ABSTRACT
Thiobencarb is a thiocarbamate herbicide used in rice paddies worldwide. Microbial degradation plays a crucial role in the dissipation of thiobencarb in the environment. However, the physiological and genetic mechanisms underlying thiobencarb degradation remain unknown. In this study, a novel thiobencarb degradation pathway was proposed in Acidovorax sp. strain T1. Thiobencarb was oxidized and cleaved at the C—S bond, generating diethylcarbamothioic S-acid and 4-chlorobenzaldehyde (4CDA). 4CDA was then oxidized to 4-chlorobenzoic acid (4CBA) and hydrolytically dechlorinated to 4-hydroxybenzoic acid (4HBA). The identification of catabolic genes suggested further hydroxylation to protocatechuic acid (PCA) and finally degradation through the protocatechuate 4,5-dioxygenase pathway. A novel two-component monooxygenase system identified in the strain, TmoAB, was responsible for the initial catabolic reaction. TmoA shared 28 to 32% identity with the oxygenase components of pyrimidine monooxygenase from Agrobacterium fabrum, alkanesulfonate monooxygenase from Pseudomonas savastanoi, and dibenzothiophene monooxygenase from Rhodococcus sp. TmoB shared 25 to 37% identity with reported flavin reductases and oxidized NADH but not NADPH. TmoAB is a flavin mononucleotide (FMN)-dependent monooxygenase and catalyzed the C—S bond cleavage of thiobencarb. Introduction of tmoAB into cells of the thiobencarb degradation-deficient mutant T1m restored its ability to degrade and utilize thiobencarb. A dehydrogenase gene, tmoC, was located 7,129 bp downstream of tmoAB, and its transcription was clearly induced by thiobencarb. The purified TmoC catalyzed the dehydrogenation of 4CDA to 4CBA using NAD+ as a cofactor. A gene cluster responsible for the complete 4CBA metabolic pathway was also cloned, and its involvement in thiobencarb degradation was preliminarily verified by transcriptional analysis.
IMPORTANCE Microbial degradation is the main factor in thiobencarb dissipation in soil. In previous studies, thiobencarb was degraded initially via N-deethylation, sulfoxidation, hydroxylation, and dechlorination. However, enzymes and genes involved in the microbial degradation of thiobencarb have not been studied. This study revealed a new thiobencarb degradation pathway in Acidovorax sp. strain T1 and identified a novel two-component FMN-dependent monooxygenase system, TmoAB. Under TmoAB-mediated catalysis, thiobencarb was cleaved at the C—S bond, producing diethylcarbamothioic S-acid and 4CDA. Furthermore, the downstream degradation pathway of thiobencarb was proposed. Our study provides the physiological, biochemical, and genetic foundation of thiobencarb degradation in this microorganism.
KEYWORDS: thiobencarb, degradation pathway, two-component FMN-dependent monooxygenase system, C—S bond cleavage, Acidovorax sp. T1
INTRODUCTION
Thiobencarb (S-4-chlorobenzyl N,N-diethylthiocarbamate) is a highly effective thiocarbamate herbicide that is widely used in rice paddies worldwide to control annual grasses and broadleaf weeds (1). However, the long-term application of thiobencarb has led to many negative effects on farmland ecosystems and aquatic environments (2–4) because thiobencarb is highly toxic to invertebrates and moderately toxic to fish (5–7). Thiobencarb also inhibits the growth of microbes, reducing their number and diversity (8). In addition, its reductive dechlorination product, deschlorothiobencarb, is 10 times more toxic than thiobencarb (9), often causing severe rice dwarfing with subsequent yield loss (10).
Aerobic and anaerobic microorganisms play crucial roles in the dissipation of thiobencarb in soil (11). To date, two aerobic bacterial strains capable of degrading thiobencarb have been reported, including an Aspergillus niger strain (12) and a Corynebacterium sp. strain (13). However, the thiobencarb metabolic pathway and related genes in these strains have not been elucidated. No anaerobic thiobencarb transformation strain has been reported. Thiobencarb degradation pathways mediated by soil microbes (11, 14) have been proposed via the identification of metabolites. In aerobic soil, thiobencarb degradation is initiated through three mechanisms: N-deethylation to produce deethylated thiobencarb, followed by further N-deethylation to generate 4-cholorobenzyl thiocarbamate; sulfoxidation to produce thiobencarb sulfoxide; and hydroxylation of the benzene ring to generate hydroxyl-thiobencarb (14). In anaerobic soil, thiobencarb is dechlorinated to deschlorothiobencarb by anaerobic bacteria (15). However, these hypothetical thiobencarb degradation pathways in soil have not been physiologically or genetically confirmed using pure cultures.
In this study, an aerobic thiobencarb-utilizing bacterium, Acidovorax sp. strain T1, was isolated from soil. Acidovorax sp. T1 cleaved the C—S bond of thiobencarb to release diethylcarbamothioic S-acid and 4-chlorobenzaldehyde (4CDA), which was further dehydrogenated to 4-chlorobenzoic acid (4CBA). A monooxygenase gene cluster, tmoAB, which was responsible for the C—S cleavage of thiobencarb, and a dehydrogenase gene, tmoC, which was responsible for 4CDA dehydrogenation, were identified. This study provides a biochemical and genetic foundation for a novel microbial thiobencarb degradation pathway.
RESULTS
Identification of metabolites during thiobencarb degradation by Acidovorax sp. T1.
Strain T1 was isolated from soil from a pesticide manufacturer and utilized thiobencarb as a sole carbon, nitrogen, and sulfur source for growth and degraded 95.4% of 0.4 mM thiobencarb within 36 h of incubation (see Fig. S1 in the supplemental material). Furthermore, Acidovorax sp. T1 could degrade and utilize 4CBA, 4-hydroxybenzoic acid (4HBA), and protocatechuic acid (PCA) as sole carbon sources for growth (data not shown). The 16S rRNA gene sequence of strain T1 shared 99.2%, 99.1%, and 98.2% identity with those of Acidovorax radicis N35T, Acidovorax defluvii BSB411T, and Acidovorax delafieldii ATCC 17505T, respectively. Thus, strain T1 was preliminarily identified as Acidovorax sp.
During the degradation of thiobencarb by Acidovorax sp. T1, two peaks were detected by high-performance liquid chromatography (HPLC) in samples taken between 6 h and 18 h, and the two peaks disappeared as incubation progressed, indicating further degradation of the two metabolites. The retention times for the two metabolites were 3.82 and 5.02 min, which were equal to the retention times of the authentic 4CBA (3.84 min) and 4CDA (5.02 min) standards (see Fig. S2 in the supplemental material). In gas chromatography-mass spectrometry (GC-MS) analysis, two metabolites with retention times of 7.56 min and 9.57 min were detected (Fig. 1A). The mass spectrum of the 7.56-min metabolite was typical for a molecule containing one chlorine, with a prominent protonated molecular ion at m/z 139.9/142.1 and fragments at m/z 139.0/141.0 (loss of a hydrogen atom at —CHO) and 111.0/113.1 (loss of —CHO). This metabolite and its fragments showed two peaks because chlorine atoms have two isotopes (35Cl and 37Cl), and their relative intensities were consistent with the natural abundances of 76% 35Cl and 24% 37Cl (Fig. 1B). Thus, based on the HPLC and GC-MS analyses, the metabolite at 7.56 min was identified as 4CDA. The mass spectrum of the 9.57-min metabolite was also typical for a molecule containing one chlorine, with a protonated molecular ion at m/z 228.1/230.1 [4CBA with a group of —Si(CH3)3] and prominent ion fragments at 213.1/215.0 (loss of —CH3), 139.0/140.9 [loss of —OSi(CH3)3], and 111.0/113.0 [loss of —COOSi(CH3)3] (Fig. 1C). Thus, the metabolite at 9.57 min was identified as 4CBA.
FIG 1.
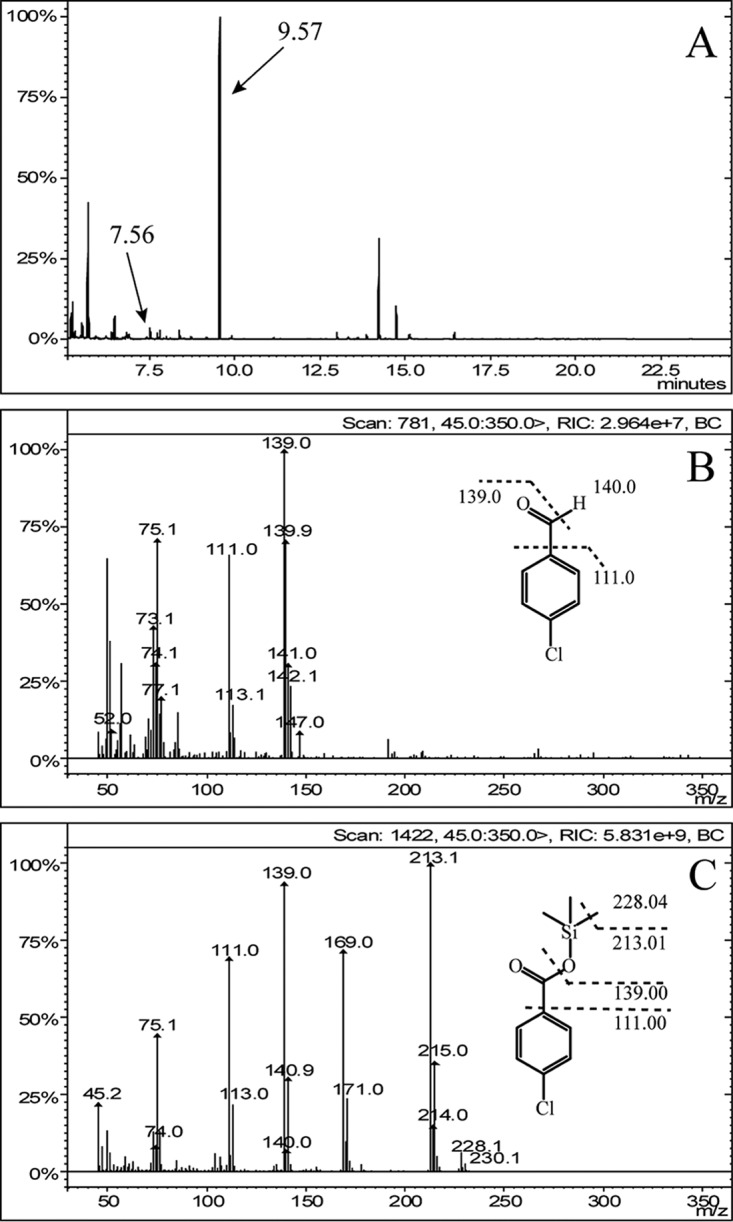
GC-MS spectra of the silanized derivatives of the intermediates during thiobencarb degradation by Acidovorax sp. T1. (A) GC spectrum of the silanized derivatives of the intermediates. (B) Mass spectrum of the product at GC retention time 7.56 min. (C) Mass spectrum of the product at GC retention time 9.57 min.
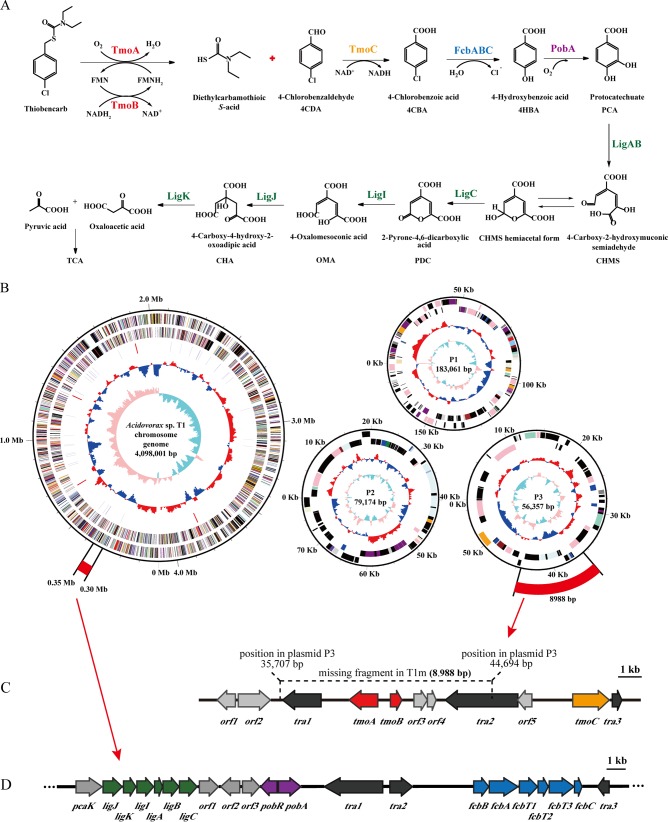
Based on the metabolite identification, we proposed a novel thiobencarb degradation pathway in Acidovorax sp. T1 (Fig. 2A). The initial step in this pathway was the cleavage of the C—S bond to form diethylcarbamothioic S-acid and 4CDA. 4CDA was then dehydrogenated to 4CBA, and 4CBA was completely degraded. In this study, the monooxygenase responsible for the C—S bond cleavage of thiobencarb was designated Tmo (thiobencarb monooxygenase).
FIG 2.
Degradation mechanism of thiobencarb in Acidovorax sp. T1. (A) Degradation pathway for thiobencarb. (B) Genome of Acidovorax sp. T1, containing one circular chromosome and three circular plasmids (P1, P2, and P3). The scale for each replicon is marked on the black scale circle. The positions of gene clusters for thiobencarb degradation and 4CBA degradation are marked by the outermost red arcs. The first and second circles from the center indicate the GC skew (G−C/G+C) and G+C content: for GC skew, the parts greater than and less than zero are colored pink and light blue, respectively; for G+C content, the parts greater than and less than zero are colored red and blue, respectively. The third circle represents rRNA (red) and tRNA (gray). The fourth and fifth circles show CDSs (coding sequences) on the negative and positive DNA strands, respectively. (C) Organization of the tmoA, tmoB, and tmoC genes in plasmid P3. (D) Organization of the lig, pob, and fcb operons in the chromosome.
Screen of the thiobencarb degradation-deficient mutant strain T1m.
Colonies of strain T1 grown on 1/5 Luria-Bertani (LB) (i.e., 0.2 times the concentration of LB) agar plates supplemented with 0.4 mM thiobencarb, which has low solubility, produced transparent circles due to thiobencarb degradation. Interestingly, very few mutant colonies (approximately 0.5%) lost the ability to produce transparent circles. There were no obvious differences in morphology and growth curves between these mutant strains and the wild-type strain in 1/5 LB medium. However, whole-cell transformation experiments showed that these mutants lost the ability to degrade thiobencarb, although they rapidly and completely degraded the thiobencarb metabolites 4CDA and 4CBA. These results indicated that the gene responsible for the initial step (C—S bond cleavage) in thiobencarb degradation was unstable and was lost without selective pressure from thiobencarb. One of the thiobencarb degradation-deficient mutants was designated T1m.
Identification of a thiobencarb monooxygenase gene via comparative genomic analysis.
The complete genome of strain T1 and the draft genome of T1m were sequenced on an Illumina HiSeq 2500 system. The complete genome (4.417 Mb) of strain T1 revealed four replicons consisting of one circular chromosome (4.098 Mb) and three circular plasmids (P1, 183.1 kb; P2, 79.2 kb; and P3, 56.4 kb) (Fig. 2B). The draft genome sequence of mutant T1m contained 233 contigs, representing a total size of 4.215 Mb. Based on a pairwise comparison, mutant T1m lost an 8,988-bp fragment located between bp 35707 and 44694 on the 56.4-kb plasmid (Fig. 2B). The loss of this DNA fragment was further confirmed by PCR (data not shown). In addition, as shown in Fig. 2C and Table 1, the fragment was flanked by two transposase genes, tra1 and tra2. This gene arrangement indicated that the 8,988-bp fragment was located on a mobile genetic element, which likely contributed to its instability in Acidovorax sp. T1.
TABLE 1.
Deduced functions of ORFs within or near the missing 8,988-bp fragment sequence
| Gene name | Position in plasmid P3/no. of amino acids | Homologous protein | Source | UniProtKB accession no. | Identity (%) |
|---|---|---|---|---|---|
| orf1 | 33174–33938/254 | Putative ATP-binding protein | Mycobacterium tuberculosis H37Rv | Q50701.1 | 29 |
| orf2 | 34061–35419/452 | Acetylglutamate kinase | Mycobacterium leprae TN | Q9CC13.2 | 32 |
| tra1 | 35901–37517/538 | Putative transposase y4qJ | Sinorhizobium fredii NGR234 | P55631.1 | 26 |
| tmoA | 38683–39894/403 | Pyrimidine monooxygenase RutA | Agrobacterium fabrum strain C58 | P58759.1 | 32 |
| tmoB | 40418–40921/167 | FMN reductase (NADH) NtaB | Aminobacter aminovorans | P54990.2 | 37 |
| orf3 | 41339–41998/199 | Biotin synthase | Rhodopseudomonas palustris CGA009 | Q6N859.1 | 30 |
| orf4 | 41973–42362/129 | Transmembrane channel-like protein 1 | Mus musculus | Q8R4P5.1 | 55 |
| tra2 | 42721–45750/1009 | Transposase for transposon Tn3926 | Escherichia coli | P13694.1 | 23 |
| orf5 | 45747–46340/197 | Serine recombinase gin | Escherichia phage D108 | Q38199.2 | 49 |
| tmoC | 48050–49597/515 | Benzaldehyde dehydrogenase | Pseudomonas putida | P43503.1 | 48 |
| tra3 | 49702–50133/143 | Transposase for IS200 | Salmonella enterica | P59696.1 | 43 |
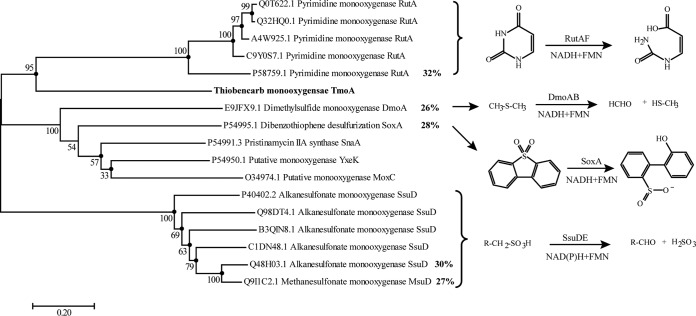
A complete physical map of the 8,988-bp fragment and its upstream and downstream genes is presented in Fig. 2C. Six putative genes were predicted, and the putative function of each gene is listed in Table 1. Among these genes, one gene, designated tmoA, was homologous to the genes encoding the oxygenase component of several two-component flavin mononucleotide (FMN)-dependent monooxygenase systems. TmoA consisted of 403 amino acids and shared 32% identity with RutA, the oxygenase component of pyrimidine monooxygenase (16) from Agrobacterium fabrum strain C58; 30% identity with SsuD, the oxygenase component of alkanesulfonate monooxygenase (17) from Pseudomonas savastanoi pv. phaseolicola 1448A; and 28% identity with SoxA, the oxygenase component of dibenzothiophene monooxygenase from Rhodococcus sp. strain IGTS8 (18). As alkanesulfonate monooxygenase and dibenzothiophene monooxygenase catalyzed the cleavage of C—S bonds, TmoA was potentially the oxygenase component of the thiobencarb monooxygenase. A flavin reductase gene, designated tmoB, was located 523 bp upstream of tmoA. TmoB consisted of 167 amino acid residues and shared 37% identity with the reductase component of nitrilotriacetate monooxygenase NtaAB from Aminobacter aminovorans (19), 34% identity with the reductase component of flavin-dependent monooxygenase HsaAB (20) from Rhodococcus jostii RHA1, 30% identity with the reductase component of 4-hydroxyphenylacetate 3-monooxygenase from Acinetobacter baumannii (21), and 29% identity with the reductase component of styrene monooxygenase StyAB from Pseudomonas fluorescens ST (22), suggesting that TmoB was the reductase component of the thiobencarb monooxygenase.
Functional complementation of tmoA and tmoB in T1m.
The fragments containing tmoA or tmoB or both (tmoAB) were ligated into the broad-host-range vector pBBR1MCS-2. The recombinant vectors were introduced into Escherichia coli DH5α and mutant T1m, respectively. Whole-cell transformation experiments revealed that tmoAB or tmoA, but not tmoB, restored the ability of mutant T1m to degrade and utilize thiobencarb (Fig. 3). Interestingly, E. coli DH5α carrying either tmoAB or tmoA also acquired the ability to degrade thiobencarb but was unable to utilize thiobencarb as a sole carbon source for growth, possibly because E. coli DH5α lacks the downstream thiobencarb degradation pathway (data not shown). These results indicated that tmoAB was responsible for the C—S bond cleavage of thiobencarb in Acidovorax sp. T1.
FIG 3.
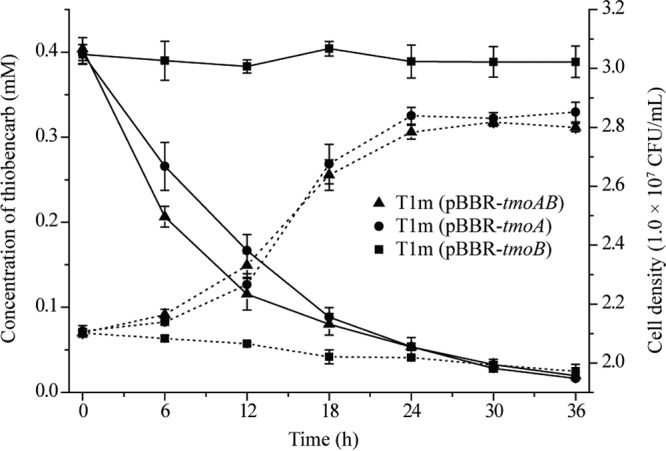
Degradation of thiobencarb by recombinant strains T1m(pBBR-tmoAB), T1m(pBBR-tmoA), and T1m(pBBR-tmoB). Shown are the time course of thiobencarb degradation by recombinant T1m(pBBR-tmoAB), T1m(pBBR-tmoA), and T1m(pBBR-tmoB) and their growth in MSM with thiobencarb as the sole source of carbon, nitrogen, and sulfur. The solid and dotted lines indicate the residual thiobencarb and strain growth, respectively. The data were derived from three independent measurements, and the error bars indicate standard deviations.
Expression of tmoA and tmoB, purification of TmoA and TmoB, and reconstruction of the thiobencarb monooxygenase in vitro.
The tmoA and tmoB genes were individually overexpressed in E. coli BL21(DE3) using the pET24b(+) expression system. The fusion proteins with a His6 tag at the C terminus were purified using a Co2+ chelating column. According to SDS-PAGE analysis, the molecular masses of the recombinant proteins His6-TmoA and His6-TmoB were approximately 46 and 20 kDa, respectively (see Fig. S3 in the supplemental material), corresponding to the deduced molecular masses of the proteins (46.67 and 19.22 kDa, respectively). TmoB oxidized NADH but not NADPH in the presence of either FMN or flavin adenine dinucleotide (FAD). When FMN was used as a cofactor, NADH oxidation activity of 13.83 U mg−1 was observed, and when FMN was replaced by FAD, only 1.93 U mg−1 activity was detected, suggesting that FMN was a more favored electron acceptor than FAD for TmoB. The Km value of TmoB for NADH with FMN as a cofactor was 33.71 ± 1.86 μM, and the catalytic efficiency (kcat/Km) was 31.65 ± 1.44 min−1 μM−1.
Individually, TmoA and TmoB did not exhibit thiobencarb-transforming activity, whereas the mixture of TmoA and TmoB transformed thiobencarb, indicating that the thiobencarb monooxygenase consisted of two components. Interestingly, the mixture of TmoA and MeaY, the reductase component of 2-methyl-6-ethylaniline monooxygenase MeaXY (23), also displayed thiobencarb-transforming activity. The results indicated that TmoA had low specificity for its partner, flavin reductase. Two products were detected in the GC-MS analysis during thiobencarb conversion by TmoAB (Fig. 4). The product with a retention time of 7.56 min was identified as 4CDA (Fig. 4B), and the amount of accumulated 4CDA (0.171 mM) was approximately equivalent to the amount of transformed thiobencarb (0.175 mM), indicating a nearly stoichiometric formation of 4CDA from thiobencarb. Another product, with a retention time of 8.52 min, was detected after silylation. This 8.52-min product was identified as diethylcarbamothioic S-acid with a —Si(CH3)3 group at the S atom based on its molecular ion peak at m/z 205.1 with fragments at m/z 147.1 (loss of two —C2H5) and m/z 133.0 [loss of —N(C2H5)2] (Fig. 4C). These results indicated that TmoAB was a two-component monooxygenase that catalyzed the cleavage of the C—S bond of thiobencarb to generate 4CDA and diethylcarbamothioic S-acid.
FIG 4.
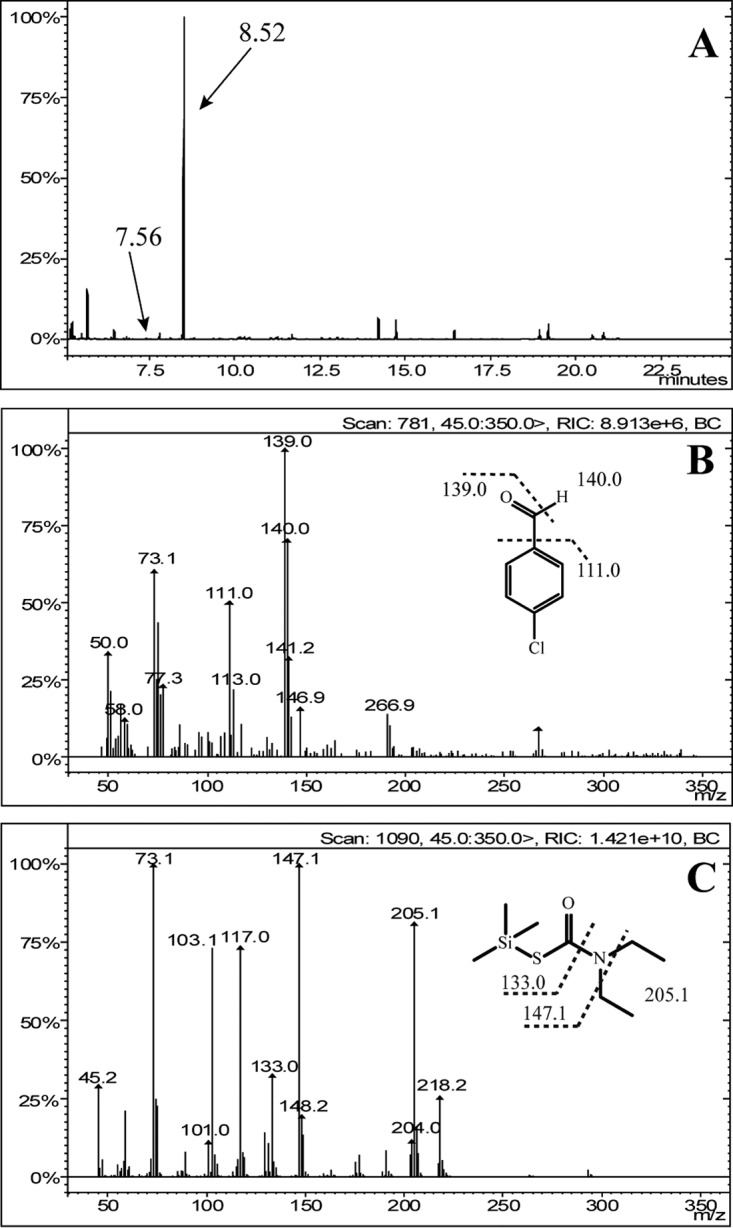
GC-MS spectra of the silanized derivatives of the metabolites generated during thiobencarb transformation by purified TmoAB. (A) GC spectrum of the silanized derivative of the metabolites. (B) Mass spectrum of the product at GC retention time 7.56 min. (C) Mass spectrum of the product at GC retention time 8.52 min.
Characterization of TmoAB.
TmoAB displayed thiobencarb monooxygenase activity in the presence of FMN but not FAD, indicating that TmoAB is an FMN-dependent monooxygenase. TmoAB was unable to transform other thiocarbamate herbicides, such as molinate, vernolate, and asulam, as well as other nitrogen and sulfur compounds, such as dimethyl sulfide, alkanesulfonates, dibenzothiophene, dibenzothiophene sulfone, and pyrimidine. Thiobencarb monooxygenase activity gradually increased with increasing concentrations of TmoB, reaching maximal activity (0.45 U mg−1) when TmoB was 0.05 μM (see Fig. S4 in the supplemental material), indicating that TmoA and TmoB had an optimal molar ratio at 20:1. The Km value of TmoAB for thiobencarb was 95.28 ± 10.81 μM, and the kcat/Km value was 0.17 ± 0.01 μM−1 min−1. TmoAB exhibited the highest catalytic activity at 20°C and pH 7.4 in 50 mM Tris-HCl (see Fig. S5 and S6 in the supplemental material). The effects of metal ions and potential enzyme inhibitors on TmoAB activity are shown in Fig. S7 in the supplemental material. TmoAB activity was severely inhibited by 1 mM Hg2+, Ni2+, Cd2+, Zn2+, and Cu2+ and moderately inhibited by 1 mM Ba2+, Mn2+, and Co2+, as well as 5 mM dithiothreitol (DTT) or EDTA. In particular, TmoAB activity increased 2.62-fold in the presence of catalase compared with that in a reaction without catalase, possibly because catalase removed the H2O2 produced in the reaction.
tmoC encodes a 4CDA dehydrogenase.
The second step in the thiobencarb degradation pathway was the dehydrogenation of 4CDA to 4CBA. Open reading frame (ORF) analysis revealed that a putative dehydrogenase gene, designated tmoC, was located 7,129 bp downstream of tmoAB. TmoC consisted of 515 amino acid residues and exhibited 48% and 42% amino acid sequence similarities with the benzaldehyde dehydrogenase XylC from Pseudomonas putida (24) and the 4-hydroxybenzaldehyde dehydrogenase PHBDD from Pseudomonas putida NCIMB 9866 (25), respectively. As 4CDA had structural similarity to both benzaldehyde and 4-hydroxybenzaldehyde, tmoC potentially encoded the 4CDA dehydrogenase. TmoC was overexpressed in E. coli BL21(DE3) as a C-terminally His6-tagged fusion protein and was purified using a Co2+ chelating column. SDS-PAGE analysis showed that the molecular mass of His6-TmoC was approximately 56 kDa (see Fig. S3 in the supplemental material), matching the calculated mass of the tagged protein (56.19 kDa). Purified TmoC rapidly transformed 4CDA to 4CBA (12.80 U/mg) using NAD+ as a cofactor (see Fig. S8 in the supplemental material), suggesting that tmoC encoded the 4CDA dehydrogenase. The Km values of TmoC for NAD+ and 4CDA were 28.44 ± 0.54 μM and 103.50 ± 11.76 μM, respectively, and the kcat/Km values for NAD+ and 4CDA were 17.98 ± 0.73 μM−1 min−1 and 10.18 ± 0.87 μM−1 min−1, respectively.
Verification of gene transcriptional expression.
Real-time quantitative PCR (RT-qPCR) was performed to compare the mRNA levels of the genes tmoA, tmoB, and tmoC in cells grown on thiobencarb or glucose. The data in Fig. 5 show that there were no distinct differences in the transcription levels of tmoA and tmoB between cells grown on thiobencarb versus glucose, suggesting that tmoA and tmoB were constitutively transcribed. The transcription of tmoC in cells grown on thiobencarb increased 7.8-fold compared with cells grown on glucose (Fig. 5). Genome analysis revealed the presence of an approximately 36-kb DNA fragment containing three metabolic operons in the chromosome: an fcb operon responsible for the conversion of 4CBA to 4HBA (26), a pob operon responsible for the conversion of 4HBA to PCA (27), and a lig operon responsible for PCA catabolism (28) (Fig. 2B and D and Table 2). These three metabolic operons, which were highly conserved in sequence and function, formed a complete 4CBA degradation pathway (Fig. 2A). To investigate whether the three operons were involved in thiobencarb degradation, transcriptional analyses were carried out. The RT-qPCR results showed that the relative expression levels of fcbB, pobA, and ligB under thiobencarb-induced conditions were 37.3-, 5.5-, and 3.6-fold higher than those under uninduced conditions (Fig. 5). These results indicated that the three operons were involved in thiobencarb degradation. Furthermore, the induction effect of 4CDA, the cleavage product of thiobencarb, was also studied. The results showed that the transcription levels of tmoC, fcbB, and pobA in cells grown on 4CDA were 5.5-, 48.2-, and 7.1-fold higher than those of glucose-grown cells (see Fig. S9 in the supplemental material). Considering that thiobencarb is a xenobiotic and is not the direct substrate of the enzymes involved in the downstream pathway, the increase in the transcription level of downstream metabolic genes in thiobencarb-grown cells was most likely due to the induction effect of 4CDA or other intermediates.
FIG 5.
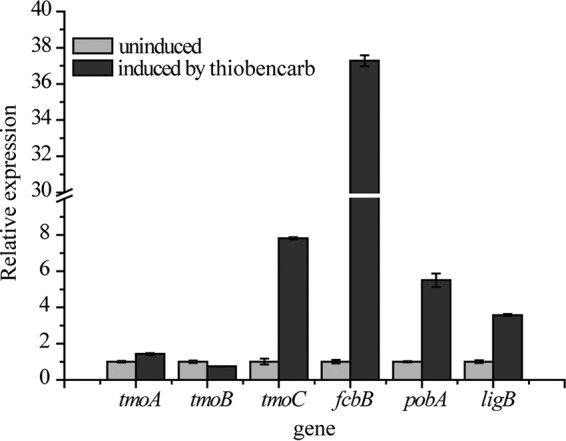
Transcriptional levels of tmoA, tmoB, tmoC, fcbB, pobA, and ligB in Acidovorax sp. T1 with or without thiobencarb induction. Relative expression levels were calculated using the 2−ΔΔCT threshold cycle (CT) method with the 16S rRNA gene used as the reference gene. The data were derived from three independent measurements, and the error bars indicate standard deviations.
TABLE 2.
Deduced function of each ORF within the bp 307519–343106 fragment (GenBank accession no. MF189566) of the T1 chromosome
| Gene name | Gene position in fragment MF189566/no. of amino acids | Homologous protein | Source | UniProtKB accession no. | % identity |
|---|---|---|---|---|---|
| pcaK | 4040–5416/458 | 4-Hydroxybenzoate transporter PcaK | Acinetobacter sp. strain ADP1 | Q43975.3 | 46 |
| ligJ | 5447–6475/342 | 2-Amino-3-carboxymuconate-6-semialdehyde decarboxylase | Rattus norvegicus (Norway rat) | Q8R5M5.1 | 22 |
| ligK | 6505–7188/227 | Oxaloacetate decarboxylase | Pseudomonas straminea NGJ1 | Q9AQI0.1 | 92 |
| ligI | 7202–8119/305 | 2-Pyrone-4,6-dicarboxylate hydrolase | Comamonas testosteroni BR6020 | Q93PS7.1 | 90 |
| ligA | 8122–8565/147 | Protocatechuate 4,5-dioxygenase alpha chain | Pseudomonas paucimobilis | P22635.1 | 65 |
| ligB | 8566–9432/288 | Protocatechuate 4,5-dioxygenase beta chain | P. paucimobilis | P22636.1 | 65 |
| ligC | 9429–10388/319 | 4-Carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase | P. paucimobilis | Q9KWL3.1 | 77 |
| orf1 | 10478–11491/337 | Probable aldo-keto reductase | Arabidopsis thaliana (thale cress) | Q84M96.1 | 44 |
| orf2 | 11580–12581/333 | UPF0065 protein BP0148 | Bordetella pertussis Tohama I | O30446.1 | 30 |
| orf3 | 12720–13655/311 | Nitrogen assimilation regulatory protein Nac | Klebsiella aerogenes | Q08597.1 | 85 |
| pobR | 13671–14519/282 | p-Hydroxybenzoate hydroxylase transcriptional activator | Acinetobacter sp. strain ADP1 | Q43992.1 | 50 |
| pobA | 14601–15776/391 | 4-Hydroxybenzoate 3-monooxygenase | Pseudomonas fluorescens | P00438.2 | 67 |
| tra1 | 16973–20053/1026 | Transposase for insertion sequence element IS1071 in transposon Tn5271 | Comamonas testosteroni | Q04222.1 | 99 |
| tra2 | 20413–21618/401 | Putative transposase y4bL/y4kJ/y4tB | Sinorhizobium fredii NGR234 | P55379.1 | 31 |
| fcbB | 24792–25601/269 | 4-Chlorobenzoyl-CoAa dehalogenase | Pseudomonas sp. strain CBS3 | A5JTM5.1 | 86 |
| fcbA | 25613–27130/505 | 4-Chlorobenzoyl-CoA ligase | Pseudomonas sp. CBS3 | A5JTM6.1 | 58 |
| fcbT1 | 27175–28155/326 | Solute-binding protein Bpro 4736 | Polaromonas sp. strain JS666 | Q122C7.1 | 26 |
| fcbT2 | 28152–28709/185 | Putative TRAP transporter small permease protein HI 1030 | Haemophilus influenzae Rd KW20 | P44994.1 | 27 |
| fcbT3 | 28722–30041/439 | Putative TRAP transporter large permease protein HI 0050 | Haemophilus influenzae Rd KW20 | P44483.1 | 28 |
| fcbC | 30044–30472/142 | 4-Hydroxybenzoyl-CoA thioesterase | Pseudomonas sp. CBS3 | P56653.1 | 65 |
| tra3 | 31255–31869/204 | Probable transposase for insertion element | Shigella flexneri | Q79CE8.1 | 64 |
CoA, coenzyme A.
DISCUSSION
Previous studies have demonstrated that the dissipation of thiobencarb in soil occurs mainly through microbial degradation, and four hypothetical degradation mechanisms have been proposed: N-deethylation, sulfoxidation, and hydroxylation under aerobic conditions and dechlorination under anaerobic conditions. However, these hypothetical degradation mechanisms have not yet been confirmed in pure culture. In this study, we proposed a novel thiobencarb degradation pathway in an isolated strain, Acidovorax sp. T1. Unlike the four proposed degradation mechanisms, the initial step of thiobencarb degradation in the strain was the cleavage of the C—S bond, producing diethylcarbamothioic S-acid and 4CDA.
The phenomenon where degradative strains lose the ability to degrade target compounds has been extensively reported (23, 29–31). The most likely reason is that the degradation gene is located on a degradative plasmid or on a mobile genetic element, which are prone to loss when cells are subcultured on complete medium without selective pressure. In our study, we found it was easy to screen thiobencarb degradation-deficient mutants; approximately 0.5% of colonies lost the ability to produce transparent circles on a 1/5 LB agar plate supplemented with thiobencarb. Genome analysis revealed that tmoAB was located on an 8,988-bp mobile genetic element in plasmid P3. In mutant T1m, the lack of tmoAB was due to the loss of the 8,988-bp mobile genetic element rather than the whole plasmid. Our unpublished data showed that in seven other thiobencarb degradation-deficient mutants, the 8,988-bp mobile genetic element was also lost, whereas the rest of plasmid P3 remained. This evidence indicated that the reason why Acidovorax sp. T1 was prone to losing the ability to degrade thiobencarb was most likely the loss of the transposable element containing tmoAB rather than the complete loss of the degradative plasmid.
The two-component FMN-dependent monooxygenase system catalyzes a diverse range of reactions, such as the hydroxylation of aromatic compounds and long-chain alkanes, the biosynthesis of antibiotics and bioluminescence, and the desulfurization of sulfonated compounds. In this system, the oxygenase component utilizes the reduced flavin supplied by the reductase component to oxidize the substrate. The two-component FMN-dependent monooxygenase system is characterized by its specificity for FMN and the formation of a TIM barrel fold consisting of eight α-helices and eight parallel β-strands. In this study, a monooxygenase system that catalyzed the cleavage of the C—S bond of thiobencarb was identified in Acidovorax sp. T1 and characterized. The thiobencarb monooxygenase consisted of two components: an oxygenase (TmoA) and a flavin reductase (TmoB) that used FMN, but not FAD, as a cofactor. Sequence alignment and structure prediction results suggested TmoA had a classical TIM barrel structure and contained the conserved sites for flavin binding (Val164, Arg185, Tyr186, and Ser238) (32–34) (see Fig. S10 in the supplemental material). Furthermore, TmoB has the conserved SXXPP and GDH motifs of the HpaC-like flavin reductase subfamily (35, 36) (see Fig. S11 in the supplemental material). This evidence indicated that TmoAB was a two-component FMN-dependent monooxygenase system. TmoA shared homology with the oxygenase components of certain two-component FMN-dependent monooxygenases that catalyzed the cleavage of N—C (16, 37, 38) or S—C (39–41) bonds in nitrogen or sulfur heterocyclic compounds, such as pyrimidine, alkanesulfonate, and dibenzothiophene. However, the identities between TmoA and these oxygenases were very low (less than 32%), and TmoA formed a separate branch in the neighbor-joining (NJ), maximum-likelihood (ML), and minimum evolution (ME) trees constructed based on the oxygenase components of related monooxygenases (Fig. 6). Furthermore, TmoAB was unable to transform pyrimidine, alkanesulfonate, and dibenzothiophene, which are the substrates of related monooxygenases. Thus, TmoAB appears to be a novel two-component FMN-dependent monooxygenase system.
FIG 6.
Phylogenetic tree constructed based on the alignment of TmoA with the oxygenase components of certain related two-component flavin-dependent monooxygenases. Multiple-alignment analysis was performed with ClustalX v2.0, and the phylogenetic tree was constructed with the NJ, ML, and ME algorithms using MEGA 7.0; bootstrap values (based on 1,200 replications) are indicated at branch nodes. The solid circles indicate that the corresponding branches were also recovered using the ML and ME algorithms. Bar, 0.20 substitutions per nucleotide position. Each item was arranged in the following order: UniProtKB accession number and protein name.
Bioinformatic analysis revealed that the genome of Acidovorax sp. T1 contained tmoAB; tmoC; and the fcb, pob, and lig operons, which together formed a complete thiobencarb degradation pathway in silico (Fig. 2A). RT-qPCR results indicated that the transcription of tmoC, fcbB, pobA, and ligB was significantly increased in thiobencarb-grown cells of Acidovorax sp. T1. Furthermore, Acidovorax sp. T1 could utilize 4CBA, 4HBA, and PCA. Based on the above-mentioned evidence, we proposed a complete thiobencarb degradation pathway in Acidovorax sp. T1: thiobencarb is cleaved at the C—S bond, generating diethylcarbamothioic S-acid and 4CDA, which is further dehydrogenated to 4CBA; 4CBA is subsequently transformed to 4HBA and further degraded via the PCA 4,5-cleavage pathway to pyruvic acid and oxaloacetic acid, which are metabolized through the citric acid cycle (Krebs cycle). The diethylcarbamothioic S-acid moiety contained a nitrogen atom and a sulfur atom, and structurally, the nitrogen atom was located in the center of the diethylcarbamothioic S-acid. Acidovorax sp. T1 utilized thiobencarb as the sole carbon, nitrogen, and sulfur source for growth, indicating that the generated diethylcarbamothioic S-acid was completely decomposed and the nitrogen and sulfur were released as mineral nutrients. Further study is needed to elucidate the metabolism of diethylcarbamothioic S-acid in Acidovorax sp. T1.
MATERIALS AND METHODS
Chemicals and media.
Thiobencarb (96% purity) was purchased from J&K Scientific Ltd. (Shanghai, China). 4CDA (99% purity), 4CBA (99% purity), and silylation reagent {99% BSTFA [bis(trimethylsilyl)trifluoroacetamide] plus 1% TMCS [chlorotrimethylsilane]} were obtained from Aladdin Co., Ltd. (Shanghai, China). Methanol (chromatography grade) and acetic acid (analytical grade) were purchased from Shanghai Chemical Reagent Co., Ltd. (Shanghai, China). 1/5 Luria-Bertani (LB) medium contained 2 g liter−1 tryptone, 2 g liter−1 NaCl, and 1 g liter−1 yeast extract. Mineral salts medium (MSM) contained the following components (per liter): 1 g of NaCl, 1.5 g of K2HPO4, 0.5 g of KH2PO4, and 0.2 g of MgCl2, pH 7.0.
Strains, vectors, and culture conditions.
The bacterial strains and vectors used in this study are listed in Table 3, and the primers are listed in Table 4. Strain T1 was grown at 30°C in 1/5 LB or MSM containing 0.4 mM thiobencarb. E. coli strains were grown at 37°C in LB broth supplemented with appropriate antibiotics, as indicated in Table 3.
TABLE 3.
Strains and vectors used in this study
| Straina or vector | Relevant characteristic(s)b | Source or reference |
|---|---|---|
| Strains | ||
| Acidovorax sp. T1 (= CCTCC M2016205) | Degrades thiobencarb | This study |
| Acidovorax sp. T1m | Mutant of T1; degrades 4CBA and 4CDA, but not thiobencarb | This study |
| E. coli DH5α | Host strain for cloning vector | TaKaRa |
| E. coli BL21(DE3) | Host strain for expression vector | TaKaRa |
| Vectors | ||
| pMD19-T | TA clone vector; Ampr | TaKaRa |
| pBBR-tmoA | pBBR1MCS-2 derivative carrying tmoA; Kmr | This study |
| pBBR-tmoAB | pBBR1MCS-2 derivative carrying tmoAB; Kmr | This study |
| pBBR-tmoB | pBBR1MCS-2 derivative carrying tmoB; Kmr | This study |
| pET24b(+) | Expression vector; Kmr | Novagen |
| pET-tmoA | pET24b(+) derivative carrying tmoA; Kmr | This study |
| pET-tmoB | pET24b(+) derivative carrying tmoB; Kmr | This study |
| pET-tmoC | pET24b(+) derivative carrying tmoC; Kmr | This study |
CCTCC, China Center for Type Culture Collection.
Kmr, kanamycin resistant; Ampr, ampicillin resistant.
TABLE 4.
PCR primers used in this study
| Primer | DNA sequence (5′–3′)a | Purpose |
|---|---|---|
| 27F | AGAGTTTGATCCTGGCTC AG | Amplification of 16S rRNA |
| 1492R | TACGGCTACCTTGTTACGACT T | |
| pBBR-KpnI-tmoA-F | GGGGTACCTCCACTGAATTGTCAGTCAAAG | Amplification of tmoA for gene functional verification |
| pBBR-XbaI-tmoA-R | GCTCTAGAAATCACATCGCGCAGGCCGCGTTCA | |
| pBBR-KpnI-tmoAB-F | GGGGTACCGCACTCCCATTCCTATTTCG | Amplification of fragment containing gene tmoA and tmoB for gene functional verification |
| pBBR-XbaI-tmoAB-R | GCTCTAGAGACCTGGCGTAGCTCATCC | |
| pBBR-KpnI-tmoB-F | GGGGTACCACAACAATTGAGCTTCAGCCTG | Amplification of tmoB for gene functional verification |
| pBBR-XbaI-tmoB-R | GCTCTAGATGCGGTGCTGAGTTGGCGGAATGAG | |
| pET-NdeI-tmoA-F | GGAATTCCATATGTCCACTGAATTGTCAGTCAAAG | Amplification of tmoA for expression in E. coli BL21 |
| pET-SalI-tmoA-R | ACGCGTCGACAATCACATCGCGCAGGCCGCGTTCA | |
| pET-NdeI-tmoB-F | GGAATTCCATATGACAACAATTGAGCTTCAGCCTG | Amplification of tmoB for expression in E. coli BL21 |
| pET-SalI-tmoB-R | ACGCGTCGACTGCGGTGCTGAGTTGGCGGAATGAG | |
| pET-NdeI-tmoC-F | GGAATTCCATATGTTTTTCATCGCAGCCAATGACG | Amplification of tmoC for expression in E. coli BL21 |
| pET-XhoI-tmoC-R | CCGCTCGAGCAGTGGGTAGGCCGGTGCTTCA | |
| RT-16S-F | GCGAACGGGTGAGTAATACATC | Amplification of a 159-bp fragment of 16S rRNA by RT-qPCR |
| RT-16S-R | TCCCACCAACTACCTAATCTGC | |
| RT-tmoA-F | AGCCCAGCGAAGTCAAGAT | Amplification of a 101-bp fragment of tmoA by RT-qPCR |
| RT-tmoA-R | GTCCCACAACTGTTTCACCAT | |
| RT-tmoB-F | CCGATGAGCATGTCAAGGC | Amplification of a 150-bp fragment of tmoB by RT-qPCR |
| RT-tmoB-R | CAGGGAATGTCTGATAGATGGAAC | |
| RT-tmoC-F | TTTTCGAACCGACCGTGCTC | Amplification of a 114-bp fragment of tmoC by RT-qPCR |
| RT-tmoC-R | CAACGGCCTCTTCGTCGGTA | |
| RT-fcbB-F | TGTGAAGAGGCCGGTGTTGG | Amplification of a 158-bp fragment of fcbB by RT-qPCR |
| RT-fcbB-R | TGTAGCTCGTCGCCGTGTCATT | |
| RT-pobA-F | GCATGAACTCACGGGTGGCA | Amplification of a 178-bp fragment of pobA by RT-qPCR |
| RT-pobA-R | CTGCTCGCCGTCCTTGTGATAG | |
| RT-ligB-F | GTTTGCGGTCAACGTGGTGC | Amplification of a 104-bp fragment of ligB by RT-qPCR |
| RT-ligB-R | TGAGGTCCTCGTCATAGCTTTCC |
Restriction sites are underlined.
Isolation and identification of the thiobencarb-degrading strain and its mutant.
The thiobencarb-degrading strain was isolated using the conventional enrichment technique described by Zhang et al. (42) with minor modifications. The soil sample used as the initial inoculant was collected from Zhenjiang Jiangsu Pesticide Chemical Co., Ltd. (32°10′23.27″N, 119°36′22.16″E) in Jiangsu Province, China. A 1.0-g soil sample was added to a 250-ml Erlenmeyer flask containing 50 ml MSM supplemented with 0.4 mM thiobencarb. After incubation at 180 rpm and 30°C for 5 days, 5 ml of the enrichment culture was transferred to another 50 ml fresh enrichment medium and incubated for another 5 days. After four rounds of transfer, the enrichment culture was serially diluted and spread on a 1/5 LB agar plate supplemented with 100 mg liter−1 thiobencarb. The strain that produced a transparent zone around the colony was purified via repeated plate streaking and tested for its ability to degrade thiobencarb. The isolate was characterized and identified by 16S rRNA gene sequence analysis according to the method described by Chu et al. (43).
Biotransformation and intermediate identification.
The thiobencarb-degrading strain was cultured in 1/5 LB broth, and cells were harvested at mid-exponential phase by centrifugation (2,600 × g; 10 min) at 4°C, washed twice using fresh MSM to remove nutrients, and resuspended in MSM as the inoculum. The cells were then inoculated into 50 ml of MSM supplemented with 0.4 mM thiobencarb and incubated at 30°C and 180 rpm on a rotary shaker. A 2-ml culture aliquot was sampled every 6 h to monitor cell growth and the concentrations of thiobencarb and its metabolites. Cell growth was detected by the colony-counting method, and the thiobencarb concentration was determined by HPLC as described in “Analytical methods” below. The metabolites were identified by GC-MS. Each treatment was performed in triplicate. Control experiments without inoculation or thiobencarb were performed under the same conditions.
Sequencing, assembly, annotation, and genome comparison.
DNA manipulation was performed according to standard protocols as described by Sambrook and Russell (44). To sequence the genomes of strain T1 and mutant T1m, 300- to 500-bp shotgun libraries were constructed and sequenced on an Illumina HiSeq system, and 8- to 10-kb shotgun libraries were constructed and sequenced on a PacBio RS system. Sequencing reads were assembled using SOAP de novo software (version 2.04) (http://soap.genomics.org.cn/soapdenovo.html). De novo gene prediction was performed using the Glimmer (version 3.0) system (http://ccb.jhu.edu/software/glimmer/index.shtml). Functional annotation was accomplished by performing BLAST analysis of protein sequences in the UniProtKB/Swiss-Prot, nonredundant protein (NR), KEGG, and COG databases. To identify the missing DNA fragment in mutant T1m, an all-versus-all genome alignment between strains T1 and T1m was performed using Mauve software (version 1.2.3) (45).
Functional study of tmoAB in the mutant T1m and E. coli.
Fragments containing tmoA, tmoB, or tmoAB were amplified from the genomic DNA of strain T1 using PrimeStar polymerase (TaKaRa, Dalian, China) and the primer pairs listed in Table 4. The restriction sites KpnI (in the forward primer) and XbaI (in the reverse primer) were introduced to ensure the correct orientation of gene insertion. The amplified fragments were digested and ligated into the corresponding sites of the broad-host-range vector pBBR1MCS-2 (46). The resulting recombinants, pBBR-tmoA, pBBR-tmoB, and pBBR-tmoAB, were transformed into E. coli DH5α and mutant T1m by electrotransformation. Gene function was confirmed by whole-cell transformation experiments according to the method described by Wang et al. (30).
Gene expression and purification of the recombinant TmoA, TmoB, and TmoC.
tmoA, tmoB, and tmoC were amplified from the genomic DNA of strain T1 using PrimeStar polymerase and the primers listed in Table 4. The amplified products were digested with NdeI and SalI and ligated into the corresponding site of pET24b(+) to generate pET-tmoA, pET-tmoB, and pET-tmoC. The recombinant vectors were then transformed into E. coli BL21(DE3). Recombinant E. coli BL21(DE3) isolates harboring pET-tmoA, pET-tmoB, and pET-tmoC were cultured at 37°C in LB with 50 mg liter−1 kanamycin to an optical density at 600 nm (OD600) of 0.5 and then induced with 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 8 h at 16°C (47). The induced cells were harvested, washed, and resuspended in 50 mM phosphate-buffered saline (PBS) buffer (pH 7.4) (1 mM dithiothreitol was added for BL21 harboring pET-tmoC), sonicated (UH-650B ultrasonic processor; Auto Science; 40% intensity) for 10 min, and then centrifuged at 4°C and 14,940 × g for 30 min to remove undisrupted cells and cell debris. Crude extracts were loaded on 1-ml His-Trap cobalt affinity columns (His GraviTrap Talon; GE), which had been equilibrated with binding buffer. The target proteins were eluted with 50 mM imidazole for His6-TmoA and His6-TmoB and 100 mM imidazole for His6-TmoC after elution of the nontarget proteins with 5 mM imidazole. The proteins were dialyzed overnight at 4°C to remove imidazole using dialysis membranes (10 kDa) against 50 mM phosphate buffer (pH 7.4) for His6-TmoA and His6-TmoB and 40 mM HEPES-KOH buffer (pH 8.0) (25) for His6-TmoC. Protein concentrations were quantified using the Bradford method (48). Calibration curves were prepared from standard solutions of bovine serum albumin. The molecular weights of the proteins were determined by SDS-PAGE.
Enzyme activity assays.
The flavin reductase activity of TmoB was determined in a reaction mixture containing 50 mM PBS buffer (pH 7.4), 3 μM FMN (or FAD), 0.5 mM NADH (or NADPH), and 0.5 μM purified TmoB in a total volume of 1 ml at 30°C. A reaction mixture without TmoB was used as a control. The reactions were initiated by the addition of NADH; the enzyme activity was immediately measured using a Uvikon 752 spectrophotometer. One unit of TmoB activity was defined as the amount of TmoB required to oxidize 1 μmol NADH per minute, calculated from the absorbance decrease rate at 340 nm using a ε340 nm (the molar extinction coefficient of the substance at 340 nm) value of 6.22 mM−1 cm−1 for NADH.
Thiobencarb monooxygenase activity and the optimum molar concentration ratio of TmoA and TmoB were determined in a reaction mixture containing 50 mM Tris-HCl buffer (pH 7.4), 0.2 mM thiobencarb, 3 μM FMN, 0.5 mM NADH, 175 U of catalase (from bovine liver; Sigma), 1 μM TmoA, and 0 to 0.06 μM TmoB in a total volume of 1 ml. The reaction was initiated by the addition of TmoA. After incubation at 30°C for 60 min, the reaction was terminated by the addition of 50 μl HCl (0.5% [vol/vol]). Thiobencarb degradation was measured by HPLC, and the products were identified by GC-MS as described below. One unit of monooxygenase activity was defined as the amount of enzyme required to oxidize 1 μmol thiobencarb per minute. The optimal temperature range of the monooxygenase was assayed from 5 to 50°C, and the relative activity was calculated by assuming that the activity at 20°C was 100%. The optimal pH range was determined at pH values from 3.0 to 8.8, with the activity observed at pH 7.4 in PBS buffer set as 100%. Three buffers were used: 50 mM citric acid buffer (pH 3.0 to 6.0), 50 mM PBS buffer (pH 6.0 to 8.0), and 50 mM Tris-HCl buffer (7.0 to 8.8). To investigate the effects of potential inhibitors on monooxygenase activity, various chemical agents (urea, DTT, and EDTA; final concentration, 5 mM) and metal ions (Ca2+, Fe3+, Al3+, Li+, Zn2+, Mg2+, Co2+, Cd2+, Cu2+, Hg2+, and Mn2+; final concentration, 1.0 mM) were individually added, and the reactions were performed at 20°C for 60 min.
4CDA dehydrogenase activity of TmoC was detected by the absorbance increase rate at 340 nm. The reaction system consisted of 40 mM HEPES-KOH buffer (pH 8.0), 0.5 mM NAD+, and 2.75 μg purified TmoC in a total volume of 1 ml at 30°C (25). One unit of dehydrogenase activity was defined as the amount of TmoC required to catalyze the reduction of 1 μmol NAD+ per minute.
To investigate the kinetic parameters of TmoAB, TmoB, and TmoC, various concentrations of each substrate (0.02 to 0.2 mM thiobencarb for TmoAB, 0.01 to 0.1 mM NADH for TmoB, and 0.05 to 0.3 mM NAD+ and 0.05 to 0.2 mM 4CDA for TmoC) were added to the corresponding reaction mixtures as described above. Kinetic parameters were obtained by nonlinear regression analysis via the Michaelis-Menten equation.
RNA preparation and transcription analysis.
Cells were precultured in 1/5 LB and harvested at the mid-exponential phase, washed twice with MSM, and resuspended in MSM. The cell suspension was transferred to a 50-ml Erlenmeyer flask containing 20 ml MSM supplemented with 0.4 mM glucose or thiobencarb or 4CDA. The initial OD600 of the cultures was adjusted to 0.6. After incubation at 30°C and 180 rpm for 6 h, the cells were harvested. Total RNA was isolated using an RNA isolation kit (TaKaRa). Reverse transcription-PCR was carried out with a PrimeScript RT reagent kit (TaKaRa), and RT-qPCR was performed according to the procedure described by Yao et al. (49); the primers used for RT-qPCR are listed in Table 4. All samples were run in triplicate. Relative expression levels were calculated using the cycle threshold (2−ΔΔCT) method, and the 16S rRNA gene was used as the reference gene.
Analytical methods.
For the determination of residual thiobencarb and metabolite identification, culture and enzymatic samples were acidified to pH 2.0 with 0.5% HCl and extracted twice with ethyl acetate and dichloromethane separately. The extracts were dried over anhydrous sodium sulfate, evaporated to dryness using a nitrogen-blowing instrument, and then redissolved in methanol. For HPLC analysis, a Thermo Scientific Syncronis C18 column (5 μm; 250 by 4.6 mm) was used; the mobile phase was a mixture of methanol and 0.02 M ammonium acetate (pH 4.0; 80:20 [vol/vol]) at a flow rate of 1 ml min−1. The detection wavelengths were 220 nm and 238 nm, and the injection volume was 20 μl. The silylation of the sample was carried out under stringent anhydrous conditions; 100 μl silylation reagent (99% BSTFA plus 1% TMCS) was added to a vial containing nitrogen-dried sample, which was sealed and reacted at 75°C for 30 min. The silanized sample was directly analyzed by GC-MS. GC-MS was performed on a Bruker 320GC-MS/MS system linked to a BR-5MS column (Bruker; 30 m by 2.5 mm by 2.5 μm). The oven temperature was programmed to remain at 50°C for 3 min, followed by a linear increase to 180°C at a rate of 20°C min−1, a continuing linear increase to 280°C at a rate of 10°C min−1, and hold at 280°C for 5 min.
Accession number(s).
The GenBank accession numbers of the 16S rRNA gene of strain T1; the chromosome genome of strain T1; the plasmids P1, P2, and P3; the 36-kb gene fragment containing the fcb, pob, and lig operons; and the draft genome of mutant T1m are KX162718, CP021648, CP021649, CP021650, CP021651, MF189566, and NIAZ00000000, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Key R&D Project of China (2016YFD0801102), the National Natural Science Foundation of China (31500041), the Science and Technology Project of Jiangsu Province (BE2016374), the Project of University-Industry Collaboration of Guangdong Province (grant 2013B090500017), and the Natural Science Foundation of Jiangsu Province (no. BK20141366).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01490-17.
REFERENCES
- 1.Tanetani Y, Kaku K, Ikeda M, Shimizu T. 2013. Action mechanism of a herbicide, thiobencarb. J Pestic Sci 38:39–43. doi: 10.1584/jpestics.D12-047. [DOI] [Google Scholar]
- 2.Quayle WC, Oliver DP, Zrna S. 2006. Field dissipation and environmental hazard assessment of clomazone, molinate, and thiobencarb in Australian rice culture. J Agric Food Chem 54:7213–7220. doi: 10.1021/jf061107u. [DOI] [PubMed] [Google Scholar]
- 3.Finlayson BJ, Faggella GA. 1986. Comparison of laboratory and field observations of fish exposed to the herbicides molinate and thiobencarb. T Am Fish Soc 115:882–890. doi:. [DOI] [Google Scholar]
- 4.Sapari P, Ismail BS. 2012. Pollution levels of thiobencarb, propanil, and pretilachlor in rice fields of the muda irrigation scheme, Kedah, Malaysia. Environ Monit Assess 184:6347–6356. doi: 10.1007/s10661-011-2424-9. [DOI] [PubMed] [Google Scholar]
- 5.Fernández VC, Sancho E, Ferrando MD, Andreu E. 2002. Thiobencarb-induced changes in acetylcholinesterase activity of the fish Anguilla anguilla. Pestic Biochem Phys 72:55–63. doi: 10.1006/pest.2001.2581. [DOI] [Google Scholar]
- 6.Cashman JR, Olsen LD, Nishioka RS, Gray ES, Bern HA. 1990. S-oxygenation of thiobencarb (Bolero) in hepatic preparations from striped bass (Morone saxatilis) and mammalian systems. Chem Res Toxicol 3:433–440. doi: 10.1021/tx00017a008. [DOI] [PubMed] [Google Scholar]
- 7.Villalobos SA, Hamm JT, Teh SJ, Hinton DE. 2000. Thiobencarb-induced embryotoxicity in medaka (Oryzias latipes): stage-specific toxicity and the protective role of chorion. Aquat Toxicol 48:309–326. doi: 10.1016/S0166-445X(99)00032-6. [DOI] [PubMed] [Google Scholar]
- 8.Jena PK, Adhya TK, Rao VR. 1990. Nitrogen-fixing bacterial populations as influenced by butachlor and thiobencarb in rice soils. Zentralbl Mikrobiol 145:469–474. [Google Scholar]
- 9.Palumbo AJ, Tenbrook PL, Phipps A, Tjeerdema RS. 2004. Comparative toxicity of thiobencarb and deschlorothiobencarb to rice (Oryza sativa). Bull Environ Contam Toxicol 73:213–218. doi: 10.1007/s00128-004-0415-z. [DOI] [PubMed] [Google Scholar]
- 10.Gunasekara AS, Tenbrook PL, Palumbo AJ, Johnson CS, Tjeerdema RS. 2005. Influence of phosphate and copper on reductive dechlorination of thiobencarb in California rice field soils. J Agric Food Chem 53:10113–10119. doi: 10.1021/jf051656k. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura Y, Ishikawa K, Kuwatsuka S. 1977. Degradation of benthiocarb in soils as affected by soil conditions. J Pestic Sci 2:7–16. doi: 10.1584/jpestics.2.7. [DOI] [Google Scholar]
- 12.Torra RM, Yajima M, Yamanaka S, Kodama T. 2004. Degradation of the herbicides thiobencarb, butachlor and molinate by a newly isolated Aspergillus niger. J Pestic Sci 29:214–216. doi: 10.1584/jpestics.29.214. [DOI] [Google Scholar]
- 13.Miwa N, Takeda Y, Kuwatsuka S. 1988. Plasmid in the degrader of the herbicide thiobencarb (benthiocarb) isolated from soil: a possible mechanism for enrichment of pesticide degraders in soil. J Pestic Sci 13:291–293. doi: 10.1584/jpestics.13.291. [DOI] [Google Scholar]
- 14.Ishikawa K, Nakamura Y, Kuwatsuka S. 1976. Degradation of benthiocarb herbicide in soil. J Pestic Sci 1:49–57. doi: 10.1584/jpestics.1.49. [DOI] [Google Scholar]
- 15.Moon YH, Kuwatsuka S. 1985. Characterization of microbes causing dechlorination of benthiocarb (thiobencarb) in diluted soil suspension. J Pestic Sci 10:541–547. doi: 10.1584/jpestics.10.541. [DOI] [Google Scholar]
- 16.Kim KS, Pelton JG, Inwood WB, Andersen U, Kustu S, Wemmer DE. 2010. The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems. J Bacteriol 192:4089–4102. doi: 10.1128/JB.00201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichhorn E Jr, van der Ploeg JR, Leisinger T. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem 274:26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- 18.Denome SA, Oldfield C, Nash LJ, Young KD. 1994. Characterization of the desulfurization genes from Rhodococcus sp. strain IGTS8. J Bacteriol 176:6707–6716. doi: 10.1128/jb.176.21.6707-6716.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knobel HR, Egli T, van der Meer JR. 1996. Cloning and characterization of the genes encoding nitrilotriacetate monooxygenase of Chelatobacter heintzii ATCC 29600. J Bacteriol 178:6123–6132. doi: 10.1128/jb.178.21.6123-6132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dresen C, Lin LY, D'Angelo I, Tocheva EI, Strynadka N, Eltis LD. 2010. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J Biol Chem 285:22264–22275. doi: 10.1074/jbc.M109.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sucharitakul J, Chaiyen P, Entsch B, Ballou DP. 2005. The reductase of p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii requires p-hydroxyphenylacetate for effective catalysis. Biochemistry 44:10434–10442. doi: 10.1021/bi050615e. [DOI] [PubMed] [Google Scholar]
- 22.Beltrametti F, Marconi AM, Bestetti G, Colombo C, Galli E, Ruzzi M, Zennaro E. 1997. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol 63:2232–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng MG, Meng Q, Yang YJ, Chu CW, Chen Q, Li Y, Cheng D, Hong Q, Yan X, He J. 2017. The two-component monooxygenase MeaXY initiates the downstream pathway of chloroacetanilide herbicide catabolism in Sphingomonads. Appl Environ Microbiol 83:e03241–e03216. doi: 10.1128/AEM.03241-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue J, Shaw JP, Rekik M, Harayama S. 1995. Overlapping substrate specificities of benzaldehyde dehydrogenase (the xylC gene product) and 2-hydroxymuconic semialdehyde dehydrogenase (the xylG gene product) encoded by TOL plasmid pWW0 of Pseudomonas putida. J Bacteriol 177:1196–1201. doi: 10.1128/jb.177.5.1196-1201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YF, Chao H, Zhou NY. 2014. The catabolism of 2,4-xylenol and p-cresol share the enzymes for the oxidation of para-methyl group in Pseudomonas putida NCIMB 9866. Appl Microbiol Biotechnol 98:1349–1356. doi: 10.1007/s00253-013-5001-z. [DOI] [PubMed] [Google Scholar]
- 26.Chae JC, Zylstra GJ. 2006. 4-Chlorobenzoate uptake in Comamonas sp. strain DJ-12 is mediated by a tripartite ATP-independent periplasmic transporter. J Bacteriol 188:8407–8412. doi: 10.1128/JB.00880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JY, Zhou L, Chen B, Sun S, Zhang W, Li M, Tang H, Jiang BL, Tang JL, He YW. 2015. A functional 4-hydroxybenzoate degradation pathway in the phytopathogen Xanthomonas campestris is required for full pathogenicity. Sci Rep 5:18456. doi: 10.1038/srep18456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara H, Masai E, Katayama Y, Fukuda M. 2000. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J Bacteriol 182:6950–6957. doi: 10.1128/JB.182.24.6950-6957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, Wang CH, Deng SK, Wu YD, Li Y, Yao L, Jiang JD, Yan X, He J, Li SP. 2014. Novel three-component Rieske non-heme iron oxygenase system catalyzing the N-dealkylation of chloroacetanilide herbicides in sphingomonads DC-6 and DC-2. Appl Environ Microbiol 80:5078–5085. doi: 10.1128/AEM.00659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang CH, Chen Q, Wang R, Shi C, Yan X, He J, Hong Q, Li SP. 2014. A novel angular dioxygenase gene cluster encoding 3-phenoxybenzoate 1′,2′-dioxygenase in Sphingobium wenxiniae JZ-1. Appl Environ Microbiol 80:3811–3818. doi: 10.1128/AEM.00208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong WL, Wang F, Huang F, Wang Y, Zhou J, Ye X, Li ZK, Hou Y, Huang Y, Ma J, Jiang M, Cui ZL. 2016. Metabolic pathway involved in 6-chloro-2-benzoxazolinone degradation by Pigmentiphaga sp. strain DL-8 and identification of the novel metal-dependent hydrolase CbaA. Appl Environ Microbiol 82:4169–4179. doi: 10.1128/AEM.00532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jun SY, Lewis KM, Youn B, Xun L, Kang CH. 2016. Structural and biochemical characterization of EDTA monooxygenase and its physical interaction with a partner flavin reductase. Mol Microbiol 100:989–1003. doi: 10.1111/mmi.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okai M, Lee WC, Guan LJ, Ohshiro T, Izumi Y, Tanokura M. 2017. Crystal structure of dibenzothiophene sulfone monooxygenase BdsA from Bacillus subtilis WU-S2B. Proteins 85:1171–1177. doi: 10.1002/prot.25267. [DOI] [PubMed] [Google Scholar]
- 34.Eichhorn E, Davey CA, Sargent DF, Leisinger T, Richmond TJ. 2002. Crystal structure of Escherichia coli alkanesulfonate monooxygenase SsuD. J Mol Biol 324:457–468. doi: 10.1016/S0022-2836(02)01069-0. [DOI] [PubMed] [Google Scholar]
- 35.Galán B, Díaz E, Prieto MA, García JL. 2000. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new flavin:NAD(P)H reductase subfamily. J Bacteriol 182:627–636. doi: 10.1128/JB.182.3.627-636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CY, Lohman JR, Cao H, Tan K, Rudolf JD, Ma M, Xu W, Bingman CA, Yennamalli RM, Bigelow L. 2016. Crystal structures of SgcE6 and SgcC, the two-component monooxygenase that catalyzes hydroxylation of a carrier protein-tethered substrate in biosynthesis of the enediyne antitumor antibiotic C-1027 in Streptomyces globisporus. Biochemistry 55:5142–5154. doi: 10.1021/acs.biochem.6b00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uetz T, Schneider R, Snozzi M, Egli T. 1992. Purification and characterization of a two-component monooxygenase that hydroxylates nitrilotriacetate from “Chelatobacter” strain ATCC 29600. J Bacteriol 174:1179–1188. doi: 10.1128/jb.174.4.1179-1188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bohuslavek J, Payne JW, Liu Y, Bolton H Jr, Xun L. 2001. Cloning, sequencing, and characterization of a gene cluster involved in EDTA degradation from the bacterium BNC1. Appl Environ Microbiol 67:688–695. doi: 10.1128/AEM.67.2.688-695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellis HR. 2011. Mechanism for sulfur acquisition by the alkanesulfonate monooxygenase system. Bioorg Chem 39:178–184. doi: 10.1016/j.bioorg.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Boden R, Borodina E, Wood AP, Kelly DP, Murrell JC, Schafer H. 2011. Purification and characterization of dimethylsulfide monooxygenase from Hyphomicrobium sulfonivorans. J Bacteriol 193:1250–1258. doi: 10.1128/JB.00977-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldfield C, Pogrebinsky O, Simmonds J, Olson ES, Kulpa CF. 1997. Elucidation of the metabolic pathway for dibenzothiophene desulphurization by Rhodococcus sp. strain IGTS8 (ATCC 53968). Microbiology 143:2961–2973. doi: 10.1099/00221287-143-9-2961. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, Zheng JW, Liang B. 2011. Biodegradation of chloroacetamide herbicides by Paracoccus sp. FLY-8 in vitro. J Agric Food Chem 59:4614–4621. doi: 10.1021/jf104695g. [DOI] [PubMed] [Google Scholar]
- 43.Chu CW, Yuan CS, Liu X, Yao L, Zhu JC, He J, Kwon SW, Huang X. 2015. Phenylobacterium kunshanense sp. nov., isolated from the sludge of a pesticide manufacturing factory. Int J Syst Evol Microbiol 65:325–330. doi: 10.1099/ijs.0.063644-0. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.Darling AE, Treangen TJ, Messeguer X, Perna NT. 2007. Analyzing patterns of microbial evolution using the Mauve genome alignment system. Methods Mol Biol 396:135–152. doi: 10.1007/978-1-59745-515-2_10. [DOI] [PubMed] [Google Scholar]
- 46.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 47.Fang T, Zhou NY. 2014. Purification and characterization of salicylate 5-hydroxylase, a three-component monooxygenase from Ralstonia sp. strain U2. Appl Microbiol Biotechnol 98:671–679. doi: 10.1007/s00253-013-4914-x. [DOI] [PubMed] [Google Scholar]
- 48.Bradford MM. 1976. A rapid method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 49.Yao L, Yu LL, Zhang JJ, Xie XT, Tao Q, Yan X, Hong Q, Qiu JG, He J, Ding DR. 2016. A tetrahydrofolate-dependent methyltransferase catalyzing the demethylation of dicamba in Sphingomonas sp. strain Ndbn-20. Appl Environ Microbiol 82:5621–5630. doi: 10.1128/AEM.01201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.