Abstract
Transmembrane helix M6 of the sarco(endo)plasmic reticulum Ca2+ ATPase (SERCA) has been shown to form a site of interaction with phospholamban (PLN). Site-directed mutagenesis was carried out in the cytoplasmic loop (L67) between M6 and M7 in SERCA1a to detect other SERCA-PLN binding sites. Mutants N810A, D813A, and R822A had diminished ability to interact functionally with PLN, but only D813A and R822A had reduced physical interaction with PLN. PLN mutants R25A, Q26A, N27A, L28A, Q29A, and N30A had enhanced physical interaction with wild-type (wt) SERCA1a, but physical interaction of these PLN mutants with SERCA1a mutants D813A and R822A was reduced about 2.5 fold (range 1.44–2.82). Exceptions were the interactions of PLN N27A and N30A with SERCA1a D813A, which were reduced by 7.3- and 5.8-fold, respectively. A superinhibitory PLN deletion mutant, PLNΔ21–29, had strong physical interactions with SERCA1a and with SERCA1a mutant D813A. Physical interactions with SERCA1a and mutant D813A were sharply diminished, however, for the PLN deletion mutant, PLNΔ21–30, lacking PLN N30. Physical interactions between SERCA1a and a PLN-cytochrome b5 chimera containing PLN residues 1–29 were much stronger than those between a PLN-cytochrome b5 chimera containing PLN residues 1–21 and lacking N27. These results suggest that a SERCA1-PLN interaction site occurs between L67 of SERCA1a and domain IB of PLN, which involves SERCA1a D813 and PLN N27 and N30.
Muscle contraction is regulated by Ca2+ release from the sarcoplasmic reticulum through Ca2+ release channels. Relaxation is regulated by the subsequent return of Ca2+ to the lumen of the sarcoplasmic reticulum through the action of Ca2+ pumps, the 110-kDa transmembrane proteins referred to as sarco(endo)plasmic reticulum Ca2+ ATPases or SERCAs (1). The energy for Ca2+ transport is provided by ATP; intermediate steps involve the formation and hydrolysis of a phosphoenzyme intermediate and several conformational states have been identified. The crystal structure of SERCA1a at 2.6 Å (2) confirms earlier predictions that the protein contains three distinct cytosolic domains supported on a stalk and 10 helices forming a transmembrane domain (1, 3). Two moles of Ca2+ are bound side-by-side in the center of a cluster of transmembrane helices M4, M5, M6, and M8, but M4 and M6 helices unwind near their centers to accommodate the binding of Ca2+. The phosphorylation and nucleotide binding domains resemble the major and subdomains of the haloacid dehalogenase (4). Unexpected features that are of obvious functional significance are the association of two helices at the N terminus of the molecule with a cytosolic β-strand domain between helices M2 and M3 to form an actuator domain and a cytosolic loop (L67) between M6 and M7 (F809-G831), which winds around and links together several elements of the stalk sector. These two features set up linkages that might involve virtually all of the transmembrane helices in the conformational movements that drive Ca2+ transport.
The role of amino acid residues in the L67 loop in Ca2+ transport has been investigated through site-directed mutagenesis (5, 6). Mutations K819A and R822A inhibit Ca2+ ATPase activity, diminish phosphorylated intermediate formation, and diminish Ca2+ binding affinity slightly. The triple mutation, D813N-D815N-D818N interferes with Ca2+-dependent activation of the ATPase, and most of this effect is due to mutation of D813. Mutations in this region of the protein have the potential to disrupt the network of hydrogen bonds formed between L67 and other elements of the protein.
Phospholamban (PLN) is a 52-aa integral membrane protein, highly expressed in cardiac and slow-twitch skeletal muscle fibers, that interacts with and, at low Ca2+ concentrations, reversibly inhibits the activity of SERCA2a by lowering its apparent affinity for Ca2+ (7). Studies of mice in which PLN is ablated or replaced by highly inhibitory forms of PLN demonstrate that PLN is a modulator of the activity of the Ca2+ pump and a major regulator of the dynamics of cardiac contractility (8–11). It has been proposed that PLN contains three domains. Domain IA, amino acids 1–20, contains sites of regulatory phosphorylation by protein kinase A at S16 and by Ca2+-calmodulin kinase at T17; domain IB, amino acids 21–30, contains a high proportion of amidated residues; domain II, amino acids 31–52, forms a transmembrane domain. A structure of PLN in trifluoroethanol, determined by NMR spectroscopy, demonstrates that the protein forms two α-helices connected by a turn made up of I18, E19, M20, and P21 (12). This turn, at the end of domain IA, could provide flexibility in the orientation of the two helices relative to each other. A structure of [C41F]PLN1–52 in chloroform-methanol shows the same features, but the β-turn begins at T17 (13).
In previous papers, two different sites in SERCA that interact with PLN have been identified: a cytosolic interaction site involves the sequence Lys-Asp-Asp-Lys-Pro-Val402 (14); a transmembrane interaction site involves residues V795 and L802 in the face of transmembrane helix M6, which is directed toward the lipid bilayer (15). Complementary studies with PLN show that charged and hydrophobic residues in domain IA interact with the cytosolic domains of SERCA (16) and that the SERCA-interacting face of PLN domain II is on the opposite side to the face that interacts with other PLN molecules to form a pentameric structure (17). Mutations in PLN domain IB cause both loss and gain of inhibitory function, the mutation N27A being the most inhibitory yet observed (18). Although interactions between domain IB and SERCA are likely, it is not known where these interaction sites might lie in SERCA. The fact that domain IB forms part of a helix that is likely to extend directly above the site of PLN domain II interaction with M6 (12) suggests that the L67 is a candidate region for interaction with PLN domain IB. However, helices M2 and M9 flank the proposed site where PLN interacts with M6 (2), so that M2 and M9 are also candidate interaction sites. In this study, we focused on L67, examining the question of whether this region might be involved in PLN-SERCA interactions and whether residues in this loop might interact with residues in PLN domain IB.
Materials and Methods
Materials.
Enzymes for DNA manipulation were obtained from New England Biolabs and Amersham Pharmacia. G-Sepharose and a chemiluminescence kit for measurement of coimmunoprecipitation were purchased from Pierce.
Oligonucleotide-Directed Mutagenesis and Expression.
Site-directed mutagenesis of SERCA1a and PLN was carried out as described (15, 17). The coexpression of various combinations of wild-type (wt) or mutant PLN and wt or mutant SERCA1a in HEK-293 cells and preparation of microsome fractions were described in earlier publications (14–17).
Coimmunoprecipitation of SERCA1a and PLN from Microsomal Fractions.
Coimmunoprecipitation of SERCA1a and PLN was carried out as described (15), using anti-PLN monoclonal antibody 1D11, specific for an epitope between PLN residues 7 and 17 (15) and anti-SERCA1a monoclonal antibody A52, specific for an epitope between residues 657 to 672 in SERCA1a (15). Immunoprecipitates, separated by SDS/PAGE, were transferred to either 0.05- or 0.45-μm nitrocellulose membranes. Signals were detected with an enhanced chemiluminescence kit (Pierce Super Signal). The relative amounts of SERCA1a and PLN in each lane in exposed films were quantified by scanning densitometry by using a Kodak X-omat Processor. Protein expression levels in all experiments were estimated by immunoblotting using antibodies A52, and 1D11.
Ca2+ Transport Activity.
Measurements of Ca2+ transport activity in microsomal fractions were carried out as described (14–17).
Results
Functional Interactions Between PLN and SERCA1a Mutated in L67.
Earlier, we showed that PLN interacts with and regulates both SERCA2a and SERCA1a (14). The loop between M6 and M7 (L67) is well conserved between SERCA1a and SERCA2a, differing at (SERCA1a numbering) D818 = N, R819 = K, and S823 = N (19), and might be a site of interaction with PLN. To test for functional alterations, we mutated all of the amino acids in SERCA1a between F809 and P827 to Ala and measured the effect on apparent Ca2+ affinities of their coexpression with wt PLN. Results for F809A, P811A, and P812A have been published (15). For mutants L814A, I816A, M817A, or D818A, activity was so low that we were unable to evaluate functional interactions with PLN.
Functional data describing the apparent Ca2+ affinity of wt and mutant SERCA1a expressed in the presence and absence of wt PLN are presented in Table 1. The KCa value for SERCA1a expressed in the absence of PLN was 6.38 pCa units (pCa = −log10[Ca2+]) and in the presence of PLN was 6.04 pCa units. Thus, the ΔKCa attributed to the inhibitory interaction with wt PLN was −0.34 pCa units (Table 1). For the mutants D815A, R819A, P820A, P821A, S823A, P824A, K825A, E826A, and P827A, KCa, ranging from pCa 6.40 to 6.63, and changes because of interaction with PLN (ΔKCa values) ranging from −0.25 to −0.43 pCa units, were comparable to wt. The apparent Ca2+ affinities for mutants N810A, D813A, and R822A were lower than wt, ranging from pCa 5.67 to pCa 6.01, and ΔKCa values for this group of mutants were as follows: N810A, −0.10; D813A, −0.08, and R822, −0.09, all of which were significantly different from the wt ΔKCa value of −0.34.
Table 1.
KCa of wt SERCA1a and mutants expressed in HEK-293 cells in the presence and absence of PLN
| Mutation | KCa − PLN | KCa + PLN | ΔKCa | n |
|---|---|---|---|---|
| wt | 6.38 ± 0.03 | 6.04 ± 0.03 | −0.34 ± 0.01 | 20 |
| N810A | 5.75 ± 0.11 | 5.65 ± 0.11 | −0.10 ± 0.03* | 3 |
| D813A | 5.67 ± 0.06 | 5.60 ± 0.02 | −0.08 ± 0.11* | 3 |
| D815A | 6.40 ± 0.02 | 6.15 ± 0.04 | −0.25 ± 0.05 | 3 |
| R819A | 6.41 ± 0.03 | 6.00 ± 0.10 | −0.41 ± 0.14 | 3 |
| P820A | 6.63 ± 0.06 | 6.21 ± 0.04 | −0.42 ± 0.05 | 3 |
| P821A | 6.45 ± 0.06 | 6.08 ± 0.04 | −0.36 ± 0.05 | 3 |
| R822A | 5.98 ± 0.08 | 5.96 ± 0.06 | −0.09 ± 0.10* | 5 |
| S823A | 6.43 ± 0.01 | 6.11 ± 0.07 | −0.32 ± 0.13 | 3 |
| P824A | 6.36 ± 0.04 | 6.00 ± 0.06 | −0.35 ± 0.04 | 3 |
| K825A | 6.42 ± 0.06 | 6.14 ± 0.05 | −0.28 ± 0.04 | 3 |
| E826A | 6.55 ± 0.04 | 6.12 ± 0.05 | −0.43 ± 0.02 | 3 |
| P827A | 6.50 ± 0.03 | 6.17 ± 0.07 | −0.33 ± 0.06 | 3 |
, P < 0.05 against wt SERCA1a.
Although there appears to be a correlation between low intrinsic Ca2+ affinity and reduced inhibition by PLN in these mutants, our earlier studies (15) showed that such a correlation does not hold. Thus, SERCA1a mutants I307A, I761A, and N914A, with intrinsic KCa values of 5.74, 6.07, and 6.07 pCa units, respectively, had ΔKCa values of −0.36, −0.28, and −0.28 pCa units, when expressed with PLN. These values were not significantly different from wt.
Physical Interactions Between PLN and SERCA1a Carrying Mutations in L67.
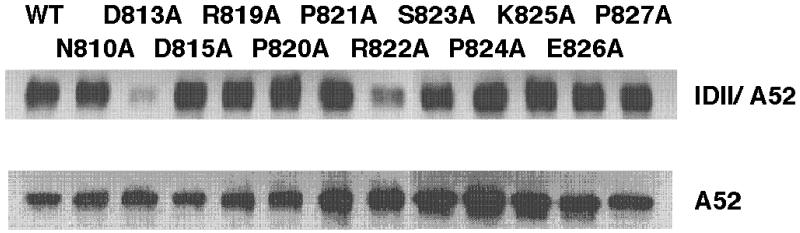
Earlier (15, 20), we showed that there is a reasonable correlation between loss or gain of PLN-SERCA functional interactions and loss or gain of PLN-SERCA physical interactions, as judged by coimmunoprecipitation of SERCA molecules with antibodies against PLN. Expression and coimmunoprecipitation data for 12 SERCA1a mutants not described previously are presented in Fig. 1. In the Lower lanes of Fig. 1, Western blotting with antibody A52, specific for SERCA1a, shows that all mutants were expressed as well as, or better than, wt. In the Upper lanes, microsomes were dissolved in Tween-20, and PLN was immunoprecipitated by antibody IDII. The immunoprecipitate was separated by SDS/PAGE and the gel was stained with antibody A52 to detect coimmunoprecipitated SERCA1a. Mutants D813A and R822A showed a loss of physical interaction that corresponded with their loss of functional interaction, but mutant N810A, which lost the ability to interact functionally with PLN, retained full physical interaction by comparison with wt. Data presented in line 1, Table 2 show that the ability of mutant D813A to interact with PLN was reduced to 32% of wt and of mutant R822A to 64% of wt.
Figure 1.
Coimmunoprecipitation of wt PLN with L67 mutants of SERCA1a. Microsomal fractions from HEK-293 cells coexpressing wt PLN and SERCA1a L67 mutants were dissolved in Tween 20, specific proteins were coimmunoprecipitated with anti-PLN antibody IDII, and the precipitate was separated by 8% SDS/PAGE. After electrophoresis, the proteins were transferred to a nitrocellulose membrane and stained with anti-SERCA1a antibody, A52. The Upper row was stained with the A52 antibody against SERCA1 and defines the relative amount of SERCA1a that was coimmunoprecipitated with antibody ID11 against PLN. The Lower row shows staining of 10 μg of microsomes from each expressed sample with the A52 antibody, to show that each mutant SERCA1 was well expressed.
Table 2.
Relative physical interactions between PLN domain IB mutants and SERCA1a L67 mutants D813A and R822A
| wt
|
D813A
|
R822A
|
||||||
|---|---|---|---|---|---|---|---|---|
| Mean ± SE | n | Mean ± SE | wt/D813A | n | Mean ± SE | wt/R822A | n | |
| wtPLN | 1 | 3 | 0.32 ± 0.07* | 3.1 | 4 | 0.64 ± 0.07* | 1.6 | 4 |
| R25A | 1.10 ± 0.05 | 3 | 0.54 ± 0.13* | 2.0 | 4 | 0.76 ± 0.12* | 1.4 | 4 |
| Q26A | 1.61 ± 0.18* | 3 | 0.86 ± 0.09 | 1.9 | 4 | 1.05 ± 0.12 | 1.5 | 4 |
| N27A | 2.64 ± 0.18* | 3 | 0.36 ± 0.06* | 7.3 | 4 | 1.03 ± 0.15 | 2.6 | 4 |
| L28A | 2.31 ± 0.25* | 3 | 0.84 ± 0.16 | 2.7 | 4 | 0.82 ± 0.20 | 2.8 | 4 |
| Q29A | 2.42 ± 0.28* | 3 | 1.00 ± 0.07 | 2.4 | 4 | 0.95 ± 0.08 | 2.6 | 4 |
| N30A | 2.95 ± 0.19* | 3 | 0.51 ± 0.06* | 5.8 | 4 | 1.15 ± 0.35 | 2.6 | 4 |
| I40A | 4.34 ± 0.64* | 3 | 1.60 ± 0.15* | 2.7 | 4 | 2.02 ± 0.65* | 2.2 | 4 |
The density of bands of wt SERCA1a or L67 mutants D813A and R822A coimmunoprecipitated with PLN domain IB mutants was measured, and the mean ± SE was calculated for each sample. Statistical analysis was carried out using Student's unpaired t test.
, P < 0.05 vs. coexpression of wt PLN and SERCA1a.
Physical Interactions Between SERCA1a D813A and R822A Mutants and PLN Domain IB Mutants.
To characterize the SERCA1a D813A and R822A mutants more completely and, in an attempt to determine whether there might be interaction between residues in SERCA1a L67 and PLN domain IB, we analyzed the physical interactions between wt and D813A and R822A mutant SERCA1a molecules and mutants R25A, Q26A, N27A, L28A, Q29A, and N30A in PLN domain IB and I40A in domain II. As shown in Table 2, PLN mutants Q26A, N27A, L28A, Q29A, N30A, and I40A had enhanced physical interactions with SERCA1a, in line with earlier studies showing enhanced functional and/or physical interactions for N27A, Q29A, N30A, and I40A (15, 18). Mutants Q26A and L28A, however, have been shown previously to have diminished functional interactions with SERCA1a (15, 18). When each of these PLN mutants was expressed with the SERCA1a mutants D813A and R822A, there was an almost uniform reduction in physical interaction, ranging from 1.4-fold to 3.1-fold (Table 2). However, the interaction of PLN mutant N27A with SERCA1a mutant D813A was reduced by 7.3-fold, and the interaction of PLN mutant N30A with SERCA1a mutant D813A was reduced by 5.8-fold, suggesting that specific interactions occur among SERCA1a D813 and PLN N27 and N30.
Effect of Deletion of PLN Domain IB on Physical Interactions with SERCA1a.
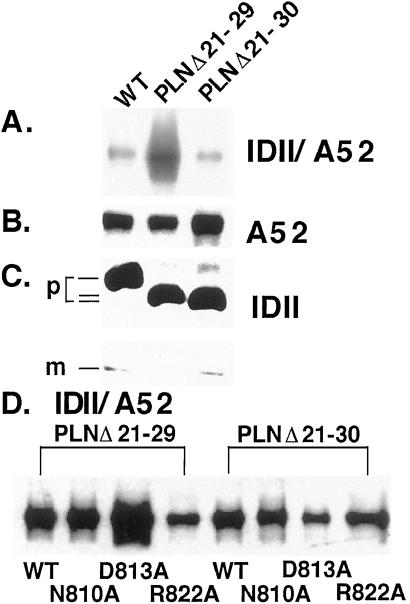
We investigated the role of N30 in PLN association with SERCA1a by using deletion analysis. Earlier (21), we observed that the PLN deletion mutant, PLNΔ21–29, was highly inhibitory. Data presented in Fig. 2A show that physical association of PLNΔ21–29 with wt SERCA1a was enhanced 4.5-fold over wt PLN. By contrast, there was no increase in physical association of PLNΔ21–30 with wt SERCA1a. When PLNΔ21–29 was coexpressed with SERCA1a mutants N810A, D813A, and R822A (Fig. 2D Left), no difference in physical interaction was observed with SERCA1a mutant N810A, but the interaction with mutant D813A was enhanced 1.7-fold, whereas interaction with R822A was decreased by 50%. When we tested the physical association of PLNΔ21–30 with both wt and mutant SERCA1a (Fig. 2D Right), physical association was diminished with wt SERCA1a and with all three SERCA1a mutants, N810A, D813A, and R822A.
Figure 2.
Coimmunoprecipitation of SERCA1a and SERCA1a mutants with PLNΔ21–29 and PLNΔ21–30. (A) Microsomal fractions from HEK-293 cells coexpressing SERCA1a with either PLNΔ21–29 or PLNΔ21–30 were dissolved in Tween 20, and specific proteins were coimmunoprecipitated with anti-PLN antibody IDII. The immunoprecipitates were separated by SDS/PAGE, transferred to nitrocellulose membranes, and stained with anti-SERCA1a antibody A52. The interaction between SERCA1a and the PLN mutant PLNΔ21–29, but not PLNΔ21–30, is enhanced 4.5-fold. The relative levels of expression of SERCA1a (B) or of PLN and PLN deletion mutants (C) are shown. m, monomer; p, pentamer. (D). Microsomal fractions from HEK-293 cells coexpressing SERCA1a mutants N810A, D813A, or R822A with either PLNΔ21–29 or PLNΔ21–30 were analyzed in the same way. The interaction between SERCA1a mutant D813A and the PLN mutant PLNΔ21–29, but not PLNΔ21–30, is enhanced 1.7-fold.
Interactions Between SERCA1a and PLN-Cytochrome b5 Chimeric Proteins.
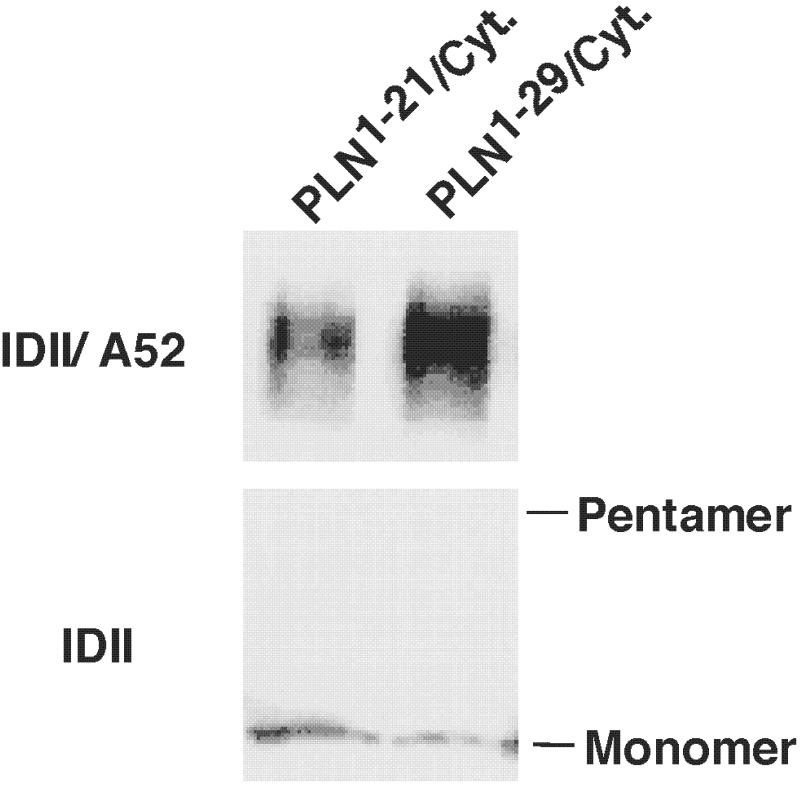
Earlier (22), we showed that PLN domain I-Cytochrome b5 transmembrane domain chimeras PLN1–21/cytb5 and PLN1–29/cytb5, do not inhibit the activity of SERCA2a. Data presented in Fig. 3 show that physical interactions between SERCA1a and PLN1–29/cytb5 were 1.6-fold higher than those between PLN1–21/cytb5 lacking a key PLN residue, N27. These studies, combined with other studies described above, support the view that a dynamic interaction occurs between D813 in SERCA1a and N27 and N30 in PLN.
Figure 3.
Coimmunoprecipitation of SERCA1a with PLN-cytochrome b5 chimeras. Microsomal fractions from HEK-293 cells coexpressing SERCA1a with either chimera PLN1–21/cytb5101–134 or PLN1–29/cytb5109–134 were dissolved in Tween 20, and specific proteins were coimmunoprecipitated with anti-PLN antibody IDII. The immunoprecipitates were separated by SDS/PAGE, transferred to nitrocellulose membranes, and stained with anti-SERCA1a antibody A52 (Upper).The relative level of expression of chimeras PLN1–21/cytb5101–134 and PLN1–29/cytb5109–134 is illustrated by their levels of staining with anti-PLN antibody IDII (Lower). Physical interactions between SERCA1a and PLN1–29/cytb5 were 1.6-fold higher than those between PLN1–21/cytb5 lacking a key PLN residue, N27.
Discussion
Physical and Functional Probing of Sites of Interaction Between PLN Domain IB and SERCA1a L67.
In previous papers, we showed that PLN interacts with the cytoplasmic sequence Lys-Asp-Asp-Lys-Pro-Val402, which forms an exposed loop on the surface of the nucleotide binding domain in the crystal structure of SERCA1a (2), and with residues V795 and L802 on a face in transmembrane helix M6 that is oriented away from the Ca2+ binding sites (2). Our objective in this study was to use mutagenesis to evaluate whether the trajectory of PLN from the top of M6 to the cytoplasmic site might interact with residues in L67, a loop that encircles and interacts with several elements in the stalk region of SERCA1a. This goal has proven to be difficult, probably because L67 of SERCA1a is linked to so many elements in the protein that mutation has complex effects on the structure of the molecule. Nonetheless, we have, through a series of approaches, highlighted a probable interaction among N27 and N30 in PLN and D813 in SERCA1a.
Our initial evaluation of functional interactions, showed that mutants N810A, D813A and R822A had diminished ability to interact functionally with PLN. We then evaluated physical interactions defined by coimmunoprecipitation of SERCA1a molecules with an antibody directed against PLN. The N810 mutant retained full physical interaction, even though it had reduced functional interaction. Physical interactions between mutant D813A and PLN, however, were reduced to 32% of wt SERCA1a, in line with reduced functional interactions, and PLN interaction with SERCA1a mutant R822A was reduced to 64% of wt. These studies focused particular attention on SERCA1a residue D813.
We then turned our attention to sites in PLN domain IB, the region in PLN most likely to interact with L67 in SERCA1a. Earlier (17), we found that mutant N27A was among the most highly inhibitory mutants known and that mutants N29A and N30A were also superinhibitory, whereas mutants R25A, Q26A, and L28A had diminished functional interactions. These observations had already highlighted this sequence as forming a potential interaction site with SERCA1a. We found that mutants Q26A, N27A, L28A, N29A, and N30A in domain IB and mutant I40A all had enhanced physical interaction with wt SERCA1a. The level of physical interaction was reduced almost uniformly by about 2.3-fold (range 1.4–3.1) for the interaction of all of the PLN mutants with SERCA1a R822A and for most PLN mutants with SERCA1a D813A. The interaction of PLN mutant N27A with SERCA1a D813A, however, was reduced by 7.3-fold and of PLN mutant N30A by 5.8-fold. These observations support the view that N27 and N30, presumably on the same helical face of PLN domain IB, interact rather specifically with D813 in SERCA1a L67.
We attempted, without success, to measure cross-linking within a group of mutant proteins—N810C, D813C, D818C, and R822C—in SERCA1a and a pair of PLN mutants—N27C and N30—in PLN, by using protocols that were successful in studies of cross-linking of residues in transmembrane helices M4 and M6 in SERCA1a (23). Our lack of success was not surprising, because our attempts to form cross-links among several residues shown to be proximal in the crystal structure of SERCA1a (2) have also been unsuccessful.
Earlier (21), we showed that the internal deletion mutant, PLNΔ21–29, was highly inhibitory, and, in experiments described here, we observed that the physical association of this mutant with wt SERCA1a was enhanced 4.5-fold. Enhanced physical association was not observed for PLNΔ21–30, which lacked N30, highlighting once again the importance of this residue for PLN interaction with SERCA1a. Physical association between PLNΔ21–29 and SERCA1a mutant D813A was enhanced 1.7-fold over wt SERCA1a, whereas physical associations were only slightly altered or decreased for SERCA1a mutations N810A and R822A. This observation might be interpreted to indicate that an interaction between SERCA D813 and PLN N30 is highly favored in wt proteins, but that the conformation of PLNΔ21–29 favors an interaction between PLN N30 and SERCA D813A. In this light, the experiment, although providing a different answer than expected, might again be highlighting the close association between SERCA D813 and PLN N30.
Chimeric molecules in which the C terminus of PLN was replaced with a cytochrome b5 transmembrane segment do not interact functionally with SERCA1 (22). It was, therefore, interesting to observe that PLN1–29/cytb5 had strong physical interactions with SERCA1a, but these interactions were reduced by 40% when PLN1–29/cytb5 was replaced with PLN1–21/cytb5. These studies provide further evidence that residues 22–29, containing N27, are important for interactions between PLN and SERCA1a. When combined with other studies described above, they support the view that a dynamic interaction occurs between D813 in SERCA1a and N27 and N30 in PLN.
Structural Implications of PLN-SERCA1a Interaction Sites.
PLN has been shown by NMR to be composed of two helices separated by a β-bend involving (Thr)-Ile-Glu-Met-Pro21 (12, 13, 24). The crystal structure of the E1[Ca2+]2 conformation of SERCA1a, at a resolution of 2.6 Å (2), shows that phosphorylation, nucleotide binding, and actuator sequences form distinct cytosolic domains and that 10 transmembrane helices form the Ca2+ binding and translocation domain. The site of cytoplasmic interaction between PLN and SERCA1 occurs in an exposed loop in the nucleotide binding domain.
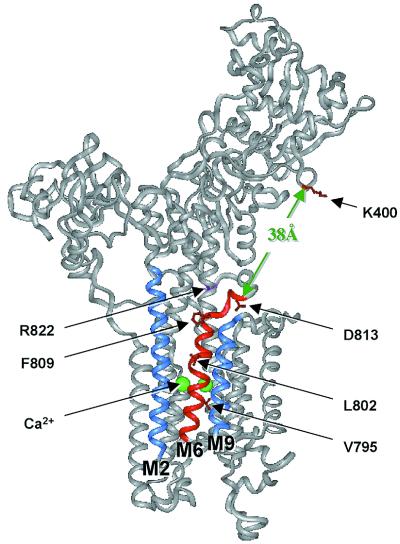
The structure would, in principle, allow modeling of the PLN structure into the E1[Ca2+]2, conformation of SERCA1a by orienting the PLN transmembrane region into a groove that is formed between M2 and M9 on the transmembrane surface of SERCA1a and is backed by M6 (Fig. 4). Such a model would be consistent with our results obtained by mutagenesis (15). This orientation of PLN domain II in the membrane domain of the PLN/SERCA1a complex would place PLN N27 and N30 near SERCA1a D813 and would place the N terminus of PLN near K400, in accord with the results of both cross-linking and mutagenesis (14, 25). Unfortunately, the E1[Ca2+]2 conformation, for which crystal structure is known, does not interact with PLN. An E2 structure, which is capable of interacting with PLN (the E2[V10] conformation) is known only at low resolution (26). Comparison of the two structures at low resolution (27–29) showed that formation of E2[V10] from E1[Ca2+]2 would require minor reorientation of transmembrane helices, a movement of L67 away from the membrane and a partial closure of the N-domain onto the P-domain by a rotation about a hinge near to Asp-Pro-Pro-Arg604.
Figure 4.
Sites in the crystal structure of SERCA1a where PLN is proposed to interact. A version of the crystal structure of SERCA1a (2) was generated in the insight ii program (Molecular Simulations, Waltham, MA). PLN domain IA interacts with SERCA1a residues K397-V402 (red) in the nucleotide binding domain. PLN domain II binds to transmembrane helix M6 (red), lying in a groove between helices M2 and M9 (blue). Two Ca2+ ions (green) bind to the opposite face of M6. If PLN domain II is located in the groove formed by helices M2, M6, and M9 and if it is also associated with K400 in the cytosolic domain, then PLN domain IB must cross the region from the top of the M6 helix (G808) to D813 located in L67. This distance is about 10 Å. It must then cross a gap of about 38 Å to reach the cytosolic interaction site identified as K400. Evidence for interactions among residues N27 and N30 in PLN domain IB and residue D813 in SERCA1a L67 is fully consistent with this model for PLN interaction with SERCA1a. The crystal structure of SERCA1a was reprinted by permission from Nature (2), copyright 2000 Macmillan Magazines Ltd.
In the crystal structure of the E1[Ca2+]2 conformation of SERCA1a (2), the distance between the α-carbons of G808, marking the top of transmembrane helix M6, and D813 is 10 Å and the distance between D813 and the amino group of K400 is 38 Å. The 38 Å between D813 and K400 might be extended to 45–50 Å, after removal of Ca2+, but even this distance could be bridged by the N-terminal 26 residues of PLN, especially since there is unwinding of the PLN helix between residues 17 and 21.
The PLN domain IB-II helix is predicted to be oriented approximately parallel to M6. Since D813 is offset relative to the top of the M6 helix, either a bend in the N27-N30 region of PLN or a movement of D813 in the E2 conformation relative to the E1[Ca2+]2 conformation would be required for interaction of PLN N27 or N30 with D813 in SERCA1a. Although there is no independent evidence for a bend in PLN from the NMR results, these results were obtained in the helix-enhancing solvent trifluoroethanol or in chloroform-methanol. Because this portion of the sequence of PLN is polar, it might be partially uncoiled in an aqueous environment. It is likely to be more relevant, however, that the position of L67 changes in the transition to the E2 conformation, following the position of domain P (2, 28). Its position in E2 is much higher from the membrane surface than in the presence of Ca2+. This might locate D813 closer to that portion of the PLN helix which contains N27 and N30.
In the structure of L67 in the Ca2+ bound form of SERCA1a, D813 lies above M6 and forms a hydrogen bond with N755 in helix M5. By contrast, R822 lies behind M5, in a position that would be inaccessible to a PLN helix. The mutation R822A would break up the interaction of this residue with E340 on M4 and T247 on M3, and this destabilization could diminish the interaction of SERCA1a with PLN, which we observed, without a requirement for a direct interaction of R822 with PLN. The possibility that PLN might interact with all of D813, R822, and K400 is obviated because the combined distances between D813, R822, and K400 are greater than the potential length of PLN domains IA and IB. Thus, it is most likely that PLN interacts only with D813, which is close to M6, stabilizing its interaction with SERCA1a. However, because the crystal structure is derived from a conformation of SERCA1a that does not bind to PLN, D813 and R822 may lie in different positions in the Ca2+-free conformation.
The physical interactions of SERCA1a with PLN is not disrupted by PLN phosphorylation(20), even though PLN does not inhibit SERCA1a after phosphorylation (7). It will be interesting to discover how PLN phosphorylation can blunt functional interactions without disrupting physical interactions. PLN phosphorylation might break up physical interactions occurring between PLN domain IB and SERCA1a L67, leaving intact the interactions between the N terminus of PLN and the SERCA1a binding site near K400.
Acknowledgments
We thank Dr. Chikashi Toyoshima, University of Tokyo, for helpful discussions. We also thank Dr. Robert G. Johnson and Merck Research Laboratories for the gift of the anti-PLN antibody 1D11 and Dr. R. J. Kaufman, Genetics Institute, Boston, for the gift of the pMT2 vector. This work was supported by grants to D.H.M. from the Heart and Stroke Foundation of Ontario and by Grant MT-12545 from the Medical Research Council of Canada. M.A. was a postdoctoral fellow of the Heart and Stroke Foundation of Canada.
Abbreviations
- SERCA
sarco(endo)plasmic reticulum Ca2+ ATPase
- PLN
phospholamban
- wt
wild type
- pCa
−log10[Ca2+]
References
- 1.MacLennan D H, Rice W J, Green N M. J Biol Chem. 1997;272:28815–28818. doi: 10.1074/jbc.272.46.28815. [DOI] [PubMed] [Google Scholar]
- 2.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Nature (London) 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 3.MacLennan D H, Brandl C J, Korczak B, Green N M. Nature (London) 1985;316:696–700. doi: 10.1038/316696a0. [DOI] [PubMed] [Google Scholar]
- 4.Stokes D L, Green N M. Biophys J. 2000;78:1765–1776. doi: 10.1016/s0006-3495(00)76727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menguy T, Corre F, Bouneau L, Deschamps S, Moller J V, Champeil P, le Maire M, Falson P. J Biol Chem. 1998;273:20134–20143. doi: 10.1074/jbc.273.32.20134. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Lewis D, Sumbilla C, Inesi G, Toyoshima C. J Biol Chem. 2001;276:15232–15239. doi: 10.1074/jbc.M010813200. [DOI] [PubMed] [Google Scholar]
- 7.Simmerman H K, Jones L R. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 8.Luo W, Grupp I L, Harrer J, Ponniah S, Grupp G, Duffy J J, Doetschman T, Kranias E G. Circ Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 9.Zvaritch E, Backx P H, Jirik F, Kimura Y, de Leon S, Schmidt A G, Hoit B D, Lester J W, Kranias E G, MacLennan D H. J Biol Chem. 2000;275:14985–14991. doi: 10.1074/jbc.275.20.14985. [DOI] [PubMed] [Google Scholar]
- 10.Zhai J, Schmidt A G, Hoit B D, Kimura Y, MacLennan D H, Kranias E G. J Biol Chem. 2000;275:10538–10544. doi: 10.1074/jbc.275.14.10538. [DOI] [PubMed] [Google Scholar]
- 11.Haghighi K, Schmidt A G, Hoit B D, Brittsan A G, Yatani A, Lester J W, Zhai J, Kimura Y, Dorn G W, 2nd, MacLennan D H, Kranias E G. J Biol Chem. 2001;27:27. doi: 10.1074/jbc.M102403200. [DOI] [PubMed] [Google Scholar]
- 12.Pollesello P, Annila A, Ovaska M. Biophys J. 1999;76:1784–1795. doi: 10.1016/S0006-3495(99)77339-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamberth S, Schmid H, Muenchbach M, Vorherr T, Krebs J, Carafoli E, Griesinger C. Helv Chim Acta. 2000;83:2141–2152. [Google Scholar]
- 14.Toyofuku T, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1994;269:22929–22932. [PubMed] [Google Scholar]
- 15.Asahi M, Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1999;274:32855–32862. doi: 10.1074/jbc.274.46.32855. [DOI] [PubMed] [Google Scholar]
- 16.Toyofuku T, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1994;269:3088–3094. [PubMed] [Google Scholar]
- 17.Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- 18.Kimura Y, Asahi M, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1998;273:14238–14241. doi: 10.1074/jbc.273.23.14238. [DOI] [PubMed] [Google Scholar]
- 19.Burk S E, Lytton J, MacLennan D H, Shull G E. J Biol Chem. 1989;264:18561–18568. [PubMed] [Google Scholar]
- 20.Asahi M, McKenna E, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 2000;275:15034–15038. doi: 10.1074/jbc.275.20.15034. [DOI] [PubMed] [Google Scholar]
- 21.Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1996;271:21726–21731. doi: 10.1074/jbc.271.36.21726. [DOI] [PubMed] [Google Scholar]
- 22.Kimura Y, Asahi M, Kurzydlowski K, Tada M, MacLennan D H. FEBS Lett. 1998;425:509–512. doi: 10.1016/s0014-5793(98)00151-3. [DOI] [PubMed] [Google Scholar]
- 23.Rice W J, Green N M, MacLennan D H. J Biol Chem. 1997;272:31412–31419. doi: 10.1074/jbc.272.50.31412. [DOI] [PubMed] [Google Scholar]
- 24.Mortishire-Smith R J, Pitzenberger S M, Burke C J, Middaugh C R, Garsky V M, Johnson R G. Biochemistry. 1995;34:7603–7613. doi: 10.1021/bi00023a006. [DOI] [PubMed] [Google Scholar]
- 25.James P, Inui M, Tada M, Chiesi M, Carafoli E. Nature (London) 1989;342:90–92. doi: 10.1038/342090a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Toyoshima C, Yonekura K, Green N M, Stokes D L. Nature (London) 1998;392:835–839. doi: 10.1038/33959. [DOI] [PubMed] [Google Scholar]
- 27.Stokes D L, Zhang P, Toyoshima C, Yonekura K, Ogawa H, Lewis M R, Shi D. Acta Physiol Scand Suppl. 1998;643:35–43. [PubMed] [Google Scholar]
- 28.Ogawa H, Stokes D L, Sasabe H, Toyoshima C. Biophys J. 1998;75:41–52. doi: 10.1016/S0006-3495(98)77493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice W J, Young H S, Martin D W, Sachs J R, Stokes D L. Biophys J. 2001;80:2187–2197. doi: 10.1016/S0006-3495(01)76191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]