Abstract
Silver-Russell syndrome (SRS) is a growth retardation syndrome in which loss of methylation on chromosome 11p15 (11p15 LOM) and maternal uniparental disomy for chromosome 7 [UPD(7)mat] explain 20–60% and 10% of the syndrome, respectively. To search for a molecular cause for the remaining SRS cases, and to find a possible common epigenetic change, we studied DNA methylation pattern of more than 450 000 CpG sites in 44 SRS patients. Common to all three SRS subgroups, we found a hypomethylated region at the promoter region of HOXA4 in 55% of the patients. We then tested 39 patients with severe growth restriction of unknown etiology, and found hypomethylation of HOXA4 in 44% of the patients. Finally, we found that methylation at multiple CpG sites in the HOXA4 promoter region was associated with height in a cohort of 227 healthy children, suggesting that HOXA4 may play a role in regulating human growth by epigenetic mechanisms.
Introduction
Silver-Russell syndrome is an imprinting disorder characterized by severe pre-and postnatal growth retardation and typical clinical features, including relative macrocephaly, protruding forehead, body asymmetry and feeding difficulties1. Imprinting disorders affect the function of genes that are normally expressed predominantly or solely from one parental chromosome. Chromosomal abnormalities or changes in DNA methylation level can alter imprinted gene dosage and lead to phenotypes affecting growth or neurological functioning2. Opposite changes in the same imprinted genomic regions can lead to opposite growth phenotypes, such as growth restriction in SRS and overgrowth in Beckwith-Wiedemann syndrome (BWS), caused by hypo- and hypermethylation, respectively, of the imprinted H19-IGF2 region in 11p15 (H19/IGF2:IG-DMR) in a proportion of the cases3. SRS is a heterogeneous syndrome both in phenotype and in molecular etiology, but severe growth retardation is a central feature shared by the patients.
SRS patients can be divided into three subgroups based on their molecular etiology: maternal uniparental disomy of chromosome 7 (UPD(7)mat), loss of methylation on chromosome 11p15, (11p15 LOM; also known as hypomethylation of H19/IGF2:IG-DMR or H19 hypomethylation), and clinical SRS without any of these molecular findings. Approximately 10% of SRS patients have UPD(7)mat and 20–60% of SRS patients have 11p15 LOM4–10. Duplications and deletions at 11p15 affect <1% of SRS patients and even other molecular changes, such as UPD of chromosome 11 have been detected in single cases3. The remaining cases can be designated clinical SRS, which make up 30–70% of the SRS patients.
As all SRS patients share the same clinical diagnosis, while a substantial amount of patients test negative for molecular diagnosis, we hypothesized that there might occur further epigenetic changes that all subgroups of SRS share in common. To search for such genes or regions, we analyzed 44 SRS patients for methylation changes by the genome-wide Illumina Infinium HumanMethylation450K BeadChip assay that provides quantitative methylation data at more than 450 000 single CpG sites. In addition, we evaluated associations between the found methylation changes and severe growth restriction of unknown etiology (SGR), as well as growth in healthy children.
Results
Genome-wide methylation analysis pinpointed HOXA4 hypomethylation as a common epigenetic change among SRS subgroups
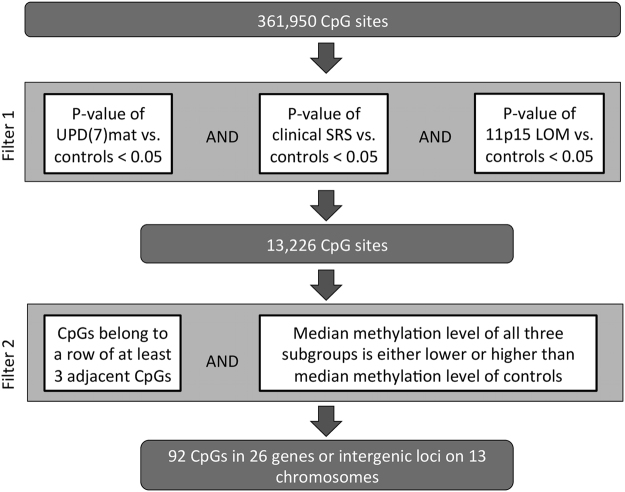
We used the Infinium HumanMethylation450K BeadChip (Illumina) to identify shared methylation changes in the three different SRS subgroups. We used a filtering approach (Fig. 1) to search for areas that were significantly differentially methylated in all subgroups compared to controls over a stretch of consecutive CpGs. In the first stage of filtering (filter 1), altogether 13,226 CpGs were found, in which the differential methylation between each of the three SRS groups compared to controls reached the empirical Bayes significance (nominal p-value < 0.05). We then chose regions that had at least three consecutive significant CpGs for all subgroups and discarded CpGs in which the methylation level of different groups varied to opposite directions in comparison to the controls (filter 2). This filtering process substantially narrowed down the potential areas and resulted in 92 CpGs located in 26 genes or intergenic loci on 13 different chromosomes (Supplementary Table 1). Of these CpGs, 66 showed only small differences in median Beta-value (<0.02) between each of the subgroups and controls. Altogether 16 CpGs showed median Beta-value differences >0.05 (5% absolute methylation level) for all three subgroups. Two of the 16 CpGs were located in an intergenic region on 5p15.33 and 14 were located in the HOXA4 gene region on 7p15.2. HOXA4 was the only area that showed at least 5% median methylation difference for all subgroups vs. controls for at least three consecutive significant CpGs. In total, HOXA4 showed 12 consecutive probes with such differences. The differences for the individual groups in these 12 CpGs were also larger than with any other CpGs, ranging from 6.8–25% in UPD(7)mat, 7.1–21% in clinical SRS and 5.0–11% in 11p15 LOM patients. As a result of the filtering process of genome-wide methylation data, HOXA4 emerged as a specific candidate for further study.
Figure 1.
Filtering for common differentially methylated regions in SRS patients.
HOXA4 methylation levels differ among subgroups and are lowest near the HOXA4 transcription start site
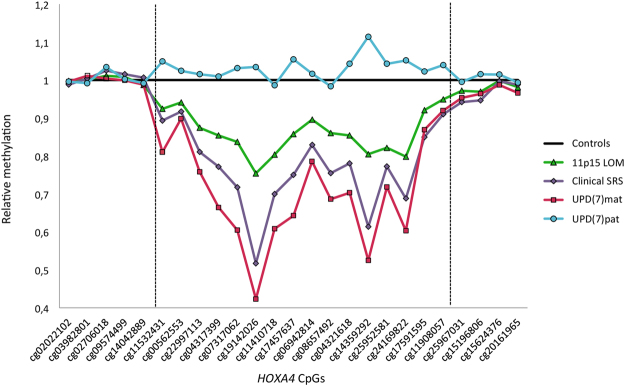
All three SRS subgroups showed hypomethylation throughout the HOXA4 region identified by filtering when comparing median methylation levels relative to controls (Fig. 2). This differentially methylated area included two stretches of consecutive CpGs: from cg11532431 to cg14359292 (chr7:27169674–27170892 in GRCh37/hg19 assembly), and from cg24169822 to cg11908057 (chr7:27170994–27171154). Localized between these stretches, cg25952581 (at chr7:27170961) did not reach the empirical Bayes significance for differential methylation between controls and 11p15 LOM group (p = 0.082), but clinical SRS and UPD(7)mat had significant methylation differences (p = 0.013 and p < 0.001 respectively), and the methylation pattern was similar to the pattern of other CpGs in the region. P-values for differential methylation between subgroups and controls for all HOXA4 CpGs included in our filtering are shown in Table 1. For the majority of the CpGs, the median methylation level was lowest in the UPD(7)mat group, followed by clinical SRS, 11p15 LOM, and controls in ascending order. The methylation level of the single UPD(7)pat sample was higher than controls for most of the CpGs. Figure 3 shows examples of methylation levels at four individual CpG sites, where methylation level of most SRS patients fell below -2 SD and UPD(7)pat remained within the range of the control group’s methylation level. Some SRS patients showed extremely low hypomethylation patterns and were hypomethylated even as low as 8 SD below the mean of the control group at cg22997113.
Figure 2.
HOXA4. Median methylation levels of SRS subgroups relative to controls (1). Boundaries of the region in which HOXA4 CpGs are hypomethylated in SRS groups relative to controls are marked with two dashed vertical lines.
Table 1.
P-values for differential methylation for SRS subgroups (11p15 LOM, clinical SRS, UPD(7)mat) compared to controls for all HOXA4 CpGs that passed quality control and were included in the filtering process of Illumina 450 K BeadChip assay data. Regulatory feature group as indicated by Illumina is shown in column “Regulatory”: “PROMOTER” = Promoter_Associated, “CELL TYPE” = Unclassified_Cell_type_specific. Relation to UCSC CpG island chr7:27169572-27170638 is shown in column “CGI”.
| Coordinates in hg19 (chr 7) | CpG ID | Regulatory | CGI | 11p15 LOM | clinical SRS | UPD(7)mat |
|---|---|---|---|---|---|---|
| 27168609 | cg02022102 | N_Shore | 0,1337 | 0,3379 | 0,0345 | |
| 27168688 | cg03982801 | N_Shore | 0,4355 | 0,2119 | 0,8381 | |
| 27168780 | cg02706018 | N_Shore | 0,9538 | 0,3056 | 0,5738 | |
| 27168962 | cg09574499 | N_Shore | 0,8489 | 0,2095 | 0,4154 | |
| 27169208 | cg14042889 | N_Shore | 0,6536 | 0,8850 | 0,6749 | |
| 27169674 | cg11532431 | CELL TYPE | Island | 0,0011 | 0,0013 | 8,2E-06 |
| 27169740 | cg00562553 | Island | 0,0052 | 0,0037 | 0,0002 | |
| 27170241 | cg22997113 | PROMOTER | Island | 0,0006 | 0,0001 | 1,5E-06 |
| 27170313 | cg04317399 | PROMOTER | Island | 0,0309 | 0,0030 | 4,1E-05 |
| 27170388 | cg07317062 | PROMOTER | Island | 0,0073 | 0,0014 | 8,4E-06 |
| 27170394 | cg19142026 | PROMOTER | Island | 0,0061 | 0,0005 | 7,3E-06 |
| 27170412 | cg11410718 | PROMOTER | Island | 0,0037 | 0,0004 | 3,3E-06 |
| 27170717 | cg17457637 | PROMOTER | S_Shore | 0,0349 | 0,0011 | 1,4E-05 |
| 27170819 | cg06942814 | PROMOTER | S_Shore | 0,0116 | 0,0004 | 1,9E-05 |
| 27170832 | cg08657492 | PROMOTER | S_Shore | 0,0063 | 0,0001 | 4,4E-06 |
| 27170880 | cg04321618 | PROMOTER | S_Shore | 0,0172 | 0,0009 | 9,7E-06 |
| 27170892 | cg14359292 | PROMOTER | S_Shore | 0,0125 | 0,0001 | 9,8E-07 |
| 27170961 | cg25952581 | PROMOTER | S_Shore | 0,0818 | 0,0125 | 0,0002 |
| 27170994 | cg24169822 | PROMOTER | S_Shore | 0,0120 | 0,0005 | 5,4E-06 |
| 27171051 | cg17591595 | PROMOTER | S_Shore | 0,0371 | 0,0006 | 0,0033 |
| 27171154 | cg11908057 | S_Shore | 0,0108 | 0,0001 | 0,0031 | |
| 27171203 | cg25967031 | S_Shore | 0,0804 | 0,0035 | 0,0167 | |
| 27171213 | cg15196806 | S_Shore | 0,0526 | 0,0173 | 0,0674 | |
| 27171391 | cg15624376 | S_Shore | 0,8751 | 0,8147 | 0,2402 | |
| 27171401 | cg20161965 | S_Shore | 0,0634 | 0,2399 | 0,0240 |
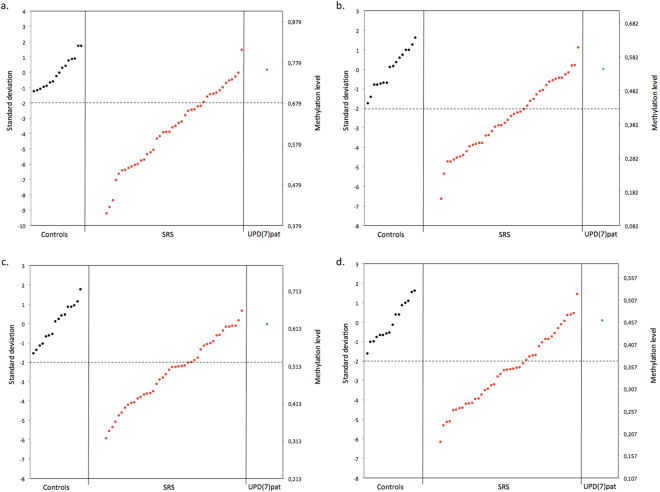
Figure 3.
Methylation level at HOXA4 cg22997113 (a), cg11410718 (b), cg08657492 (c) and cg04321618 (d). Individuals within each group are represented in ascending order of methylation. Dashed line represents -2 standard deviations of the arithmetic mean of control group methylation level.
The identified differentially methylated area of HOXA4 was located in the promoter region and co-localized with the CpG island at chr7:27169573–27170638. Approximately half of the differentially methylated CpGs were located within the CpG island and the rest within the proximal CpG island shore. According to SwitchGear Genomics transcription start sites, the HOXA4 TSS is at chr7:27170399 and according to FANTOM5, at chr7:27170364–27170377 in the hg19 build (GRCh37/hg19 assembly). Cg19142026 at position chr7:27170394, the CpG with lowest relative median methylation of the HOXA4 differentially methylated area for all SRS subgroups, was within 5-17 bp, respectively, of the TSS.
HOXA4 hypomethylation frequency was highest for UPD(7)mat, followed by clinical SRS and 11p15 LOM patients
While group results showed the trend that UPD(7)mat patients had the lowest median level of methylation for CpGs in the HOXA4 differentially methylated area, followed by the clinical SRS and 11p15 LOM groups, the methylation level varied between individuals within each subgroup. Figure 4(a) shows hypomethylation of individual SRS patients for CpGs in the HOXA4 differentially methylated area. The frequency of HOXA4 hypomethylation was highest for UPD(7)mat patients at 80%, followed by clinical SRS at 62% and 11p15 LOM group at 38%, when hypomethylation for an individual patient was defined by at least two-thirds of the CpGs being hypomethylated (<−2 SD). In addition to the patients that were hypomethylated in majority of the CpGs in the HOXA4 area, three 11p15 LOM patients and two clinical SRS patients had hypomethylation in at least one-third of the HOXA4 CpGs, demonstrating a partial deviation from normal methylation status in the area.
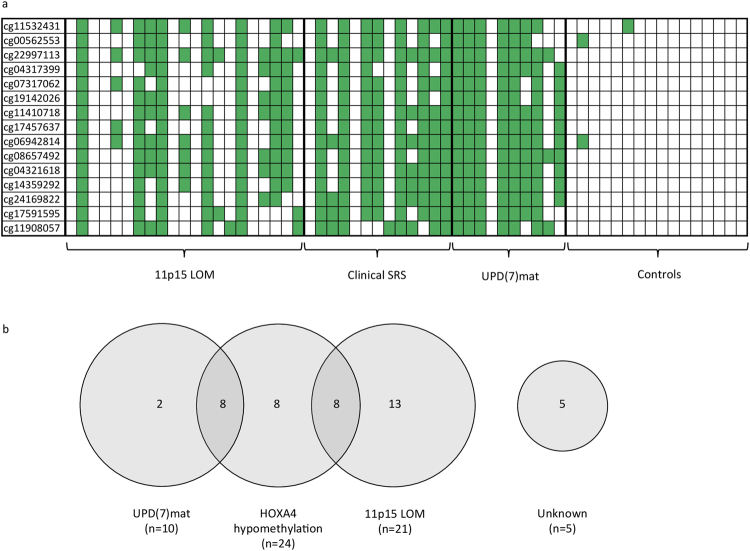
Figure 4.
(a) HOXA4 CpG methylation status represented by individuals in each SRS group. Each column represents one patient. Green color indicates methylation level below -2SD of the controls. (b) Molecular changes of SRS patients, n = 44. HOXA4 hypomethylation is present in 55% of the patients.
Shared molecular etiology for SRS patients
Analysis of the methylation data of HOXA4 region studied by the Illumina assay revealed that altogether 24 (55%) of the SRS patients were hypomethylated in at least two-thirds of the CpG sites at HOXA4 differentially methylated area. Often both UPD(7)mat and HOXA4 hypomethylation, or HOXA4 hypomethylation and 11p15 LOM occurred in the same individual, as seen in Fig. 4(b). Either 11p15 LOM, UPD(7)mat, HOXA4 hypomethylation or a combination of two changes was found in 39 out of 44 SRS patients (89%). Only 5 patients (11%) were left without a molecular change defined by the chosen criteria. In two of these patients, we observed partial hypomethylation in the HOXA4 differentially methylated area.
Frequency of 11p15 LOM was higher than previously tested
After initial analysis of our 44 SRS patients, we observed that more individuals had 11p15 LOM than had previously been identified. 15 of our 21 11p15 LOM SRS patients had been previously found hypomethylated with a restriction site-specific methylation method11. Of the six patients that had previously not been found hypomethylated, two had not been tested and four had been tested but found negative. Supplementary Table 2 summarizes our result, indicating that 21 of our patients had clear methylation changes in this area with at least two-thirds of the 36 CpGs hypomethylated (below -2 SD of the controls) in the region.
EpiTYPER methylation assay revealed HOXA4 hypomethylation in patients with severe growth restriction of unknown etiology (SGR)
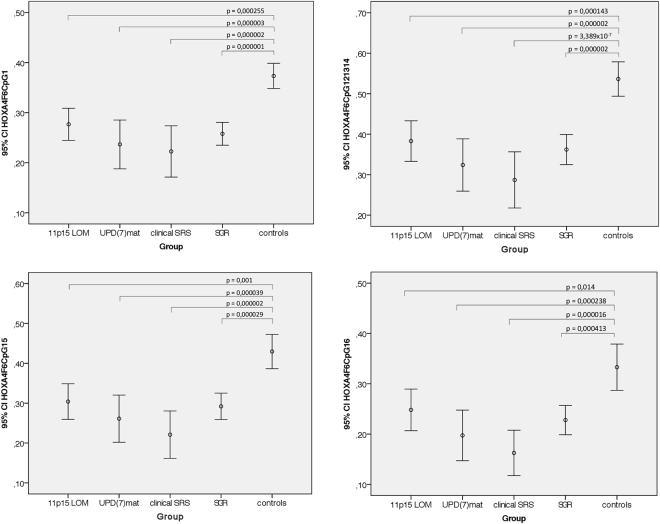
We used the Sequenom EpiTYPER method to assay the methylation level of the HOXA4 promoter region in order to validate results from the Illumina genome-wide methylation study and to study a group of children with SGR. The participants included 44 SRS patients, 16 controls and 39 children with SGR. The EpiTYPER assay spanned chr7:27170191–27170313, overlapping the area with significant methylation differences in the genome-wide methylation study. One CpG, cg04317399 at chr7:27170313, was covered by both Illumina 450 K and EpiTYPER, here named HOXA_F6_CpG_16. Analysis showed a strong correlation between the two methods, R2 = 0.86. (Supplementary Figure 1). Methylation levels of four different CpG sites (HOXA4F6CpG1, HOXA4F6CpG12.13.14, HOXA4F6CpG15, HOXA4F6CpG16) were measured for all participants. Statistically significant differences between the groups were determined by one-way ANOVA (HOXA4F6CpG1: F(4,92) = 10.831, p < 0.001; HOXA4F6CpG12.13.14: F(4, 91) = 11.549, p < 0.001; HOXA4F6CpG15: F(4,92) = 9.268, p < 0.001; HOXA4F6CpG16: F(4, 92) = 7.556, p < 0.001). Post hoc comparisons using 2-sided Dunnett t-tests indicated that the mean methylation levels of 11p15 LOM, UPD(7)mat, clinical SRS and SGR groups were statistically significantly different from the mean of the controls at all four studied sites (Fig. 5). The largest mean difference (I-J = −0.249, p = 3.389 × 10−7) was observed between clinical SRS (I) and controls (J) at HOXA4F6CpG12.13.14. Also, an analysis of individual SGR patients showed that 44% (17/39) of the patients were hypomethylated for at least two-thirds of the CpGs in the HOXA4 promoter region at chr7: 27170191–27170313. Supplementary Table 3 shows individual methylation status of all SRS and SGR patients studied with the EpiTYPER assay. Thus, HOXA4 hypomethylation was not limited to SRS patients, but also implicated in patients with SGR.
Figure 5.
Methylation levels measured at four EpiTYPER sites for 11p15 LOM (n = 21), UPD(7)mat (n = 10), clinical SRS (n = 13), and SGR (n = 38, for HOXA4_F6_CpG_12.13.14 n = 37) patients compared to controls (n = 15). Comparisons made by 2-sided Dunnett t-tests.
Methylation level of HOXA4 cg11908057 correlates with height in healthy children
We studied correlation of methylation levels of HOXA4 and height of altogether 227 healthy children from the BAMSE (Swedish abbreviation for Children, Allergy, Milieu, Stockhom, Epidemiology) cohort, a birth cohort of children born between 1994 and 1996 in Stockholm, Sweden. Methylation levels were measured with Infinium HumanMethylation450K BeadChip (Illumina)12. Sixteen CpGs of the HOXA4 differentially methylated area were studied. We observed a statistically significant correlation between the methylation level of one of the HOXA4 differentially methylated CpGs, cg11908057, and height at four years of age (r = 0.142, n = 227, p = 0.033), eight years of age (r = 0.148, n = 225 p = 0.026) and sixteen years of age (r = 0.164, n = 200, p = 0.020) by computing Pearson correlation coefficient. As methylation data for some of the CpGs did not meet criteria for normal distribution, additional analysis using all CpGs by computing Spearman correlations revealed statistically significant positive correlation between methylation level and height at sixteen years of age for 5 additional CpGs: cg04317399 (r = 0.146, n = 200, p = 0.040), cg19142026 (r = 0.139, n = 200, p = 0.050), cg04321618 (r = 0.149, n = 200, p = 0.036), cg14359292 (r = 0.140, n = 200, p = 0.047) and cg25952581 (r = 0.145, n = 200, p = 0.040). Cg11908057 demonstrated similar results as above: positive correlation was observed between methylation level and height at eight years of age (r = 0.140, n = 225, p = 0.036) and sixteen years of age (r = 0.147, n = 200, p = 0.038). Figure 6 demonstrates that the methylation level of cg11908057 in healthy children at eight years of age ranges from 0.69 to 0.91. This variation in methylation corresponds to a difference of 0.75 SD (or 4.5 cm) in height. In the same individuals, we studied correlation of height at age eight with some of the most strongly height-associated SNPs13,14 (Supplementary Table 4). In the tested SNPs, there was no significant association between height and genotype (Supplementary Figure 2). The results indicate that in our dataset of healthy-school aged children from the BAMSE cohort, the methylation level of cg11908057 at HOXA4 was better in explaining height variation at age eight than some of the most strongly height-associated SNPs. We also used regulomeDB15 to study whether cg11908057 is localized in a potential regulatory protein-binding site. There was some evidence of binding of an enhancer of zeste homolog 2 (EZH2), a methyltransferase enzyme encoded by the EZH2 gene on 7q36.1. Mutations of EZH2 cause Weaver syndrome16, a pre- and postnatal overgrowth syndrome resulting in tall adult stature.
Figure 6.
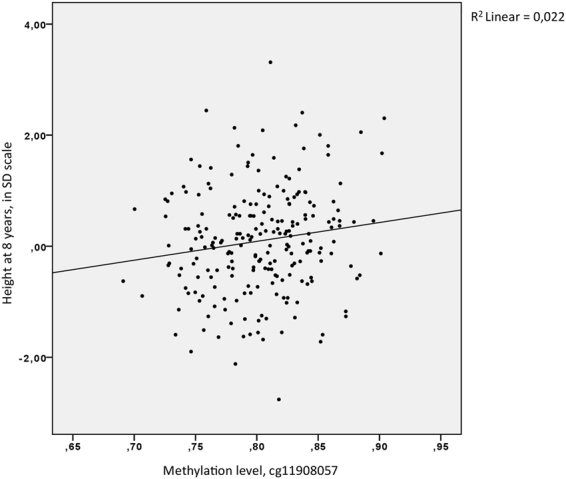
Height measurements (in SD scale) of BAMSE individuals (n = 225) at age 8 plotted against meathylation level of cg11908057 measured by Illumina 450 K BeadChip assay.
Methylation level of HOXA4 promoter region correlates with expression level of HOXA4 in blood
We used data previously published in Hannula-Jouppi et al.17, to study the correlation of HOXA4 promoter region CpG methylation level and expression level of HOXA4 in blood in nine UPD(7)mat patients, one individual with UPD(7)pat and ten controls. The average methylation level of the HOXA4 region in SD scale correlated negatively with the expression level of HOXA4 in blood, as demonstrated in Fig. 7. In all differentially methylated CpGs of HOXA4, we observed a statistically significant negative correlation between expression and methylation level by Spearman correlation analysis. The negative correlation was strongest for cg22997113 (r = −0.81, n = 20, p = 1.40 × 10−5). Data for other CpGs is listed in Supplementary Table 5.
Figure 7.
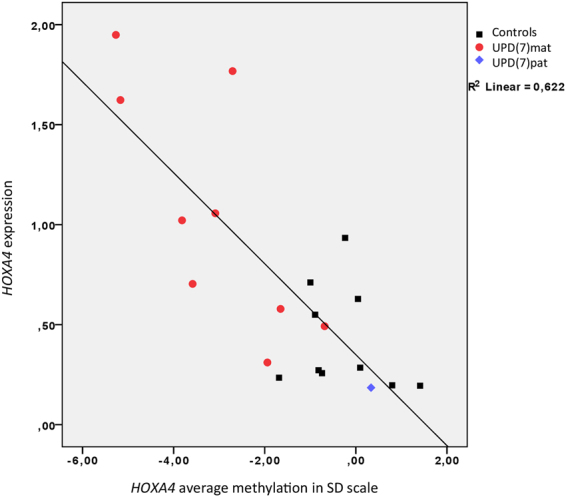
HOXA4 expression level in blood measured by qPCR vs. average methylation level of HOXA4 CpGs in SD scale. Average methylation was calculated from all HOXA4 CpGs that showed significant differential methylation between all SRS groups and controls with Illumina 450 K analysis.
Discussion
We conducted a genome-wide methylation study, determining the methylation status of more than 450,000 CpG sites for UPD(7)mat, 11p15 LOM and clinical SRS patients, and aimed the data analysis at finding common methylation changes among these three groups. Initial analysis showed such changes in 26 genes or intergenic loci, and further analysis pinpointed HOXA4 promoter hypomethylation as a major defect in SRS. HOXA4 hypomethylation occurred in 55% of SRS patients, and was thus more common in our cohort than 11p15 LOM (48%) or UPD(7)mat (23%), and specifically, was found in eight 11p15 LOM (38%), eight UPD(7)mat (80%), and eight patients without any prior molecular diagnosis (62%). Furthermore, targeted methylation analysis revealed that HOXA4 hypomethylation was also present in children with severe growth-restriction of unknown etiology. Further analysis of the HOXA4 region identified multiple CpG sites where lower methylation level was associated with lower height in healthy school-aged children.
HOXA4 belongs to the cluster of HOXA genes located on 7p15.2. The HOXA4 hypomethylated region was located at chr7:27169674–27171154, with most CpGs classified as promoter-associated, and half of the CpGs located within the overlapping CpG island and the rest in the proximal island shore. CpG islands in somatic cells are usually unmethylated, and methylated promoter CpG islands are involved in long-term repression of, for example, imprinted genes18. Methylated CGIs are often observed near developmentally important genes, such as the HOX genes, and it is thought that the large numbers of potential CGI promoters within HOX loci may contribute to their regulation19. Relative HOXA4 methylation level was lowest for all the SRS subgroups at cg19142026, in close proximity to the functionally relevant transcription start site. Lower methylation level is often related to transcriptional activation of genes, while hypermethylation can lead to transcriptional repression16,20. We observed that hypomethylation of the HOXA4 promoter region was correlated with increased expression of HOXA4 in blood, when using expression data from UPD(7)mat, UPD(7)pat and controls from our previously published study17. Other reports have shown that hypermethylation of HOXA4 promoter region promotes inactivation of gene expression21,22. Altered methylation levels of the HOXA4 promoter therefore seem to have functional relevance by affecting gene expression.
Several genes within the HOXA gene cluster (HOXA2, HOXA3, HOXA4, HOXA5, HOXA6 and HOXA11) are predicted as maternally expressed imprinted genes23. In our previous study, HOXA4 was monoallelically expressed and the expression of HOXA4 was increased in whole blood of the UPD(7)mat group relative to controls17, supporting the prediction of HOXA4 as a maternally expressed imprinted gene. In UPD(7)mat patients, HOXA4 hypomethylation can therefore be explained by the occurrence of two maternal copies of chromosome 7, although the hypomethylation was not observed in all of the UPD(7)mat patients in our study. In 11p15 LOM patients, HOXA4 hypomethylation could potentially be due to multi-locus imprinting disturbance (MLID). MLID is reported to be present in 15–38% of 11p15 LOM patients1.
The HOX/Hox genes in human and other species are highly interesting in the context of growth disorders. They encode a family of transcription factors that regulate early developmental morphogenetic processes and are important regulators of anterior-posterior axis development24. Ten of the 39 HOX genes have been associated with human disorders25, such as HOXA13 with Hand-foot-genital syndrome (HFGS). Human phenotypes of the loss-of-function mutations in genes of the HOXA cluster include facial dysmorphisms, limb anomalies, cardiac defects and urogenital malformations. As the Hox genes are highly conserved, human phenotypes are very similar to their mouse orthologs. Hox gene knockouts in mouse affect the skeleton, primary vertebral column and slightly less frequently the bones of the forelimb and hindlimb. Mouse Hoxa4 loss-of-function homozygote phenotype showed skeletal abnormalities including partial anterior transformation of C3, posterior transformation of C7 to T1, presence of a C7 cervical rib and malformation of the sternum26,27. The reported Hox gene defects that often affect bone morphology suggest that disturbance in the normal pattern of HOXA4 methylation may also have relevance considering skeletal malformations in SRS patients. Interestingly, in 3-M syndrome, a growth disorder with a phenotype very similar to SRS, downregulation of IGF2 and upregulation of several HOX genes has been reported28.
Our study also included two rare samples, a UPD(7)pat and a UPD(7q31-qter)mat, in order to enable further investigation of the potential findings29. The methylation level of the UPD(7)pat sample was higher than the median methylation level of controls in most differentially methylated CpGs of HOXA4, but still within normal range (+/−2SD). It is interesting to note that there is no growth abnormality in this individual or other reported UPD(7)pat cases29–31. The segmental UPD(7q31-qter)mat was hypomethylated at 10 of the HOXA4 CpGs, even though HOXA4 (at 7p15.2) is located outside of the segmental UPD(7q31-qter)mat region. Further studies are required to better understand this observation.
Unexpectedly, we found that some of the patients that had been tested negative for 11p15 LOM previously, were actually positive according to our results from the genome-wide methylation study. Prickett et al. reported similar findings, where methylation-sensitive RFLP PCR method failed to detect 11p15 LOM patients while 11p15 LOM was found by the Infinium Human Methylation 450 K BeadChip array32. These findings suggest that the genome-wide methylation assay may be more sensitive at detecting hypomethylated patients than some other methods.
Methylation levels may vary in different tissues and time in development. Our results are limited to methylation levels present at the time of sampling in accessible whole blood tissue. Variation in the methylation level of the different white blood cell populations in whole blood can complicate interpretation of whole blood methylation profiles33. In the HOXA4 region, such variation between blood cell types was not observed, and therefore potential differences in blood cell populations of the participants should not affect our results. Also genome-wide methylation studies of human blood have reported age-related modifications in specific CpGs during early childhood34, adulthood35 and from newborn to elderly in a meta-analysis36. Our patients included both children and adults, while control samples were all adults. CpGs in the HOXA4 region were, however, not among the reported age-modified loci, and therefore age of the participants is not expected to significantly affect our results.
Our genome-wide study of SRS patients found hypomethylation at multiple adjacent CpGs in the promoter region of the developmentally important HOXA4 gene. Our results indicated that HOXA4 hypomethylation was not specific to SRS, but was also present in children with severe growth restriction of unknown etiology. Our study in healthy children indicated an effect in the same direction, showing the association of hypomethylation of multiple HOXA4 CpGs with short stature. It is also worth mentioning that in HOXA3, a gene adjacent to HOXA4, an association between birth weight and methylation has been observed in two CpGs37, demonstrating that the region may be important in regulating body size. We hypothesize that HOXA4 plays a role in growth-regulating pathways affecting SRS, related growth disorders, and also stature in general. Remarkably, the effect size of HOXA4 methylation in our healthy child cohort was larger than found for any of nine highly significantly height-associated SNPs. As growth disorders share many diagnostic features, it is plausible that underlying genetic or epigenetic defects are shared across closely related phenotypes. Our results suggest a new candidate region potentially relevant in the pathogenesis of SRS, other growth disorders and growth in general.
Methods
Patients and controls
We studied 44 SRS patients. The patients included three subgroups based on molecular findings: 21 patients with 11p15 LOM, 10 patients with UPD(7)mat, and 13 patients negative for both 11p15 LOM and UPD(7)mat, assigned as clinical SRS. One paternal uniparental disomy 7 (UPD(7)pat)27 sample without growth retardation was also included for comparison. One of the patients in UPD(7)mat had segmental UPD(7q31-qter)mat. SRS diagnoses were made based on criteria for SRS diagnosis in use at the time of evaluation. Information of birth weight, birth length, postnatal growth, body asymmetry, protruding forehead, relative macrocephaly and feeding difficulties are listed in Supplementary Table 6. Birth length and weight for gestational age in SD scale are according to Finnish growth references38. Ten control blood samples were obtained from adult volunteers of normal height. Six additional adult control samples were from healthy blood donors31. 39 patients with severe growth restriction of unknown etiology (SGR) were included in a targeted analysis of HOXA4 hypomethylation. SGR patients had severe growth restriction but did not fulfill criteria for SRS diagnosis at the time of evaluation. Information on birth length and birth weight for gestational age, and postnatal growth is listed in Supplementary Table 7. The majority of the SGR patients were born small for gestational age (SGA) and all of them had postnatal growth restriction. Methylation data from the BAMSE study12 were used to test for correlation with height in healthy children. The study was approved by the Ethical Review Board of the Hospital for Children and Adolescents, Helsinki University Central Hospital, Helsinki, Finland (SRS and SGR patients and controls) and the Ethical Review Board North at Karolinska Institutet (BAMSE, blood donor controls). Informed consent was obtained from all study participants. Clinical investigations have been conducted according to the Declaration of Helsinki.
Genome-wide methylation analysis with Illumina 450K BeadChip assay
Methylation analysis was carried out on 44 SRS patients, one individual with UPD(7)pat, and 10 control samples. DNA from EDTA blood samples was extracted using the FlexiGene DNA Kit (Qiagen) according to manufacturer’s instructions. 500 ng of DNA was bisulfite converted with the EZ-96 Methylation Kit (Zymo research corporation) according to the manufacturer’s instructions. Samples from 27 patients, UPD(7)pat and 10 control samples were analyzed at the Karolinska Institutet Bioinformatic and Expression Analysis (BEA) Core Facility, and additional 17 samples at the Mutation Analysis Core Facility (MAF, www.maf.ki.se). Bisulfite-treated DNA was amplified, fragmented and hybridized to the HumanMethylation450 BeadChip (Illumina) according to standard Illumina protocol and imaging was performed using Illumina iScan scanner. Additional data from similarly performed methylation analysis of six whole blood control samples31 produced at MAF were integrated into the analysis. Data produced at different facilities were comparable (data not shown). Methylation data for the BAMSE cohort had also been produced at MAF.
Bioinformatics analysis of the Illumina 450K BeadChip assay for SRS patients and controls
The.idat files were imported in R and analyzed using the minfi package39. After quality check, the data were normalized using the subset-quantile within array normalization (SWAN) method40. The probes overlapping with known SNPs as well as probes prone to cross-hybridization problems were removed, rendering 361,948 CpG sites for further analysis. The M-values were extracted and the batch effects removed using the ComBat method41. Differentially methylated CpG sites were obtained using linear models (y~subgroup + gender) and pairwise comparisons with empirical Bayes as implemented in the limma package42. The differentially methylated CpG sites with p-value < 0.05 after Benjamini and Hochberg correction for multiple testing were considered for further analysis.
Filtering process for common methylation differences in genome-wide methylation data
We analyzed 361,948 CpGs for significant differential methylation of the UPD(7)mat, 11p15 LOM, and clinical SRS groups in comparison to controls. At the first stage of filtering (filter 1), we selected CpGs that showed significant differential methylation with a nominal p-value < 0.05 (empirical Bayes significance) for each of the three patient groups when compared to controls. Out of these common differentially methylated CpGs we selected those that fulfilled two criteria: 1) there were at least three consecutive CpGs probes with a significant difference, 2) the median methylation level of all of the three subgroups was either lower or higher than that of the controls (filter 2). Consecutive CpGs were defined as occurring next to each other based on their genomic coordinates; they may not be representative of true consecutive CpGs in the genome, as those CpGs not included in the array as well as those discarded in the quality control steps were not considered. The filtering process is shown in Fig. 1.
The output list of significant CpGs that passed the filtering process was subjected to further selection by comparing the median methylation level of each of the subgroups to controls (Supplementary Table 1). We set a threshold of 5% methylation difference to further narrow down significant probes. The HOXA4 differentially methylated area was defined by the CpGs that passed the filtering process, comprising altogether 12 + 3 consecutive CpG sites.
DNA methylation analysis of HOXA4 region with EpiTYPER
We measured the methylation level of CpGs in the HOXA4 differentially methylated area using Sequenom EpiTYPER (Sequenom, San Diego, CA, USA). The analysis was carried out on 44 SRS patients, one individual with UPD(7)pat, 39 SGR patients, and 16 controls. Bisulfite conversion with the EZ-96 Methylation Kit (Zymo research corporation), subsequent sample processing, and quality control were carried out at the Karolinska Institutet Mutation Analysis Core Facility. All assays were performed in technical duplicates, and mean of the duplicate measurements was used in the analyses. If one of the duplicate samples failed, then data from the one informative sample only was used. One control sample, one SGR sample and the duplicate of one 11p15 LOM SRS sample were excluded due to experimental failure. Normality of the data was tested using Shapiro-Wilk test of normality and homogeneity of variances by Levene’s statistic test. Data were further analyzed by one-way ANOVA in SPSS. Dunnett t-tests, which compare all patient groups against the control group, were performed post hoc.
Individual methylation level analysis and hypomethylated status
For each CpG in the HOXA4 differentially methylated area, and each EpiTYPER site, standard deviation was calculated from the methylation level of the 16 control samples. Normal distribution of the control group data for each CpG (or a combination of CpG sites in Epityper site HOXA4_F6_CpG_12.13.14) was tested by Shapiro-Wilk normality test. Individual SD values for the SRS patients were calculated based on the standard deviation of the controls. A CpG site was considered hypomethylated when the methylation level was below -2 SD. We considered an individual patient generally hypomethylated at HOXA4 when the methylation level was below -2 SD for at least two-thirds of the CpG sites. For the Illumina assay, the treshold was 10 out of 15 CpG sites (67%), and for the EpiTYPER assay 3 our of 4 sites (75%).
Similarly, the patients were tested for 11p15 LOM in preliminary stages of the data analysis. We considered an individual patient having 11p15 LOM when at least two-thirds or 24 out of 36 (67%) CpG sites were hypomethylated at chr11:2019090–2023450 in hg19 build.
Correlation analysis of HOXA4 methylation level and height in healthy children
Illumina 450 K methylation data from 227 healthy children from the BAMSE cohort were used to study methylation level of each of the HOXA4 differentially methylated area CpGs in relation to their height measurements. Both height measurements and DNA samples for methylation analysis were obtained at approximately age 8. Additionally, records of birth length, height at 4 y and height at 16 y were obtained43. Height measurements were converted to units of standard deviation (SD) for exact age and gender to perform correlation analysis with methylation values. Pearson product-moment correlation coefficient was computed for CpGs with normally distributed data. Normality of the data was tested with Kolmogorov-Smirnov test of normality. Additional correlation analyses were performed by computing Spearman rank-order correlation coefficient for all CpGs in the HOXA4 region of interest. The analysis was performed for each of the CpGs in comparison to birth length and height measurements at each age in SPSS.
Correlation analysis of HOXA4 expression and methylation level
Expression data was taken from our previous publication, where expression of HOXA4 in blood was studied by qPCR17. The expression data from nine UPD(7)mat SRS patients, one individual with UPD(7)pat and ten controls were analyzed for correlation with each of the 16 HOXA4 CpGs listed in Supplementary Table 5. Normality of the data was tested with Shapiro-Wilk test of normality. As expression data was not normally distributed, Spearman rank-order correlation analysis was performed. To calculate average methylation level of the 16 HOXA4 CpGs for each individual, the methylation levels were first converted to SD scale, based on the standard deviation of the 10 controls.
Data availability
The Illumina 450 K BeadChip methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus44 and are accessible through GEO Series accession number GSE104451 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104451).
Electronic supplementary material
Acknowledgements
We thank the patients and families for participation and all referring physicians for collaboration. We thank Eira Leinonen and Auli Saarinen for excellent assistance. This study was supported by the Sigrid Jusélius Foundation (Finland), Swedish Research Council, Swedish Foundation for Strategic Research (the Epigene study), Karolinska Institutet Distinguished Professor Award (to J.K.), the Knut and Alice Wallenberg Foundation, and Doctoral Programme in Biomedicine. The BAMSE birth cohort was supported by grants from the Swedish Foundation for Strategic Research, the Swedish Research Council, Swedish Heart-Lung Foundation, the Stockholm County Council and the Swedish Foundation for Health Care Sciences and Allergy Research, and the European Union. Illumina BeadChip and Sequenom EpiTyper analyses were performed at Karolinska Institutet Mutation Analysis Facility. J.K. is a recipient of The Royal Society Wolfson Research Excellence Award.
Author Contributions
M.M. designed and performed experiments, interpreted results and drafted the manuscript. K.H.-J. collected data, designed experiments and interpreted the results. L.E.R. designed and performed experiments, and interpreted results. C.S. performed data analysis. S.K.M. performed data analysis. A.B., E.M., G.P. contributed the BAMSE cohort. M.L.-N. collected and analyzed/interpreted clinical data. D.G. designed experiments, performed data analysis, interpreted results. J.K. conceived the study, designed experiments, interpreted results. All authors read, edited and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16070-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wakeling EL, et al. Diagnosis and management of Silver-Russell syndrome: first international consensus statement. Nat. Rev. Endocrinol. 2017;13(2):105–24. doi: 10.1038/nrendo.2016.138. [DOI] [PubMed] [Google Scholar]
- 2.Ishida M, Moore GE. The role of imprinted genes in humans. Mol. Aspects Med. 2013;34(4):826–40. doi: 10.1016/j.mam.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Eggermann K, et al. EMQN best practice guidelines for the molecular genetic testing and reporting of chromosome 11p15 imprinting disorders: Silver–Russell and Beckwith–Wiedemann syndrome. Eur. J. Hum. Genet. 2016;24(10):1377–87. doi: 10.1038/ejhg.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotzot D, et al. Uniparental disomy 7 in Silver-Russell syndrome and primordial growth retardation. Hum. Mol. Genet. 1995;4:583–7. doi: 10.1093/hmg/4.4.583. [DOI] [PubMed] [Google Scholar]
- 5.Preece MA, et al. Maternal uniparental disomy 7 in Silver-Russell syndrome. J. Med. Genet. 1997;34(1):6–9. doi: 10.1136/jmg.34.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eggermann T, et al. Molecular studies in 37 Silver-Russell syndrome patients: frequency and etiology of uniparental disomy. Hum. Genet. 1997;100:415–9. doi: 10.1007/s004390050526. [DOI] [PubMed] [Google Scholar]
- 7.Gicquel C, et al. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat. Genet. 2005;37:1003–7. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 8.Eggermann T, et al. Epigenetic mutations in 11p15 in Silver-Russell syndrome are restricted to the telomeric imprinting domain. J. Med. Genet. 2006;43:615–6. doi: 10.1136/jmg.2005.038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bliek J, et al. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am. J. Hum. Genet. 2006;78:604–14. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schönherr N, et al. Epi)mutations in 11p15 significantly contribute to Silver-Russell syndrome: but are they generally involved in growth retardation? Eur. J. Med. Genet. 2006;49:414–8. doi: 10.1016/j.ejmg.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Bruce S, Hannula-Jouppi K, Lindgren CM, Lipsanen-Nyman M, Kere J. Restriction Site-Specific Methylation Studies of Imprinted Genes with Quantitative Real-Time PCR. Clin. Chem. 2008;54(3):491–9. doi: 10.1373/clinchem.2007.098491. [DOI] [PubMed] [Google Scholar]
- 12.Gruzieva O, et al. Epigenome-Wide Meta-Analysis of Methylation in Children Related to Prenatal NO2 Air Pollution Exposure. Environ. Health Perspect. 2017;125(1):104–110. doi: 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lettre G, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40(5):584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40(5):575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson WT, et al. Mutations in EZH2 cause Weaver syndrome. Am. J. Hum. Genet. 2012;90:110–118. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannula-Jouppi K, et al. Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7. Epigenetics. 2014;9(3):351–65. doi: 10.4161/epi.27160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 19.Illingworth R, et al. A Novel CpG Island Set Identifies Tissue-Specific Methylation at Developmental Gene Loci. PLoS Biol. 2008;6(1):e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 21.Zangenberg M, et al. The combined expression of HOXA4 and MEIS1 is an independent prognostic factor in patients with AML. Eur. J. Haematol. 2009;83(5):439–48. doi: 10.1111/j.1600-0609.2009.01309.x. [DOI] [PubMed] [Google Scholar]
- 22.Strathdee G, Sim A, Parker A, Oscier D, Brown R. Promoter hypermethylation silences expression of the HoxA4 gene and correlates with IgVh mutational status in CLL. Leukemia. 2006;20(7):1326–9. doi: 10.1038/sj.leu.2404254. [DOI] [PubMed] [Google Scholar]
- 23.Luedi PP, et al. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–30. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis EB. Clusters of master control genes regulate the development of higher organisms. JAMA. 1992;267:1524–1531. doi: 10.1001/jama.1992.03480110100042. [DOI] [PubMed] [Google Scholar]
- 25.Quinonez SC, Innis JW. Human HOX gene disorders. Mol. Genet. Metab. 2014;111(1):4–15. doi: 10.1016/j.ymgme.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Horan GS, Wu K, Wolgemuth DJ, Behringer RR. Homeotic transformation of cervical vertebrae in Hoxa-4 mutant mice. Proc. Natl. Acad. Sci. USA. 1994;91(26):12644–8. doi: 10.1073/pnas.91.26.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostic D, Capecchi MR. Targeted disruptions of the murine Hoxa-4 and Hoxa-6 genes result in homeotic transformations of components of the vertebral column. Mech. Dev. 1994;46(3):231–47. doi: 10.1016/0925-4773(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 28.Murray PG, et al. 3-M syndrome: a growth disorder associated with IGF2 silencing. Endocr. Connect. 2013;2(4):225–35. doi: 10.1530/EC-13-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Höglund P, Holmberg C, de la Chapelle A, Kere J. Paternal isodisomy for chromosome 7 is compatible with normal growth and development in a patient with congenital chloride diarrhea. Am. J. Hum. Genet. 1994;55:747–52. [PMC free article] [PubMed] [Google Scholar]
- 30.Pan Y, et al. Paternal isodisomy of chromosome 7 associated with complete situs inversus and immotile cilia. Am. J. Hum. Genet. 1998;62:1551–5. doi: 10.1086/301857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Caignec C, et al. Third case of paternal isodisomy for chromosome 7 with cystic fibrosis: a new patient presenting with normal growth. Am. J. Med. Genet. 2007;143A:2696–9. doi: 10.1002/ajmg.a.31999. [DOI] [PubMed] [Google Scholar]
- 32.Prickett AR, et al. Genome-wide methylation analysis in Silver-Russell syndrome patients. Hum. Genet. 2015;134(3):317–32. doi: 10.1007/s00439-014-1526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinius LE, et al. Differential DNA Methylation in Purified Human Blood Cells: Implications for Cell Lineage and Studies on Disease Susceptibility. PLoS ONE. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acevedo N, et al. Age-associated DNA methylation changes in immune genes, histone modifiers and chromatin remodeling factors within 5 years after birth in human blood leukocytes. Clin. Epigenetics. 2015;7(1):34. doi: 10.1186/s13148-015-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannum G, et al. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell. 2013;9(2):359–67. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bacalini MG, et al. A meta-analysis on age-associated changes in blood DNA methylation: results from an original analysis pipeline for Infinium 450k data. Aging. 2015;7(2):97–109. doi: 10.18632/aging.100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpkin AJ, et al. Longitudinal analysis of DNA methylation associated with birth weight and gestational age. Hum. Mol. Genet. 2015;24(13):3752–63. doi: 10.1093/hmg/ddv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saari A, et al. New Finnish growth references for children and adolescents aged 0 to 20 years: Length/height-for-age, weight-for-length/height, and body mass index-for-age. Ann Med. 2011;43(3):235–48. doi: 10.3109/07853890.2010.515603. [DOI] [PubMed] [Google Scholar]
- 39.Aryee MJ, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13(6):R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 42.Smyth, G. K. Limma: linear models for microarray data in Bioinformatics and Computational Biology Solutions using R and Bioconductor (ed. Gentleman, R., Carey, V., Dudoit, S., Irizarry, R., Huber, W.) 397–420 (Springer, 2005).
- 43.Ekström S, et al. Maternal body mass index in early pregnancy and offspring asthma, rhinitis and eczema up to 16 years of age. Clin. Exp. Allergy. 2015;45(1):283–91. doi: 10.1111/cea.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina 450 K BeadChip methylation data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus44 and are accessible through GEO Series accession number GSE104451 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104451).