ABSTRACT
Almost all animals possess gut microbial communities, but the nature of these communities varies immensely. For example, in social bees and mammals, the composition is relatively constant within species and is dominated by specialist bacteria that do not live elsewhere; in laboratory studies and field surveys of Drosophila melanogaster, however, gut communities consist of bacteria that are ingested with food and that vary widely among individuals and localities. We addressed whether an ecological specialist in its natural habitat has a microbiota dominated by gut specialists or by environmental bacteria. Drosophila nigrospiracula is a species that is endemic to the Sonoran Desert and is restricted to decaying tissues of two giant columnar cacti, Pachycereus pringlei (cardón cactus) and Carnegiea gigantea (saguaro cactus). We found that the D. nigrospiracula microbiota differs strikingly from that of the cactus tissue on which the flies feed. The most abundant bacteria in the flies are rare or completely absent in the cactus tissue and are consistently abundant in flies from different cacti and localities. Several of these fly-associated bacterial groups, such as the bacterial order Orbales and the genera Serpens and Dysgonomonas, have been identified in prior surveys of insects from the orders Hymenoptera, Coleoptera, Lepidoptera, and Diptera, including several Drosophila species. Although the functions of these bacterial groups are mostly unexplored, Orbales species studied in bees are known to break down plant polysaccharides and use the resulting sugars. Thus, these bacterial groups appear to be specialized to the insect gut environment, where they may colonize through direct host-to-host transmission in natural settings.
IMPORTANCE Flies in the genus Drosophila have become laboratory models for microbiota research, yet the bacteria commonly used in these experiments are rarely found in wild-caught flies and instead represent bacteria also present in the food. This study shows that an ecologically specialized Drosophila species possesses a distinctive microbiome, composed of bacterial types absent from the flies' natural food but widespread in other wild-caught insects. This study highlights the importance of fieldwork-informed microbiota research.
KEYWORDS: Drosophila, Dysgonomonas, gut microbiota, Orbales, Serpens
INTRODUCTION
In the genetic model organism Drosophila melanogaster, gut bacteria have been shown to have beneficial effects on fitness in various laboratory trials (1–5) and have been proposed to influence mating preferences and reproductive isolation (6). Despite its apparent importance to host biology, the composition of the microbiota of D. melanogaster varies widely, even among laboratories using the same genetic strain and the same diet, suggesting that variations in the rearing environments affect community composition (7, 8). Indeed, the bacteria retrieved from laboratory-reared D. melanogaster flies reflect the bacteria that live in the food source (9), and the size of the D. melanogaster gut community depends on bacterial titers in the food (10–12). Wild-caught Drosophila specimens show even greater variations in microbiota composition than do laboratory cultures, possibly reflecting variations in their environments (7, 8, 13–15).
The genus Drosophila includes a diversity of species, with different life histories, diets, and ecological associations (16), and thus provides a model for diversification and adaptation. While D. melanogaster is a cosmopolitan species that feeds on fermenting fruit and is associated with anthropogenic habitats, many other Drosophila species have more restricted geographic ranges and more specialized feeding habits. Studies of gut bacteria in wild-caught Drosophila species have revealed that the microbiotas of laboratory-reared Drosophila flies are not taxonomically or compositionally representative of those of wild flies (7, 8, 17). The most common bacteria in many wild-caught Drosophila specimens are from the recently described order Orbales (17), a group that has repeatedly been sampled from the guts of insects, including corbiculate bees, butterflies, darkling beetles, red palm weevils, and tephritid flies (18–20). Whereas the composition of laboratory-reared D. melanogaster microbiomes is heavily dependent on direct ingestion of bacteria with food (10, 12), it is not known whether this is also the case for wild populations of D. melanogaster or for other, ecologically diverse Drosophila species.
Prominent examples of ecological specialization in the genus Drosophila are the cactophilic species, for which larval development is restricted to the rotting tissue of particular cactus species indigenous to North and South American deserts. We investigated the microbiome of Drosophila nigrospiracula, a Drosophila repleta group species endemic to the Sonoran Desert, where it feeds and breeds exclusively in saguaro cactus (Carnegiea gigantea) or cardón cactus (Pachycereus pringlei), both of which are large columnar cacti (21–23). The restriction of D. nigrospiracula to these hosts, which was originally revealed in the rearing records from necrotic cacti and from collections of adults from rotting tissue (21), is attributed to the flies' ability to tolerate the specific alkaloids gigantine and carnegine that are found in the plant tissues (24). Although both cacti are used by D. nigrospiracula, they differ chemically and thus might be expected to harbor different microbiotas, with potential consequences for the microbiotas of the flies. Our study addresses the relationship between the composition of the host-associated microbiotas and microbes present in the environment; specifically, we focus on wild D. nigrospiracula flies and the necrotic cacti on which they feed.
RESULTS
Sample reads and OTU composition.
A 291-bp region of the 16S rRNA gene was amplified from 90 wild-collected cactus or fly tissue samples. After sequence quality trimming and rarefaction cutoffs, 21 cactus samples (7 saguaro cactus samples and 14 cardón cactus samples) and 63 D. nigrospiracula individuals were included in microbial community comparisons (see Data Set S1a in the supplemental material). Rarefaction analysis revealed that both cactus and D. nigrospiracula microbiotas were thoroughly sampled (Fig. S2). Subsampling at 30,000 sequences/sample across all samples resulted in a total of 894 bacterial operational taxonomic units (OTUs) (≥97% sequence similarity). Individual D. nigrospiracula flies harbored an average of 373 ± 12 OTUs (mean ± standard error [SE]), but only 50 ± 4 OTUs made up 90% of the total sequences across all flies. Cactus tissue samples had an average of 290 ± 10 OTUs, and 45 ± 4 OTUs accounted for 90% of the total sequences from cacti.
Variation in microbiota compositions.
The factor explaining the most variation in community composition, for both flies and cactus tissue, was the individual cactus of origin, which accounted for 19 to 35% of variation for flies and 30 to 53% for cactus samples (Table 1). The amount of variation explained was generally greater for presence/absence metrics than for metrics weighted by relative abundances. Further, microbiotas did not differ significantly between male and female flies (Table 1). Sampling at each locality and for each cactus species was too limited to make conclusions about an effect of locality or cactus species on microbiota composition.
TABLE 1.
Comparison of microbial community compositions with Adonisa
| Data included and comparison category | Weighted UniFrac |
Unweighted UniFrac |
Bray-Curtis dissimilarity |
Jaccard index |
||||
|---|---|---|---|---|---|---|---|---|
| P | r2 | P | r2 | P | r2 | P | r2 | |
| Flies and cacti | ||||||||
| Fly or cactus | 0.001 | 0.126 | 0.001 | 0.209 | 0.001 | 0.144 | 0.001 | 0.189 |
| Locality | 0.052 | 0.025 | 0.002 | 0.053 | 0.002 | 0.037 | 0.002 | 0.045 |
| Cactus individual | 0.001 | 0.134 | 0.001 | 0.234 | 0.001 | 0.156 | 0.001 | 0.188 |
| Only flies | ||||||||
| Cactus species | 0.001 | 0.099 | 0.001 | 0.173 | 0.001 | 0.079 | 0.001 | 0.125 |
| Cactus individual | 0.001 | 0.196 | 0.001 | 0.348 | 0.001 | 0.193 | 0.001 | 0.261 |
| Locality | 0.07 | 0.031 | 0.004 | 0.06 | 0.002 | 0.042 | 0.001 | 0.057 |
| Sex | 0.294 | 0.018 | 0.348 | 0.016 | 0.055 | 0.027 | 0.385 | 0.016 |
| Only cacti | ||||||||
| Cactus species | 0.002 | 0.244 | 0.042 | 0.097 | 0.002 | 0.188 | 0.022 | 0.121 |
| Cactus individual | 0.001 | 0.532 | 0.001 | 0.303 | 0.001 | 0.453 | 0.001 | 0.35 |
| Locality | 0.007 | 0.209 | 0.007 | 0.137 | 0.002 | 0.155 | 0.005 | 0.151 |
| Rot type | 0.001 | 0.352 | 0.001 | 0.291 | 0.001 | 0.336 | 0.001 | 0.329 |
Analysis of variance using distance matrices (Adonis function) was performed using 999 permutations, F tests, and 97% OTUs.
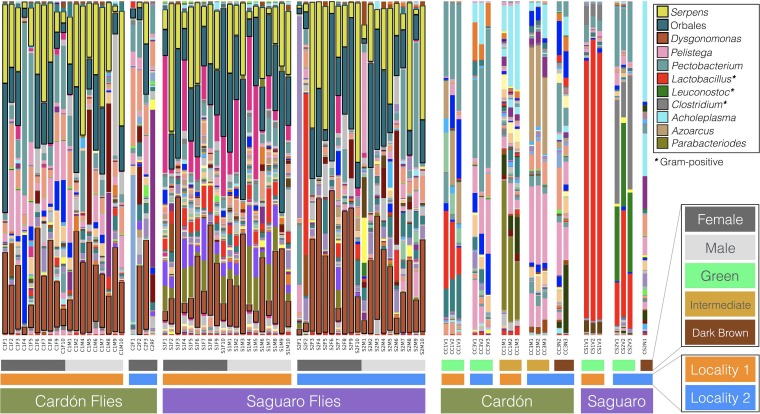
Cactus tissue samples displayed considerable variation in microbiotas (Fig. 1 and 2). Cactus tissues harbored similar numbers of bacterial OTUs regardless of their decay state (i.e., green, intermediate, or dark brown) (Data Set S1a). The taxonomic composition of less decayed samples (i.e., green) tended to have relatively high levels of representation of Firmicutes, especially Lactobacillus (Gram positive), whereas more decomposed tissues tended to have greater relative abundances of Enterobacteriaceae (Pectobacterium), Bacteroidetes (Dysgonomonas), Mollicutes (Acholeplasma), and Burkholderiales (Pelistega) (Fig. 2).
FIG 1.
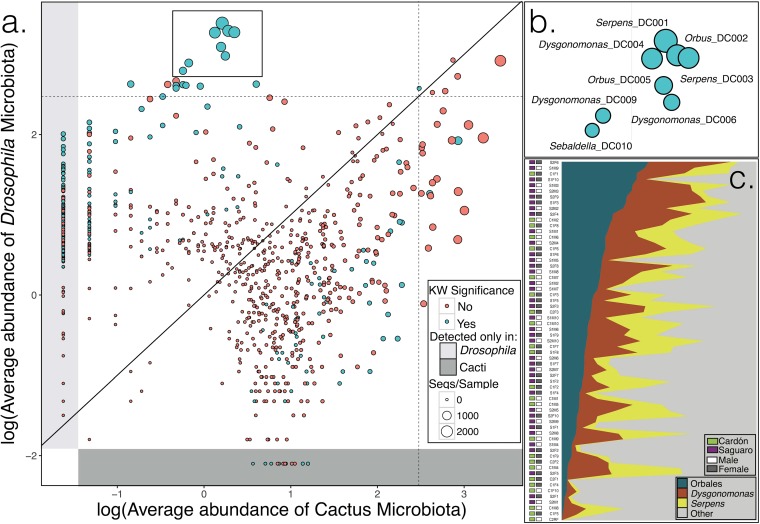
Abundance of bacteria in flies and cacti. (a) Average abundance of each OTU in Drosophila nigrospiracula individuals and in cactus tissues. Bacteria present at equal abundances in flies and cacti would fall near the diagonal line. OTUs that differ significantly in their distributions are colored green. Dashed lines indicate 1% of the average microbiota (subsampled at 30,000 sequences [Seqs]/sample) in flies and cacti (log10 scale). The plot includes all samples of flies and cacti. KW, Kruskal-Wallis. (b) Genus names and OTU identifications for highly abundant OTUs in D. nigrospiracula. (c) Relative abundances of bacteria in Orbales, Dysgonomonas, and Serpens across D. nigrospiracula individuals (OTUs were pooled by genus; samples were rarefied to 30,000 sequences/sample).
FIG 2.
Bacterial genera identified in D. nigrospiracula and cactus samples. Bars are for individual D. nigrospiracula flies or individual cactus tissue samples.
Comparison of microbiotas from cacti and Drosophila flies.
To test whether bacteria in the cactus food directly colonize the Drosophila gut, we compared the microbial communities of paired cactus tissue and fly individuals from the same cactus sample (Fig. S3). The composition of the D. nigrospiracula microbiota differed strongly from the communities in their cactus food with respect to membership and relative abundances (Kruskal-Wallis tests) (Table 1; also see Data Set S1d). The differences were more pronounced with presence/absence metrics that clearly differentiated cacti from flies (unweighted UniFrac analyses) (Fig. S4), indicating that low-abundance OTU membership differed in flies and cacti (Fig. 2). The unweighted pair group method with arithmetic mean (UPGMA) dendrograms of the microbiotas showed that cactus tissue bacterial communities generally clustered separately from D. nigrospiracula microbiotas (Fig. S5). Furthermore, the fly gut microbiotas not only were distinct but also were largely dominated by the OTUs that were absent or nearly absent from cactus tissues (Fig. 1).
Fly microbiotas, on average, had more bacterial OTUs (mean, 373 OTUs) than did cactus tissue (mean, 290 OTUs) [t(70.73) = 5.34; P < 0.0001], and this difference held if OTUs that were never >1% of any sample were removed [fly mean, 92 OTUs; cactus mean, 80 OTUs; t(36.30) = 3.79; P < 0.0003]. The majority of bacterial OTUs (670/894 OTUs) were shared at similar abundances and did not have significantly different distributions between flies and cactus tissues (Fig. 1), but there were many more OTUs overrepresented in flies than overrepresented in cactus samples. Among the significantly different OTUs, 74% (165/224 OTUs) were overrepresented in flies (Kruskal-Wallis test) (Data Set S1d). Nearly 20% of all OTUs (184/894 OTUs) were present only in flies and were undetected in cacti, whereas 10/894 OTUs were exclusive to cactus samples. The core microbiome analysis found 18 OTUs (including Orbales, Dysgonomonas, and Serpens) that were present in all D. nigrospiracula individuals and an additional 39 OTUs that were found in 90% of individuals (Data Set S1f). Many of the most abundant bacterial OTUs in the D. nigrospiracula microbiota were positively correlated with each other (Fig. S6). The bacterial OTUs that were enriched in flies were also broadly distributed across D. nigrospiracula individuals (with 57 OTUs present in >90% of the flies and 18 OTUs present in 100%), across cactus species, and across localities.
Widespread bacteria in wild Drosophila species.
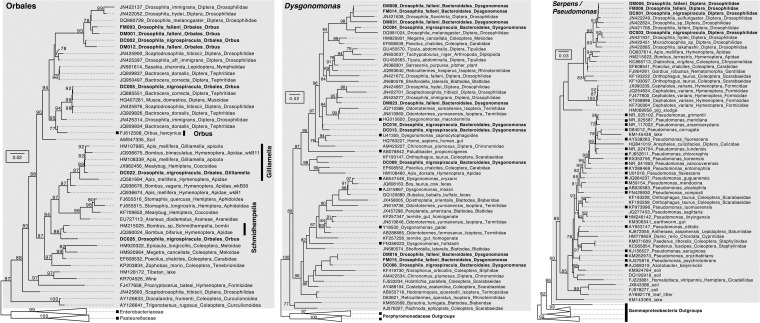
Among the 40 most abundant OTUs in D. nigrospiracula, nearly one-quarter (9/40 OTUs) had a 100% identical match in the top 40 OTUs in a recent study of the mycophagous (mushroom-feeding) Drosophila microbiota (263-bp region in the 16S rRNA gene) (Fig. S7). The mushroom-feeding Drosophila and the cactus-feeding Drosophila share Orbales, Dysgonomonas, and Serpens as gut-biased bacteria not detected in their mushroom or cactus food (Fig. 1 and 2; also see Fig. S3). Alignment with the previously published sequences from the microbiota of wild-caught Drosophila flies revealed that many of the abundant OTUs were closely related, often with >97% sequence similarity. Phylogenetic analyses showed that clades of Orbales, Dysgonomonas, and Serpens/Pseudomonas bacteria were associated with wild-collected Drosophila flies and also are found in many diverse insects (Fig. 3), suggesting that these represent insect gut specialists.
FIG 3.
Phylogenies of Orbales, Dysgonomonas, and Serpens/Pseudomonas sequences, showing that many sequences are found in association with insects. Sequences from cactophilic Drosophila nigrospiracula and mycophagous Drosophila falleni are in bold; representative bacterial species are marked with black squares; nodes have bootstrap support values. The sequences were collected from GenBank and RDP.
DISCUSSION
The D. nigrospiracula microbiota differs sharply from the bacterial communities in the flies' cactus food (Fig. 1) and largely consists of insect-specialized bacterial types. Individual flies consistently harbored identical bacterial OTUs across collection localities and host cactus species. Many of these consistently associated bacterial types were abundant members of the fly microbiota but were rare or undetected in the corresponding cactus tissue. Cactus microbiotas varied more than the fly microbiotas and appeared to differ greatly among individual cactus plants and among different states of decomposition. Several of the major bacterial taxa found in D. nigrospiracula were identified previously in distantly related and ecologically diverse Drosophila species and other insects. The widespread observation of these OTUs in Drosophila and their rarity apart from insects support the hypothesis that these bacterial taxa are specialized residents of insect guts.
In laboratory-based studies of D. melanogaster microbiotas, the food and the flies have similar microbiota compositions, dominated by Acetobacter and Lactobacillus species (7–11). These findings suggest that Drosophila adults that arrive in new food patches could seed them with microorganisms capable of metabolizing or detoxifying compounds that could then be utilized as food by developing flies, as has been shown for yeasts (25–30). In this scenario, the microbial communities of the food would be driven by Drosophila adults acting as vectors of microorganisms. However, we found a very different pattern for wild-caught D. nigrospiracula flies; we observed large compositional differences between the microbiotas of cactus tissues and flies. This finding parallels the recent discovery of differences between the microbiotas of mushrooms and those of mycophagous Drosophila species (17). In contrast, laboratory-reared Drosophila species harbor less microbial diversity and lack many of the most abundant microbiota members of their wild counterparts (7, 8). Although it is difficult to entirely rule out the presence of the gut-biased bacterial OTUs in the food tissue at extremely low abundance, we note that our methods can retrieve extremely rare taxa and that the cactus samples harbored fewer OTUs than did the D. nigrospiracula samples (Data Set S1a).
The bacterial OTUs that were generally abundant in the D. nigrospiracula microbiota were also broadly distributed across the flies surveyed and were concentrated in three distinctive taxonomic groups, Orbales, Dysgonomonas, and Serpens (Fig. 1b and c and 2; also see Fig. S3). These taxa were consistently present (Data Set S1f) and constituted large proportions of the microbiotas of individual Drosophila flies (Fig. 1c). These microorganisms are generally positively correlated with each other (Fig. S6) and may have metabolic interdependencies that reinforce their coexistence. Database searches and phylogenetic analyses show that these microbes are widespread in insects (Fig. 3); however, the 16S rRNA gene does not differentiate closely related sequences, and future work should use more phylogenetically informative genes. The current findings are aligned with the idea of a within-species core microbiota that is influenced by host ecology and the consequent transient microbes. Overall, our findings support the occurrence of one set of taxa that is determined by the external environment and diet and a second set of taxa, including members of these three distinctive groups, that is governed by internal gut community processes, which is consistent with some observations for the human gut microbiota (31).
Although flies undoubtedly encounter diverse bacteria throughout their lives, only a few phylogenetic groups dominate colonization of the Drosophila gut (7, 8, 17). Social interactions, broadly defined, can provide a route for microorganisms to colonize new hosts and may lead to widespread bacterium-host associations. Among honeybees, Gilliamella (a member of the order Orbales) is socially transmitted among colony members and forms a biofilm on the hindgut lining (32). Drosophila-associated Orbales species may be similarly transmitted among flies that cooccur at feeding, mating, oviposition, and defecation sites. Alternatively, gut conditions (e.g., low oxygen levels, host immune responses, and pH) may eliminate bacteria not specialized for the gut. Drosophila nigrospiracula larvae and adults are present only in actively necrotic (i.e., dark brown) tissue, whereas prenecrotic (i.e., green and intermediate) tissues largely lacked the fly-specific bacterial OTUs. Thus, the D. nigrospiracula-associated bacteria appear to be absent from the plant tissue until colonization by flies and then they are present in only low relative abundance. Defecation by the flies in the cactus tissue most likely is the source for the small number of fly-specific sequences retrieved from necrotic tissue; an alternative is that these bacteria, although rare, are a signature of a community that correlates with suitable breeding conditions for D. nigrospiracula.
Individual cactus samples had unique microbial communities, suggesting that microbiotas may be stochastically assembled from the local environment. The large compositional range of microbial communities among cacti may be due to the vast diversity of bacteria present in the soil environment that can structure microbial rhizosphere communities in saguaro and cardón cacti (33, 34). Bacterial communities within cacti were very similar among replicate samples of a tissue, but there were large compositional differences between tissues in different states of necrosis, even within the same cactus individual (Fig. 2). Decomposition greatly affected the bacterial communities; in a single cactus, Gram-positive species (Lactobacillus, Leuconostoc, and Clostridium) were common in early decomposition, whereas Gram-negative species (Acholeplasma, Azoarcus, and Parabacteroides) increased in abundance in later states of decay (Fig. 2). However, our limited sampling of cactus individuals prevents us from drawing broad conclusions about their microbiotas.
Many insect gut communities are dominated by environmentally derived microbes, with large variations among individuals (35–37). Exceptions are some species with social behavior (e.g., termites and corbiculate social bees such as Apis and Bombus) or conspicuous transmission mechanisms (e.g., egg smearing). A recent survey of gut communities in noncorbiculate social bees (Ceratina and Megalopta) revealed that, similar to findings for Drosophila and Apis, the microbiotas of guts were very different from those of food and Orbales species were much more abundant in the gut (38). Ecological conditions and not just sociality appear to be important in determining whether a core microbiota is present. For example, all ant species are social but only certain lifestyles appear to possess a core gut microbiota (39). The repeated retrieval of the same or closely related bacterial species from diverse insects (e.g., Drosophila, Apis, Bombus, and other species from Diptera, Hymenoptera, Coleoptera, and Lepidoptera) but not from environmental food sources suggests that some microbial groups might have been overlooked as insect specialists, particularly among insect species that aggregate at a shared food source (e.g., carrion or rotting plants or fruit). These common insect-associated bacteria are likely to be biologically relevant to their hosts. For example, the Apis gut associate Gilliamella apicola, a close relative of the Orbales species found in Drosophila, provides its host with fatty acids by fermenting plant polysaccharides and can detoxify sugars that can be poisonous to insects (40). Given the centrality of insects for ecosystem functioning, agriculture, and human disease transmission, elucidating the diversity, colonization mechanisms, and functional consequences of these bacteria may be useful in, for example, the conservation of pollinators and the biocontrol of pests.
Laboratory-reared Drosophila adults differ from wild counterparts in the taxonomic composition and overall diversity of their microbiotas. Much of the microbiota research using the model organism Drosophila melanogaster has focused on bacterial associates in the genera Acetobacter and Lactobacillus, but the ecological relevance of these associations has largely not been borne out in wild-caught individuals of Drosophila melanogaster or other Drosophila species (7, 8, 14). Indeed, Acetobacter and Lactobacillus are largely absent in wild-caught individuals of mushroom-feeding Drosophila species but are present in these species when they are reared in the laboratory (17). Similarly, this study shows that they are largely absent from wild-caught D. nigrospiracula individuals. Several explanations for these different distributions of gut-restricted bacteria and environmental bacteria are possible. Potentially, certain organisms, such as Acetobacter and Lactobacillus species, are less able to disperse among transient food sources in nature and thus are rarely picked up by Drosophila hosts. Additionally, Drosophila species may differ in immune responses, resulting in different characteristic gut microbiotas. The genus Drosophila contains over 1,300 species, representing highly diverse lifestyles and diets (16); of these, very few species have been surveyed for gut microbiota composition. Based on current sampling, it is possible that distinctive gut-restricted bacteria occur more often in ecologically specialized species, such as D. nigrospiracula and the mycophagous Drosophila species, than in ecologically generalized species, such as D. melanogaster.
In conclusion, our survey reveals a select community of bacteria associated with D. nigrospiracula and not with its food sources. Although wild D. nigrospiracula flies have a portion of their microbiota that appears to be environmentally variable and derived from the diet, the most abundant gut bacteria are undetected or extremely rare in the food. These gut-biased bacterial groups have now been identified in 10 Drosophila species that differ in ecology and geography (7, 14, 17); further, members of these same groups have been found in diverse insects (i.e., honeybees, bumblebees, cockroaches, termites, dung beetles, and house flies). In the debate about microbial biogeography, animal gut specialists do not seem to follow the “everything is everywhere” paradigm; instead, host movement, host aggregation, and fecal-oral transmission may enable gut-restricted bacteria to migrate to new resources. Meaningful microbiota research needs to focus on natural host-microbe interactions, informed by ecological interactions observed in nature.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Sampling was conducted in September and October 2014 in Bahia de Kino, Sonora, Mexico (see Fig. S1 in the supplemental material). At two localities, located 1 km apart, D. nigrospiracula adults and plant tissues were sampled at a decaying cardón cactus and a decaying saguaro cactus. Collection coordinates were as follows: 28°50′21.6″N, 111°47′41.8″W (locality 1) and 28°49′51.6″N, 111°48′03.6″W (locality 2). Cacti were 65 m and 223 m apart at locality 1 and locality 2, respectively. Flies were collected with an insect net directly from necrotic cactus tissue and were placed in empty sterile vials, where they spent no more than 2 h before being keyed to species and sex. Individual flies were stored in 1.5-ml tubes with 95% ethanol. Tissue samples of the cactus from which the flies were feeding were collected (in triplicate), with sterile tools, into 1.5-ml tubes with 95% ethanol and were stored until further processing.
DNA extraction was performed for each whole fly using the DNeasy blood and tissue kit (Qiagen, Valencia, CA), following the manufacturer's protocol, with the modification that flies were initially ground with sterile pestles in 1.5-ml tubes with animal tissue lysis (ATL) buffer and proteinase K and were incubated for 60 min at 56°C to increase the DNA yield. For decaying cactus samples, total DNA extraction was performed using the PowerPlant DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA), following the manufacturer's protocol and adding the phenolic separation solution. DNA extracts were normalized to ≤50 ng/μl. PCR assays for each sample and an extraction control were performed using the bacterial 16S rRNA primers 515f and 806r, to confirm the presence of bacterial DNA, before sequencing using the conditions described by Caporaso et al. (41). The extraction controls had no amplification.
Amplicon sequencing of bacterial 16S rRNA gene.
We used amplicon sequencing of the variable V4 region of the 16S rRNA gene to obtain microbiota profiles of flies and of the necrotic cactus tissue on which the flies were collected. PCR was performed as described by Caporaso et al. (41), using the barcoded primers 515f and 806r. Amplicon primer barcodes for individual samples are summarized in Data Set S1a. PCR assays and sequencing were performed at the High-Throughput Genome Analysis Core at the Argonne National Laboratory. Pooled amplicons from triplicate PCRs were prepared for each sample. Multiplexed, paired-end sequencing was performed (forward, 151 bp; reverse, 151 bp) using an Illumina MiSeq system. Sample processing, sequencing, and core amplicon data analysis were performed by the Earth Microbiome Project (http://www.earthmicrobiome.org) (42), and all amplicon sequence data and metadata have been made public through the Qiita data portal (qiita.microbio.me/emp). PCRs, library construction, and sequencing protocols were as detailed by Caporaso et al. (41).
Sequence assembly and quality control.
Barcode removal, sequence quality filtering, paired-read merging, OTU construction (97% sequence similarity), and chimeric screening were performed using QIIME (41), with default settings. Representative OTU sequences were assigned a taxonomic identity using the Greengenes and RDP (26 October 2016) databases (43, 44). To remove potential sequencing artifacts, OTUs that were present at <1% in all samples were removed prior to downstream analyses (45). To determine an appropriate subsampling depth, rarefaction and completeness curves for each sample were constructed with iNEXT in R (46, 47), using 50 bootstrap replicates. Samples with small numbers of reads (<2,500 reads) were not used in further analyses (Table S1). Rarefaction to 30,000 sequences/sample was performed to enable even-sampling comparisons. Raw and rarefied OTU abundance tables are presented in Data Set S1b and c, and representative sequences are presented in Data Set S1e. Representative OTUs were aligned with PyNAST (48), and phylogenies were created with FastTree (49) for phylogenetic diversity metrics.
Community diversity analyses.
Alpha diversity, richness, and coverage for each sample were estimated with QIIME (Data Set S1a). Pairwise dissimilarity (beta diversity) was measured using both relative abundance and presence/absence methods for phylogenetic metrics (weighted and unweighted UniFrac metrics) and nonphylogenetic metrics (Bray-Curtis dissimilarity and Jaccard index). Principal-coordinate analysis (PCoA) was performed with QIIME, using rarefied OTU tables (Data Set S1c). Microbial communities were hierarchically clustered with UPGMA analysis in QIIME, and node support was computed with 999 jackknife resamplings (30,000 sequences/sample). Dendrograms of UPGMA results were visualized with GraPhlAn (50).
Compositional differences among flies and cacti were tested for significance using the Adonis function (analysis of variance using distance metrics, also called nonparametric multivariate analysis of variance) (51). Pairwise dissimilarity matrices from different diversity metrics were used for Adonis analysis, and significance was based on F tests of permutations in QIIME. Core microbiome analysis was performed with QIIME for rarefied D. nigrospiracula individuals. Comparisons of the mean abundances of individual OTUs across D. nigrospiracula and cactus samples were performed using a nonparametric Kruskal-Wallis test with the Bonferroni correction, using a significance level of 0.05. OTUs were grouped by taxonomic assignment at the order level, to visualize differences in composition between flies and cacti. Pairwise correlations (Kendall's tau) were computed for the top 30 abundant OTUs among D. nigrospiracula individuals using the vegan package in R; OTUs were ordered with hierarchical clustering (i.e., hclust-complete), and results were visualized with corrplot (52).
Comparison of D. nigrospiracula microbiotas to previous surveys of wild Drosophila microbiotas.
Representative sequences for the top 40 abundant OTUs identified in mycophagous Drosophila species (17) were aligned to the top 40 OTUs we found in D. nigrospiracula with Infernal, in the RDP pipeline (53, 54). Regions that did not overlap were trimmed from the alignment, and a pairwise distance matrix was exported. A heatmap of the pairwise distances was created in superheat (https://github.com/rlbarter/superheat). BLAST searches in GenBank and the RDP Hierarchy Browser were used to collect sequences closely related to Orbales, Dysgonomonas, and Serpens/Pseudomonas. Sequences were aligned with Infernal, and the phylogeny was constructed with FastTree (49).
Accession number(s).
Molecular sequence data reported in this paper have been deposited in the NCBI Sequence Read Archive as part of BioProject PRJNA385203.
Supplementary Material
ACKNOWLEDGMENTS
Funding was provided by Mexico Consejo Nacional de Ciencia y Tecnología (CONACYT) grant 180385 to T.A.M., a CONACYT graduate fellowship to J.C.-P., and U.S. National Institutes of Health grant RO1GM108477-02 to N.A.M.
J.C.-P., N.A.M., and T.A.M. designed the survey, J.C.-P. collected data, and V.G.M. and J.C.-P. performed analyses. All authors wrote the paper and contributed substantially to revisions.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01551-17.
REFERENCES
- 1.Broderick NA, Buchon N, Lemaitre B. 2014. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. mBio 5:e01117-14. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brummel T, Ching A, Seroude L, Simon AF, Benzer S. 2004. Drosophila lifespan enhancement by exogenous bacteria. Proc Natl Acad Sci U S A 101:12974–12979. doi: 10.1073/pnas.0405207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaston JM, Dobson AJ, Newell PD, Douglas AE. 2016. Host genetic control of the microbiota mediates the Drosophila nutritional phenotype. Appl Environ Microbiol 82:671–679. doi: 10.1128/AEM.03301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 5.Wong AC, Dobson AJ, Douglas AE. 2014. Gut microbiota dictates the metabolic response of Drosophila to diet. J Exp Biol 217:1894–1901. doi: 10.1242/jeb.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. 2010. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci U S A 107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet 7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staubach F, Baines JF, Künzel S, Bik EM, Petrov DA. 2013. Host species and environmental effects on bacterial communities associated with Drosophila in the laboratory and in the natural environment. PLoS One 8:e70749. doi: 10.1371/journal.pone.0070749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong AC, Luo Y, Jing X, Franzenburg S, Bost A, Douglas AE. 2015. The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl Environ Microbiol 81:6232–6240. doi: 10.1128/AEM.01442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blum JE, Fischer CN, Miles J, Handelsman J. 2013. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 4:e00860-13. doi: 10.1128/mBio.00860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamps JA, Yang LH, Morales VM, Boundy-Mills KL. 2012. Drosophila regulate yeast density and increase yeast community similarity in a natural substrate. PLoS One 7:e42238. doi: 10.1371/journal.pone.0042238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada R, Deshpande SA, Bruce KD, Mak EM, Ja WW. 2015. Microbes promote amino acid harvest to rescue undernutrition in Drosophila. Cell Rep 10:865–872. doi: 10.1016/j.celrep.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broderick NA, Lemaitre B. 2012. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, Promislow DEL. 2007. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl Environ Microbiol 73:3470–3479. doi: 10.1128/AEM.02120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong AC, Chaston JM, Douglas AE. 2013. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J 7:1922–1932. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markow TA. 2015. The secret lives of Drosophila flies. eLife 4:e06793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinson VG, Douglas AE, Jaenike J. 2017. Community structure of the gut microbiota in sympatric species of wild Drosophila. Ecol Lett 20:629–639. doi: 10.1111/ele.12761. [DOI] [PubMed] [Google Scholar]
- 18.Hammer TJ, McMillan WO, Fierer N. 2014. Metamorphosis of a butterfly-associated bacterial community. PLoS One 9:e86995. doi: 10.1371/journal.pone.0086995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JY, Lee J, Shin NR, Yun JH, Whon TW, Kim MS, Jung MJ, Roh SW, Hyun DW, Bae JW. 2013. Orbus sasakiae sp. nov., a bacterium isolated from the gut of the butterfly Sasakia charonda, and emended description of the genus Orbus. Int J Syst Evol Microbiol 63:1766–1770. doi: 10.1099/ijs.0.041871-0. [DOI] [PubMed] [Google Scholar]
- 20.Kwong WK, Moran NA. 2013. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: Snodgrassella alvi gen. nov., sp. nov., a member of the Neisseriaceae family of the Betaproteobacteria; and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the Enterobacteriales order of the Gammaproteobacteria. Int J Syst Evol Microbiol 63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 21.Fellows DP, Heed WB. 1972. Factors affecting host plant selection in desert-adapted cactiphilic Drosophila. Ecology 53:850–858. doi: 10.2307/1934300. [DOI] [Google Scholar]
- 22.Heed WB. 1978. Ecology and genetics of Sonoran Desert Drosophila, p 109–126. In Brassard PF. (ed), Ecological genetics: the interface. Springer-Verlag, New York, NY. [Google Scholar]
- 23.Heed WB. 1982. The origin of Drosophila in the Sonoran Desert, p 65–80. In Barker JSF, Starmer WT (ed), Ecological genetics and evolution: the cactus-yeast-Drosophila model system. Academic Press, Sydney, Australia. [Google Scholar]
- 24.Danielson PB, Frank MR, Fogleman JC. 1994. Comparison of larval and adult P-450 activity levels for alkaloid metabolism in desert Drosophila. J Chem Ecol 20:1893–1906. doi: 10.1007/BF02066231. [DOI] [PubMed] [Google Scholar]
- 25.Giglioli I. 1897. Insects and yeasts. Nature 56:575–577. [Google Scholar]
- 26.Gilbert DG. 1980. Dispersal of yeasts and bacteria by Drosophila in a temperate forest. Oecologia 46:135–137. doi: 10.1007/BF00346979. [DOI] [PubMed] [Google Scholar]
- 27.Starmer WT, Aberdeen V. 1990. The nutritional importance of pure and mixed cultures of yeasts in the development of Drosophila mulleri larvae in Opuntia tissues and its relationship to host plant shifts, p 145–160. In Barker JSF, Starmer WT, MacIntyre RJ (ed), Ecological and evolutionary genetics of Drosophila. Springer, New York, NY. [Google Scholar]
- 28.Starmer WT, Barker JSF, Phaff HJ, Fogleman JC. 1986. Adaptations of Drosophila and yeasts: their interactions with the volatile 2-propanol in the cactus microorganism Drosophila model system. Aust J Biol Sci 39:69–77. [PubMed] [Google Scholar]
- 29.Starmer WT, Peris F, Fontdevila A. 1988. The transmission of yeasts by Drosophila buzzatii during courtship and mating. Anim Behav 36:1691–1695. doi: 10.1016/S0003-3472(88)80109-X. [DOI] [Google Scholar]
- 30.Starmer WT, Phaff HJ, Bowles JM, Lachance MA. 1988. Yeasts vectored by insects feeding on decaying saguaro cactus. Southwest Nat 33:362–363. doi: 10.2307/3671766. [DOI] [Google Scholar]
- 31.Gibbons SM, Kearney SM, Smillie CS, Alm EJ. 2017. Two dynamic regimes in the human gut microbiome. PLoS Comput Biol 13:e1005364. doi: 10.1371/journal.pcbi.1005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honeybee worker. Appl Environ Microbiol 78:2830–2840. doi: 10.1128/AEM.07810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrew DR, Fitak RR, Munguia-Vega A, Racolta A, Martinson VG, Dontsova K. 2012. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl Environ Microbiol 78:7527–7537. doi: 10.1128/AEM.01459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonseca-Garcia C, Coleman-Derr D, Garrido E, Visel A, Tringe SG, Partida-Martinez LP. 2016. The cacti microbiome: interplay between habitat-filtering and host-specificity. Front Microbiol 7:150. doi: 10.3389/fmicb.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones RT, Sanchez LG, Fierer N. 2013. A cross-taxon analysis of insect-associated bacterial diversity. PLoS One 8:e61218. doi: 10.1371/journal.pone.0061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yun J-H, Roh SW, Whon TW, Jung M-J, Kim M-S, Park D-S, Yoon C, Nam Y-D, Kim Y-J, Choi J-H. 2014. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl Environ Microbiol 80:5254–5264. doi: 10.1128/AEM.01226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP, Fierer N. 2017. Caterpillars lack a resident gut microbiome. Proc Natl Acad Sci U S A 114:9641–9646. doi: 10.1073/pnas.1707186114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graystock P, Rehan SM, McFrederick QS. 2017. Hunting for healthy microbiomes: determining the core microbiomes of Ceratina, Megalopta, and Apis bees and how they associate with microbes in bee collected pollen. Conserv Genet 18:701–711. doi: 10.1007/s10592-017-0937-7. [DOI] [Google Scholar]
- 39.Sanders JG, Łukasik P, Frederickson ME, Russell JA, Koga R, Knight R, Pierce NE. 2017. Dramatic differences in gut bacterial densities correlate with diet and habitat in rainforest ants. Integr Comp Biol 57:705–722. doi: 10.1093/icb/icx088. [DOI] [PubMed] [Google Scholar]
- 40.Zheng H, Nishida A, Kwong WK, Koch H, Engel P, Steele MI, Moran NA. 2016. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio 7:e01326–16. doi: 10.1128/mBio.01326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilbert JA, Jansson JK, Knight R. 2014. The Earth Microbiome Project: successes and aspirations. BMC Biol 12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan YM, Wang Q, Cole JR, Rosen GL. 2012. Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS One 7:e32491. doi: 10.1371/journal.pone.0032491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chao A, Gotelli NJ, Hsieh TC, Sander EL, Ma KH, Colwell RK, Ellison AM. 2014. Rarefaction and extrapolation with Hill numbers: a framework for sampling and estimation in species diversity studies. Ecol Monogr 84:45–67. doi: 10.1890/13-0133.1. [DOI] [Google Scholar]
- 47.Hsieh TC, Ma KH, Chao A. 2013. iNEXT online: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7:1451–1456. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 48.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. 2015. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 52.Wei T. 2015. CORRPLOT, visualization of a correlation matrix, v0.73. https://cran.r-project.org/web/packages/corrplot/corrplot.pdf.
- 53.Cole JR, Wang Q, Fish JA, Chai BL, McGarrell DM, Sun YN, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.