Abstract
Human identity testing is critical to the fields of forensics, paternity, and hematopoietic stem cell transplantation. Most bone marrow (BM) engraftment testing currently uses microsatellites or short tandem repeats that are resolved by capillary electrophoresis. Single-nucleotide polymorphisms (SNPs) are theoretically a better choice among polymorphic DNA; however, ultrasensitive detection of SNPs using next-generation sequencing is currently not possible because of its inherently high error rate. We circumvent this problem by analyzing blocks of closely spaced SNPs, or haplotypes. As proof-of-principle, we chose the HLA-A locus because it is highly polymorphic and is already genotyped to select proper donors for BM transplant recipients. We aligned common HLA-A alleles and identified a region containing 18 closely spaced SNPs, flanked by nonpolymorphic DNA for primer placement. Analysis of cell line mixtures shows that the assay is accurate and precise, and has a lower limit of detection of approximately 0.01%. The BM from a series of hematopoietic stem cell transplantation patients who tested as all donor by short tandem repeat analysis demonstrated 0% to 1.5% patient DNA. Comprehensive analysis of the human genome using the 1000 Genomes database identified many additional loci that could be used for this purpose. This assay may prove useful to identify hematopoietic stem cell transplantation patients destined to relapse, microchimerism associated with solid organ transplantation, forensic applications, and possibly patient identification.
CME Accreditation Statement: This activity (“JMD 2014 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint sponsorship of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2014 CME Program in Molecular Diagnostics”) for a maximum of 48 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Myeloablative conditioning and allogeneic stem cell transplantation have historically been limited to the treatment of lethal hematological malignancies in children or young adults. More recently, with the advent of highly immunosuppressive, nonmyeloablative regimens, the clinical use of allogeneic stem cell transplantation has expanded to include older, less fit patients with hematological malignancies and patients with nonmalignant disorders, such as sickle-cell disease.1, 2, 3, 4 Nonmyeloablative conditioning regimens offer the additional safeguard of recovery of autologous hematopoiesis in the event of graft rejection and may be a safer option in patients at risk for immune-mediated rejection of the donor graft.
Chimerism testing at set intervals is an effective method for detecting graft rejection or recurrence of the original hematopoietic neoplasm after allogeneic hematopoietic stem cell transplantation (HSCT) [with either bone marrow (BM) or peripheral blood stem cells]. Decades ago, BM engraftment (BME) monitoring was performed using Southern blot analysis and minisatellite or variable number of tandem repeats loci.5 Today, short tandem repeat (STR) or microsatellite loci are most commonly used for this purpose.6, 7, 8 STRs are composed of 10 to 60 tandemly repeated units, in which each unit is 1 to 6 bases in length. They are widely distributed throughout the human genome and highly variable between individuals; therefore, they allow for excellent differentiation between individuals, including patient and donor, even if they are closely related. Most laboratories use multiplex PCR-based kits, originally developed for forensics analysis using Combined DNA Index System loci.7, 9, 10 STR analysis most commonly involves PCR amplification using fluorescently labeled primers, followed by amplicon separation by capillary electrophoresis.
Other polymorphic DNAs that could be used to monitor BME include single-nucleotide polymorphisms (SNPs).11, 12, 13 SNPs are theoretically superior to STR-based analyses because analysis of STR loci by capillary electrophoresis is relatively insensitive [limit of detection (LD), 1% to 5%] and microsatellite alleles of varying length amplify with different efficiencies, thus making them inherently biased. STR amplification can also be difficult in the setting of highly degraded DNA. However, SNPs are less attractive as targets because of their inherently lower informativity (eg, only two possible bases for a bi-allelic SNP versus ≥10 alleles for some microsatellites), requiring many more SNPs to be tested to identify those that distinguish donor from recipient. For example, we previously estimated that one would need to screen >20 to 30 individual SNPs to confidently identify one SNP where the donor is homozygous for one allele and an unrelated recipient is homozygous for the other allele.11 Fewer would need to be included if heterozygotes were included, but more would have to be analyzed for related individuals.
Recently emerging next-generation sequencing (NGS) technologies, along with their decreasing costs, are now feasible for clinical testing. However, all NGS technologies currently have high error rates, in the range of 0.04% to 1% at each base,14 which precludes their use for ultrasensitive detection of one SNP. One solution to this problem is sequencing blocks of closely spaced SNPs (ie, haplotypes). Haplotypes are regions of the genome, where polymorphic areas are sufficiently close that they are inherited together, including either genes (eg, HLA-A and HLA-B) within a locus or multiple SNPs within a region of DNA.
Herein, we first used the HLA-A locus as proof-of-principle to demonstrate that this approach permits high sensitivity, precision, and accuracy. We then studied BM samples from a cohort of patients who engrafted after HSCT and tested as all donors by STRs, and found that low-level patient DNA is commonly present. To identify additional loci that could be used for this purpose, we comprehensively analyzed the human genome and identified other regions with highly informative haplotypes. We discuss additional situations where routine haplotyping patient samples could improve patient safety.
Materials and Methods
Sample Collection and Preparation
Cell lines HCT116 (A*01:01:01 and A*02:01:01) and DLD-1 (A*02:01:01 and A*24:02:01:01) were chosen because of their available HLA-A haplotypes, and we confirmed them in the Immunogenetics Laboratory of the Johns Hopkins University (Baltimore, MD), as well as their DNA fingerprint profile using the AmpFlSTR Profiler Plus PCR Amplification Kit (Life Technologies, Carlsbad, CA). Many cells were expanded to permit making cell dilutions, because we have found that cell dilutions are generally more accurate then DNA dilutions (data not shown). Cell-to-cell dilutions were made, where an appropriate number of HCT116 cells was added to 10 million DLD-1 cells for each dilution. DNA was extracted from each cell pellet using a DNeasy Blood and Tissue kit (Qiagen, Valencia, CA). Extracted DNA was quantified by Quantifiler (Applied Biosystems, Carlsbad, CA) and stored at −20°C.
BME Samples
Samples from 18 patients who underwent allogeneic HSCT were obtained from the Molecular Diagnostic Laboratory at the Johns Hopkins Hospital on an Institutional Review Board–approved protocol. Samples were selected on the basis of the disease type, the ability to distinguish patient from donor alleles, their classification as donor by STR analysis, and the fact that at least 600 ng (100,000 genomes) of DNA was available to test (Table 1). Each BM sample was prepared for sequencing, as described below, and sequenced on the Ion Torrent Personal Genome Machine (PGM) to ensure approximately 100,000 reads were obtained.
Table 1.
BMT Patient Characteristics and Patient DNA Detected
| Patient no. | Disease | Patient HLA-A | Donor HLA-A | Difference∗ | Patients (%) |
|---|---|---|---|---|---|
| BME14 | AML | A*02:05:01/A*03:01:01:01 | A*02:05:01/A*01:01:01:01 | 11,6 | 0.000 |
| BME20 | MDS | A*29:02:01:01/A*26:01:01 | A*01:01:01:01/A*26:01:01 | 12,6 | 0.001 |
| BME18 | AML | A*03:01:01:01/A*30:01:01 | A*11:01:01/A*30:01:01 | 7,5 | 0.019 |
| BME10 | AML | A*32:01:01/A*34:02:01 | A*01:01:01:01/A*34:02:01 | 12,3 | 0.030 |
| BME19 | AML | A*24:02:01:01/A*68:01:01:01 | A*32:01:01/A*68:01:01:01 | 6,7 | 0.052 |
| BME25 | ALL | A*01:01:01:01/A*03:01:01:01 | A*33:01:01/A*03:01:01:01 | 12,11 | 0.060 |
| BME30 | AML | A*03:01:01:01/A*24:02:01:01 | A*03:01:01:01/A*01:01:01:01 | 7,6 | 0.136 |
| BME27 | AML | A*02:01:01:01/A*24:02:01:01 | A*02:01:01:01/A*02:01:01:01 | 7 | 0.140 |
| BME16 | AML | A*02:XX/A*26:XX | A*02:xx/A*03:xx | 6,9 | 0.189 |
| BME11 | AML | A*30:02:01/A*74:01 | A*68:02:01:01/A*74:01 | 8,2 | 0.190 |
| BME4 | AML-M6 | A*02:01:01:01/A*29:02:01:01 | A*02:01:01:01/A*68:01:01:01 | 6,5 | 0.312 |
| BME26 | ALL | A*11:01:01/A*30:01:01 | A*25:01:01/A*30:01:01 | 6,9 | 0.328 |
| BME1 | MM | A*02:01:01:01/A*23:01:01 | A*30:xx/A*02:xx | 4,7 | 0.416 |
| BME22 | CLL | A*02:01:01:01/A*66:01 | A*01:01:01:01/A*66:01 | 11,9 | 0.640 |
| BME21 | MCL | A*68:02:01:01/A*29:02:01:01 | A*30:01:01/A*29:02:01:01 | 8,6 | 0.750 |
| BME29 | AML | A*11:01:01/A*26:01:01 | A*11:01:01/A*11:01:01 | 6 | 0.863 |
| BME23 | DLBCL | A*02:01:01:01/A*26:01:01 | A*02:01:01:01/A*02:01:01:01 | 9 | 1.110 |
| BME24 | HD | A*01:01:01:01/A*29:02:01:01 | A*30:02:01/A*29:02:01:01 | 13,12 | 1.470 |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; HD, Hodgkin disease; MCL, mantle cell lymphoma; MDS, myelodysplastic syndrome; MM, multiple myeloma.
Number of SNP differences between unique alleles (bold) and between the unique patient and shared alleles.
HLA-A PCR Amplification
Forward and reverse primers were ordered with Ion Torrent–specific adaptors (A- and P1-adaptors) added at their 5′ ends. Briefly, the first round PCR included 600 ng (100,000 genomes) of DNA, 200 nmol/L forward and reverse primers, in Platinum PCR SuperMix High Fidelity (Invitrogen, Carlsbad, CA) in a total 100-μL reaction volume. The forward primer was as follows: 5′-CCATCTCATCCCTGCGTGTCTCCGACtcagAGGACCTGCGCTCTTGGAC-3′ [A-adaptor (underlined letters), library key (lowercase letters), and HLA-A primer (italicized letters)]. The reverse primer was as follows: 5′-CCTCTCTATGGGCAGTCGGTGATCGTTCTCCAGGTATCTGCGGA-3′ [P1-adaptor (underlined letters) and HLA-A reverse (italicized letters)]. The expected size of the full-length PCR product was 298 bp, including adapters. After amplification, adaptor containing PCR product was visualized by gel electrophoresis (Novex TBE Gels; Life Technologies) and quantified using high-sensitivity double-stranded DNA reagents and a Qubit 2.0 fluorometer version 3.10 (Invitrogen). With the desire to paint a single molecule on each bead for emulsion PCR, samples were diluted to a 0.03 nmol/L working concentration. During emulsion PCR, each single molecule produces a clone of progeny molecules (estimated to be approximately 500,000) for sequencing.
Ion Torrent PGM Library Preparation and Sequencing
Emulsion PCR using 20 to 25 μL working stock, NGS, and mapping to hg19 were all done per the manufacturer’s protocol (Life Technologies). Assessment of the percentage amplicon-containing beads was performed per the manufacturer’s protocol (Life Technologies) and measured with a Qubit 2.0 fluorometer (Invitrogen). Amplicon-coated beads were analyzed on 314 and 316 chips using the Ion PGM Sequencing 200 kit on the Life Technologies’ Ion Torrent PGM, semiconductor sequencer, which detects dNTP incorporation using the hydrogen ion that is released (along with pyrophosphate) when a dNTP is incorporated into an elongating DNA strand.15, 16
Microsatellite Analysis
PCR amplification of nine microsatellites (AmpFlSTR Profiler kit; Applied Biosystems) or 15 microsatellites (Identifiler; Applied Biosystems) was performed according to the manufacturer’s instructions. Amplicons were resolved on a capillary electrophoresis ABI3130xl Genetic Analyzer (Applied Biosystems).
Bioinformatics/Analysis
Initial processing of the data was done using the Ion Torrent platform-specific pipeline software Torrent Suite version 3.2.1 to generate sequence reads, trim adapter sequences, and remove poor quality reads. Resulting sequence files were aligned to an hg19 reference sequence. Further analysis of Fastq files was done using Geneious Pro 5.5.7, where perfectly matched reads were counted. Some samples were log10 transformed and graphed (GraphPad Prism version 5.04; GraphPad Software Inc., San Diego, CA).
Analysis of molecular specificity was done with homozygous HLA-A*01:01:01:01 and HLA-A*02:01:01:01 samples (hereafter simply A*01 and A*02, respectively). Each sample was analyzed for the other allele, followed by the introduction of the other allele’s base at 11 SNP positions. For example, homozygous A*01 was analyzed for perfectly matched A*01 reads, followed by perfect A*02 reads. On this, one A*02-specific base was introduced into the A*01 sequence at each of the 11 SNP positions and analyzed. After introducing one A*02-specific base at all of the possible positions, two A*02-specific bases were introduced, repeating the process for 11 possible combinations. The process was continued all of the way to substituting 10 A*02-specific bases. The number of reads found, for each combination, was averaged and the percentage of reads found was graphed.
The BM transplant samples were analyzed for the amount of patient (patient unique allele) in the sample. After eliminating reads representing a haplotype shared by both individuals, we calculated percentage patient DNA (patient/patient + donor). For cases where three alleles were shared, the equation [(2 × unique patient)/(unique patient + shared patient − donor)] was used.
Bioinformatics Analysis of 1000 Genomes Database
By use of 1000 Genomes release version 3_20110521 from build hg18 (http://www.1000genomes.org, last accessed July 3, 2014), we identified regions that were polymorphic and flanked by constant regions. We required a minimum of nine variants with a minor allele frequency of ≥9% in three of the major populations: CEU (CEPH, Utah residents with Northern and Western European ancestry); JPTCHB (a combined Asian population including JPT, Japanese in Tokyo, Japan, and CHB, Han Chinese in Beijing, China); and YRI (Yoruba in Ibadan, Nigeria) within 300 bp. Size was chosen so that it could be amplified and sequenced in a single read, well within the limits of current NGS technology. We required potential regions to be flanked by constant regions of at least 20 bp for primer placement.
Polymorphic regions were converted from hg18 to hg19 using University of California, Santa Cruz’s LiftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver, last accessed March 28, 2014). A custom script was written to convert phased variants from 1000 Genomes variant call files (both IMPUTE217 and SHAPEIT218 versions; http://www.1000genomes.org) into IMPUTE reference panel format haplotypes at every region. A custom program was then used to assess all possible haplotype combinations between two theoretical individuals for informativity and probability of occurrence. Particular haplotype combinations were defined as informative if at least one haplotype of individual B (patient) had at least two SNP differences from both haplotypes of individual A (donor). Once informative haplotype combinations were determined, the probabilities of these combinations occurring in two unrelated individuals were calculated (on the basis of the 1000 Genomes frequencies; http://www.1000genomes.org) and summed. Results from the two methods of phasing the chromosomes were highly correlated (data not shown), so only the data from SHAPEIT2 phased haplotypes are presented. Thus, the calculated probability score reflects the informativity of the region for two unrelated individuals.
Results
Overall Strategy
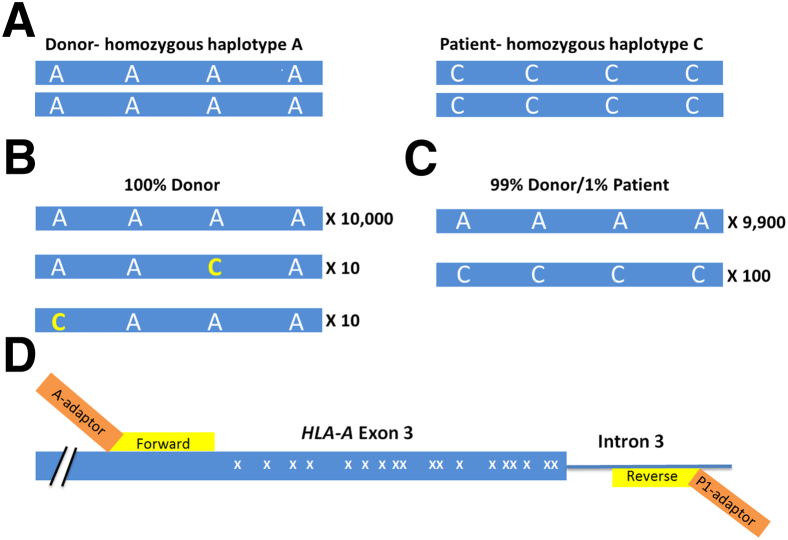
The overall strategy is demonstrated in Figure 1. Imagine a region of a gene that contains four SNPs and two individuals: a donor who is homozygous for adenine at all four SNPs (designated homozygous haplotype A) and a patient (recipient) who is homozygous for cytosine at all four SNPs (homozygous haplotype C) (Figure 1A). In a sample that is pure donor (top bar, 10,000 reads), because of the high error rates of NGS, mutations will be seen by NGS, some of which will coincidentally occur at SNP positions (10 reads each) (Figure 1B). Because these involve only one of the four SNPs, these can be attributed as PCR errors and discarded because these molecules do not match either the donor or patient alleles. If the strategy used only one SNP, instead of a haplotype, one would be unable to distinguish between 10 true patient reads and 10 PCR error reads. In contrast, if 100 molecules are detected in the post-transplant sample with cytosine at all four SNPs, this would indicate the presence of true patient-specific DNA (Figure 1C).
Figure 1.
Haplotype counting strategy. A: Shown is a theoretical locus containing four possible SNPs, where the donor has homozygous adenine at each of the SNPs and the patient (recipient, right) has homozygous cytosine. B: In a post-transplant sample, 10,000 reads that perfectly match the donor genotype, 10 reads with a cytosine (yellow) at the third SNP, and 10 reads with a cytosine at the first SNP (yellow) are detected. The 20 reads with a single cytosine can be interpreted as PCR errors because they do not perfectly match either donor or patient alleles. Accordingly, these results would be interpreted as 100% donor. C: Another post-transplant sample contains 9900 reads that perfectly match the donor haplotype, whereas 100 reads perfectly match the patient haplotype. Accordingly, this sample is 1% patient. D: The distal region of HLA-A exon 3 (blue) and intron 3 (blue line) containing 18 potential SNPs (indicated with X) and two HLA-A PCR primers (yellow). The PCR primers are tailed with adapters (orange) for the DNA sequencing primer to bind (A-adapter) or to covalently anchor the amplicon onto the bead (P1-adapter). The total length of the HLA-targeted region is 245 bp, and the total length of the amplicon is 298 bp, including the adapters. Not shown is the four-base library key located between the A-adaptor and the forward HLA-A primer (as described in Materials and Methods).
Analysis of HLA-A Alleles
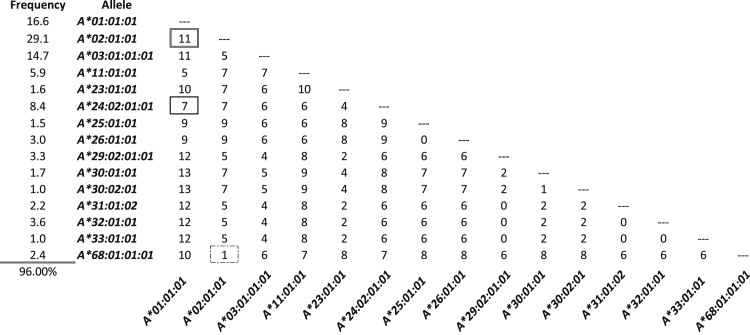
We performed alignments of common HLA-A alleles in the European-origin population using the Major Histocompatibility Complex database, a publically accessible platform for DNA and clinical data related to the human major histocompatibility complex (http://www.ncbi.nlm.nih.gov/gv/mhc/main.cgi?cmd=init, last accessed March 28, 2014). Regions with a high density of SNPs and flanked by nonpolymorphic DNA were identified. One region, HLA-A exon 3, contained 18 possible SNPs and at least 15 major alleles in the European-origin population. We tested a series of primers surrounding this region and selected the best pair on the basis of amplification efficiency and specificity (as described in Materials and Methods) (Figure 1D). The number of SNP differences in this region between the most common alleles of the European-origin population is given (Figure 2); some combinations of HLA-A alleles are easily differentiated, whereas others are more difficult. For example, 11 SNPs differentiate allele A*01 from A*02, so that even with a relatively high base substitution error rate (eg, 1% per base), a sample homozygous for A*01 should not contain any false-positive A*02 allele reads. In contrast, A*02 and HLA-A*68:01:01:01 (hereafter A*68) have only one SNP difference, so a pure sample homozygous for A*02 will likely contain reads matching A*68 because of the relatively high error rate intrinsic to current NGS technologies.
Figure 2.
Numbers of discriminating SNPs between common HLA-A alleles. Common European-origin HLA-A alleles were collected and aligned, and the number of discriminating SNPs between them was determined. The homozygous genotypes analyzed later in Figure 3, A*01 and A*02, differ by 11 SNPs (double box). The genotypes from the dilution series later in Figure 4 are A*01/A*02 into A*02/A*24, where the unique alleles vary by seven SNPs (bold box). Other combinations vary by fewer SNPs, such as A*68 versus A*02 (dashed box). Included are only alleles that occur at ≥1% in the population of European origin.
Specificity of the Assay for SNPs
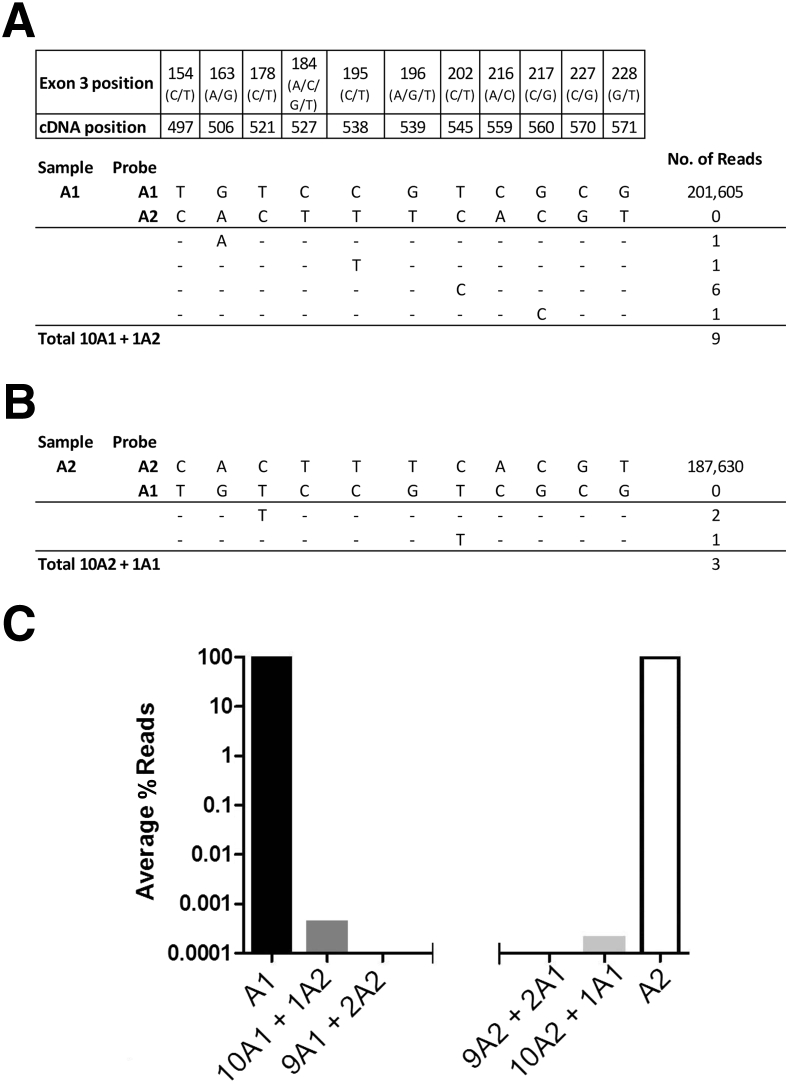
To test the possible cross talk between molecules that vary by 11 SNPs, we sequenced two samples, one homozygous for A*01 and another homozygous for A*02, and analyzed each for the other allele (Figure 3). The A*01 and A*02 samples contained approximately 200,000 perfect matching reads. Neither pure sample contained any perfect reads of the other haplotype. We then examined the A*01 sample files for reads containing a single A*02 SNP at each of the 11 positions and found an average of 0.3% to 0.8% reads that contained a single error from the perfect haplotype (Figure 3A). When two SNPs of the opposite haplotype were searched, no reads were obtained. Similar results were obtained with the pure A*02 sample (Figure 3B). A double-waterfall plot shows that when enough discriminating SNPs between two individuals’ alleles exist, the assay is highly specific (Figure 3C and Supplemental Tables S1 and S2).
Figure 3.
Molecular specificity. Samples homozygous for the A*01 allele (A) and the A*02 allele (B) were PCR amplified and NGS sequenced. Each was analyzed for the presence of the other allele and for hybrid molecules containing one or more SNPs of the other allele. C: Waterfall plot of the percentage reads for each combination of the 11 SNP differences for the two samples. For example, the 10A1 + 1A2 bar reflects the mean number of erroneous reads (9/11 = 0.82), in the pure A*01 sample where any one of the 11 SNP positions matches the A*02 allele (0.82/201,605, from A). Likewise, the 9A1 + 2A2 bar reflects the lack of any reads in the pure A*01 sample, where two adjacent SNPs match the A*02 allele at any of the positions.
HLA-A Dose-Response Curve, Accuracy, Precision, and LD
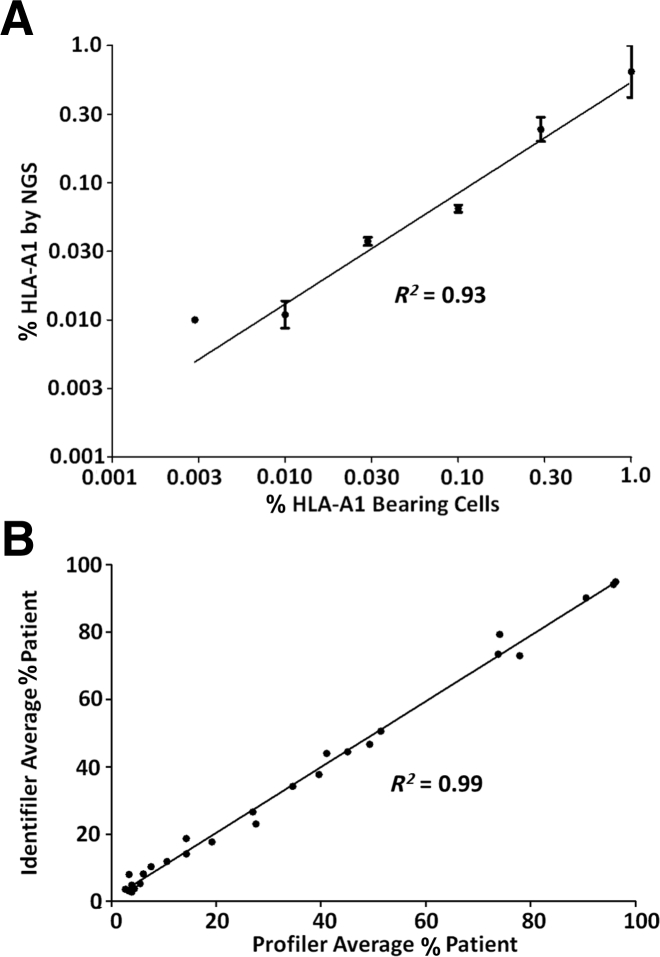
To assess the accuracy and LD, we generated a dilution series from two cell lines with known HLA-A genotypes. These samples were chosen because the two alleles of interest (A*01 and A*02) vary from one another by 11 SNPs, and both vary from the commonly shared allele (A*24) by seven SNPs (Figure 2). Dilutions were made with cell mixes varying from 1 in 1 million (0.0001%) to 1 in 100 (1%) using a total of 10 million cells for each dilution. DNA was isolated and PCR was performed using 600 ng of DNA. We chose this relatively large amount of DNA on the basis of the desire to achieve an LD of at least 1:10,000 (0.01%) and to exceed that target LD by using 10× excess DNA. This relatively high DNA input reflects approximately 100,000 genomes (on the basis of approximately 6 pg/haploid genome) and was chosen to prevent bottlenecking and resultant allele dropout. For example, if DNA representing only 100 genomes were analyzed to 100,000 × depth of coverage, the LD is input DNA limited, not depth of coverage limited, and is at best 1%. A sample with a minor allele frequency of 0.1% would likely not be detected. Each sample was sequenced at least twice, and results were graphed as the percentage HLA-A*01–bearing cells (Figure 4A). Least-squares analysis generated a straight line demonstrating excellent accuracy (R2 = 0.93). To determine precision, we performed additional replicates at 0.1% and 0.01% dilutions (four total) (Supplemental Table S3). The assay was highly precise at 0.1% cell mix (mean, 0.065%; coefficient of variation, 0.12%), but less so at the 0.01% cell mix, as expected (mean, 0.012%; coefficient of variation, 0.40%). Because we detected the minor HLA-A allele in four of four replicates at 0.01%, but only one of two replicates at 0.003%, we concluded that 0.01% is the lower LD for this assay (Supplemental Table S3). These results compare favorably with those of microsatellites, which are highly linear and generally accurate, but with a much poorer LD of only approximately 3% (Figure 4B).
Figure 4.
Linearity NGS haplotyping and microsatellite analysis. Linearity of the SNP haplotyping approach. A*01/A*02 cells were serially diluted into A*02/A*24 cells, and DNA was isolated and tested by HLA-A NGS. A: NGS HLA-A*01 counts were expressed as a percentage, as a function of the percentage of HLA-A*01 bearing cells. Error bars: ±1 SEM. The error bar for 0.1% bearing cells is too small to be seen. The error bar for 0.003% is absent because only half the replicates were amplified. The NGS haplotyping data begin at 1% and are serially diluted down from there. B: Post-transplant patient samples with various percentage of patient DNA were analyzed using the Profiler and Identifiler kits. Note the lack of the ability to detect DNA lower than the LD of 3%.
SNP Haplotype Assay Detects Patient DNA in BM Samples That Tested All Donors by STR Analysis
We selected 18 patients whose donor-patient HLA genotypes varied by at least four SNPs (Table 1). The allele unique to donor varied from that unique to patient by 4 to 13 SNPs. To further prevent false-positive results, the unique patient allele varied from the shared allele by 2 to 12 SNPs. We also required that 600 ng be available for testing, so that the number of genomes (approximately 100,000) exceeded by an order of magnitude the desired LD. Several samples were excluded that could not meet these criteria.
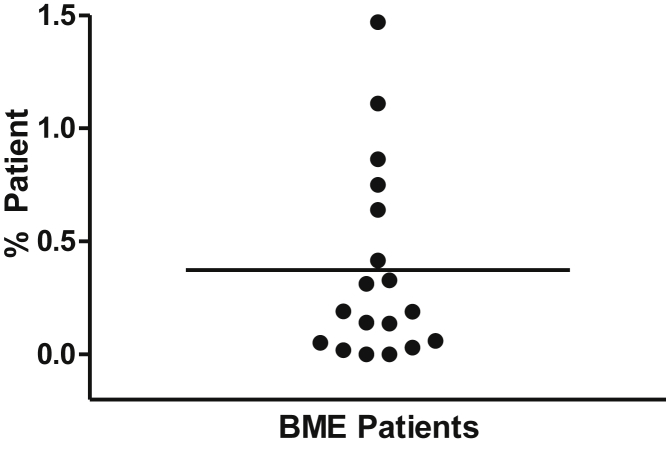
All samples tested positive for some level of patient DNA, except for one sample. The positives ranged from 0.001% to 1.47% patient DNA (mean, 0.373) (Figure 5), confirming that the haplotype-based assay was more sensitive. We also noted that of the four samples with the highest levels of patient DNA, three of them were from patients with lymphoma.
Figure 5.
Percentage of patient DNA in BM samples. Unique patient DNA was detected in BM samples, which demonstrated all donors by conventional short tandem repeat assays, after hematopoietic stem cell transplantation.
The Human Genome Contains Clusters of SNPs That Can Be Amplified to Give Information on Haplotypes
We used variant calls from the 1000 Genomes project (http://www.1000genomes.org) to identify regions containing at least nine SNPs in all three populations (Europeans, Asians, and Africans; as described in Materials and Methods) within 300 bp. We further required these regions to be flanked by at least 20 bp of nonpolymorphic DNA. The nonpolymorphic regions provide potential primer sites, whereas the variable regions are suitable for haplotype counting. We identified 4349 such loci across the genome (Supplemental Figure S1), requiring that nine or more variants have minor allele frequencies >9% in all three populations (Supplemental Table S4). One concern is that a cluster of SNPs may not be that informative because it simply represents a high-frequency haplotype. For example, a haplotype that exists within a population at a 2% level and containing 20 SNPs, in conjunction with a second haplotype with the other base at each of the 20 positions present in 98% of the population, is a relatively uninformative marker. In contrast, a locus with 20 SNPs with 10 different haplotypes, each of which is present at 10% in the population, is highly informative.
By using the 4349 regions identified as SNP dense, we analyzed the 1000 (1092) Genomes project (http://www.1000genomes.org) phased-variant call files to calculate overall probability that two unrelated individuals would be different at each locus (as described in Materials and Methods). Briefly, at each locus, a file was generated that contained all of the SNPs for each of the 2184 (2 × 1092) alleles. We then determined the number of distinct alleles and the number of times each of them was represented. We then calculated all possible combinations of alleles within donors and recipients, and finally determined the combined probability that two unrelated individuals would be informative (Table 2 and Supplemental Table S5). These putative alternate loci will need to be experimentally validated in large cohorts of patients, and experiments are in progress for this validation.
Table 2.
Alternate Loci for Haplotype Counting
| Chromosome∗ | Gene† | Start‡ | Stop | Length (bp) | No. of SNPs | No. of haplotypes | CEU | JPT | YRI | Mean |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | — | 238855189 | 238855428 | 240 | 9 | 7 | 0.63 | 0.46 | 0.63 | 0.57 |
| 1 | — | 238416454 | 238416673 | 220 | 9 | 11 | 0.39 | 0.46 | 0.43 | 0.43 |
| 2 | — | 106142506 | 106142791 | 286 | 10 | 8 | 0.63 | 0.61 | 0.66 | 0.63 |
| 2 | VIT | 36927840 | 36928071 | 232 | 9 | 6 | 0.58 | 0.59 | 0.61 | 0.59 |
| 3 | ST6GAL1 | 186654745 | 186655022 | 278 | 9 | 12 | 0.72 | 0.67 | 0.73 | 0.70 |
| 3 | — | 97907092 | 97907374 | 283 | 10 | 13 | 0.58 | 0.42 | 0.64 | 0.55 |
| 4 | SORCS2 | 7447136 | 7447434 | 299 | 9 | 16 | 0.60 | 0.46 | 0.65 | 0.57 |
| 4 | — | 66995978 | 66996254 | 277 | 9 | 8 | 0.63 | 0.45 | 0.60 | 0.56 |
| 5 | — | 125498526 | 125498753 | 228 | 10 | 19 | 0.67 | 0.58 | 0.67 | 0.64 |
| 5 | — | 178259629 | 178259892 | 264 | 13 | 23 | 0.69 | 0.56 | 0.64 | 0.63 |
| 6 | HLA-DRB1 | 32551493 | 32551765 | 273 | 9 | 306 | 0.98 | 0.95 | 0.96 | 0.96 |
| 6 | HLA-B | 31319390 | 31319668 | 279 | 10 | 27 | 0.93 | 0.91 | 0.88 | 0.91 |
| 7 | — | 64895079 | 64895355 | 277 | 10 | 12 | 0.59 | 0.61 | 0.66 | 0.62 |
| 7 | — | 9111002 | 9111297 | 296 | 9 | 11 | 0.60 | 0.64 | 0.59 | 0.61 |
| 8 | — | 6160141 | 6160442 | 302 | 10 | 13 | 0.56 | 0.67 | 0.63 | 0.62 |
| 8 | CSMD1 | 3478267 | 3478568 | 302 | 10 | 13 | 0.63 | 0.51 | 0.66 | 0.60 |
| 9 | — | 132197723 | 132197947 | 225 | 11 | 11 | 0.66 | 0.45 | 0.53 | 0.55 |
| 9 | — | 95691312 | 95691534 | 223 | 10 | 14 | 0.39 | 0.35 | 0.46 | 0.40 |
| 10 | — | 133376146 | 133376435 | 290 | 9 | 14 | 0.74 | 0.74 | 0.78 | 0.75 |
| 10 | — | 123095093 | 123095377 | 285 | 10 | 18 | 0.74 | 0.78 | 0.69 | 0.74 |
| 11 | CNTN5 | 99491238 | 99491538 | 301 | 9 | 7 | 0.62 | 0.50 | 0.60 | 0.57 |
| 11 | — | 5078880 | 5079122 | 243 | 10 | 15 | 0.65 | 0.40 | 0.61 | 0.55 |
| 12 | — | 82460264 | 82460531 | 268 | 9 | 7 | 0.65 | 0.66 | 0.66 | 0.66 |
| 12 | — | 17884472 | 17884719 | 248 | 9 | 10 | 0.67 | 0.61 | 0.66 | 0.65 |
| 13 | FARP1 | 99084002 | 99084259 | 258 | 9 | 17 | 0.63 | 0.70 | 0.71 | 0.68 |
| 13 | — | 33553507 | 33553784 | 278 | 9 | 29 | 0.68 | 0.45 | 0.65 | 0.59 |
| 14 | — | 107094021 | 107094317 | 297 | 10 | 25 | 0.53 | 0.51 | 0.66 | 0.57 |
| 14 | TRA | 22736235 | 22736521 | 287 | 9 | 11 | 0.51 | 0.57 | 0.61 | 0.56 |
| 15 | — | 34750054 | 34750330 | 277 | 9 | 8 | 0.51 | 0.57 | 0.61 | 0.56 |
| 15 | — | 25047393 | 25047681 | 289 | 10 | 25 | 0.51 | 0.44 | 0.55 | 0.50 |
| 16 | — | 56576389 | 56576687 | 299 | 11 | 46 | 0.76 | 0.82 | 0.80 | 0.79 |
| 16 | — | 84540653 | 84540932 | 280 | 9 | 16 | 0.69 | 0.66 | 0.74 | 0.70 |
| 17 | TBCD | 80804085 | 80804372 | 288 | 9 | 33 | 0.70 | 0.70 | 0.74 | 0.71 |
| 17 | RBFOX3 | 77148074 | 77148367 | 294 | 10 | 15 | 0.38 | 0.60 | 0.52 | 0.50 |
| 18 | — | 76597079 | 76597332 | 254 | 9 | 14 | 0.40 | 0.40 | 0.40 | 0.40 |
| 18 | CLUL1 | 631210 | 631400 | 191 | 9 | 12 | 0.35 | 0.27 | 0.38 | 0.33 |
| 19 | CCDC61 | 46500080 | 46500360 | 281 | 9 | 14 | 0.41 | 0.38 | 0.43 | 0.41 |
| 19 | — | 57525179 | 57525424 | 246 | 13 | 10 | 0.37 | 0.39 | 0.41 | 0.39 |
| 20 | PROKR2 | 5289890 | 5290161 | 272 | 9 | 7 | 0.64 | 0.51 | 0.59 | 0.58 |
| 20 | SIRPA | 1895467 | 1895674 | 208 | 11 | 24 | 0.49 | 0.58 | 0.56 | 0.54 |
| 21 | — | 20548907 | 20549196 | 290 | 9 | 6 | 0.44 | 0.35 | 0.42 | 0.40 |
| 21 | UMODL1 | 43528819 | 43529079 | 261 | 9 | 10 | 0.33 | 0.38 | 0.39 | 0.36 |
—, No data.
Chromosome 22 is absent because of the lack of good polymorphic loci. X and Y were excluded.
Gene, if present.
Hg19 coordinates.
Discussion
Herein, we demonstrated that we can detect mixtures of human DNA down to an LD of 0.01% (1 in 10,000) using haplotype counting by NGS, 100-fold more sensitive than current STR-based methods. False positives from NGS are avoided by using haplotypes because if they vary from one another by enough SNPs, they should demonstrate no cross talk, even with a mutation frequency of 0.1% to 1% per base. Although we have chosen HLA-A19, 20 as a proof-of-concept in this assay, we report other haplotype loci from the 1000 Genomes project21 (http://www.1000genomes.org) that could be used for this purpose. Selection of suitable loci may be influenced by the patient’s ethnic background, number of discriminating SNPs, and ease of primer placement. The set of haplotypes used for this purpose ideally would be suitable for transplant analysis of all patient ethnicities. Patient DNA is consistently detected in BM samples that test all donor by the conventional STR assay.
Detection of minimal residual disease in cases such as acute promyelocytic leukemia and subsequent early intervention has been associated with significantly higher survival.22, 23 The haplotype counting-based assay may prove valuable to detect relapse in HSCT patients earlier than the existing microsatellite-based assays. One of the other non-chromosome 6 (containing human HLA) loci (Table 2) might be better for this purpose, because one mechanism that leukemic cells can escape donor anti-leukemic T cells is through the loss of the mismatched HLA allele, estimated to occur in 29.4% to 66.7% of such patients.24, 25 Another limitation of the HLA-A haplotype approach is that some transplant donors are HLA identical, and loci other than HLA would be required to monitor such patients.
HSCT has traditionally been used to treat malignant and nonmalignant hematological disorders. In addition, with the transplantation of solid organs, some amount of lymphoid tissue may be transferred by cell migration from the donor organs, thereby generating a chimerism in the patient.26 In contrast, intentional induction of a microchimerism, by injecting donor BM, is a strategy used to generate donor-specific tolerance in extremity transplantation.27, 28, 29 The development and persistence of a donor-recipient microchimerism may be associated with the acceptance of transplanted organs.26 Using NGS to detect low levels of donor cells may provide an opportunity to document such a microchimerism (or lack thereof) and possibly manage immunosuppressive regimens to optimize engraftment.
The NGS of highly polymorphic regions may have applications outside of transplantation medicine. A microchimerism resulting from bidirectional exchange of cells between mother and fetus has been detected in women long after pregnancy using Y chromosome fluorescence in situ hybridization and PCR for the SRY gene.30, 31, 32 Our assay could be applied for such studies with the additional benefit of also being able to detect exchanged cells between a mother and a daughter. NGS haplotyping might also aid in the detection of rare tissue regenerative cells in the heart transplant setting. In sex-mismatched heart transplant and using Y chromosome fluorescence in situ hybridization, studies have shown a cardiac chimerism caused by migration of recipient cells to the grafted heart.33, 34
The field of forensics has relied on STR loci for identification of suspects and human remains. In theory, testing haplotypes could allow one to identify the presence of suspect in a large mixture of DNAs, such as in polysuspect cases.35, 36 In addition to forensic applications, tools for quality control and sample tracking in a large inventory of human DNA samples would be valuable. Such tools have previously been reported using panels of SNPs,37 and the NGS-haplotyping assay could be applied in similar situations. Similar markers could be developed to distinguish among species.
To ensure patient identity and exclude the possibility of a mislabeled specimen, haplotyping to uniquely define the patient could easily be included in any NGS-based genetic test. They could also be used for any absolutely critical test, such as ABO typing of blood products,38 and matched to the intended patient’s genotype encoded in an implanted microchip immediately before transfusion. Although rare, wrong-patient adverse events occur in hospital settings, especially when patients share the same name or patients have similar appearances.39 To circumvent these preventable errors, a biological identifier, such as those described herein, could be implemented to unequivocally distinguish patients.
NGS technologies, because of their declining costs and improved read lengths and base calling, have been making their way into the clinical setting. Haplotype counting using NGS described herein may become a valuable tool to analyze DNA mixes and perform identity testing because of its high sensitivity, precision, and accuracy.
Note Added in Proof
A group recently published that circulating DNA of donor origin can be used to detect rejection of the transplanted heart.40
Acknowledgments
We thank Drs. Ephraim J. Fuchs, Richard J. Jones, Dwight Oliver, Allison Klein, Javier Bolanos-Meade, Bert Vogelstein, Alexis Norris, Soonweng Cho, Elliot Z. Chen, Laura Wood, Eric Stevens, Alexis Carter, Stacy Mosier, Rosie Jiang, and Jennifer Meyers for helpful discussions.
Footnotes
Supported in part by The Sol Goldman Foundation.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2014.04.003.
Supplemental Data
Relative densities of minor allele frequency-filtered variants across the human autosomes. The densities of variants with a minor allele frequency >9% in the CEU, JPTCHB, and YRI populations along each autosome were calculated in 1-kb windows using the IRanges R package (http://www.r-project.org).
References
- 1.Slavin S., Nagler A., Naparstek E., Kapelushnik Y., Aker M., Cividalli G., Varadi G., Kirschbaum M., Ackerstein A., Samuel S., Amar A., Brautbar C., Ben-Tal O., Eldor A., Or R. Nonmyeloablative stem cell transplantation and cell therapy as an alternative to conventional bone marrow transplantation with lethal cytoreduction for the treatment of malignant and nonmalignant hematologic diseases. Blood. 1998;91:756–763. [PubMed] [Google Scholar]
- 2.Khouri I.F., Saliba R.M., Giralt S.A., Lee M.S., Okoroji G.J., Hagemeister F.B., Korbling M., Younes A., Ippoliti C., Gajewski J.L., McLaughlin P., Anderlini P., Donato M.L., Cabanillas F.F., Champlin R.E. Nonablative allogeneic hematopoietic transplantation as adoptive immunotherapy for indolent lymphoma: low incidence of toxicity, acute graft-versus-host disease, and treatment-related mortality. Blood. 2001;98:3595–3599. doi: 10.1182/blood.v98.13.3595. [DOI] [PubMed] [Google Scholar]
- 3.Brodsky R.A., Luznik L., Bolanos-Meade J., Leffell M.S., Jones R.J., Fuchs E.J. Reduced intensity HLA-haploidentical BMT with post transplantation cyclophosphamide in nonmalignant hematologic diseases. Bone Marrow Transplant. 2008;42:523–527. doi: 10.1038/bmt.2008.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolanos-Meade J., Fuchs E.J., Luznik L., Lanzkron S.M., Gamper C.J., Jones R.J., Brodsky R.A. HLA-haploidentical bone marrow transplantation with posttransplant cyclophosphamide expands the donor pool for patients with sickle cell disease. Blood. 2012;120:4285–4291. doi: 10.1182/blood-2012-07-438408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeffreys A.J., Wilson V., Thein S.L. Individual-specific “fingerprints” of human DNA. Nature. 1985;316:76–79. doi: 10.1038/316076a0. [DOI] [PubMed] [Google Scholar]
- 6.Scharf S.J., Smith A.G., Hansen J.A., McFarland C., Erlich H.A. Quantitative determination of bone marrow transplant engraftment using fluorescent polymerase chain reaction primers for human identity markers. Blood. 1995;85:1954–1963. [PubMed] [Google Scholar]
- 7.Thiede C., Florek M., Bornhauser M., Ritter M., Mohr B., Brendel C., Ehninger G., Neubauer A. Rapid quantification of mixed chimerism using multiplex amplification of short tandem repeat markers and fluorescence detection. Bone Marrow Transplant. 1999;23:1055–1060. doi: 10.1038/sj.bmt.1701779. [DOI] [PubMed] [Google Scholar]
- 8.Schichman S.A., Suess P., Vertino A.M., Gray P.S. Comparison of short tandem repeat and variable number tandem repeat genetic markers for quantitative determination of allogeneic bone marrow transplant engraftment. Bone Marrow Transplant. 2002;29:243–248. doi: 10.1038/sj.bmt.1703360. [DOI] [PubMed] [Google Scholar]
- 9.Mills K.A., Even D., Murray J.C. Tetranucleotide repeat polymorphism at the human alpha fibrinogen locus (FGA) Hum Mol Genet. 1992;1:779. doi: 10.1093/hmg/1.9.779. [DOI] [PubMed] [Google Scholar]
- 10.Huang N.E., Schumm J., Budowle B. Chinese population data on three tetrameric short tandem repeat loci–HUMTHO1, TPOX, and CSF1PO–derived using multiplex PCR and manual typing. Forensic Sci Int. 1995;71:131–136. doi: 10.1016/0379-0738(94)01646-m. [DOI] [PubMed] [Google Scholar]
- 11.Oliver D.H., Thompson R.E., Griffin C.A., Eshleman J.R. Use of single nucleotide polymorphisms (SNP) and real-time polymerase chain reaction for bone marrow engraftment analysis. J Mol Diagn. 2000;2:202–208. doi: 10.1016/S1525-1578(10)60638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochberg E.P., Miklos D.B., Neuberg D., Eichner D.A., McLaughlin S.F., Mattes-Ritz A., Alyea E.P., Antin J.H., Soiffer R.J., Ritz J. A novel rapid single nucleotide polymorphism (SNP)-based method for assessment of hematopoietic chimerism after allogeneic stem cell transplantation. Blood. 2003;101:363–369. doi: 10.1182/blood-2002-05-1365. [DOI] [PubMed] [Google Scholar]
- 13.Gineikiene E., Stoskus M., Griskevicius L. Single nucleotide polymorphism-based system improves the applicability of quantitative PCR for chimerism monitoring. J Mol Diagn. 2009;11:66–74. doi: 10.2353/jmoldx.2009.080039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bragg L.M., Stone G., Butler M.K., Hugenholtz P., Tyson G.W. Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput Biol. 2013;9:e1003031. doi: 10.1371/journal.pcbi.1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W., Davey M. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 16.Merriman B., Ion Torrent R&D Team. Rothberg J.M. Progress in ion torrent semiconductor chip based sequencing. Electrophoresis. 2012;33:3397–3417. doi: 10.1002/elps.201200424. [DOI] [PubMed] [Google Scholar]
- 17.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delaneau O., Zagury J.F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 19.Gabriel C., Furst D., Fae I., Wenda S., Zollikofer C., Mytilineos J., Fischer G.F. HLA typing by next-generation sequencing: getting closer to reality. Tissue Antigens. 2014;83:65–75. doi: 10.1111/tan.12298. [DOI] [PubMed] [Google Scholar]
- 20.De Santis D., Dinauer D., Duke J., Erlich H.A., Holcomb C.L., Lind C., Mackiewicz K., Monos D., Moudgil A., Norman P., Parham P., Sasson A., Allcock R.J. 16(th) IHIW: review of HLA typing by NGS. Int J Immunogenet. 2013;40:72–76. doi: 10.1111/iji.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke L., Zheng-Bradley X., Smith R., Kulesha E., Xiao C., Toneva I., Vaughan B., Preuss D., Leinonen R., Shumway M., Sherry S., Flicek P., 1000 Genomes Project Consortium The 1000 Genomes Project: data management and community access. Nat Methods. 2012;9:459–462. doi: 10.1038/nmeth.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteve J., Escoda L., Martin G., Rubio V., Díaz-Mediavilla J., González M., Rivas C., Alvarez C., Gonzalez San Miguel J.D., Brunet S., Tomás J.F., Tormo M., Sayas M.J., Sanchez Godoy P., Colomer D., Bolufer P., Sanz M.A., Spanish Cooperative Group PETHEMA Outcome of patients with acute promyelocytic leukemia failing to front-line treatment with all-trans retinoic acid and anthracycline-based chemotherapy (PETHEMA protocols LPA96 and LPA99): benefit of an early intervention. Leukemia. 2007;21:446–452. doi: 10.1038/sj.leu.2404501. [DOI] [PubMed] [Google Scholar]
- 23.Chendamarai E., Balasubramanian P., George B., Viswabandya A., Abraham A., Ahmed R., Alex A.A., Ganesan S., Lakshmi K.M., Sitaram U., Nair S.C., Chandy M., Janet N.B., Srivastava V.M., Srivastava A., Mathews V. Role of minimal residual disease monitoring in acute promyelocytic leukemia treated with arsenic trioxide in frontline therapy. Blood. 2012;119:3413–3419. doi: 10.1182/blood-2011-11-393264. [DOI] [PubMed] [Google Scholar]
- 24.Vago L., Perna S.K., Zanussi M., Mazzi B., Barlassina C., Stanghellini M.T., Perrelli N.F., Cosentino C., Torri F., Angius A., Forno B., Casucci M., Bernardi M., Peccatori J., Corti C., Bondanza A., Ferrari M., Rossini S., Roncarolo M.G., Bordignon C., Bonini C., Ciceri F., Fleischhauer K. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med. 2009;361:478–488. doi: 10.1056/NEJMoa0811036. [DOI] [PubMed] [Google Scholar]
- 25.Villalobos I.B., Takahashi Y., Akatsuka Y., Muramatsu H., Nishio N., Hama A., Yagasaki H., Saji H., Kato M., Ogawa S., Kojima S. Relapse of leukemia with loss of mismatched HLA resulting from uniparental disomy after haploidentical hematopoietic stem cell transplantation. Blood. 2010;115:3158–3161. doi: 10.1182/blood-2009-11-254284. [DOI] [PubMed] [Google Scholar]
- 26.Starzl T.E., Demetris A.J., Trucco M., Murase N., Ricordi C., Ildstad S., Ramos H., Todo S., Tzakis A., Fung J.J., Nalesnik M., Zeevi A., Rudert W.A., Kocova M. Cell migration and chimerism after whole-organ transplantation: the basis of graft acceptance. Hepatology. 1993;17:1127–1152. [PMC free article] [PubMed] [Google Scholar]
- 27.Foster R.D., Fan L., Neipp M., Kaufman C., McCalmont T., Ascher N., Ildstad S., Anthony J.P., Niepp M. Donor-specific tolerance induction in composite tissue allografts. Am J Surg. 1998;176:418–421. doi: 10.1016/s0002-9610(98)00248-7. [DOI] [PubMed] [Google Scholar]
- 28.Foster R.D., Ascher N.L., McCalmont T.H., Neipp M., Anthony J.P., Mathes S.J. Mixed allogeneic chimerism as a reliable model for composite tissue allograft tolerance induction across major and minor histocompatibility barriers. Transplantation. 2001;72:791–797. doi: 10.1097/00007890-200109150-00009. [DOI] [PubMed] [Google Scholar]
- 29.Schneeberger S., Gorantla V.S., Brandacher G., Zeevi A., Demetris A.J., Lunz J.G., Metes D.M., Donnenberg A.D., Shores J.T., Dimartini A.F., Kiss J.E., Imbriglia J.E., Azari K., Goitz R.J., Manders E.K., Nguyen V.T., Cooney D.S., Wachtman G.S., Keith J.D., Fletcher D.R., Macedo C., Planinsic R., Losee J.E., Shapiro R., Starzl T.E., Lee W.P. Upper-extremity transplantation using a cell-based protocol to minimize immunosuppression. Ann Surg. 2013;257:345–351. doi: 10.1097/SLA.0b013e31826d90bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bianchi D.W., Zickwolf G.K., Weil G.J., Sylvester S., DeMaria M.A. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A. 1996;93:705–708. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cirello V., Perrino M., Colombo C., Muzza M., Filopanti M., Vicentini L., Beck-Peccoz P., Fugazzola L. Fetal cell microchimerism in papillary thyroid cancer: studies in peripheral blood and tissues. Int J Cancer. 2010;126:2874–2878. doi: 10.1002/ijc.24993. [DOI] [PubMed] [Google Scholar]
- 32.Chan W.F., Gurnot C., Montine T.J., Sonnen J.A., Guthrie K.A., Nelson J.L. Male microchimerism in the human female brain. PLoS One. 2012;7:e45592. doi: 10.1371/journal.pone.0045592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quaini F., Urbanek K., Beltrami A.P., Finato N., Beltrami C.A., Nadal-Ginard B., Kajstura J., Leri A., Anversa P. Chimerism of the transplanted heart. N Engl J Med. 2002;346:5–15. doi: 10.1056/NEJMoa012081. [DOI] [PubMed] [Google Scholar]
- 34.Hocht-Zeisberg E., Kahnert H., Guan K., Wulf G., Hemmerlein B., Schlott T., Tenderich G., Korfer R., Raute-Kreinsen U., Hasenfuss G. Cellular repopulation of myocardial infarction in patients with sex-mismatched heart transplantation. Eur Heart J. 2004;25:749–758. doi: 10.1016/j.ehj.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 35.Nurit B., Anat G., Michal S., Lilach F., Maya F. Evaluating the prevalence of DNA mixtures found in fingernail samples from victims and suspects in homicide cases. Forensic Sci Int Genet. 2011;5:532–537. doi: 10.1016/j.fsigen.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt C., Benecke M. Five cases of forensic short tandem repeat DNA typing. Electrophoresis. 1997;18:690–694. doi: 10.1002/elps.1150180506. [DOI] [PubMed] [Google Scholar]
- 37.Pakstis A.J., Speed W.C., Fang R., Hyland F.C., Furtado M.R., Kidd J.R., Kidd K.K. SNPs for a universal individual identification panel. Hum Genet. 2010;127:315–324. doi: 10.1007/s00439-009-0771-1. [DOI] [PubMed] [Google Scholar]
- 38.Dzik W.H., Corwin H., Goodnough L.T., Higgins M., Kaplan H., Murphy M., Ness P., Shulman I.A., Yomtovian R. Patient safety and blood transfusion: new solutions. Transfus Med Rev. 2003;17:169–180. doi: 10.1016/s0887-7963(03)00017-8. [DOI] [PubMed] [Google Scholar]
- 39.Seiden S.C., Barach P. Wrong-side/wrong-site, wrong-procedure, and wrong-patient adverse events: are they preventable? Arch Surg. 2006;141:931–939. doi: 10.1001/archsurg.141.9.931. [DOI] [PubMed] [Google Scholar]
- 40.De Vlaminck I., Valantine H.A., Snyder T.M., Strehl C., Cohen G., Luikart H., Neff N.F., Okamoto J., Bernstein D., Weisshaar D., Quake S.R., Khush K.K. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77. doi: 10.1126/scitranslmed.3007803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative densities of minor allele frequency-filtered variants across the human autosomes. The densities of variants with a minor allele frequency >9% in the CEU, JPTCHB, and YRI populations along each autosome were calculated in 1-kb windows using the IRanges R package (http://www.r-project.org).