Abstract
MicroRNAs are small non-coding RNAs, which are critical regulators of carcinogenesis and tumor progression. Previous studies have identified that microRNA-20b (miR-20b) acts as an oncogene in numerous cancers. However, the role of miR-20b in prostate cancer remains unclear. The present study aimed to investigate the expression of miR-20b in prostate cancer and to examine whether modulating miR-20b expression impacts prostate cancer cellular proliferation and migration. It was revealed that miR-20b was strongly expressed in prostate cancer tissues compared with adjacent normal prostate tissues (P<0.05). Knockdown of miR-20b expression by miR-20b inhibitor inhibited VCaP and PC-3 cell growth and migration. Through bioinformatics analysis, phosphatase and tensin homolog (PTEN) was predicted as a target gene of miR-20b in prostate cancer cells, which was validated by dual-luciferase reporter assay and western blot analysis. In addition, restoration of PTEN expression levels did not affect endogenous miR-20b expression in prostate cancer cells. In conclusion, the present study indicated that miR-20b promotes cellular proliferation and migration by directly regulating PTEN in prostate cancer.
Keywords: micro RNAs, microRNA-20b, prostate cancer, phosphatase and tensin homolog
Introduction
Prostate cancer is the second most common type of malignancy in the male population worldwide, and is the third leading cause of cancer-associated mortality in male patients (1). Surgery and chemotherapy are the main treatment strategies for prostate cancer (2–4). Due to the malignant migration and invasion potential of human prostate cancer cells, the current therapy for this type of cancer remains unsatisfactory (5). In addition, the molecular mechanism behind the progression of prostate cancer remains largely unclear.
MicroRNAs (miRNAs) are a class of small non-coding RNAs, which control protein-coding gene expression by directly binding the 3′-untranslated regions (3′-UTRs) of these genes (6,7). Accumulating evidence has indicated that miRNAs are important regulatory factors involved in various biological processes, including cell differentiation, growth, metastasis and apoptosis (8,9). Aberrant expression of miRNAs has been identified in numerous cancers, including lung and breast cancer and hepatocellular carcinoma (10–15). Numerous miRNAs may be regarded as potential and novel biomarkers for the diagnosis, therapy and prognosis of numerous types of cancer (16,17).
A large number of miRNAs have been identified to be aberrantly expressed in human prostate cancer, including miR-22, miR-23b/27b, miR-103, miR-135b, miR-181, miR-192 and miR-613 (18–24). These miRNAs perform tumor-suppressive or oncogenic roles in the regulation of prostate cancer cell growth, invasion, migration and apoptosis. The mature sequence of miR-20b is located at chromosome Xq26.2, which is a fragile chromosomal region associated with numerous types of human cancer (25). However, the role of miR-20b in prostate cancer remains unknown.
The present study investigated the potential effect of miR-20b on prostate cancer. Firstly, the expression of miR-20b was examined in prostate cancer tissues and adjacent normal prostate tissues. MTT and wound-healing assays were then used to detect the effect of miR-20b on prostate cancer cell growth and migration. Finally, phosphatase and tensin homolog (PTEN) was identified as a potential target gene of miR-20b by bioinformatics analysis, dual-luciferase reporter assay and western blot analysis.
Materials and methods
Clinical specimens
A total of 35 pairs of prostate cancer tissues and adjacent normal prostate tissues were obtained from the Institute of Urology, First Affiliated Hospital of Nanchang University (Nanchang, China). The patients with prostate cancer did not receive chemotherapy or radiotherapy prior to prostatectomy. All samples were immediately snap-frozen in liquid nitrogen until RNA extraction. The histological diagnosis of each sample was confirmed simultaneously by two pathologists using hematoxylin and eosin staining. Written consent forms were obtained from each patient, and the present study was approved by the Ethics Committee of First Affiliated Hospital of Nanchang University (Nanchang, China).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of each sample was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), following the manufacturer's protocol. Total RNA (1 µg) was treated with DNase to remove contaminating DNA. Subsequently, the RNA was reverse transcribed into first-strand complementary DNA (cDNA) using M-MLV reverse transcriptase (Promega Corporation, Madison, WI, USA) for mRNA expression analysis. The miScript reverse transcription kit (Qiagen GmbH, Hilden, Germany) was used to reverse total RNA into cDNA for miRNA expression analysis. RT-qPCR reaction was performed with the Power SYBR-Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol on the Roche LightCycler 480 Real-Time PCR Machine (Roche Diagnostics, Basel, Switzerland). The qPCR conditions consisted of a uracil-N-glycosylase carry-over protection step of 55°C for 2 min, 10 min of DNA polymerase activation at 95°C, followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. The gene or miRNA primers were obtained from Invitrogen (Thermo Fisher Scientific, Inc.). The sequences of primers are shown in Table I. U6 and GAPDH were used as the internal controls. The relative gene and miRNA expression levels were calculated using the 2−ΔΔCq method (26). Each experiment was conducted at least three times.
Table I.
Primers for reverse transcription-quantitative polymerase chain reaction.
| Gene | Forward (5′-3′) | Reverse (5′-3′ |
|---|---|---|
| miR-20b | GAGGACGGAACCGGAAAC | Universal primer |
| PTEN | GTGCAGATAATGACAAG | GATTTGACGGCTCCTCT |
| U6 | CTCGCTTCGGCAGCAC | ACGCTTCACGAATTTGC |
| GAPDH | CAAGGTCATCCATGACAA | GTCCACCACCCTGTTGCTG |
PTEN, phosphatase and tensin homolog; miR-20b, microRNA-20b.
Cell culture and transfection
Two human prostate cancer cell lines (VCaP and PC-3) were purchased from Shanghai Institute of Biological Sciences, Chinese Academy of Science (Shanghai, China). All cells were cultured in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
A total of four pairs of inhibitors (inhibitor 1#-4#) and corresponding inhibitor controls (inhibitor control 1#-4#) of miR-20b were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequences were as follows: Inhibitor 1#, 5′-UGCUCAUAGUGCAGGUAGUU-3′ and inhibitor control 1#, 5′-GAUGAGCAACAUUGAGGACU-3′; inhibitor 2#, 5′-GCAGGUAGUUUUGGCAUGAC-3′ and inhibitor control 2#, 5′-AGCCCGGAUUACCUGUAGCU-3′; inhibitor 3#, 5′-UCUACUGUAGUAUGGGCACU-3′ and inhibitor control 3#, 5′-GACCCUCAUUCCACGCAUC-3′; inhibitor 4#, 5′-UAUGGGCACUUCCAGUACU-3′ and inhibitor control 4#, 5′-CAUACAUUACCCGAAGUCUA-3′. The inhibitors and inhibitor controls were transfected into VCaP and PC-3 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at a concentration of 50 nM, according to the manufacturer's protocol.
Cell proliferation assay
Cell proliferation was examined using an MTT assay. Briefly, ~8,000 VCaP and PC-3 cells/well were seeded onto 96-well plates. After 24 h, cells were transfected with miR-20b inhibitor and inhibitor control for 48 h and then treated with 20 ml/well MTT (Sigma-Aldrich; Merck KGaA; 5 mg/ml). Following incubation for 4 h at 37°C, the free supernatant of cells was discarded and the formazan products were dissolved by 150 µl dimethyl sulfoxide. Finally, the optical density (OD) at 450 nm was detected by a microplate reader (Synergy™ HT Multi-Mode Microplate Reader; BioTek Instruments, Inc., Winooski, VT, USA).
Cell migration assay
A wound-healing assay was used to assess prostate cancer cell migration ability of miR-20b. VCaP and PC-3 cells were plated onto 12-well plates and transfected with miR-20b inhibitor and inhibitor control. Following transfection for 24 h at 37°C in a humidified atmosphere with 5% CO2, wounds were made with a yellow pipette tip among cells in each well. Images were captured in five random visual fields (magnification, ×100) at 0 and 12 h by a Leica DMI 6000B microscope (Leica Microsystems, Inc., Buffalo Grove, IL, USA) subsequent to wounding in order to determine the width of wound healing.
Western blot analysis
The protein of cells was extracted using lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. 45 µg of protein samples in each group was subjected to 10% SDS-PAGE, and then transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). Subsequent to blocking with 5% non-fat milk for 2 h at 37°C, the membrane was incubated with the rabbit monoclonal anti-PTEN antibodies (dilution, 1:500; cat. no. ab32199; Abcam, Cambridge, UK) overnight at 4°C, followed by anti-rabbit horseradish peroxidase-linked secondary antibodies (dilution, 1:2,000; cat. no. ab6721; Abcam) for 1 h at 37°C. The bands were obtained using AnalySIS 3.0 image analysis system (Soft Imaging System GmbH, Münster, Germany), and the protein density was quantified with Odyssey v1.2 software (LI-COR Biosciences, Lincoln, NE, USA).
Bioinformatics analysis
The potential target of miR-20b was created by combining three public algorithms, which were TargetScan (http://www.targetscan.org/), miRanda (http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/). The putative genes that were predicted by the three algorithms were accepted and the candidates were chosen based on the gene function.
Dual-luciferase reporter assay
The mRNA 3′-UTR of PTEN containing the predicted binding region or mutated binding region was sub-cloned into a psiCHECK-2 luciferase reporter vector (Promega Corporation). The psiCHECK-2 vector containing wild-type (WT) or mutant (MUT) mRNA 3′-UTRs of PTEN with the miR-20b inhibitors or inhibitor controls were co-transfected into VCaP and PC-3 cells using Lipofectamine 2000 at 37°C in a humidified atmosphere with 5% CO2. Luciferase activities were detected by the Dual-Luciferase Reporter Assay System (Promega Corporation) 48 h post-transfection, according to the manufacturer's protocol.
Statistical analysis
Each experiment was performed in triplicate for biological repeat. Statistical analysis was performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation. Statistical differences were analyzed using Student's t-test or one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-20b is significantly upregulated in prostate cancer tissues
The expression level of miR-20b in 35 pairs of prostate cancer tissues and adjacent normal prostate tissues was analyzed by RT-qPCR. The results revealed that the miR-20b expression was significantly upregulated in prostate cancer tissues, compared with adjacent normal prostate tissues (P<0.05; Fig. 1). The data indicated that the upregulation of miR-20b may be involved in the development of human prostate cancer.
Figure 1.
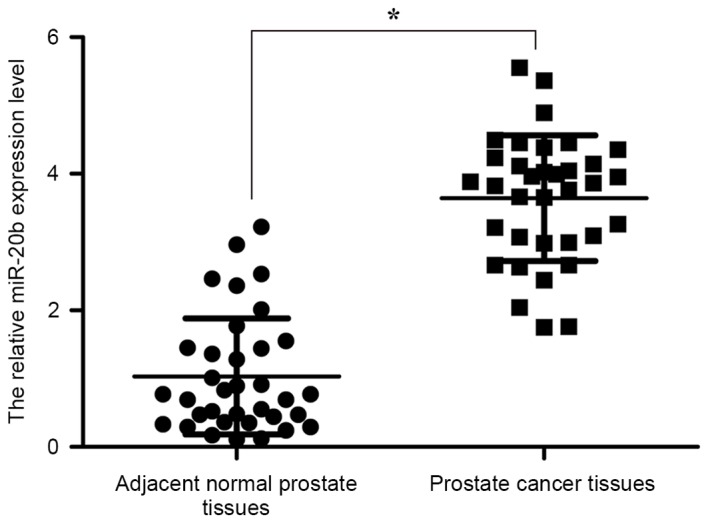
miR-20b is significantly upregulated in prostate cancer tissues. Expression levels of miR-20b in 35 pairs of prostate cancer tissues and adjacent normal prostate tissues was measured by reverse transcription-quantitative polymerase chain reaction. U6 was used as an internal control. *P<0.05. miR-20b, microRNA-20b.
Knockdown of miR-20b inhibits prostate cancer cell proliferation
A total of four pairs of inhibitors and inhibitor controls of the miR-20b were transfected into VCaP and PC-3 cells. The transfection efficiency was analyzed by RT-qPCR at 24 h post-transfection. As shown in Fig. 2, the transfection efficiency of cells was the highest in the inhibitor 2# group compared with other groups (P<0.05). Therefore, the miR-20b inhibitor 2# group was selected for subsequent experiments.
Figure 2.
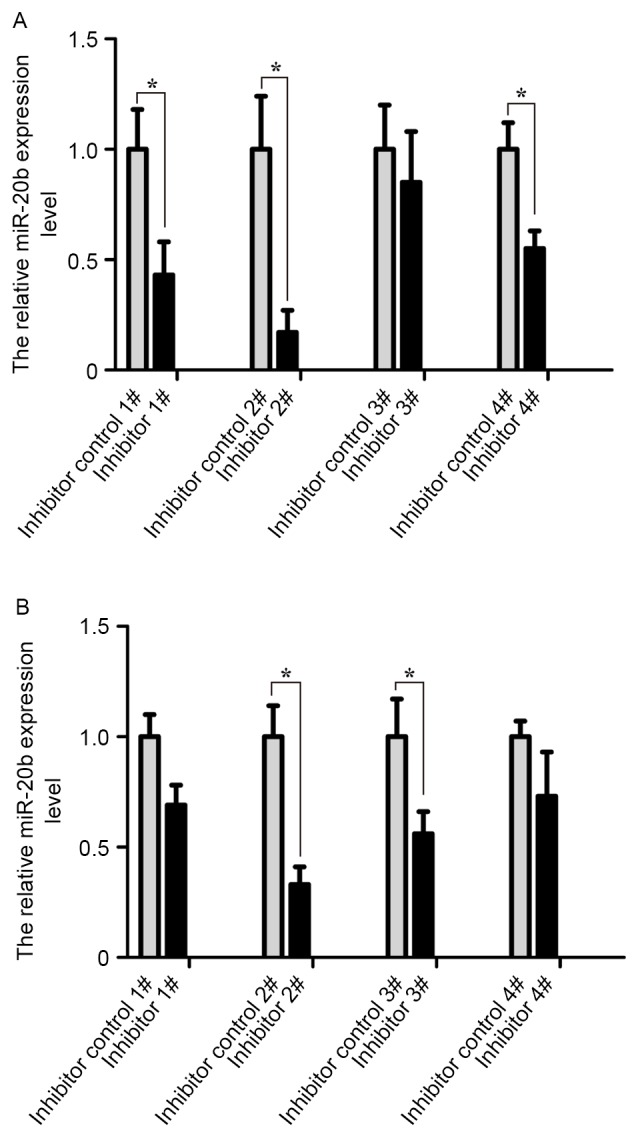
Analysis of transfection efficiency of four pairs of inhibitors and inhibitor controls of miR-20b. The fold-changes of miR-20b expression in (A) VCaP and (B) PC-3 cells treated with miR-20b inhibitors and inhibitor controls were analyzed by reverse transcription-quantitative polymerase chain reaction following transfection for 24 h. *P<0.05. miR-20b, microRNA-20b.
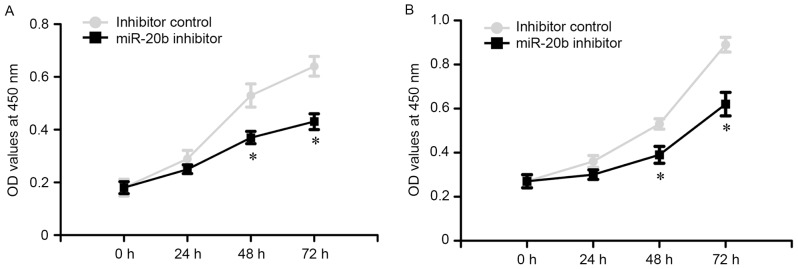
To evaluate the potential role of miR-20b on the proliferation of prostate cancer cells, a MTT assay was performed. The OD values of VCaP and PC-3 cells were measured at 0, 24, 48 and 72 h following transfection. The results revealed that miR-20b inhibitor reduced the growth of VCaP and PC-3 cells, as compared with the inhibitor control (Fig. 3; P<0.05).
Figure 3.
Knockdown of miR-20b inhibits prostate cancer cell proliferation. (A) Effect of miR-20b on prostate cancer cell proliferation was measured using an MTT assay following miR-20b inhibitor or inhibitor control infection in VCaP cells at 0, 24, 48 and 72 h. (B) Effect of miR-20b on prostate cancer cell proliferation was measured by MTT assay following miR-20b inhibitor or inhibitor control infection in PC-3 cells at 0, 24, 48 and 72 h. *P<0.05. miR-20b, microRNA-20b; OD, optical density.
Knockdown of miR-20b suppresses prostate cancer cell migration
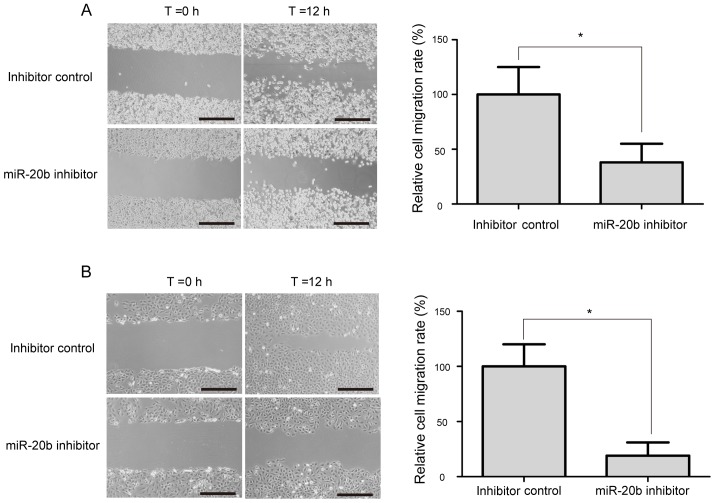
The wound-healing assay was performed to evaluate the potential role of miR-20b on the migration of prostate cancer cell. As shown in Fig. 4, a significant decrease in cell migration ability was observed in miR-20b inhibitor-treated VCaP and PC-3 cells, compared with inhibitor control-transfected cells (P<0.05).
Figure 4.
Knockdown of miR-20b inhibits prostate cancer cell migration. (A) Images of the VCaP cells transfected with miR-20b inhibitor or inhibitor control at 0 and 12 h after the scratches were made at the same point. (B) Images of the PC-3 cells transfected with miR-20b inhibitor or inhibitor control at 0 and 12 h after the scratches were made at the same point. Scale bar=200 µm; *P<0.05. miR-20b, microRNA-20b.
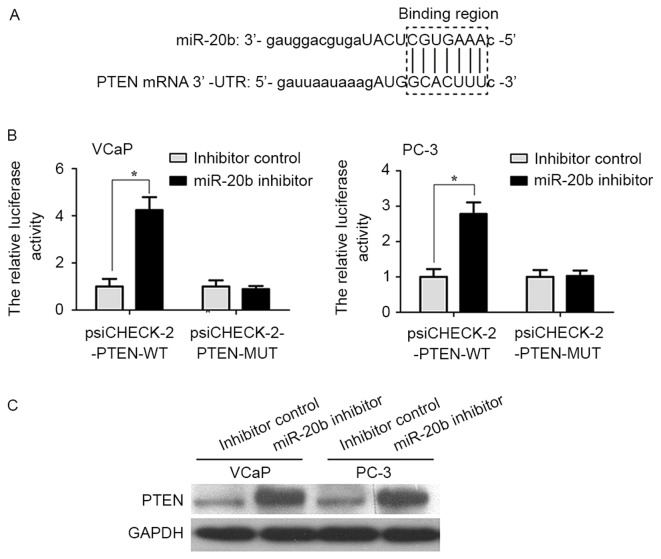
PTEN is a direct target gene of miR-20b in prostate cancer cells
Bioinformatics analysis predicted that PTEN was a target gene of miR-20b (Fig. 5A). In order to provide additional direct evidence for the association between PTEN and miR-20b, the binding region of miR-20b in the mRNA 3′UTR of PTEN was characterized by the dual-luciferase reporter assay. As shown in Fig. 5B, the relative luciferase activity of the psiCHECK-2-PTEN-WT vector in VCaP and PC-3 cells was increased compared with in the psiCHECK-2-PTEN-MUT vector (P<0.05). In addition, it was examined whether miR-20b inhibits endogenous PTEN expression in VCaP and PC-3 cells. Compared with inhibitor control, endogenous PTEN protein expression levels were significantly increased in VCaP and PC-3 cells transfected with miR-20b inhibitor compared with that in inhibitor control-transfected cells (Fig. 5C; P<0.05).
Figure 5.
PTEN is a direct target gene of miR-20b in prostate cancer cells. (A) Presentation of the miR-20b binding region within the mRNA 3′-untranslated region of PTEN. (B) VCaP and PC-3 cells were co-transfected with psiCHECK-2-PTEN-WT or psiCHECK-2-PTEN-MUT and miR-20b inhibitor or inhibitor control for 48 h, and the luciferase activities were assessed to determine the direct interaction between miR-20b and PTEN. *P<0.05. (C) Western blot analysis detected PTEN expression in VCaP and PC-3 cells transfected with miR-20b inhibitor or inhibitor control. GAPDH was used as an internal control. PTEN, phosphatase and tensin homolog; WT, wild-type; MUT, mutant; miR-20b, microRNA-20b.
Accumulating evidence has indicated that certain target genes may also reverse regulate expression of miRNA. In order to investigate whether PTEN may regulate miR-20b expression in prostate cancer cells, PTEN overexpression plasmid (PTEN-pCDNA3.1/HA) was synthesized (Invitrogen; Thermo Fisher Scientific, Inc.). The PTEN-pCDNA3.1/HA and pCDNA3.1/HA empty vectors were transfected into VCaP and PC-3 cells using Lipofectamine 2000, according to the manufacturer's protocol. The results demonstrated that restoration of PTEN expression in VCaP and PC-3 cells did not alter the expression level of miR-20b (Fig. 6).
Figure 6.
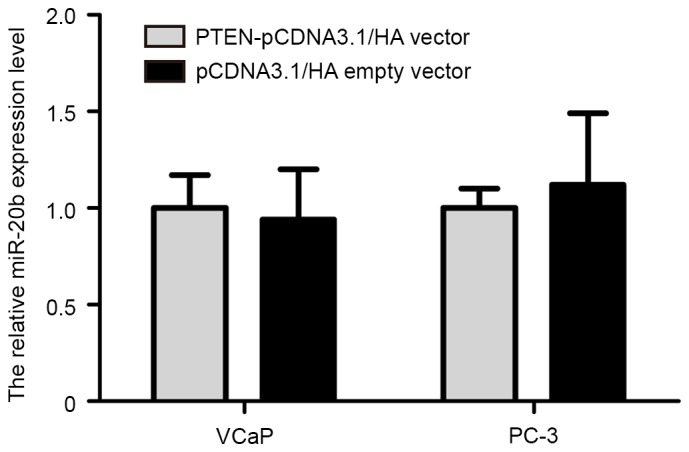
The effect of PTEN on miR-20b expression in prostate cancer cells. Reverse transcription-quantitative polymerase chain reaction demonstrated that restoration of PTEN expression in VCaP and PC-3 cells did not alter the expression level of miR-20b. PTEN, phosphatase and tensin homolog; miR-20b, microRNA-20b; pcDNA, pseudo-complementary DNA.
Discussion
Previous studies have reported that miR-20b is upregulated in gastric and cervical cancer (27,28). Studies have indicated that miR-20b functions as an oncogene in numerous types of cancer. miR-20b promotes breast cancer cell growth, partly by targeting the PTEN gene (29). miR-20b suppresses PTEN expression resulting in B7-H1 overexpression in advanced colorectal cancer (30). miR-20b presents high expression level and has oncogenic potential in human T-cell leukemia (31). However, the expression and function of miR-20b has not been documented in prostate cancer.
In the present study, miR-20b appeared to be markedly upregulated in prostate cancer tissues compared with adjacent normal prostate tissues, which is consistent with previous studies (27–29,31). The proliferation and migration abilities of human prostate cancer cells transfected with miR-20b inhibitor were significantly reduced compared with the inhibitor control, which indicated that miR-20b acts as an oncogene in prostate cancer cells. These results were also similar to observations in colorectal and breast cancer (32,33), in which miR-20b expression was upregulated, and knockdown of miR-20b inhibited cellular proliferation and migration.
The PTEN gene is a classic tumor suppressor gene in various human cancers, which is located at chromosome 10q23.31 (34). PTEN functions as a negative regulator of the phosphoinositide 3-kinase/Akt pathway through dephosphorylation of phosphatidylinositol 3,4,5 trisphosphate, and is involved in regulation of cellular proliferation, metastasis and apoptosis during progression of cancers (34). PTEN has been reported to be regulated by numerous miRNAs in multiple cancers, including colorectal carcinoma, glioma, ovarian and breast cancer (29,35–37). In the present study, it was also determined that miR-20b inhibited PTEN expression by directly binding to the mRNA 3′-UTR of PTEN in VCaP and PC-3 cells. Additionally, restoration of PTEN expression did not affect endogenous miR-20b expression in VCaP and PC-3 cells. Above all, the present study indicated that miR-20b serves as an important oncogene in promoting prostate cancer cell growth and migration by directly regulating PTEN.
In conclusion, the present study identified that miR-20b expression was significantly upregulated in prostate cancer tissues. Notably, knockdown of miR-20b expression exhibits an anti-tumor effect in vitro. miR-20b acts as an oncogene that performs a critical role in the growth and migration of prostate cancer cell by targeting PTEN. Therefore, miR-20b may serve as a novel biomarker and potential therapeutic target in prostate cancer.
Acknowledgements
The present study was supported by the Natural Science Foundation of Jiangxi province (grant no. 20151BAB205017).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Silvestri I, Cattarino S, Aglianò AM, Collalti G, Sciarra A. Beyond the immune suppression: The immunotherapy in prostate cancer. Biomed Res Int. 2015;2015:794968. doi: 10.1155/2015/794968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose JN, Crook JM. The role of radiation therapy in the treatment of metastatic castrate-resistant prostate cancer. Ther Adv Urol. 2015;7:135–145. doi: 10.1177/1756287215576647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nazim SM, Abbas F. Role of surgery in locally advanced prostate cancer. Pak J Med Sci. 2015;31:710–716. doi: 10.12669/pjms.313.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Marzo AM, DeWeese TL, Platz EA, Meeker AK, Nakayama M, Epstein JI, Isaacs WB, Nelson WG. Pathological and molecular mechanisms of prostate carcinogenesis: Implications for diagnosis, detection, prevention, and treatment. J Cell Biochem. 2004;91:459–477. doi: 10.1002/jcb.10747. [DOI] [PubMed] [Google Scholar]
- 6.Mo YY. MicroRNA regulatory networks and human disease. Cell Mol Life Sci. 2012;69:3529–3531. doi: 10.1007/s00018-012-1123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 8.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 10.Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He Y, Chen G, Zhou Q, Wang W, Zhou X. Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J Exp Clin Cancer Res. 2016;35:7. doi: 10.1186/s13046-016-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim G, An HJ, Lee MJ, Song JY, Jeong JY, Lee JH, Jeong HC. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer. 2016;91:15–22. doi: 10.1016/j.lungcan.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T, Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, et al. Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene. 2016;35:801. doi: 10.1038/onc.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinnerthaler G, Hackl H, Gampenrieder SP, Hamacher F, Hufnagl C, Hauser-Kronberger C, Zehentmayr F, Fastner G, Sedlmayer F, Mlineritsch B, Greil R. miR-16-5p is a stably-expressed housekeeping microRNA in breast cancer tissues from primary tumors and from metastatic sites. Int J Mol Sci. 2016;17:pii: E156. doi: 10.3390/ijms17020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, Han S, Yuan Q, Yang M. MiRNA-192 [corrected] and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, Sun X, Wang L, Tang S, Yan C. Downregulation of MicroRNA-145 caused by Hepatitis B virus X protein promotes expression of CUL5 and contributes to pathogenesis of Hepatitis B virus-associated hepatocellular carcinoma. Cell Physiol Biochem. 2015;37:1547–1559. doi: 10.1159/000438522. [DOI] [PubMed] [Google Scholar]
- 16.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruan K, Fang X, Ouyang G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 18.Pasqualini L, Bu H, Puhr M, Narisu N, Rainer J, Schlick B, Schäfer G, Angelova M, Trajanoski Z, Börno ST, et al. miR-22 and miR-29a are members of the androgen receptor cistrome modulating LAMC1 and Mcl-1 in prostate cancer. Mol Endocrinol. 2015;29:1037–1054. doi: 10.1210/me.2014-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun T, Yang M, Chen S, Balk S, Pomerantz M, Hsieh CL, Brown M, Lee GM, Kantoff PW. The altered expression of MiR-221/−222 and MiR-23b/−27b is associated with the development of human castration resistant prostate cancer. Prostate. 2012;72:1093–1103. doi: 10.1002/pros.22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu X, Zhang W, Su Y, Lu L, Wang D, Wang H. MicroRNA-103 suppresses tumor cell proliferation by targeting PDCD10 in prostate cancer. Prostate. 2016;76:543–551. doi: 10.1002/pros.23143. [DOI] [PubMed] [Google Scholar]
- 21.Aakula A, Leivonen SK, Hintsanen P, Aittokallio T, Ceder Y, Børresen-Dale AL, Perälä M, Östling P, Kallioniemi O. MicroRNA-135b regulates ERα, AR and HIF1AN and affects breast and prostate cancer cell growth. Mol Oncol. 2015;9:1287–1300. doi: 10.1016/j.molonc.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong SJ, Liu J, Wang X, Qu LX. microRNA-181 promotes prostate cancer cell proliferation by regulating DAX-1 expression. Exp Ther Med. 2014;8:1296–1300. doi: 10.3892/etm.2014.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Fan Z, Lu S, Yang J, Hao T, Huo Q. MiR-192 suppresses the tumorigenicity of prostate cancer cells by targeting and inhibiting nin one binding protein. Int J Mol Med. 2016;37:485–492. doi: 10.3892/ijmm.2016.2449. [DOI] [PubMed] [Google Scholar]
- 24.Ren W, Li C, Duan W, Du S, Yang F, Zhou J, Xing J. MicroRNA-613 represses prostate cancer cell proliferation and invasion through targeting Frizzled7. Biochem Biophys Res Commun. 2016;469:633–638. doi: 10.1016/j.bbrc.2015.12.054. [DOI] [PubMed] [Google Scholar]
- 25.Saleiban A, Faxälv L, Claesson K, Jönsson JI, Osman A. miR-20b regulates expression of proteinase-activated receptor-1 (PAR-1) thrombin receptor in melanoma cells. Pigment Cell Melanoma Res. 2014;27:431–441. doi: 10.1111/pcmr.12217. [DOI] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Xue TM, Tao LD, Zhang M, Xu GC, Zhang J, Zhang PJ. miR-20b overexpression is predictive of poor prognosis in gastric cancer. Onco Targets Ther. 2015;8:1871–1876. doi: 10.2147/OTT.S85236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li MY, Hu XX. Meta-analysis of microRNA expression profiling studies in human cervical cancer. Med Oncol. 2015;32:510. doi: 10.1007/s12032-015-0510-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhou W, Shi G, Zhang Q, Wu Q, Li B, Zhang Z. MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN) Cell Biosci. 2014;4:62. doi: 10.1186/2045-3701-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, Chen L, Zou L, Yang P, Wu R, Mao Y, Zhou H, Li R, Wang K, Wang W, et al. MiR-20b, −21 and −130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum Immunol. 2014;75:348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi T, Iijima T, Wakaume R, Takahashi K, Matsumoto H, Nakano D, Nakayama Y, Mori T, Horiguchi S, Miyaki M. Underexpression of miR-126 and miR-20b in hereditary and nonhereditary colorectal tumors. Oncology. 2014;87:58–66. doi: 10.1159/000363303. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad A, Ginnebaugh KR, Sethi S, Chen W, Ali R, Mittal S, Sarkar FH. miR-20b is up-regulated in brain metastases from primary breast cancers. Oncotarget. 2015;6:12188–12195. doi: 10.18632/oncotarget.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 35.Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, Tan W, Wei G, Zhou Y. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013;12:30. doi: 10.1186/1476-4598-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: MiR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]