ABSTRACT
The use of phages as antibacterial agents is limited by their generally narrow host ranges. The aim of this study was to make a T4-like phage, WG01, obtain the host range of another T4-like phage, QL01, by replacing its host-determinant gene region with that of QL01. This process triggered a direct expansion of the WG01 host range. The offspring of WG01 obtained the host ranges of both QL01 and WG01, as well as the ability to infect eight additional host bacteria in comparison to the wild-type strains. WQD had the widest host range; therefore, the corresponding fragments, named QD, could be used for constructing a homologous sequence library. Moreover, after a sequencing analysis of gene 37, we identified two different mechanisms responsible for the expanded host range: (i) the first generation of WG01 formed chimeras without mutations, and (ii) the second generation of WG01 mutants formed from the chimeras. The expansion of the host range indicated that regions other than the C-terminal region may indirectly change the receptor specificity by altering the supportive capacity of the binding site. Additionally, we also found the novel means by which subsequent generations expanded their host ranges, namely, by exchanging gene 37 to acquire a wider temperature range for lysis. The method developed in this work offers a quick way to change or expand the host range of a phage. Future clinical applications for screening phages against a given clinical isolate could be achieved after acquiring more suitable homologous sequences.
IMPORTANCE T4-like phages have been established as safe in numerous phage therapy applications. The primary drawbacks to the use of phages as therapeutic agents include their highly specific host ranges. Thus, changing or expanding the host range of T4-like phages is beneficial for selecting phages for phage therapy. In this study, the host range of the T4-like phage WG01 was expanded using genetic manipulation. The WG01 derivatives acquired a novel means of expanding their host ranges by acquiring a wider temperature range for lysis. A region was located that had the potential to be used as a sequence region for homologous sequence recombination.
KEYWORDS: Escherichia coli, T4-like phages, gp37, homologous recombination, host range expansion
INTRODUCTION
Escherichia coli is an important infectious pathogen in both humans and animals (1–5). Antibiotics are commonly used to treat colibacillosis; however, antibiotic resistance among bacteria is increasingly recognized as a problem in both the veterinary and medical fields (6–8). Moreover, strains from farm animals exhibiting antibiotic resistance may pose a zoonotic risk, creating adverse health effects for the consumer (9, 10). Increased antibiotic resistance, particularly of multidrug-resistant strains, has led to the reconsideration of phages as alternative therapeutic agents (11–13). Phages have multiple advantages over antibiotics, including a ubiquitous presence (14) and specificity for target bacteria, thereby reducing the extent of damage to the normal flora (15).
The primary drawbacks to the use of phages as therapeutic agents include their narrow and specific host ranges (16, 17). To fix this problem, two possible methods that can be used are (i) to employ a cocktail to cover an extended host range (18) and (ii) to perform genetic manipulation to change or expand the host range and perform an oriented induction for screening phages of a given clinical isolate. T4-like phages are a type of virulent phage that has already been described to be safe in numerous phage therapy applications (19). In T4-like phages, the gp37 C-terminal region of the T4 family of phages and gp38 of the T2 phage family may be important regions for the determination of receptor specificity (20, 21). To change the host range of a given phage, extensive effort has been made to change the sequence of the long tail fiber genes. For example, gene 37 and gene 38 from the T2 genome were replaced with those of IP008 or PP01, each of which enabled T2 to acquire the same host range as that of the donor phages (22, 23); exchanging the 382-bp segment of gene 37 of T4 allowed phage TuIb to infect E. coli B (20). These experiments focused on results in which the modified phage obtained the same lysis capability as the donor phage; however, to our knowledge, no further research has been performed regarding an expansion of the phage host range.
In the T4-like phages we isolated in this study, WG01 exhibited a substantial difference in host range compared to another phage, QL01. Interestingly, these two phages exhibited a high homology on the level of the entire genome but low similarity regarding the sequence of gp37. We attempted to change and expand the host range of WG01 using two steps: (i) we located the receptor specificity determinant region of gp37, and (ii) we changed this region using homologous recombination with numerous homologous sequences.
RESULTS AND DISCUSSION
Phage isolation and morphology.
T4-like phages were isolated from duck feces, and two that exhibited different host ranges, WG01 and QL01 (Table 1), were chosen for further study. The morphological characterization revealed that the WG01 phage possessed an icosahedral head and a contractile tail (see Fig. S1A in the supplemental material), as did the QL01 phage (Fig. S1B).
TABLE 1.
Escherichia coli strains used to determine the host ranges
| Strain | QL01 | WG01 | Strain | QL01 | WG01 | Strain | QL01 | WG01 | Strain | QL01 | WG01 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DE001 | +a | −b | DE061 | − | − | DE167 | − | − | DE365 | + | − |
| DE002 | − | − | DE064 | − | − | DE169 | + | − | DE373 | − | − |
| DE003 | + | − | DE065 | − | − | DE172 | − | − | DE376 | − | − |
| DE005 | − | − | DE069 | − | + | DE182 | − | − | DE384 | + | − |
| DE007 | − | − | DE070 | − | − | DE183 | − | − | DE388 | − | − |
| DE008 | + | − | DE071 | − | − | DE186 | + | − | DE389 | + | − |
| DE010 | − | − | DE072 | + | + | DE192 | + | + | DE400 | + | − |
| DE011 | − | − | DE074 | − | − | DE197 | − | − | DE402 | + | − |
| DE013 | − | − | DE075 | − | − | DE205B | + | + | DE404 | + | − |
| DE015 | + | − | DE077 | − | − | DE207 | − | − | DE407 | + | − |
| DE017 | + | + | DE083 | − | − | DE209 | − | − | DE414 | + | − |
| DE018 | − | − | DE085 | − | − | DE217 | + | + | DE419 | + | − |
| DE019 | − | − | DE094 | − | − | DE235 | − | − | DE426 | − | − |
| DE020 | − | − | DE098 | − | − | DE241 | − | + | DE432 | − | − |
| DE021 | + | + | DE101 | + | + | DE242 | − | − | DE452 | + | + |
| DE022 | − | − | DE102 | + | + | DE248 | + | − | DE458 | − | − |
| DE023 | + | − | DE104 | + | − | DE257 | − | − | DE464 | − | − |
| DE028 | + | − | DE119 | + | − | DE278 | + | − | XM | + | + |
| DE032 | + | − | DE120 | + | − | DE283 | + | − | HX01 | + | − |
| DE034 | − | + | DE123 | + | − | DE295 | + | − | HX04 | − | − |
| DE037 | − | − | DE127 | − | − | DE296 | + | − | NT−1 | + | + |
| DE041 | − | − | DE129 | − | − | DE301 | + | − | MC1061 | − | + |
| DE044 | − | + | DE132 | − | − | DE302 | + | − | MG1655 | − | − |
| DE046 | − | − | DE137 | − | − | DE303 | + | − | O157:H7 | − | − |
| DE049 | − | − | DE144 | − | − | DE312 | + | − | RS218 | + | + |
| DE050 | − | − | DE147 | − | + | DE316 | + | − | 5155 | + | + |
| DE054 | − | − | DE148 | + | + | DE322 | + | − | |||
| DE056 | − | − | DE158 | − | + | DE327 | + | − | |||
| DE060 | − | − | DE160 | + | − | DE353 | − | + |
+, clear lysis.
−, no reaction.
Comparative genomic analysis.
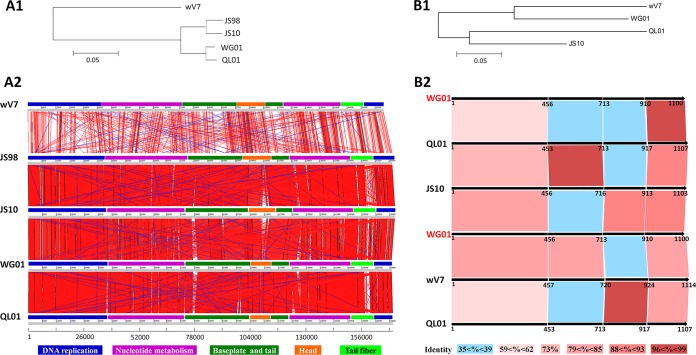
The genome of WG01 (NC_031928.1) consists of linear double-stranded DNA 169,936 bp in size with a 60-bp redundancy at each end. The comparative genomic analysis of WG01 revealed a high homology with other T4-like phages, including QL01 (NC_028847.1), Bp7 (NC_019500), JS98 (NC_010105), IME08 (NC_014260), JS10 (NC_012741), and vB_EcoM_VR5 (KP007359) (see Table S2). The high sequence homology (Fig. 1A1 and A2; see also Table S2) indicated that WG01 belonged to the T4-like virus Myoviridae family of the order Caudovirales (24) and the JS98 subgroup (25). The highest similarity throughout the WG01 genome was observed with QL01, covering 94% (identity, 98%). The majority of the WG01 proteins exhibited strong sequence similarity with those of QL01, and more than 90% shared high amino acid sequence identity (>90%) (see Table S3).
FIG 1.
Comparisons of the whole-genome and amino acid sequences of gp37. (A1) Neighbor-joining tree based on the alignment of the entire genomes of five T4-like phages derived from the “distance tree results” in the BLASTN search of the NCBI database. The results revealed that WG01 shared close phylogenic relationships with JS98, QL01, and JS10 but exhibited a distant phylogenic relationship with wV7. (A2) Comparisons of the entire genome sequences were carried out using the Artemis Comparison Tool (ACT). Different functional modules are represented by different colors, and the color legend is shown at the bottom. The different colored lines represent different similarities of the genes. The red lines represent the highest similarity and the blue lines represent the lowest similarity. (B1) Neighbor-joining tree analysis and bootstrap analysis (1,000 bootstrap replicates) based on the alignment of the gp37 amino acid sequences of four T4-like phages were carried out using ClustalX and MEGA. WG01 shared a closer phylogenic relationship with wV7 than with QL01 and JS10. (B2) The different amino acids domains (1 to 456, 457 to 713, 714 to 910, and 911 to 1,100) of WG01 gp37 were compared with the corresponding protein sequences of QL01, JS10, and wV7. The color keys representing the different similarities are presented at the bottom. In A1 and B1, the scales at the bottom represent the evolutionary distances.
Phylogenetic analyses were performed for 33 T4-like phages from the NCBI database based on the amino acid sequences of the major capsid protein (gp23) and tail fiber protein (gp37) with MEGA6 using the neighbor-joining algorithm (see Fig. S2). These 33 phages were derived from different hosts with distant relationships on separate branches of a trunk, as displayed in the distance tree. The phylogenetic analyses of gp23 and gp37 revealed different evolutionary relationships. In the distance tree of gp23 (Fig. S2A), WG01, Bp7, and QL01 displayed a close evolutionary relationship; however, in the distance tree of gp37 (Fig. S2B), WG01, Bp7, JS98, and vB_EcoM-VR26 shared a close evolutionary relationship, and QL01 and JS10 shared a close evolutionary relationship.
Comparison of the gp37 amino acid sequence.
A comparison of the gp37 amino acid sequences between WG01 and QL01 revealed a sequence conservation of 59% identity for the entire sequence, and different parts of gp37 from WG01 and QL01 exhibited different similarities. In particular, there was a high sequence identity (100%) in the first 64 amino acids (aa) of the N-terminal domain and the first 183 aa of the C-terminal domain; however, only trace homology was exhibited in the other regions (see Fig. S3 and S4A).
The sequence similarity between the target sequence (the sequences of gp37 of WG01, QL01, JS10, and wV7) and the template sequence (the sequence of the experimentally solved three-dimensional structure of T4 gp37) dropped below 30%. The model of the gp37 sequence of these four phages could not be predicted for analyzing which segments of DNA fragments could be selected for genetic manipulation experiments.
A comparison between the gp37 amino acid sequences of WG01, QL01, JS10, and wV7 showed that the amino acid site 714 of WG01 gp37 was special. The sequences on either side of amino acid site 714 exhibited different similarity results compared with their homologous sequences. According to the diverse similarities of the different regions, four regions (amino acids 1 to 456 [Fig. S3A], 457 to 713 [Fig. S3B], 714 to 910 [Fig. S3C], and 911 to 1,100 [Fig. S3D]) of the WG01 gp37 amino acid sequences were divided and compared with the corresponding regions of QL01, JS10, and wV7. Amino acids 457 to 713 of WG01 gp37 exhibited a higher sequence homology with the corresponding regions of wV7, with an identity of 80%, and exhibited a lower homology with the corresponding parts of QL01 and JS10, with an identity of 39% (Fig. 1B2; see also Fig. S3B). However, amino acids 714 to 910 of WG01 gp37 exhibited a higher homology with the corresponding parts of JS10, with an identity of 88%, and exhibited a lower homology with the corresponding parts of QL01 and wV7, with identities of 37% and 36%, respectively (Fig. 1B2; see also Fig. S3C).
These comparisons revealed that amino acid 714 may be an important dividing point; therefore, the DNA fragments for homologous recombination were selected at different sites before or after amino acid 714 (nucleotide 2139) (see Fig. S4).
Homologous recombination of different parts of gene37 between WG01 and QL01.
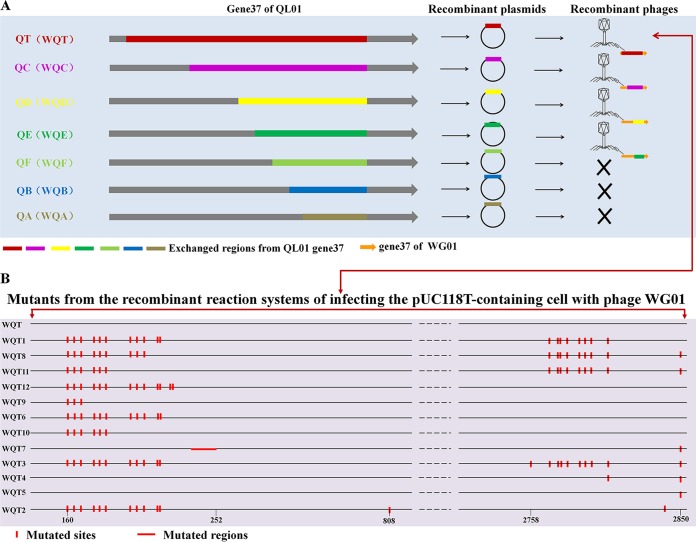
The rationale for the homologous recombination of WG01 is displayed in Fig. 2. When performing a plaque assay with E. coli strain DE205B, the host of QL01, four types of recombinant phages (WQT1, WQC1, WQD1, and WQE1) (Fig. 3A) were isolated and able to form clear plaques. The sequencing analysis of gene 37 for WQT1, WQC1, WQD1, and WQE1 confirmed that they had originated from QL01 (see Table S4). The chimeric phages possessed the body of WG01 and different portions of gene 37 from QL01. The sequencing analysis of the recombinant phages demonstrated that no nucleic acid mutations had occurred in WQE1. Moreover, the nucleic acid mutation that occurred in WQC1 and WQD1 did not cause an amino acid substitution; however, the nucleic acid mutation which occurred in WQT1 did cause an amino acid substitution.
FIG 2.
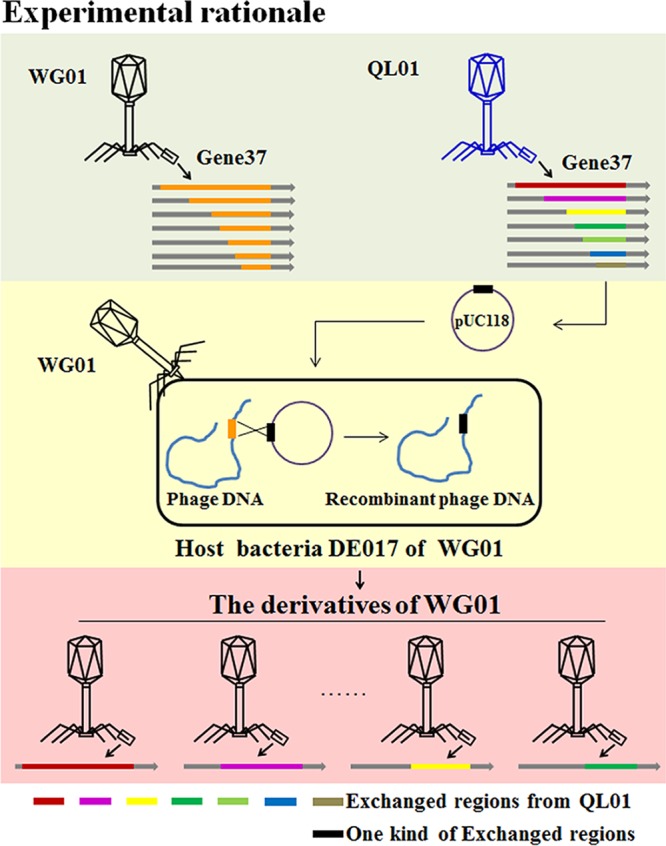
Rationale for the homologous recombination of WG01. The different parts from gene 37 of the QL01 phage were digested and inserted into pUC118 to produce different recombinant plasmids. DE017 was transformed with the recombinant plasmid. Then, the transformant cells were infected by WG01. When the WG01 genome was injected into the bacteria and nucleic acid was synthesized, homologous recombination of gene 37 between QL01 and WG01 occurred.
FIG 3.
Sketch map of gene 37 positions of QL01 for WG01 recombination, results of the isolated chimeras, and mutation sites of the mutants derived from the same WQT chimera. (A) Sketch map of gene 37 positions of QL01 for WG01 recombination and results of the chimeras isolated with the DE205B host. Different positions from the end indicate the DNA fragments for WG01 recombination; these fragments were named QT, QC, QD, QE, QF, QB, and QA. Four types of recombinant phages, namely, WQT1, WQC1, WQD1, and WQE1, were isolated from DE205B, the host of QL01. The Xs indicate that no chimeric phages were isolated with DE205B by infecting the plasmid-containing cell with phage WG01. (B) The mutation sites of mutants derived from the same WQT chimera. There are 12 types of mutants that originate from DE017 cells carrying plasmids containing the QT inserts and that were infected with WG01, including single point mutations, multipoint mutations, and DNA fragment deletions. The sequences of the 12 types of mutants were compared with that of WQT. The mutation sites are marked with red lines. The numbering scale at the bottom represents the nucleotide positions.
Mutations occurring in recombinant phages derived from the same WQT chimera.
The sequencing analysis of gene 37 of the mutants derived from the same WQT chimera indicated that there were 11 types of phages in the 31 clear phage plaques (WQT2, WQT3, WQT4, WQT5, WQT6, WQT7, WQT8, WQT9, WQT10, WQT11, and WQT12) (Fig. 3B). The sequencing results for gene 37 of these phages confirmed that they had originated from QL01. The mutants possessed the body of WG01 and gene 37 of QL01. All 12 mutants were called WQTs. Twelve types of mutants originated from DE017 cells carrying plasmids containing the QT inserts and infected with WG01, including single point mutations, multipoint mutations, and DNA fragment deletions (Fig. 3B; see also Table S4). The mutations were focused in the regions of nucleic acids 162 to 209 and 2,760 to 2,850.
Spontaneous mutations could be selected when the phage filtrate was plated directly on the bacteria that the phages did not infect, making it possible for the phages of the T4 family to expand the host range (21, 26–28). According to the results in this section, we speculated that the bacterium-phage combination carrying plasmids containing the homologous sequences encouraged the occurrence of mutations, which also improved the opportunity to produce phages with different host ranges.
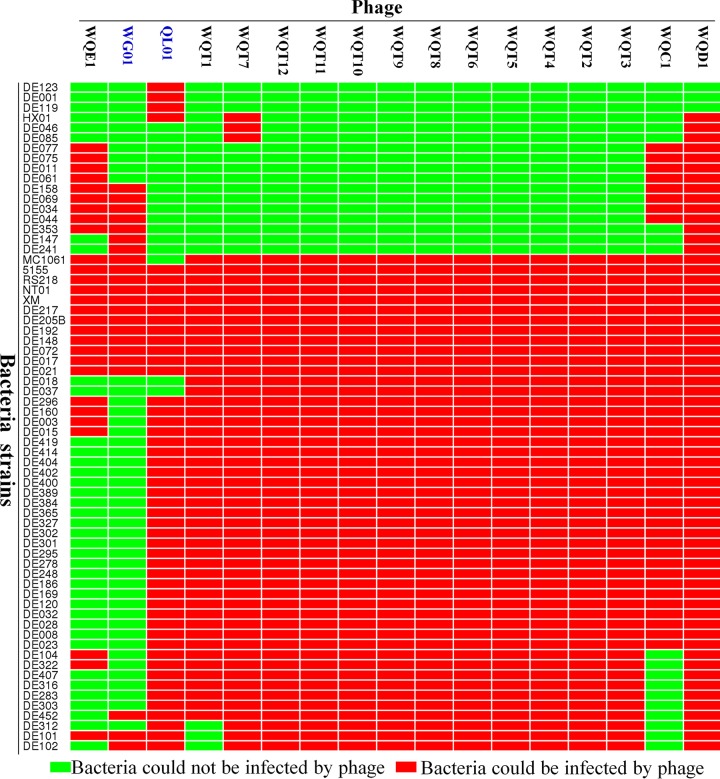
Host range analysis of QL01, WG01, and WG01 derivatives: two different methods for expanding phage host range.
The host ranges of the phages WG01 and QL01, as well as the 15 WG01 derivatives, were tested using 113 E. coli strains. The results of the host range analysis are displayed in Fig. 4 and Table S1. QL01, WG01, WQC1, WQD1, and WQE1 lysed 50, 22, 47, 63, and 28 of the 113 E. coli strains, respectively. In the mutants derived from the same chimera of WQT, WQT7 was able to lyse the largest number of strains (52 strains).
FIG 4.
Host range results of WG01, QL01, and WG01 derivatives. The heat map of host range results was produced using the OmicShare platform. The red color represents that the bacteria could be infected by the phages. The light green color represents that the bacteria could not be infected by the phages. QL01, WG01, WQC1, WQD1, and WQE1 lysed 50, 22, 47, 63, and 28 of the 113 E. coli strains, respectively. The mutants derived from the same chimeras of WQT, WQT2, WQT3, WQT4, WQT5, WQT6, WQT8, WQT9, WQT10, WQT11, and WQT12 had the same host ranges, each of which could lyse 49 strains. WQT7 was able to lyse the largest number of strains (52 strains).
Each of the WG01 derivatives had a wider host spectrum than that of WG01, and its derivatives were able to lyse a total of 63 E. coli strains (Fig. 4; see also Table S1). Moreover, the derivatives could lyse 41 bacterial strains that WG01 could not lyse, 16 bacterial strains that QL01 could not lyse, 33 bacterial strains that QL01 could lyse but WG01 could not, 8 bacterial strains that WG01 could lyse but QL01 could not, and 8 bacterial strains that neither WG01 nor QL01 could lyse (Fig. 5). WQD1 exhibited the widest host range of the 15 WG01 derivative phages, which demonstrated the ability to lyse 63 strains. In WQD1, the starting position of the exchange fragment was the site of amino acid 714 (nucleotide 2,139), which was a dividing point of WG01 gp37 (Fig. S4B).
FIG 5.
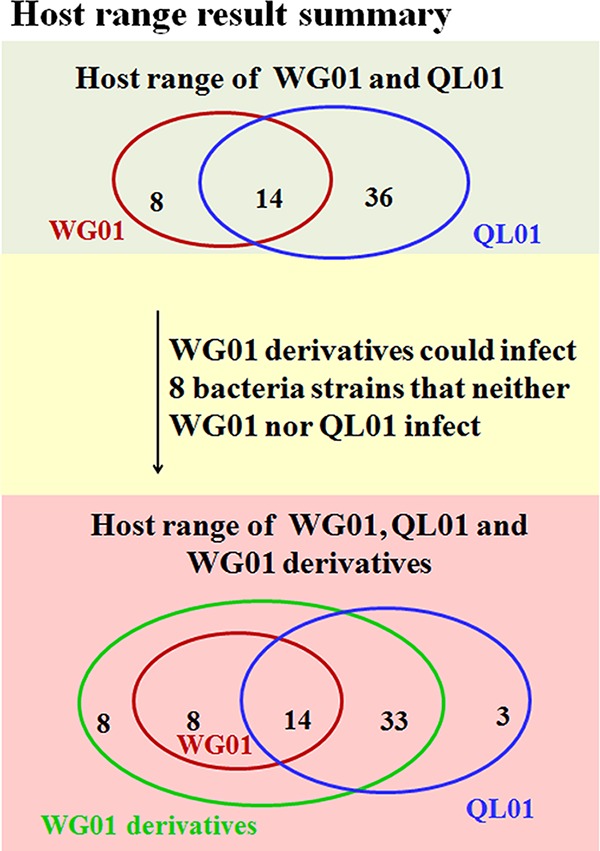
Summary of the final experimental host range results. (Top) The experimental results of host range analysis showed that in 113 E. coli strains used for the host spectrums testing, WG01 could lyse 22 E. coli strains and QL01 could lyse 50 E. coli strains. There were 14 strains that WG01 and QL01 could lyse. (Bottom) There were 22 strains that WG01 and WG01 derivatives could lyse, 47 strains that QL01 and WG01 derivatives could lyse, and 3 strains that only QL01 could lyse. All the WG01 derivatives could lyse 63 E. coli strains, including 41 strains that WG01 could not lyse, 16 strains that QL01 could not lyse, 33 strains that QL01 could lyse and WG01 could not, and 8 strains that WG01 could lyse and QL01 could not. Moreover, the derivative phages lysed 8 bacterial strains that neither WG01 nor QL01 lysed.
According to the experimental results described above, the host range of WG01 was expanded by simply exchanging the gene 37 sequence with that of QL01, and there are two different methods that can be used to expand the host range after chimeric phages have been constructed, namely, (i) from the chimeras themselves and (ii) from the mutants derived from the same chimera. These two different approaches were pursued as follows.
The first means of expanding the host range of WG01 was via the chimeras themselves. The WG01 chimeric phages acquired the ability to infect eight additional host bacteria that neither of the parent phages could infect. Moreover, the chimeric phages exhibited some interesting phenomena. One interesting phenomenon was that the four types of chimeric phages, namely, the WQTs, WQC1, WQD1, and WQE1, had in common the region of 183 aa in the C-terminal domain of gp37, which might determine the receptor specificity (20, 29). Therefore, the chimeric phages should have the same host ranges. However, they showed dramatically different host ranges, indicating that the receptor specificity of the phages may not be decided only by a constant fragment as reported by Montag et al. (20). We speculated that one of the explanations for this was that besides the direct binding region, other regions could indirectly change the receptor specificity by altering the supportive capacity of the binding site and changing the conformation of the tail fiber protein. For example, in the case of adenovirus fiber protein, receptor binding is influenced by both the C-terminal knob domain and the central shaft region (30–32). Another interesting phenomenon was that WQD1 had a wider host range than those of the other three kinds of derivatives, namely, the WQTs, WQC1, and WQE1. The tail fiber sequences of the WQTs and WQC1 covered that of WQD1, indicating that they should have the same host range as WQD1 or a wider one. We speculated that one possible reason for this feature was that the C-terminal domain from amino acid 714 to the end of QL01 might facilitate the formation of an optimal three-dimensional protein conformation in WG01, which could generate a phage with greater lysis ability.
The second method of host range expansion was unexpected. The C-terminal domain of gp37 has been considered to be an important factor for the determination of receptor specificity (20, 29); however, mutations that occurred in the N-terminal domain also influenced the host range. Moreover, compared to WQT and other WQTs, the 12 missing amino acids in the N-terminal domain created the WQT7 derivative with the widest host range. This finding indicated that changes in amino acid regions other than those in the C-terminal domain also influenced the receptor specificity, as previously described for adenovirus (30–32), likely via an altered ability to support the direct receptor binding site.
Growth characteristics of the WG01, QL01, and WG01 derivatives: a unique means of expanding phage host range.
The growth characteristic results for WG01, QL01, WQT1, WQC1, WQD1, and WQE1 are displayed in Fig. 6 and Fig. S5. On the DE205B strain lawn, QL01, WQT1, WQC1, and WQD1 all formed plaques at 28°C, 16°C, and 4°C, rather than at 37°C; WQE1 only formed plaques at 28°C. Moreover, WG01 could not infect DE205B and was able to form clear plaques at 37°C and 28°C with the host DE017. All of the phages were adapted to grow at temperatures lower than 28°C rather than within the range of poultry gut temperatures (none of the phages could form plaques at 42°C, which is not displayed), although phages WG01 and QL01 were isolated from duck feces.
FIG 6.
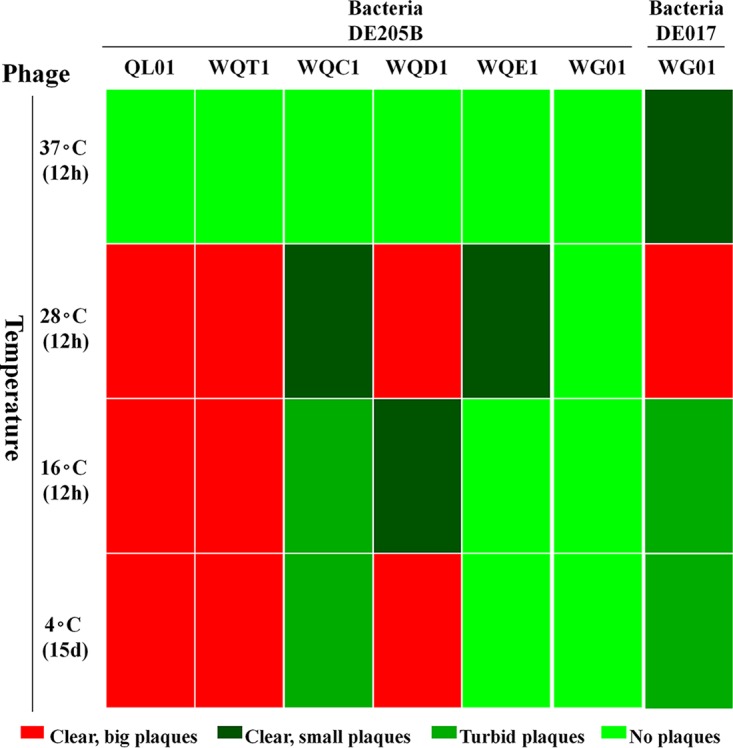
Growth characteristics of the WG01, QL01, and WG01 derivatives. The heat map of the growth characteristics was produced using the OmicShare platform. The growth characteristics of WG01, QL01, WQT1, WQC1, WQD1, and WQE1 are displayed. On a lawn of DE205B cells, QL01 and WQT1 formed large clear plaques at 28°C, 16°C, and 4°C; WQC1 formed small clear plaques at 28°C and turbid and small plaques at 16°C and 4°C; WQD1 formed large clear plaques at 28°C and 4°C and small clear plaques at 16°C; WQE1 formed small clear plaques at 28°C. WG01 could not infect DE205B but could form large clear plaques at 28°C, small clear plaques at 37°C, and small turbid plaques at 16°C and 4°C with the DE017 host.
The WG01qlE phage formed relatively smaller (<0.5 mm) plaques on a lawn of strain DE205B than those of the other four phages, namely, QL01, WQT1, WQC1, and WQD1. An alteration of the different parts of the gp37 tail fiber protein of QL01 enabled the WG01 derivatives to infect DE205B and, furthermore, enabled the WG01 derivatives of WQT1, WQC1, and WQD1 to grow well at low temperatures, such as 4°C.
According to the published literature, the failure of low-temperature T4-like coliphages to thrive at high temperatures is attributed to the impact on the multiplication (and/or the injection) step of the phages (33, 34). However, in our study, the adsorption of phages could influence growth characteristics, because the WG01 growth characteristic was altered just by changing the sequence of gene 37. The homologous recombination methods of the T4-like phage provided a tool that can be used to construct phages with the ability to grow well at wider temperature ranges (e.g., 4°C). This may make it possible to utilize phages to kill pathogenic bacteria on fresh and minimally processed produce during storage at 4°C (33, 35). Furthermore, alternative phages can be generated to kill pathogenic bacteria in a variety of environments exhibiting different temperatures.
Conclusion.
Our study expanded the host range of WG01 by altering different parts of gene 37 within the chimeras themselves and the mutants derived from the chimeras. Additionally, the temperature range of WG01 was expanded to infect bacteria at different temperatures, even at 4°C. This method provides a tool that can be used to change and expand the host range of phages rapidly. Future clinical applications for screening phages of given clinical isolates can be developed following the acquisition of a greater number of suitable homologous sequences. It may also be possible to modify the genomes themselves to directly change or expand the host ranges of other bacteriophages using this method.
MATERIALS AND METHODS
Bacterial strains and medium.
E. coli strains DE017 and DE205B were used as the hosts of phages WG01 and QL01, respectively, for propagation. A total of 104 avian E. coli strains used in these experiments were isolated from the brains of ducks with clinical signs of septicemia and neurological symptoms at different times and in different areas within the eastern region of China, as previously described (36–38). The pathogenic avian E. coli strain, IMT5155, was kindly provided by Lothar H. Wieler and Christa Ewers (39). The RS218 strain is a neonatal meningitis E. coli (NMEC) strain purchased from the Control Institute of Veterinary Bioproducts and Pharmaceuticals of China. All of the E. coli strains used in this study are listed in Table S1 in the supplemental material. All bacterial strains were grown in Luria-Bertani-Miller (LB-Miller) liquid medium or on LB-Miller agar plates at 37°C. For the dilution and preservation of the phages, an SM buffer (10 mM MgSO4, 100 mM NaCl, 0.01% gelatin, and 50 mM Tris-HCl [pH 7.5]) was used.
Isolation and propagation of bacteriophages.
Duck feces were collected from a poultry market in Nanjing, Jiangsu Province, China. SM buffer was added to the fecal samples, which were then centrifuged twice at 4,000 × g for 10 min at 4°C, and the supernatants were filtered through 0.22-μm Millipore filters (Merck Millipore) and stored at 4°C. Phage detection and isolation were performed via the double-layer agar plate method, and samples were stored at 4°C (40).
Electron microscopy.
The phage filtrates were applied to copper grids coated with a carbon support film before negative staining with phosphotungstic acid (PTA; 2% [wt/vol]). Electron micrographs were observed using an H_7650 (Hitachi, Japan) transmission electron microscope (TEM).
DNA isolation, genome sequencing, function prediction, and comparative genomic analysis.
The phage DNA isolation of WG01 was performed according to the protocol described previously (41). The phage supernatant was collected by centrifugation at 10,000 × g for 10 min and filtered through a 0.22-μm Millipore filter. Chloroform was added to reach a final concentration of 0.5%. The phage DNA was extracted from the treated lysates using an Abigen λ phage DNA extraction kit (catalog no. AB1141). Genomic sequencing was performed using Illumina MiSeq, and reads were assembled into contigs using Newbler (version 2.8). The open reading frames (ORFs) were predicted using GeneMarkS (http://topaz.gatech.edu/GeneMark/genemarks.cgi). The functions of the putative ORFs were analyzed in BLASTP searches of the NCBI database. The phylogenetic analysis and comparisons of amino acid sequences were carried out with ClustalX (version 2.1) and MEGA (version 6.06) (42). The whole-genome sequences and amino acid sequences were compared using BLASTN and BLASTP searches, respectively, of the NCBI database. The comparisons of the whole-genome sequences were carried out with the Artemis Comparison Tool (ACT) (43, 44) (http://www.webact.org). The model was predicted by employing the SWISS-MODEL server (http://swissmodel.expasy.org) (45).
Selection of different DNA fragments of gene 37 for gene manipulation experiments.
The C-terminal region of gp37 of the T4 family of phages may be an important region for the determination of receptor specificity (20, 29); thus, we can make comparisons of gp37 between different T4-like phages and perform a genetic manipulation of gene 37 in which the manipulated phage and the donor phage were WG01 and QL01, respectively. The gp37 amino acid sequences of 33 T4-like phages (Table S2) were downloaded from the NCBI database, and comparisons of the amino acid sequences were carried out with ClustalX (version 2.1) (data not shown). The results revealed that the sequence of the gp37 C-terminal domain of WG01 exhibited a high similarity with that of JS10. In addition, the sequence of the gp37 C-terminal domain of QL01 exhibited a high similarity with those of e11/2 and wV7. Thus, the sequences of gp37 from WG01, QL01, JS10, and wV7 were selected for the comparative analysis, and the three-dimensional structures of these from the four phages were predicted by the SWISS-MODEL server to analyze which segments of DNA fragments could be selected for genetic manipulation experiments.
Construction of the plasmids for altering different parts of WG01 gene 37.
Construction of the plasmid for the alteration of gene 37 was performed according to the protocol described previously (22) with some modifications. On the basis of the comparison of the results mentioned above, seven segments of gene 37 DNA fragments (i.e., QT, QC, QD, QE, QF, QB, and QA) were selected and amplified by PCR or fusion PCR using the corresponding primers (Table 2) and WG01 and/or QL01 phage genomic DNA as the templates. The PCR fragments were digested with BamHI and SphI and inserted into the plasmid pUC118 with ampicillin resistance (TaKaRa) to construct different recombinant plasmids. Seven types of recombinant plasmids with these fragments were constructed and named pUC118T, pUC118C, pUC118D, pUC118E, pUC118F, pUC118B, and pUC118A.
TABLE 2.
Primers used in this study for altering different parts of gene 37 of WG01
| Primer | Sequence (5′ to 3′)a |
|---|---|
| WQT | F, CGCGGATCCCCAGTTGGCTGAAGGCGAACTGGC |
| R, ACATGCATGCCCTGTTTCAGTCCACAAGGCACCAGTGG | |
| WQA | F1, CGCGGATCCGGGACCCAAGGTCATACATC |
| R1, GTAGCATCTCCGCCCAATCTACGCATGCCTAATTCAACAG | |
| F2, CTGTTGAATTAGGCATGCGTAGATTGGGCGGAGATGCTAC | |
| R2, ACATGCATGCGTGTGCTCATTGATTGCAGC | |
| WQB | F1, CGCGGATCCGGGACCCAAGGTCATACATC |
| R1, GGCTTTTTCCTTTGATAATTGGATAATAACTGTCTTCACCCACGC | |
| F2, GCGTGGGTGAAGACAGTTATTATCCAATTATCAAAGGAAAAAGCC | |
| R2, ACATGCATGCGTGTGCTCATTGATTGCAGC | |
| WQC | F1, CGCGGATCC GGCACAAGAACTTTCCTGAGCG |
| R1, CCAGCCAGTAGCATCAGATGTTTCCATTACGTTTTTACGGTCACC | |
| F2, GGTGACCGTAAAAACGTAATGGAAACATCTGATGCTACTGGCTGG | |
| R2, ACATGCATGCGGTCGCCTTCAACCAATTCAGG | |
| WQD | F1, CGCGGATCC GCTGGTAATGCGGTCAATGCTC |
| R1, GCCTCTTTTAACGATTAATTTGCCTGTCATCGTGTCACCTTC | |
| F2, GAAGGTGACACGATGACAGGCAAATTAATCGTTAAAAGAGGC | |
| R2, ACATGCATGCGGTCGCCTTCAACCAATTCAGG | |
| WQE | F1, CGCGGATCC GCTGGTAATGCGGTCAATGCTC |
| R1, GAAATTATAAAATGAAGCAGTGTCATCAGCACCACCATTACCTACATACC | |
| F2, GGTATGTAGGTAATGGTGGTGCTGATGACACTGCTTCATTTTATAATTTC | |
| R2, ACATGCATGCGGTCGCCTTCAACCAATTCAGG | |
| WQF | F1, CGCGGATCCGCTGGTAATGCGGTCAATGCTC |
| R1, CCTTGAGTAGCTGTCCACTGTCTACCGTTCATATAGATGCGGTC | |
| F2, GACCGCATCTATATGAACGGTAGACAGTGGACAGCTACTCAAGG | |
| R2, ACATGCATGCGGTCGCCTTCAACCAATTCAGG | |
| Fcheckb | AATCAGGTCGTGTATTCAGTC |
| Rcheckb | ACCGACCGTTCATATTACTCA |
Primers were for amplifying fragments for recombination. F, forward; R, reverse.
Control primer.
Homologous recombination of different gene 37 sections from WG01 with the corresponding sections from QL01.
Homologous recombination was carried out as described previously (22) with some modifications. Strain DE017 was transformed with the recombinant plasmid, which contained different parts of gene 37 of QL01. The transformant cells were incubated in 10 ml of LB broth with 50 μg/ml ampicillin at 28°C with shaking (120 rpm). WG01 phage infection was performed at an optical density at 600 nm (OD600) of 0.4. After a 4-h incubation, the culture was centrifuged to remove cell debris. The lysate was mixed with DE205B in 0.5% agar and overlaid on an LB plate at 28°C. The single clear plaque was transferred into the SM buffer and used for the plaque assay. The same procedure was repeated three times to purify the recombinant phage.
The sequencing analysis of gene 37 from the recombinant phages was performed with the corresponding primers, Fcheck, and Rcheck (Table 2).
Isolation of recombinant phages from the same chimeric parent of WQT.
The sequencing analysis of the recombinant phages WQT1, WQC1, WQD1, and QE1 revealed that nucleic acid mutations that occurred in the WQT1 recombinant phage caused amino acid substitutions. Thus, the other 31 phage isolates from the DE017 cells carrying plasmids containing the QT insert and that were infected with WG01 were selected and used to perform the plaque assay, purification, and sequencing. The sequences of the mutants were compared with that of WQT to observe which nucleic acid sites generated mutations.
Comparison of the host ranges of two wild-type strains (WG01 and QL01) and WG01 derivatives.
A total of 113 isolates (Table S1) were used to test the host ranges of phages in this study according to a previous article (37) with some modifications. Briefly, the bacterial cultures during log phase (100 μl) were spread on an LB agar plate and allowed to dry. Phage suspensions (10 μl at 109 PFU/ml) were dropped into the center of each square and allowed to dry. Following an 8-h incubation at 28°C, the plates were examined for lysis.
Comparison of the growth characteristics of two wild-type strains (WG01 and QL01) and WG01 derivatives.
On the lawn of DE205B cells, the WG01, QL01, and WG01 derivatives were mixed with DE205B (simultaneously, WG01 was mixed with DE017) in 0.5% agar and overlaid on LB plates at 42°C, 37°C, 28°C, 16°C, and 4°C. The phage plaque formation and phage sizes were observed.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (no. 31322054), the Central University Basic Scientific Research Fund “Animal pathogenic bacteria” (KJYQ201402), the Priority Academic Program Development of Jiangsu Higher Education Institutions, the Natural Science Foundation of Jiangsu Province (no. BK20141363), and the Special Fund for Public Welfare Industry of Chinese Ministry of Agriculture (no. 201303041).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01576-17.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Newitt S, MacGregor V, Robbins V, Bayliss L, Chattaway MA, Dallman T, Ready D, Aird H, Puleston R, Hawker J. 2016. Two linked enteroinvasive Escherichia coli outbreaks, Nottingham, UK, June 2014. Emerg Infect Dis 22:1178–1184. doi: 10.3201/eid2207.152080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim KS. 2012. Current concepts on the pathogenesis of Escherichia coli meningitis: implications for therapy and prevention. Curr Opin Infect Dis 25:273–278. doi: 10.1097/QCO.0b013e3283521eb0. [DOI] [PubMed] [Google Scholar]
- 5.Ghunaim H, Abu-Madi MA, Kariyawasam S. 2014. Advances in vaccination against avian pathogenic Escherichia coli respiratory disease: potentials and limitations. Vet Microbiol 172:13–22. doi: 10.1016/j.vetmic.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Diarra MS, Malouin F. 2014. Antibiotics in Canadian poultry productions and anticipated alternatives. Front Microbiol 5:282. doi: 10.3389/fmicb.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Kraker ME, Davey PG, Grundmann H. 2011. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med 8:e1001104. doi: 10.1371/journal.pmed.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fruth A, Prager R, Tietze E, Rabsch W, Flieger A. 2015. Molecular epidemiological view on Shiga toxin-producing Escherichia coli causing human disease in Germany: diversity, prevalence, and outbreaks. Int J Med Microbiol 305:697–704. doi: 10.1016/j.ijmm.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Mellata M. 2013. Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog Dis 10:916–932. doi: 10.1089/fpd.2013.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobsen L, Spangholm DJ, Pedersen K, Jensen LB, Emborg HD, Agerso Y, Aarestrup FM, Hammerum AM, Frimodt-Moller N. 2010. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int J Food Microbiol 142:264–272. doi: 10.1016/j.ijfoodmicro.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Brüssow H. 2012. What is needed for phage therapy to become a reality in Western medicine? Virology 434:138–142. doi: 10.1016/j.virol.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Keen EC. 2012. Phage therapy: concept to cure. Front Microbiol 3:238. doi: 10.3389/fmicb.2012.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittebole X, De Roock S, Opal SM. 2014. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5:226–235. doi: 10.4161/viru.25991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson RP, Gyles CL, Huff WE, Ojha S, Huff GR, Rath NC, Donoghue AM. 2008. Bacteriophages for prophylaxis and therapy in cattle, poultry and pigs. Anim Health Res Rev 9:201–215. doi: 10.1017/S1466252308001576. [DOI] [PubMed] [Google Scholar]
- 15.Clark JR, March JB. 2006. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol 24:212–218. doi: 10.1016/j.tibtech.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Haq IU, Chaudhry WN, Akhtar MN, Andleeb S, Qadri I. 2012. Bacteriophages and their implications on future biotechnology: a review. Virol J 9:9. doi: 10.1186/1743-422X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermoso JA, Garcia JL, Garcia P. 2007. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr Opin Microbiol 10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Chan BK, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 19.Brüssow H. 2005. Phage therapy: the Escherichia coli experience. Microbiology 151:2133–2140. doi: 10.1099/mic.0.27849-0. [DOI] [PubMed] [Google Scholar]
- 20.Montag D, Hashemolhosseini S, Henning U. 1990. Receptor-recognizing proteins of T-even type bacteriophages: the receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb. J Mol Biol 216:327–334. doi: 10.1016/S0022-2836(05)80324-9. [DOI] [PubMed] [Google Scholar]
- 21.Riede I, Degen M, Henning U. 1985. The receptor specificity of bacteriophages can be determined by a tail fiber modifying protein. EMBO J 4:2343–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y. 2005. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J Biotechnol 115:101–107. doi: 10.1016/j.jbiotec.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Mahichi F, Synnott AJ, Yamamichi K, Osada T, Tanji Y. 2009. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol Lett 295:211–217. doi: 10.1111/j.1574-6968.2009.01588.x. [DOI] [PubMed] [Google Scholar]
- 24.Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol 9:224. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuber S, Ngom-Bru C, Barretto C, Bruttin A, Brussow H, Denou E. 2007. Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. J Bacteriol 189:8206–8214. doi: 10.1128/JB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashemolhosseini S, Montag D, Kramer L, Henning U. 1994. Determinants of receptor specificity of coliphages of the T4 family. A chaperone alters the host range. J Mol Biol 241:524–533. [DOI] [PubMed] [Google Scholar]
- 27.Drexler K, Dannull J, Hindennach I, Mutschler B, Henning U. 1991. Single mutations in a gene for a tail fiber component of an Escherichia coli phage can cause an extension from a protein to a carbohydrate as a receptor. J Mol Biol 219:655–663. doi: 10.1016/0022-2836(91)90662-P. [DOI] [PubMed] [Google Scholar]
- 28.Tétart F, Repoila F, Monod C, Krisch HM. 1996. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol 258:726–731. doi: 10.1006/jmbi.1996.0281. [DOI] [PubMed] [Google Scholar]
- 29.Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ. 2010. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci U S A 107:20287–20292. doi: 10.1073/pnas.1011218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu EY, Pache L, Von Seggern DJ, Mullen T, Mikyas Y, Stewart PL, Nemerow GR. 2003. Flexibility of the adenovirus fiber is required for efficient receptor interaction. J Virol 77:7225–7235. doi: 10.1128/JVI.77.13.7225-7235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura T, Sato K, Hamada H. 2003. Reduction of natural adenovirus tropism to the liver by both ablation of fiber-coxsackievirus and adenovirus receptor interaction and use of replaceable short fiber. J Virol 77:2512–2521. doi: 10.1128/JVI.77.4.2512-2521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bokiej M, Ogden KM, Ikizler MR, Reiter DM, Stehle T, Dermody TS. 2012. Optimum length and flexibility of reovirus attachment protein σ1 are required for efficient viral infection. J Virol 86:10270–10280. doi: 10.1128/JVI.01338-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaliniene L, Zajanckauskaite A, Simoliunas E, Truncaite L, Meskys R. 2015. Low-temperature bacterial viruses VR—a small but diverse group of E. coli phages. Arch Virol 160:1367–1370. doi: 10.1007/s00705-015-2388-0. [DOI] [PubMed] [Google Scholar]
- 34.Kaliniene L, Klausa V, Truncaite L. 2010. Low-temperature T4-like coliphages vB_EcoM-VR5, vB_EcoM-VR7 and vB_EcoM-VR20. Arch Virol 155:871–880. doi: 10.1007/s00705-010-0656-6. [DOI] [PubMed] [Google Scholar]
- 35.Sharma M. 2013. Lytic bacteriophages: potential interventions against enteric bacterial pathogens on produce. Bacteriophage 3:e25518. doi: 10.4161/bact.25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J, Sun M, Bao Y, Pan Z, Zhang W, Lu C, Yao H. 2013. Genetic diversity and features analysis of type VI secretion systems loci in avian pathogenic Escherichia coli by wide genomic scanning. Infect Genet Evol 20:454–464. doi: 10.1016/j.meegid.2013.09.031. [DOI] [PubMed] [Google Scholar]
- 37.Chen M, Xu J, Yao H, Lu C, Zhang W. 2016. Isolation, genome sequencing and functional analysis of two T7-like coliphages of avian pathogenic Escherichia coli. Gene 582:47–58. doi: 10.1016/j.gene.2016.01.049. [DOI] [PubMed] [Google Scholar]
- 38.Chen MM, Zhang L, Xin SP, Yao HC, Lu CP, Zhang W. 2017. Inducible prophage mutant of Escherichia coli can lyse new host and the key sites of receptor recognition identification. Front Microbiol 8:147. doi: 10.3389/fmicb.2017.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai J, Wang S, Guerlebeck D, Laturnus C, Guenther S, Shi Z, Lu C, Ewers C. 2010. Suppression subtractive hybridization identifies an autotransporter adhesin gene of E. coli IMT5155 specifically associated with avian pathogenic Escherichia coli (APEC). BMC Microbiol 10:236. doi: 10.1186/1471-2180-10-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams MH. 1959. Bacteriophages. Interscience Publishers, New York, NY. [Google Scholar]
- 41.Xu J, Chen M, He L, Zhang S, Ding T, Yao H, Lu C, Zhang W. 2016. Isolation and characterization of a T4-like phage with a relatively wide host range within Escherichia coli. J Basic Microbiol 56:405–421. doi: 10.1002/jobm.201500440. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 44.Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. 2005. WebACT–an online companion for the Artemis Comparison Tool. Bioinformatics 21:3665–3666. doi: 10.1093/bioinformatics/bti601. [DOI] [PubMed] [Google Scholar]
- 45.Arnold K, Bordoli L, Kopp J, Schwede T. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.