ABSTRACT
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:H− strains, first identified in Germany, have emerged as important pathogens throughout Europe. Besides chromosomally encoded Shiga toxin 2a (the major virulence factor), several putative virulence loci, including the hly, etp, and sfp operons, encoding EHEC hemolysin, type II secretion system proteins, and Sfp fimbriae, respectively, are located on the 121-kb plasmid pSFO157 in German strains. Here we report novel SF EHEC O157:H− strains isolated from patients in the Czech Republic. These strains share the core genomes and chromosomal virulence loci encoding toxins (stx2a and the cdtV-ABC operon) and adhesins (eae-γ, efa1, lpfAO157OI-141, and lpfAO157OI-154) with German strains but differ essentially in their plasmids. In contrast to all previously detected SF EHEC O157:H− strains, the Czech strains carry two plasmids, of 79 kb and 86 kb. The 79-kb plasmid harbors the sfp operon, but neither of the plasmids contains the hly and etp operons. Sequence analyses demonstrated that the 79-kb plasmid (pSFO157 258/98-1) evolved from pSFO157 of German strains by deletion of a 41,534-bp region via homologous recombination, resulting in loss of the hly and etp operons. The 86-kb plasmid (pSFO157 258/98-2) displays 98% sequence similarity to a 92.7-kb plasmid of an extraintestinal pathogenic E. coli bloodstream isolate. Our finding of this novel plasmid composition in SF EHEC O157:H− strains extends the evolutionary history of EHEC O157 plasmids. Moreover, the unique molecular plasmid characteristics permit the identification of such strains, thereby facilitating further investigations of their geographic distribution, clinical significance, and epidemiology.
IMPORTANCE Since their first identification in Germany in 1989, sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H− (nonmotile) strains have emerged as important causes of the life-threatening disease hemolytic-uremic syndrome in Europe. They account for 10 to 20% of sporadic cases of this disease and have caused several large outbreaks. The strains isolated throughout Europe share conserved chromosomal and plasmid characteristics. Here we identified novel sorbitol-fermenting enterohemorrhagic E. coli O157:H− patient isolates in the Czech Republic which differ from all such strains reported previously by their unique plasmid characteristics, including plasmid number, composition of plasmid-carried virulence genes, and plasmid origins. Our findings contribute substantially to understanding the evolution of E. coli O157 strains and their plasmids. In practical terms, they enable the identification of strains with these novel plasmid characteristics in patient stool samples and thus the investigation of their roles as human pathogens in other geographic areas.
KEYWORDS: EHEC O157, outbreaks, Sfp fimbriae, enterohemorrhagic E. coli, hemolytic-uremic syndrome, plasmid analysis
INTRODUCTION
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:H− (nonmotile) strains, first identified in Germany in 1989 (1), have emerged as important human pathogens throughout Europe (2–10). In Germany and the Czech Republic, SF EHEC O157:H− strains account for ∼20% and ∼13%, respectively, of all EHEC strains isolated from patients with hemolytic-uremic syndrome (HUS) (10, 11). In the Czech Republic, SF EHEC O157:H− strains cause HUS as frequently as classical non-sorbitol-fermenting (NSF) EHEC O157:H7 strains do (10). SF EHEC O157:H− strains have caused several outbreaks in Europe (1, 8, 12–19), the three largest of which consisted of 38, 28, and 25 HUS cases and occurred in Germany (12, 13, 19). The outbreaks caused by SF EHEC O157:H− were characterized by an accumulation of HUS cases (“HUS outbreaks”) without a parallel increase of surrounding cases of diarrhea (12–14, 16–19), which is typical for outbreaks caused by NSF EHEC O157:H7 (20). This indicates a high rate of progression of SF EHEC O157:H− infections to HUS, which, together with the high case fatality rate (8.8% in the three largest outbreaks) (12, 13, 19), suggests that SF EHEC O157:H− might be more virulent than NSF EHEC O157:H7 (13–16). Although the reasons for this are still unknown, differences in virulence factor spectra between these two groups of EHEC O157 strains (15, 21–23) might contribute, at least partially, to the apparently increased virulence of SF EHEC O157:H−.
Besides their defining characteristics, i.e., nonmotility and the ability to ferment sorbitol and produce β-d-glucuronidase, SF EHEC O157 strains differ from NSF EHEC O157:H7 strains by a spectrum of molecular and phenotypic features (2). The major chromosomal differences include the following: (i) the presence of the cdtV-ABC operon, encoding cytolethal distending toxin V (CdtV), in most SF EHEC O157:H− strains, compared to its rare occurrence in NSF EHEC O157:H7 strains (23); (ii) the presence of a complete efa1 locus, encoding the EHEC factor for adherence (Efa1), in SF EHEC O157:H− strains, in contrast to a truncated efa1 locus in NSF EHEC O157:H7 strains (24); (iii) the absence of the ter operon, encoding tellurite resistance (25), the ure operon, encoding urease (26), and iha, encoding the iron-regulated gene A homologue adhesin (Iha) (27), in SF EHEC O157:H− strains, in contrast to the regular presence of these loci in NSF EHEC O157:H7 strains (25–27); and (iv) the presence of intact operons encoding type 1 fimbriae and curli fimbriae and expression of the respective adhesins in SF EHEC O157:H− strains (15, 28) but not in NSF EHEC O157:H7 strains, in which these loci are deleted or mutated (15, 28, 29). In addition to these chromosomal differences, SF and NSF EHEC O157 strains also differ in their plasmid contents and plasmid gene compositions. Specifically, SF EHEC O157:H− strains carry a 121-kb plasmid (pSFO157) (30, 31) in lieu of the 92-kb plasmid of EHEC O157:H7 (pO157) (32, 33). The pSFO157 plasmid typically contains the hlyCABD operon, encoding EHEC hemolysin (30, 34), an important EHEC virulence factor (35), and the etp operon, which encodes a type II secretion system (30, 36). However, espP and katP, encoding the serine protease EspP and catalase peroxidase, respectively, which are regularly found together with hlyCABD and etp on pO157 (32, 33), are absent from pSFO157 (30). In their stead, the sfpAHCDJFG operon, encoding Sfp fimbriae (37), a putative adhesin of SF EHEC O157:H− (22), is present (30). This plasmid virulence gene composition is conserved among SF EHEC O157:H− strains isolated in Germany and other countries (2, 4, 6, 7, 18, 38). We isolated SF EHEC O157:H− strains from patients in the Czech Republic that differ from the typical German SF EHEC O157:H− strains in their plasmid profile and plasmid virulence gene composition. In the present study, we compared the phylogenies, molecular characteristics, and corresponding phenotypes of these unusual SF EHEC O157:H− strains to those of prototype German SF EHEC O157:H− strains. We provide sequences of the novel plasmids harbored by the Czech SF EHEC O157:H− strains and portray their evolutionary history.
RESULTS
Phylogenetic relationships of Czech and German SF EHEC O157:H− strains.
The novel SF EHEC O157:H− Czech strains 258/98 and 269/98 with the unique features were isolated from patients with HUS and diarrhea, respectively (Table 1). Similar to prototype German SF EHEC O157:H− strains 493/89 and 3072/96, the Czech strains belonged to sequence type (ST) 11 and clonal complex (CC) 11 by multilocus sequence typing (MLST) (Table 1). The same was true for NSF EHEC O157:H7 strains EDL933 and Sakai, which were used for comparison (Table 1). To gain deeper insight into the phylogenetic relationships of the strains investigated, we performed whole-genome sequencing (WGS) of strains 258/98 and 3072/96 and compared the core genome coding regions with published WGS data for strains 493/89, EDL933, and Sakai. Altogether, 2,299 genes were present in all strains (see Table S1 in the supplemental material). Analysis of clonal relationships based on the allelic profiles derived from sequences of these 2,299 genes demonstrated that the Czech and German SF EHEC O157:H− strains are closely related to each other but differ by almost 600 alleles from NSF EHEC O157:H7 (Fig. 1).
TABLE 1.
Phylogenies, chromosomal virulence loci, and corresponding phenotypes of Czech and German SF EHEC O157:H− strains
| Strain | Reference | Disease or origin, country of origina | Serotype [fliC type] | ST (CC)b | stx locus (or loci) | Presence of chromosomal virulence locus |
Phenotypeg |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cdtVc | eae | efa1d | lpfAO157 OI-141 | lpfAO157 OI-154 | iha | tere | uref | Stx type (LA) | Stx titer (Vero cells) | CdtV titer (CHO cells) | Growth on CT-SMAC | SF/GUD phenotype | ||||||
| 258/98 | 3 | HUS, CR | O157:H− [H7] | 11 (11) | stx2a | + | γ | + | + | + | − | − | − | 2a | 1:1,024 | 1:8 | − | +/+ |
| 269/98 | 3 | D, CR | O157:H− [H7] | 11 (11) | stx2a | + | γ | + | + | + | − | − | − | 2a | 1:512 | 1:8 | − | +/+ |
| 493/89 | 70 | HUS, G | O157:H− [H7] | 11 (11) | stx2a | + | γ | + | + | + | − | − | − | 2a | 1:512 | 1:4 | − | +/+ |
| 3072/96 | 30, 37 | HUS, G | O157:H− [H7] | 11 (11) | stx2a | + | γ | + | + | + | − | − | − | 2a | 1:1,024 | 1:4 | − | +/+ |
| EDL933 | 71 | Outbreak, USA | O157:H7 [H7] | 11 (11) | stx1a + stx2a | − | γ | − | + | + | + | + | + | 1a + 2a | 1:2,048 | <1:2 | + | −/− |
| Sakai | 72 | Outbreak, J | O157:H7 [H7] | 11 (11) | stx1a + stx2a | − | γ | − | + | + | + | + | + | 1a + 2a | 1:2,048 | <1:2 | + | −/− |
HUS, hemolytic-uremic syndrome; D, diarrhea; G, Germany; CR, Czech Republic; USA, United States; J, Japan.
ST, sequence type; CC, clonal complex.
+, all components of the cdtV operon (cdtV-A, cdtV-B, and cdtV-C) were present; −, all these genes were absent.
+, complete efa1 was detected by PCR; −, only the 5′ fragment of efa1 was present.
+, all genes of the ter operon (terZABCDEF) were present; −, all these genes were absent.
+, all genes of the ure operon (ureABCDEFG) were present; −, all these genes were absent.
Shiga toxin (Stx) and cytolethal distending toxin V (CdtV) titers are the highest dilutions of culture supernatants that caused typical morphological effects in 50% of cells. LA, latex agglutination assay; CHO, Chinese hamster ovary cells; CT-SMAC, cefixime-tellurite sorbitol MacConkey agar; SF, sorbitol fermentation; GUD, β-d-glucuronidase.
FIG 1.
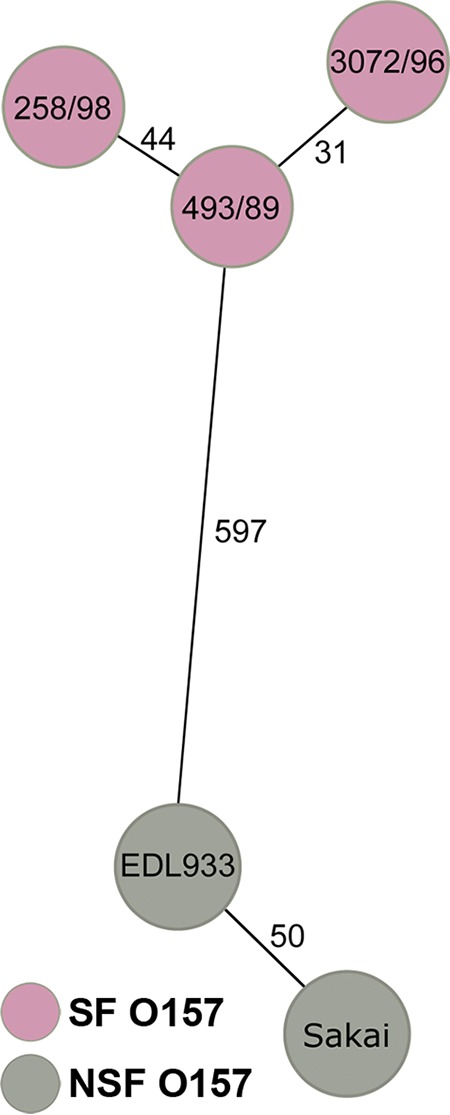
Whole-genome relationships of sorbitol-fermenting (SF) EHEC O157:H− Czech (258/98) and German (493/89 and 3072/96) strains in comparison to non-sorbitol-fermenting (NSF) EHEC O157:H7 strains. The minimum spanning tree is based on allelic profiles derived from sequences of 2,299 core genome genes present in all strains (shown in Table S1 in the supplemental material). Each circle represents a given allelic profile based on these 2,299 target genes. SF and NSF EHEC O157 strains are distinguished by the colors of the circles. The numbers on the connecting lines illustrate the numbers of alleles by which the respective strains differ.
Czech and German SF EHEC O157 strains share chromosomal virulence loci and phenotypes.
Similar to prototype German SF EHEC O157:H− strains 493/89 and 3072/96, the Czech SF EHEC O157 strains 258/98 and 269/98 were nonmotile (Table 1). Molecular subtyping of the flagellar subunit-encoding fliC gene demonstrated that all SF EHEC O157:H− strains possessed fliCH7, as did NSF EHEC O157:H7 strains EDL933 and Sakai (Table 1). The presence of O157 lipopolysaccharide (LPS) was confirmed for all strains by a PCR assay targeting rfbEO157. Chromosomal virulence loci and the respective phenotypes were identical for Czech and German SF EHEC O157:H− strains. Specifically, all strains possessed stx2a but not stx1a, expressed only Stx2a in a latex agglutination assay, and produced similar cytotoxicity titers on Vero cells (Table 1). Moreover, all possessed the cdtV-ABC operon, encoding CdtV, and expressed biologically active CdtV as demonstrated by the ability of culture supernatants to cause a progressive distension of Chinese hamster ovary (CHO) cells (Table 1), which is a hallmark of Cdt biological activity (23). All SF O157:H− strains shared several adhesin-encoding genes, including eae (subtype γ), encoding the major EHEC adhesin intimin (39), efa1, encoding Efa1, and lpfAO157OI-141 and lpfAO157OI-154, encoding major subunits of EHEC O157 long polar fimbriae (LPF) on O island (OI) 141 (LPF-1) and OI 154 (LPF-2), respectively (40, 41). In contrast, none of the strains carried iha, encoding Iha (Table 1). Furthermore, none of the SF EHEC O157:H− strains harbored the terZABCDEF operon, encoding tellurite resistance, and accordingly, none of them grew on cefixime-tellurite sorbitol MacConkey agar (CT-SMAC). Similarly, the ureDABCEFG operon, encoding urease, was absent from all SF EHEC O157:H− strains (Table 1). The majority of the chromosomal virulence characteristics of the Czech and German SF EHEC O157:H− strains differed from those of NSF EHEC O157:H7 strains EDL933 and Sakai (Table 1).
Czech and German SF EHEC O157 strains differ in their plasmid profiles and plasmid-carried virulence loci.
In contrast to the conserved chromosomal virulence characteristics, striking differences were found in plasmid profiles and plasmid gene compositions between the Czech and German SF EHEC O157:H− strains. Plasmid profiling demonstrated that Czech strains 258/98 and 269/98 contained two large plasmids, of ∼79 kb and ∼86 kb, whereas a single plasmid of 121 kb was present in German strains 493/89 and 3072/96 (Table 2; Fig. 2). In contrast to the German strains, which carried EHEC-hlyA and etpD on their plasmids, the Czech strains did not carry these genes as determined by both PCR and Southern blot hybridization (Table 2). However, sfpA, encoding the major subunit of Sfp fimbriae (37), which is a typical molecular feature of SF EHEC O157:H− (42), was present in each of the Czech strains and was located on a SmaI plasmid fragment of the same size as that for sfpA in German strains (Table 2). Hybridization of undigested plasmids with the sfpA probe demonstrated that sfpA is located on the 79-kb plasmid, but not on the 86-kb plasmid, in Czech strains and on the 121-kb plasmid in German strains (Table 2; Fig. 2). The plasmid profiles and plasmid gene compositions of Czech and German SF EHEC O157:H− strains differed from those of NSF EHEC O157:H7 strains EDL933 and Sakai (Table 2; Fig. 2); the latter strains harbored, in accordance with published data (32, 33), 92-kb plasmids containing the EHEC-hlyA, katP, espP, and etpD genes but not sfpA (Table 2). In contrast to NSF EHEC O157:H7 strains that produced EHEC hemolysin, neither EHEC-hlyA-negative Czech SF EHEC O157:H− strains nor EHEC-hlyA-positive German SF EHEC O157:H− strains displayed an enterohemolytic phenotype (Table 2). The lack of EHEC hemolysin production is a common feature of EHEC-hlyA-harboring SF EHEC O157:H− strains from different countries (2, 4, 6, 7, 10), but this could not be explained by analysis of the pSFO157 sequence (30). Unlike the results for EHEC hemolysin, both Czech and German SF EHEC O157:H− strains expressed Sfp fimbriae as demonstrated by immunoblotting with anti-SfpA antibody (Table 2).
TABLE 2.
Plasmid profiles, plasmid-borne virulence genes, and corresponding phenotypes of Czech and German SF EHEC O157:H− strains
| Strain | Serotype | Country of origina | Plasmid size (kb)/sfpA hybridizationb | Presence of plasmid-borne virulence genec |
Phenotype |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| EHEC-hlyA | katP | espP | etpD | sfpA | EHEC hemolysind | Sfp fimbriaee | ||||
| 258/98 | O157:H− | CR | 79/+, 86/− | −/− | −/− | −/− | −/− | +/4.9 | − | + |
| 269/98 | O157:H− | CR | 79/+, 86/− | −/− | −/− | −/− | −/− | +/4.9 | − | + |
| 493/89 | O157:H− | G | 121/+ | +/15.0 | −/− | −/− | +/3.9; 1.9 | +/4.9 | − | + |
| 3072/96 | O157:H− | G | 121/+ | +/15.0 | −/− | −/− | +/3.9; 1.9 | +/4.9 | − | + |
| EDL933 | O157:H7 | USA | 92/− | +/12.0 | +/9.0 | +/7.5 | +/3.9; 1.9 | −/− | + | − |
| Sakai | O157:H7 | J | 92/− | +/12.0 | +/9.0 | +/7.5 | +/3.9; 1.9 | −/− | + | − |
CR, Czech Republic; G, Germany; USA, United States; J, Japan.
Hybridization of undigested plasmids with an sfpA probe. +, positive result; −, no signal obtained.
Detection of genes was performed by PCR/Southern blot hybridization with the respective probe. +, positive result; −, no signal obtained. Plasmid DNA was digested with BamHI before hybridization with the EHEC-hlyA, espP, and etpD probes and with SmaI before hybridization with the katP and sfpA probes. Sizes of hybridizing fragments (in kilobases) are shown.
Production of EHEC hemolysin was sought on enterohemolysin agar. +, hemolysis present; −, no hemolysis (SF EHEC O157:H− strains usually do not produce EHEC hemolysin even though they possess the hlyCABD operon).
Sfp fimbriae were detected by immunoblotting with an anti-SfpA antibody. +, signal present; −, signal absent.
FIG 2.
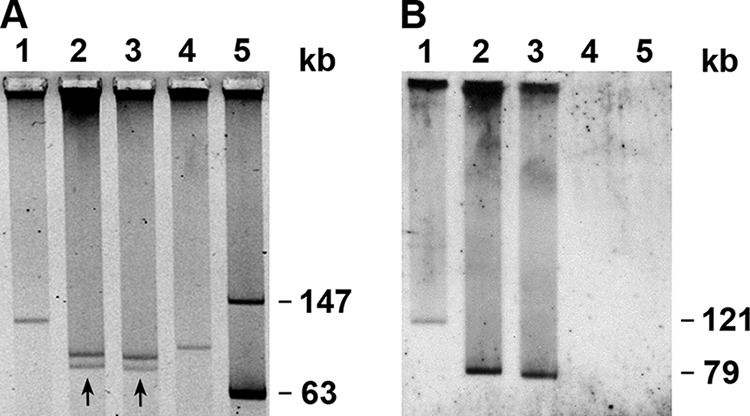
Plasmid profiles (A) and plasmid hybridization with an sfpA probe (B) of SF EHEC O157:H− Czech and German strains and EHEC O157:H7 strain EDL933. Lanes 1, strain 3072/96 (SF EHEC O157:H−, Germany); lanes 2, strain 258/98 (SF EHEC O157:H−, Czech Republic); lanes 3, strain 269/98 (SF EHEC O157:H−, Czech Republic); lanes 4, strain EDL933 (NSF EHEC O157:H7, United States); lanes 5, molecular mass marker (plasmid 39R861) (69). The plasmids of strains 258/98 and 269/98 that hybridized with the sfpA probe are marked by arrows in panel A, and the sizes of the sfpA-hybridizing plasmids are indicated in panel B.
Sequence analyses and origins of plasmids of Czech SF EHEC O157:H− strain 258/98.
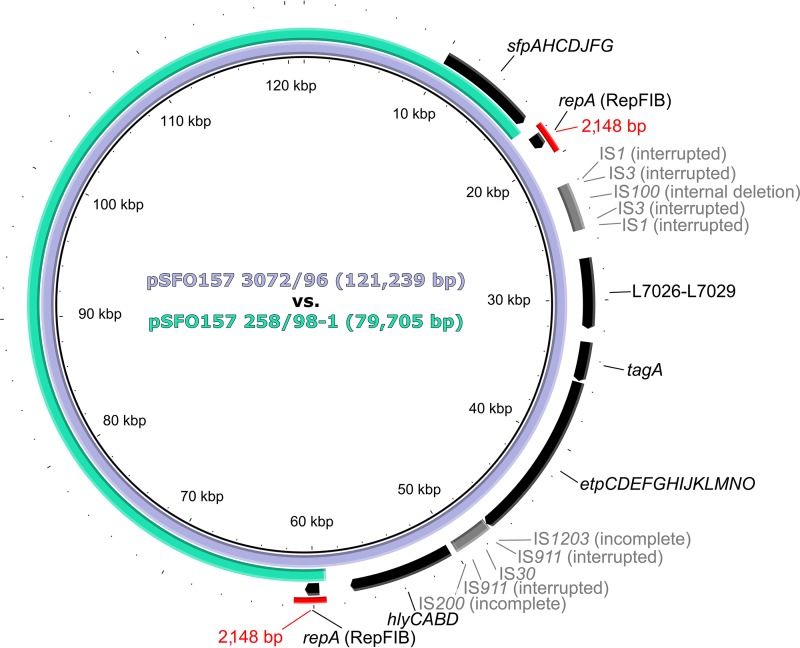
To gain a deeper insight into the differences between the plasmids of the Czech and German SF EHEC O157:H− strains, we sequenced plasmid DNA from strain 258/98 and compared the sequence to the published sequence of pSFO157 from strain 3072/96 (GenBank accession no. AF401292) (30). Sequencing of the 258/98 plasmid DNA resulted in 175,092 reads; of these, 65,933 mapped to the reference sequence of pSFO157 3072/96, resulting in an average 123-fold sequencing depth. The mapped reads of the 258/98 plasmid DNA displayed a perfect match, without any variations, for 67 of the 96 annotated open reading frames (ORFs) of the reference sequence. The remaining ORFs were not detected. In-depth analysis of the mapping results identified a deletion of ∼41 kb in the 258/98 plasmid, spanning the region from ORF w0013 to ORF w0041 in the reference sequence. This deletion was further specified by use of the de novo assembly data as being 41,534 bp, which corresponds to nucleotide positions 17,615 to 59,148 in the reference sequence (Fig. 3). We confirmed the gap by mapping the de novo contigs. The gap was flanked by two nearly identical (99%) regions of 2,148 bp, containing the identical ORFs w0013 and w0041, encoding the IncFIB replication protein RepA (Fig. 3). This indicated that this pSFO157 258/98 plasmid (which we term pSFO157 258/98-1) evolved from pSFO157 3072/96 via deletion of a 41,534-bp region by homologous recombination between the 2,148-bp fragments, resulting in a plasmid size of 79,705 bp (Fig. 3). The sequence of the pSFO157 258/98-1 plasmid, including the Sfp fimbria-encoding sfp operon located on this plasmid (Fig. 3), was 100% identical in the overlapping region to the pSFO157 3072/96 reference sequence (Fig. 3). The sequencing of the pSFO157 258/98-1 plasmid corroborated the absence of etpD and EHEC-hlyA in strain 258/98 determined by PCR and Southern blot hybridization (Table 2), as these genes are located in the plasmid region that was deleted in comparison to the pSFO157 3072/96 sequence (Fig. 3).
FIG 3.
Comparison of pSFO157 3072/96 from German SF EHEC O157:H− strain 3072/96 (reference plasmid) (GenBank accession no. AF401292) and pSFO157 258/98-1 from Czech SF EHEC O157:H− strain 258/98 (consensus sequence derived by combined de novo assembly and read mapping to the reference). The sequences share 100% identity over the overlapping region (modified BRIG image) (63). Plasmid pSFO157 258/98-1 evolved from the reference plasmid by homologous recombination between 2,148-bp fragments (depicted in red; 99% identity) resulting in a deletion of 41,534 bp. The sizes of the plasmids are shown inside the scheme. Annotations are specified only for the sfp operon and the deleted region (ORFs/operons are shown in black, and insertion sequence [IS] elements are shown in gray). sfp, plasmid-borne genes encoding sorbitol-fermenting EHEC O157 fimbriae; repA, gene encoding replication protein A; L7026-L7029, genes encoding hypothetical proteins; tagA, gene encoding ToxR-regulated lipoprotein; etp, genes encoding type II secretion system-related proteins; hly, operon encoding EHEC hemolysin and its secretion machinery.
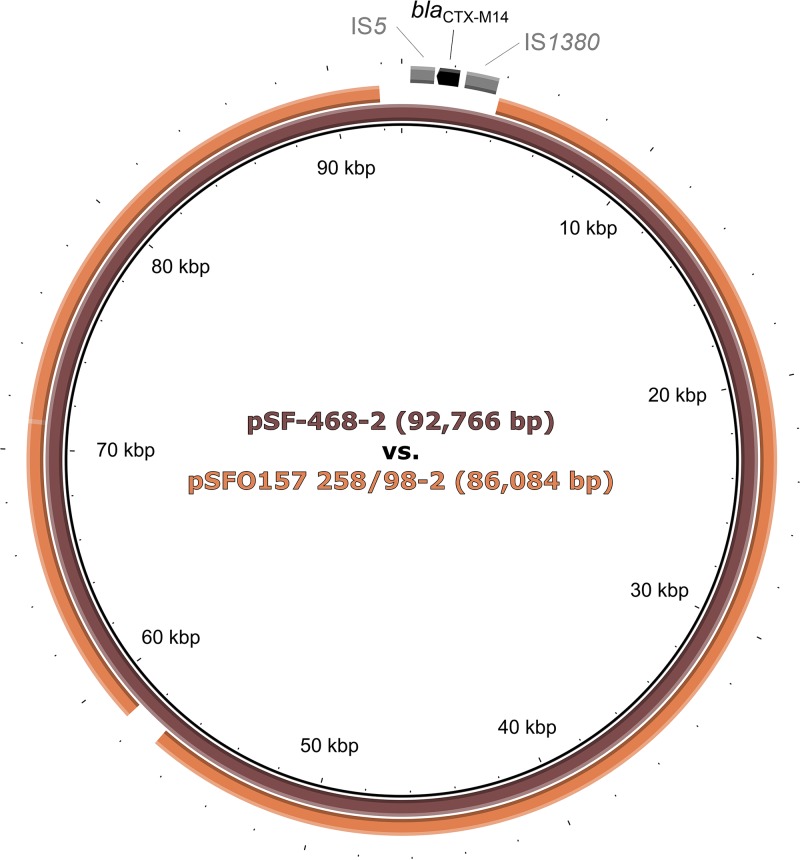
As the Czech strains also contain a second large plasmid, of 86 kb (Fig. 2; Table 2) (termed pSFO157 258/98-2), we performed BLAST queries of the de novo-assembled contigs of 258/98 plasmid DNA that did not map to pSFO157 3072/96. Interestingly, two large contigs, of 40.9 kb and 31.9 kb, showed 99% similarity to a 92.7-kb plasmid (pSF-468-2) (GenBank accession no. CP012627) of an extraintestinal pathogenic E. coli (ExPEC) strain isolated from a bloodstream infection (43). Subsequent mapping of the consensus sequence of pSFO157 258/98-2 to the reference sequence of pSF-468-2 demonstrated that pSFO157 258/98-2 covered 92% of the reference plasmid, with an identity of 98% in the overlapping region (Fig. 4). The most prominent difference between these two plasmids was a deletion of a 4,779-bp region (nucleotide positions 91,881 to 3,893 in the reference sequence) in pSFO157 258/98-2, covering the blaCTX-M14 extended-spectrum-β-lactamase (ESBL) gene of pSF-468-2 (43) and two adjacent insertion sequences (Fig. 4). In addition, a region of 1,518 bp at nucleotide positions 56,979 to 58,496 of the reference plasmid, containing an ORF with a hypothetical protein product (locus tag AN206_27055), was replaced by a fragment of 38 bp in pSFO157 258/98-2 (Fig. 4). In both cases, the mechanistic background of these events remains unknown, since sequence analyses revealed no motifs within the plasmids that are required for site-specific or homologous recombination. The pSFO157 258/98-2 plasmid contains the complete IncI conjugal transfer region present in pSF-468-2. However, neither of these plasmids carries ExPEC virulence genes encoding toxins, adhesins, or iron uptake systems (44).
FIG 4.
Comparison of pSF-468-2 from ExPEC strain SF-468 (reference plasmid) (GenBank accession no. CP012627) and pSFO157 258/98-2 from Czech SF EHEC O157:H− strain 258/98 (consensus sequence derived by combined de novo assembly and read mapping to the reference). The latter covers 92% of the reference plasmid, with 98% identity over the overlapping region (modified BRIG image) (63). Plasmid pSFO157 258/98-2 differs from the reference plasmid by a deletion of 4,779 bp (positions 91,881 to 3,893 in the reference plasmid) covering the blaCTX-M14 gene and two insertion sequences in pSF-468-2. A region of 1,518 bp (positions 56,979 to 58,496 in the reference plasmid) that contains an ORF encoding a hypothetical protein (locus tag AN206_27055) is replaced by a 38-bp fragment in pSFO157 258/98-2. Sizes of the plasmids are shown inside the scheme. Annotations are specified only for the deleted region (the ORF is shown in black, and IS elements are shown in gray). bla, gene encoding a beta-lactamase; IS, insertion sequence.
Antibiotic susceptibilities of Czech and German SF EHEC O157:H− strains.
To confirm the absence of the blaCTX-M14 ESBL gene in plasmid DNA of strain 258/98 on a phenotypic level, we tested the susceptibilities of strains 258/98 and 3072/96 (used for comparison) to ampicillin, piperacillin, cefuroxime, cefotaxime, cefpodoxime, ceftazidime, cefepime, piperacillin-tazobactam, tigecycline, imipenem, ertapenem, meropenem, gentamicin, amikacin, trimethoprim-sulfamethoxazole, ciprofloxacin, nitrofurantoin, and fosfomycin. We also specifically investigated the phenotypic presence of ESBLs by using the MAST ESBL detection disc set. Both strains were susceptible to all antibiotics tested and were phenotypically negative for ESBLs.
DISCUSSION
We report novel SF EHEC O157:H− strains isolated from patients in the Czech Republic. These strains are closely related to German SF EHEC O157:H− strains by their core genomes and share identical chromosomal virulence characteristics with them, but they differ essentially from German strains in their unique plasmid composition. In contrast to the German strains, which harbor a single, 121-kb pSFO157 plasmid, the Czech strains carry two plasmids, of 79 kb (pSFO157 258/98-1) and 86 kb (pSFO157 258/98-2). Sequence analyses of these plasmids demonstrated that neither of them contains the hlyCABD and etpCDEFGHIJKLMNO operons that are present on pSFO157 of German strains (30, 31), but the 79-kb plasmid pSFO157 258/98-1 harbors an sfpAHCDJFG operon that is 100% identical to that carried on pSFO157 3072/96. Notably, these two pSFO157 258/98 plasmids have different evolutionary origins. The 79-kb pSFO157 258/98-1 plasmid evolved from the 121-kb pSFO157 plasmid of German strains (represented here by pSFO157 3072/96) by homologous recombination between two nearly identical 2,148-bp regions, containing ORFs w0013 and w0041, which resulted in a 41,534-bp deletion (Fig. 3). This deletion led to the loss of the hlyCABD and etp operons but retained the sfpAHCDJFG operon (Fig. 3). In contrast to pSFO157 258/98-1, the 86-kb plasmid pSFO157 258/98-2 showed no homology to pSFO157 3072/96. This in turn indicates the lack of its relatedness to plasmids of NSF EHEC O157:H7 strains, including pO157 of strains EDL933 and Sakai (32, 33) and pO157_2 of strain G5101 (31), all of which share >99% overall similarities with pSFO157 3072/96 (31). Surprisingly, pSFO157 258/98-2 was highly (98%) similar to a 92.7-kb plasmid of an ExPEC bloodstream isolate belonging to a globally distributed ST95 ExPEC clonal lineage (43, 45). However, the blaCTX-M14 ESBL gene carried by ExPEC plasmid pSF-468-2 (43, 45) was absent from pSFO157 258/98-2 (Fig. 4). Accordingly, similar to strain 3072/96, strain 258/98 was susceptible to all antimicrobials tested and did not display an ESBL phenotype.
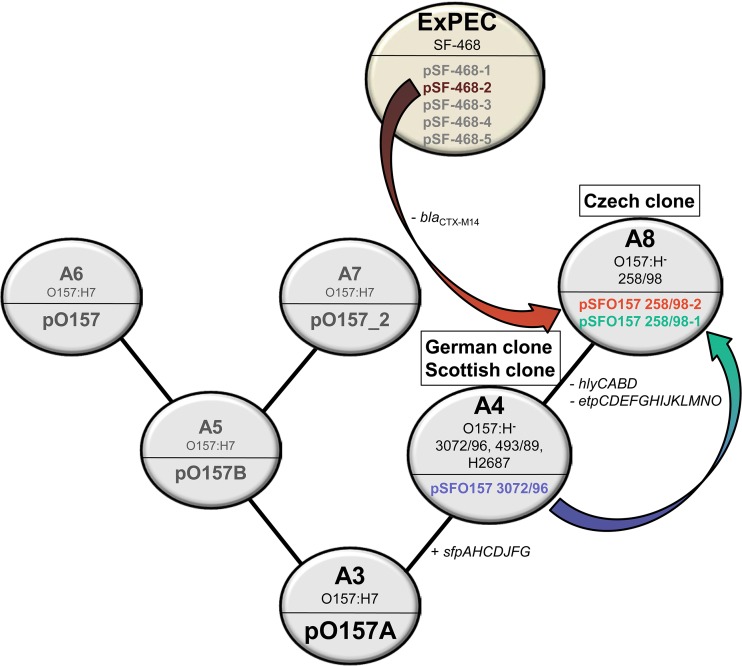
Our data contribute to elucidating the evolutionary history of EHEC O157 plasmids. Previous studies indicated that both NSF EHEC O157:H7 and SF EHEC O157:H− plasmids are nonconjugative because they lack a number of transfer genes of the plasmid F required for conjugation (30, 31, 33, 46). Therefore, their recent introduction into EHEC O157 by conjugation is unlikely (31). It was hypothesized that these plasmids evolved together with their bacterial hosts from a hypothetical ancestral plasmid carried by the O157 intermediate strain of clonal complex A3 (31, 47, 48), through a process of multiple structural changes due to acquisitions and losses of various genes via insertion/transposition events, and possibly plasmid fusion (30, 31). This hypothesis is supported by comparative sequence analyses of pO157 and pO157_2 of NSF EHEC O157:H7 strains Sakai and G5101, respectively, and pSFO157 of SF EHEC O157:H− strain 3072/96 (30, 31). Our sequence analyses of the two 258/98 plasmids confirm this hypothesis and, moreover, bring new insights into the evolutionary history of EHEC O157 plasmids proposed by Rump et al. (31), based on the stepwise evolutionary model of EHEC O157 (47, 48). In this extended plasmid evolutionary scenario that includes our data (Fig. 5), the Czech SF EHEC O157:H− strains represent the novel clonal complex A8 (termed the “Czech clone”). One plasmid of these strains (pSFO157 258/98-1) evolved from pSFO157 of the “German clone” (the A4 clonal complex, which also contains Scottish SF EHEC O157 strains) by deletion of a large plasmid region that led to the loss of the hly and etp operons. The second plasmid (pSFO157 258/98-2), which is highly similar to the ExPEC plasmid pSF-468-2, might have been acquired by SF EHEC O157:H− from ExPEC, plausibly by conjugation (as supported by the presence of the complete IncI conjugal transfer machinery in pSF-468-2) in the human intestine, which is the niche for SF EHEC O157:H− during infection and the primary source of ExPEC (44). This demonstrates a novel mechanism in the evolution of EHEC O157 plasmids. The absence of the blaCTX-M14 gene harbored by pSF-468-2 in pSFO157 258/98-2 might be due to the loss of this gene before the plasmid acquisition by SF EHEC O157:H− or within its new host. Alternatively, blaCTX-M14 might have been acquired in the lineage that led to pSF-468-2 after it had diverged from an ancestor that also gave rise to pSFO157 258/98-2.
FIG 5.
Extended evolutionary scenario for SF EHEC O157 plasmids based on sequence analyses of the plasmids of SF EHEC O157:H− strain 258/98. The pSFO157 plasmid of the A4 clonal complex (SF EHEC O157:H− strains of the German clone and the Scottish clone), represented by pSFO157 3072/96, evolved from a common O157 ancestor plasmid of a hypothetical intermediate strain of the A3 clonal complex by acquisition of the sfpAHCDJFG operon. From pSFO157 of the A4 complex, pSFO157 258/98-1 (79 kb) (A8 clonal complex; Czech clone) evolved via deletion of a 41,534-bp region by homologous recombination, leading to loss of the hlyCABD and etpCDEFGHIJKLMNO operons but retention of the sfpAHCDJFG operon. In addition, the A8 clonal complex strains acquired an EHEC O157-unrelated plasmid, pSFO157 258/98-2 (86 kb), which is 98% similar to pSF-468-2 (92.7 kb) (GenBank accession no. CP012627) from an ExPEC strain. The clonal complexes A5, A6, and A7 represent the evolution of NSF EHEC O157:H7 plasmids and were previously described in detail (31). (Adapted from reference 31 with permission.)
Except for the Czech Republic, where the novel SF EHEC O157 strains account for 20% of all SF EHEC O157:H− strains isolated from patients with HUS (10), the distribution and frequencies of these strains in other geographic regions, as well as the epidemiology of these infections, are currently unknown. In the Czech Republic, a SF EHEC O157:H− strain with these unique plasmid characteristics was isolated from a cow epidemiologically associated with an HUS case (3), suggesting that cattle can be a reservoir for these novel strains, as also demonstrated for the “classical” SF EHEC O157:H− strains belonging to the German clone (6, 17, 18). Notably, SF E. coli O157:H− (fliCH7) strains that share chromosomal (presence of cdtV-ABC and eae-γ) and plasmid (presence of sfpA and absence of EHEC-hlyA and etpD) characteristics with the novel SF EHEC O157:H− strains but lack stx2a were isolated from patients with HUS (n = 1) or diarrhea (n = 2) in the Czech Republic (M. Marejková and M. Bielaszewska, unpublished data). Since SF EHEC O157:H− strains can frequently lose their stx2a genes via loss of stx2a-harboring phages (38, 49), these strains plausibly represent stx-negative variants of the novel SF EHEC O157:H− clone. Similar strains, i.e., stx-negative SF E. coli O157:H− (fliCH7) harboring sfpA but lacking EHEC-hlyA, were recently reported for patients with HUS or diarrhea in Germany (50). These strains clustered with SF EHEC O157:H− in the WGS-based analysis (50), corroborating their descent from SF EHEC O157:H− by stx2a loss (50). Altogether, these findings indicate that a subset of SF EHEC O157:H− strains of the novel Czech clone may occur as stx-negative variants. As a consequence, such strains in patient stools are missed by diagnostic approaches that rely solely on the detection of stx and/or Stx. This in turn hampers the determination of their etiological role in human diseases. In order to reliably identify such strains, we propose the testing of all SF E. coli O157:H− (fliCH7) isolates, regardless of whether they possess or lack stx2a, for the unique plasmid molecular characteristics of the novel SF EHEC O157:H− clone (presence of sfpA but absence of EHEC-hlyA and etpD). This will enable further investigations of the geographic distribution of such strains, their role as human pathogens, and the epidemiology of these infections. This is particularly important considering the propensity of the stx-negative SF EHEC O157:H− variants to again become EHEC via transduction by stx2a-harboring phages (49).
In conclusion, we identified and characterized novel plasmids in SF EHEC O157:H− strains isolated from patients. Our data extend the evolutionary history of EHEC O157 plasmids and demonstrate an important role for plasmids in the evolution of SF EHEC O157:H− pathogens. The unique molecular characteristics of these novel plasmids enable the identification of such strains, thereby facilitating further investigations of their geographic distribution, clinical significance, and epidemiology.
MATERIALS AND METHODS
Strains and serotyping.
The origins and serotypes of the strains used are listed in Table 1. Conventional serotyping was performed as described previously (51). Motility was determined using soft (0.5%) agar (10). The presence of O157 LPS was confirmed by PCR targeting rfbEO157 (52), and the fliC gene was subtyped using HhaI restriction fragment length polymorphism (RFLP) analysis (53).
Detection of virulence genes.
Chromosomal virulence loci encoding toxins (stx and the cdtV operon), adhesins (eae, efa1, lpfAO157OI-141, lpfAO157OI-154, and iha), and other characteristics (ter and ure operons) and plasmid-borne putative virulence genes (EHEC-hlyA, katP, espP, etpD, and sfpA) were detected using established PCR protocols (4, 23–26, 34, 36, 37, 54); stx and eae genes were subtyped as described previously (55, 56).
Phenotypes.
Sorbitol fermentation was detected on sorbitol MacConkey agar (SMAC) (Oxoid, Hampshire, United Kingdom), β-d-glucuronidase activity on nutrient agar with 4-methylumbelliferyl-β-d-glucuronide (MUG) (Becton Dickinson, Heidelberg, Germany), production of EHEC hemolysin on enterohemolysin agar (Sifin, Berlin, Germany), and tellurite resistance on CT-SMAC (Oxoid, Hampshire, United Kingdom). Stx production was determined using a latex agglutination assay (verotoxin-producing E. coli reverse passive latex agglutination [VTEC-RPLA] assay; Denka Seiken Co. Ltd., Tokyo, Japan) according to the manufacturer's instructions. Vero cell cytotoxicity assay and CHO assay were performed as described previously (23, 57). Stx and CdtV titers were defined as the highest dilutions of culture supernatants that caused Vero cell detachment after 3 days and CHO distension after 4 days of incubation, respectively, in 50% of cells. Expression of Sfp fimbriae was determined using immunoblotting with an anti-SfpA antibody (produced by Davids Biotechnologie, Regensburg, Germany) and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Dianova, Hamburg, Germany) as described previously (22).
Plasmid profiles and Southern blot hybridization.
Plasmid profiles were determined as described earlier (58, 59). For Southern blot hybridization, plasmid-extracted DNAs (Plasmid Midi kit; Qiagen, Hilden, Germany), either undigested or digested with BamHI or SmaI (New England BioLabs, Frankfurt, Germany), were separated in a 0.6% agarose gel, blotted to a nylon membrane (Roche Molecular Biochemicals, Mannheim, Germany), and hybridized with digoxigenin-labeled (DIG High Prime kit; Roche Molecular Biochemicals) sfpA, EHEC-hlyA, katP, espP, and etpD probes prepared as described earlier (60). Labeled probes were detected using a DIG Luminescent detection kit (Roche Molecular Biochemicals).
Sequence analyses of plasmids from SF EHEC O157 strain 258/98.
To determine the sequences of 258/98 plasmids, we first extracted plasmid DNA by using the method of Barton et al. (61) to detect and size large plasmids by pulsed-field gel electrophoresis (PFGE) (61), performed with the following modifications: 1% agarose gel, initial switch of 1 s, final switch of 25 s, and run time of 21 h. Subsequently, the linearized plasmids were manually excised from the PFGE gel, and DNA was eluted using a GFX PCR DNA and gel band purification kit (GE Healthcare, Freiburg, Germany). Finally, the plasmid DNA was tested for the presence of sfpA by PCR (37). Subsequent library preparation using Nextera XT chemistry (Illumina Inc., San Diego, CA, USA) and 250-bp paired-end sequencing on an Illumina MiSeq machine (Illumina) were done as described recently (62). After quality trimming of the resulting reads using CLC Bio genomics workbench software, version 10 (Qiagen, Aarhus, Denmark), with default parameters, reads were mapped to the reference sequence of pSFO157 of strain 3072/96 (GenBank accession no. AF401292) (30; http://www.ncbi.nlm.nih.gov/nucleotide). Default parameters within the CLC Bio genomics workbench software were used, with the exception of the parameters “length fraction = 0.8” and “similarity fraction = 0.95.” In parallel, we created two de novo assemblies. First, we assembled all reads originating from the 258/98 plasmid DNA and compared the resulting contigs with the sequence of pSFO157 3072/96. Second, we assembled all plasmid reads that were not mapped to the reference sequence of pSFO157 3072/96 and queried all resulting contigs of >5 kb by using NCBI Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn). Finally, we compared all de novo-assembled contigs that did not map to the pSFO157 3072/96 sequence with the reference sequence of ExPEC plasmid pSF-468-2 (GenBank accession no. CP012627) (43; http://www.ncbi.nlm.nih.gov/nucleotide). Differences between the sequences were extracted from the reference mapping results within the CLC Bio genomics workbench software and visualized using BRIG (63).
MLST and WGS-based analysis of the core genomes.
MLST was performed as described previously (11), and sequence types (STs) were assigned in accordance with the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). Related STs were grouped into clonal complexes (CCs) in accordance with the MLST website.
For the whole-genome sequencing of strains 258/98 and 3072/96, the genomic DNAs were purified by use of a MagAttract HMW DNA kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Subsequent library preparation and sequencing were done as described above for the plasmid DNA. Sequence quality trimming, de novo assembly, and extraction of coding regions were done as described recently (62), using SeqSphere+ software, version 2.0 beta (Ridom GmbH, Münster, Germany). For the gene-by-gene core genome analysis, we included all core genome genes present in all strains analyzed (see Table S1 in the supplemental material) and displayed them in a minimum spanning tree by using SeqSphere+ software. For comparison, we used published genome sequences of SF EHEC O157:H− strain 493/89 (GenBank accession no. AETY00000000) (64) and NSF EHEC O157:H7 strains EDL933 (GenBank accession no. NZ_CP008957) (65) and Sakai (GenBank accession no. NC_002695) (66) (http://www.ncbi.nlm.nih.gov/nucleotide).
Antimicrobial susceptibility testing.
Susceptibilities of strains 258/98 and 3072/96 to antimicrobials were tested by the disc diffusion method, using clinical breakpoints established by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI) (for fosfomycin) (67, 68). In addition, the presence of ESBLs was investigated phenotypically by using the MAST ESBL detection disc set (Mast Group Ltd., Bootle, United Kingdom).
Accession number(s).
All generated raw reads, including those derived from either plasmid DNA of strain 258/98 or whole genomic DNAs of strains 258/98 and 3072/96, were deposited at the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena/) under study accession no. PRJEB21607.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a grant from the German Research Foundation (grant ME3205/2-1 to A.M.) and by a funding project of the Ministry of Health of the Czech Republic (“Conceptual Development of Research Organization” [National Institute of Public Health]) (grant NIPH 75010330).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Ulrich Dobrindt (Institute for Hygiene, Münster, Germany) and Craig M. Stephens (Santa Clara University, Santa Clara, CA, USA) for fruitful discussions during preparation of the manuscript. The technical assistance of Ralph Fischer, Isabell Höfig, Andrea Lagemann, Thomas Böking, and Ursula Keckevoet (Münster) and of Ute Siewert und Ute Strutz (Wernigerode) is greatly appreciated.
A.B. performed WGS analyses and cell culture assays. M.M. carried out serotyping, genotypic, and phenotypic analyses of Czech strains. R.P. analyzed plasmid profiles and performed Southern blot hybridization. A.M. and B.M.-B. sequenced 258/98 plasmids and analyzed the data. A.K. performed antibiotic susceptibility tests, and W.Z. performed phenotypic analyses of German strains. H.K. and A.M. participated in study design, supervised the study, and raised funding. M.B. participated in study design, data analysis, and drafted the manuscript, together with A.B. and A.M. All authors read, edited, and approved the final version of the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01454-17.
REFERENCES
- 1.Karch H, Wiss R, Gloning H, Emmrich P, Aleksić S, Bockemühl J. 1990. Hemolytic-uremic syndrome in infants due to verotoxin-producing Escherichia coli. Dtsch Med Wochenschr 115:489–495. doi: 10.1055/s-2008-1065036. [DOI] [PubMed] [Google Scholar]
- 2.Karch H, Tarr PI, Bielaszewska M. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol 295:405–418. doi: 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Bielaszewska M, Schmidt H, Liesegang A, Prager R, Rabsch W, Tschäpe H, Cízek A, Janda J, Bláhová K, Karch H. 2000. Cattle can be a reservoir of sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains and a source of human diseases. J Clin Microbiol 38:3470–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eklund M, Bielaszewska M, Nakari UM, Karch H, Siitonen A. 2006. Molecular and phenotypic profiling of sorbitol-fermenting Escherichia coli O157:H− human isolates from Finland. Clin Microbiol Infect 12:634–641. doi: 10.1111/j.1469-0691.2006.01478.x. [DOI] [PubMed] [Google Scholar]
- 5.Jakubczak A, Szych J, Januszkiewicz K. 2008. Characterization of first sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strain isolated in Poland. Med Dosw Mikrobiol 60:173–181. [PubMed] [Google Scholar]
- 6.Orth D, Grif K, Dierich MP, Würzner R. 2006. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157: indications for an animal reservoir. Epidemiol Infect 134:719–723. doi: 10.1017/S0950268805005467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buvens G, Piérard D, Hachimi-Idrissi S, Lauwers S. 2009. First sorbitol-fermenting verocytotoxin-producing Escherichia coli O157:H− isolated in Belgium. Acta Clin Belg 64:59–64. doi: 10.1179/acb.2009.011. [DOI] [PubMed] [Google Scholar]
- 8.Pollock KG, Locking ME, Beattie TJ, Maxwell H, Ramage I, Hughes D, Cowieson J, Allison L, Hanson M, Cowden JM. 2010. Sorbitol-fermenting Escherichia coli O157, Scotland. Emerg Infect Dis 16:881–882. doi: 10.3201/eid1605.091919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandal LT, Løbersli I, Stavnes TL, Wester AL, Lindstedt BA. 2012. First report of the Shiga toxin 1 gene in sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol 50:1825–1826. doi: 10.1128/JCM.06435-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marejková M, Bláhová K, Janda J, Fruth A, Petráš P. 2013. Enterohemorrhagic Escherichia coli as causes of hemolytic uremic syndrome in the Czech Republic. PLoS One 8:e73927. doi: 10.1371/journal.pone.0073927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellmann A, Bielaszewska M, Köck R, Friedrich AW, Fruth A, Middendorf B, Harmsen D, Schmidt MA, Karch H. 2008. Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerg Infect Dis 14:1287–1290. doi: 10.3201/eid1408.071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ammon A, Petersen LR, Karch H. 1999. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H−. J Infect Dis 179:1274–1277. doi: 10.1086/314715. [DOI] [PubMed] [Google Scholar]
- 13.Alpers K, Werber D, Frank C, Koch J, Friedrich AW, Karch H, An DER, Heiden M, Prager R, Fruth A, Bielaszewska M, Morlock G, Heissenhuber A, Diedler A, Gerber A, Ammon A. 2009. Sorbitol-fermenting enterohaemorrhagic Escherichia coli O157:H− causes another outbreak of haemolytic uraemic syndrome in children. Epidemiol Infect 137:389–395. doi: 10.1017/S0950268808001465. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen S, Frank C, Fruth A, Spode A, Prager R, Graff A, Plenge-Bönig A, Loos S, Lütgehetmann M, Kemper MJ, Müller-Wiefel DE, Werber D. 2011. Desperately seeking diarrhoea: outbreak of haemolytic uraemic syndrome caused by emerging sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H−, Germany, 2009. Zoonoses Public Health 58:567–572. doi: 10.1111/j.1863-2378.2011.01405.x. [DOI] [PubMed] [Google Scholar]
- 15.Rosser T, Dransfield T, Allison L, Hanson M, Holden N, Evans J, Naylor S, La Ragione R, Low JC, Gally DL. 2008. Pathogenic potential of emergent sorbitol-fermenting Escherichia coli O157:NM. Infect Immun 76:5598–5607. doi: 10.1128/IAI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haugum K, Lindstedt BA, Løbersli I, Kapperud G, Brandal LT. 2012. Identification of the anti-terminator qO111:H− gene in Norwegian sorbitol-fermenting Escherichia coli O157:NM. FEMS Microbiol Lett 329:102–110. doi: 10.1111/j.1574-6968.2012.02505.x. [DOI] [PubMed] [Google Scholar]
- 17.King LA, Loukiadis E, Mariani-Kurkdjian P, Haeghebaert S, Weill FX, Baliere C, Ganet S, Gouali M, Vaillant V, Pihier N, Callon H, Novo R, Gaillot O, Thevenot-Sergentet D, Bingen E, Chaud P, de Valk H. 2014. Foodborne transmission of sorbitol-fermenting Escherichia coli O157:[H7] via ground beef: an outbreak in northern France, 2011. Clin Microbiol Infect 20:O1136–O1144. doi: 10.1111/1469-0691.12736. [DOI] [PubMed] [Google Scholar]
- 18.Jaakkonen A, Salmenlinna S, Rimhanen-Finne R, Lundström H, Heinikainen S, Hakkinen M, Hallanvuo S. 2017. Severe outbreak of sorbitol-fermenting Escherichia coli O157 via unpasteurized milk and farm visits, Finland 2012. Zoonoses Public Health 64:468–475. doi: 10.1111/zph.12327. [DOI] [PubMed] [Google Scholar]
- 19.Vygen-Bonnet S, Rosner B, Wilking H, Fruth A, Prager R, Kossow A, Lang C, Simon S, Seidel J, Faber M, Schielke A, Michaelis K, Holzer A, Kamphausen R, Kalhöfer D, Thole S, Mellmann A, Flieger A, Stark K. 2017. Ongoing haemolytic uraemic syndrome (HUS) outbreak caused by sorbitol-fermenting (SF) Shiga toxin-producing Escherichia coli (STEC) O157, Germany, December 2016 to May 2017. Euro Surveill 22:30541. doi: 10.2807/1560-7917.ES.2017.22.21.30541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg Infect Dis 11:603–609. doi: 10.3201/eid1104.040739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bielaszewska M, Sinha B, Kuczius T, Karch H. 2005. Cytolethal distending toxin from Shiga toxin-producing Escherichia coli O157 causes irreversible G2/M arrest, inhibition of proliferation, and death of human endothelial cells. Infect Immun 73:552–562. doi: 10.1128/IAI.73.1.552-562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müsken A, Bielaszewska M, Greune L, Schweppe CH, Müthing J, Schmidt H, Schmidt MA, Karch H, Zhang W. 2008. Anaerobic conditions promote expression of Sfp fimbriae and adherence of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM to human intestinal epithelial cells. Appl Environ Microbiol 74:1087–1093. doi: 10.1128/AEM.02496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janka A, Bielaszewska M, Dobrindt U, Greune L, Schmidt MA, Karch H. 2003. Cytolethal distending toxin gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect Immun 71:3634–3638. doi: 10.1128/IAI.71.6.3634-3638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janka A, Bielaszewska M, Dobrindt U, Karch H. 2002. Identification and distribution of the enterohemorrhagic Escherichia coli factor for adherence (efa1) gene in sorbitol-fermenting Escherichia coli O157:H−. Int J Med Microbiol 292:207–214. doi: 10.1078/1438-4221-00206. [DOI] [PubMed] [Google Scholar]
- 25.Bielaszewska M, Tarr PI, Karch H, Zhang W, Mathys W. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J Clin Microbiol 43:452–454. doi: 10.1128/JCM.43.1.452-454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich AW, Köck R, Bielaszewska M, Zhang W, Karch H, Mathys W. 2005. Distribution of the urease gene cluster among and urease activities of enterohemorrhagic Escherichia coli O157 isolates from humans. J Clin Microbiol 43:546–550. doi: 10.1128/JCM.43.2.546-550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarr PI, Bilge SS, Vary JC Jr, Jelacic S, Habeeb RL, Ward TR, Baylor MR, Besser TE. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun 68:1400–1407. doi: 10.1128/IAI.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaikh N, Holt NJ, Johnson JR, Tarr PI. 2007. Fim operon variation in the emergence of enterohemorrhagic Escherichia coli: an evolutionary and functional analysis. FEMS Microbiol Lett 273:58–63. doi: 10.1111/j.1574-6968.2007.00781.x. [DOI] [PubMed] [Google Scholar]
- 29.Uhlich GA, Keen JE, Elder RO. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol 67:2367–2370. doi: 10.1128/AEM.67.5.2367-2370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunder W, Karch H, Schmidt H. 2006. Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H− strain 3072/96. Int J Med Microbiol 296:467–474. doi: 10.1016/j.ijmm.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Rump LV, Meng J, Strain EA, Cao G, Allard MW, Gonzalez-Escalona N. 2012. Complete DNA sequence analysis of enterohemorrhagic Escherichia coli plasmid pO157_2 in β-glucuronidase-positive E. coli O157:H7 reveals a novel evolutionary path. J Bacteriol 194:3457–3463. doi: 10.1128/JB.00197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, Blattner FR. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res 26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han CG, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res 5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt H, Beutin L, Karch H. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun 63:1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bielaszewska M, Rüter C, Kunsmann L, Greune L, Bauwens A, Zhang W, Kuczius T, Kim KS, Mellmann A, Schmidt MA, Karch H. 2013. Enterohemorrhagic Escherichia coli hemolysin employs outer membrane vesicles to target mitochondria and cause endothelial and epithelial apoptosis. PLoS Pathog 9:e1003797. doi: 10.1371/journal.ppat.1003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt H, Henkel B, Karch H. 1997. A gene cluster closely related to type II secretion pathway operons of gram-negative bacteria is located on the large plasmid of enterohemorrhagic Escherichia coli O157 strains. FEMS Microbiol Lett 148:265–272. doi: 10.1111/j.1574-6968.1997.tb10299.x. [DOI] [PubMed] [Google Scholar]
- 37.Brunder W, Khan AS, Hacker J, Karch H. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H−. Infect Immun 69:4447–4457. doi: 10.1128/IAI.69.7.4447-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedrich AW, Zhang W, Bielaszewska M, Mellmann A, Köck R, Fruth A, Tschäpe H, Karch H. 2007. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin Infect Dis 45:39–45. doi: 10.1086/518573. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Kaper JB. 1992. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]
- 40.Torres AG, Giron JA, Perna NT, Burland V, Blattner FR, Avelino-Flores F, Kaper JB. 2002. Identification and characterization of lpfABCC′DE, a fimbrial operon of enterohemorrhagic Escherichia coli O157:H7. Infect Immun 70:5416–5427. doi: 10.1128/IAI.70.10.5416-5427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB. 2004. Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol Lett 238:333–344. [DOI] [PubMed] [Google Scholar]
- 42.Friedrich AW, Nierhoff KV, Bielaszewska M, Mellmann A, Karch H. 2004. Phylogeny, clinical associations, and diagnostic utility of the pilin subunit gene (sfpA) of sorbitol-fermenting, enterohemorrhagic Escherichia coli O157:H−. J Clin Microbiol 42:4697–4701. doi: 10.1128/JCM.42.10.4697-4701.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens CM, Skerker JM, Sekhon MS, Arkin AP, Riley LW. 2015. Complete genome sequences of four Escherichia coli ST95 isolates from bloodstream infections. Genome Announc 3:e01241-15. doi: 10.1128/genomeA.01241-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leimbach A, Hacker J, Dobrindt U. 2013. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 358:3–32. doi: 10.1007/82_2012_303. [DOI] [PubMed] [Google Scholar]
- 45.Stephens CM, Adams-Sapper S, Sekhon M, Johnson JR, Riley LW. 2017. Genomic analysis of factors associated with low prevalence of antibiotic resistance in extraintestinal pathogenic Escherichia coli sequence type 95 strains. mSphere 2:e00390-16. doi: 10.1128/mSphere.00390-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frost LS, Ippen-Ihler K, Skurray RA. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58:162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol 187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng PC, Monday SR, Lacher DW, Allison L, Siitonen A, Keys C, Eklund M, Nagano H, Karch H, Keen J, Whittam TS. 2007. Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg Infect Dis 13:1701–1706. doi: 10.3201/eid1311.070381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mellmann A, Bielaszewska M, Karch H. 2009. Intrahost genome alterations in enterohemorrhagic Escherichia coli. Gastroenterology 136:1925–1938. doi: 10.1053/j.gastro.2008.12.072. [DOI] [PubMed] [Google Scholar]
- 50.Kossow A, Zhang W, Bielaszewska M, Rhode S, Hansen K, Fruth A, Rüter C, Karch H, Mellmann A. 2016. Molecular characterization of human atypical sorbitol-fermenting enteropathogenic Escherichia coli O157 reveals high diversity. J Clin Microbiol 54:1357–1363. doi: 10.1128/JCM.02897-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prager R, Strutz U, Fruth A, Tschäpe H. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar (H)-antigens: serotyping versus fliC polymorphisms. Int J Med Microbiol 292:477–486. doi: 10.1078/1438-4221-00226. [DOI] [PubMed] [Google Scholar]
- 52.Nagano I, Kunishima M, Itoh Y, Wu Z, Takahashi Y. 1998. Detection of verotoxin-producing Escherichia coli O157:H7 by multiplex polymerase chain reaction. Microbiol Immunol 42:371–376. doi: 10.1111/j.1348-0421.1998.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Mellmann A, Sonntag AK, Wieler L, Bielaszewska M, Tschäpe H, Karch H, Friedrich AW. 2007. Structural and functional differences between disease-associated genes of enterohaemorrhagic Escherichia coli O111. Int J Med Microbiol 297:17–26. doi: 10.1016/j.ijmm.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Toma C, Martínez Espinosa E, Song T, Miliwebsky E, Chinen I, Iyoda S, Iwanaga M, Rivas M. 2004. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J Clin Microbiol 42:4937–4946. doi: 10.1128/JCM.42.11.4937-4946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang WL, Köhler B, Oswald E, Beutin L, Karch H, Morabito S, Caprioli A, Suerbaum S, Schmidt H. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J Clin Microbiol 40:4486–4492. doi: 10.1128/JCM.40.12.4486-4492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karmali MA, Petric M, Lim C, Fleming PC, Arbus GS, Lior H. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis 151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 58.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang W, Bielaszewska M, Kunsmann L, Mellmann A, Bauwens A, Köck R, Kossow A, Anders A, Gatermann S, Karch H. 2013. Lability of the pAA virulence plasmid in Escherichia coli O104:H4: implications for virulence in humans. PLoS One 8:e66717. doi: 10.1371/journal.pone.0066717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bielaszewska M, Prager R, Vandivinit L, Müsken A, Mellmann A, Holt NJ, Tarr PI, Karch H, Zhang W. 2009. Detection and characterization of the fimbrial sfp cluster in enterohemorrhagic Escherichia coli O165:H25/NM isolates from humans and cattle. Appl Environ Microbiol 75:64–71. doi: 10.1128/AEM.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 62.Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, Prior K, Harmsen D. 2016. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 54:2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rump LV, Strain EA, Cao G, Allard MW, Fischer M, Brown EW, Gonzalez-Escalona N. 2011. Draft genome sequences of six Escherichia coli isolates from the stepwise model of emergence of Escherichia coli O157:H7. J Bacteriol 193:2058–2059. doi: 10.1128/JB.00118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Latif H, Li HJ, Charusanti P, Palsson BØ, Aziz RK. 2014. A gapless, unambiguous genome sequence of the enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Genome Announc 2:e00821-14. doi: 10.1128/genomeA.00821-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 67.European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters, version 7.1. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf Accessed 22 June 2017.
- 68.Matuschek E, Brown DF, Kahlmeter G. 2014. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect 20:O255–O266. doi: 10.1111/1469-0691.12373. [DOI] [PubMed] [Google Scholar]
- 69.Threlfall EJ, Rowe B, Ferguson JL, Ward LR. 1986. Characterization of plasmids conferring resistance to gentamicin and apramycin in strains of Salmonella typhimurium phage type 204c isolated in Britain. J Hyg (Lond) 97:419–426. doi: 10.1017/S0022172400063609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karch H, Böhm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. 1993. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol 31:1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 72.Watanabe H, Wada A, Inagaki Y, Itoh K, Tamura K. 1996. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet 348:831–832. doi: 10.1016/S0140-6736(05)65257-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.