Abstract
Individuals with 22q11.2 microdeletion syndrome (22q11.2 DS) show cognitive and behavioral dysfunctions, developmental delays in childhood and risk of developing schizophrenia and autism. Despite extensive previous studies in adult animal models, a possible embryonic root of this syndrome has not been determined. Here, in neurons from a 22q11.2 DS mouse model (Lgdel +/−), we found embryonic-premature alterations in the neuronal chloride cotransporters indicated by dysregulated NKCC1 and KCC2 protein expression levels. We demonstrate with large-scale spiking activity recordings a concurrent deregulation of the spontaneous network activity and homeostatic network plasticity. Additionally, Lgdel +/− networks at early development show abnormal neuritogenesis and void of synchronized spontaneous activity. Furthermore, parallel experiments on Dgcr8 +/− mouse cultures reveal a significant, yet not exclusive contribution of the dgcr8 gene to our phenotypes of Lgdel +/− networks. Finally, we show that application of bumetanide, an inhibitor of NKCC1, significantly decreases the hyper-excitable action of GABAA receptor signaling and restores network homeostatic plasticity in Lgdel +/− networks. Overall, by exploiting an on-a-chip 22q11.2 DS model, our results suggest a delayed GABA-switch in Lgdel +/− neurons, which may contribute to a delayed embryonic development. Prospectively, acting on the GABA-polarity switch offers a potential target for 22q11.2 DS therapeutic intervention.
Introduction
The 22q11.2 deletion syndrome (22q11.2 DS), also known as DiGeorge Syndrome (DGS), confers the most significant genetic risk for developing schizophrenia (25–30%) in early adulthood1, autism spectrum disorders (20–25%)2 and predisposition to different psychiatric disorders including cognitive, learning, and behavioral impairments (>60%)3,4. The syndrome occurs in 1 of every 4000 live-births5 and caused by hemizygous deletion of 1.5–3 megabases (Mb) within the region q11.2 of the human chromosome 224,6.
Neuropathology studies have suggested that neurodevelopmental abnormalities occurring in the early brain formation may play a vital role in the pathogenesis of the complex neuropsychiatric phenotypes of this syndrome. Additionally, a wealth of data has demonstrated that the distribution and function of gamma-aminobutyric acid (GABA)-ergic-interneurons are perturbed in the brains of adult 22q11.2 DS patients7 and that impaired GABAergic-signaling is involved in several neurodevelopmental disorders such as fragile×8, autism9, schizophrenia10, Rett Syndrome9, and Down Syndrome11. GABA is the primary inhibitory neurotransmitter in the adult central nervous system (CNS), but in fetal life and early postnatal development, it acts mostly as excitatory, exerting a vital trophic and regulatory role for the maturation of neurons12. Alterations in the GABA-polarity switch can cause a delay in the functional maturation of developing neural circuits13, consequences for network excitability14 and can give rise to neuropsychiatric phenotypes. However, impairments associated with the GABAergic-signaling and correlated network dysfunctions at the early onset of brain circuit development have not yet been reported in the 22q11.2 DS.
During early brain development, immature neurons have a high intracellular chloride concentration [Cl−]int., thus their GABAA receptors (GABAARs)-mediated-signaling sets for depolarization and action potential generation, leading to activation of NMDA receptors (NMDARs) and hence inducing an elevation of Ca2+ concentration15. This excitatory-depolarizing property of GABA is mediated by developmental changes in the expression of neuronal cation-chloride cotransporters (CCCs) that regulate the neuronal chloride homeostasis16. Therefore, in embryonic and early postnatal life, the expression of accumulating-chloride Na+-K+-Cl− cotransporter (NKCC1) is high, while on later developmental stages the expression of extruding-chloride K+-Cl− cotransporter (KCC2) increases17,18. Thus, this dynamic change in the expression of NKCC1 to KCC2 sets an accurate developmental switch of GABAergic-signaling from excitatory-to-inhibitory.
The switch of GABAergic-signaling is one of the drivers that modify the spontaneous activity in early neuronal development. This spontaneous activity plays a fundamental role during critical stages of early brain circuits’ development and in their orchestrated activity-dependent formation19,20. On the other hand, several lines of evidence indicate the essential role of neuronal electrical activity to regulate the synaptic strength and to refine the maturation of developing neural circuits within a neuronal network20. Additionally, in very early brain development, the abnormal neuronal activity can alter the critical homeostatic balance between excitatory (Glutamatergic) and inhibitory (GABAergic) neurotransmission and may contribute to many neurological disorders such as autism, schizophrenia, and epilepsy21,22. GABAergic synapses and receptors are initiated before Glutamatergic ones in the hippocampus and other regions of the brain23; hence, GABAergic-signaling plays an important role for processing and storage of information by controlling signaling cascades that regulate maturation, survival, and differentiation and promote synaptic stabilization and connection, and neuronal network wiring19. Interestingly, the fraction of GABAergic-neurons in rats is already established during the embryonic period from E10.5 through E18.5 and several days after birth24, and the activity oscillation of these interneurons sets the melody for the early network activity25. In turn, this evidence suggests that altered GABA-ergic signaling might be a key factor in the pathophysiology of neural deficits observed in 22q11.2 DS. Several studies performed in adult brains have provided compelling evidence on the neuronal impairments and brain dysfunctions associated with the 22q11.2 microdeletion, but the pathogenesis at early onset of brain development has remained to be investigated. To access the molecular, cellular, synaptic, and circuit-level changes underlying the accurate human phenotype of this syndrome, several mouse models were designed to carry chromosomal defects, which are syntenic to the human 22q11.2 deletion. Two of these multigene deletion models, Df(16A +/−)26 and Lgdel +/− 27, carry a hemizygous deletion in the mouse chromosome 16 that is syntenic to the 1.5 Mb microdeletion of the human chromosome 22q11.2. Notably, these mouse models have become standard models for studying schizophrenia due to their common facets with the 22q11 deletion in schizophrenia. In addition, mouse models carrying single-gene mutations were also generated, including a heterozygous mouse model for the Dgcr8 +/− 28, which carries a deletion in the DGS chromosomal region 8 (dgcr8) gene, one of the deleted genes of the 22q11.2 microdeletion. This gene is a key component of the Microprocessor complex required for biogenesis of microRNA (miRNA)29 and holds a fundamental role in the molecular control of brain development30. Effects of the dgcr8 gene deletion were directly associated with the pathophysiology and cognitive phenotypes of the 22q11.2 DS26,31.
Here, we investigated the role of the switch of GABAergic-signaling in developing neuronal networks prepared from Lgdel +/− and Dgcr8 +/− mouse embryos (Fig. 1a). Specifically, we performed fluorescence high-content imaging (HCI) microscopy and high-resolution bioelectrical extracellular signal recordings using 4096-electrodes complementary metal-oxide semiconductor-multielectrode arrays (CMOS-MEAs) on cultured networks (Fig. 1b,c). Notably, the GABA-polarity switch has been reported to occur at ~10 days-in vitro (DIVs)18 in embryonic rat hippocampal neuronal cultures, and within the second postnatal week ~P932 in rat hippocampal slices. Here, hippocampal neurons from embryonic wild-type (WT), Lgdel +/− and Dgcr8 +/− mice (Fig. 1a and Supplementary Figure S5) were plated at a time when GABA was excitatory (E18.5, 2.5 days before birth)18. The WT refers to the phenotype of the typical form of a mouse as it occurs in nature. Therefore, the WT has a normal genotype, in contrast to the mutant genotypes in Lgdel +/− and Dgcr8 +/− mice; hence, it is considered the control mouse model in our experiments. The expression of NKCC1 and KCC2 as well as the spontaneous electrical activity were monitored and quantified over 26 DIVs. We found that diminished 22q11.2 gene dosage in an embryonic 22q11.2 DS Lgdel +/− mouse culture exhibited a noticeable delay in the regular excitatory-to-inhibitory switch of GABA-signaling during in vitro development, and showed abnormally high levels of NKCC1 even after three weeks in culture. Correlated with this delayed GABA-polarity switch observed in Lgdel +/−, we found an altered spontaneous network activity, lack of synchronization and widespread homeostatic plasticity defects. Concurrent experiments performed on Dgcr8 +/− mouse cultures allowed us to evaluate the contribution of dgcr8 single-gene deletion to the GABA-polarity switch and early network dysfunctions on the multigene deletion of neuronal networks from Lgdel +/− mouse. Finally, the treatment of Lgdel +/− cultures with NKCC1 inhibitor bumetanide rescued the hyper-excitability of GABA-signaling and restored the homeostatic plasticity to levels seen in WT neuronal networks.
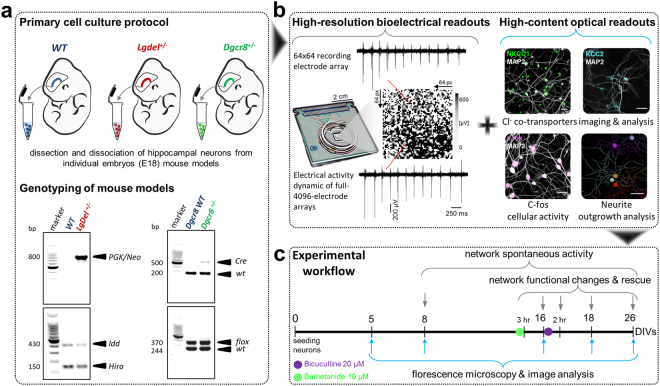
Figure 1.
Schematic of experimental implementation of the 22q11.2 DS model. (a) Single-embryo primary cell culture preparation of hippocampal neurons from WT, Lgdel +/− and Dgcr8 +/− mice and their genotyping. Each cropped gel image reports two representative embryos from groups of animals genotyped for PGK/Neo (top left) and Idd/Hira alleles (bottom left) in (WT and Lgdel +/− embryos) or Cre (top right) and flox alleles (bottom right) in (Dgcr8 WT and Dgcr8 +/− embryos); “full-length gels are presented in Supplementary Figure S5”. (b) Overview of the experimental readouts used in this study, combining electrical measures and optical HCI. Scale bars represent 30 μm. (c) Workflow timeline of the experimental implementation.
Results
Altered expression of NKCC1 and KCC2 in Lgdel+/− and Dgcr8+/− embryonic hippocampal neurons
To investigate whether the GABA-signaling was disrupted by the 22q11.2 microdeletion in Lgdel +/− mouse neuronal culture, we prepared primary hippocampal neurons from embryonic WT and Lgdel +/− mice that were grown in 96 wells imaging-microplates for up to 26 DIVs using optimized cell culture conditions (see Methods). In the same way, we also prepared cultures from Dgcr8 +/− mouse embryos to characterize the effects of the dgcr8 gene deletion. Then, by immunofluorescence and HCI, we measured the expression of the two chloride cotransporters NKCC1 and KCC2 at 8, 16, 26 DIVs. As shown in (Fig. 2a,b and Supplementary Figure S1a), the analysis of the fluorescence intensity for NKCC1 at 8 DIVs showed a significantly higher expression in Lgdel +/− (138.9 ± 4.67%; p < 0.05, ANOVA) compared to Dgcr8 +/− (112.1 ± 4.67%) and to the WT samples. At 16 DIVs, NKCC1 significantly decreased in Lgdel +/− (112.5 ± 1.1%) compared to its expression at 8 DIVs, but it remained significantly higher than WT (88.4 ± 2.86%; p < 0.05, ANOVA) and slightly higher than Dgcr8 +/− (102.3 ± 4.64%). Further, although at 26 DIVs the NKCC1 expression reached its lowest level in Lgdel +/− (105.5 ± 3.78%), its value remained significantly higher compared to Dgcr8 +/− (91.8 ± 2.16%; p < 0.05, ANOVA) and WT (79.6 ± 2.5%; p < 0.05, ANOVA), respectively. On the other hand, as shown in (Fig. 2c,d and Supplementary Figure S1b), the analysis of the fluorescence intensity for KCC2 at 8 DIVs showed the lowest level across the developmental time points, with no significant differences for both Lgdel +/− (90.68 ± 2.23%) and Dgcr8 +/− (87.6 ± 1.49%) compared to the WT samples. At 16 DIVs, KCC2 expression significantly increased in WT cultures (134.5 ± 1.96%) compared to its expression at 8 DIVs, which was also significantly higher compared to Lgdel +/− (91.5 ± 5.64%; p < 0.05, ANOVA) and Dgcr8 +/− (100.29 ± 3.81%; p < 0.05, ANOVA) cultures, respectively. At 26 DIVs, WT cultures showed the highest level of KCC2 expression (214.13 ± 1.88%), while Lgdel +/− and Dgcr8 +/−cultures, albeit their increased KCC2 expression from 16 DIVs, displayed a significantly lower level of expression compared to WT (141.86 ± 4.82%; and 127.6 ± 1.78%; p < 0.05, ANOVA), respectively.
Figure 2.
Altered expression of NKCC1 and KCC2 in Lgdel +/− and Dgcr8 +/− neurons. (a) Fluorescence micrographs showing intensity expression of NKCC1 in cultures from WT, Lgdel +/− and Dgcr8 +/− (n = 5), at 8, 16, 26 DIVs. Scale bar represents 30 μm. (b) Quantification of NKCC1 intensity shows a significantly higher expression in Lgdel +/− neurons (n = 5) compared to WT neurons (n = 7) across all developmental ages (*p < 0.05, ANOVA) and slightly higher than Dgcr8 +/− neurons (n = 5) (ns denotes not significant). (c) Fluorescence micrographs showing the changes of intensity expression of KCC2 in cultures from the three animal genotypes at 8, 16, 26 DIVs. Scale bar represents 30 μm. (d) Quantification of KCC2 intensity at 8 DIVs shows the non-significant difference between genotypes (ns denotes not significant), but displaying a significantly lower expression in Lgdel +/− and Dgcr8 +/− neurons compared to WT neurons at 16 and 26 DIVs (*p < 0.05, ANOVA).
To assess whether the altered protein levels of NKCC1 and KCC2 were accompanied by changes in their transcript levels, we performed quantitative real-time polymerase chain reaction (qRT-PCR) on samples from WT, Lgdel +/− and Dgcr8 +/− at the same time points as in previous experiments (8, 16 and 26 DIVS), by using specific primers for NKCC1 and KCC2 genes. We found a significant alteration of these transcripts only in Lgdel +/− cultures at 8 DIVs compared to WT cultures (Supplementary Figure S2a–d). Importantly, given that we quantified the cotransporters’ expression exclusively for neuronal populations, which is a void resolution in qRT-PCR readouts; this could account for the observed differences between protein and transcript levels in Lgdel +/− cultures compared to WT. On the other hand, the discordance between protein expression and transcripts may suggest a possible alteration in the post-transcriptional control of the cotransporters expression in Lgdel +/− cultures33.
Furthermore, the dgcr8 gene (one of the deleted genes in the 22q11.2 microdeletion), is essential for the biogenesis of miRNAs. Thus, we also attempted to assess the miRNA expression profiles in cultured hippocampal neurons from WT and Lgdel +/− mice, albeit these characterizations are beyond the scope of the current work. We quantified the read counts for all genome-aligned miRNA sequences during embryonic developmental phases (8, 16, and 26 DIVs). We found an incremental delay in the expression level of miRNAs in Lgdel +/− neurons over developmental ages, compared to WT neurons, albeit the increase of read counts from 8 to 26 DIVs in both Lgdel +/− and WT neurons (Supplementary Figure S2e). Further, the analysis of small RNA deep sequencing data from 8 to 26 DIVs in Lgdel +/− and WT samples showed a progressive delay in the increase of miRNAs global expression in Lgdel +/− compared to WT. Interestingly, among the most downregulated miRNAs in LgDel +/− samples, in our experimental attempt, we found miR-340–5p, a candidate regulator of NKCC1 expression, as it was highlighted by different prediction software to possibly target Nkcc1 mRNA. Additionally, we also found miR-101a-3p downregulated in Lgdel +/− compared to WT samples. This is in accordance with a recent study that reported the role of miR-101 to orchestrate early postnatal network development and also to target NKCC1 to facilitate the GABA-signaling switch34.
Taken together, these results reveal the higher NKCC1 and the lower KCC2 expressions in embryonic Lgdel +/− neurons compared to WT neurons. Thus, the deregulation of NKCC1 and KCC2 suggests a delayed GABA-switch in Lgdel +/− neurons that may lead to a developmental delay and altered network formation. Given that NKCC1 and KCC2 expressions were also found significantly altered in Dgcr8 +/− compared to WT cultures, this may indicate a contribution of the dgcr8 gene, i.e., inferred by the altered miRNA expression level, to modulate the GABA-switch delay in the early embryonic development of Lgdel +/− neuronal networks.
Delayed GABAergic-signaling in Lgdel+/− and Dgcr8+/− embryonic hippocampal neurons correlates with an altered spontaneous network activity
Spontaneous electrical neuronal activity is associated with the early development of neural circuits20,25,35. Therefore, we examined whether the expression levels of the chloride cotransporters in WT, Lgdel +/− and Dgcr8 +/− cultures could be correlated with the spontaneous network activity. In particular, we aimed to pinpoint the developmental GABA-polarity switch during network maturation. We first characterized the biochemical profile of the chloride cotransporters NKCC1 and KCC2 (Fig. 3a–c) and the electrophysiological network activity profile (Fig. 3d) at 8, 16 and 26 DIVs. First, based on the quantitative expressions of immature chloride cotransporters that were described earlier in this study, we generated a single-plot showing the expression of NKCC1 and KCC2 in the different genotypes. In turn, this allowed us to estimate the intersection point that confers the GABA-polarity switch time-point. Next, we correlated the occurrence of the switch with the similar spontaneous network firing activity, as obtained from high-resolution extracellular electrical recordings.
Figure 3.
Altered GABAergic-signaling in Lgdel +/− and Dgcr8 +/− neurons is correlated with excitable neuronal network firing activity. (a) In WT neurons, the quantification of the fluorescence intensities of NKCC1 and KCC2 allows estimating an intersection point that indicates the GABA-polarity switch time point at 8 DIVs. Cartoon (right up) illustrates the physiological arrangement (size) of the chloride cotransporters derived from data at 16 DIVs, indicating low and high expression levels of NKCC1 and KCC2, respectively. (b) As in (a), but for Lgdel +/− neurons, the intersection point corresponding to the GABA-polarity switch is delayed with respect to WT, and it is estimated at ~20.5 DIVs. Cartoon (right middle) illustrates the high expression level of NKCC1 and the low expression level of KCC2 observed at 16 DIVs. (c) As in (a) and (b), but for Dgcr8 +/− neurons, the GABA-polarity switch is delayed with respect to WT, and it is estimated at ~17.5 DIVs. Cartoon (right down) illustrates the arrangement of NKCC1 and KCC2 cotransporters at 16 DIVs. (d) The MFR computed from the spontaneous activity of WT, Lgdel +/− and Dgcr8 +/− embryonic hippocampal neuronal networks at 8, 16 and 26 DIVs. At 16 DIVs, compared to WT, the hyper-excitable Lgdel +/− and Dgcr8 +/− networks showing evident correlation with the expression level of the chloride cotransporters and with the delayed GABA-polarity switches. The hyper-excitable state is indicated by a significantly higher MFR in Lgdel +/− and Dgcr8 +/− networks compared to WT networks (**denotes p < 0.01 Lgdel +/− vs. WT,++ denotes p < 0.01 Dgcr8 +/− vs. WT, ANOVA).
To perform network activity recordings from WT, Lgdel +/− and Dgcr8 +/− neuronal cultures at 8, 16, and 26 DIVs, we used CMOS-MEAs36–38 with cell-culture and substrate functionalization conditions optimized for mouse primary neuronal cultures (see Methods). These devices provide 4096 closely spaced electrodes for simultaneously recording the spontaneous electrical activity of several thousands of single hippocampal neurons within the network. To characterize the network-wide activity levels we computed the mean firing rate (MFR), and we used this parameter to correlate the activity with the estimated timing of the GABA-polarity switch in the different cultures. In WT cultures, we found a dynamic switch in the action of GABA from excitatory-to-inhibitory that occurred at 8 DIVs, as shown by the intersection of NKCC1 and KCC2 expressions in (Fig. 3a). This GABA-polarity switch time-point was correlated with the increase of the MFR that followed the temporal maturation of the neural network from 8 to 26 DIVs until the complete maturation of the glutamatergic synapses was reached39 (Fig. 3d; blue line). On the other hand, it is intriguing to remark that the intersection of NKCC1 and KCC2 in Lgdel +/− cultures was delayed and occurred after 16 DIV (estimated at ~20.5 DIVs) (Fig. 3b). This delayed switch was correlated with the observed excitable network activity indicated by the pinnacle of MFR at 16 DIVs (Fig. 3d; red line). Afterward, the activity in Lgdel +/− cultures progressively declined over 26 DIVs. Finally, results from the intersection of NKCC1 and KCC2 expression levels of Dgcr8 +/− cultures elucidated a GABA-polarity switch that also occurred after 16 DIV (estimated at ~17.5 DIVs) (Fig. 3c), slightly before the switch estimated for Lgdel +/− cultures. The switch time-point of Dgcr8 +/− cultures was correlated with the pinnacle of MFR at 16 DIVs (Fig. 3d; green line), which is indicative of an excitable network behavior as observed in Lgdel +/− cultures, but minimally subsided to reach a similar level of activity as in WT cultures at 26 DIVs.
Altogether, these results suggest significant developmental delays in networks of embryonic Lgdel +/− and Dgcr8 +/− neurons compared to WT. For both Lgdel +/− and Dgcr8 +/− cell cultures, this functional derangement in the network activity is correlated with an altered expression of the chloride cotransporters, as indicated by the delayed decrease of NKCC1 expression and the delayed increased of KCC2 expression (Fig. 3, cartoon at 16 DIVs). Our results also unveil the contribution of the dgcr8 gene deletion to the altered GABA-polarity switch and the electrophysiological network activity observed in the Lgdel +/− cultures.
Network homeostatic plasticity is impaired in Lgdel+/− and attenuated in Dgcr8+/− embryonic hippocampal cultures
Network homeostatic plasticity is a key mechanism that allows maintaining a stable, functional state in neural circuits when the network is subdued by developmental or input-driven changes40. Homeostatic responses are an essential property of healthy networks. In contrast, the disability of a network to restore similar initial activity levels upon mild extrinsic perturbations (induced by pharmacological manipulations), or intrinsic modifications (neurodevelopmental changes), is an indicator of pathological network impairments. Notably, previous studies reported an age-dependent alteration of synaptic plasticity in adult Df(16)A +/− and Dgcr8 +/−mice28,41,42. We thus asked whether Lgdel +/− and Dgcr8 +/− neuronal networks exhibited plasticity deficits also at early neurodevelopmental phases, which in turn, might affect the neuronal maturation.
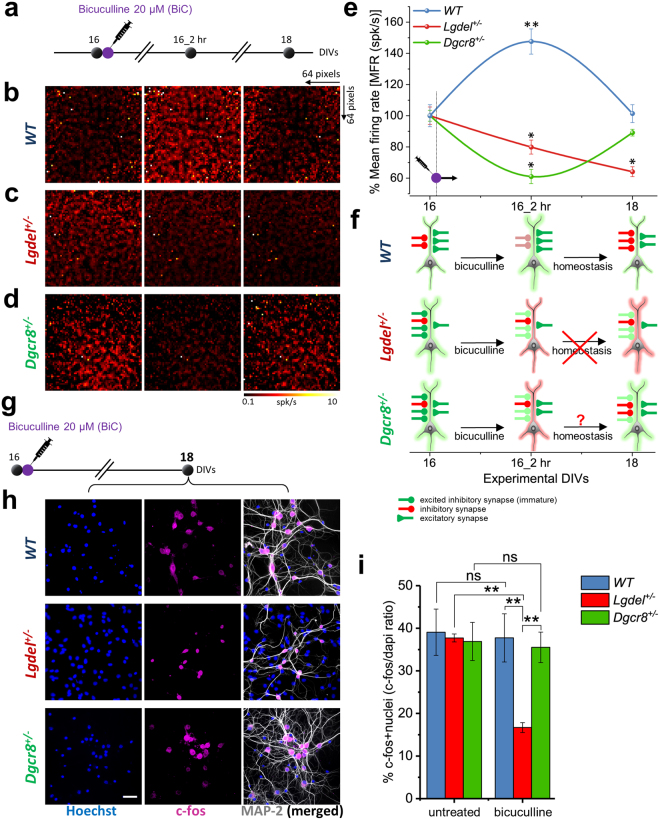
To characterize homeostatic plasticity responses, hippocampal neuronal cultures from WT, Lgdel +/− and Dgcr8 +/− mouse embryos were treated at 16 DIVs with 20 μM bicuculline and the network-wide firing activity was then monitored for 48 hr after bicuculline-treatment with CMOS-MEAs recordings (Fig. 4a). As it was previously demonstrated40; here, we aimed at using this mild pharmacological manipulation to temporarily destabilize the network by slightly elevating the network firing rate through a small reduction of the GABAergic network inhibition. As expected, WT cultures showed a significant increase in the mean firing activity of the network after 2 hr bicuculline-treatment (147.58 ± 10.05% spk/s; p < 0.01, ANOVA) and a return nearly to its baseline after 48 hr (101.38 ± 0.7% spk/s) (Fig. 4b,e blue). In contrast, in Lgdel +/− cultures (Fig. 4c,e red), the network firing activity progressively declined after 2 hr bicuculline-treatment (79.88 ± 4.48% spk/s; p < 0.05, ANOVA) and this diminution continued over 48 hr after bicuculline-treatment (64.11 ± 3.13% spk/s; p < 0.05, ANOVA) with respect to its baseline firing activity. Remarkably, Dgcr8 +/− cultures showed a promiscuous combination of the network firing behavior observed in WT and Lgdel +/− cultures (Fig. 4d,e green). Indeed, Dgcr8 +/− cultures responded with a significant decay in activity after 2 hr bicuculline-treatment (61.01 ± 1.56% spk/s; p < 0.05, ANOVA), as observed in LgDel +/− networks, and a restored firing activity returning to the baseline after 48 hr bicuculline-treatment (89.12 ± 2.15% spk/s; p < 0.05, ANOVA), as yielded in the WT networks. These results reveal the homeostatic dysregulation of Lgdel +/− and Dgcr8 +/− networks with respect to WT.
Figure 4.
Electrical and optical evidence of dysregulated network homeostatic plasticity in Lgdel +/− and Dgcr8 +/− networks. (a) Schematic illustration of the experimental protocol used to test network homeostatic plasticity responses upon 20 µM bicuculline-treatment. (b) Snapshots showing the homogenous averaged MFR maps (64 × 64 electrode pixels) for a WT network at the different experimental time points and each map is computed for a recording of 10 minutes. (c) As in (b), but for a Lgdel +/− network. At 16 DIVs the network manifests an already higher averaged MFR than WT. Upon bicuculline-treatment the maps at 16_2 hr and 18 DIVs indicate a decrease of the network firing activity without the homeostatic restoration to the initial condition. (d) As in (b) and (c), but for a Dgcr8 +/− network. (e) Quantification of the MFR shows after bicuculline-treatment (16_2 hr), a significantly decrease in Lgdel +/− and Dgcr8 +/− networks, and a significantly increase in WT from the initial baseline (**p < 0.01 WT, *p < 0.05 Lgdel +/−, *p < 0.05 Dgcr8 +/−, ANOVA; n = 5). At 18 DIVs, the MFR continuously decreases in Lgdel +/− networks, while it returns to the baseline value in Dgcr8 +/− networks and sets to a nearly similar MFR of WT. (f) Cartoon illustrates the homeostatic regulation of the excitation-inhibition balance in WT, Lgdel +/− and Dgcr8 +/− neurons at the different time points. (g) Schematic of the experimental protocol used to assay the cellular neuronal activity with c-fos immunofluorescence after 48 hr from bicuculline-treatment. (h) Fluorescence micrographs showing the c-fos expression 48 hr after bicuculline-treatment in WT, Lgdel +/− and Dgcr8 +/− networks and confirming previous electrical read-outs. (i) Quantification of c-fos-positive nuclei ratio 48 hr after bicuculline-treatment. Data showing a significant decrease in expression in Lgdel +/− neurons compared to WT, and Dgcr8 +/− neurons (**p < 0.01, ANOVA; n = 5), as well as compared to the expression in Lgdel +/− neurons before bicuculline-treatment (untreated) (**p < 0.01, ANOVA, n = 5).
Our results are delineated in (Fig. 4f) showing in WT, the homeostatically regulated firing activity by the cooperation of excitatory (green) and inhibitory (red) synapses. At 16 DIVs Lgdel +/− already holds higher immature excited inhibitory synapses than excitatory synapses, which leads to a reduction of the network firing activity after bicuculline-treatment due to dysregulated homeostatic plasticity. Although Dgcr8 +/− at 16 DIVs possess more immature excited inhibitory synapses than excitatory synapses as Lgdel +/−, Dgcr8 +/− succeeds to return to the baseline network firing activity. This happens because the GABA-polarity switch in Dgcr8 +/− neurons occurs earlier than in Lgdel +/− neurons.
Further, to validate at the cellular level these altered homeostatic plasticity responses observed with electrical recordings, we measured by HCI the level of c-fos expression in neurons from WT, Lgdel +/− and Dgcr8 +/− cultures at 16 DIVs (baseline) and 18 DIVs (after 48 hr bicuculline-treatment) (Fig. 4g–i and Supplementary Figure S3a,b). Quantifications of these optical readouts revealed the changes in the cellular neuronal activity (Fig. 4i). At 16 DIVs, before bicuculline-treatment, we found similar c-fos-positive nuclei ratios under untreated conditions in WT, Lgdel +/− and Dgcr8 +/− cultures (39.06 ± 5.44%, 37.72 ± 0.92% and 36.9 ± 4.48%), respectively (Fig. 4h and Supplementary Figure S3a,b). After 48 hr bicuculline-treatment, the c-fos-positive nuclei ratios in WT and Dgcr8 +/− cultures showed a restoration to the untreated baseline condition (37.73 ± 5.65% and 35.51 ± 3.58%), respectively (Fig. 4h,i). In contrast, Lgdel +/− cultures exhibited a significant reduction of c-fos-positive nuclei ratio (16.68 ± 1.16%; p < 0.01, ANOVA) compared to their baseline untreated condition (Fig. 4h,i and Supplementary Figure S3a,b). Thus, this significant cellular loss of c-fos-positive neurons in Lgdel +/− cultures confirms the decayed network activity after 48 hr bicuculline-treatment observed with our electrical recordings.
Next, we tested whether homeostatic plasticity deficits in Lgdel +/− and Dgcr8 +/− cultures yielded a functional derangement of the network dynamics based on the particular firing frequency of neuronal populations in the network. To do so, we exploited our large-scale electrical recordings to analyze the multiunit distributions of neuronal firing rates using lognormal-like quantification, as previously reported in in vivo 43 and in vitro 44 studies. Although there is no discernable boundary between the two sides of a lognormal distribution, a biological notion can be resolved based on the firing frequencies associated with a given neuronal population. Indeed, under physiological conditions, the right-tail of the firing distribution denotes a minority of highly active neurons (interneurons). Given the strong connections of this fast-firing minority population in a network confer stability and transformation of neural information. On the other hand, the left tail of the distribution indicates the firing activity of slow-discharge neurons that may provide excitation and set plastic features to a neural network45. In turn, this can provide a balance mechanism to maintain the mean activity level of a self-organized network and to ensure inhibition by counteracting the effects of the fast-firing minority population. We found that after 2 hr bicuculline-treatment, the lognormal-like distribution of WT networks was skewed toward high frequencies compared to the distribution at 16 DIVs, as indicated by the mean parameter of the Gaussian fit (Supplementary Figure S4a,b,d blue). After 48 hr bicuculline-treatment, the distribution of WT networks shifted toward left (low firing frequencies), thus indicating that networks were driven by homeostatic plasticity to stabilize the firing activity to a baseline state similar to the one observed at 16 DIVs (Supplementary Figure S4c,d blue). The peaks of the Gaussian distributions in Lgdel +/− and Dgcr8 +/− networks at 16 DIVs already yielded higher frequencies with respect to WT networks (Supplementary Figure S4a,d red, and green). On the contrary to the WT, we found that in Lgdel +/− and Dgcr8 +/− networks after 2 hr bicuculline-treatment, the lognormal-like distributions shifted to the left, toward lower frequencies (Supplementary Figure S4b,d red, and green). After 48 hr bicuculline-treatment, the distribution of Lgdel +/− networks shifted further left toward low frequencies, while Dgcr8 +/− networks returned nearly to the initial state observed at 16 DIVs (Supplementary Figure S4c,d red, and green). Therefore, right tails of the distributions of Lgdel +/− and Dgcr8 +/− networks confirm that GABA is excitatory and in turn, by blocking this neuronal population with bicuculline, the firing activity is decreased, and the distributions shift toward the left.
In sum, these results demonstrate the abnormal firing dynamics of Lgdel +/− and Dgcr8 +/− networks associated with dysregulated homeostatic plasticity to set the appropriate synaptic rule to counter the contribution of excitable GABAergic neurons. Further, these responses can be interpreted as a consequence of the altered GABA-polarity switch that we have found in the neuronal populations of Lgdel +/− and Dgcr8 +/− networks.
Synchronous activity is altered in Lgdel+/− embryonic hippocampal networks
Synchronous spiking activity and changes in its dynamics are fundamental for brain circuits development46 and count for the critical establishment of synaptic connections47 and activity-dependent network formation48. Synchronous activity arises from dynamically connected neural assemblies has been postulated to render processing of neuronal information during early developmental phases49. A wealth of data suggests that in vitro spontaneous events of synchronous spikes (or bursts) expressed by networks formed by embryonic neurons may retain some properties of the early synchronous activity observed in vivo 50. Here, we investigated whether burst properties varied during the formation of Lgdel +/− and Dgcr8 +/− neuronal networks, compared to WT networks (Fig. 5a). In order to characterize the onset of bursts and their properties during development from CMOS-MEAs recordings obtained from these neuronal cultures, we quantified the mean bursting rate (MBR) (Fig. 5b), the average count of network-wide burst events (NBs) (Fig. 5c) and the distributions of inter-burst intervals (IBIs) (Fig. 5d). The MBR was computed from all single active electrodes that were recording bursts during sessions of 10 minutes; the NBs confer the level of temporal synchronization over a 100 ms time-window, and the IBIs characterize the mean time-distance between each subsequent NB event.
Figure 5.
Desynchronization and deregulation of spontaneous bursting activity in Lgdel +/− networks over developmental phases. (a) Graphs display the network-wide averaged firing rate (AFR) and raster plots of the spiking activity from recordings in WT, Lgdel +/− and Dgcr8 +/− networks (n = 5) at 8, 16 and 26 DIVs. (b) MBR indicates the mean bursting rate computed for single electrodes. At 8 DIVs, the MBR displays a significantly lower value in Lgdel +/− network compared to WT and Dgcr8 +/− networks (**denotes p < 0.01 to WT, ++ denotes p < 0.01 to Dgcr8 +/−, ANOVA). During development, Lgdel +/− networks manifest a tendency to increase the MBR and reach the highest value at 26 DIVs, with a similar MBR to Dgcr8 +/− networks, but significantly lower than WT networks (**denotes p < 0.01, ANOVA). (c) The quantification of NBs shows the lack of Lgdel +/− networks to express these events at 8 DIVs compared to WT and Dgcr8 +/− networks. In all networks NBs increase over developmental phases. Lgdel +/− networks reach nearly similar NB counts of WT at 16 DIVs but manifest a significantly lower NB counts than Dgcr8 +/− network (*p < 0.01, ANOVA). At 26 DIVs Lgdel +/− networks manifest significantly lower NB counts than WT and Dgcr8 +/− networks (*p < 0.01, ANOVA). (d) Distribution of IBIs yielded from the temporal classification of bursts in the selected time bins from WT networks. (e) As in (d), but for Lgdel +/− networks. (f) As in (d) and (e), but for Dgcr8 +/− networks.
In WT and Dgcr8 +/− networks, we observed the first network-wide synchronous spiking events (NBs) at 8 DIVs, whilst Lgdel +/− networks did not show any NB event at this age (Fig. 5a and Supplementary movies), albeit minimal single-electrode bursting activity as indicated by the MBR value (Fig. 5b). During later stages of development, the spiking rate increased in all networks, and the dynamics of their activity changed, showing a different degree of burstiness, in addition to the temporal clustering of the burst events51. At 16 DIVs, regular network-wide bursts were observed in WT, Lgdel +/− and Dgcr8 +/− cultures (Fig. 5a). Notably, at 26 DIVs WT and Dgcr8 +/− cultures showed a particular change in their bursting pattern and expressed NBs that occurred over short-time periods separated by prolonged silent time windows (Fig. 5a). This dynamic was not observed in Lgdel +/− cultures that remained expressing consistent bursting patterns, but with a lower spiking rate compared to events at earlier DIVs (Fig. 5a). As shown in (Fig. 5b,c), the quantification of the burst parameters at 8 DIVs in Dgcr8 +/− networks showed very similar MBR (88.72 ± 5.86% burst/min) and NBs (45 ± 16.89%) compared to WT networks. In contrast, Lgdel +/− networks showed a significantly lower MBR (33.16 ± 6.7% burst/min; p < 0.01, ANOVA) compared to WT and Dgcr8 +/− networks and lacked the NBs. At 16 DIVs, Lgdel +/− networks showed nearly similar MBR (53.24 ± 5.87% burst/min) compared to WT and Dgcr8 +/− networks, as well as similar NBs to WT, but 1.6-fold lower NBs compared to Dgcr8 +/− networks. At 26 DIVs, the MBR increased significantly for WT networks compared to Lgdel +/− and Dgcr8 +/− networks (93.7 ± 5.03% burst/min; p < 0.05, ANOVA), while the NBs for WT and Dgcr8 +/− networks significantly increased compared to Lgdel +/− networks. Noticeably, the IBIs changed over development. At early network developmental phases, the IBIs were marked with widely temporally distributed bursts, while at older stages they were characterized by bursting dynamics that changed into more narrowly distributed events (Fig. 5d). At 26 DIVs, the network-wide synchronized events recorded from WT and Dgcr8 +/− cultures appeared as “superburst-like patterns” (Fig. 5a). Interestingly, these bursting patterns were previously associated with a higher level of network maturation in rodent neuronal cultures51. In contrast, Lgdel +/− networks did not show any superburst-like events and remained expressing fixed-size bursts; thus, indicating a lower level of maturation on both WT and Dgcr8 +/− cultures (Fig. 5a).
Taken together, these results indicate that networks formed by Lgdel +/− embryonic neurons show a significant delay in the expression of synchronous spiking activity and the developmental changes of the dynamics of these events with respect to WT and Dgcr8 +/− networks. Importantly, this might indicate the presence of severe alterations in the activity-dependent formation of neuronal networks in Lgdel +/− models during their early development.
Morphological alterations in Lgdel+/− and Dgcr8+/− embryonic hippocampal neurons
Evidence in adult animals has been found for abnormalities in spine morphogenesis that ascribed to the deficits of structural synaptic plasticity and neuronal morphological alterations in the 22q11.2 microdeletion mouse model52,53. Here, we aimed at assessing whether, at very early embryonic developmental stages, Lgdel +/− and Dgcr8 +/− networks exhibited structural deficits in features of neurite outgrowth that could be associated with the observed functional impairments. We performed morphological measurements of neurite processes by using an automated fluorescence HCI system on WT, Lgdel +/− and Dgcr8 +/− embryonic hippocampal cultures at 5 DIVs. The acquired images were quantified based on the detection of two fluorescence channels (Fig. 6a). We quantified the neuritogenesis parameters and generated a series of measures on the neurite outgrowth.
Figure 6.
Neurite outgrowth measurements unveil alteration in neuromorphogenesis in Lgdel +/− and Dgcr8 +/− neurons. (a) Micrographs showing the raw image of WT neurons (MAP-2) and the segmentation mask for cell bodies (Hoechst) that are used by the neurite outgrowth algorithm to obtain the final cell body and neurite segmentation masks. Scale bar represents 30 µm. (b) Micrographs of WT, Lgdel +/− and Dgcr8 +/− neurons before segmentation (raw images) and after final segmentation. Scale bar is 30 µm. (c) Quantifications of neurite outgrowth measurements from WT, Lgdel +/− and Dgcr8 +/− neurons (n = 5 for). Statistical comparisons confirm that all parameters’ values in Lgdel +/− and Dgcr8 +/− neurons are significantly lower than WT neurons (**p < 0.01, ANOVA). (d) Signal traces of typical spontaneous spikes extracellularly recorded from WT, Lgdel +/− and Dgcr8 +/− cultures at 8 DIVs. (e) Quantifications of the negative peaks of the spontaneous extracellular spikes recorded from WT, Lgdel +/− and Dgcr8 +/− cultures (n = 5) at different time points (8, 12, 16, 18 DIVs). This analysis reveals the non-significant difference of the spike amplitudes between the genotypes.
Our results showed that all measures of the averaged neurite outgrowth parameters were significantly reduced in both Lgdel +/− and Dgcr8 +/− cultures compared to WT cultures (p < 0.01, ANOVA) (Fig. 6b,c). Theoretical and experimental approaches have suggested that neuritogenesis plays a role in modulating synaptic efficacy and in the generation of spiking activity54,55. Thus, we next examined whether alterations in neurite morphology at early onset (5 DIVs) was accompanied by changes in the amplitude of the spontaneous firing activity in later developing phases (8, 12, 16 and 18 DIVs). To do so, we analyzed the averaged extracellular signals-amplitudes that were recorded from WT, Lgdel +/− and Dgcr8 +/− networks. This analysis was performed with a highly conservative thresholding method that was assessed by pre-selecting three different signal amplitude values. Further, the average of signal amplitudes was computed for all extracellular signals that were larger than (−250 μV). However, despite the observed morphological differences, the signal amplitude analysis showed no significant difference between WT, Lgdel +/− and Dgcr8 +/− (Fig. 6d,e). On the whole, these results suggest that the structural deficits observed in Lgdel +/− and Dgcr8 +/− cultures might be a consequence of the network activity dysfunctions, resulting from dysregulated expression of the chloride cotransporters, rather than being their source.
Bumetanide restores physiological network behavior and rescues homeostatic deficits in Lgdel+/− embryonic hippocampal neurons
Recently, a growing body of evidence suggests that the NKCC1 antagonist bumetanide, an already FDA approved drug56, attenuates the intracellular chloride concentration [Cl−]int. during the excitatory action of GABA responses in immature neurons, thus producing a negative shift in reversal potential of the GABAA receptor (EGABA). Noticeably, when bumetanide is administrated in a concentration range of 2–10 μM, it selectively inhibits NKCC1 without affecting KCC257. Therefore, we evaluated whether 10 μM of bumetanide could consistently suppress the high activity of NKCC1 observed in Lgdel +/− immature networks. The rescuing capability of bumetanide was tested both on the spontaneous network-wide activity and to the homeostatic network response induced by bicuculline.
In the first set of experiments (Fig. 7a), we applied bumetanide at 16 DIVs to the cell culture media and evaluated its effect on the network-wide spontaneous activity after 3 hr. We found the MFR reduced by 2-fold in bumetanide-treated Lgdel +/− cultures (74.53 ± 7.69%; p < 0.01, ANOVA) compared to untreated Lgdel +/− cultures (147.51 ± 5.58%). In WT cultures treated with bumetanide, the spiking activity (88.17 ± 2.46%) remained similar to one of the bumetanide-untreated (89.98 ± 12.86%) samples. At 18 DIVs, the same Lgdel +/− cultures that were previously treated with bumetanide showed 1.67-fold lower mean firing rate (83.76 ± 8.05%; p < 0.01, ANOVA) than untreated Lgdel +/− samples (139.53 ± 1.03%). In WT cultures we observed nearly identical mean spiking activity levels both for bumetanide-treated (87.80 ± 4.14%) and untreated (88.21 ± 14.38%) samples, respectively. At 26 DIVs, bumetanide-untreated Lgdel +/− cultures showed a dramatic decrease in activity (83.76 ± 8.05%) compared to WT networks, while bumetanide-treated Lgdel +/− cultures displayed 1.47-fold significant recovery (113.4 ± 8.45; p < 0.05, ANOVA) compared to Lgdel +/− untreated cultures and a MFR comparable to WT cultures.
Figure 7.
Bumetanide rescues the abnormally excitable Lgdel +/− neuronal network and restores physiological homeostatic plasticity. (a) Schematic summary of the experimental protocol used to test the effect of 10 µM bumetanide (administrated 3 hr before the baseline recordings at 16 DIVs) to rescue the spontaneous hyper-excitable activity in Lgdel +/− networks. (b) MFR indicates the spontaneous firing activity of WT and Lgdel +/− networks across the developmental phases from 16 to 26 DIVs. After 3 hr from bumetanide-treatment, Lgdel +/− cultures at 16 DIVs exert 2-fold decrease of MFR compared to untreated Lgdel +/− cultures (**p < 0.01, ANOVA; n = 5) and express a nearly analogous MFR as WT cultures. At 18 DIVs, the MFR in bumetanide-treated Lgdel +/− cultures remains significantly low (1.67-fold) compared to the untreated cultures (**p < 0.01, ANOVA; n = 5). At 26 DIVs, bumetanide-untreated Lgdel +/− cultures exert a steep decrease in the MFR, while the BUM-treated Lgdel +/− cultures display 1.47-fold significant recovery (**p < 0.05, ANOVA; n = 5). (c) Schematic summary of the experimental protocol used to test the effect of 10 µM bumetanide (administrated 3 hr before the baseline recordings at 16 DIVs) to rescue dysregulated homeostatic plasticity response in Lgdel +/− networks upon 2 hr of 20 µM bicuculline-treatment. (d) At 16 DIVs, 5 hr after bumetanide-treatment and 2 hr after bicuculline, Lgdel +/− networks display 1.67-fold significant increase of MFR compared to bumetanide-untreated bicuculline-treated Lgdel +/− cultures (**p < 0.01, ANOVA; n = 5), and showing a nearly analogous MFR as WT cultures. At 18 DIVs, bumetanide-treated Lgdel +/− cultures display a complete restoration to the baseline activity recorded at 16 DIVs (prior bicuculline-treatment) compared to bumetanide-untreated Lgdel +/− cultures (*p < 0.01, ANOVA; n = 5).
In the second set of experiments (Fig. 7b), we evaluated the effects of bumetanide to rescue response of homeostatic Lgdel +/− networks. To do so, at 16 DIVs and immediately after baseline spiking activity recordings, 10 μM bumetanide was added to Lgdel +/− and WT cultures and left in the bath for 3 hr. Then, 20 μM bicuculline was co-administrated for 2 hr, and the network activity was recorded. Similar to WT networks, we found that the firing activity in Lgdel +/− cultures treated with bumetanide and bicuculline increased 1.67-fold (133.55 ± 6.19%; p < 0.01, ANOVA) compared to bumetanide-untreated Lgdel +/− cultures (79.88 ± 4.48%). We observed ostensibly similar spiking activity in bumetanide-treated (140.06 ± 8.74%) and bumetanide-untreated (147.58 ± 0.05%) WT networks under bicuculline, respectively. Remarkably, at 18 DIVs the network activity of bumetanide-treated Lgdel +/− was completely returned to the baseline activity recorded at 16 DIVs (94.95 ± 11.09%; p < 0.01, ANOVA), similarly as bumetanide-treated (96.7 ± 6.35%) and bumetanide-untreated (101.38 ± 1.7%) WT samples. On the other hand, bumetanide-untreated Lgdel +/− cultures showed a persistent decrease in the spiking activity that reached a 1.5-fold lower level than bumetanide-treated Lgdel +/− cultures.
In summary, by regulating the activity of the chloride cotransporter NKCC1, our results on bumetanide-treated Lgdel +/− cultures suggest that the altered expression level of NKCC1 is the molecular mechanism responsible for the delayed network activity development and impaired network homeostatic plasticity observed in Lgdel +/−networks. This delineates the potential therapeutic effect of bumetanide to restore normal embryonic development in 22q11.2 DS.
Discussion
Several studies on adult 22q11.2 DS patients have suggested neurodevelopmental abnormalities playing an important role in the pathogenesis of this syndrome. Additionally, the last decade has witnessed a surge of interest in investigating the role of GABAergic-signaling in several neurodevelopmental disorders8–10,58, including schizophrenia, which is a neuropsychiatric disorder associated with a high genetic risk for 22q11.2 DS. Importantly, defects of GABAergic-neurotransmission observed in adult patients with the 22q11.2 DS may also be related to aggravation of GABAergic-signaling during early development, but this hypothesis was not yet examined at embryonic developmental stages. Interestingly, GABAergic-interneurons distribution was reported to be significantly altered in adult Lgdel +/− mouse brain circuits59.
In this work, we examined GABAergic-signaling in in vitro networks prepared from embryonic hippocampal neurons from Lgdel +/− and Dgcr8 +/− mice embryos. We monitored the level of expression of the cation-chloride cotransporters NKCC1 and KCC2, the associated electrophysiological developmental profile, and we characterized homeostatic network responses to mild perturbations induced by small doses of bicuculline.
Our results reveal for the first time a critical developmental delay in embryonic networks of Lgdel +/− and Dgcr8 +/−neurons. We observed alterations in the expression of NKCC1 and KCC2 cotransporters and the association of these expression levels with a hyper-excitability of the network. These deregulated expressions reveal an altered timing of the GABA-polarity switch and correlate with a delay in the functional maturation of the networks. Our data pinpoint specific time switches occurring at ~20.5 DIVs for Lgdel +/− neuronal networks and at ~17.5 DIVs for Dgcr8 +/− neuronal networks, thus occurring significantly later compared to WT networks, switching earlier than 8 DIVs. Alterations in the GABA-polarity switch have previously been documented to impact the balance between GABAergic and Glutamatergic transmission in neural networks60. Central to these findings is the indication on the early depolarization of GABA-releasing synapses to drive the development of Glutamatergic synapses61, where the latter was shown to be reduced in Df(16)A +/− mouse model (i.e., a multigene deletion mouse model that spans a segment syntenic to the 1.5-Mb human 22q11.2 microdeletion and encompasses 27 genes)52. Within this line of evidence, our results explicitly elucidate the analogues deregulation in the network-wide spontaneous activity mandated by hyper-excitable Lgdel +/− and Dgcr8 +/− networks compared to the WT ones. We also found additional evidence on the delayed developmental profile of Lgdel +/− networks by analyzing the spontaneous synchronous network activity. Indeed, Lgdel +/− networks at 8 DIVs express asynchronous spikes, while WT and Dgcr8 +/− networks already exert synchronous activity at this early stage of network development. At older developmental stages (26 DIVs), Lgdel +/− networks also express synchronous activity, but display dynamics corresponding to an early stage of maturation compared to WT and Dgcr8 +/− networks. We postulate that these results on Lgdel +/− networks likely arise from the overall delayed NKCC1 and KCC2 expression profile that induces an immature balance of excitation-inhibition62.
As shown in our results, the delayed GABA-polarity switch also has implications on establishing homeostatic plasticity properties during early phases of network development. An extensive literature highlights the essential role of homeostatic plasticity to adjust the network firing rate dynamically and to maintain neuronal network stability40. While WT networks at 16 DIVs exerted an increased excitation in response to bicuculline, we observed an impaired network homeostatic plasticity in Lgdel +/− cultures. These networks are unable to reset to the baseline activity levels and to counteract the dysfunction of inhibitory circuits that still operate as excitatory due to the chloride cotransporters alterations. On the other hand, the homeostatic response of Dgcr8 +/− networks was found altered with respect to WT, but it was not completely disrupted as for Lgdel +/− networks. The alteration is likely due to a slightly earlier GABA-polarity switch in Dgcr8 +/− networks than Lgdel +/− networks. Cellular quantifications of the c-fos-positive nuclei and the analysis of the dynamical shift of lognormal-like distributions of the neuronal firing frequencies further confirm our results on the electrically characterized homeostatic response.
Given the compelling evidence from some converging experiments on the 22q11.2 mouse model that suggested deficits in the synaptic connectivity and spine morphogenesis52,53, we also assessed the presence of abnormal neuritogenesis in Lgdel +/− and Dgcr8 +/− networks. In both conditions, we found these abnormalities by multi-parameter analysis obtained with unbiased automated image-based assays. Interestingly, these structural alterations may arise from the network dysfunction per se rather than being the cause of these malfunctions63. To investigate this, we compared the amplitude of the spikes over development (8, 16, 18 DIVs) for WT, Lgdel +/−, and Dgcr8 +/− networks. Our data show a non-significant change in the spiking amplitude, despite the neuritic structure abnormalities. These results suggest that abnormal neuritogenesis in Lgdel +/− and Dgcr8 +/− networks might be a consequence of the network dysfunction.
To attenuate the excitable network-wide spontaneous activity and to attempt rescuing the neuronal chloride homeostatic deficits, we treated the Lgdel +/− immature networks with 10 μM bumetanide14. Our results demonstrate that this treatment can rescue both the activity levels and homeostatic responses of Lgdel +/− networks with respect to WT. The treatment effect does not only confirm that our results on the network dysfunctions are essentially due to the alteration of the GABAergic-signaling switch but importantly, it also indicates the first time the possible therapeutic effects of blocking the high activity of NKCC1 in early developing networks of the 22q11.2 DS. Remarkably, the bumetanide concentration used in our study selectively inhibits NKCC1 hyperactivity in Lgdel +/− networks and shows no effects on the network activity and homeostatic response of WT neuronal networks. In turn, this indicates the GABA-dependent hyper-excitability mechanism of bumetanide.
Our study also reveals a significant, yet not exclusive contribution of the dgcr8 gene haploinsufficiency in the GABA-polarity switch and electrophysiological phenotypes observed in the Lgdel +/− neuronal networks. Dgcr8 gene (one of the deleted genes in the 22q11.2 microdeletion) is essential for the biogenesis of miRNAs, and it exerts miRNA-independent gene-regulatory functions29. Noticeably, miRNAs fulfills various functions in neuronal development64, plasticity, and neurological diseases65. In this context, compelling evidence identified altered miRNA biogenesis associated with the 22q11.2 microdeletion in Df(16)A +/− adult mice (8 week old), which was shown to rise due to dgcr8 gene deficiency26. Although no study has shown such alterations during embryonic development in Lgdel +/− neuronal networks, we attempted to assess the miRNA expression profiles in cultured hippocampal neurons from WT and Lgdel +/− mice. Our results showed an incremental delay in the expression level of miRNAs in Lgdel +/− neurons over developmental ages, compared to WT neurons, albeit the increase of read counts from 8 to 26 DIVs in both Lgdel +/− and WT neurons. In turn, the altered expression of miRNAs may play a role, at least partly, in the functional phenotypes observed in our study. Indeed, such as a post-transcriptional regulation could explain the distinct alteration in NKCC1 and KCC2 protein levels rather than in their mRNA levels. Also, given that Dgcr8 +/− cultures display intermediate phenotypes between WT and Lgdel +/− cultures, it is tempting to speculate, at least partly, the causative effects of dgcr8 deletion, and in turn, the miRNAs downregulation, on the delayed timing of the GABA-switch observed in Lgdel +/− cultures. This notion is further corroborated by our recent study that reported the evidence of influential alternative miRNA-independent functions of dgcr8 during brain development on developmental-related gene expression66. Thus, the dgcr8 haploinsufficiency may dysregulate the gene expression of key genes necessary for the developmental excitatory-to-inhibitory GABA-polarity switch in embryonic Lgdel +/− networks. To this end, our results on Dgcr8 +/− neurons reveal the contribution of the dgcr8 gene haploinsufficiency to the GABAergic-signaling deficits in Lgdel +/− networks, but also indicate the need for extensive future work to characterize the contribution of other deleted genes in Lgdel +/− model. Further, a next challenge will be to identify a possible link of GABA-signaling dysregulation to neuronal activity-regulated transcriptional genes67 in Lgdel +/− networks.
Lumping together, the implications of our findings on the delay of GABA-signaling, electrophysiological developmental profile and homeostatic plasticity defects that occur in the early embryonic developmental phases in Lgdel +/− and Dgcr8 +/− networks, are tempting to speculate that these deficits might operate the kernel of functional and structural abnormalities that affect adult animal models of the 22q11.2 DS. Prospectively, our in vitro study suggests a possible direction for further in vivo studies to assess the potential rescue of bumetanide and the contribution of different deleted genes in the 22q11.2 DS to the GABA-signaling properties of neuronal systems. Additionally, 22q11.2 DS on-CMOS-MEA chips combined with HCI may offer an excellent in vitro model and readouts to accelerate the probing of the genetic, biology, and of the electrical physiology of the 22q11.2 DS-associated neurodevelopmental disorders, before in vivo studies and clinical investigation.
Methods
CMOS-MEAs and acquisition system
We performed all electrical recordings using a custom-built acquisition platform based on components of the BioCam system (3Brain GmbH). We grew mouse neuronal hippocampal cultures on CMOS-MEAs (BioChip 4096S+ from 3Brain GmbH) that integrate an array of 4096 recording electrodes (21 × 21 μm2 in size, 42 μm pitch) on an active area of (2.67 × 2.67 mm2) centered in a working area of (6 × 6 mm2). We used the BrainWave software (3Brain GmbH) for data recording and spike detection.
Preparation and coating of CMOS-MEAs
We sterilized CMOS-MEA devices thoroughly outside the cell culture chamber ring using tissue moistened with EtOH 96%. We placed each device into a sterile 100 × 20 mm Petri dish (Corning) and sterilized the entire culture chamber by filling it with 70% EtOH. After 20 minutes, we rinsed the CMOS-MEAs 4x with sterile double-distilled water (DDW) and dried them under a sterile laminar flow hood. Chips were pre-conditioned by overnight incubation at 37 °C and 5% CO2 with Complete Neurobasal Media (CNM) containing 2% B-27 1% penicillin/streptomycin and 1% GlutaMax supplements (all reagents from Life Technologies). The next day, CNM was aspirated, and chips were immediately coated with 50 μg/ml poly-DL-ornithine (PDLO) (Sigma-Aldrich) and incubated overnight at 37 °C and 5% CO2. We filled a 35 × 10 mm Petri dish (Corning) with sterile DDW and placed it inside the Petri dish (besides the chip) both to maintain proper humidity and to avoid evaporation of the coating reagents overnight. The next day, CMOS-MEAs were rinsed 4x with sterile DDW and left to dry under the hood before cell seeding.
Mouse lines and genotyping
Mice were housed under standard conditions in the animal facility at the Fondazione Istituto Italiano di Tecnologia (IIT). All experiments and procedures were approved by the Italian Ministry of Health and IIT Animal Use Committee, by the Guide for the Care and Use of Laboratory Animals of the European Community Council Directives. Lgdel +/− mice (DGS model27) carrying a hemizygous deletion from Idd to Hira (Lgdel +/−), were maintained on a C57BL/6 J (Charles River) background. Lgdel +/− mice were crossed with C57BL/6 J mice. The yielded WT (control) and Lgdel +/− embryos were used for the experiments. The conditional heterozygous ablation of dgcr8 gene was performed by crossing dgcr8 flox/flox mice with Emx1 Cre/wt knock-in mice66. Embryos yielded from these crossings were used for the experiments: dgcr8 flox/wt/Emx1 wt/wt (WT, controls), dgcr8 flox/wt/Emx1 Cre/wt (dgcr8 +/−conditional dgcr8 heterozygous). All primers used for the genotyping procedures are listed in Table 1.
Preparation of primary hippocampal cultures from mouse embryos
We obtained primary hippocampal neurons from brain tissues of embryos of WT, Lgdel +/−, and Dgcr8 +/− mice at day 18 (E18) using a similar previously described method on rat cultures68. All work with primary cultures was performed in accordance with the Italian guidelines and regulations, and all animal procedures carried out in this work were approved by the institutional (IIT) Ethics Committee and by the Italian Ministry of Health and Animal Care (Authorization No. 057/2013 and 214/2015). Briefly, embryos were removed and decapitated, and the brains were extracted from the skulls and placed in cold Hanks Balanced Salt Solution (HBSS). We dissected hippocampi of WT and Lgdel +/− embryos (C57/BL6J mouse background) and dgcr8 WT and Dgcr8 +/−. We placed hippocampi in 0.125% trypsin-EDTA and incubated them for 30 minutes at 37 °C in a water bath to dissociate the tissue. We then blocked the trypsin activity by CNM, supplemented with 10% fetal bovine serum (FBS) and centrifuged the tubes for 5 minutes at 1200 rpm. The supernatant was discarded, and fresh CNM and 10% FBS were added. The hippocampi were dissociated gently by pipetting, and then the solution was filtered through a cell strainer and centrifuged for 10 minutes at 700 rpm. The supernatant was discarded, and cells were resuspended in CNM and counted using trypan blue and a hemocytometer. We seeded 100.000 cells/chip on the previously coated active area and then cultures were incubated at 37 °C with 5% CO2 and 95% humidity. After 1.5 hr, we added 1.6 ml of CNM to the culture chamber and incubated under the same conditions. On the other hand, we seeded 10.000 cells/well in BD Falcon 96-well imaging plates (Corning) for HCI experiments (BD Pathway 855, BD Biosciences). For maintenance and cell culture growth, one-third of the medium was routinely replaced with a fresh CNM every four days. All reagents were obtained, unless indicated differently, from Life Technologies.
RNA extraction, cDNA preparation, and qRT-PCR
We extracted total RNA from WT, Lgdel +/−, dgcr8 WT, and Dgcr8 +/− primary hippocampal cultures at 8, 16 and 26 DIVs (three embryos per condition) using QIAzol lysis reagent (Qiagen). Synthesis of cDNA from total RNA was performed with ImProm-IITM Reverse Transcription System (Promega). We performed qRT–PCR by using Quantifast SYBR Green method (Qiagen) with the following probes11; NKCC1 (Slc12a2, NM_009194)-Fwd 5′-CCACCAGGAAACCATACCA-3′ and NKCC1-Rev 5′-AAGGCAGGCAAGTCTACC-3′, KCC2 (Slc12a5, NM_020333)-Fwd 5′-GGACCACTAGCTGACCTC-3′ and KCC2-Rev 5′-CACCTGAGCCGTTTGATG-3′, GAPDH (NM_008084)-Fwd 5′-GAACATCATCCCTGCATCCA and GAPDH-Rev 5′-CCAGTGAGCTTCCCGTTCA-3′. Expression analysis was performed using the comparative cycle threshold (Ct) method.
Immunofluorescence protocol
We performed immunostaining on mouse hippocampal cultures that were grown on CMOS-MEAs, and microplates using a previously described protocol44. Briefly, we removed the culture medium and washed using phosphate-buffered saline (PBS-1X) (Life Technologies) at 37 °C, and then we fixed samples in paraformaldehyde (4% in PBS-1X) for 15 minutes at room temperature (RT). We washed the sample 4x with PBS-1X, then cells were permeabilized with 0.1% Triton X-100 in PBS-1X (PBST) for 10 minutes, and then cells were blocked with 5% normal goat serum (NGS) (EuroClone) for 1 hr before incubation with primary antibodies. Cells were then incubated overnight at 4 °C in primary antibodies diluted in the NGS blocking buffer. The following primary antibodies were used: guinea pig anti-MAP-2 (Synaptic Systems; 1:1000), rabbit anti-c-fos (Calbiochem Millipore; 1:500), and rabbit anti-NKCC1 (Millipore; 1:100), and KCC2 (Millipore; 1:500). We performed three successive washing steps in PBST for five minutes per wash, and cells were incubated for 1 hr in the dark at RT with the corresponding secondary antibodies, including Alexa Fluor 488, Alexa Fluor 546, and Alexa Fluor 647 (all from Life Technologies; 1:1000). Subsequently, the cells were washed 4x with PBST and incubated for 15 minutes in the dark at RT with the nuclear marker Hoechst 33342 (Thermo Fisher Scientific; 1:500) diluted in PBS-1X. We visualized and acquired all images with 20X and 25X objective lenses using a BD Pathway 855 (BD Biosciences), and a Leica SP5 upright confocal microscope (Leica Microsystems), respectively.
Drug treatments
Bicuculline methiodide
We prepared a fresh solution of bicuculline for each experiment by dissolving 8 mg/ml (2503-Tocris) in DDW. The final working concentration used for all experiments was 20 µM.
Bumetanide
We prepared a fresh solution of bumetanide for each experiment by dissolving 20 mg/ml (B3023-Sigma-Aldrich) in dimethyl sulfoxide (DMSO). The final working concentration used for all experiments was 10 µM.
Extracellular recordings and analyses of spiking, bursting and dynamic changes
We performed multiple extracellular recordings from hippocampal neuronal networks grown on CMOS-MEAs at different developmental time points (DIVs). Each recording session lasted 10 minutes, and neural activity was acquired at a full-frame resolution of 7.8 kHz/electrode and from the whole array of 4096 electrodes (acquisition rate of 60 Mbyte/s). The time stamps of the spikes were detected using the precise timing spike detection (PTSD) algorithm of the Brainwave software application (3Brain GmbH). As in previous works, we adopted a threshold of 9x the standard deviation for spike detection, whereas burst events were identified if we detected at least five consecutive spikes in an inter-spike interval (ISI) lower than 100 ms. On the other hand, NBs were detected and analyzed as previously reported69. Only electrodes that recorded spikes with rates between 0.1 and 10 events/sec (active electrodes) were considered in our analysis. All spike trains were exported from BrainWave software in a MATLAB file format and were analyzed using custom Python scripts (Python Software Foundation). The analysis included first-order statistics of network-wide mean activity parameters (MFR and MBR), in addition to further analyses of the neuronal firing frequency distributions (lognormal-like distributions), NBs, and the distribution of IBIs. Lognormal-like distributions and fit statistics were analyzed as previously described44. Briefly, we assigned each active electrode to its corresponding averaged MFR value, then we normalized the occurrence of these active electrodes as a proportion of their corresponding spiking activity divided by the total number of electrodes in the array, which is 4096. For acceptance of NBs within a time-window of 150 msec (within a given temporal bin of 15 msec), the value of firing threshold was set approximately 15% of the total network spiking activity, and events were detected whenever the number of spiking activity exceeded this specific threshold. Finally, we quantified the distribution of IBIs as a bimodal temporal clustering of bursts that were binned using a Gaussian window with a 5% bin size of the burst duration, as previously reported51.
Image analyses
To acquire images from neurons grown in 96 wells microplates, we used the BD Pathway 855 Bioimager in the non-confocal mode using 20x/0.75 NA objective (BD Biosciences) in the form of 3 × 3 montage (9 image fields/well).
c-fos
To quantify the neuronal activity through cellular c-fos expression, we grew neuronal cultures on two different cell culture platforms. First, neuronal networks grown on CMOS-MEAs were acquired on a Leica SP5 upright confocal microscope using a 25x/0.95 NA objective (Leica Microsystems). Second, to consolidate the cellular readouts of c-fos expression in high-throughput experimentation, we performed multiplexed HCI by growing neurons in 96 wells microplates (Corning). Consequently, the quantification of c-fos expression for all these data was performed by counting the c-fos-positive nuclei. Images acquired from CMOS-MEAs were quantified using the object count tool of the NIS-Element Advance Research Software AR (Nikon), which allowed us to perform automated object detection and counting by using image thresholding based on area, circularity, and EqDiameter parameters (Nikon NIS-Elements Advanced Research User’s Guide V.4). Images were acquired from at least nine fields in each confocal image from four different preparations and different animals. For data acquired with the HCI platform, we quantified the c-fos-positive nuclei ratio using BD AttoVision software v1.6 (BD Biosciences), which allowed us to apply the polygon segmentation methods to generate polygon-shaped ROIs from a single dye (c-fos and Hoechst for the nuclei); then, the total number of ROIs indicated the positive expression of c-fos with respect to their nuclei.
NKCC1 and KCC2
We quantified the average intensity expressions of the chloride cotransporters to investigate the GABAergic-signaling switch in Lgdel +/− and dgcr8 +/−. Neurons from WT, Lgdel +/−, and dgcr8 +/− embryos were grown in 96 wells microplates (Corning). We measured the expression intensity of NKCC1 and KCC2 by using multiplexed immunofluorescence HCI. The level of chloride cotransporters was analyzed using BD AttoVision software v1.6 (BD Bioscience) by quantifying the gray level in the segmented polygon-shaped ROIs. Given that NKCC1 is a nonspecific neuronal protein, we performed a segmentation method based on two dyes (channel A) for NKCC1, and (channel B) for MAP-2, thus resulting in two output ROIs (one for each channel). The sum of the averaged intensity in ROIs from NKCC1 (channel A) was selected for quantification only if they matched the segmented ROIs of the neuronal MAP-2 expression (channel B). Under this condition, the averaged intensity in ROIs from NKCC1 was considered as the final expression of NKCC1 at its associated time-point. On the other hand, KCC2 protein is a neuronal-specific marker, thus the polygon segmentation methods were used to generate polygon shaped ROIs from a single dye (nucleus), and the sum of the averaged intensity values of these individual ROIs was computed as the final expression of KCC2 at a precisely measured time-point.
Neurite Outgrowth
We performed morphometric measurements of neurite processes on WT, Lgdel +/− and Dgcr8 +/− mouse embryonic hippocampal cultures at 5 DIVs grown in 96 wells microplates. The images were analyzed using BD’s neurite outgrowth algorithms (BD Bioscience). The neurite outgrowth analysis required two dye channels; (channel A) for the nucleus and (channel B) for the neurites of a neuron. Therefore, the Hoechst (channel A) was used to generate segmentation masks for the nuclei and to create cell body boundaries around them. Next, MAP-2 (channel B) was used to define the structure of the neuronal network. After analysis, image masks were generated as illustrated in (Fig. 7a). Finally, neurite outgrowth parameters (Table 2, see also BD AttoVision User’s Guide V1.6), including neurite total and maximum length per cell, the number of roots/neurites emerging per cell, neurite extremities (primary, secondary, and tertiary branching if exist), were generated and exported as text files for further quantifications.
Statistical analyses and experimental parameters
All experimental data reported in this work are expressed as the mean ± standard error of the mean (SEM). Differences between groups were examined for statistical significance, where appropriate using one-way analysis of variance (ANOVA) or two-way ANOVA followed by Tukey’s posthoc testing. A non-parametric Kruskal-Wallis method was used for data that were not normally distributed. A non-parametric Kolmogorov-Smirnov test was used to assess differences between distributions and p < 0.05 was considered significant. The statistical tests used for each specific experiment are depicted in the Figures’ legends.
To ensure the consistency and reproducibility of our results, we conducted at least three repeated trials for each experimental condition. These tests were performed in different cell culture preparations and from at least three different animals. The number of samples used in each experiment is depicted in the Figures’ legends.
Electronic supplementary material
Acknowledgements
We thank Prof. BE. Morrow (A. Einstein College of Medicine, NY, USA) and Prof. R. Kucherlapati (Harvard Medical School, Boston, USA) for Lgdel mice and Dr. E. Fuchs (Rockefeller University, NY, USA) for Dgcr8-flox and Dr. S. Itohara (RIKEN, Wako, Japan) for Emx1-Cre mouse line. We also thank Dr. MJ. Marzi, and Dr. F. Nicassio (CGS@SEMM Milano, Italy) for their support on the deep-sequencing data.
We thank Dr. A. Maccione for initial experimental trials, and the technical support of Dr. M. Nanni, Dr. R. Pelizzoli, Dr. E. Albanesi, and Dr. M. Pesce (IIT-NBT). We would also like to acknowledge the Animal Facility of IIT Genoa (F. Piccardi, D. Cantatore, Dr. R. Navone, S. Morando, T. Luchetta and Dr. M. Morini) for their assistance.
This study was partially supported by the European Commission for Research within the Seventh Framework Program for the NAMASEN (FP7-264872) Marie-Curie ITN.
Author Contributions
H.A. and L.B. wrote the manuscript. H.A. planned and performed the experiments, analyzed the data, and prepared all Figures. F.M. contributed to the experimental planning, co-performed the cell culture experiments, and performed and analyzed qRT-PCR readouts. D.D.P.T. suggested and provided the mouse models for the study. D.D.P.T and L.B. are co-last authors. All authors revised, reviewed and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Hayder Amin and Federica Marinaro contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15793-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Drew LJ, et al. The 22q11.2 microdeletion: fifteen years of insights into the genetic and neural complexity of psychiatric disorders. Int. J. Dev. Neurosci. 2011;29:259–281. doi: 10.1016/j.ijdevneu.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niklasson L, Rasmussen P, Óskarsdóttir S, Gillberg C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res. Dev. Disabil. 2009;30:763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 4.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat. Rev. Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robin NH, Shprintzen RJ. Defining the clinical spectrum of deletion 22q11.2. J. Pediatr. 2005;147:90–6. doi: 10.1016/j.jpeds.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Shaikh TH, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 7.Mori T, et al. Neuroradiological and Neurofunctional Examinations for Patients with 22q11.2 Deletion. Neuropediatrics. 2011;42:215–221. doi: 10.1055/s-0031-1295479. [DOI] [PubMed] [Google Scholar]
- 8.He Q, Nomura T, Xu J, Contractor A. The developmental switch in GABA polarity is delayed in fragile X mice. J. Neurosci. 2014;34:446–50. doi: 10.1523/JNEUROSCI.4447-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coghlan S, et al. GABA system dysfunction in autism and related disorders: From synapse to symptoms. Neurosci. Biobehav. Rev. 2012;36:2044–2055. doi: 10.1016/j.neubiorev.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyde, T. M. et al. Expression of GABA Signaling Molecules KCC2, NKCC1, and GAD1 in Cortical Development and Schizophrenia. J. Neurosci. 31 (2011). [DOI] [PMC free article] [PubMed]
- 11.Deidda G, et al. Reversing excitatory GABAAR signaling restores synaptic plasticity and memory in a mouse model of Down syndrome. Nat. Med. 2015;21:318–326. doi: 10.1038/nm.3827. [DOI] [PubMed] [Google Scholar]
- 12.Represa A, Ben-Ari Y. Trophic actions of GABA on neuronal development. Trends in Neurosciences. 2005;28:278–283. doi: 10.1016/j.tins.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Owens DF, Kriegstein AR. Is there more to gaba than synaptic inhibition? Nat. Rev. Neurosci. 2002;3:715–727. doi: 10.1038/nrn919. [DOI] [PubMed] [Google Scholar]
- 14.Dzhala VI, et al. NKCC1 transporter facilitates seizures in the developing brain. Nat. Med. 2005;11:1205–1213. doi: 10.1038/nm1301. [DOI] [PubMed] [Google Scholar]
- 15.Khazipov R, et al. Early Development of Neuronal Activity in the Primate Hippocampus In Utero. J. Neurosci. 2001;21:9770–9781. doi: 10.1523/JNEUROSCI.21-24-09770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raimondo JV, Richards BA, Woodin MA. Neuronal chloride and excitability - the big impact of small changes. Curr. Opin. Neurobiol. 2017;43:35–42. doi: 10.1016/j.conb.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Rivera C, Voipio J, Kaila K. Two developmental switches in GABAergic signalling: the K+ −Cl− cotransporter KCC2 and carbonic anhydrase CAVII. J. Physiol. 2005;562:27–36. doi: 10.1113/jphysiol.2004.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguly K, Schinder AF, Wong ST, Poo M. GABA itself promotes the developmental switch of neuronal GABAergic responses from excitation to inhibition. Cell. 2001;105:521–32. doi: 10.1016/S0092-8674(01)00341-5. [DOI] [PubMed] [Google Scholar]
- 19.Cellot G, Cherubini E. Functional role of ambient GABA in refining neuronal circuits early in postnatal development. Front. Neural Circuits. 2013;7:136. doi: 10.3389/fncir.2013.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitzer NC. Electrical activity in early neuronal development. Nature. 2006;444:707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- 21.Rubenstein JLR, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes, Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183X.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möhler H, Gaba A. Receptors in Central Nervous System Disease: Anxiety, Epilepsy, and Insomnia. J. Recept. Signal Transduct. 2006;26:731–740. doi: 10.1080/10799890600920035. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Ari Y. Excitatory actions of gaba during development: the nature of the nurture. Nat. Rev. Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- 24.Sahara, S., Yanagawa, Y., O’Leary, D. D. M. & Stevens, C. F. The Fraction of Cortical GABAergic Neurons Is Constant from Near the Start of Cortical Neurogenesis to Adulthood. J. Neurosci. 32 (2012). [DOI] [PMC free article] [PubMed]
- 25.Ben-Ari Y, Khalilov I, Represa A, Gozlan H. Interneurons set the tune of developing networks. Trends Neurosci. 2004;27:422–427. doi: 10.1016/j.tins.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Stark KL, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 27.Merscher S, et al. TBX1 Is Responsible for Cardiovascular Defects in Velo-Cardio-Facial/DiGeorge Syndrome. Cell. 2001;104:619–629. doi: 10.1016/S0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 28.Earls LR, et al. Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 2012;32:14132–14144. doi: 10.1523/JNEUROSCI.1312-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burger K, Gullerova M. Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nat. Rev. Mol. Cell Biol. 2015;16:417–430. doi: 10.1038/nrm3994. [DOI] [PubMed] [Google Scholar]
- 30.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 32.Rivera C, et al. The K+/Cl− co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature. 1999;397:251–5. doi: 10.1038/16697. [DOI] [PubMed] [Google Scholar]
- 33.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippi G, et al. MicroRNA-101 Regulates Multiple Developmental Programs to Constrain Excitation in Adult Neural Networks. Neuron. 2016;92:1337–1351. doi: 10.1016/j.neuron.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blankenship AG, Feller MB. Mechanisms underlying spontaneous patterned activity in developing neural circuits. Nat. Rev. Neurosci. 2010;11:18–29. doi: 10.1038/nrn2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berdondini, L. et al. High-density microelectrode arrays for electrophysiological activity imaging of neuronal networks. In ICECS2001. 8th IEEE International Conference on Electronics, Circuits and Systems (Cat. No. 01EX483) 3, 1239–1242 (IEEE, 2001).
- 37.Eversmann B, et al. A 128 x 128 cmos biosensor array for extracellular recording of neural activity. IEEE J. Solid-State Circuits. 2003;38:2306–2317. doi: 10.1109/JSSC.2003.819174. [DOI] [Google Scholar]
- 38.Franks, W. et al. CMOS monolithic microelectrode array for stimulation and recording of natural neural networks. in TRANSDUCERS ’03. 12th International Conference on Solid-State Sensors, Actuators and Microsystems. Digest of Technical Papers (Cat. No. 03TH8664) 2, 963–966 (IEEE, 2003).
- 39.Keefer EW, Gramowski A, Gross GW. NMDA receptor-dependent periodic oscillations in cultured spinal cord networks. J. Neurophysiol. 2001;86:3030–42. doi: 10.1152/jn.2001.86.6.3030. [DOI] [PubMed] [Google Scholar]
- 40.Turrigiano G. Stabilizing Neuronal Function Homeostatic Synaptic Plasticity: Local and Global Mechanisms for Homeostatic Synaptic Plasticity: Local and Global Mechanisms for Stabilizing Neuronal Function. Cold Spring Harb Perspect Biol. 2011;4:a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Earls LR, et al. Dysregulation of presynaptic calcium and synaptic plasticity in a mouse model of 22q11 deletion syndrome. J. Neurosci. 2010;30:15843–15855. doi: 10.1523/JNEUROSCI.1425-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fénelon K, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. USA. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizuseki K, Buzsáki G. Preconfigured, skewed distribution of firing rates in the hippocampus and entorhinal cortex. Cell Rep. 2013;4:1010–21. doi: 10.1016/j.celrep.2013.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amin, H. et al. Electrical responses and spontaneous activity of human iPS-derived neuronal networks characterized for 3-month culture with 4096-electrode arrays. Front. Neurosci. 10 (2016). [DOI] [PMC free article] [PubMed]
- 45.Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- 46.Kirkby LA, Sack GS, Firl A, Feller MB. A Role for Correlated Spontaneous Activity in the Assembly of Neural Circuits. Neuron. 2014;81:218. doi: 10.1016/j.neuron.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saint-Amant L, Drapeau P. Synchronization of an Embryonic Network of Identified Spinal Interneurons Solely by Electrical Coupling. Neuron. 2001;31:1035–1046. doi: 10.1016/S0896-6273(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 48.Ben-Ari Y. Developing networks play a similar melody. Trends Neurosci. 2001;24:353–360. doi: 10.1016/S0166-2236(00)01813-0. [DOI] [PubMed] [Google Scholar]
- 49.Pouget A, Dayan P, Zemel R. Information processing with population codes. Nat. Rev. Neurosci. 2000;1:125–132. doi: 10.1038/35039062. [DOI] [PubMed] [Google Scholar]
- 50.Rolston JD, Wagenaar DA, Potter SM. Precisely timed spatiotemporal patterns of neural activity in dissociated cortical cultures. Neuroscience. 2007;148:294–303. doi: 10.1016/j.neuroscience.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagenaar DA, Pine J, Potter SM. An extremely rich repertoire of bursting patterns during the development of cortical cultures. BMC Neurosci. 2006;7:11. doi: 10.1186/1471-2202-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukai J, et al. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat. Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meechan DW, Tucker ES, Maynard TM, LaMantia A-S. Diminished dosage of 22q11 genes disrupts neurogenesis and cortical development in a mouse model of 22q11 deletion/DiGeorge syndrome. Proc. Natl. Acad. Sci. USA. 2009;106:16434–16445. doi: 10.1073/pnas.0905696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verzi, D. W., Rheuben, M. B. & Baer, S. M. Impact of Time-Dependent Changes in Spine Density and Spine Shape on the Input – Output Properties of a Dendritic Branch: A Computational Study. 2073–2089, 10.1152/jn.00373.2004 (2005). [DOI] [PubMed]
- 55.Palmer LM, Stuart GJ. Membrane potential changes in dendritic spines during action potentials and synaptic input. J. Neurosci. 2009;29:6897–6903. doi: 10.1523/JNEUROSCI.5847-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maa EH, Kahle KT, Walcott BP, Spitz MC, Staley KJ. Diuretics and epilepsy: Will the past and present meet? Epilepsia. 2011;52:1559–1569. doi: 10.1111/j.1528-1167.2011.03203.x. [DOI] [PubMed] [Google Scholar]
- 57.Payne JA, Rivera C, Voipio J, Kaila K. Cation–chloride co-transporters in neuronal communication, development and trauma. Trends Neurosci. 2003;26:199–206. doi: 10.1016/S0166-2236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- 58.Costa ACS, Scott-McKean JJ. Prospects for Improving Brain Function in Individuals with Down Syndrome. CNS Drugs. 2013;27:679–702. doi: 10.1007/s40263-013-0089-3. [DOI] [PubMed] [Google Scholar]
- 59.Meechan DW, et al. Cognitive ability is associated with altered medial frontal cortical circuits in the LgDel mouse model of 22q11.2DS. Cereb. Cortex. 2015;25:1143–1151. doi: 10.1093/cercor/bht308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfeffer CK, et al. NKCC1-Dependent GABAergic Excitation Drives Synaptic Network Maturation during Early Hippocampal Development. J. Neurosci. 2009;29:3419–3430. doi: 10.1523/JNEUROSCI.1377-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben-Ari Y, Gaiarsa J-L, Tyzio R, Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol. Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- 62.Gibson JR, Bartley AF, Hays SA, Huber KM. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J. Neurophysiol. 2008;100:2615–26. doi: 10.1152/jn.90752.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schobel SA, et al. Imaging Patients with Psychosis and a Mouse Model Establishes a Spreading Pattern of Hippocampal Dysfunction and Implicates Glutamate as a Driver. Neuron. 2013;78:81–93. doi: 10.1016/j.neuron.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schratt G. microRNAs at the synapse. Nat. Rev. Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 65.Geaghan M, Cairns MJ. MicroRNA and Posttranscriptional Dysregulation in Psychiatry. Biol. Psychiatry. 2015;78:231–239. doi: 10.1016/j.biopsych.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 66.Marinaro, F. et al. MicroRNA‐independent functions of DGCR8 are essential for neocortical development and TBR1 expression. EMBO Rep. e201642800, 10.15252/embr.201642800 (2017). [DOI] [PMC free article] [PubMed]
- 67.West AE, Greenberg ME. Neuronal Activity-Regulated Gene Transcription in Synapse Development and Cognitive Function. Cold Spring Harb. Perspect. Biol. 2011;3:a005744–a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amin H, Nieus T, Lonardoni D, Maccione A, Berdondini L. High-resolution bioelectrical imaging of Aβ-induced network dysfunction on CMOS-MEAs for neurotoxicity and rescue studies. Sci. Rep. 2017;7:2460. doi: 10.1038/s41598-017-02635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barkan U, Horn D. Spatiotemporal clustering of synchronized bursting events in neuronal networks. Neurocomputing. 2006;69:1108–1111. doi: 10.1016/j.neucom.2005.12.121. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.