromycin incorporation into nascent protein was also significantly reduced in SeV-infected MEFs from Nlrx1−/− versus wild-type mice (Supplementary Fig. 4g). Thus, the negative impact of NLRX1 deficiency on protein synthesis in virus-infected cells is not limited to human hepatocytes.
To directly characterize the impact of NLRX1 deficiency on mRNA translation by ribosomes, we used sucrose density gradient fractionation to profile polysome formation following SeV infection of NLRX1-T3 versus control cells (Fig. 4f). NLRX1 deletion resulted in a significant shift in the gradient distribution of IRF1, IRF3 and ACTB mRNAs from SeV-infected cells, with a substantially lower proportion of each of the mRNAs associated with translationally-active polysomes (fractions 7–13) (Fig. 4g). These differences were statistically significant in replicate independent experiments. Moreover, the formation of translationally competent 80S ribosomes was reduced in SeV-infected NLRX1-T3 versus control cells, as evidenced by a sharp reduction in the 80S/40S subunit ratio (Fig. 4h, Supplementary Fig. 4h), suggesting a defect in initiation of translation. Collectively, these three separate measures of protein synthesis, [35S]-Met/Cys incorporation, puromycin labeling, and polysome profiling, indicate that mRNA translation is globally suppressed by the absence of NLRX1 expression in SeV-infected cells.
NLRX1 limits PKR-mediated global translational shutdown
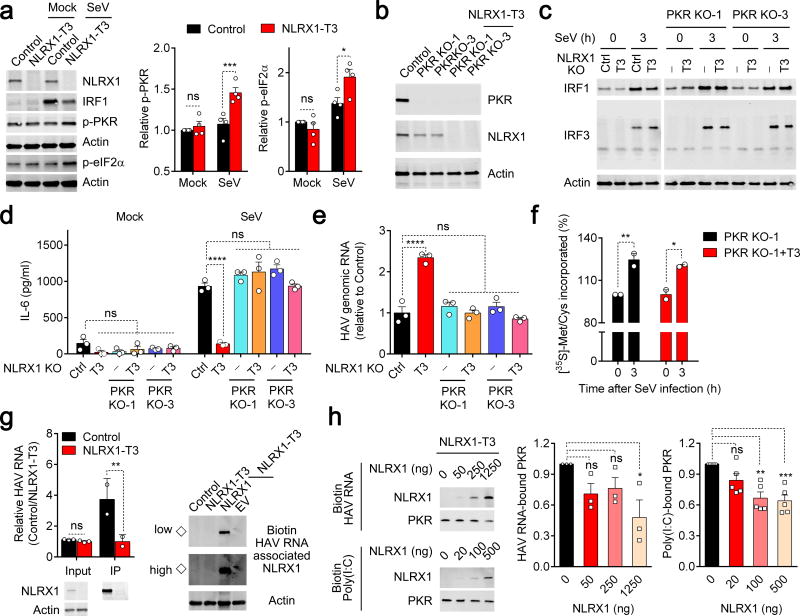
The double-stranded RNA-activated kinase PKR is triggered by virus infection to phosphorylate eIF2α, thereby globally shutting down host cell translation26. To ascertain whether NLRX1 influences this PKR response, we characterized PKR activation in NLRX1-deficient cells. NLRX1 deficiency enhanced SeV-induced PKR autophosphorylation, as well as phosphorylation of the PKR substrate, eIF2α (Fig. 5a, Supplementary Fig. 5a). To determine whether NLRX1 facilitates virus-induced increases in IRF1 protein synthesis by suppressing PKR responses, we generated NLRX1-PKR double-deficient PH5CH8 cells (Fig. 5b). The negative impact of NLRX1 deletion on SeV-triggered IRF1 responses was completely abolished by PKR deficiency (Fig. 5c). PKR deficiency also fully restored the SeV-triggered immediate IL-6 protein response (Fig. 5d), and abrogated the beneficial effect conferred on HAV replication by NLRX1 deficiency (Fig. 5e). We also assessed the impact of SeV infection on nascent protein synthesis in NLRX1-PKR double-deficient cells. In contrast to the lack of an increase in [35S]-Met/Cys or puromycin incorporation in NLRX1-deficient cells, protein synthesis was increased in response to SeV infection in both NLRX1-PKR double-deficient cells and PKR-deficient cells (Fig. 5f, Supplementary Fig. 5b).
Figure 5.
NLRX1 reduces PKR activation and subsequent inhibition of protein synthesis by competing for viral RNA binding. (a) Immunoblots of PKR-eIF2α signaling in NLRX1-deficient PH5CH8 cells 4.5 h after SeV infection. Quantification (infrared fluorescence intensity) of p-PKR or p-eIF2α was normalized to actin (n=4 experiments). (b) Immunoblots of PKR and NLRX1 in PKR-deficient and NLRX1-PKR double-deficient PH5CH8 cells. (c) Immunoblots of SeV-induced IRF1 expression and IRF3 dimerization in NLRX1-deficient and NLRX1-PKR double-deficient PH5CH8 cells. The image was generated from 2 separate gels in one experiment; data are representative of 2 experiments. (d,e) ELISA for SeV-triggered IL-6 protein production (d) and qRT-PCR for HAV RNA (e) in control, NLRX1-deficient and NLRX1-PKR double-deficient PH5CH8 cells. Data are representative of 2–3 experiments, each with 3 technical replicates. (f) [35S]-Met/Cys incorporation as measurement of global protein synthesis in SeV- versus mock-infected NLRX1-PKR double-deficient PH5CH8 cells. Data are from 4 technical replicates of 2 experiments. (g) Bidirectional assessment of NLRX1-HAV RNA association. (left) qRT-PCR of HAV RNA co-immunoprecipitating with NLRX1. HAV RNA was transfected as in Fig. 1b, followed by immunoprecipitation with anti-NLRX1 at 3 h. Immunoblots of input and immunoprecipitated (IP) NLRX1 are shown at the bottom. Data are representative of 2 experiments (n=2–3). (right) Immunoblots of NLRX1 co-precipitating with biotin-tagged HAV RNA in cell lysates. Immunoblots are representative of 3 experiments (◇ indicates endogenous NLRX1 associated with HAV RNA). “High” and “Low” indicate detection intensity. (h) Impact of NLRX1 reconstitution on PKR association with biotin-tagged HAV RNA or poly(I:C) in NLRX1-T3 cell lysates. Immunoblots are representative of 3 (HAV) or 5 [poly(I:C)] experiments. Quantification of PKR binding is summarized on the right. Data are shown as mean ± S.E.M. ns = nonsignificant, *p< 0.05; **p < 0.01; ***p< 0.001 by two-way ANOVA (a,d,f,g) and one-way ANOVA (e,h).
To explore the mechanism by which NLRX1 suppresses PKR responses, we investigated whether NLRX1 physically interacts with PKR in hepatocytes. However, no co-immunoprecipitation of PKR protein was detectable with either endogenous or overexpressed NLRX1 in PH5CH8 cells (Supplementary Fig. 5c,d). Since NLRX1 binds both viral RNA and poly(I:C), a common surrogate for viral dsRNA27,28, we reasoned that NLRX1 might suppress PKR activation by competing for viral RNA in infected cells. To test this hypothesis, we first confirmed that NLRX1 binds genomic HAV RNA, which is known to have extensive secondary structure29, in bidirectional pull down assays (Fig. 5g). Next, we designed a cell-free competition assay in which purified recombinant NLRX1 protein and biotin-tagged HAV RNA or poly(I:C) were added to lysates of NLRX1-deficient cells as a source of PKR. The addition of increasing amounts of recombinant NLRX1 protein led to a progressive reduction in PKR binding to viral RNA and poly(I:C) (Fig. 5h), indicating that NLRX1 competes with PKR for binding to viral RNA. Taken together, these results demonstrate that NLRX1 suppresses PKR activation, in part by competitively binding viral RNA, thereby preventing PKR-mediated translational shutdown that would otherwise attenuate IRF1-mediated antiviral responses (Supplementary Fig. 5e).
DISCUSSION
Although NLRX1 has been reported to be a negative regulator of innate immunity3,6,8,30, others have not confirmed this9,31 or even suggested that NLRX1 enhances antiviral responses10. Consistent with the latter, we found NLRX1 to be an antiviral factor and a positive regulator of early innate immune responses in human hepatocytes. HAV and HCV, both common causes of viral hepatitis in humans, are unrelated, positive-strand hepatotropic RNA viruses that share many similarities in their replication cycles and in their interactions with innate immunity12. Both produce double-stranded RNA replication intermediates that induce RIG-I and TLR3 signaling, thereby activating NF-κB and IRF family members leading to antiviral defense12,32. To counter these host defenses, both HAV and HCV express proteases that target for degradation MAVS and TRIF, key adaptor proteins in the RIG-I and TLR3 signaling pathways33–36. Adding to these similarities, we show here that NLRX1 acts to restrict both HAV and HCV replication.
Surprisingly, we observed that NLRX1 had clear, opposing effects on SeV-induced activation of two members of the IRF family. NLXR1 was required for maximum increases in IRF1 abundance, yet suppressed IRF3 dimerization.As key factors in inducing cytokine transcription, IRF family members are triggered by various stimuli in all cell types. Whereas IRF3 and IRF7 are activated by virus infection in most cell types37,38, other family members, including IRF1 as well as IRF5, 8 and 9, are selectively activated by different stimuli in a cell-type dependent manner39–44. This complexity in IRF activation provides the diversity of responses required for effective host defense. However, it also allows for a single regulatory molecule, such as NLRX1, to have different functional consequences when expressed in different cell types or pathogen contexts or, potentially, at different points of time in the response to an invading virus. The opposing regulatory activities of NLRX1 on early IRF3 and IRF1 responses may account for much of the controversy surrounding this protein7,8,10. We found NLRX1 positively regulates IRF1 signaling in both BMDMs and primary MEFs. Yet, NLRX1 is known to attenuate innate immune control of viral replication in these cell types6–8. We infer from this that IRF1 does not play a dominant role in determining the outcome of infection in these cells, at least not under the conditions tested, whereas our data show it does in hepatocytes. Thus, whether NLRX1 functions as a pro-viral or antiviral factor, or neither9,31, likely reflects which IRFs dominate in inducing antiviral responses.
Although previous studies indicate NLRX1 negatively regulates LPS/TLR4-triggered NF-κB signaling in macrophages and HEK293T cells3,7, we found that NLRX1 has a slight, positive effect on RIG-I-mediated NF-κB signaling in hepatocytes. However, this effect is not critical for the promotion of early cytokine responses by NLRX1, as our data show that NLRX1 regulates the IL6 response downstream of the influence of NF-κB.
Negative regulation of MAVS-mediated IRF3 responses by NLRX1 has been described previously6, but how NLRX1 impacts IRF1 responses has not been studied. We found that depleting NLRX1 did not alter SeV-induced IRF1 mRNA transcription in PH5CH8 cells, but it significantly reduced the synthesis of IRF1 protein. Reductions in protein synthesis were not specific to IRF1, but rather reflected a global shutdown of protein synthesis due to enhanced PKR activation, a phenomenon that would likely have its greatest impact on proteins that turn over rapidly, such as IRF1 that has a half-life of about 60 min.
Emerging evidence is increasingly linking translational control to regulation of immediate innate immune responses. Both IRF7 and IRF8 appear to be controlled via global, eIF4E-dependent regulation of translation45–47. PKR restricts protein synthesis globally by phosphorylating eIF2α, thereby blocking the production of virus48. However, PKR activation also reduces the synthesis of effector proteins encoded by interferon-stimulated genes49, and, as we show here, the production of IRF1 driven by an early, NF-κB-mediated response to SeV. PKR is thus a double-edged sword, potentially weakening the cell’s antiviral defenses as well as presenting a hurdle to be overcome by the virus. The negative impact of PKR activation on host defense may be particularly important when cells are infected with viruses such as HCV that can continue to translate their RNAs despite PKR activation49. Our data suggest NLRX1 moderates the potentially negative consequences of PKR activation by competing with it for binding to viral RNA. This is distinct from how other NLR family members, including NLRP3, NLRP1 and NLRC4, physically interact with PKR to maximize inflammasome activation50.
We speculate that the opposing actions of NLRX1 on early IRF1 and IRF3 antiviral responses may underlie much of the controversy surrounding the biological function of this NLR family member. Loss of NLRX1 expression may result in contrary effects on outcome of infections by different viruses in different tissues, depending on which IRF dominates in driving the antiviral response.
METHODS
Detailed information concerning experimental design and reagents used in this study can be found in the Life Sciences Reporting Summary that accompanies this publication.
Reagents and antibodies
Puromycin and blasticidin were from Invitrogen. 3-methyladenine (3-MA), tauroursodeoxycholic acid (TUDCA), N-acetyl-L-cysteine (NAC), ruxolitinib, tofacitinib, cycloheximide and actinomycin-D were from Sigma-Aldrich. PSI-7977 (Sofosbuvir) was from Chemscene. Poly(I:C) (high molecular weight, InvivoGen) was either added to cell culture supernatants at a final concentration of 50 µg/ml, or transfected at a concentration of 2 µg/ml.
Antibodies used in this study included anti-NLRX1 (1:5,000 dilution; this antibody was described previously6); anti-IRF1 (1:1,000 dilution; clone D5E4, #8478), anti-phospho-NF-κB p65 (anti-p-RELA; 1:1,000 dilution; clone 93H1, #3033), anti-NF-κB p65 (anti-RELA; 1:1,000 dilution; clone D14E12, #8242), anti-p50 (1:1,000 dilution; clone 5D10D11, #13681), anti-phospho-IκBα (1:500 dilution; clone 14D4, #2859), anti-IκBα (1:1,000 dilution; clone L35A5, #4814), anti-PKR (1:1,1000 dilution; clone D7F7, #12297) and anti-phospho-eIF2α (1:1,000 dilution; clone S51, #9721) from Cell Signaling; anti-IRF3 (1:500 dilution; clone FL-425, sc-9082), anti-eIF2α (1:500 dilution; clone FL-315, sc-11386) and anti-Lamin A/C (1:1,000 dilution; clone N-18, sc-6215) from Santa Cruz; anti-phospho-PKR (1:1,000 dilution; clone E120, ab32036) and mouse IgG1 (ab18448) from Abcam; anti-Cardif (anti-MAVS; 1:2,000 dilution; clone AT107, #ALX-210-929) from Enzo Life Sciences; anti-actin (1:40,000 dilution; clone AC-74, A2228) from Sigma, and anti-GAPDH (1:50,000 dilution; AM4300) from Ambion; anti-puromycin (1: 20,000 dilution for immunoblot and 1:10,000 dilution for immunofluorescence; clone 12D10, MABE343) from EMD Millipore. Goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody (1:300 dilution; A-11001) was from ThermoFisher. IRDye 680 or 800 secondary antibodies included #926-32211, #926-32212, #926-32214, #926-68020 and #926-68073 (1:12,000) from LI-COR Biosciences.
Cells
Tissues for preparation of fetal human liver cells were from random donors, and were provided by the accredited nonprofit corporation Advanced Biosciences Resources, Inc. (ABR) and obtained from fetuses between 19–21 weeks gestation during elective termination of pregnancy. Tissues were collected with written informed consent from all donors and in accordance with the US Food and Drug Administration CFR Part 1271 Good Tissue Practices regulations. Tissue processing, and hepatoblast isolation and culture were as previously described51. The use of commercially procured fetal liver cells was reviewed by the University of North Carolina at Chapel Hill Office of Human Research Ethics, and was determined to be exempt from review by the University of North Carolina at Chapel Hill Institutional Review Board.
293FT cells and human hepatocyte-derived cell lines including Huh-7.552 and PH5CH813 were mycoplasma-free and cultured as previously described51. Murine bone marrow-derived macrophages (BMDMs) and mouse embryo fibroblasts (MEFs) were generated and cultured as previously described8.
Mice and HAV RNA inoculation
Age- and sex-matched wild-type and Nlrx1−/− mice were previously described7. Animals were stratified by sex prior to randomization. Experiments were performed in a non-blinded fashion in accordance with the NIH Guide for the Care and Use of Laboratory Animals and with the approval of the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill. Thirteen-week-old mice were inoculated intravenously with PBS or 15 µg in vitro-transcribed HAV RNA in a volume of PBS equivalent to ~5% body weight. Mice were sacrificed 3 h after injection, and liver tissue collected for analysis of cytokine mRNAs by qRT-PCR.
Virus
Sendai virus (SeV, Cantell strain) was obtained from Charles River Laboratory. Cultures were exposed to SeV at a concentration of 600 HA units/ml. High-titer HAV (HM175/18f strain53) stock was prepared as described previously54. HAV infection was carried out at m.o.i = 10 for PH5CH8 cells and m.o.i = 1 for Huh-7.5 cells. Infectious molecular clones for HCV (pJFH1-QL, containing the cell culture–adaptive mutation Q221L in the NS3 helicase)55, a reporter virus expressing Gaussia princeps luciferase (pJFH1-QL/GLuc)51, and HAV (pT7-18f)56 were described previously.
Plasmids and oligonucleotides
Firefly luciferase reporter vectors including pIFNB1-Luc, pIRF3-responsive (4*PRD(I/III))-Luc and pPRDII-Luc as well as the Renilla luciferase control reporter vector, pRL-TK, were described previously57. The IL6 3′UTR construct was kindly provided by B. Glaunsinger (University of California, Berkeley), and was sub-cloned to psiCHECK2. Plasmids for overexpressing NLRX1 and PKR were previously described6. An expression vector for reconstitution of NLRX1 was generated by removal of the TurboGFP-2A region from pLOC-RFP58, producing pLOC∆GFP which retains the RFP open reading frame (ORF) (empty vector, EV). The RFP ORF was then replaced by sequence encoding the NLRX1 ORF to produce pLOC-NLRX1T3 using a PCR-based strategy as previously described59. Oligonucleotides used in this study are listed in Supplementary Table 2. All constructs were verified by DNA sequencing.
In vitro transcription and transfection
In vitro transcription and transfection of viral RNA was performed as previously described51. For biotin labeling of HAV RNA, transcription mixes were supplemented with 1× concentrated Biotin RNA Labeling Mix (Roche). siLentfect Lipid Reagent (Bio-Rad) was used for transfection of siRNAs (siRNAs: NLRX1 5′-AUCCCGACGGAAGAUGUGC-3′ and control #2, both from Dharmacon; NFKB1 from Santa Cruz). Fugen HD Reagent (Promega) was used for DNA transfections.
Lentivirus production and transduction
For sgRNA CRISPR/Cas9 lentivirus production, individual sgRNA expressing vectors and the 3rd Generation Packaging Mix vectors (ABM, Inc., Canada) were co-transfected into 293FT cells. Transduction was facilitated by the addition of 8 µg/ml polybrene, and the resulting cells subjected to selection with puromycin (6 µg/ml) for single-deleted cells, and puromycin (6 µg/ml) and blasticidin (10 µg/ml) for double-deleted cells.
RNA extraction and quantitative RT-PCR
Total RNA extraction was carried out with the RNeasy Kit (Qiagen), followed by a two-step quantitative RT-PCR analysis of indicated genes with the SuperScript® III First-Strand Synthesis System (Invitrogen) and iTaq SYBR Green Supermix (Bio-Rad). The abundance of indicated genes was normalized to that of ACTB (human) or Actb (mouse). HAV and HCV RNA abundance was quantified as previously described51,60.
Nuclear protein extraction
Extraction of nuclear proteins was carried out as previously described61, with minor modification. Briefly, the washed nuclei pellet was re-suspended in 50 µl of nuclear protein extraction buffer (20 mM Tris-HCl, pH 7.5, 400 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA) supplemented with complete protease inhibitor and PhoSTOP (Roche).
Protein-protein/RNA interactions
Cell lysates were prepared from 10-cm dish cultures (1.5~2 × 106 cells) with lysis buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 250 mM NaCl, 10% Glycerol, 5 mM EDTA, 1× complete protease inhibitor, 1× PhoSTOP) supplemented with detergents as follows. For protein-protein interaction assays, 0.2% Triton X-100 and 0.3% NP-40 were added. The resulting lysates were used for immunoprecipitation as previously described62. For protein-RNA interaction, 0.5% Triton X-100 with 0.1% NP-40 was added. Recombinant NLRX1 was generated as previously described63. Lysates were mixed with an equal volume of 2× incubation buffer (300 mM KCl, 40 mM HEPES, pH7.5, 10% glycerol, 200 Units/ml RNaseOUT (Thermo Fisher), 10 mM magnesium acetate, 2mM DTT). For precipitation, 500 ng biotin-labeled HAV RNA, 100 ng biotin-labeled poly(I:C) (InvivoGen) or 1 µl polyclonal anti-NLRX1 was used in each reaction. All reactions were gently rotated at 4 °C for 2 h, followed by the addition of magnetic streptavidin T1 beads (Invitrogen) for another 30 min rotation or protein G-Sepharose (GE Healthcare) for another 1 h rotation. After 5 intense washes with 1× incubation buffer, the final product was analyzed by qRT-PCR and/or immunoblotting.
EMSA, native PAGE, immunoblots and visualization
For EMSA, IRDye 700-labelled oligonucleotides containing a consensus NF-κB binding site (5′- AGTTGAGGGGACTTTCCCAGGC-3′; LI-COR Biosciences) were incubated with nuclear extracts according to the manufacturer’s protocol (Odyssey Infrared EMSA kit). Electrophoresis of the resultant products was carried out with 5% TBE gel (Bio-Rad). Native PAGE for analysis of IRF3 dimerization was performed as previously described64. Immunoblotting was carried out using standard methods. An Odyssey Imaging System (Li-COR Biosciences) was used for visualization and signal (infrared fluorescence) intensity analysis.
Polysome profiling, [35S]-metabolic labeling, and puromycin incorporation assay
Control and NLRX1-depleted PH5CH8 cells (2 × 107) were infected with SeV (3 × 104 HA units) for 2 h. Cells were then harvested and polysome gradients prepared and analyzed as previously described65. For metabolic labeling, 3 × 105 cells were infected with SeV (1.5 × 103 HA units) for 2 h 15 min. After 15 min starvation in methionine/cysteine-free medium, newly synthesized proteins were pulse-labeled with [35S]-labeled methionine and cysteine, 125 µCi [35S]/ml, for 30 min as described66. Protein synthesis was assessed in cells infected by SeV under similar conditions by monitoring puromycin incorporation into nascent protein using a method described previously67. In brief, cells grown on glass slides were pulsed with puromycin (10 µg/ml) for 10 min, then fixed with 4% paraformaldehyde followed by staining with anti-puromycin and DAPI. Slides were imaged by laser-scanning confocal microscopy at a fixed gain in an Olympus FV1000 instrument at 40× magnification, and recorded images analyzed with ImageJ (Fiji). Individual cells were identified by DAPI staining of nuclei, and scored for intensity of the puromycin signal greater than an arbitrary threshold set with mock-infected control cells in each experiment. Alternatively, cells were lysed and puromycin-labeled proteins identified by immunoblotting, and quantified by assessing infra-red fluorescence intensity with the Odyssey Imaging System.
Luciferase reporter assays
Luciferase assays, including secreted Gaussia luciferase (GLuc) analysis of HCV replication and dual luciferase assays to analyze promoter activation, were carried out as described previously57,68.
ELISA
IL-6 concentrations were determined with the human IL-6 ELISA Kit (eBioscience) following the manufacturer’s protocol.
Statistical analysis
Unless noted otherwise, all between-group comparisons were carried out by one-way or two-way ANOVA or two-sided t test. Calculations were made using Prism 6.0 software (GraphPad Software, Inc.).
Data availability
Data that support the findings of this study are either shown directly in figures accompanying the main text or in supplementary figures, or are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank B. Glaunsinger (University of California, Berkeley) for the human IL6 3′UTR expression vector, and L. Hensley, W. Lovell, M. Deng, and M. Chua for technical support. This work was supported by NIH grants U19-AI109965 (S.M.L., J.A.D. and J.P-Y.T.), R01-AI103083 (S.M.L.) and R01-AI131685 (S.M.L. and J.K.W.), R01-AI103311 (N.J.M.), R21-AI117575 (J.K.W.), and the University of North Carolina Cancer Research Fund (S.M.L. and J.P-Y.T.)
Footnotes
AUTHOR CONTRIBUTIONS
H.F. and S.M.L. designed the experiments; H.F., E.M.L., D.Y., D.R.M., O.G-L. and I.M performed experiments; E.W., H.G., J.M., J.K.W., N.J.M., L.M.R., J.P-Y.T. and J.A.D. provided critical reagents and intellectual input; S.M.L. supervised the study; H.F. and S.M.L. analyzed the data and wrote the manuscript; all authors reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider M, et al. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol. 2012;13:823–831. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia X, et al. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity. 2011;34:843–853. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185:1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- 5.Allen IC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore CB, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 7.Allen IC, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–865. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, et al. NLRX1 Sequesters STING to negatively regulate the interferon response, thereby facilitating the replication of HIV-1 and DNA viruses. Cell Host Microbe. 2016;19:515–528. doi: 10.1016/j.chom.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebsamen M, et al. NLRX1/NOD5 deficiency does not affect MAVS signalling. Cell Death Differ. 2011;18:1387. doi: 10.1038/cdd.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaworska J, et al. NLRX1 prevents mitochondrial induced apoptosis and enhances macrophage antiviral immunity by interacting with influenza virus PB1-F2 protein. Proc Natl Acad Sci U S A. 2014;111:E2110–2119. doi: 10.1073/pnas.1322118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tattoli I, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu L, Lemon SM. Hepatitis A and hepatitis C viruses: divergent infection outcomes marked by similarities in induction and evasion of interferon responses. Semin Liver Dis. 2010;30:319–332. doi: 10.1055/s-0030-1267534. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda M, et al. Human hepatocyte clonal cell lines that support persistent replication of hepatitis C virus. Virus Res. 1998;56:157–167. doi: 10.1016/s0168-1702(98)00063-x. [DOI] [PubMed] [Google Scholar]
- 14.Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 15.Yamane D, et al. Differential hepatitis C virus RNA target site selection and host factor activities of naturally occurring miR-122 3 variants. Nucleic Acids Res. 2017;45:4743–4755. doi: 10.1093/nar/gkw1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Z, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdul-Sater AA, et al. Enhancement of reactive oxygen species production and chlamydial infection by the mitochondrial Nod-like family member NLRX1. J Biol Chem. 2010;285:41637–41645. doi: 10.1074/jbc.M110.137885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei Y, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–946. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soares F, et al. The mitochondrial protein NLRX1 controls the balance between extrinsic and intrinsic apoptosis. J Biol Chem. 2014;289:19317–19330. doi: 10.1074/jbc.M114.550111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumpter R, Jr, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda K, et al. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc Natl Acad Sci U S A. 2013;110:9409–9414. doi: 10.1073/pnas.1307419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odendall C, et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 24.Mboko WP, et al. Interferon regulatory factor 1 restricts gammaherpesvirus replication in primary immune cells. J Virol. 2014;88:6993–7004. doi: 10.1128/JVI.00638-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita T, Kimura Y, Miyamoto M, Barsoumian EL, Taniguchi T. Induction of endogenous IFN-alpha and IFN-beta genes by a regulatory transcription factor, IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 26.Dauber B, Wolff T. Activation of the antiviral kinase PKR and viral countermeasures. Viruses. 2009;1:523–544. doi: 10.3390/v1030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong M, Yoon SI, Wilson IA. Structure and functional characterization of the RNA-binding element of the NLRX1 innate immune modulator. Immunity. 2012;36:337–347. doi: 10.1016/j.immuni.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unger BL, et al. Nod-like receptor X-1 is required for rhinovirus-induced barrier dysfunction in airway epithelial cells. J Virol. 2014;88:3705–3718. doi: 10.1128/JVI.03039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown EA, Zajac AJ, Lemon SM. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5' nontranslated region of hepatitis A virus RNA: Comparison with the IRES of encephalomyocarditis virus. J Virol. 1994;68:1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barouch DH, et al. Rapid inflammasome activation following mucosal SIV infection of Rhesus monkeys. Cell. 2016;165:656–667. doi: 10.1016/j.cell.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soares F, et al. NLRX1 does not inhibit MAVS-dependent antiviral signalling. Innate Immun. 2013;19:438–448. doi: 10.1177/1753425912467383. [DOI] [PubMed] [Google Scholar]
- 32.Hirai-Yuki A, et al. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science. 2016;353:1541–1545. doi: 10.1126/science.aaf8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, et al. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu L, et al. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011;7:e1002169. doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 38.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tailor P, et al. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007;27:228–239. doi: 10.1016/j.immuni.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li P, et al. IRF8 and IRF3 cooperatively regulate rapid interferon-beta induction in human blood monocytes. Blood. 2011;117:2847–2854. doi: 10.1182/blood-2010-07-294272. [DOI] [PubMed] [Google Scholar]
- 41.Lazear HM, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9:e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss G, et al. MyD88 drives the IFN-beta response to Lactobacillus acidophilus in dendritic cells through a mechanism involving IRF1, IRF3, and IRF7. J Immunol. 2012;189:2860–2868. doi: 10.4049/jimmunol.1103491. [DOI] [PubMed] [Google Scholar]
- 43.Ousman SS, Wang J, Campbell IL. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J Virol. 2005;79:7514–7527. doi: 10.1128/JVI.79.12.7514-7527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherbik SV, Stockman BM, Brinton MA. Differential expression of interferon (IFN) regulatory factors and IFN-stimulated genes at early times after West Nile virus infection of mouse embryo fibroblasts. J Virol. 2007;81:12005–12018. doi: 10.1128/JVI.01359-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colina R, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 47.Lee MS, Kim B, Oh GT, Kim YJ. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat Immunol. 2013;14:346–355. doi: 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- 48.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu B, et al. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamane D, et al. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med. 2014;20:927–935. doi: 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemon SM, et al. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol. 1991;65:2056–2065. doi: 10.1128/jvi.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng Z, et al. Human pDCs preferentially sense enveloped hepatitis A virions. J Clin Invest. 2015;125:169–176. doi: 10.1172/JCI77527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimakami T, et al. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology. 2011;140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi M, Lemon SM. Replication of subgenomic hepatitis A virus RNAs expressing firefly luciferase is enhanced by mutations associated with adaptation of virus to growth in cultured cells. J Virol. 2002;76:1171–1180. doi: 10.1128/JVI.76.3.1171-1180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dansako H, et al. Class A scavenger receptor 1 (MSR1) restricts hepatitis C virus replication by mediating toll-like receptor 3 recognition of viral RNAs produced in neighboring cells. PLoS Pathog. 2013;9:e1003345. doi: 10.1371/journal.ppat.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saeed M, et al. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature. 2015;524:471–475. doi: 10.1038/nature14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48:463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lanford RE, et al. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc Natl Acad Sci U S A. 2011;108:11223–11228. doi: 10.1073/pnas.1101939108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: A two minute cell fractionation method. BMC Res Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Masaki T, Shimakami T, Lemon SM. hnRNP L and NF90 interact with hepatitis C virus 5'-terminal untranslated RNA and promote efficient replication. J Virol. 2014;88:7199–7209. doi: 10.1128/JVI.00225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mo J, et al. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J Biol Chem. 2012;287:23057–23067. doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Q, et al. Role of double-stranded RNA pattern recognition receptors in rhinovirus-induced airway epithelial cell responses. J Immunol. 2009;183:6989–6997. doi: 10.4049/jimmunol.0901386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Masaki T, et al. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe. 2015;17:217–228. doi: 10.1016/j.chom.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziehr B, Vincent HA, Moorman NJ. Human Cytomegalovirus pTRS1 and pIRS1 Antagonize Protein Kinase R To Facilitate Virus Replication. J Virol. 2016;90:3839–3848. doi: 10.1128/JVI.02714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 68.Shimakami T, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are either shown directly in figures accompanying the main text or in supplementary figures, or are available from the corresponding author upon request.