ABSTRACT
Heterotrophic bacteria have recently been reported to oxidize sulfide to sulfite and thiosulfate by using sulfide:quinone oxidoreductase (SQR) and persulfide dioxygenase (PDO). In chemolithotrophic bacteria, both SQR and PDO have been reported to function in the periplasmic space, with SQR as a peripheral membrane protein whose C terminus inserts into the cytoplasmic membrane and PDO as a soluble protein. Cupriavidus pinatubonensis JMP134, best known for its ability to degrade 2,4-dichlorophenoxyacetic acid and other aromatic pollutants, has a gene cluster of sqr and pdo encoding C. pinatubonensis SQR (CpSQR) and CpPDO2. When cloned in Escherichia coli, the enzymes are functional. Here we investigated whether they function in the periplasmic space or in the cytoplasm in heterotrophic bacteria. By using sequence analysis, biochemical detection, and green fluorescent protein (GFP)/PhoA fusion proteins, we found that CpSQR was located on the cytoplasmic side of the membrane and CpPDO2 was a soluble protein in the cytoplasm with a tendency to be peripherally located near the membrane. The location proximity of these proteins near the membrane in the cytoplasm may facilitate sulfide oxidation in heterotrophic bacteria. The information may guide the use of heterotrophic bacteria in bioremediation of organic pollutants as well as H2S.
IMPORTANCE Sulfide (H2S, HS−, and S2−), which is common in natural gas and wastewater, causes a serious malodor at low levels and is deadly at high levels. Microbial oxidation of sulfide is a valid bioremediation method, in which chemolithotrophic bacteria that use sulfide as the energy source are often used to remove sulfide. Heterotrophic bacteria with SQR and PDO have recently been reported to oxidize sulfide to sulfite and thiosulfate. Cupriavidus pinatubonensis JMP134 has been extensively characterized for its ability to degrade organic pollutants, and it also contains SQR and PDO. This paper shows the localization of SQR and PDO inside the cytoplasm in the vicinity of the membrane. The information may provide guidance for using heterotrophic bacteria in sulfide bioremediation.
KEYWORDS: sulfide:quinone oxidoreductase, persulfide dioxygenase, Cupriavidus pinatubonensis JMP134, subcellular localization, rhodanese, heterotrophic bacteria, sulfide oxidation
INTRODUCTION
Hydrogen sulfide (H2S) is a toxic gas at high levels but may have physiological functions at low levels. Under aerobic conditions, hydrogen sulfide is produced mainly from cysteine and iron-sulfur proteins in various organisms (1, 2). It functions as a “gastrotransmitter” (3), participating in a variety of important physiological processes in animals (4–6), and acts as a defense against antibiotics in bacteria (7). Since H2S is toxic at high levels (8), its concentration is usually maintained in a range, with excess H2S being metabolized in cells. A new sulfide oxidation pathway has been discovered from animal mitochondria (1, 9), and the pathway is also present in heterotrophic bacteria (10–12). In bacteria, sulfide:quinone oxidoreductase (SQR) oxidizes sulfide to polysulfide, rhodanese speeds up the reaction of polysulfide with glutathione (GSH) to produce glutathione persulfide (GSSH), and persulfide dioxygenase (PDO) oxidizes GSSH to sulfite, which spontaneously reacts with polysulfide to produce thiosulfate (12).
The discovery of SQR can be traced back to the 1970s, when Knaff showed that ubiquinone is a cosubstrate in the reaction (13). SQR has been identified and studied in a variety of anaerobic phototrophic bacteria that use sulfide as the reducing power for photosynthesis and chemolithotrophic bacteria that oxidize sulfide to conserve energy for growth (14, 15). Heterotrophic bacteria carrying SQR and PDO have been reported to oxidize sulfide (H2S, HS−, and S2−) to sulfite and thiosulfate (12), showing potential to be used in H2S bioremediation, which has been done with chemolithotrophic bacteria (16, 17).
In spite of lacking a signal peptide, SQR is thought to be a membrane-associated protein on the periplasmic side (18, 19). To date, studies with SQR have been focused mainly on its biochemical functions (1, 19–23) and crystal structures (24–26). Marcia et al. (26) have determined the X-ray structure of the Aquifex aeolicus SQR, showing that it is an integral monotopic membrane protein with an amphipathic motif of a helix-turn-helix inserting about 12 Å into the lipid bilayer. Further, SQR displays a diffusion behavior on the lipid bilayer, different from the case for transmembrane proteins (27). However, these studies do not reveal whether SQR is inserted into the membrane on the cytoplasmic side or the periplasmic side. The only evidence that suggests the periplasmic localization of SQR is the whole-cell assay of a Rhodobacter capsulatus strain carrying a fusion protein of alkaline phosphatase (PhoA) and its SQR, showing a 10-fold increase in PhoA activity after sulfide induction (19). Since PhoA is active only when translocated into the periplasmic space (28, 29), the increased PhoA activity suggests the PhoA-SQR fusion is in the periplasmic space of R. capsulatus. Further, green fluorescent protein (GFP) is also frequently used to check the location of a protein within a bacterial cell, as it is normally fluorescent only in the cytoplasm (28, 29). Thus, the PhoA fusion and GFP fusion are often used to check the location of a protein in bacteria.
According to structural and sequence analyses, SQRs are divided into six types (18). The type I SQRs play a role in sulfide-dependent energy conservation for chemolithotrophic or phototrophic growth (20, 30). The periplasmic R. capsulatus SQR is a type I SQR (19). Type II SQRs were recently identified from animal mitochondria and bacteria, and they may play a detoxification role (4, 12). Cupriavidus pinatubonensis (ex. C. necator) JMP134 has been extensively studied for its ability to degrade a variety of organic pollutants, such as 2,4-dichlorophenoxyacetic acid and 2,4,6-trichlorophenol (31, 32), and it contains an SQR (CpSQR) that is a fusion of a rhodanese domain and a type II SQR domain (12). The sqr gene is next to a pdo gene within the chromosome, coding for CpPDO2. When both genes are expressed in Escherichia coli, the recombinant E. coli oxidizes sulfide to sulfite and thiosulfate (12). Further, C. pinatubonensis JMP134 contains another PDO, CpPDO1 (10). The localization of PDOs in chemolithotrophic Acidithiobacillus spp. is controversial. PDOs were initially identified with crude enzyme extracts and were proposed to be in the periplasmic space (33). However, the genes coding for PDOs from two Acidithiobacillus spp. have recently been identified, and the proteins are likely in the cytoplasm due to the lack of signaling peptides (12). Here we show evidence that CpSQR is a membrane protein active on the cytoplasmic side and that CpPDO2 is a soluble protein in the cytoplasm with a tendency to be peripherally located near the cytoplasmic membrane.
RESULTS AND DISCUSSION
The membrane association of CpSQR.
CpSQR does not have an apparent signal peptide. To test whether CpSQR was a membrane-associated protein, Cpsqr was tagged with the GFP gene in pBBR2-Cpsqr-gfp, which was transformed into E. coli BL21(DE3) and C. pinatubonensis JMP134 cells. The green fluorescence appeared bright at the periphery of E. coli and C. pinatubonensis cells containing pBBR2-Cpsqr-gfp (Fig. 1) or pBBR2-MhpT-GFP, which encodes a fusion of the membrane protein MhpT tagged with GFP (34). The control of E. coli BL21(DE3) with a cytoplasmic protein GAPDH (glyceraldehyde-3-phosphate dehydrogenase)-GFP fusion showed fluorescence evenly distributed in the cytoplasm (Fig. 1).
FIG 1.
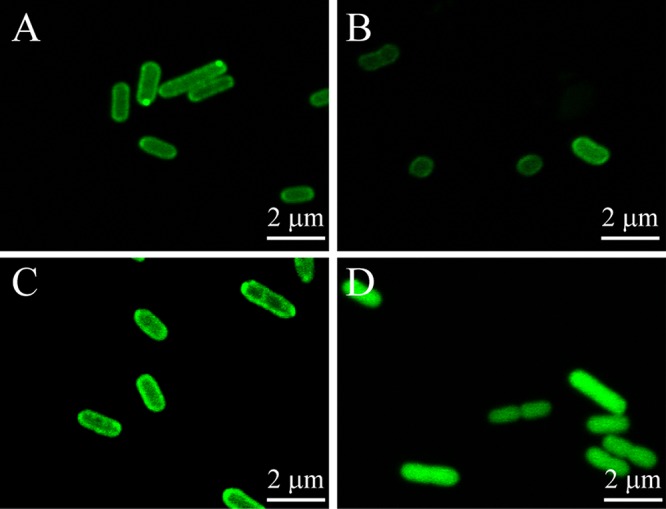
GFP analysis of CpSQR localization in E. coli BL21(DE3) and C. pinatubonensis JMP134. (A) E. coli(pBBR2-Cpsqr-gfp); (B) C. pinatubonensis(pBBR2-Cpsqr-gfp); (C) E. coli(pBBR2-mhpT-gfp) (positive control); (D) E. coli(pBBR2-gapA-gfp) (negative control). Confocal fluorescence microscopic images are shown.
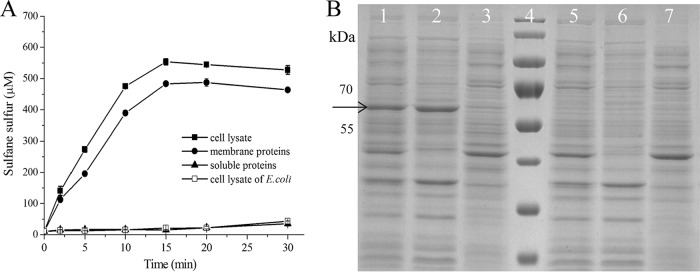
The membrane association of CpSQR was further tested with E. coli BL21(DE3)(pET30-Cpsqr) cells and cell fractions. The cells were cultured, induced, harvested, and resuspended in 50 mM Tris buffer, pH 8. The resuspended cells at an optical density at 600 nm (OD600) of 5 completely oxidized 1,000 μM sulfide within 5 min of incubation. When the cell suspension was disrupted and fractionated via ultracentrifugation with the membrane pellet resuspended in the same volume of the buffer, the whole-cell lysate and the membrane fraction of E. coli BL21(DE3)(pET30-Cpsqr) cells also completely oxidized 1 mM sulfide within 15 min, with the production of about 500 μM sulfane sulfur, while the soluble proteins in the supernatant after ultracentrifugation did not show apparent sulfide oxidation and generated a small amount of polysulfide (Fig. 2A). Using the first data points at 2 min (Fig. 2A), the calculated rates of polysulfide production were 423, 1,061, and 82 nmol min−1 mg−1 of protein with the cell lysate, the membrane fraction, and the soluble proteins, respectively. The overproduced CpSQR was clearly detectable as a dominant band in an SDS-polyacrylamide gel (Fig. 2B). The E. coli BL21(DE3) control without CpSQR did not have SQR activity (Fig. 2A) and did not show a CpSQR band (Fig. 2B). The results were consistent with the fluorescent imaging (Fig. 1), indicating that CpSQR is membrane associated.
FIG 2.
Detection of CpSQR in cellular fractions. E. coli(pET-Cpsqr) was used to isolate the cellular components. The whole-cell lysate (832 μg of protein ml−1), the membrane component (266 μg ml−1), and the soluble proteins (470 μg ml−1) in Tris buffer (pH 8) were obtained. The whole-cell lysate (665 μg of protein ml−1) of E. coli BL21 was used as the negative control. (A) CpSQR activities were shown as sulfane sulfur production. All the fractions were diluted 1:5, and Na2S was added to 1,000 μM to initiate the reaction. (B) SDS-PAGE analysis with cellular components. Lanes 1 to 3, E. coli (pET30-Cpsqr). Lane 1, whole-cell lysate; lane 2, membrane component; lane 3, soluble proteins. Lane 4, marker. Lanes 5 to 7, E. coli control. Lane 5, whole-cell lysate; lane 6, membrane component; lane 7, soluble proteins. Ten micrograms of proteins of each sample was loaded. The arrow points to the position of CpSQR.
Our results are consistent with previous reports that SQRs are membrane associated (19, 24, 26, 35); however, the orientation of SQRs on the cell membrane has been investigated only in R. capsulatus (19). Because R. capsulatus uses sulfide as the reducing power for photosynthesis and deposits zero-valence elemental sulfur outside the cells, it is likely that SQR is attached to the periplasmic side of the cytoplasmic membrane. Since the report of R. capsulatus SQR, SQRs have been considered to be peripheral membrane proteins on the periplasmic side (19, 26, 27, 30). For C. pinatubonensis JMP134 and heterotrophic bacteria, sulfide is oxidized to sulfite and thiosulfate by the concerted actions of SQR and PDO (12). The process is likely for detoxification instead of energy conservation for growth, because PDO oxidizes sulfane sulfur to sulfite without ATP production (1, 10), while autotrophic bacteria are likely to use the Sox system or the reverse sulfite dissimilatory reductase to oxidize sulfane sulfur, which is linked to ATP production (36, 37). Thus, for heterotrophic bacteria that may use SQR for intracellular sulfide detoxification, a cytoplasmic localization would be ideal. This possibility was further investigated with CpSQR.
Sequence analysis-guided construction of CpSQR fragment-PhoA/GFP fusions.
Potential transmembrane regions of CpSQR were analyzed by using the function “Prediction of Transmembrane Segments” at the Biology Workbench website of the San Diego Supercomputer Center. Three segments were predicted to possibly go through the cytoplasmic membrane: amino acids (aa) 147 to 175 (29 aa), 213 to 229 (17 aa), and 343 to 358 (16 aa). Other programs also predicted that amino acid residues 150 to 170 might be a transmembrane region (Table 1). Since the SQR domain starts from the amino acid residue 158, we constructed five fragmentx-PhoA/GFP fusion proteins (where x = 157, 176, 230, 359, or 450), in which PhoA without the signal peptide or GFP was linked to the C-terminally truncated CpSQR at the x sites.
TABLE 1.
Predicted transmembrane regions
| Software | Transmembrane region (aa) | Length (aa) | Scorea |
|---|---|---|---|
| Biology Workbench | 147–175 | 29 | 1.5 |
| 213–229 | 17 | 1.3 | |
| 343–358 | 16 | 1.3 | |
| PRED-TMR | 151–170 | 20 | |
| DAS transmembrane prediction server | 151–168 | 18 | 1.7 |
| 153–163 | 11 | 2.2 | |
| HMMTOP | 148–170 | 23 | |
| TMpred | 151–170 | 20 | ++ |
| 334–359 | 26 | + | |
| SMART | 151–165 | 15 | |
| 448–468 | 21 | ||
| SPLIT SERVER | 147–171 | 25 | |
| TMHMM | ∼150–169 | 18 |
All the scores listed are not significant.
The sequence analysis of the N-terminal DUF442 domain predicted that there was an atypical α helix before amino acid residues 30, 62, 84, 107, and 125, and we constructed fragmentx-PhoA/GFP fusions (where x = 26, 49, 71, 83, 107, 115, or 128) in which the first 128 amino acid residues constitute the full length of the DUF442 domain (12).
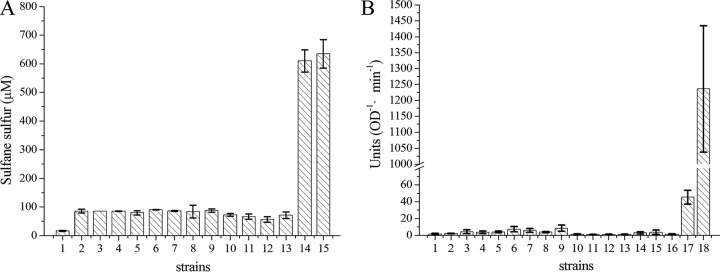
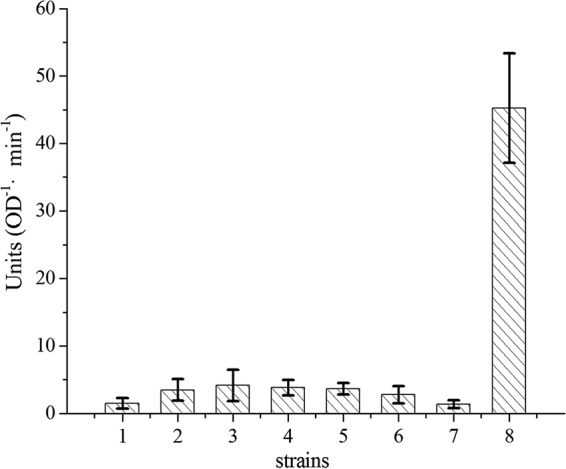
Cells with the fusion of CpSQR and PhoA had SQR activity; other fusions with SQR fragments had no SQR activity (Fig. 3A). Full-length PhoA and TolB-PhoA were used as the positive controls. TolB, the periplasmic component of the Tol-Pal transenvelope protein complex in Gram-negative bacteria, was chosen because it has been reported to localize in the periplasmic space without a typical signal peptide (38) and it has a molecular weight similar to that of CpSQR. Whole cells with TolB-PhoA at an OD600 of 1 had a PhoA activity of about 45 units, while cells with the full-length PhoA showed an activity of 1,230 units. As the expression of a fused protein may be different from that of a single protein, we used TolB-PhoA to compare with CpSQR-PhoA fusion proteins. Whole-cell analysis did not show any significant PhoA activity with any of the fusions (Fig. 3B), but all fragmentx-GFP fusions had fluorescence (data not shown).
FIG 3.
Whole-cell analysis of SQR and PhoA activities with fusion proteins. (A) Detection of SQR activity in E. coli with fragmentx-PhoA fusion proteins. Cells were resuspended in 50 mM Tris buffer (pH 8) at an OD600 of 5, and Na2S was added to 1,000 μM to initiate the reaction at 30°C. Sulfane sulfur was detected after incubation for 5 min, and SQR activity was represented by the amount of sulfane sulfur generated (μM). (B) Detection of PhoA activity in E. coli with fragmentx-PhoA fusion proteins. Cells were resuspended in 50 mM Tris buffer (pH 8) at an OD600 of 1 for PhoA activity detection. Bars 1, E. coli; bars 2 to 13, E. coli(pBBR2-fragmentx-phoA) (x = 26, 49, 71, 83, 106, 115, 128, 157, 176, 230, 359, 450, respectively); bars 14, E. coli(pBBR2-Cpsqr-phoA); bars 15, E. coli(pBBR2-Cpsqr); bar 16, E. coli(pBBR2-phoA with no signal peptide); bar 17, E. coli(pBBR2-tolB-phoA); bar 18, E. coli(pBBR2-signal-phoA). Error bars indicate standard deviations.
Our data for CpSQR-PhoA and CpSQR-GFP fusions showed that CpSQR is associated with the membrane on the cytoplasmic side. For inner membrane proteins, fused GFP is fluorescent when attached to their cytoplasmic domains, and PhoA is active when attached to the periplasmic domains (39–41). Thus, the reporters tagged to the C terminus of a protein in the cytoplasm will have low PhoA activity and high GFP fluorescence. In contrast, a tagged protein with its C terminus in the periplasmic space will have high PhoA activity and no GFP fluorescence (28). Our results (Fig. 1 and 3) for CpSQR-GFP and CpSQR-PhoA indicate that the C terminus of CpSQR is located in the cytoplasm. Since none of the CpSQR fragment-PhoA fusions (fragmentx-PhoA) showed apparent PhoA activity in E. coli, no transmembrane segment of the protein was identified. Further, the whole cells were fluorescent for E. coli with the CpSQR fragment-GFP fusions. Since the fusions did not have SQR activity, we did not check all of them with confocal microscopy. The CpSQR fragment 128-GFP is evenly distributed within the cell (Fig. 4A). These results are in agreement with the structural data showing that SQRs are peripheral membrane proteins with the C terminus inserted into the cytoplasmic membrane (26). CpSQR also has an amphipathic helix-turn-helix tripodal motif at its C terminus, which should be the motif for membrane insertion (26). The cytoplasmic localization of CpSQR is consistent with the sulfide detoxification role proposed for the enzyme in C. pinatubonensis JMP134.
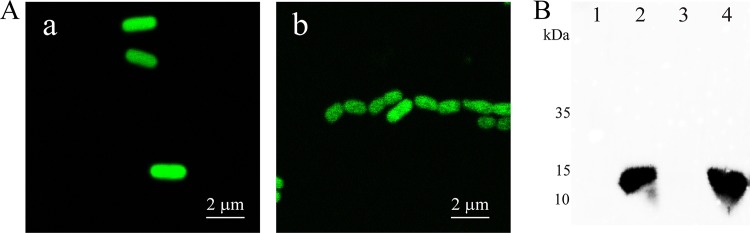
FIG 4.
GFP fusion and Western blot analyses of cellular localization of CpDUF442. (A) GFP fluorescence of E. coli BL21(DE3)(pBBR2-Cpduf442-gfp) (a) and C. pinatubonensis JMP134(pBBR2-Cpduf442-gfp) (b). (B) Western blotting of CpDUF442 in cellular fractions. An anti-His tag polyclonal antibody was used as the primary antibody. Lane 1, E. coli BL21(DE3) whole-cell lysate (12 μg of protein). Lanes 2 to 4, E. coli BL21(DE3)(pET30-Cpduf442). Lane 2, soluble fraction (8.8 μg of protein); lane 3, membrane fraction (4.7 μg of protein); lane 4, whole-cell lysate (17 μg of protein).
The DUF442 domain does not bind to the membrane.
The DUF442 domain of CpSQR is soluble when cloned and expressed in E. coli (12), and the fluorescence of CpDUF442-GFP in both E. coli BL21(DE3) and C. pinatubonensis JMP134 showed an even distribution in the cytoplasm (Fig. 4A). After cell disruption and the separation of the soluble proteins from the membrane proteins, Western blotting showed that CpDUF442 was present only in the soluble fraction and not in the membrane fraction (Fig. 4B), indicating that the CpDUF442 domain of CpSQR, when produced as an individual protein, is not bound to the membrane. However, as a domain of CpSQR, CpDUF442 is associated with the membrane.
Subcellular localization of CpPDO2.
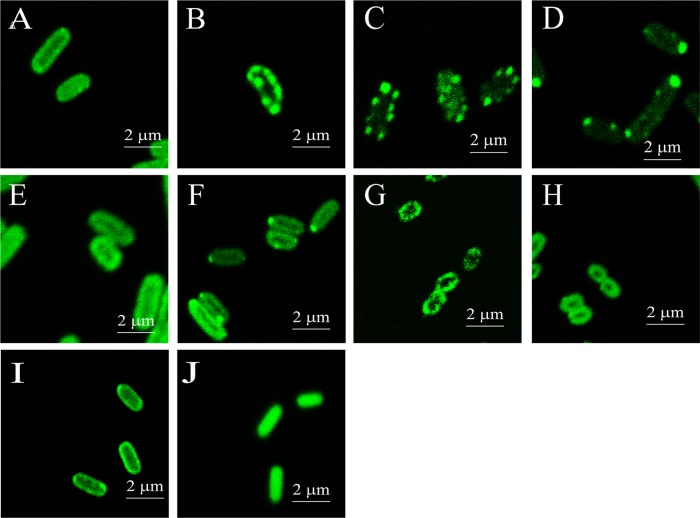
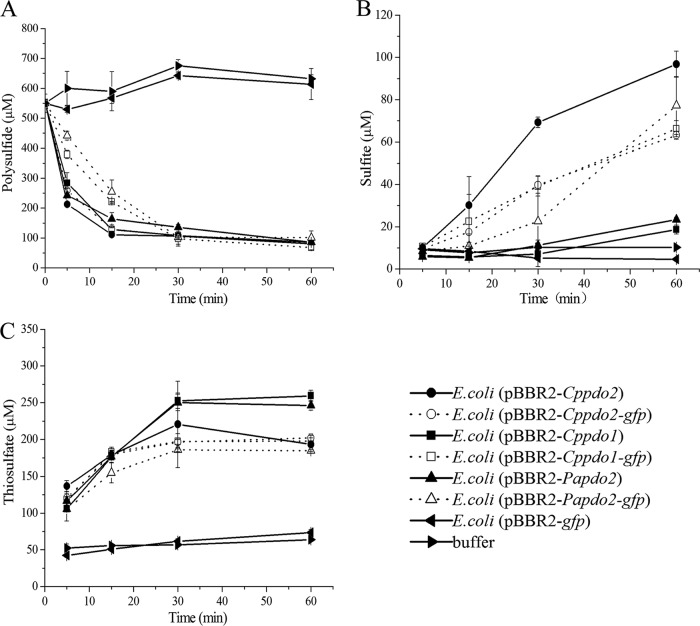
The position of PDO in the cell was also investigated. CpPDO2 is likely a cytoplasmic protein because it does not have a signal peptide for membrane translocation and it uses ferrous ion (Fe2+) for catalysis (10). Recombinant E. coli BL21(DE3) cells with pBBR2-Cppdo2-phoA, producing the CpPDO2-PhoA fusion, did not have any apparent PhoA activity (Fig. 5). When E. coli BL21(DE3) and C. pinatubonensis JMP134 Δsqr Δpdo2 and Δpdo2 mutants were transformed with pBBR2-Cppdo2-gfp, we detected green fluorescence around the cell surface (Fig. 6). Since the overproduced CpPDO2 in E. coli BL21(DE3) was soluble (10), the membrane association of CpPDO2-GFP was unexpected. We then constructed several GFP fusions with other PDOs, including CpPDO1, Pseudomonas aeruginosa PDO2 (PaPDO2), Agrobacterium tumefaciens Blh (AtBlh), and Myxococcus xanthus PDO1b (MxPDO1b), as well as a glyoxalase II (E. coli GloB1 [EcGloB1]). All of them belong to the metallo-β-lactamase superfamily. Surprisingly, all the tested proteins with GFP showed membrane association in E. coli BL21(DE3) (Fig. 6A to F), while the control, GAPDH-GFP, did not (Fig. 6J). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is a soluble enzyme of glycolysis in E. coli. Since some PDO-GFP fusions were shown as spots along the cell membrane, whether they formed inclusion bodies was checked. SDS-PAGE and Western blot analysis showed that the majority of expressed PDO-GFPs existed as soluble proteins, and none of the PDO-GFPs was found in inclusion bodies, which would be in the debris fraction pelleted after high-speed centrifugation (Fig. 7). Whether the PDO-GFP fusions had PDO activity was also tested. Recombinant E. coli cells with PDOs are known to oxidize polysulfide (12). Recombinant E. coli with several PDO-GFP fusions and the positive control with the corresponding PDO rapidly oxidized polysulfide to sulfite and thiosulfate, showing that PDO-GFP fusions have PDO activity (Fig. 8).
FIG 5.
PhoA activities in E. coli cells with PDO-PhoA fusions. The method was the same as for Fig. 3B. Bar 1, E. coli BL21(DE3). Bars 2 to 8, E. coli BL21(DE3) with PDO-PhoA fusions. Bar 2, pBBR2-Cppdo2-phoA; bar 3, pBBR2-Cppdo1-phoA; bar 4, pBBR2-Papdo2-phoA; bar 5, pBBR2-EcgloB1-phoA; bar 6, pBBR2-Mxpdo1a-phoA; bar 7, pBBR2-Atblh-phoA; bar 8, pBBR2-tolB-phoA. Error bars indicate standard deviations.
FIG 6.
GFP fluorescence analysis of cellular localization of PDO-GFP fusions in E. coli. (A to F) E. coli BL21(DE3) with pBBR2-Cppdo2-gfp (A), pBBR2-Cppdo1-gfp (B), pBBR2-Papdo2-gfp (C), pBBR2-EcgloB1-gfp (D), pBBR2-Mxpdo1a-gfp (E), or pBBR2-Atblh-gfp (F). (G) C. pinatubonensis JMP134 Δsqr Δpdo2(pBBR2-Cppdo2-gfp). (H) C. pinatubonensis JMP134 Δpdo2(pBBR2-Cppdo2-gfp). (I) E. coli BL21(DE3)(pBBR2-mhpT-gfp) (positive control). (J) E. coli BL21(DE3)(pBBR2-gapA-gfp) (negative control). The cloned genes were induced with IPTG before analysis.
FIG 7.
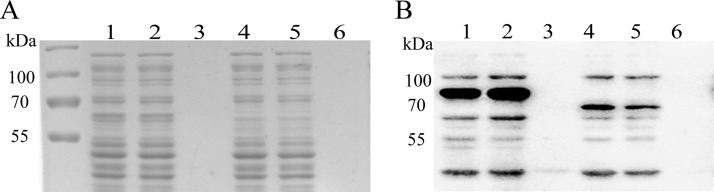
SDS-PAGE and Western blotting of PDO-GFP fusions in E. coli BL21(DE3). The cells were suspended in 50 mM Tris-HCl buffer (pH 8.0) and disrupted; the lysate was centrifuged at 12,000 × g for 10 min to separate supernatant and cell debris, which was resuspended with the same buffer to the same volume. (A) SDS-PAGE. (B) Western blotting. Anti-GFP polyclonal antibody was used as the first antibody. Lanes 1 to 3, cell lysate (597.7 μg of protein ml−1), supernatant (560.1 μg ml−1), and debris resuspension (15.33 μg ml−1), respectively, of E. coli BL21(DE3)(pBBR2-Cppdo2-gfp). Lanes 4 to 6, cell lysate (552.8 μg ml−1), supernatant (533.4 μg ml−1), and debris resuspension (12.2 μg ml−1), respectively, of E. coli BL21(DE3)(pBBR2-Papdo2-gfp). Lanes 7 to 9, cell lysate (528.5 μg ml−1), supernatant (504 μg ml−1) and debris resuspension (21.8 μg ml−1) of E. coli BL21(DE3)(pBBR2-mhpT-gfp). The sample volume was 15 μl for each well.
FIG 8.
Recombinant E. coli with PDO-GFP fusions oxidizes polysulfide. Cells were resuspended in 50 mM Tris-HCl buffer (pH 8.0) at an OD600 of 2. Polysulfide was added to 550 μM to initiate the reaction. Polysulfide (A), sulfite (B), and thiosulfate (C) were consumed and produced by E. coli BL21(DE3) with pBBR2-Cppdo2-gfp, pBBR2-Cppdo1-gfp, pBBR2-Papdo2-gfp, pBBR2-Cppdo2, pBBR2-Cppdo1, pBBR2-Papdo2, or pBBR2-gfp or the buffer, as indicted. All data are averages for at least three samples with standard deviations (error bars).
The PhoA/GFP fusion analyses also revealed that CpPDO2 and several tested bacterial PDOs are all cytoplasmic proteins that are peripherally located. The localization of CpPDO2 close to the membrane in the cytoplasm is not CpSQR dependent but is an intrinsic property of CpPDO2 and related proteins, as CpPDO2-GFP was associated with the membrane in both C. pinatubonensis JMP134 Δsqr Δpdo2 and Δpdo2 mutants (Fig. 6G and H). A significant number of cytoplasmic proteins may interact with membranes via amphipathic α-helixes or hydrophobic patches; however, bioinformatics analysis cannot predict which cytoplasmic protein will interact with the membrane, because most cytoplasmic proteins contain these motifs (42). Only the cytoplasmic proteins that form stable interactions with the membrane can be isolated with the membrane fraction, such as l-lactate dehydrogenase (43). CpPDO2 and other PDOs are likely to form weak interactions with the membrane, as they are mainly in the soluble fraction when overexpressed in E. coli (10). The cytoplasmic location of CpPDO2 is also consistent with the proposed localization of PDOs in chemolithotrophic Acidithiobacillus spp. (44). The CpDUF442 domain by itself was a soluble cytoplasmic protein that did not localize to the vicinity of the membrane (Fig. 4A). Since CpDUF442 is a domain of CpSQR, its native location is expected to be associated with the membrane on the cytoplasmic side. The role of CpDUF442 is to speed up the reaction between polysulfide and GSH to produce GSSH that is oxidized by CpPDO2 (12).
In summary, CpSQR and CpPDO2 are involved in sulfide oxidation in C. pinatubonensis JMP134, and they function inside the cytoplasm, which is different from the proposed location of SQR in the periplasmic spaces of phototrophic and chemolithotrophic bacteria (19, 20, 26, 27). The location of the two proteins with three enzymatic activities, SQR, rhodanese, and PDO, close to the membrane may facilitate sulfide oxidation via metabolic channeling, enabling the bacterium to rapidly detoxify sulfide (33). This information is critical to understand how heterotrophic bacteria oxidize sulfide, which may be helpful in guiding the use of these bacteria in H2S bioremediation.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains, plasmids, and recombinant cells used in this study are listed in Table 2. All primers are listed in Table 3. C. pinatubonensis JMP134 was cultivated in lysogeny broth (LB) at 30°C. E. coli BL21(DE3) was used to produce the recombinant proteins in LB at 37°C. Kanamycin (50 mg/liter) was added as required. pET30 Ec/Lic (Novagen) was used as an overexpression vector for protein production and purification, and the low-copy-number plasmid pBBR1MCS2 (45) was used as an expression vector for physiological analysis.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| C. pinatubonensis JMP134 | Wild type | Ron L. Crawford |
| E. coli BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm met (DE3) | Invitrogen |
| Plasmids | ||
| pET30 Ec/Lic | Kmr, expression vector | Invitrogen |
| pBBR1MCS2 | Kmr, expression vector | Invitrogen |
| pET30-Cpsqr | pET30 Ec/Lic containing Cpsqr gene from C. pinatubonensis JMP134 | This study |
| pET30-Cpduf442 | pET30 Ec/Lic containing Cpduf442 gene from C. pinatubonensis JMP134 | 40 |
| pBBR2-signal-phoA | pBBR1MCS2 containing phoA encoding the full-length PhoA with its signal peptide from E. coli BL21(DE3) | This study |
| pBBR2-phoA | pBBR1MCS2 containing a phoA gene fragment that encodes PhoA without its signal peptide | This study |
| pBBR2-gapA-gfp | pBBR1MCS2 containing gapA of E. coli MG1655 fused with the GFP gene | This study |
| pBBR2-mhpT-gfp | pBBR1MCS2 containing mhpT (41) of E. coli MG1655 fused with the GFP gene | This study |
| pBBR2-tolB-phoA | pBBR1MCS2 containing tolB (14) of E. coli MG1655 fused with phoA | This study |
| pBBR2-Cpsqr-gfp | pBBR1MCS2 containing Cpsqr fused with the GFP gene | This study |
| pBBR2-Cpsqr-phoA | pBBR1MCS2 containing Cpsqr fused with phoA | This study |
| pBBR2-fragmentx-gfp | pBBR1MCS2 containing a Cpsqr fragment encoding the N-terminal x amino acid residues fused with the GFP gene | This study |
| pBBR2-fragmentx-phoA | pBBR1MCS2 containing a Cpsqr fragment encoding the N-terminal x amino acid residues fused with phoA | This study |
| pBBR2-Cppdo2-phoA | pBBR1MCS2 containing Cppdo2 fused with phoA | This study |
| pBBR2-Cppdo2-gfp | pBBR1MCS2 containing Cppdo2 fused with the GFP gene | This study |
| pBBR2-Cppdo1-gfp | pBBR1MCS2 containing Cppdo1 fused with the GFP gene | This study |
| pBBR2-Cppdo1-phoA | pBBR1MCS2 containing Cppdo1 fused with phoA | This study |
| pBBR2-Papdo2-gfp | pBBR1MCS2 containing Papdo2 (21) of Pseudomonas aeruginosa PAO1 fused with the GFP gene | This study |
| pBBR2-Papdo2-phoA | pBBR1MCS2 containing Papdo2 fused with phoA | This study |
| pBBR2-Mxpdo1a-gfp | pBBR1MCS2 containing Mxpdo1a (21) of Myxococcus xanthus DK 1622 fused with the GFP gene | This study |
| pBBR2-Mxpdo1a-phoA | pBBR1MCS2 containing Mxpdo1a fused with phoA | This study |
| pBBR2-Atblh-gfp | pBBR1MCS2 containing Atblh (21) of Agrobacterium tumefaciens C58 fused with the GFP gene | This study |
| pBBR2-Atblh-phoA | pBBR1MCS2 containing Atblh fused with phoA | This study |
| pBBR2-EcgloB-gfp | pBBR1MCS2 containing EcgloB (21) of E. coli MG1655 fused with the GFP gene | This study |
| pBBR2-EcgloB-phoA | pBBR1MCS2 containing EcgloB fused with phoA | This study |
TABLE 3.
Oligonucleotide primers used for plasmid construction
| Primer | Nucleotide sequencea | Target protein |
|---|---|---|
| gfp-F | CACACAGGAAACAGCTATGAGTAAAGGAGAAGAAC | GFP |
| gfp-R | TAACAAAATATTAACGCTTATTTGTATAGTTCATACAT | |
| phoA-F | CACACAGGAAACAGCTATGCGGGCACCAGAAATGC | PhoA |
| phoA-R | TATTTGTACAAAATATTAACGCTTATTTCAGCCCCAGAGCG | |
| Signal-phoA-F | CACACAGGAAACAGCTATGAAACAAAGCACTATTGCAC | Signal-PhoA |
| Cpsqr-F | CACACAGGAAACAGCTATGCAACCCCGCGCCCTCACC | CpSQR |
| Cpsqr-R | AACAAAATATTAACGCTTATCCCTGCAACTCGGGT | |
| Cpsqr-phoA-R | TCCTGCTGCACTTCCTGCACTTCCTCCCTGCAACTCGGGTG | CpSQR-linker |
| Cpsqr-phoA-F | CAGCAGGAAGTGGAGAATTCCGGGCACCAGAAATGCCTG | Linker-PhoA |
| Cpsqr-gfp-F | CAGCAGGAAGTGGAGAATTCATGAGTAAAGGAGAAGAACT | Linker-GFP |
| Cpduf-gfp-R | TGCTGCACTTCCTGCACTTCCTAGATCATAGCCGGCGGCC | CpDUF442-linker |
| Linker-F | GGAAGTGCAGGAAGTGCAGCAGGAAGTG | Linker-PhoA/GFP |
| Fragment26-phoA-R | TTCCTGCTGCACTTCCTGCACTTCCGGCCGCCAGAGCGTGC | FragmentR6-linker |
| Fragment49-phoA-R | TCCTGCTGCACTTCCTGCACTTCCTTCCTGGAAGTTTGGCT | Fragment49-linker |
| Fragment71-phoA-R | TTCCTGCTGCACTTCCTGCACTTCCGACCTTGCCCGATTCC | Fragment71-linker |
| Fragment83-phoA-R | CCTGCTGCACTTCCTGCACTTCCCAGGAGTTGGCCGAACG | Fragment83-linker |
| Fragment106-phoA-R | TTCCTGCTGCACTTCCTGCACTTCCCAGCGCCCACAGCGTC | Fragment106-linker |
| Fragment115-phoA-R | CCTGCTGCACTTCCTGCACTTCCCGGCAGTGGCCTGATCAC | Fragment115-linker |
| Fragment157-phoA-R | TTCCTGCTGCACTTCCTGCACTTCCGCCGCCAATGATGACC | Fragment157-linker |
| Fragment176-phoA-R | CTTCCTGCTGCACTTCCTGCACTTCCATCCAGCCCCGGC | Fragment176-linker |
| Fragment230-phoA-R | CTTCCTGCTGCACTTCCTGCACTTCCCTCGGGCTCGAAGG | Fragment230-linker |
| Fragment359-phoA-R | CTTCCTGCTGCACTTCCTGCACTTCCGACGTACTGCATCA | Fragment359-linker |
| Fragment450-phoA-R | CTTCCTGCTGCACTTCCTGCACTTCCGTTGCCGGCGTCGC | Fragment450-linker |
| tolB-phoA-F | CACACAGGAAACAGCTATGAAGCAGGCATTACGAG | TolB-linker |
| tolB-phoA-R | CCTGCTGCACTTCCTGCACTTCCCAGATACGGCGACCAGG | |
| mhpT-gfp-F | CACACAGGAAACAGCTATGTCGACTCGTACCCCTT | MhpT-linker |
| mhpT-gfp-R | TGCTGCACTTCCTGCACTTCCGGCATCGGCGCACGGCTGTA | |
| Cppdo1-gfp-F | ACACAGGAAACAGCTATGCAAACCTTCTAT | CpPDO1-linker |
| Cppdo1-gfp-R | ACTTCCTGCACTTCCGGCGCCATGCGGCACG | |
| Cppdo2-gfp-F | CACACAGGAAACAGCTATGACACCGACCATGCCAAG | CpPDO2-linker |
| Cppdo2-gfp-R | TGCTGCACTTCCTGCACTTCCGAGGGCGTTGAGGGGAATCT | |
| Papdo2-gfp-F | ACACAGGAAACAGCTATGTTGAAACCCGAC | PaPDO2-linker |
| Papdo2-gfp-R | TGCACTTCCTGCACTTCCGAACAGATCCAGCGGGA | |
| Atblh-gfp-F | ACACAGGAAACAGCTATGAAGGCCGTAAGG | AtBlh-linker |
| Atblh-gfp-R | ACTTCCTGCACTTCCCCATGTGCTGCCCTCCAG | |
| Mxpdo1a-gfp-F | ACACAGGAAACAGCTATGCTCTTCCGCCAG | MxPDO1a-linker |
| Mxpdo1a-gfp-R | ACTTCCTGCACTTCCATGTGTGAAGCTGCCTC | |
| gloB-gfp-F | ACACAGGAAACAGCTATGAATCTTAACAGT | GloB-linker |
| gloB-gfp-R | ACTTCCTGCACTTCCGAACCTATCTTTCTTTG | |
| gapA-gfp-F | CACACAGGAAACAGCTATGACTATCAAAGTAGGTAT | GAPDH-linker |
| gapA-gfp-R | CACTTCCTGCACTTCCTTTGGAGATGTGAGCGAT | |
| (pET)Cpsqr-F | TAAGAAGGAGATATACATATGCAACCCCGCGCCCT | CpSQR |
| (pET)Cpsqr-R | GTGGTGGTGCTCGAGTCCCTGCAACTCGGGTGT |
Underlining indicates sequences overlapping with the linker.
Recombinant construction.
Each gene was amplified by PCR from the genomic DNA and cloned into pET30 Ec/Lic or pBBR1MCS2 by using the In-Fusion HD cloning kit (Clontech, USA). A reporter gene, phoA, of E. coli lacking the signal sequence, the first 59 amino acid residues at the N terminus, or the GFP gene was PCR amplified and fused to the C termini of CpSQR (WP_011299713.1), truncated CpSQR, CpPDO2 (WP_041680387.1), and CpPDO1 (AAZ62692.1) of C. pinatubonensis JMP134, PaPDO2 (NP_251605) of Pseudomonas aeruginosa PAO1, MxPDO1a (YP_633997.1) of Myxococcus xanthus DK 1622, AtBlh (AAK89929.1) (10) of Agrobacterium tumefaciens C58, and EcGloB (NP_414748.1) of E. coli MG1655 via a linker (5′-GGAAGTGCAGGAAGTGCAGCAGGAAGTGGAGAATTC-3′) (46). The truncated CpSQR contained the N-terminal fragmentx, where x stands for the number of amino acid residues from the N terminus (x = 26, 49, 71, 83, 107, 115, 128, 157, 176, 230, 359, or 450; CpDUF442 [rhodanese], x = 128). The membrane protein 3-hydroxyphenylpropionic acid transporter (MhpT) (NP_414887.2) (34) and the cytoplasmic protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (NP_416293.1) were linked with GFP as controls for cellular localization; periplasmic protein TolB (NP_415268.1) (38) was linked with PhoA as a control. The PCR products were cloned into pBBR1MCS2 under control of the lac promoter (45). The recombinant plasmid was transferred into E. coli BL21(DE3) or C. pinatubonensis JMP134 by electroporation. In C. pinatubonensis, the lac promoter is constitutively expressed due to the lack of the repressor LacI.
Whole-cell analysis.
E. coli and its recombinant cells were cultivated at 30°C with shaking, induced with 0.15 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) when the OD600 reached 0.4, and further cultivated at 30°C for 4 h. C. pinatubonensis JMP134 and its recombinant cells were cultivated at 30°C with shaking for 6 h. The cells were harvested by centrifugation (5,000 × g, 10 min) and resuspended in 50 mM Tris-HCl buffer (pH 8.0) to the desired OD600. Na2S or polysulfide was added to the cell suspension, followed by sampling at indicated the incubation times for detection of the substrate, intermediates, and products. Sulfide and polysulfide were detected with colorimetric assays (12). Thiosulfate and sulfite were detected after monobromobimane (mBBr) derivatization by using high-pressure liquid chromatography (HPLC) (12, 47). Polysulfide was produced by mixing 13 mg of sulfur power and 70 mg of sodium sulfide in 5 ml of anoxic distilled water under argon gas, as previous described (12).
Alkaline phosphatase activity was measured according to a reported method by using a cell suspension at an OD600 of 1 (19, 29), and one unit was defined as 1,000 times the absorbance at 420 nm of the reaction solution in 1 min (units · OD−1 · min−1). Cells with green fluorescence protein were fixed on a glass coverslip with 0.4% agarose and visualized by using a Zeiss confocal fluorescence microscope (LSM 780; Zeiss, Germany).
Isolation of cellular components.
E. coli BL21(DE3) and its recombinant cells were cultivated and induced as described above. Cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl (pH 8) to an OD600 of 5, and disrupted with a pressure cell homogenizer (Stansted Fluid Power Ltd., Harlow, Essex, UK). The lysate was centrifuged at 12,000 × g for 10 min to remove cell debris. The supernatant was then ultracentrifuged at 270,000 × g for 60 min to pellet the cell membranes. The supernatant contained soluble proteins; the pellet contained the membrane component, which was resuspended in the same buffer to the original volume. The procedures were done at 4°C.
Fractionation, SDS-PAGE, and Western analysis of recombinant proteins.
The proteins from the lysate, membrane fraction, and soluble fraction were separated through a 12% SDS-polyacrylamide gel and stained with Coomassie brilliant blue stain. An unstained gel was transferred (60 min, 100 V) onto a polyvinylidene fluoride (PVDF) membrane (Sangon, Shanghai, China), and the recombinant proteins were detected by using anti-His or anti-GFP antibody conjugated with horseradish peroxidase (Sangon, Shanghai, China) and an enhanced chemiluminescence reagent (Sangon, Shanghai, China); fluorescence intensity was detected by using a transilluminator (Fluor Chem Q; Protein Simple, San Jose, CA).
ACKNOWLEDGMENTS
This work was financially supported by grants from the National Natural Science Foundation of China (21477062), the Natural Science Foundation of Shandong Province (ZR2014CM003 and ZR2016CM03), and the State Key Laboratory of Microbial Technology at Shandong University.
REFERENCES
- 1.Hildebrandt TM, Grieshaber MK. 2008. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J 275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- 2.Kimura H. 2002. Hydrogen sulfide as a neuromodulator. Mol Neurobiol 26:13–19. doi: 10.1385/MN:26:1:013. [DOI] [PubMed] [Google Scholar]
- 3.Kimura H, Nagai Y, Umemura K, Kimura Y. 2005. Physiological roles of hydrogen sulfide: synaptic modulation, neuroprotection, and smooth muscle relaxation. Antioxidants Redox Signal 7:795. doi: 10.1089/ars.2005.7.795. [DOI] [PubMed] [Google Scholar]
- 4.Jackson MR, Melideo SL, Jorns MS. 2015. Role of human sulfide:quinone oxidoreductase in H2S metabolism. Methods Enzymol 554:255–270. doi: 10.1016/bs.mie.2014.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Shahak Y, Hauska G. 2008. Sulfide oxidation from cyanobacteria to humans: sulfide–quinone oxidoreductase (SQR), p 319–335. In Hell R, Dahl C, Knaff D, Leustek T (ed), Sulfur metabolism in phototrophic organisms. Advances in photosynthesis and respiration, vol 27 Springer, Dordrecht, The Netherlands. [Google Scholar]
- 6.Zhang X, Bian JS. 2014. Hydrogen sulfide: a neuromodulator and neuroprotectant in the central nervous system. ACS Chem Neurosci 5:876. doi: 10.1021/cn500185g. [DOI] [PubMed] [Google Scholar]
- 7.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 8.Wang R. 2012. Physiological implications of hydrogen sulfide: a whiff exploration that blossomed. Physiol Rev 92:791. doi: 10.1152/physrev.00017.2011. [DOI] [PubMed] [Google Scholar]
- 9.Goubern M, Andriamihaja M, Nubel T, Blachier F, Bouillaud F. 2007. Sulfide, the first inorganic substrate for human cells. FASEB J 21:1699–1706. doi: 10.1096/fj.06-7407com. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Xin Y, Xun L. 2014. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl Environ Microbiol 80:1799–1806. doi: 10.1128/AEM.03281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luebke JL, Shen J, Bruce KE, Kehl-Fie TE, Peng H, Skaar EP, Giedroc DP. 2015. The CsoR-like sulfurtransferase repressor (CstR) is a persulfide sensor in Staphylococcus aureus. Mol Microbiol 94:1343–1360. doi: 10.1111/mmi.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin Y, Liu H, Cui F, Liu H, Xun L. 2016. Recombinant Escherichia coli with sulfide: quinone oxidoreductase and persulfide dioxygenase rapidly oxidizes sulfide to sulfite and thiosulfate via a new pathway. Environ Microbiol 18:5123–5136. doi: 10.1111/1462-2920.13511. [DOI] [PubMed] [Google Scholar]
- 13.Knaff DB. 1975. The effect of o-phenanthroline on the midpoint potential of the primary electron acceptor of photosystem II. Biochim Biophys Acta 376:583–587. doi: 10.1016/0005-2728(75)90180-2. [DOI] [PubMed] [Google Scholar]
- 14.Klatt JM, Haas S, Yilmaz P, De BD, Polerecky L. 2015. Hydrogen sulfide can inhibit and enhance oxygenic photosynthesis in a cyanobacterium from sulfidic springs. Environ Microbiol 17:3301. doi: 10.1111/1462-2920.12791. [DOI] [PubMed] [Google Scholar]
- 15.Reinartz M, Tschape J, Bruser T, Truper HG, Dahl C. 1998. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol 170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 16.Gerrity S, Kennelly C, Clifford E, Collins G. 2016. Hydrogen sulfide oxidation in novel horizontal-flow biofilm reactors dominated by an Acidithiobacillus and a Thiobacillus species. Environ Technol 37:2252–2264. doi: 10.1080/09593330.2016.1147609. [DOI] [PubMed] [Google Scholar]
- 17.Pokorna D, Zabranska J. 2015. Sulfur-oxidizing bacteria in environmental technology. Biotechnol Adv 33:1246–1259. doi: 10.1016/j.biotechadv.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Marcia M, Ermler U, Peng G, Michel H. 2010. A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78:1073–1083. doi: 10.1002/prot.22665. [DOI] [PubMed] [Google Scholar]
- 19.Schütz M, Maldener I, Griesbeck C, Hauska G. 1999. Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic localization, and extension of gene sequence analysis. J Bacteriol 181:6516–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nübel T, Klughammer C, Huber R, Hauska G, Schütz M. 2000. Sulfide:quinone oxidoreductase in membranes of the hyperthermophilic bacterium Aquifex aeolicus (VF5). Arch Microbiol 173:233–244. doi: 10.1007/s002030000135. [DOI] [PubMed] [Google Scholar]
- 21.Theissen U, Martin W. 2008. Sulfide:quinone oxidoreductase (SQR) from the lugworm Arenicola marina shows cyanide- and thioredoxin-dependent activity. FEBS J 275:1131–1139. doi: 10.1111/j.1742-4658.2008.06273.x. [DOI] [PubMed] [Google Scholar]
- 22.Vande Weghe JG, Ow DW. 1999. A fission yeast gene for mitochondrial sulfide oxidation. J Biol Chem 274:13250–13257. doi: 10.1074/jbc.274.19.13250. [DOI] [PubMed] [Google Scholar]
- 23.Wakai S, Mei K, Kanao T, Kamimura K. 2004. Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, NASF-1. Biosci Biotechnol Biochem 68:2519–2528. doi: 10.1271/bbb.68.2519. [DOI] [PubMed] [Google Scholar]
- 24.Brito JA, Sousa FL, Stelter M, Bandeiras TM, Vonrhein C, Teixeira M, Pereira MM, Archer M. 2009. Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry 48:5613. doi: 10.1021/bi9003827. [DOI] [PubMed] [Google Scholar]
- 25.Chen ZW, Koh M, Van Driessche G, Van Beeumen JJ, Bartsch RG, Meyer TE, Cusanovich MA, Mathews FS. 1994. The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium. Science 266:430–432. doi: 10.1126/science.7939681. [DOI] [PubMed] [Google Scholar]
- 26.Marcia M, Ermler U, Peng G, Michel H. 2009. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci U S A 106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harb F, Prunetti L, Giudici-Orticoni MT, Guiral M, Tinland B. 2015. Insertion and self-diffusion of a monotopic protein, the Aquifex aeolicus sulfide quinone reductase, in supported lipid bilayers. Eur Phys J E Soft Matter 38:110. doi: 10.1140/epje/i2015-15110-8. [DOI] [PubMed] [Google Scholar]
- 28.Fontaine F, Fuchs RT, Storz G. 2011. Membrane localization of small proteins in Escherichia coli. J Biol Chem 286:32464–32474. doi: 10.1074/jbc.M111.245696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manoil C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol 34:61. doi: 10.1016/S0091-679X(08)61676-3. [DOI] [PubMed] [Google Scholar]
- 30.Arieli B, Shahak Y, Taglicht D, Hauska G, Padan E. 1994. Purification and characterization of sulfide-quinone reductase, a novel enzyme driving anoxygenic photosynthesis in Oscillatoria limnetica. J Biol Chem 269:5705–5711. [PubMed] [Google Scholar]
- 31.Louie TM, Webster CM, Xun L. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J Bacteriol 184:3492–3500. doi: 10.1128/JB.184.13.3492-3500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pérez-Pantoja D, Rodrigo DLI, Pieper DH, González B. 2008. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol Rev 32:736–794. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Bercoff A, McLysaght A, Conant GC. 2011. Patterns of indirect protein interactions suggest a spatial organization to metabolism. Mol Biosyst 7:3056–3064. doi: 10.1039/c1mb05168g. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Chen B, Chao H, Zhou NY. 2013. mhpT encodes an active transporter involved in 3-(3-hydroxyphenyl)propionate catabolism by Escherichia coli K-12. Appl Environ Microbiol 79:6362–6368. doi: 10.1128/AEM.02110-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangold S, Valdés J, Holmes DS, Dopson M. 2011. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front Microbiol 2:17. doi: 10.3389/fmicb.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregersen LH, Bryant DA, Frigaard NU. 2011. Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front Microbiol 2:116. doi: 10.3389/fmicb.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenk S, Arnds J, Zerjatke K, Musat N, Amann R, Mussmann M. 2011. Novel groups of Gammaproteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ Microbiol 13:758. doi: 10.1111/j.1462-2920.2010.02380.x. [DOI] [PubMed] [Google Scholar]
- 38.Isnard M, Rigal A, Lazzaroni JC, Lazdunski C, Lloubes R. 1994. Maturation and localization of the TolB protein required for colicin import. J Bacteriol 176:6392. doi: 10.1128/jb.176.20.6392-6396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drew D, Sjöstrand D, Nilsson J, Urbig T, Chin CN, de Gier JW, Von HG. 2002. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc Natl Acad Sci U S A 99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drew DE, Von HG, Nordlund P, de Gier JW. 2001. Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett 507:220–224. doi: 10.1016/S0014-5793(01)02980-5. [DOI] [PubMed] [Google Scholar]
- 41.Duffy EB, Barquera B. 2006. Membrane topology mapping of the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J Bacteriol 188:8343–8351. doi: 10.1128/JB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papanastasiou M, Orfanoudaki G, Koukaki M, Kountourakis N, Sardis MF, Aivaliotis M, Karamanou S, Economou A. 2013. The Escherichia coli peripheral inner membrane proteome. Mol Cell Proteomics 12:599–610. doi: 10.1074/mcp.M112.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Futai M, Kimura H. 1977. Inducible membrane-bound l-lactate dehydrogenase from Escherichia coli. Purification and properties. J Biol Chem 252:5820–5827. [PubMed] [Google Scholar]
- 44.Wang H, Liu S, Liu X, Li X, Wen Q, Lin J. 2014. Identification and characterization of an ETHE1-like sulfur dioxygenase in extremely acidophilic Acidithiobacillus spp. Appl Microbiol Biotechnol 98:7511–7522. doi: 10.1007/s00253-014-5830-4. [DOI] [PubMed] [Google Scholar]
- 45.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Nd RR, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Zaro JL, Shen WC. 2013. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65:1357. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura Y, Toyofuku Y, Koike S, Shibuya N, Nagahara N, Lefer D, Ogasawara Y, Kimura H. 2015. Identification of H2S3 and H2S produced by 3-mercaptopyruvate sulfurtransferase in the brain. Sci Rep 5:14774. doi: 10.1038/srep14774. [DOI] [PMC free article] [PubMed] [Google Scholar]