ABSTRACT
Methylglyoxal (MG) is a cytotoxic, nonenzymatic by-product of glycolysis that readily glycates proteins and DNA, resulting in carbonyl stress. Glyoxalase I and II (GloA and GloB) sequentially convert MG into d-lactic acid using glutathione (GSH) as a cofactor. The glyoxalase system is essential for the mitigation of MG-induced carbonyl stress, preventing subsequent cell death, and recycling GSH for maintenance of cellular redox poise. All pathogenic liberibacters identified to date are uncultured, including “Candidatus Liberibacter asiaticus,” a psyllid endosymbiont and causal agent of the severely damaging citrus disease “huanglongbing.” In silico analysis revealed the absence of gloA in “Ca. Liberibacter asiaticus” and all other pathogenic liberibacters. Both gloA and gloB are present in Liberibacter crescens, the only liberibacter that has been cultured. L. crescens GloA was functional in a heterologous host. Marker interruption of gloA in L. crescens appeared to be lethal. Key glycolytic enzymes were either missing or significantly downregulated in “Ca. Liberibacter asiaticus” compared to (cultured) L. crescens. Marker interruption of sut, a sucrose transporter gene in L. crescens, decreased its ability to take up exogenously supplied sucrose in culture. “Ca. Liberibacter asiaticus” lacks a homologous sugar transporter but has a functional ATP/ADP translocase, enabling it to thrive both in psyllids and in the sugar-rich citrus phloem by (i) avoiding sucrose uptake, (ii) avoiding MG generation via glycolysis, and (iii) directly importing ATP from the host cell. MG detoxification enzymes appear to be predictive of “Candidatus” status for many uncultured pathogenic and environmental bacteria.
IMPORTANCE Discovered more than 100 years ago, the glyoxalase system is thought to be present across all domains of life and fundamental to cellular growth and viability. The glyoxalase system protects against carbonyl stress caused by methylglyoxal (MG), a highly reactive, mutagenic and cytotoxic compound that is nonenzymatically formed as a by-product of glycolysis. The uncultured alphaproteobacterium “Ca. Liberibacter asiaticus” is a well-adapted endosymbiont of the Asian citrus psyllid, which transmits the severely damaging citrus disease “huanglongbing.” “Ca. Liberibacter asiaticus” lacks a functional glyoxalase pathway. We report here that the bacterium is able to thrive both in psyllids and in the sugar-rich citrus phloem by (i) avoiding sucrose uptake, (ii) avoiding (significant) MG generation via glycolysis, and (iii) directly importing ATP from the host cell. We hypothesize that failure to culture “Ca. Liberibacter asiaticus” is at least partly due to its dependence on host cells for both ATP and MG detoxification.
KEYWORDS: ATP/ADP translocase, carbonyl stress, citrus greening, culturability, glycolysis, glyoxalase, huanglongbing, liberibacter, methylglyoxal, sugar transporter
INTRODUCTION
Huanglongbing (HLB [or citrus greening]) has emerged as the single most devastating citrus disease worldwide (1). HLB is associated with three fastidious and to date, uncultured, alphaproteobacteria; “Candidatus Liberibacter asiaticus” in Asia and the Americas, “Ca. Liberibacter americanus” in Brazil, and “Ca. Liberibacter africanus” in Africa. “Ca. Liberibacter asiaticus” and “Ca. Liberibacter americanus” are primarily vectored and transmitted among citrus and some other Rutaceae species by the Asian citrus psyllid Diaphorina citri Kuwayama (Sternorrhyncha: Psyllidae) (2). “Ca. Liberibacter asiaticus” growth in planta is phloem limited, causing perturbed assimilate partitioning, leaf chlorosis, loss in productivity, and eventual decline and death of infected trees. However, the bacterium is systemic and circulative in its psyllid host, causing only localized apoptosis in the midgut tissue and without causing any overt disease symptoms (3, 4).
In addition to the three citrus-associated species, liberibacters are emerging as a versatile group of plant pathogens capable of infecting a wide range of plant hosts. The most economically important of the latter is “Ca. Liberibacter solanacearum,” which causes economically serious diseases of solanaceous crops, for example, the “zebra chip” disease of potato (5). Attempts to culture any of the pathogenic liberibacters in axenic media have remained unsuccessful except transiently (6, 7). The lack of axenic culture methods has severely restricted functional genomic analyses, molecular characterization of host-pathogen interactions, and subsequent development of chemical control methods for HLB (8–10).
Only a single strain of a single liberibacter species, L. crescens strain BT-1 (NC_019907.1), has been cultured in vitro (11). BT-1 was originally isolated from the sap of Babaco mountain papaya (Carica stipulata × C. pubescens) and has no known insect host. Consistent failures in several labs to reinoculate BT-1 into Babaco and other plants have led to speculation that BT-1 may not be pathogenic. L. crescens appears to be the most basal Liberibacter lineage with a genome size of 1.5 Mb, diverging from other Liberibacter spp. early during evolution of the genus (12). All sequenced strains of the pathogenic liberibacters, e.g., “Ca. Liberibacter asiaticus” strain gxpsy (NC_020549.1) (13). “Ca. Liberibacter americanus” strain Sao Paulo (CP006604.1) (14), “Ca. Liberibacter africanus” strain PTSAPSY (NZ_CP004021.1) (15), and “Ca. Liberibacter solanacearum” strain CLso-ZC1 (NC_014774.1) (5) have further reduced genome sizes of about 1.2 Mb. Comparative genomics and metabolic pathway analyses have revealed a trend for the reduction or complete absence of several biosynthetic pathways, metabolic enzymes, and secretion systems in these genomes (14).
In silico analyses of all available Liberibacter genomes (5, 13–15) revealed the conspicuous absence of a functional glyoxalase pathway in all pathogenic (and uncultured) liberibacters, but the pathway is present in L. crescens (11). The two-enzyme glyoxalase pathway is an evolutionary conserved system for the mitigation of methylglyoxal (MG)-induced carbonyl stress and is ubiquitous across all domains of life, including bacteria (16). MG (2-oxopropanal or pyruvaldehyde [CH3COCHO]) is an α-ketoaldehyde originating as a by-product of several metabolic pathways, including glycolysis, ketone metabolism, lipid peroxidation, oxidative degradation of glucose, and fragmentation of glycated proteins (16). The nonenzymatic formation of MG as a consequence of the flux of triose phosphate intermediates, glyceraldehyde 3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP), is an intrinsic feature of the glycolysis pathway and unavoidable under physiological conditions (17). Nevertheless, MG is a potent electrophile, highly cytotoxic and mutagenic, since it readily glycates proteins via the Maillard reaction, forms cross-linked adducts with amino groups of adenine and guanine nucleotides, and results in the peroxidation of free and membrane-bound lipids (18).
The glyoxalase system, consisting of glyoxalase I (GloA; lactoylglutathione lyase; EC 4.4.1.5) and glyoxalase II (GloB; hydroxyacylglutathione hydrolase; EC 3.1.2.6), provides the primary cellular defense against proteome glycation in both prokaryotes and eukaryotes (16–18). Potential alternative MG detoxification enzymes have been described. For example, a metal ion- and glutathione (GSH)-independent route for direct detoxification of MG to d-lactic acid is catalyzed by glyoxalase III (GloIII; EC 4.2.1.130) in Escherichia coli (19), Schizosaccharomyces pombe (20), and rice (21). Nonglyoxalase enzymes such as aldehyde reductase (AR; EC 1.1.1.2), aldehyde dehydrogenase (AD; EC 1.2.1.3), and aldo/keto reductases (AKR superfamily; EC 1.1) can also potentially detoxify MG (22). However, none of these enzymes have been predicted to exist in the uncultured liberibacters.
GloA catalyzes isomerization of hemithioacetal formed by the spontaneous combination of MG and GSH leading to the formation of S-lactoylglutathione. GloB hydrolyzes S-lactoylglutathione to d-lactic acid, recycling GSH in the process, and thus maintains cellular redox poise (16, 17). The importance of the glyoxalase system has been well established in multiple developmental, physiological, and pathological processes in both prokaryotes and eukaryotes, including organismal longevity, cell proliferation, embryogenesis, and cell death (23, 24). Both plant glyoxalase enzymes exist as multigene families, and both are required for regulation of cell division, mitigation of biotic and abiotic stress, reproductive and morphogenetic cognizance, signaling cascades, and the regulation of protein turnover (25).
In an effort to understand why pathogenic liberibacters have remained uncultured, we discovered that the pathogenic liberibacters (i) are deficient in the biologically fundamental MG detoxification pathway (i.e., gloA is missing), (ii) lack a functional sucrose importer, (iii) circumvent MG generation by lack of a functional glycolytic pathway, and (iv) directly scavenge ATP from the host cell by virtue of a functional ATP/ADP translocase. We also demonstrate that, in contrast, an intact glyoxalase system (both gloA and gloB) is present in cultured L. crescens and that L. crescens gloA expresses a functional enzyme required for its culture. In addition, L. crescens also expresses a functional sucrose importer and has a functional glycolytic pathway. We hypothesize that MG detoxification is a necessary requirement for culturing liberibacters.
RESULTS
L. crescens has a functional glyoxalase system.
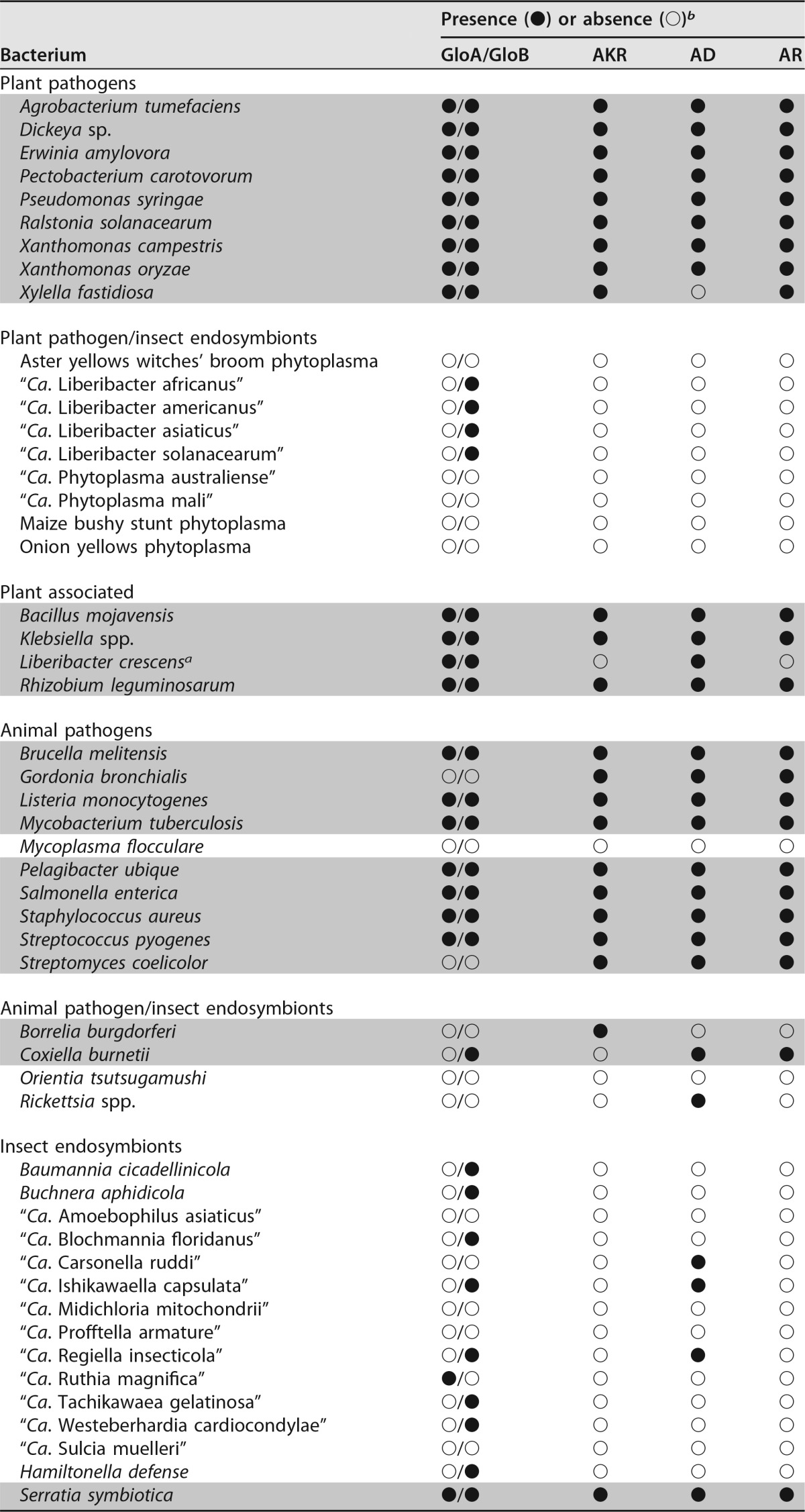
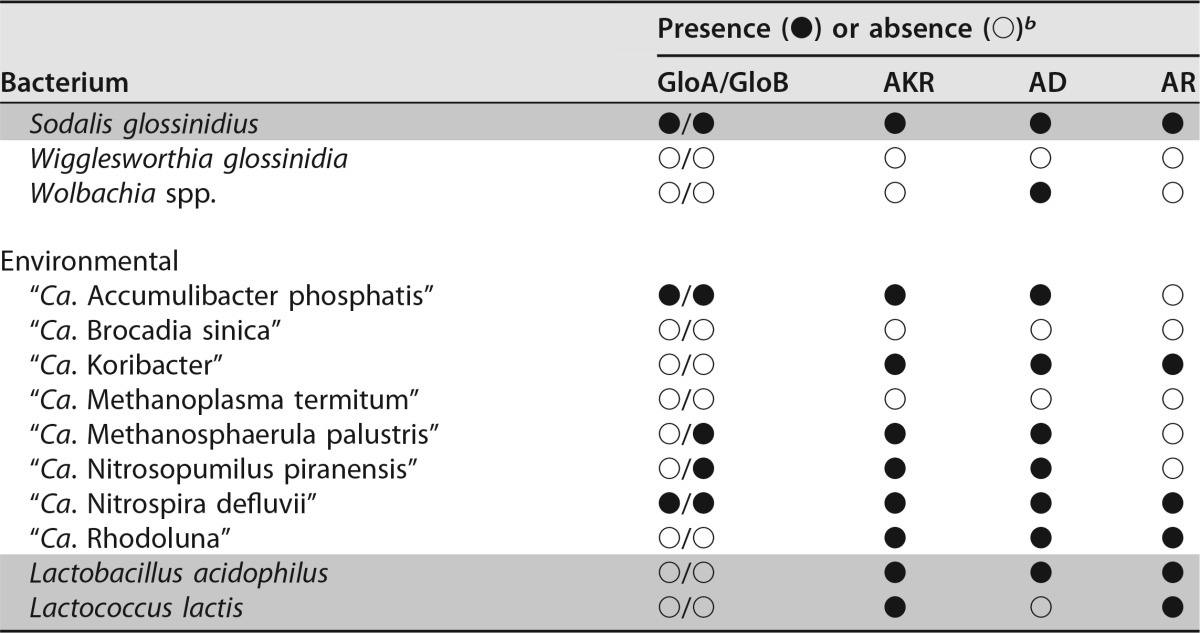
Both enzymes of the glyoxalase pathway, GloA and GloB (locus tags B488_RS02175 and B488_RS05485, respectively), are annotated in the cultured L. crescens strain BT-1 genome (GenBank accession number NC_019907). L. crescens GloA appeared to have all the expected features of a functional GloA, including the hallmark dimerization, glutathione, and divalent metal-binding domains. Similarly, GloB had the conventional metallohydrolase domain (see Fig. S1 and S2 in the supplemental material). However, no evidence of a gloA gene was found in any other liberibacter (Table 1). In addition, the predicted GloB enzymes in “Ca. Liberibacter asiaticus” (CLIBASIA_02350; GenBank accession number NC_012985), “Ca. Liberibacter americanus” (LAM_RS04885; GenBank accession number NC_022793), “Ca. Liberibacter africanus” (G293_RS03490; GenBank accession number NZ_CP004021), and “Ca. Liberibacter solanacearum” (CKC_RS00125; GenBank accession number NC_014774) had only limited identity (53 to 56%) to the L. crescens GloB enzyme.
TABLE 1.
Survey of glyoxalase and nonglyoxalase methylglyoxal detoxification enzymes present in representative plant- and human-pathogenic, symbiotic, and environmental bacteria
a Plant and/or insect hosts are currently unknown for L. crescens.
b GloA, glyoxalase I (EC 4.4.1.5); GloB, glyoxalase II (EC 3.1.2.6); AKR, aldo/keto reductase superfamily (EC 1.1); AD, aldehyde dehydrogenase (EC 1.2.1.3); AR, aldehyde reductase (EC 1.1.1.2). The “●” and “○” symbols represent the presence or absence of either the glyoxalase or alternate (putative) methylglyoxal detoxification enzymes. They are marked as present in the subject species if any of the three query sequences used for each enzyme yielded values of >70% coverage with >30% identity. Gray-shaded rows denote culturability (defined as replicative growth) in axenic media.
L. crescens proved to be readily tractable to marker interruption (26), which was successfully used to knock out several genes, including the sucrose transporter gene sut (locus tag B488_RS00965), as described in detail below. However, three separate attempts to inactivate L. crescens gloA by marker interruption failed, despite success in each experiment to achieve targeted insertions of a similarly sized homology region in a nonessential locus, lcrI (B488_RS03405, encoding restriction subunit R of an annotated type I restriction-modification system) (data not shown). This suggested that GloA is essential for free-living growth of L. crescens BT-1.
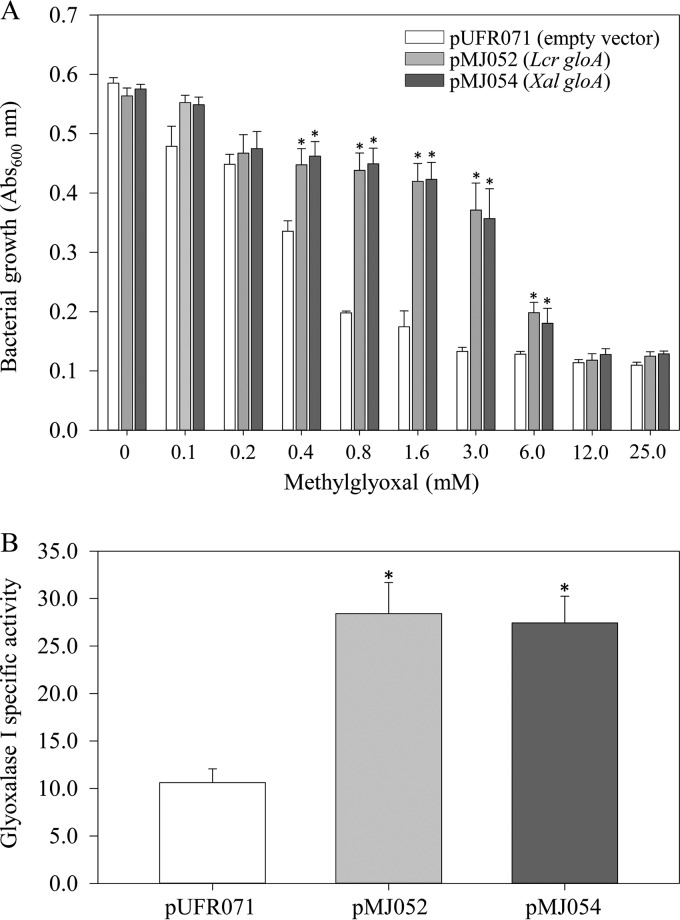
Given the difficulty in obtaining gloA deletion mutants, heterologous expression of L. crescens gloA in wild-type Xanthomonas albilineans was used for functional validation. The gloA genes from both L. crescens (B488_RS02175) and X. albilineans (XALXAFL071_RS04055; GenBank accession number NZ_JZIL01000006) were individually cloned into the wide-host-range (repW) shuttle vector pUFR071 (27) under the control of the lacZ promoter. X. albilineans cells carrying either pMJ052 (lacZ::B488_RS02175) or pMJ054 (lacZ::XALXAFL071_RS04055) exhibited significantly higher growth rates in the presence of exogenously supplied MG (at levels ranging from 0.4 to 6.0 mM) compared to the empty vector control (Fig. 1A). Episomal expression of either L. crescens or X. albilineans gloA in X. albilineans resulted in equivalent levels of alleviation of MG-induced cytotoxicity. Significantly higher levels of MG (∼7-fold) were required for 50% growth inhibition (i.e., the half-maximal inhibitory concentration [IC50]) of X. albilineans expressing L. crescens gloA (IC50 = 4.3 mM) or the native enzyme (IC50 = 4.5 mM) compared to X. albilineans cells carrying the empty vector (IC50 = 0.6 mM).
FIG 1.
Functional validation of L. crescens gloA in the heterologous host X. albilineans. (A) X. albilineans cells carrying L. crescens gloA (pMJ052), X. albilineans gloA (pMJ054), or empty vector (pUFR071) were grown for 4 days in modified Wilbrink's medium supplemented with 0 to 25 mM methylglyoxal. (B) Extractable glyoxalase I enzyme activity (S-lactoylglutathione, min−1 mg of protein−1) in X. albilineans cells carrying L. crescens gloA (pMJ052), X. albilineans gloA (pMJ054), or empty vector (pUFR071). The bars represent averages ± the standard errors (A, n = 8; B, n = 4). Asterisks represent significant differences at P < 0.05.
The enzymatic function of L. crescens GloA was further corroborated by quantifying formation of S-lactoylglutathione in a spectrophotometric assay. X. albilineans cells transformed with either pMJ052 or pMJ054 displayed 2.7- and 2.6-fold higher GloA activity compared to pUFR071-transformed cells (28.41 ± 2.81, 27.44 ± 3.28, and 10.62 ± 1.45 μmol of S-lactoylglutathione formed min−1 mg of protein−1, respectively) (Fig. 1B).
Uncultured liberibacters lack MG detoxification mechanisms.
The MG detoxification enzyme repertoire of 64 representative pathogenic, symbiotic, and environmental bacteria was predicted using the NCBI database of finished genome projects. All uncultured pathogenic liberibacters that are classified as “Candidatus” are deficient in not only gloA but also in all other reported alternative MG detoxification enzymes, including GloIII, AKR, AD, and AR. Interestingly, absence of the glyoxalase system and any alternative MG detoxification enzyme appeared to be predictive of inability to achieve replicative growth in culture (Table 1). The ability to detoxify MG seems to be necessary, but not sufficient, for culturability.
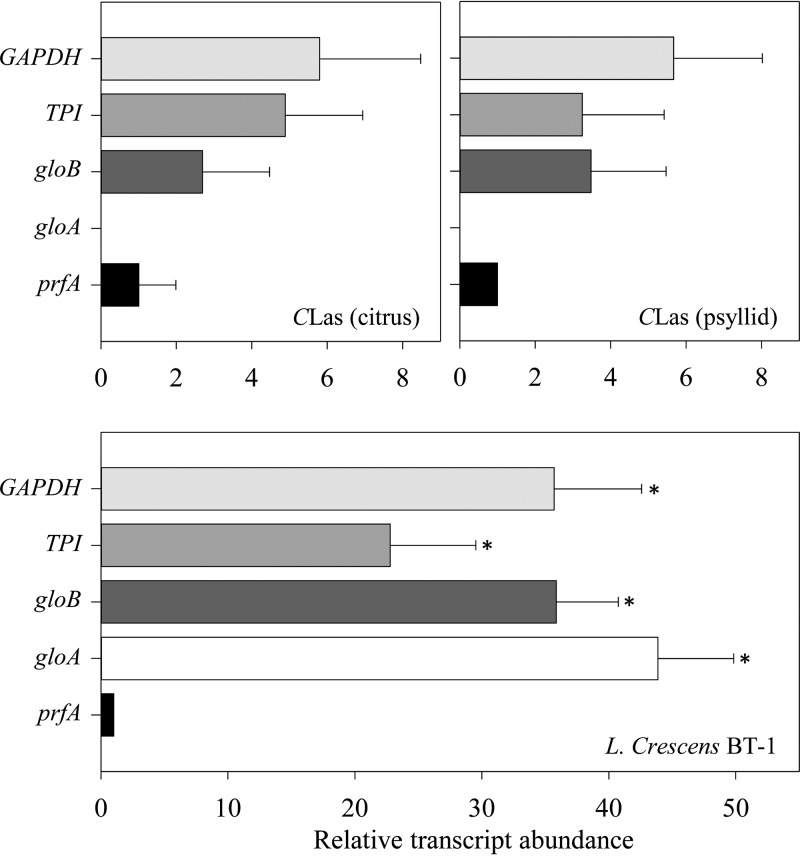
Quantitative reverse transcription-PCR (RT-PCR) analyses were used to examine the expression of both glyoxalase genes in culture-grown L. crescens and of gloB in “Ca. Liberibacter asiaticus” (gloA is missing) in both hosts (Fig. 2). Both gloA (P = 0.000232) and gloB (P = 0.000137) were highly expressed compared to the housekeeping control gene prfA (B488_RS00360) during growth of L. crescens in culture. In contrast in “Ca. Liberibacter asiaticus,” gloB expression was negligible in both citrus and psyllid hosts. All transcript data were normalized against gyrB (L. crescens [B488_RS06700] or “Ca. Liberibacter asiaticus” [CLIBASIA_03525]) within each sample.
FIG 2.
Relative expression of glyoxalase and key glycolysis pathway genes in L. crescens grown in culture and “Ca. Liberibacter asiaticus” in both its citrus and psyllid hosts. The transcript abundance of each gene was normalized against the expression levels of the chromosomal reference gene gyrB within each sample. Bars represent averages ± the standard errors of the means (n = 3). Asterisks represent significant differences (P < 0.05) in the transcript abundance with respect to prfA within each set.
“Ca. Liberibacter asiaticus” does not import or utilize sucrose via glycolysis.
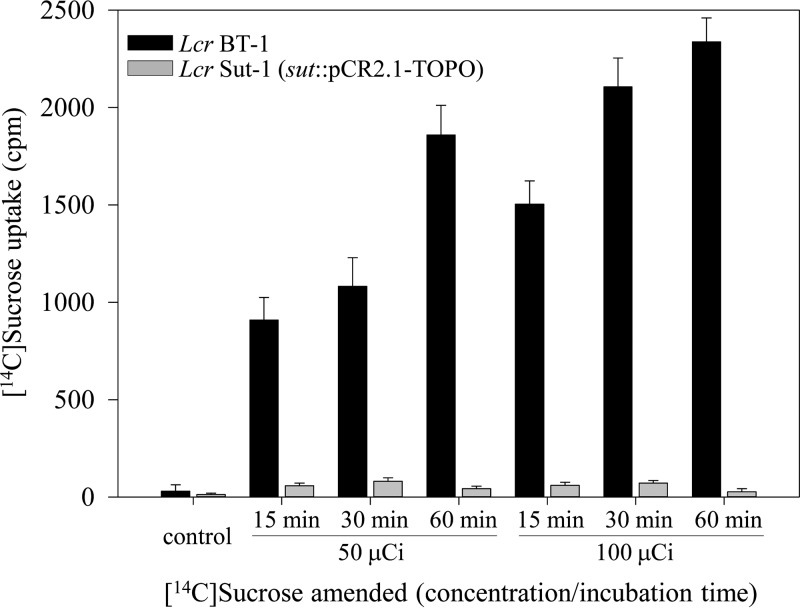
Among liberibacters sequenced to date, only the cultured L. crescens strain BT-1 carries a sugar transporter protein, Sut, encoded by B488_RS00965, belonging to the major facilitator superfamily (MFS) of transporter proteins. A partial internal fragment of sut was PCR amplified, cloned into suicide vector pCR2.1-TOPO, and used for marker interruption of the target in BT-1 cells. Inactivation of sut in BT-1 resulted in significantly diminished capacity by the mutant strain, Sut-1, to take up exogenously supplied isotopic sucrose compared to the wild-type parent (Fig. 3). Sucrose uptake was discernible as early as 15 min by BT-1 cells, which also accumulated significantly higher levels of radiolabel (15.9- to 43.5-fold at 50 μCi and 25.2- to 86.5-fold at 100 μCi exogenously supplied [14C]sucrose) compared to Sut-1. No orthologs of sut (B488_RS00965) were identified in the genomes of any of the uncultured pathogenic liberibacters.
FIG 3.
Functional characterization of L. crescens sucrose transporter sut (B488_RS00965). Five-day-old wild-type BT-1 and interruption mutant Sut-1 cells (sut::pCR2.1-TOPO) were incubated in the presence of [14C]sucrose at room temperature. At the indicated time points, 500-μl culture aliquots were withdrawn, washed twice to remove external [14C]sucrose, and resuspended in PBS and scintillation fluid. The amount of radioactive sucrose taken up by the cells was expressed as counts per minute (cpm). Bars represent average data ± standard errors of at least three independent experiments.
A key glycolysis/gluconeogenesis enzyme, phosphoglucose isomerase (EC 5.3.1.9), catalyzing interconversion of glucose-6-phosphate and fructose-6-phosphate, is missing in “Ca. Liberibacter asiaticus” but is present in L. crescens strain BT-1 (B488_RS06225). The transcriptional activities of two additional rate-limiting enzymes of the glycolysis pathway, glyceraldehyde-3-phosphate dehydrogenase (GAPDH; EC 1.2.1.12) and triosephosphate isomerase (TPI; EC 5.3.1.1), were compared in BT-1 cells grown in culture and in “Ca. Liberibacter asiaticus” infecting both psyllids and citrus (Fig. 2). As expected for BT-1, quantitative real-time PCR (qRT-PCR) revealed high levels of expression of both GAPDH (B488_RS05930) and TPI (B488_RS02240) (P = 0.000425 and 0.000261, respectively) compared against the housekeeping gene prfA. In contrast for “Ca. Liberibacter asiaticus” in both hosts, nearly negligible transcript levels of both GAPDH (CLIBASIA_02705) and TPI (CLIBASIA_00410) were observed.
“Ca. Liberibacter asiaticus” is an ATP scavenger.
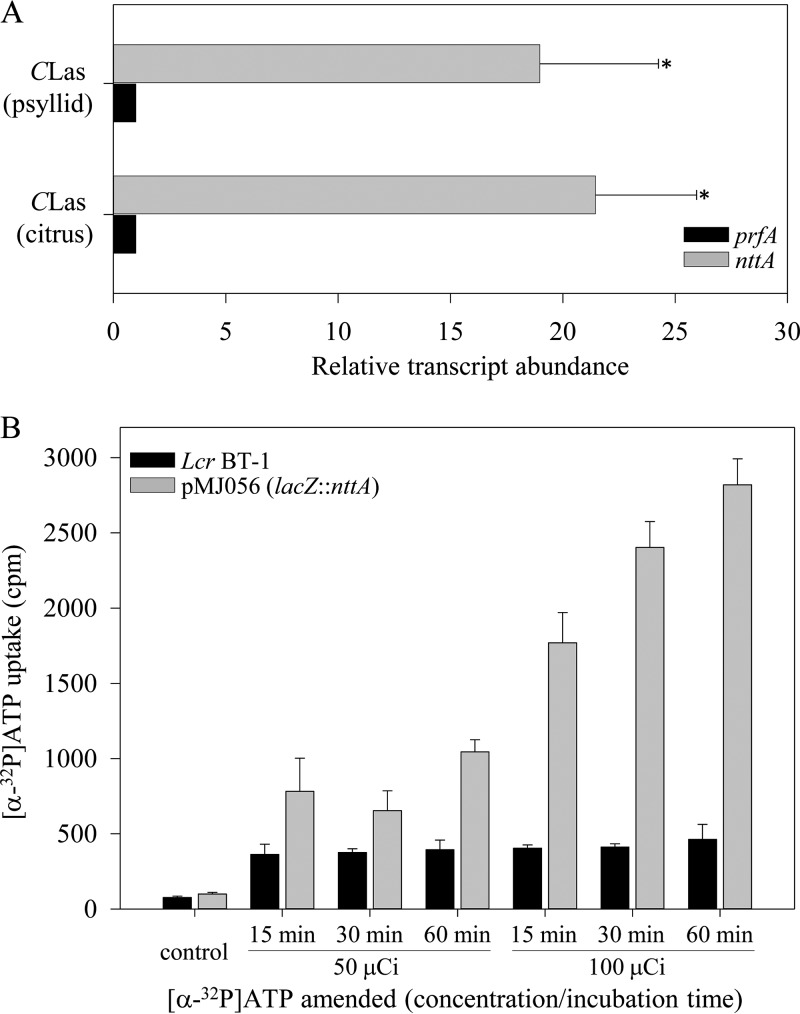
An ATP/ADP translocase gene (nttA, CLIBASIA_01040) was annotated in the “Ca. Liberibacter asiaticus” genome (28). Highly conserved orthologous nttA genes are annotated in all uncultured pathogenic liberibacters but not in the cultured L. crescens strain BT-1. Quantitative RT-PCR data revealed high levels of nttA expression in both citrus (P = 0.000639) and the psyllid host (P = 0.00532) compared to the housekeeping gene prfA (within each host) (Fig. 4A). The “Ca. Liberibacter asiaticus” ATP/ADP translocase gene (nttA) was cloned just downstream of the lacZ promoter in pUFR071, resulting in pMJ056 (lacZ::nttA). After transformation into BT-1, isotopic ATP import assays were performed. The amount of radiolabeled ATP uptake by BT-1/pMJ056 cells was measured after the addition of 50 and 100 μCi of [α-32P]ATP to the culture at the indicated time points. The levels of isotopic ATP present in NttA-expressing L. crescens cells increased 2.1- to 2.7-fold at 50 μCi and 4.4- to 6.1-fold at 100 μCi when supplied with [α-32P]ATP compared to the wild type (Fig. 4B). pMJ056 conferred the ability on L. crescens strain BT-1 to import extracellular ATP in a dose- and time-dependent manner, thus extending an earlier report that “Ca. Liberibacter asiaticus” nttA was functional in E. coli (28).
FIG 4.
(A) Relative expression of “Ca. Liberibacter asiaticus” ATP/ADP translocase (nttA [CLIBASIA_RS01005]) in both its citrus and psyllid hosts. Transcript abundances of nttA and prfA were normalized against expression levels of chromosomal reference gene gyrB within each host. Bars represent averages ± the standard errors of the means (n = 3). Asterisks represent significant differences (P < 0.05) in the transcript abundance with respect to prfA within each host. (B) Time- and dose-dependent [α-32P]ATP uptake into L. crescens cells expressing “Ca. Liberibacter asiaticus” nttA. Five-day-old wild-type BT-1 cells and transformed BT-1/pMJ056 cells expressing nttA were incubated in the presence of [α-32P]ATP at room temperature. At the indicated time points, 500-μl culture aliquots were withdrawn and washed twice to remove external [α-32P]ATP. The cells were resuspended in PBS and scintillation fluid. The amount of radioactive ATP taken up by the cells was expressed as counts per minute (cpm). Bars represent average data ± the standard errors of at least three independent experiments.
DISCUSSION
L. crescens strain BT-1 is a genetically tractable proxy for functional genomics studies of uncultured liberibacters. It has been transformed with plasmids expressing “Ca. Liberibacter asiaticus” genes and reporter constructs (8, 9; the present study) and used for Tn5 mutagenesis (29). In the present study, site-directed mutagenesis of the L. crescens sugar transporter sut (B488_RS00965) via marker interruption (Fig. 3) further expanded the list of molecular tools proven useful in L. crescens. More importantly, the relative ease of performing marker interruptions of the L. crescens sugar transporter sut and several other genes (data not shown) indicated that repeated failures to knock out gloA were due to lethality and that the glyoxalase system is essential for axenic growth of L. crescens. Taken together with circumstantial evidence from multiple bacteria (Table 1) that the presence of MG detoxification mechanisms is predictive of culturability, it seems likely that the concurrent absence of MG detoxification genes (Fig. 1 and 2 and Table 1) and the stringent energy dependence on host cell (Fig. 4) are two fundamental determinants that make “Ca. Liberibacter asiaticus” recalcitrant to axenic culturing.
In addition to the conventional two-enzyme glyoxalase system (GloA and GloB) for MG detoxification, alternative MG detoxification enzymes exist that may allow culturing of some bacteria (Table 1). Notably, AKR may provide this capacity, since it has been annotated in all the six completely sequenced bacterial genomes examined (Gordonia bronchialis, Borrelia burgdorferi, Coxiella burnetii, Sodalis glossinidius, Lactobacillus acidophilus, and Lactococcus lactis) that have been reported as axenically cultured and lack either GloA or GloB. In the case of B. burgdorferi, AKR is the sole predicted MG detoxification enzyme. GloIII, which can catalyze direct detoxification of MG to d-lactic acid (19–21), was omitted from Table 1 because GloIII (and similar proteins with a DJ-1 domain) are reportedly present at relatively low cellular concentrations and have extremely low specific activity, thereby precluding GloIII activity as a significant MG detoxification mechanism in vivo (23). Based on TBLASTN and keyword searches, all pathogenic and uncultured liberibacters sequenced to date appear to lack all previously described MG detoxification pathways.
All pathogenic liberibacters are missing a key glycolytic pathway gene, and in “Ca. Liberibacter asiaticus” two others are barely expressed (Fig. 2), findings consistent with the previously noted partial or complete loss of several metabolic pathways (14). Pathogenic liberibacters may have no need to detoxify the limited amounts of MG they produce via nonglycolysis pathways (16, 17), and therefore the absence of MG detoxification enzymes may be well tolerated. Furthermore, phloem sap and psyllid hemolymph are circulative environments that offer an unusual capability to both wash bacterial cells and detoxify any MG produced and passively excreted through liberibacter membranes. “Ca. Liberibacter asiaticus” colonization in planta is phloem limited and strictly intracellular. Preferential accumulation of plant GloA has been reported in phloem sieve elements in several species (30, 31). Interestingly, applications of zinc are known to significantly increase both GloA and GloB levels in Brassica juncea, rice, and tobacco plants (32). Zinc nutritional amendments have been extensively applied in Florida groves to help relieve symptoms of HLB infection (which resemble zinc deficiency); such amendments also resulted in unexpected increases in “Ca. Liberibacter asiaticus” titers (33). Unwittingly, such nutritional applications of zinc appear from this work to have provided “Ca. Liberibacter asiaticus” with increased capacity for MG detoxification by host cells, thereby allowing more extensive host colonization by “Ca. Liberibacter asiaticus.”
Even though occupying sugar-rich niches in both citrus phloem (34, 35) and psyllid hemolymph (36), “Ca. Liberibacter asiaticus” appears to be incapable of sugar import via known mechanisms (Fig. 3) and its utilization via primary metabolic pathways (Fig. 2). Marker interruption of L. crescens sut (B488_RS00965) demonstrated the functionality of this sole predicted sucrose importer (Fig. 3), and in silico homology searches across completed genomes of multiple pathogenic liberibacters (5, 13–15) established that this gene has been lost in all pathogenic liberibacters. L. crescens sugar transporter sut belongs to a large and diverse group of uniporter, symporter, and antiporter proteins (MFS), facilitating the transport of ions, sugar phosphates, nucleosides, amino acids, and metabolites across biological membranes, using the electrochemical potential of the transported substrates (37).
All pathogenic liberibacters have lost both sugar transport capacity and glycolysis. Notably, several glycolysis/gluconeogenesis enzymes such as phosphoglucose isomerase (EC 5.3.1.9), glucose-1-phosphatase (EC 3.1.3.10), and aldolase 1-epimerase (EC 5.1.33) were missing in “Ca. Liberibacter asiaticus.” Phosphoglucose isomerase has been shown to be essential for providing energy via central carbon metabolism and growth in E. coli (38). Besides a truncated glycolytic pathway, qRT-PCR data revealed significant transcriptional downregulation of “Ca. Liberibacter asiaticus” genes annotated as encoding the enzymatic steps involved in triose phosphate flux (GAPDH and TPI), thus circumventing nonenzymatic MG accumulation via glycolysis (17). The physiological significance of the lack of sugar uptake, glycolysis, and MG detoxification in pathogenic (and uncultured) liberibacters is supported by biochemical and molecular evidence that enhanced sugar uptake and glycolysis are directly linked to increased MG production and detoxification via an upregulated glyoxalase system in tumor cell lines (24).
An intact citric acid or tricarboxylic acid (TCA) cycle (for providing reducing equivalents) and oxidative phosphorylation apparatus for subsequent ATP synthesis are present in all liberibacters (5, 13). Despite the lack of glycolysis, in theory liberibacters could utilize pyruvic acid as the entry point into the TCA cycle (34, 35, 39). It is imperative, however, that the cyclic flux of TCA cycle intermediates must accommodate the metabolic, physiological, and energy demands of the cell. It is therefore likely that noncyclic or shunted parts of the TCA pathway could function in “Ca. Liberibacter asiaticus,” a finding consistent with the abundant availability of several TCA cycle intermediates (comprising 25% of the citrus phloem sap) and amino acids in its extracellular milieu (34–36, 39). Indeed, noncyclic variations in the TCA cycle, primarily providing biosynthetic precursors for lipids and amino acids, have been reported in bacteria grown under anaerobic or microaerophilic growth conditions (40). Similar metabolic shunts, with central carbon metabolites leaving at the key branch points, have been observed in Bradyrhizobium japonicum bacteroids without any apparent loss of nitrogen-fixing ability (41, 42). Since both the uptake and glycolytic breakdown of sucrose are inoperative in “Ca. Liberibacter asiaticus,” we speculate that the TCA cycle in “Ca. Liberibacter asiaticus” is not operative for ATP generation, making it partially or exclusively reliant on an extracellular energy source.
ATP/ADP translocases (antiporters) are integral membrane proteins that catalyze the highly specific transport of host-derived ATP across lipid membranes in exchange for bacterial ADP. ATP/ADP translocases are evolutionarily conserved ancestral proteins and have so far been identified exclusively in plastids and a few obligate intracellular bacteria belonging to Chlamydiales and the Rickettsiales (43). Since “Ca. Liberibacter asiaticus” appears to be incapable of sucrose uptake or metabolism, the ability to import ATP from the host cell (Fig. 4) confirms its status as an energy parasite sustaining an intracellular lifestyle. The presence of nttA in “Ca. Liberibacter asiaticus” that encodes an ATP/ADP translocase that is functional in E. coli (28) and L. crescens (Fig. 4) supports the observation that “Ca. Liberibacter asiaticus” lacks many enzymes needed for the metabolism of purines and pyrimidine. “Ca. Liberibacter asiaticus” is predicted to be incapable of de novo synthesis of guanine, xanthine, hypoxanthine, adenine, uracil, cytosine, or thymine and probably acquires these from the host as di- or trinucleosides (44). As expected, “Ca. Liberibacter asiaticus” infection of psyllids and citrus raises the levels of host glycolysis, TCA cycle metabolites, respiration rates (39), and ATP content (45, 46).
In summary, among all the liberibacters sequenced to date, L. crescens appears to be unique in its ability to take up and metabolize sugars for its energy requirements and in possessing a functional glyoxalase pathway for alleviating the ensuing carbonyl stress. In sharp contrast, “Ca. Liberibacter asiaticus” (and all other uncultured, pathogenic liberibacters sequenced to date) have evolved to circumvent the physiological requirements for sugar import and utilization and thus rely primarily on host cells for their energy requirements and for degradation of MG, both supporting a strict intracellular lifestyle and constraining the ability to culture these bacteria axenically. Addressing physiological requirements such as exogenously supplied trinucleotide phosphates should aid in culturing “Ca. Liberibacter” strains.
MATERIALS AND METHODS
Plant material and insect samples.
“Ca. Liberibacter asiaticus”-infected leaf samples were excised from curated citrus (Citrus paradisi) plants maintained in a quarantine greenhouse at the University of Florida, Gainesville, FL. “Ca. Liberibacter asiaticus”-infected, mixed sex adult psyllids (reared on sweet orange, C. sinensis; approximately 80% infection density) were provided by David Hall, USDA, ARS.
Bacterial growth conditions, gene cloning, and transformation.
The relevant characteristics, source, and/or reference for the bacterial strains and plasmids used in this study are listed in Table 2. The primers used for confirmation of “Ca. Liberibacter asiaticus” infection, qRT-PCR analyses, and cloning of gloA, sugar transporter (sut) and ATP/ADP translocase (nttA) genes are listed in Table 3. E. coli was grown in Luria-Bertani medium at 37°C. L. crescens and X. albilineans were cultured in BM7 medium (11) and Modified Wilbrink's medium (45), respectively, at 28°C. Genomic DNA from L. crescens and X. albilineans cells was extracted using the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich, St. Louis, MO). Full-length gloA genes from L. crescens strain BT-1 and X. albilineans strain FL07-1 were PCR amplified, cloned in pCR2.1-TOPO (Invitrogen, Carlsbad, CA), verified for sequence fidelity, and finally subcloned into a broad-host-range repW shuttle vector pUFR071 (27). Electrocompetent cells of L. crescens (9) and X. albilineans (47) were prepared and transformed as described previously.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| X. albilineans FL07-1 | Wild-type strain isolated in Florida | 47 |
| L. crescens BT-1 | Wild-type strain originally isolated from Babaco mountain papaya | 11 |
| L. crescens Sut-1 | sut::pCR2.1-TOPO derived from BT-1; Kanr | This study |
| L. crescens LcrI-1 | lcrI::pCR2.1-TOPO derived from BT-1; Kanr | This study |
| Plasmids | ||
| pCR2.1-TOPO | 3.9 kb; PCR cloning vector; Ampr Kanr | Invitrogen |
| pUFR071 | 9.4 kb; repW, ColE1, Mob+, LacZ+, Par+; Cmr Gmr | 27 |
| pMJ001 | Partial fragment of Lcr restriction subunit R lcrI (B488_RS03405) in pCR2.1-TOPO | This study |
| pMJ051 | Lcr gloA (B488_RS02175, with KpnI/HindIII ends) in pCR2.1-TOPO | This study |
| pMJ052 | KpnI/HindIII fragment from pMJ051 subcloned in pUFR071 | This study |
| pMJ053 | Xal gloA (XALXAFL071_RS04055, with EcoRI/HindIII ends) in pCR2.1-TOPO | This study |
| pMJ054 | EcoRI/HindIII fragment from pMJ053 subcloned in pUFR071 | This study |
| pMJ055 | CLas nttA (CLIBASIA_RS01005, with EcoRI/HindIII ends) in pCR2.1-TOPO | This study |
| pMJ056 | EcoRI/HindIII fragment from pMJ055 subcloned in pUFR071 | This study |
| pMJ057 | Partial fragment of Lcr gloA (B488_RS02175) in pCR2.1-TOPO | This study |
| pMJ058 | Partial fragment of Lcr sugar transporter sut (B488_RS00965) in pCR2.1-TOPO | This study |
Amp, ampicillin; Kan, kanamycin; Cm, chloramphenicol; Gm, gentamicin; Str, streptomycin. CLas, Lcr, and Xal represent “Ca. Liberibacter asiaticus,” L. crescens, and X. albilineans, respectively.
TABLE 3.
Primers used in this studya
| Target or primer | Sequence (5′–3′) | Source or reference |
|---|---|---|
| “Ca. Liberibacter asiaticus” confirmation | ||
| OI1 | GCGCGTATGCAATACGAGCGGC | 48 |
| OI2c | GCCTCGCGACTTCGCAACCCAT | 48 |
| CG03F | RGGGAAAGATTTTATTGGAG | 49 |
| CG05R | GAAAATAYCATCTCTGATATCGT | 49 |
| Quantitative RT-PCR | ||
| Lcr_gloA_F | GTTGTGTAGTCTTCAGTATCCCAG | This study |
| Lcr_gloA_R | GGGTCTCTATGAAGTTGATCGTC | This study |
| Lcr_gloB_F | CATGCGGATCATACAAAAGGC | This study |
| Lcr_gloB_R | CCTTCAGAAAGCACATGATCAAC | This study |
| CLas_gloB_F | GATTCAAACATTTCAGCATAACTATCTTC | This study |
| CLas_gloB_R | GATCATTTTCTTTGCGTCGGAG | This study |
| Lcr_TPI_F | CGGCAACATTAGTGTATTTGGC | This study |
| Lcr_TPI_R | ACAAAATCAGCTCCACAATCAAC | This study |
| CLas_TPI_F | CCGTACCAATCGCCCATATAG | This study |
| CLas_TPI_R | AGATTGTAGTTTGCCGAGTGAG | This study |
| Lcr_GAPDH_F | GTCTTATCAGCTCCTCCACAAG | This study |
| Lcr_GAPDH_R | GTGATCCAAAACAGCTTCCG | This study |
| CLas_GAPDH_F | AGTTGAGTCAAGACGCGATG | This study |
| CLas_GAPDH_R | CCTATGATTTTCACCTCTCCTGG | This study |
| CLas_nttA_F | CTACATTCCCGACCAATCCAA | This study |
| CLas_nttA_R | GGCGCTGTCATGATTAATCTTATG | This study |
| Lcr_gyrB_F | TCTTCACCAGCATCTCCAAC | This study |
| Lcr_gyrB_R | CACTTTCATCTTGGCTGCG | This study |
| CLas_gyrB_F | TTGAACAAGCTGTAATTTCTGG | 8 |
| CLas_gyrB_R | ATCTGTTTGCCAATTTAGAAGC | 8 |
| Lcr_prfA_F | AGGCTCAAGTTGGTTCAGG | This study |
| Lcr_prfA_R | ATATCACCCTCCAACATGCG | This study |
| CLas_prfA_F | TGTCTGAATCGCCTTCTGTC | 8 |
| CLas_prfA_R | GATCACCGATGACAGTATGC | 8 |
| Cloning in pUFR071 | ||
| Lcr_gloA_F | GGTACCAGGAGACATACTATGGTACGTGTTA | This study |
| Lcr_gloA_R | AAGCTTTTACCAGGTTCCTATGTTAGGCAT | This study |
| Xal_gloA_F | GAATTCAGGAGTTTCGCGATGAAATACCTGCATGCC | This study |
| Xal_gloA_R | AAGCTTTCACCATACTCCGGTATTGGGCA | This study |
| CLas_nttA_F | GAATTCAGGAGCGTTAAGATGTCGGAGGCGAAGA | This study |
| CLas_nttA_R | AAGCTTTTATTCTTTACTAATAAGCTGAGTATATTC | This study |
| Marker interruption | ||
| Lcr_gloA_internal_F | GCTCTTTGGGTTCTAGAGGATTAT | This study |
| Lcr_gloA_internal_R | CGTGTTAAAGACCTTGAGCATTC | This study |
| Lcr_lcrI_internal_F | CCATTGATCTGGTGCTGTTTATC | This study |
| Lcr_lcrI_internal_R | TTGGGCTTGGACGACTTTTGC | This study |
| Lcr_sut_internal_F | TATGCTTGGGAGCATGTTAGG | This study |
| Lcr_sut_internal_R | CAAGAACCGGAAGAGCGATAG | This study |
Abbreviations used in target and primer designations: gloA and gloB, glyoxalase I and II; TPI, triosephosphate isomerase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; gyrB, DNA gyrase subunit B; prfA, peptide chain release factor 1; nttA, ATP/ADP translocase; lcrI, restriction subunit R; sut, sugar transporter. In primer designations, CLas, Lcr, and Xal denote “Ca. Liberibacter asiaticus,” L. crescens, and X. albilineans, respectively, and “F” and “R” denote forward and reverse, respectively. Restriction sites for cloning are italicized in the corresponding sequences.
Nucleic acid extractions from psyllids and plant samples.
DNA was extracted from plant leaf discs and from whole psyllids via a DNeasy plant minikit and a blood and tissue kit, respectively (Qiagen, Valencia, CA). The presence of “Ca. Liberibacter asiaticus” in the infected citrus and psyllid samples was confirmed using conventional and nested PCR primer sets OI1/OI2c (48) and CG03F/CG05R (49) (Table 3). For the extraction of total RNA, midribs of two PCR-confirmed “Ca. Liberibacter asiaticus”-infected citrus leaves per sample and 10 frozen psyllids per sample were ground in cold mortars and pestles in 200 μl of lysis buffer RLT (provided with Qiagen RNeasy Plant minikit). RNA was extracted according to the manufacturer's protocol, diluted with nuclease-free water to 200 ng μl−1, and cleaned with a Turbo DNA-free (DNase) kit (Ambion, Austin, TX).
Site-directed mutagenesis of L. crescens via marker interruption.
Site-directed mutagenesis in L. crescens strain BT-1 cells was performed according to the strategy outlined by Castañeda et al. (26). Briefly, partial internal fragments of the L. crescens genes lcrI (B488_RS03405, encoding restriction subunit R of type I restriction-modification system) and gloA (B488_RS02175) and the sucrose transporter sut (B488_RS00965) were PCR amplified and cloned into pCR2.1-TOPO (Invitrogen). The resulting plasmids (pMJ001, pMJ057, and pMJ058, respectively) were used for electroporation of BT-1 cells. A single homologous recombination event at the target site duplicated the cloned region, integrating the vector between incomplete copies of the target gene and providing the resulting cells with kanamycin resistance. Antibiotic-resistant colonies were selected on solid BM7 medium supplemented with 4.5 μg ml−1 kanamycin sulfate. Selected colonies were subsequently grown in liquid BM7 for 5 days, cells were harvested by centrifugation, and DNA was extracted as described above and analyzed by PCR to confirm interruption of the target gene and integration of the plasmid backbone.
Glyoxalase I assay.
X. albilineans cultures were harvested at the mid-log-growth phase (A600 = 0.5) and used for protein extraction (Qproteome bacterial protein isolation kit; Qiagen). Bacterial protein extracts were quantified using a DC protein assay kit (Bio-Rad, Hercules, CA) and adjusted to 1 μg μl−1. The GloA enzyme activity was determined by measuring the production of S-lactoylglutathione (glyoxalase I activity assay kit; Sigma-Aldrich). Briefly, the assay mixture contained 2 mM MG and 2 mM GSH in 50 mM sodium phosphate buffer (pH 6.6). The reactions were read at 240 nm (Synergy HTX Multi-Mode reader; Bio Tek, Winooski, VT) before and after the addition of 1 μg μl−1 protein extract and incubation for 15 min at 25°C. One unit of GloA is defined as the amount of enzyme that will convert 1.0 μmol of S-lactoylglutathione from MG and GSH min−1 at pH 6.6 and 25°C.
Gene expression analysis using qRT-PCR.
The primer pairs used for the qRT-PCR analyses are listed in Table 3. Reverse transcription reactions were performed using 1 μg of RNA template (iScript Advanced cDNA synthesis kit; Bio-Rad). Quantitative RT-PCR analyses were performed using a CFX96 Touch Real-Time PCR detection system (Bio-Rad), as described previously (9, 10). At least three biological replicates and four technical replicates were used with no-template and no-RT controls. Relative expression levels were calculated and calibrated against prfA expression by the ΔΔCT method (50) and corrected for amplification efficiency. Data analyses and Student t tests (α = 0.05) were performed using the Bio-Rad CFX Manager Software Package 3.0.
Radiolabeled ATP and sucrose uptake assays.
[14C]sucrose and [α-32P]ATP were purchased from Perkin-Elmer (Waltham, MA). Isotopic sucrose and ATP uptake assays were performed as described previously by Vahling et al. (28) with some modifications. Five-day-old L. crescens wild-type BT-1, sugar transporter interruption mutant Sut-1, and pMJ056 (lacZ::nttA)-transformed cells (25 ml, A600 = 0.5) were collected via centrifugation and washed in 50 mM phosphate-buffered saline (PBS; pH 7.0). The bacteria were resuspended in PBS to A600 = 1.0, followed by incubation in the presence of [14C]sucrose (1 μCi 10 μl−1; specific activity, 435 mCi mmol−1) or [α-32P]ATP (10 μCi μl−1; specific activity, 3,000 Ci mmol−1) at room temperature. At the indicated time points, 500-μl culture aliquots were withdrawn, washed twice in ice-cold 50 mM PBS (pH 7.0), and resuspended in 2 ml of a solution of PBS and scintillation fluid (1:1, vol/vol) before being measured on a LS6500 multipurpose scintillation counter (Beckman Coulter, Brea, CA).
Search for MG detoxification enzymes across finished bacterial genomes.
Several fully sequenced plant pathogens, plant pathogen/insect symbionts, plant-associated pathogens, animal pathogens, animal pathogen/insect endosymbionts, insect endosymbionts, and environmental bacteria were selected to determine the presence or absence of MG detoxification enzymes via TBLASTN and confirmed manually by keyword searches. TBLASTN interrogation was performed against every selected genus, using the following protein sequences as the queries for each enzyme: GloA (E. coli, ASB80213.1; X. axonopodis, AEO43772.1; and L. crescens, AMC12618.1), GloB (E. coli, ASB77285.1; X. campestris, ALE67873.1; and L. crescens, AMC13092.1), aldo/keto reductase (X. citri, AJY98153.1; Rhizobium spp., CDM56005.1; and E. coli, NP_311718), aldehyde dehydrogenase (L. crescens, AMC12886.1; X. oryzae, AOS07057.1; and E. coli, ASB80413.1), and aldehyde reductase (E. coli, NP_310517; Marivirga tractuosa, ADR23652.1; and Pseudomonas fulva, AEF21531.1). The BLAST search was run with default parameters, and the enzyme was marked as present if any of the above query sequences yielded results of >70% coverage with >30% sequence identity. All negative results were reconfirmed manually by appropriate key word searches using each individual complete subject genome.
Supplementary Material
ACKNOWLEDGMENTS
We thank Patricia Rayside for excellent technical assistance and David Hall (ARS-USDA, Fort Pierce, FL) for providing “Ca. Liberibacter asiaticus”-infected psyllids.
This study was supported by the Florida Citrus Research and Development Foundation (CRDF) project 15-009 and by USDA-NIFA-SCRI grant 2016-70016-24844.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01670-17.
REFERENCES
- 1.Gottwald TR. 2010. Current epidemiological understanding of citrus huanglongbing. Annu Rev Phytopathol 48:119–139. doi: 10.1146/annurev-phyto-073009-114418. [DOI] [PubMed] [Google Scholar]
- 2.Grafton-Cardwell EE, Stelinski LL, Stansly PA. 2013. Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu Rev Entomol 58:413–432. doi: 10.1146/annurev-ento-120811-153542. [DOI] [PubMed] [Google Scholar]
- 3.Ammar E-D, Shatters RG Jr, Lynch C, Hall DG. 2011. Detection and relative titer of Candidatus Liberibacter asiaticus in the salivary glands and alimentary canal of Diaphorina citri (Hemiptera: Psyllidae) vector of citrus huanglongbing disease. Ann Entomol Soc Am 104:526–533. doi: 10.1603/AN10134. [DOI] [Google Scholar]
- 4.Ghanim M, Fattah-Hosseini S, Levy A, Cilia M. 2016. Morphological abnormalities and cell death in the Asian citrus psyllid (Diaphorina citri) midgut associated with Candidatus Liberibacter asiaticus. Sci Rep 6:33418. doi: 10.1038/srep33418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin H, Lou B, Glynn JM, Doddapaneni H, Civerolo EL, Chen C, Vahling CM. 2011. The complete genome sequence of “Candidatus Liberibacter solanacearum,” the bacterium associated with potato zebra chip disease. PLoS One 6:e19135. doi: 10.1371/journal.pone.0019135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis MJ, Mondal SN, Chen HQ, Rogers ME, Brlansky RH. 2008. Cocultivation of “Candidatus Liberibacter asiaticus” with actinobacteria from citrus with huanglongbing. Plant Dis 92:1547–1550. doi: 10.1094/PDIS-92-11-1547. [DOI] [PubMed] [Google Scholar]
- 7.Parker JK, Wisotsky SR, Johnson EG, Hijaz FM, Killiny N, Hilf ME, De La Fuente L. 2014. Viability of “Candidatus Liberibacter asiaticus” prolonged by addition of citrus juice to culture medium. Phytopathol 104:15–26. doi: 10.1094/PHYTO-05-13-0119-R. [DOI] [PubMed] [Google Scholar]
- 8.Fleites LA, Jain M, Zhang S, Gabriel DW. 2014. “Candidatus Liberibacter asiaticus” prophage late genes may limit host range and culturability. Appl Environ Microbiol 80:6023–6030. doi: 10.1128/AEM.01958-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain M, Fleites LA, Gabriel DW. 2015. Prophage-encoded peroxidase in “Candidatus Liberibacter asiaticus” is a secreted effector that suppresses plant defenses. Mol Plant Microbe Interact 28:1330–1337. doi: 10.1094/MPMI-07-15-0145-R. [DOI] [PubMed] [Google Scholar]
- 10.Jain M, Fleites LA, Gabriel DW. 2017. A small Wolbachia protein directly represses phage lytic cycle genes in “Candidatus Liberibacter asiaticus” within psyllids. mSphere 2:e00171-17. doi: 10.1128/mSphereDirect.00171-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard MT, Fagen JR, Davis-Richardson AG, Davis MJ, Triplett EW. 2012. Complete genome sequence of Liberibacter crescens BT-1. Stand Genomic Sci 7:271–283. doi: 10.4056/sigs.3326772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakabachi A, Nikoh N, Oshima K, Inoue H, Ohkuma M, Hongoh Y, Miyagishima SY, Hattori M, Fukatsu T. 2013. Horizontal gene acquisition of Liberibacter plant pathogens from a bacteriome-confined endosymbiont of their psyllid vector. PLoS One 8:e82612. doi: 10.1371/journal.pone.0082612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan Y, Zhou L, Hall DG, Li W, Doddapaneni H, Lin H, Liu L, Vahling CM, Gabriel DW, Williams KP, Dickerman A, Sun Y, Gottwald T. 2009. Complete genome sequence of citrus huanglongbing bacterium, “Candidatus Liberibacter asiaticus” obtained through metagenomics. Mol Plant Microbe Interact 22:1011–1020. doi: 10.1094/MPMI-22-8-1011. [DOI] [PubMed] [Google Scholar]
- 14.Wulff NA, Zhang S, Setubal JC, Almeida NF, Martins EC, Harakava R, Kumar D, Rangel LT, Foissac X, Bové JM, Gabriel DW. 2014. The complete genome sequence of “Candidatus Liberibacter americanus,” associated with citrus Huanglongbing. Mol Plant Microbe Interact 27:163–176. doi: 10.1094/MPMI-09-13-0292-R. [DOI] [PubMed] [Google Scholar]
- 15.Lin H, Pietersen G, Han C, Read DA, Lou B, Gupta G, Civerolo EL. 2015. Complete genome sequence of “Candidatus Liberibacter africanus,” a bacterium associated with citrus huanglongbing. Genome Announc 3:e00733-15. doi: 10.1128/genomeA.00733-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornalley PJ. 2003. Glyoxalase I-structure, function and a critical role in the enzymatic defence against glycation. Biochem Soc Trans 31:1343–1348. doi: 10.1042/bst0311343. [DOI] [PubMed] [Google Scholar]
- 17.Phillips SA, Thornalley PJ. 1993. The formation of methylglyoxal from triose phosphates. Eur J Biochem 212:101–105. doi: 10.1111/j.1432-1033.1993.tb17638.x. [DOI] [PubMed] [Google Scholar]
- 18.Kalapos MP. 1999. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology, and biological implications. Toxicol Lett 110:145–175. doi: 10.1016/S0378-4274(99)00160-5. [DOI] [PubMed] [Google Scholar]
- 19.Subedi KP, Choi D, Kim I, Min B, Park C. 2011. Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol Microbiol 81:926–936. doi: 10.1111/j.1365-2958.2011.07736.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Q, Su Y, Wang Z, Chen C, Wu T, Huang Y. 2014. Identification of glutathione (GSH)-independent glyoxalase III from Schizosaccharomyces pombe. BMC Evol Bio 14:86. doi: 10.1186/1471-2148-14-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh A, Kushwaha HR, Hasan MR, Pareek A, Sopory SK, Singla-Pareek SL. 2016. Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci Rep 6:18358. doi: 10.1038/srep18358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deponte M. 2014. Glyoxalase diversity in parasitic protists. Biochem Soc Trans 42:473–478. doi: 10.1042/BST20140005. [DOI] [PubMed] [Google Scholar]
- 23.Rabbani N, Mingzhan X, Thornalley PJ. 2014. Activity, regulation, copy number and function in the glyoxalase system. Biochem Soc Trans 42:419–424. doi: 10.1042/BST20140008. [DOI] [PubMed] [Google Scholar]
- 24.Bellahcène A, Nokin MJ, Castronovo V, Schalkwijk C. 2017. Methylglyoxal-derived stress: an emerging biological factor involved in the onset and progression of cancer. Semin Cancer Biol doi: 10.1016/j.semcancer.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Sankaranarayanan S, Jamshed M, Kumar A, Skori L, Scandola S, Wang T, Spiegel D, Samuel MA. 2017. Glyoxalase goes green: the expanding roles of glyoxalase in plants. Int J Mol Sci 18:898. doi: 10.3390/ijms18040898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castañeda A, Reddy JD, El-Yacoubi B, Gabriel DW. 2005. Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol Plant Microbe Interact 18:1306–1317. [DOI] [PubMed] [Google Scholar]
- 27.De Feyter R, Gabriel DW. 1991. Use of cloned DNA methylase genes to increase the frequency of transfer of foreign genes into Xanthomonas campestris pv. malvacearum. J Bacteriol 173:6421–6427. doi: 10.1128/jb.173.20.6421-6427.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vahling CM, Duan Y, Lin H. 2010. Characterization of an ATP translocase identified in the destructive plant pathogen “Candidatus Liberibacter asiaticus.” J Bacteriol 192:834–840. doi: 10.1128/JB.01279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai KK, Davis-Richardson AG, Dias R, Triplett EW. 2016. Identification of the genes required for the culture of Liberibacter crescens, the closest cultured relative of the Liberibacter plant pathogens. Front Microbiol 7:547. doi: 10.3389/fmicb.2016.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espartero J, Sanchez-Aguayo I, Pardo JM. 1995. Molecular characterization of glyoxalase-I from a higher plant: upregulation by stress. Plant Mol Biol 29:1223–1233. doi: 10.1007/BF00020464. [DOI] [PubMed] [Google Scholar]
- 31.Walz C, Giavalisco P, Schad M, Juenger M, Klose J, Kehr J. 2004. Proteomics of cucurbit phloem exudate reveals a network of defense proteins. Phytochemistry 65:1795–1804. doi: 10.1016/j.phytochem.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Singla-Pareek SL, Yadav SK, Pareek A, Reddy MK, Sopory SK. 2006. Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol 140:613–623. doi: 10.1104/pp.105.073734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang MQ, Guo Y, Powell CA, Doud MS, Yang CY, Zhou H, Duan YP. 2016. Zinc treatment increases the titre of “Candidatus Liberibacter asiaticus” in huanglongbing-affected citrus plants while affecting the bacterial microbiomes. J Appl Microbiol 120:1616–1628. doi: 10.1111/jam.13102. [DOI] [PubMed] [Google Scholar]
- 34.Hijaz F, Killiny N. 2014. Collection and chemical composition of phloem sap from Citrus sinensis L. Osbeck (sweet orange). PLoS One 9:e101830. doi: 10.1371/journal.pone.0101830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Killiny N. 2017. Metabolite signature of the phloem sap of fourteen citrus varieties with different degrees of tolerance to Candidatus Liberibacter asiaticus. Physiol Mol Plant Path 97:20–29. doi: 10.1016/j.pmpp.2016.11.004. [DOI] [Google Scholar]
- 36.Killiny N, Hijaz F, El-Shesheny I, Alfaress S, Jones SE, Rogers ME. 2017. Metabolomic analyses of the haemolymph of the Asian citrus psyllid Diaphorina citri, the vector of huanglongbing. Physiol Entomol 42:134–145. doi: 10.1111/phen.12183. [DOI] [Google Scholar]
- 37.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH. 2012. The major facilitator superfamily (MFS) revisited. FEBS J 279:2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canonaco F, Hess TA, Heri S, Wang T, Szyperski T, Sauer U. 2001. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble trans hydrogenase UdhA. FEMS Microbiol Lett 204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 39.Killiny N, Nehela Y, Hijaz F, Vincent CI. 2017. A plant pathogenic bacterium exploits the tricarboxylic acid cycle metabolic pathway of its insect vector. Virulence 8:1–11. doi: 10.1080/21505594.2017.1339008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. 2005. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of α-ketoglutarate decarboxylase. Proc Natl Acad Sci U S A 102:10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green LS, Li Y, Emerich DW, Bergersen FJ, Day DA. 2000. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J Bacteriol 182:2838–2844. doi: 10.1128/JB.182.10.2838-2844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prell J, Poole P. 2006. Metabolic changes of rhizobia in legume nodules. Trends Microbiol 14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M, Horn M. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to chlamydiae and rickettsiae. J Bacteriol 186:683–691. doi: 10.1128/JB.186.3.683-691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartung JS, Shao J, Kuykendall LD. 2011. Comparison of the “Ca. Liberibacter asiaticus” genome adapted for an intracellular lifestyle with other members of the Rhizobiales. PLoS One 6:e23289. doi: 10.1371/journal.pone.0023289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Killiny N, Hijaz F, Ebert TA, Rogers ME. 2017. A plant bacterial pathogen manipulates its insect vector's energy metabolism. Appl Environ Microbiol 83:e03005-16. doi: 10.1128/AEM.03005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pitino M, Armstrong CM, Duan Y. 2017. Molecular mechanisms behind the accumulation of ATP and H2O2 in citrus plants in response to “Candidatus Liberibacter asiaticus” infection. Hort Res 4:17040. doi: 10.1038/hortres.2017.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rott P, Fleites L, Marlow G, Royer M, Gabriel DW. 2011. Identification of new candidate pathogenicity factors in the xylem-invading pathogen Xanthomonas albilineans by transposon mutagenesis. Mol Plant Microbe Interact 24:594–605. doi: 10.1094/MPMI-07-10-0156. [DOI] [PubMed] [Google Scholar]
- 48.Jagoueix S, Bové JM, Garnier M. 1996. PCR detection of the two ‘Candidatus’ Liberibacter species associated with greening disease of citrus. Mol Cell Probe 10:43–50. doi: 10.1006/mcpr.1996.0006. [DOI] [PubMed] [Google Scholar]
- 49.Zhou LJ, Gabriel DW, Duan YP, Halbert SE, Dixon WN. 2007. First report of dodder transmission of huanglongbing from naturally infected Murraya paniculata to citrus. Plant Dis 91:227. doi: 10.1094/PDIS-91-2-0227B. [DOI] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.