Abstract
Tripartite motif–containing 29 (TRIM29) is a member of the TRIM protein family that has been implicated in hematologic and solid tumor cancers. We found that TRIM29 functions as a tumor suppressor in both the nontumorigenic MCF10A [estrogen receptor (ER)−/TRIM29+] breast cell line and the invasive MCF7 (ER+/TRIM29−) breast cell line. Silencing TRIM29 in MCF10A cells resulted in preneoplastic changes that included loss of polarity in three-dimensional culture, increased proliferation, anchorage-independent growth, and increased migration and invasion. Conversely, the introduction of TRIM29 into MCF7 cells caused reversion to a less aggressive phenotype by antagonizing the growth effect of 17β-estradiol. The interaction between TRIM29 and ER signaling in MCF7 cells was supported by a reduction in ERE binding in the presence of TRIM29 and suppression of ER-dependent gene expression of TFF1, FOS, and GREB1. By microarray analyses, we showed that younger women (<55 years of age) with early-stage, ER+ breast cancer who were given no adjuvant systemic therapy had a significantly lower risk of relapse when their tumor had high TRIM29 expression (P = 0.02). This effect was not observed in older women (>55 years of age) and thus may be due to menopause and loss of circulating estrogens. Our results suggest that loss of TRIM29 expression in normal breast luminal cells can contribute to malignant transformation and lead to progression of ER+ breast cancer in premenopausal women.
The ataxia telangiectasia group D–complementing (ATDC/TRIM29) gene was identified by its ability to partially restore the radiosensitive phenotype of cells from patients with ataxia telangiectasia,1, 2 a rare autosomal recessive disease in which patients develop immunodeficiency, neurodegenerative disorders, and cancer.3 TRIM29 encodes for a member of the tripartite motif (TRIM) protein family, which is generally defined by having an ordered series of three zinc-binding domains, a RING (R) domain, two unique B-box (BB) domains, and a coiled-coiled (CC) region. Although some members of the TRIM family may not contain all domains (eg, TRIM29 has no R domain), the order of the regions is always conserved and is the telltale sign of the TRIM motif. TRIM proteins, including TRIM29, self-interact through the CC domain, and this homo-oligomerization is necessary for appropriate localization to distinct cellular compartments that appear as cytoplasmic or nuclear “bodies.”4
TRIM29 is involved in a variety of cancers; however, its function can change, depending on the cell type, level of expression, posttranslational modification, and compartmentalization.4, 5 Although TRIM29 has been implicated as a tumor suppressor in some types of breast and bone cancers,6 it is also known to have oncogenic effects in gastric and pancreatic cancers.7, 8 Transfection of wild-type TRIM29 into osteosarcoma and breast cancer cell lines (Saos-2 and BT-549) lacking detectable mRNA and protein expression of TRIM29 results in suppression of colony-forming efficiency in soft agar,6 suggesting that TRIM29 can cause reversion of a malignant phenotype. In contrast, in vitro and in vivo studies in pancreatic cancer revealed that TRIM29 can increase proliferation and invasion through stabilization of β-catenin and activation of Disheveled 2.8 Additional evidence of the oncogenic effects of TRIM29 comes from mechanistic studies in p53+/− mouse embryonic fibroblasts, where it was discovered that TRIM29 can directly bind p53 and increase proliferation by sequestering it outside the nucleus, preventing promoter binding of p21.9 TRIM29 can also promote cell survival by inhibiting proapoptotic genes regulated by p53 (eg, NOXA).
Studies have shown that stable transfection of ERα into ER− cells, including MCF10A, gives an unexpected result of growth inhibition and/or cell death in the presence of 17β-estradiol (E2).10, 11 We present data supporting a tumor suppressor effect of TRIM29 in nontumorigenic MCF10A cells and invasive MCF7 cells. We hypothesize that TRIM29 expression is important for maintaining homeostasis in normal breast epithelium in part by balancing the stimulatory growth signals from the ER pathway as ER− luminal cells differentiate into ER+ cells and that loss of TRIM29 in ER+ breast cancer could contribute to the progression of the disease in the absence of circulating estrogens.
Materials and Methods
Cell Lines and Three-Dimensional Cell Culture
MCF10A and MCF7 cell lines were obtained from the American Type Culture Collection (Manassas, VA). MCF10A cells were maintained as monolayer in Dulbecco's modified Eagle's medium/F12 containing 5% horse serum, 20 ng/mL of epidermal growth factor, 0.5 μg/mL hydrocortisone, 10 μg/mL insulin, 50 U/mL penicillin, and 50 μg/mL streptomycin at 37°C and 5% CO2. MCF7 cells were cultured in modified Eagle's medium containing 10% fetal bovine serum (FBS), 10 μg/mL insulin, 1 mmol/L sodium pyruvate, 50 U/mL penicillin, and 50 μg/mL streptomycin at 37°C and 5% CO2. Three-dimensional Matrigel culture was performed as described previously.12
Lentiviral Constructs and Generation of Stable Cell Line
To knockdown TRIM29 expression, nucleotides 1265 to 1285 of the TRIM29 open reading frame (NM_012101) were chosen as the target sequence. Two complementary oligonucleotides strands were designed using Block-iT RNAi Designer: forward strand, 5′-CACCGGTGCATTGATGAGCAATTACCGAAGTAATTGCTCATCAATGCACC-3′ and reverse strand 5′-AAAAGGTGCATTGATGAGCAATTACTTCGGTAATTGCTCATCAATGCACC-3′ (italics shows hairpin loop sequence; underlined letters represent the four nucleotide overhangs for directional cloning). To form the short hairpin (shRNA), the forward and reversestrands of oligonucleotides were then annealed, followed by insertion into pENTR/U6 vector containing an U6 RNAi cassette to generate an entry construct expressing shRNA targeting the TRIM29 gene. The LacZ double-stranded control oligomer (forward strand, 5′-CACCGCTACACAAATCAGCGATTTCGAAAAATCGCTGATTTGTGTAG-3′, and reverse strand, 5′-AAAACTACACAAATCAGCGATTTTTCGAAATCGCTGATTTGTGTAGC-3′) supplied in the kit was also cloned as a nonsilencing control shRNA construct. The lentiviral construct was then cloned by the recombination of the U6 RNA interference (i) cassette from the pENTR/U6 entry construct into the pLenti6/BLOCK-iT-DEST vector using LR Clonase II enzyme (Invitrogen, Carlsbad, CA).
The full-length cDNA clone (inserted into gateway entry vector pDONR223) of TRIM29 was obtained from the Human ORFeome Library version 1.1 (Open Biosystems) and was recombined to lentiviral expression vector pLenti6.2/N-Lumio/V5-DEST with LR Clonase II enzyme (Invitrogen). pDONR223-TRIM29SiL was generated by introducing seven silent mutations without changing the amino acid sequence of human TRIM29 to make TRIM29SiL resistant to the continuous presence of TRIM29 shRNA. The plasmid pDONR223-TRIM29 was used as a template for site-directed mutagenesis following the QuickChange XL site-directed mutagenesis protocol (Stratagene, La Jolla, CA). The sense primer for the mutagenesis was as follows: 5′-1249GTTTCTGCAGGAATTTGGCGCTTTAATGTCCAACTATTCTCTCCCCCCACCCTG1303-3′ (italics are silently mutated). Each point mutation was checked by full DNA sequencing. TRIM29SiL was recombined to lentiviral expression vector pLenti4/V5-DEST (zeocin selection) with LR Clonase II enzyme (Invitrogen).
The ViraPower Lentiviral Expression System (Invitrogen) was used to stably express TRIM29, TRIM29 shRNA, or TRIM29SiL in cells. Lentiviral-containing supernatants were generated using 293FT cells according to the manufacturer's protocol. Transduction of MCF10A and MCF7 cells was performed in the presence of polybrene (6 μg/mL). To obtain stable transfectants, the viral supernatant was added into cells, selected using the appropriate antibiotics at 48 hours after transfection. The medium was changed every 3 to 4 days until antibiotic-resistant colonies were evident. Stable transfected cells were maintained with antibiotic-selecting medium and evaluated for changes of TRIM29 protein expression using immunoblot analysis.
Immunofluorescence Microscopy
Immunofluorescence staining of acini was conducted as described previously.13 The acinar structures were incubated with the following antibodies: rat anti-human α6-integrin (clone NKI-GoH3; Chemicon, Temecula, CA), rabbit anti–Ki-67 (Invitrogen), or rabbit anticleaved caspase-3 (Cell Signaling Technology, Danvers, MA). The slides were further incubated with goat anti-rat secondary antibody conjugated to Alexa Fluor 488 or goat anti-rabbit secondary antibody conjugated to Alexa Fluor 594 (Invitrogen). After three washes with immunofluorescent buffer, the slides were incubated with PBS containing DAPI (Sigma-Aldrich, St. Louis, MO) before being mounted with Prolong Gold anti-fade reagent (Invitrogen). Fluorescence images for Ki-67 and caspase-3 were acquired with an Olympus AX70 microscope and analyzed with AxioVision software (version 3.0; Carl Zeiss Microscopy, Thornwood, NY). Fluorescence images for α6-integrin were acquired with an Olympus 1X81-DSU spinning disk confocal microscope (Olympus, Tokyo, Japan) and processed using SlideBook™ 5.5 software (Intelligent Imaging Innovations, Inc., Denver, CO). All images were exported to Adobe Photoshope CS5 (Adobe Systems, San Jose, CA) for the production of final figures. The images presented represent three or more independent experiments.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated from cells using RNeasy kit (Qiagen, Valencia, CA), and cDNA was synthesized using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) from total RNA. Real-time PCR was performed with the LightCycler 2.0 system using the LightCycler-FastStart DNA Master SYBR Green Kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. For PCR amplification, the following primers were used: ACTB, 5′-TTCCTGGGCATGGAGTC-3′ (forward) and 5′-CAGGTCTTTGCGGATGTC-3′ (reverse); FOS, 5′-AGAATCCGAAGGGAAAGGAA-3′ (forward) and 5′-GCAGACTTCTCATCTTCTAGTTGGT-3′ (reverse); TFF1, 5′-CCCCTGGTGCTTCTATCCTAA-3′ (forward) and 5′-GATCCCTGCAGAAGTGTCTAAAA-3′ (reverse); and GREB1, 5′-CCCCTACAACGAGATCCACT-3′ (forward) and 5′-TTATTTTCATGATATAAGTCCACTCCA-3′ (reverse). Relative transcript levels were normalized to the reference gene, ACTB, and then normalized to the vehicle-treated control to determine the fold change. The results are the mean ± SE of three independent experiments performed at least in duplicate.
Immunoblot Analysis
Immunoblot analysis was performed using antibodies against TRIM29 (N-19; Santa Cruz Biotechnology, Santa Cruz, CA) and β-actin (Sigma-Aldrich). The blots were incubated with horseradish peroxidase–conjugated donkey anti-goat (TRIM29) or anti-mouse (β-actin) IgG (Santa Cruz Biotechnology), all at a dilution of 1:5000. Western blots were performed using an enhanced chemiluminescence detection system (ECL Plus) following the manufacturer's instructions (Amersham Pharmacia Biotech Inc., Piscataway, NJ). Proteins were visualized by autoradiography with Hyperfilm.
Cell Proliferation Assay
Cells were seeded (5 × 103 per well) in 96-well microtiter plates and evaluated using BrdU (5-bromo-2′-deoxyuridine) incorporation following the manufacturer's protocol (Roche Applied Science, Mannheim, Germany). Estrogen stimulation experiments were performed in MCF7 cells cultured in phenol red–free medium containing 5% charcoal-stripped fetal calf serum for 3 days before beginning the proliferation assay. A total of 10 nmol/L E2 (Sigma-Aldrich) or ethanol was added to the medium on day 1, and compound-containing medium was renewed every 24 hours.
Cell Migration and Cell Invasion Assay
Cell migration and cell invasion were analyzed with a CytoSelect 96-well cell migration assay and cell invasion assay kit, respectively, according to the manufacturer's instructions (Cell Biolabs, San Diego, CA). In both assays, 10% FBS was used as chemoattractant.
Anchorage-Independent Growth in Soft Agar
Anchorage-independent growth was assessed using a CytoSelect 96-well cell transformation assay kit (Cell Biolabs) following the instructions of the manufacturer. Briefly, cells (1 × 103) were suspended in medium containing 10% FBS with 0.4% agarose and layered on top of 0.6% agarose in medium in 96-well plates. Cultures were maintained for 7 days. Colony formation was measured by agar solubilization followed by cell lysis and quantification of cell number by the use of CyQuant GR Dye in a fluorescence plate reader. The fluorescence intensity is directly proportional to cell number.
Measurement of Estrogen-Responsive Element (ERE) Activation by ERE-Luciferase Assay
MCF7 cells were cultured in phenol red–free medium with 5% charcoal-stripped serum for 3 days and then plated in 24-well plates at a density of 2 × 105 cells per well in the same medium. Twenty-four hours after plating, MCF7 cells were transiently transfected with 1 μg of 3× ERE-TATA-luciferase reporter (Addgene, Cambridge, MA) using Lipofectamine 2000 (Invitrogen). Transfection efficiency was normalized using pRL-SV40 at 1/20 of the total DNA concentration. Twenty-four hours after transfection, MCF7 cells were cultured for an additional 24 hours in the presence of vehicle (ethanol) or E2 (10 nmol/L; Sigma-Aldrich). Cells were then harvested in 200 μL of Passive lysis buffer and analyzed using the Dual-Luciferase Reporter Assay System from Promega (Madison, WI) according to the manufacturer's instructions. Alterations in transcription from the ERE-firefly luciferase reporter plasmid as a result of the various treatments were determined after normalization using the renilla luciferase activity as control.
Gene Expression Microarray Analysis
Quick Amp Labeling Kit, two-color (Agilent Technologies, Englewood, CO), was used to generate fluorescently labeled cRNA for microarray hybridizations. Fluorescently labeled cRNA samples (825 ng each) were fragmented and combined with Agilent Hi-RPM Hybridization Buffer. Each experimental sample was assayed versus a common reference (Stratagene's Human Universal Reference) on Agilent Human A2 microarrays. Hybridizations were performed using Agilent SureHyb Hybridization chambers following a standard protocol. After incubation, the microarray slides were washed briefly, dipped in a solution of acetonitrile, and dried. Slides were scanned in an Agilent Technologies G2505C Microarray Scanner at 5-μm resolution. A Lowess normalization procedure was performed to adjust the Cy3 and Cy5 channels.14 Functional categorization of genes with a minimum of two-fold absolute changes was performed with GeneSifter software. All DNA microarray data are deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo, last accessed December 12, 2011) and its Short Read Archive, respectively. In addition, processed data from the HCI Microarray Core are available for direct loading in genome browsers using HCI's Distributed Annotation System 2 server (DAS/2; http://www.biodas.org/wiki/Main_Page, last accessed December 12, 2011).
Statistical Analyses
Publically available microarray data were used to determine the prognostic significance of TRIM29 expression in ER+ breast cancer.15, 16 Multiple probe values for TRIM29 were averaged, expression data across the tumors were split into equal tertiles, and survival was assessed by Kaplan-Meier plots. Statistical significance was determined by log-rank testing using TRIM29 as a continuous predictor. A multivariate Cox model was used to determine interactions between TRIM29 and PAM50 subtype predictions.17 All survival analysis was performed using “R” statistical computing software, version 2.8.0 (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Phenotypic Changes Resulting from TRIM29 Knockdown in Nontumorigenic MCF10A Cells
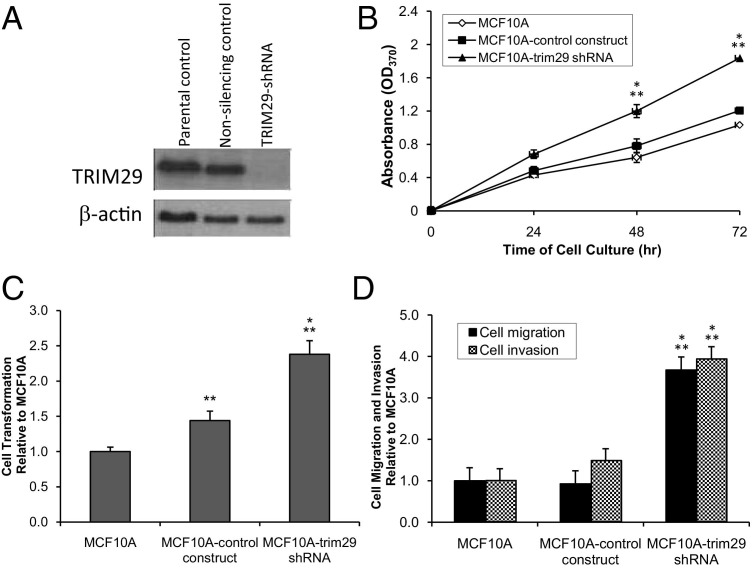
The expression of TRIM29 was silenced in MCF10A cells using shRNA, and phenotypic changes were evaluated using assays for proliferation, anchorage-independent growth, and migration and invasion (Figure 1). The immunoblots of parental and stably selected transfected cells showed high endogenous expression of TRIM29 in MCF10A cells that was effectively knocked down by the TRIM29 shRNA construct (Figure 1A). The loss of TRIM29 expression resulted in an approximately twofold increase in growth at 72 hours that was statistically significant (P < 0.05) compared with the parental or nonsilencing controls (Figure 1B). Although parental MCF10A cells and MCF10A cells transfected with shRNA control construct failed to grow in soft agar, the TRIM29 knockdown MCF10A cells increased rapidly (Figure 1C). Furthermore, in vitro basement membrane assays showed a significant increase in migration (P < 0.05) and invasion (P < 0.05) in the TRIM29 knockdown MCF10A cells compared with the parental and nonsilencing controls (Figure 1D).
Figure 1.
TRIM29 knockdown in MCF10A cells enhances proliferation, confers anchorage-independent growth, and increases migration and invasion. A: TRIM29 protein expression in MCF10A cells transfected with TRIM29 shRNA. TRIM29 and β-actin (loading control) protein were detected by Western blot analysis using anti-TRIM29 antibody N-19 or anti–β-actin antibody. B: Cell proliferation was assayed at the time points indicated using a BrdU incorporation assay. Stably transfected MCF10A cells were seeded at a density of 5000 cells per well in 96-well plates. C: Growth of MCF10A cells in soft agar was assessed using the cell transformation assay kit. Results are expressed as fold change relative to parental control. D: Migration and invasion of MCF10A cells were assayed during a 24-hour period through a Matrigel barrier in response to 10% FBS as a chemoattractant in the lower chamber. Results are expressed as fold change relative to parental control. For experiments shown in B, C, and D, data are presented as mean ± SE. *P < 0.05 versus parental control; **P < 0.05 versus nonsilencing control (one-way analysis of variance). Eight replicates for each condition were performed across three independent experiments.
Knockdown of TRIM29 Disrupts Normal Mammary Acinus Formation in Three-Dimensional Matrigel Culture
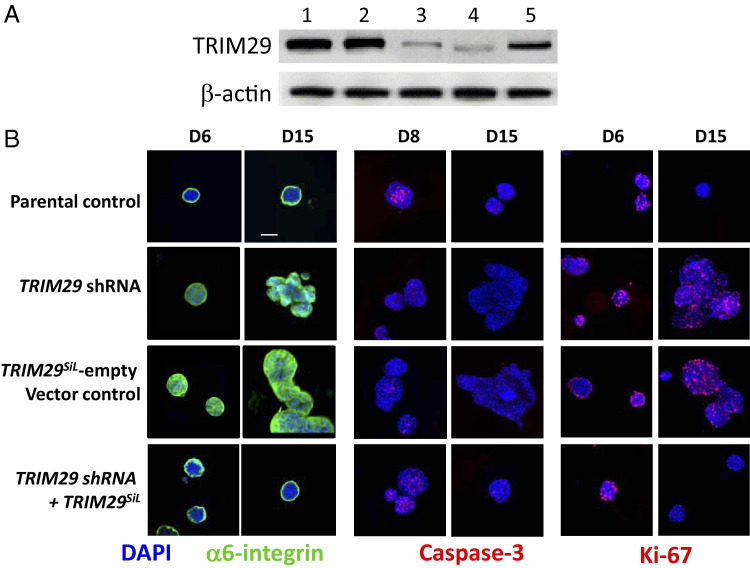
MCF10A cells grown in three-dimensional Matrigel recapitulate numerous features of breast epithelium in vivo, including the formation of acinus-like spheroids.12 Therefore, we examined whether knockdown of TRIM29 would disrupt the normal mammary acinar morphogenesis of MCF10A cells (Figure 2). To assess off-target effects that may have resulted from the short hairpin TRIM29 knockdown vector, TRIM29 was reexpressed using a cDNA with seven silent mutations (TRIM29SiL) encoding the same amino acid sequence, allowing for a functional TRIM29 protein whose expression was resistant to the TRIM29 shRNA (Figure 2A). Parental MCF10A cells grown in three-dimensional Matrigel had a well-circumscribed staining pattern of α-integrin, illustrating appropriate apico-basal polarization (Figure 2B). At day 6, the cells within the lumen of the acini continued to proliferate (Ki-67), and cell death (α-active caspase-3) was apparent by day 8. In contrast, when TRIM29 was knocked down in MCF10A cells, there was a lack of polarity (shown by diffuse α-integrin staining), increased proliferation (shown by Ki-67 staining at day 15), and reduced apoptosis (shown by caspase-3 staining at day 8) compared with the parental MCF10A cells at the same time points. To validate that the ablated acinar formation was solely due to the absence of the functional TRIM29, it was tested whether reintroduction of wild-type TRIM29, by means of a plasmid expressing the RNAi-resistant wild-type TRIM29, could reverse the phenotypic changes. The transformed phenotype caused by TRIM29 knockdown could be rescued by introducing the TRIM29SiL vector. Overall, these results provided evidence that TRIM29 is essential for the normal acinar morphogenesis of mammary epithelial cells. We also investigated whether the introduction of TRIM29 into MCF7 cells could restore the epithelioid aggregates to acini structures and found that TRIM29 alone was not sufficient to reestablish the normal morphologic characteristics (data not shown).
Figure 2.
Knockdown of TRIM29 disrupts normal mammary acinar formation in three-dimensional Matrigel culture. A: Transfection with the RNAi-resistant TRIM29 rescued the TRIM29 expression in MCF10A cells after infection with the retrovirus-based TRIM29 shRNA. Lanes: 1, Parental control; 2, shRNA nonsilencing control; 3, TRIM29 shRNA; 4, TRIM29SiL empty vector control; and 5, TRIM29SiL. The β-actin protein served as an internal loading control. B: Cells were cultured on Matrigel for the indicated number of days. D: Developing acini were immunostained with antibodies to α6-integrin subunit (green, left columns), cleaved caspase-3 (red, middle columns), or Ki-67 (red, right columns). Representative immunofluorescence images of acini are shown with the nuclei counterstained with DAPI (blue). Scale bar = 100 μm.
TRIM29 Expression Slows the Growth of MCF7 Cells in the Presence of Estrogen
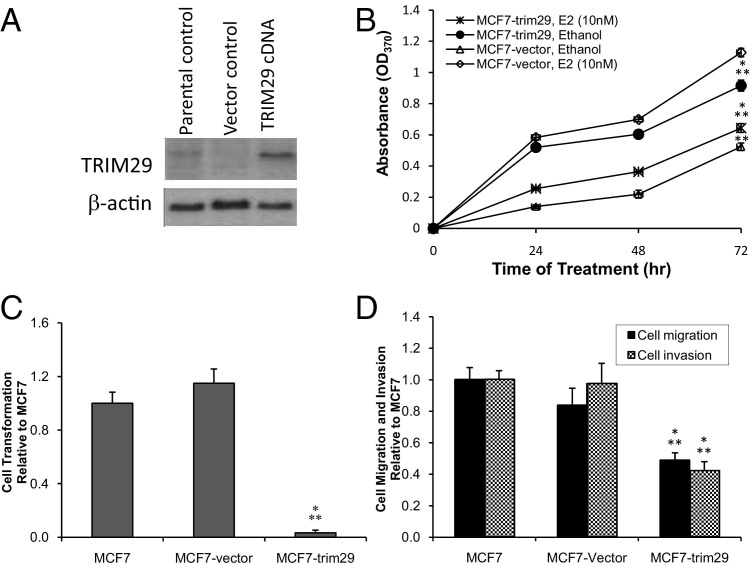
We further investigated the effect of TRIM29 expression in vitro using MCF7 breast cancer cells, which represent an invasive ER+ breast cancer that lacks TRIM29 expression (Figure 3). MCF7 cells were transfected with a lentiviral vector containing wild-type TRIM29 (Figure 3A), and the effects on proliferation (+/− estrogen; Figure 3B), anchorage-independent growth (Figure 3C), and migration and invasion (Figure 3D) were assessed. We found that MCF7 cells grown in steroid-deficient media have increased proliferation when TRIM29 is introduced, equal to the effect of adding E2 alone. However, when TRIM29 is expressed in the presence of E2, growth is decreased down to the baseline growth of parental MCF7 cells without E2. Similarly, there is a statistically significant (P < 0.05) reduction in anchorage-independent growth and migration and invasion when TRIM29 is expressed in MCF7 cells grown in E2-containing media.
Figure 3.
TRIM29 expression in MCF7 cells inhibits estradiol-induced cell proliferation, reduces anchorage-independent growth, and decreases migration and invasion. A: TRIM29 protein expression in MCF7 cells transfected with TRIM29 expression vectors. TRIM29 and β-actin (loading control) protein were detected by Western blot analysis using anti-TRIM29 antibody N-19 or anti–β-actin antibody. B: Effect of TRIM29 expression on estradiol-induced proliferation of MCF7 cells as measured by BrdU incorporation assay. MCF7 cells were seeded at a density of 5000 cells per well in 96-well plates, grown in steroid-deficient media, and then treated with either ethanol or 10 nmol/L E2, changing the medium daily during 72 hours. C: Growth of MCF7 cells in soft agar was assessed using cell transformation assay kit. Results are expressed as fold change relative to parental control. D: Migration and invasion of MCF7 cells were assayed during a 24-hour period through a Matrigel barrier in response to 10% FBS as a chemoattractant in the lower chamber. Results are expressed as fold change relative to parental control. For experiments shown in B, C, and D, data are presented as mean ± SE. *P < 0.05 versus parental control; **P < 0.05 versus vector-transfected control (one-way analysis of variance). Eight replicates for each condition were performed across three independent experiments.
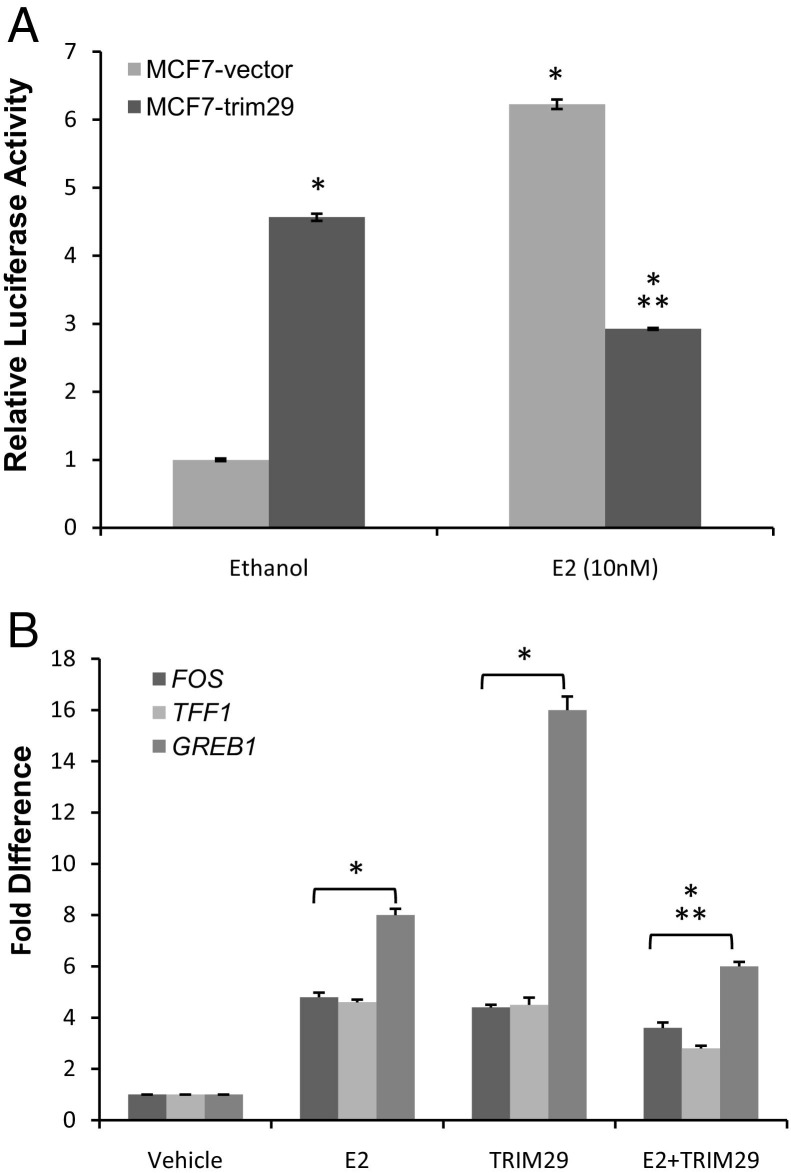
TRIM29 Expression Disrupts Estrogen-Dependent Activation of ERE-Luciferase Reporter and Attenuates Induction of Estrogen-Responsive Genes in MCF7 Cells
MCF7 cells express ERα and ERβ and are highly estrogen responsive.18 Thus, we assessed the functional consequences of TRIM29 expression on activation of the ER pathway in MCF7 cells transfected with a luciferase reporter plasmid containing the consensus ERE (Figure 4A). MCF7 cells were grown in steroid-deficient media and then treated with E2, and the luciferase activity was measured in cell extracts. Similar to the observation on proliferation, we found that ERE transcriptional activity was significantly increased when either E2 was added to the media of parental MCF7 cells or TRIM29 was expressed in the absence of E2. However, when TRIM29 was expressed in the presence of E2, the ERE transcriptional activity was ablated. We next examined whether TRIM29 expression affects the induction of endogenous estrogen-responsive genes by E2. As shown in Figure 4B, the mRNA expression levels of a number of well-characterized ER target genes, including FOS,19 TFF1,20 and GREB1,21 were increased in MCF7 cells in response to E2 treatment. The E2-dependent activation of all of these genes was markedly impaired by the presence of TRIM29. Moreover, TRIM29 expression alone induced the expression of these three genes in MCF7 cells. These results, together with the reporter assay, suggest that TRIM29 is directly involved in regulating the expression of ER target genes.
Figure 4.
Expression of TRIM29 inhibits estradiol-induced ERE activity and attenuates E2-induced transcription of estrogen-responsive genes in MCF7 cells. Each experiment was conducted twice using four replicates per condition. A: MCF7 cells transfected with 3× ERE-TATA-luciferase reporter plasmid were grown in steroid-deficient media and then treated with either ethanol or 10 nmol/L E2 for 24 hours. Cell lysates were evaluated for relative luciferase activity using the Promega dual luciferase assay system. Results are expressed as fold change relative to vehicle (ethanol) control. Data are presented as mean ± SE. *P < 0.05 versus ethanol-treated MCF7 cells; **P < 0.05 versus ethanol-treated MCF7-TRIM29 cells (one-way analysis of variance). B: MCF7 cells were grown in steroid-deficient media and then treated with either ethanol or 10 nmol/L E2 for 12 hours. Cells were harvested and mRNA levels of FOS, TFF1, and GREB1 were measured by real-time PCR and normalized to ACTB. For each gene, data are expressed as mean ± SE of fold change over control. Data for cells treated with vehicle and harvested at the starting point of E2 treatment are used as control mRNA levels. *P < 0.05 versus ethanol-treated MCF7 cells; **P < 0.05 versus E2-treated MCF7 cells (one-way analysis of variance).
Genes Differentially Expressed in MCF10A and MCF7 Cells Due to TRIM29 Expression
To assess changes in gene expression caused by TRIM29, we performed pairwise microarray analyses on both MCF10A cells transfected with the control vector or TRIM29 shRNA retrovirus and MCF7 cells transfected with the control vector or TRIM29 cDNA retrovirus. Genes that were found to be altered greater than twofold in both pairwise analyses were organized according to their involvement in biological processes by gene ontology analysis (see Supplemental Table S1 at http://ajp.amjpathol.org). Knockdown of TRIM29 in MCF10A cells caused up-regulation of cell cycle regulated genes (BTC, AXIN2, and GLP2R), and there was down-regulation of the antiproliferative gene SSTR5. In addition, positive regulators of apoptosis (DLX3 and TNFRSF11B) were down-regulated, whereas the antiapoptotic gene RUX2 was up-regulated. Components of the Wnt signaling pathway, such as WNT5A, WNT8A, and its downstream target GAD1, were also up-regulated as result of TRIM29 loss. Finally, genes involved in MAPK signaling were up-regulated, such as GNG2 and FPR1. The data support our phenotypic observation that the TRIM29-depleted MCF10A cells increase proliferation, decrease apoptosis, increase migration and invasion, and lose polarity. Introducing TRIM29 into MCF7 cells caused reciprocal changes in the expression of genes in these pathways. Moreover, the expression of ESR2, ER-related receptors β and γ, and the estrogen-responsive gene SOX5 were up-regulated from TRIM29 expression in MCF7 cells. Because of the effect that TRIM29 had on the expression of ER-regulated genes, we further identified genes containing an ERE motif that changed expression in MCF7 cells when TRIM29 was expressed (Table 1).
Table 1.
ERE Gene Changes Due to TRIM29 Expression in MCF7 Cells
| Up-regulated gene | ERE sequence⁎ | Down-regulated gene | ERE sequence⁎ |
|---|---|---|---|
| AKR1C4 | 5′-GGGTCAATTTaACCa−3′ | BDKRB2 | 5′-tGGTCtGTCTGACCT−3′ |
| ALP1 | 5′-AGGTCtGGCTGACCC−3′ | BIRC3 | 5′-AGGgCATATTGACCT−3′ |
| ASS | 5′-GGGTgACTCTGACCT−3′ | CXCL2 | 5′-AGGTCtTTATGACCT−3′ |
| CALCR | 5′-GGGTCAGTATGACag−3′ | DTNA | 5′-AGaTCAAGGTGACCT−3′ |
| CASP8 | 5′-AGGcCATCTTGACCT−3′ | HOXC12 | 5′-AGGTCtTTCTGACCT−3′ |
| CD34 | 5′-AGGTCAAATTcACCC−3′ | MATN1 | 5′-GGGTCAGGAccACCC−3′ |
| CD86 | 5′-GGGTggGAGTGACCC−3′ | NR4A3 | 5′-GGGTCAGGGcGcCCC−3′ |
| CDR1 | 5′-tGGTCACCATcACCT−3′ | PDK4 | 5′-AGGTggCCTTGACCT−3′ |
| CPM | 5′-AGGTCACTCTGACCC−3′ | PTGER3 | 5′-GGGaCAGAGTGACCT−3′ |
| ELA2A | 5′-AGGgCAGAGTGACCa−3′ | TAC1 | 5′-GGGTCACCCcGcCCC−3′ |
| FOS | 5′-AaGTCACCCTGACCT−3′ | THBD | 5′-GGGTggACATGACCC−3′ |
| GHRHR | 5′-AGGTCATGTgGACCa−3′ | TRIM31 | 5′-AGaTCAGAATGACCg−3′ |
| GREB1 | 5′-GGGTCATTCTGACCT−3′ | WISP2 | 5′-AaGTCgGAGTGACCT−3′ |
| HSPD1 | 5′-taGTCAGTGTGACCT−3′ | WNT9B | 5′-GGGTtgCACTGACCC−3′ |
| L3MBTL | 5′-GGGTCcTGTTGACCa−3′ | ||
| MTHFD2 | 5′-GGcTCACTGTGACCT−3′ | ||
| NOS3 | 5′-ctGTCACCTTGACCC−3′ | ||
| NUCB2 | 5′-GGGTtgATTTGACCC−3′ | ||
| PCHD12 | 5′-GGGTCAACTTGAgCT−3′ | ||
| PDE4C | 5′-cGGTCACACaGACCC−3′ | ||
| PHF5A | 5′-GGGaCgCCTTGACCT−3′ | ||
| PTX3 | 5′-AGGcCAGGCTGACCa−3′ | ||
| SHMT2 | 5′-AGaTCAGCCTGACCa−3′ | ||
| SLC7A5 | 5′-AaGTCAGAATGACCT−3′ | ||
| TCF2 | 5′-AGGTCAATGTGtCCg-3′ | ||
| TSHB | 5′-AGGTCAGCTTGACaT-3′ |
Sequences of previously identified EREs are shown with lower case letters denoting nonconserved bases.
Expression of TRIM29 in ER+ Breast Cancer and Prognosis
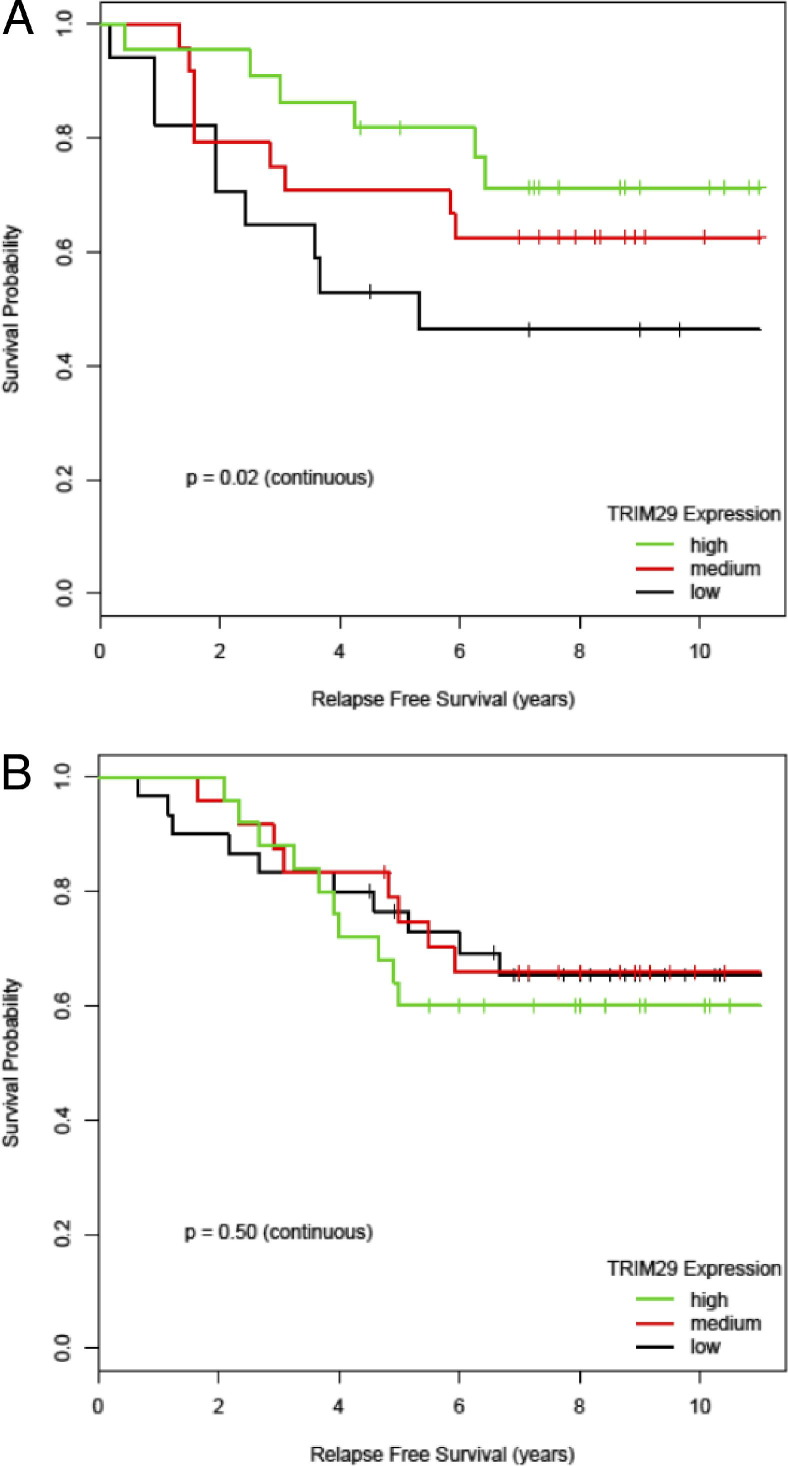
Because of our in vitro findings of TRIM29 functioning as a tumor suppressor and the interaction with the ER pathway, we wanted to assess the effects of TRIM29 tumor expression on the natural course of breast cancer progression. TRIM29 expression was analyzed from microarray for ER+, early-stage (I or II) tumors in women who did not receive any adjuvant systemic treatment.16, 22 A cut point of 55 years old was used in the data set of Wang et al to distinguish premenopausal from postmenopausal women (Figure 5). Women who were younger than 55 years with ER+ breast cancer with high TRIM29 expression had significantly longer relapse-free survival (P = 0.02; Figure 5A), whereas this effect was not observed in women older than 55 years (P = 0.5; Figure 5B). This finding was validated in an independent microarray data set from the Netherland Cancer Institute that enrolled early-stage breast cancer patients younger than 53 years (see Supplemental Figure S1 at http://ajp.amjpathol.org).15 The aggressiveness of ER+ breast cancer has also been associated with “intrinsic” subtype (ie, luminal A versus luminal B)17; however, we found no significant interaction between the different luminal subtypes and TRIM29 expression, suggesting it is an independent prognostic factor.
Figure 5.
TRIM29 expression is prognostic for relapse-free survival in young women with ER+ breast cancer given no adjuvant systemic therapy. Microarray gene expression data for TRIM29 was used from the study by Wang et al.16 Kaplan-Meier survival curves are shown for high (green), intermediate (red), and low (red) expression of TRIM29. A: Relapse-free survival for women younger than 55 years with ER+ breast cancer (n = 63). B: Relapse-free survival for women older than 55 years with ER+ breast cancer (n = 79).
Discussion
MCF10A cells were derived from a premenopausal woman diagnosed as having fibrocystic breast disease,23 which is a nonmalignant condition characterized by localized changes in breast density (ie, painful lumps) that is associated with hormonal fluctuations during the menstrual cycle. The MCF10A cell line is immortalized but retains many features of “normal” breast cells, such as forming acini, inability for anchorage-independent growth, and lack of tumor formation in mice.12, 23 As such, the cell line is often used as a model for tumor progression through additional transformations.24
It has been suggested that MCF10A cells represent a type of multipotent progenitor cell that shares gene expression profiles with many breast cell types.25 Approximately 85% to 90% of normal breast luminal epithelial cells are ER−, another feature shared with MCF10A cells. As normal ER− luminal breast cells differentiate into ER+ cells, they lose Ki-67 expression and enter senescence.26 In preinvasive and invasive ER+ breast cancer, the cells are maintained in this differentiated state and acquire additional somatic mutations (eg, loss of TP53) that promote survival and increased proliferation.27, 28
We hypothesize that TRIM29 plays a role in maintaining the balance between cell proliferation and death, accounting for its dual roles as an oncogene in some cancers and a tumor suppressor in others. Our results show that TRIM29 only had a tumor suppressor effect in MCF7 cells in the presence of E2. A previous study showed that introduction of ERα into MCF10A cells decreased the response of estrogen-regulated genes.11 A plausible role for TRIM29 in normal epithelial mammary tissue is that it is involved in luminal cell senescence in response to estrogens as cells terminally differentiate and become ER+.
Estrogens mediate cellular responses through ERα (ESR1) and ERβ (ESR2), which are encoded by separate genes.29 The ER genes regulate overlapping gene sets by binding ERE or activator protein 1 sites.30 The activator protein 1 complex is composed of the proteins c-fos and c-jun, which stimulate growth in MCF7 cells.31 We show that TRIM29 is able to induce the expression of the FOS (c-fos) gene and other ER-regulated genes (GREB1 and TFF1). Both GREB1 and TFF1 are strongly correlated with ER protein expression and are required for E2-induced proliferation and invasion of ER+ breast cancer cells.21, 32, 33 TRIM29 is able to independently induce these ER-regulated genes, but the level of induction is reduced in the presence of E2, accounting for the observed tumor suppressor effects.
In support of this tumor suppressor function of TRIM29, we found that women with early-stage, ER+ breast cancer have improved survival if the tumor has high TRIM29 expression. However, this benefit was only observed in younger women (<55 years of age), suggesting an age-related physiologic effect. Although menopausal status was not available for the cohorts studied, it is likely that many of the younger women were premenopausal or perimenopausal; thus, the presence of estrogens could account for differences in outcome, especially in the absence of any adjuvant systemic therapy. Our findings suggest that breast cancer incidence and risk of recurrence may depend on a combination of menopausal status, hormone replacement therapy, tumor ER status, and tumor TRIM29 status.
Acknowledgments
We thank the Tissue Resource and Applications Core and HCI Research Informatics at the Huntsman Cancer Center and Dr. Dawne Shelton for helpful guidance and thoughtful discussions.
Footnotes
Supported by the Huntsman Cancer Institute/Foundation. The Tissue Resource and Applications Core and the HCI Research Informatics at the Huntsman Cancer Center are shared resource facilities supported by a National Cancer Institute Cancer Center Support grant (P30 CA42014-19).
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.10.020.
Supplementary data
High expression of TRIM29 in ER+ tumors is a marker of good prognosis in young women (<53 years of age) with early-stage breast cancer given no adjuvant systemic therapy. Microarray gene expression data for TRIM29 were used from the study by van't Veer et al.15 Kaplan-Meier plots for 88 subjects show improved relapse-free survival (P < 0.05) (A) and disease-specific survival (P < 0.05) (B).
References
- 1.Kapp L.N., Painter R.B. Stable radioresistance in ataxia-telangiectasia cells containing DNA from normal human cells. Int J Radiat Biol. 1989;56:667–675. doi: 10.1080/09553008914551891. [DOI] [PubMed] [Google Scholar]
- 2.Kapp L.N., Painter R.B., Yu L.C., van Loon N., Richard C.W., 3rd, James M.R., Cox D.R., Murnane J.P. Cloning of a candidate gene for ataxia-telangiectasia group D. Am J Hum Genet. 1992;51:45–54. [PMC free article] [PubMed] [Google Scholar]
- 3.Boder E., Sedgwick R.P. Ataxia-telangiectasia; a familial syndrome of progressive cerebellar ataxia, oculocutaneous telangiectasia and frequent pulmonary infection. Pediatrics. 1958;21:526–554. [PubMed] [Google Scholar]
- 4.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P.G., Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjoblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S.D., Willis J., Dawson D., Willson J.K., Gazdar A.F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B.H., Bachman K.E., Papadopoulos N., Vogelstein B., Kinzler K.W., Velculescu V.E. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 6.Hosoi Y., Kapp L.N., Murnane J.P., Matsumoto Y., Enomoto A., Ono T., Miyagawa K. Suppression of anchorage-independent growth by expression of the ataxia-telangiectasia group D complementing gene ATDC. Biochem Biophys Res Commun. 2006;348:728–734. doi: 10.1016/j.bbrc.2006.07.115. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka Y., Inoue H., Ohmachi T., Yokoe T., Matsumoto T., Mimori K., Tanaka F., Watanabe M., Mori M. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–2549. doi: 10.1245/s10434-007-9461-1. [DOI] [PubMed] [Google Scholar]
- 8.Wang L., Heidt D.G., Lee C.J., Yang H., Logsdon C.D., Zhang L., Fearon E.R., Ljungman M., Simeone D.M. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. 2009;15:207–219. doi: 10.1016/j.ccr.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan Z., Villagra A., Peng L., Coppola D., Glozak M., Sotomayor E.M., Chen J., Lane W.S., Seto E. The ATDC (TRIM29) protein binds p53 and antagonizes p53-mediated functions. Mol Cell Biol. 2010;30:3004–3015. doi: 10.1128/MCB.01023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kushner P.J., Hort E., Shine J., Baxter J.D., Greene G.L. Construction of cell lines that express high levels of the human estrogen receptor and are killed by estrogens. Mol Endocrinol. 1990;4:1465–1473. doi: 10.1210/mend-4-10-1465. [DOI] [PubMed] [Google Scholar]
- 11.Pilat M.J., Christman J.K., Brooks S.C. Characterization of the estrogen receptor transfected MCF10A breast cell line 139B6. Breast Cancer Res Treat. 1996;37:253–266. doi: 10.1007/BF01806507. [DOI] [PubMed] [Google Scholar]
- 12.Debnath J., Muthuswamy S.K., Brugge J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 13.Muthuswamy S.K., Li D., Lelievre S., Bissell M.J., Brugge J.S. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y.H., Dudoit S., Luu P., Lin D.M., Peng V., Ngai J., Speed T.P. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van't Veer L.J., Dai H., van de Vijver M.J., He Y.D., Hart A.A., Mao M., Peterse H.L., van der Kooy K., Marton M.J., Witteveen A.T., Schreiber G.J., Kerkhoven R.M., Roberts C., Linsley P.S., Bernards R., Friend S.H. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y., Klijn J.G., Zhang Y., Sieuwerts A.M., Look M.P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M.E., Yu J., Jatkoe T., Berns E.M., Atkins D., Foekens J.A. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 17.Parker J.S., Mullins M., Cheang M.C., Leung S., Voduc D., Vickery T., Davies S., Fauron C., He X., Hu Z., Quackenbush J.F., Stijleman I.J., Palazzo J., Marron J.S., Nobel A.B., Mardis E., Nielsen T.O., Ellis M.J., Perou C.M., Bernard P.S. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroix M., Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 19.Weisz A., Rosales R. Identification of an estrogen response element upstream of the human c-fos gene that binds the estrogen receptor and the AP-1 transcription factor. Nucleic Acids Res. 1990;18:5097–5106. doi: 10.1093/nar/18.17.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown A.M., Jeltsch J.M., Roberts M., Chambon P. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci U S A. 1984;81:6344–6348. doi: 10.1073/pnas.81.20.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh M.G., Thompson D.A., Weigel R.J. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 22.van de Vijver M.J., He Y.D., van'T Veer L.J., Dai H., Hart A.A., Voskuil D.W., Schreiber G.J., Peterse J.L., Roberts C., Marton M.J., Parrish M., Atsma D., Witteveen A., Glas A., Delahaye L., van der Velde T., Bartelink H., Rodenhuis S., Rutgers E.T., Friend S.H., Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 23.Soule H.D., Maloney T.M., Wolman S.R., Peterson W.D., Jr., Brenz R., McGrath C.M., Russo J., Pauley R.J., Jones R.F., Brooks S.C. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line: MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 24.Dawson P.J., Wolman S.R., Tait L., Heppner G.H., Miller F.R. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148:313–319. [PMC free article] [PubMed] [Google Scholar]
- 25.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., Speed T., Spellman P.T., DeVries S., Lapuk A., Wang N.J., Kuo W.L., Stilwell J.L., Pinkel D., Albertson D.G., Waldman F.M., McCormick F., Dickson R.B., Johnson M.D., Lippman M., Ethier S., Gazdar A., Gray J.W. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke R.B., Howell A., Potten C.S., Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 1997;57:4987–4991. [PubMed] [Google Scholar]
- 27.Shoker B.S., Jarvis C., Clarke R.B., Anderson E., Hewlett J., Davies M.P., Sibson D.R., Sloane J.P. Estrogen receptor-positive proliferating cells in the normal and precancerous breast. Am J Pathol. 1999;155:1811–1815. doi: 10.1016/S0002-9440(10)65498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoker B.S., Jarvis C., Sibson D.R., Walker C., Sloane J.P. Oestrogen receptor expression in the normal and pre-cancerous breast. J Pathol. 1999;188:237–244. doi: 10.1002/(SICI)1096-9896(199907)188:3<237::AID-PATH343>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Enmark E., Gustafsson J.A. Oestrogen receptors - an overview. J Intern Med. 1999;246:133–138. doi: 10.1046/j.1365-2796.1999.00545.x. [DOI] [PubMed] [Google Scholar]
- 30.Webb P., Nguyen P., Valentine C., Lopez G.N., Kwok G.R., McInerney E., Katzenellenbogen B.S., Enmark E., Gustafsson J.A., Nilsson S., Kushner P.J. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13:1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- 31.Lu C., Shen Q., DuPre E., Kim H., Hilsenbeck S., Brown P.H. cFos is critical for MCF-7 breast cancer cell growth. Oncogene. 2005;24:6516–6524. doi: 10.1038/sj.onc.1208905. [DOI] [PubMed] [Google Scholar]
- 32.Rae J.M., Johnson M.D., Scheys J.O., Cordero K.E., Larios J.M., Lippman M.E. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 33.Amiry N., Kong X., Muniraj N., Kannan N., Grandison P.M., Lin J., Yang Y., Vouyovitch C.M., Borges S., Perry J.K., Mertani H.C., Zhu T., Liu D., Lobie P.E. Trefoil factor-1 (TFF1) enhances oncogenicity of mammary carcinoma cells. Endocrinology. 2009;150:4473–4483. doi: 10.1210/en.2009-0066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
High expression of TRIM29 in ER+ tumors is a marker of good prognosis in young women (<53 years of age) with early-stage breast cancer given no adjuvant systemic therapy. Microarray gene expression data for TRIM29 were used from the study by van't Veer et al.15 Kaplan-Meier plots for 88 subjects show improved relapse-free survival (P < 0.05) (A) and disease-specific survival (P < 0.05) (B).