SUMMARY
Precise genome editing via homology-directed repair (HDR) in targeted cells, particularly in vivo, provides an invaluable tool for biomedical research. However, HDR has been considered to be largely restricted to dividing cells, making it challenging to apply the technique in postmitotic neurons. Here we show that precise genome editing via HDR is possible in mature postmitotic neurons as well as mitotic cells in mice brain by combining CRISPR-Cas9-mediated DNA cleavage and the efficient delivery of donor template with adeno-associated virus (AAV). Using this strategy, we achieved efficient tagging of endogenous proteins in primary and organotypic cultures in vitro and developing, adult, aged, and pathological brains in vivo. Thus, AAV- and CRISPR-Cas9-mediated HDR will be broadly useful for precise genome editing in basic and translational neuroscience.
Keywords: Genome editing, CRISPR, Cas9, HDR, AAV, postmitotic, neuron, in vivo, SLENDR, vSLENDR
INTRODUCTION
Genome editing technologies based on the clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonuclease Cas9 enable rapid and efficient modification of endogenous genes in a variety of cell types, allowing for analysis of gene function in many organs in vivo. CRISPR-Cas9 induces DNA double strand breaks (DSBs) at single-guide RNA (sgRNA)-specific loci in the genome, which are repaired through either non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathways. While NHEJ introduces unpredictable pattern of insertions or deletions (indels) mutations, HDR directs a precise recombination event between a homologous DNA donor template and the damaged DNA site (Cong et al., 2013; Cox et al., 2015; Doudna and Charpentier, 2014; Heidenreich and Zhang, 2016; Jinek et al., 2012; Mali et al., 2013; Sander and Joung, 2014; Wang et al., 2013; Yang et al., 2013). Thus, HDR can be used to precisely introduce sequence insertions, deletions or mutations by encoding the desired changes in the donor template DNA.
While HDR-based genome editing has been demonstrated to be useful for precise genome editing, application of HDR-based genome editing has been limited to mitotically dividing cells. This is because HDR had previously been found to occur primarily in the S and G2 phases of the cell cycle in mitotically dividing cells. In fact, HDR was found to occur rarely in postmitotic cells such as neurons (Chapman et al., 2012; Cox et al., 2015; Panier and Durocher, 2013; Rothkamm et al., 2003). Thus, in the brain, HDR-based genome editing has been performed previously only in dividing cells such as neuronal progenitors in the embryo. With this strategy, we and other groups succeeded in inserting a tag sequence into a gene of interest, enabling the monitoring of the endogenous protein encoded by the gene with the tag (Mikuni et al., 2016; Uemura et al., 2016). On the other hand, tag insertion via NHEJ-based genome editing has been achieved in postmitotic cells (Auer et al., 2014; Maresca et al., 2013; Suzuki et al., 2016). However, this method lacks the precision of HDR and is subject to indel mutations. Therefore, precise genome editing via HDR has been a challenge in postmitotic cells.
A number of methods have been used to deliver genome editing machinery in vivo. In the previous studies to induce HDR in the brain in vivo, in utero electroporation (IUE) has been used to deliver HDR machinery into neuronal progenitor cells (Mikuni et al., 2016; Uemura et al., 2016). In addition, adeno-associated virus (AAV) has been used for gene delivery in a wide variety of tissues including the brain, because they can robustly infect both dividing and non-dividing cells with low immunogenicity and toxicity. It has been reported that AAV can serve as a donor template for homologous recombination at higher efficiencies than non-viral approaches, presumably due to the efficient nuclear delivery of the vector genome, the single-strand nature of the genome and the effect of inverted terminal repeats (ITRs) (Barzel et al., 2015; Gaj et al., 2016; Russell and Hirata, 1998). Moreover, the combined use of endonuclease to induce DSBs with AAV has been reported to increase the frequency of AAV-mediated gene targeting by several orders of magnitudes via unknown mechanisms (Dever et al., 2016; Gaj et al., 2016; Porteus et al., 2003). Thus, AAV-based HDR assisted by CRISPR-Cas9 may allow us to efficiently induce HDR, even in postmitotic cells such as neurons.
In the present study, we show that it is possible to perform efficient HDR-mediated genome editing not only in mitotic cells but also in postmitotic neurons in the brain by delivering HDR template with AAV. By using either Cas9 knockin mice (Chiou et al., 2015; Platt et al., 2014) or Cas9-expressing AAV, we demonstrate that efficient HDR-mediated sequence insertion can be performed in postmitotic neurons as well as in mitotic neuronal progenitors and glial cells. Through this approach, virus-mediated single-cell labeling of endogenous proteins via HDR (vSLENDR), we achieved highly efficient tagging of endogenous proteins in primary dissociated and organotypic slice cultures in vitro and various brain areas and cell-types in developing and adult brains in vivo. We further demonstrate that vSLENDR allows us to label endogenous proteins in mice models of age-related human neurological disorders at ~1 year old. Thus, HDR-mediated genome editing can be performed in virtually any cell-types, areas and ages across the brain, widely expanding the applicability of genome engineering technologies in the broad field of neuroscience.
RESULTS
HDR-Mediated Genome Editing in the Embryonic Brain
HDR occurs more efficiently in dividing cells (Mikuni et al., 2016). Thus, to develop AAV-mediated genome editing technique via HDR, we first targeted dividing neuronal progenitor cells in the embryo by intraventricular administration of AAV encoding HDR machinery. Intraventricular administration would allow widespread transgene expression throughout the brain due to the penetration of AAV into the brain parenchyma including the ventricular zone (Passini et al., 2003; Rahim et al., 2012). To test this strategy, we aimed to insert the human influenza hemagglutinin (HA) tag or monomeric EGFP (mEGFP) sequence into β-Actin, which expresses ubiquitously in most of cell types, and into CaMKIIα, which expresses exclusively in neurons (Mikuni et al., 2016).
HDR-mediated genome editing requires delivering three components in the same cells: sgRNA, donor template DNA, and Cas9. We designed single AAV vectors encoding the following two components: (1) a U6-sgRNA expression cassette targeting the vicinity of the start codon of β-Actin or CaMKIIα, and (2) an HDR template containing the HA tag or mEGFP sequence flanked by ~1 kb sequences homologous to β-Actin or CaMKIIα, respectively (termed AAV-HDR hereafter) (Figure 1A). To minimize the potential effects of gene knockdown through NHEJ, we used previously validated sgRNA targeting sequences in which CRISPR-Cas9 cleavage sites were located in 5′-UTR of β-Actin and CaMKIIα (Mikuni et al., 2016) (Figures 1C and 1F). We packaged the designed vectors with serotype 1 (AAV1), which was known to provide high transduction efficiency and tropism in both neurons and glia in vivo (Aschauer et al., 2013). As for Cas9, we utilized H11Cas9 mice (termed Cas9 mice hereafter), where Streptococcus pyogenes Cas9 (SpCas9) is constitutively expressed under control of the ubiquitous CAG promoter (Chiou et al., 2015).
Figure 1. HDR-Mediated Genome Editing in the Embryonic Brain.
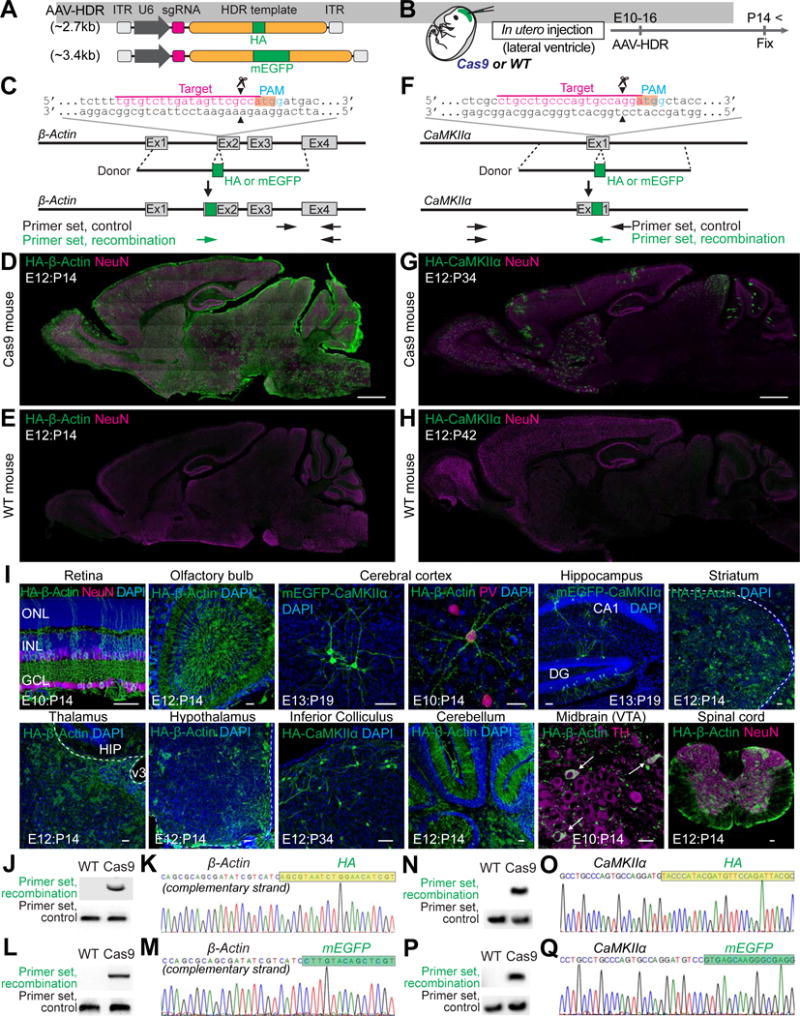
(A) Schematic of AAV-HDR for the delivery of a sgRNA and HDR donor template to insert the HA tag or mEGFP sequence. U6, human U6 Polymerase III promoter.
(B) Schematic illustration of experiments.
(C and F) Graphical representation of the mouse genomic loci of β-Actin (C) and CaMKIIα (F) showing the target sites for Cas9. The sgRNA targeting sequences are labeled in magenta. The protospacer-adjacent motif (PAM) sequences are labeled in blue. The start codons of β-Actin and CaMKIIα are marked in orange. The Cas9 cleavage sites are indicated by the black arrowheads. PCR primer sets (control and recombination) for PCR genotyping (J, L, N, and P) are indicated by the arrows.
(D, E, G, and H) Confocal microscopic images of whole sagittal brain sections of Cas9 (D and G) and wild type (E and H) mice showing immunoreactivities for NeuN (magenta) and the HA tag (green) fused to the N-terminus of endogenous β-Actin (D and E) and CaMKIIα (G and H).
(I) Confocal microscopic images of various brain regions showing the DAPI signal (blue), mEGFP-CaMKIIα fluorescence (green), and immunoreactivities for NeuN, parvalbumin (PV), or tyrosine hydroxylase (TH) (magenta) and the HA tag (green) fused to the N-terminus of endogenous β-Actin or CaMKIIα. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer (Retina). DG, dentate gyrus (Hippocampus). HIP, hippocampus; v3, third ventricle (Thalamus). VTA, ventral tegmental area; white arrows, TH positive neurons (Midbrain).
(J, L, N, and P) PCR genotyping using genomic DNAs extracted from the AAV-HDR injected brains of wild type and Cas9 mice. Recombination and control primer sets for HA-β-Actin (J), mEGFP-β-Actin (L), HA-CaMKIIα (N), and mEGFP-CaMKIIα (P) were used for PCR.
(K, M, O, and Q) DNA sequencing analysis of the PCR products for HA-β-Actin (K), mEGFP-β-Actin (M), HA-CaMKIIα (O), and mEGFP-CaMKIIα (Q). The HA tag and mEGFP sequences are marked in light green and green, respectively.
Scale bars, 1mm (D and G); 50 μm (I).
See also Figures S1–4, STAR Methods, and Table S2 and S3.
To infect dividing neuronal progenitor cells, we injected AAV-HDR for either β-Actin or CaMKIIα into the lateral ventricle of Cas9 or wild type mice brain at embryonic days (E) 10–16 (Figure 1B). We analyzed the HA signal by immunohistochemistry using anti-HA antibody, or directly imaged mEGFP fluorescence at postnatal days (P) 14–42 (Figures 1D, 1E, 1G–1I, S1, S2, and S3A–S3C). We observed strong HA staining or mEGFP fluorescence in brain slices of Cas9 mice, suggesting that the tag sequences were successfully integrated into the genome. Only background level fluorescence was observed in brain slices of wild type mice (Figures 1D, 1E, 1G, 1H, S2A, and S2B), indicating that the integration into the genome requires CRISPR-Cas9 mediated DSBs. HA- or mEGFP-β-Actin was observed in a number of NeuN-positive (neuronal) and -negative cells distributed in the entire brain regions including the olfactory bulb, retina, cerebral cortex, hippocampus, striatum, thalamus, hypothalamus, cerebellum, midbrain, and spinal cord (Figures 1I, S1, and S2). In contrast, consistent with the neuron- and region-specific expression of CaMKIIα, HA- or mEGFP-CaMKIIα was found only in neurons, but not in glia, in specific brain areas, including the olfactory bulb, cerebral cortex, hippocampus, striatum, superior/inferior colliculus, and cerebellum where CaMKIIα is expressed (Figures 1I, S1, and S2).
To test if the HA tag or mEGFP sequence was precisely inserted as intended in the genome, we performed PCR amplification of the targeted loci using primers for the HA tag or mEGFP sequence and for the genomic sequence of β-Actin or CaMKIIα outside of the homology arm (“recombination primer set” in Figures 1C, 1F, 1J–1Q). The PCR fragments were amplified from the genomic DNAs extracted from the AAV-HDR infected brains of Cas9 mice, but not from those of wild type mice (Figures 1J, 1L, 1N, and 1P). In contrast, when a primer set specific for the genomic sequence of β-Actin or CaMKIIα (“control primer set” in Figures 1C, 1F, and 1J–1Q) was used, PCR fragments were amplified both in Cas9 mice and wild type mice. Furthermore, DNA sequencing analysis of the amplified DNA fragments by the recombination primer set revealed that the HA tag or mEGFP sequence was precisely inserted at the N-terminus of β-Actin or CaMKIIα as expected (Figures 1K, 1M, 1O, and 1Q). Thus, in utero intraventricular delivery of single AAV-HDR into Cas9 mice allows for brain-wide, site-specific sequence insertion via CRISPR-Cas9-mediated HDR.
HDR-Mediated Genome Editing in Distinct Cell-Types in the Brain
Because most cell types in the brain are generated in temporally distinct phases, intraventricular delivery of viral vectors enables transgene expression in different cell types and areas in a cell birthdate-dependent manner (Sauvageot and Stiles, 2002). Indeed, we observed neurons expressing HA (or mEGFP)-CaMKIIα (only pyramidal neurons) or HA (or mEGFP)-β-Actin (both pyramidal and PV-positive inhibitory neurons) in the cortex when virus was injected at E10–13 (Figures 1I, S1, and S2). Furthermore, we observed HA-β-Actin in tyrosine hydroxylase (TH) positive dopaminergic neurons in the ventral tegmental area (VTA) in the midbrain when injected at E10, while most of HA-β-Actin positive cells in VTA were glial cells when injected at E16 (Figures 1I and S1F). In the hippocampus, HA-positive neurons were predominantly CA1 pyramidal neurons and dentate gyrus (DG) granule cells when injected at E10, while we found that most of the HA-positive neurons were dentate granule cells but only a small fraction of them was CA1 pyramidal neurons when injected at E16 (Figure S1H and Table S1). In the molecular layer of the cerebellum, we observed HA-positive, calbindin-positive Purkinje cells when injected at E10 but none when injected at E12 and E16. Instead, most of the HA positive cells were Bergman glia and stellate cells at the later stage (Figure S1I and Table S1). In retina, a large number of HA-positive ganglion cells and bipolar cells were observed when injected at E10, while no HA-positive cell was found when injected at E14 and E16 (Figure S1J and Table S1). Thus, in utero intraventricular injection of AAV-HDR enables targeting of various cell-types regulated by the timing of injection.
When AAV-HDR was injected into the lateral ventricle of P1 pups of Cas9 mice (Figure S4), most of HA-β-Actin positive cells in the hippocampus and cortex were astrocytes (Figure S4B), consistent with the peak of astrocyte proliferation during the perinatal period (Ge et al., 2012; Sauvageot and Stiles, 2002). We also observed HA-β-Actin or HA-CaMKIIα positive neurons in the olfactory bulb and DG in the hippocampus, where new neurons are continuously generated from dividing neuronal progenitors in the embryonic and adult brain (Figures S4B–S4E, and S4G) (Gage, 2000). Intriguingly, we also observed a small fraction of HA-β-Actin or HA-CaMKIIα positive cells among pyramidal neurons in the CA1 and CA3 of the hippocampus and in the cortex (Figures S4B, S4C, and S4F). These results suggest that vSLENDR enables HDR-mediated genome editing in postmitotic neurons.
HDR-Mediated Genome Editing in Postmitotic Neurons
Based on the results showing the sequence insertion in postmitotic neurons by neonatal intraventricular AAV-HDR injection, albeit low efficiency, we further examined whether vSLENDR can be used for HDR-mediated genome editing in postmitotic neurons using in vitro system. We prepared organotypic hippocampal slice cultures from Cas9 or wild type mice at P56. At day in vitro (DIV) 8, we applied AAV-HDR for HA-CaMKIIα together with AAV encoding EGFP (AAV-EGFP) as an infection marker directly onto cultures (Figure 2A). Seven days after AAV-HDR transduction, HA-CaMKIIα positive neurons were observed in the entire hippocampal slice including CA1, CA3, and DG in Cas9 mice, while only background level fluorescence was observed in wild type mice (Figure 2B). HA-CaMKIIα signals were found mostly in excitatory neurons (pyramidal neurons in CA1 and CA3 and granule cells in DG), consistent with the expression profile of CaMKIIα. The number of HA-CaMKIIα positive neurons reached at ~30% among mEGFP positive cells (CA1, HA/EGFP: 28.4% ± 3.5%, n = 497 cells/3 slices; HA/NeuN: 17.1% ± 1.5%, n = 823 cells/3 slices) when we applied 5.4*109 GC (0.5 μl of AAV-HDR at a concentration of ~1.1×1013 GC/ml; 100% in Figures 2C and 2D). The efficiency was decreased nonlinearly when we reduced the dose of AAV-HDR (Figures 2C and 2D). Similarly, when we used AAV-HDR for mEGFP-CaMKIIα or mEGFP-β-Actin, many mEGFP-CaMKIIα or mEGFP-β-Actin positive cells were observed in the entire hippocampal slice cultures in Cas9 mice, while only background level fluorescence was observed in wild type mice (Figures 2E, 2F, S5A–S5C). We again observed mEGFP-tagged proteins in cell types that are known to express the target proteins: mEGFP-CaMKIIα fluorescence was observed exclusively in excitatory neurons, while mEGFP-β-Actin fluorescence was observed in both neurons and glia (Figures 2E and 2F).
Figure 2. HDR-Mediated Genome Editing in Postmitotic Neurons in Organotypic Slice Cultures.
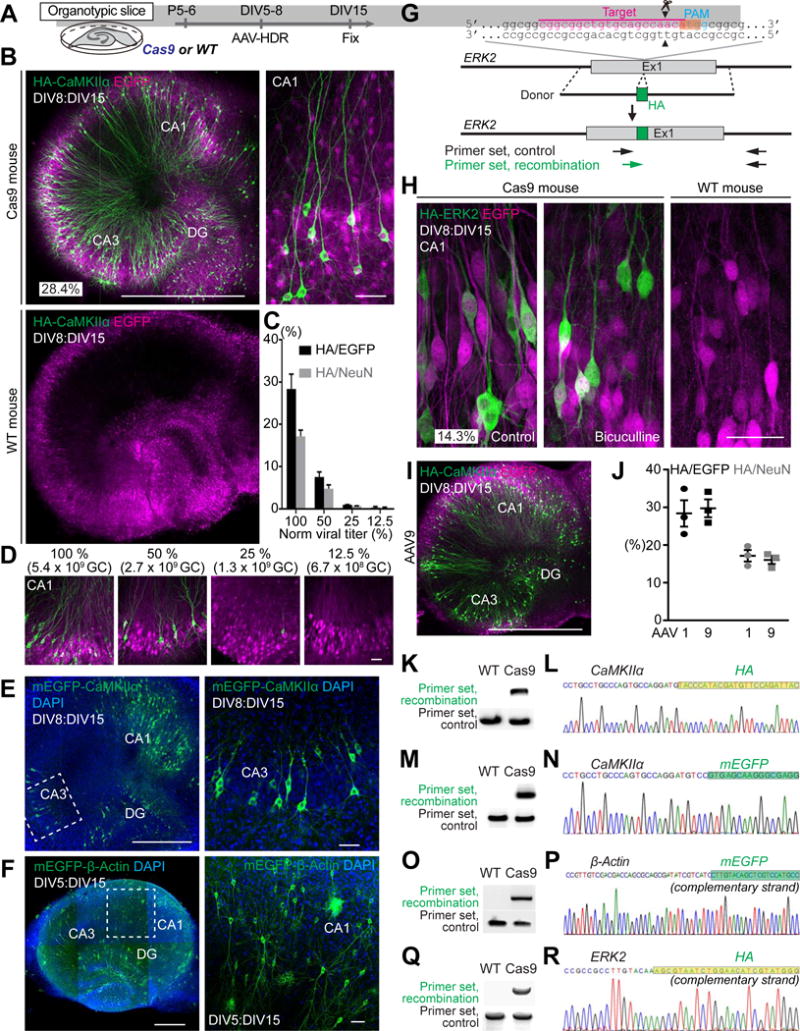
(A) Schematic illustration of experiments.
(B) Confocal microscopic images of organotypic hippocampal slice cultures of Cas9 (top) and wild type (bottom) mice transduced with AAV-HDR for HA-CaMKIIα using serotype 1, showing the EGFP fluorescence (magenta) and immunoreactivity for the HA tag (green) fused to the N-terminus of endogenous CaMKIIα.
(C and D) The effect of the copy number of AAV-HDR on HA-CaMKIIα knockin efficiency. Black and gray bars, knockin efficiency in infected cells or all neurons in CA1, respectively (the ratio of the number of HA-positive cells to that of EGFP-positive or NeuN-positive cells, respectively; 100%, n (cell) = 497 (EGFP)/823 (NeuN); 50%, n = 345/553; 25%, n = 403/602; 12.5%, n = 271/477; 3 slices for each). Representative images of slice cultures transduced with different copy number of AAV-HDR for HA-CaMKIIα are shown (D).
(E and F) Confocal microscopic images of organotypic hippocampal cultures prepared from Cas9 mice showing DAPI signal (blue) and mEGFP fluorescence (green) fused to the N-terminus of endogenous CaMKIIα (E) and β-Actin (F). Enlarged images of the white dotted squares in CA3 (E, left) and CA1 (F, left) were shown (E, right; F, right).
(G) Graphical representation of the mouse genomic loci of ERK2 showing the target sites for Cas9. The sgRNA targeting sequences are labeled in magenta. The PAM sequence is labeled in blue. The start codons of ERK2 is marked in orange. The Cas9 cleavage sites are indicated by the black arrowheads. PCR primer sets (control and recombination) for PCR genotyping (Q) are indicated by the arrows.
(H) Confocal microscopic images of organotypic hippocampal slice cultures of Cas9 (left and middle) and wild type (right) mice showing the EGFP fluorescence (magenta) and immunoreactivity for the HA tag (green) fused to the N-terminus of endogenous ERK2. The representative image of slice treated with 50 μM bicuculline for 1 h is shown (middle).
(I) Confocal microscopic images of organotypic hippocampal slice cultures of Cas9 mice transduced with AAV-HDR for HA-CaMKIIα using serotype 9 showing the EGFP fluorescence (magenta) and immunoreactivity for the HA-CaMKIIα (green).
(J) The effect of AAV serotypes on HA-CaMKIIα knockin efficiency. Black and gray squares, the knockin efficiency of AAV9-based AAV-HDR in infected cells or all neurons in CA1, respectively (the ratio of the number of HA-positive cells to that of EGFP-positive or NeuNpositive cells, respectively, n (cell) = 330 (EGFP)/604 (NeuN)/3 slices). Black and gray circles, the knockin efficiency of AAV1-based AAV-HDR (corresponding to 100% in Figure 2C) is also shown for comparison. The copy number of AAV9-based AAV-HDR is same as that of AAV1-based AAV-HDR (5.4×109 GC).
(K, M, O, and Q) PCR genotyping using genomic DNA extracted from the AAV-transduced slices of wild type and Cas9 mice. Recombination and control primer sets for HA-CaMKIIα (K), mEGFP-CaMKIIα (M), mEGFP-β-Actin (O), HA-ERK2 (Q) were used for PCR.
(L, N, P, and R) DNA sequencing analysis of the PCR products for HA-CaMKIIα (L), mEGFP-CaMKIIα (N), mEGFP-β-Actin (P), and HA-ERK2 (R). The HA or mEGFP tag sequences are marked in light green and green, respectively.
Data are represented as mean ± SEM. Scale bars, 50 μm (B; right, D, E; right, F; right, H); 1 mm (B; left, I); 500 μm (E; left, F; left).
See also Figures S5A–S5E and Table S2 and S3.
To further confirm the applicability of vSLENDR to genome editing in postmitotic cells for other genes/loci, we targeted a gene encoding a ubiquitous signaling protein ERK2. We designed and applied AAV-HDR to insert the HA sequence into the N-terminus of ERK2 in organotypic hippocampal cultures from Cas9 mice (Figure 2G). A number of HA-ERK2 positive cells were observed both in neurons and glial cells in the entire hippocampal slice in Cas9 mice, while only background level fluorescence was observed in wild type mice (Figure 2H). The number of HA-ERK2 positive neurons was ~15% among EGFP positive cells (CA1, HA/EGFP: 14.3% ± 1.9%, n = 343 cells/3 slices; HA/NeuN: 9.5% ± 1.7%, n = 532 cells/3 slices). Notably, HA-ERK2 signals were mainly found in the cytosol but accumulated into the nucleus with bicuculline treatment (Figure 2H, left and middle), consistent with previous studies showing the activity-dependent nuclear translocation of ERK2 (Zhai et al., 2013). In addition, when we injected AAV-HDR for HA-ERK2 into the lateral ventricle of P1 pups of Cas9 mice, HA-ERK2 positive neurons and astrocytes were observed in the hippocampus and cortex (Figure S4H).
While all experiments above have been performed with AAV1, AAV serotype 9 (AAV9) has been also widely used for infecting brain cells (Aschauer et al., 2013). Thus, we prepared AAV-HDR using AAV9 for HA-CaMKIIα and compared its HDR efficiency with AAV1-based AAV-HDR. We found that the efficiency of AAV9-based AAV-HDR was comparable to that of AAV1-based AAV-HDR in neurons in organotypic cultured slices (Figures 2I and 2J, AAV9, CA1, HA/EGFP: 29.7% ± 2.4%, n = 330 cells/3 slices; HA/NeuN: 16.0% ± 1.1%, n = 604 cells/3 slices). All the experiments hereafter have been performed using AAV1-based AAV-HDR.
While we designed all constructs so that the DSBs are made in the non-coding region to minimize the effects of on-target indel formation (Mikuni et al., 2016), indel-mediated knockout may occur, especially in cells that did not undergo HDR. Thus, we estimated the frequency of indel-mediated knockout in the AAV-based system, by immunostaining using the specific antibody against endogenous CaMKIIα in organotypic hippocampal slice cultures infected with AAV-HDR for CaMKIIα (Figure S5D). We found that endogenous CaMKIIα was not deleted in 98.1% of infected neurons (Figure S5E), suggesting that our strategy rarely causes knockout of the target gene. We also confirmed that the targeted HA and mEGFP sequence are inserted in the genome as designed by PCR amplification of the targeted locus of CaMKIIα, β-Actin, or ERK2, followed by DNA sequencing analysis of the amplified DNA fragments (Figures 2K–2R). Taken together, these results demonstrate that AAV-mediated HDR enables precise, targeted sequence insertion in postmitotic neurons in organotypic slice cultures and could be broadly applied to other genomic targets, although the efficiency could vary depending on the target gene.
Since the HDR efficiency in organotypic slices was a steep function of the dose of AAV-HDR, we next measured the dose-dependency of the HDR efficiency in mitotic and postmitotic cells in dissociated hippocampal culture, where the concentration of AAV-HDR can be controlled more quantitatively. We prepared dissociated hippocampal cultures from Cas9 and wild type mice at E17, and applied AAV-HDR for mEGFP-β-Actin or mEGFP-CaMKIIα together with AAV encoding mCherry (AAV-mCherry) as infection marker into the culture media at DIV10 (Figure 3A). Four days after infection, cultures were fixed and subjected to immunocytochemistry using postmitotic neuronal marker NeuN antibody. When a high dose of AAV-HDR was used (2–4×1010 GC/ml), mEGFP-β-Actin fluorescence was observed in both neurons and non-neuronal cells (NeuN-positive and negative cells), while mEGFP-CaMKIIα was observed only in NeuN-positive neurons (Figures 3B–3F). Since most neurons in hippocampal culture at the date of infection (DIV10) are postmitotic (Barnes and Polleux, 2009), these results indicate that HDR-mediated sequence insertion occurs in postmitotic neurons. When we applied different concentrations of AAV-HDR for mEGFP-β-Actin in cultures from Cas9 mice and quantified the number of mEGFP positive cells, the efficiency of mEGFP integration showed dose dependency both in neurons and non-neuronal cells (Figures 3C–3F): at a lower dose (~2×107 GC/ml), most of mEGFP-β-Actin positive cells were non-neuronal (NeuN-negative). However, at a concentration higher than ~2×109 GC/ml, mEGFP-β-Actin was observed also in neurons (NeuN-positive). These results suggested that HDR in postmitotic neurons requires ~100-fold higher concentration of AAV-HDR compared to that in mitotic cells in this system.
Figure 3. HDR-Mediated Genome Editing in Postmitotic Neurons in Primary Dissociated Cultures.
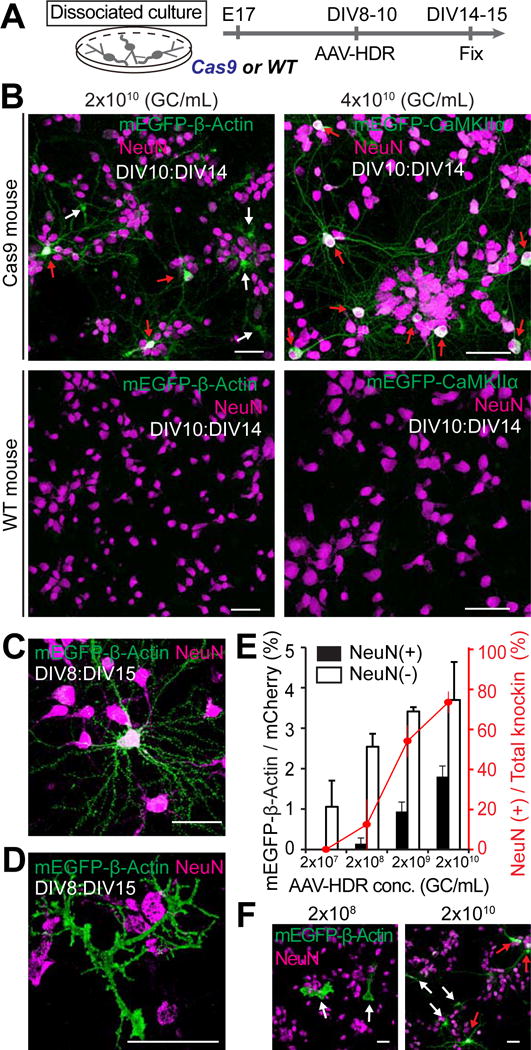
(A) Schematic illustration of experiments.
(B) Confocal microscopic images of dissociated hippocampal cultures prepared from Cas9 mice (top) and wild type mice (bottom) showing immunoreactivity for NeuN (magenta) and the HA tag (green) fused to the N-terminus of endogenous β-Actin (left; AAV-HDR, 2×1010 GC/ml) and CaMKIIα (right; AAV-HDR, 4×1010 GC/ml). Red arrows, NeuN positive neurons; white arrows, NeuN negative cells.
(C and D) Enlarged images of mEGFP-β-Actin positive neuron (C) and astrocyte (D).
(E and F) The effect of AAV-HDR concentration on mEGFP-β-Actin knockin efficiency in neurons and non-neuronal cells. Black and white bars, knockin efficiency of mEGFP-β-Actin in neurons and non-neuronal cells, respectively (the ratio of the number of mEGFP-positive and NeuN-positive or negative cells to that of mCherry-positive and NeuN-positive or negative cells, respectively; 2×1010, n (cell) = 1544/294 (mCherry and NeuN double-positive cells/mCherry-positive, NeuN-negative cells; 2×109, n = 968/203; 2×108, n = 1096/211; 2×107, n = 1276/222 from 4–6 fields of view in each of independent experiments). Red line, the fraction of neurons in total mEGFP-β-Actin knockin cells (the ratio of the number of mEGFP/NeuN double positive cells to that of total mEGFP-positive cells). Representative images of dissociated cultures transduced with AAV-HDR for mEGFP-β-Actin at a concentration of 2×108 and 2×1010 GC/ml are shown (F). Red arrows, NeuN positive neurons; white arrows, NeuN negative cells.
Data are represented as mean ± SEM. Scale bars, 50 μm (B, C, D, and F).
Dual AAV System for HDR-Mediated Genome Editing
To expand vSLENDR to animals other than Cas9 knockin mice, we next sought a system to deliver Cas9 with AAV in addition to AAV-HDR. Because AAV has a limited transgene capacity (up to ~5 kb including ITRs) (Wu et al., 2010), we employed a dual AAV system, one expressing Cas9 and the other expressing a guide RNA and the donor DNA (i.e. AAV-HDR). In order to package the large size of SpCas9 tagged with Myc (~4.2 kb) under the control of a ubiquitous promoter into AAV vectors, we utilized a short form of constitutively acting elongation factor 1α (EFS) promoter (~0.25 kb) and a minimal synthetic polyadenylation signal (48 bases) (AAV-EFS-Cas9 hereafter, ~4.8 kb in total) (Figure 4A) (Gray et al., 2011; Kostic et al., 2003; Swiech et al., 2015). We transduced AAV-EFS-Cas9 together with AAV encoding EGFP into hippocampal dissociated cultures and confirmed nuclear expression of Cas9 in all infected cells by immunocytochemistry using anti-Myc tag antibody (Figure 4B). Next, we transduced AAV-EFS-Cas9 and AAV-HDR for HA-CaMKIIα together with AAV-EGFP into hippocampal dissociated neurons prepared from wild type mice at DIV10 (Figure 4C). HA-CaMKIIα signal was observed in a population of NeuN positive neurons (Figure 4D), indicating that the dual AAV system enables efficient HDR-mediated genome editing in both mitotic cells and postmitotic neurons in vitro.
Figure 4. Dual AAV System for HDR-Mediated Genome Editing In Vitro.
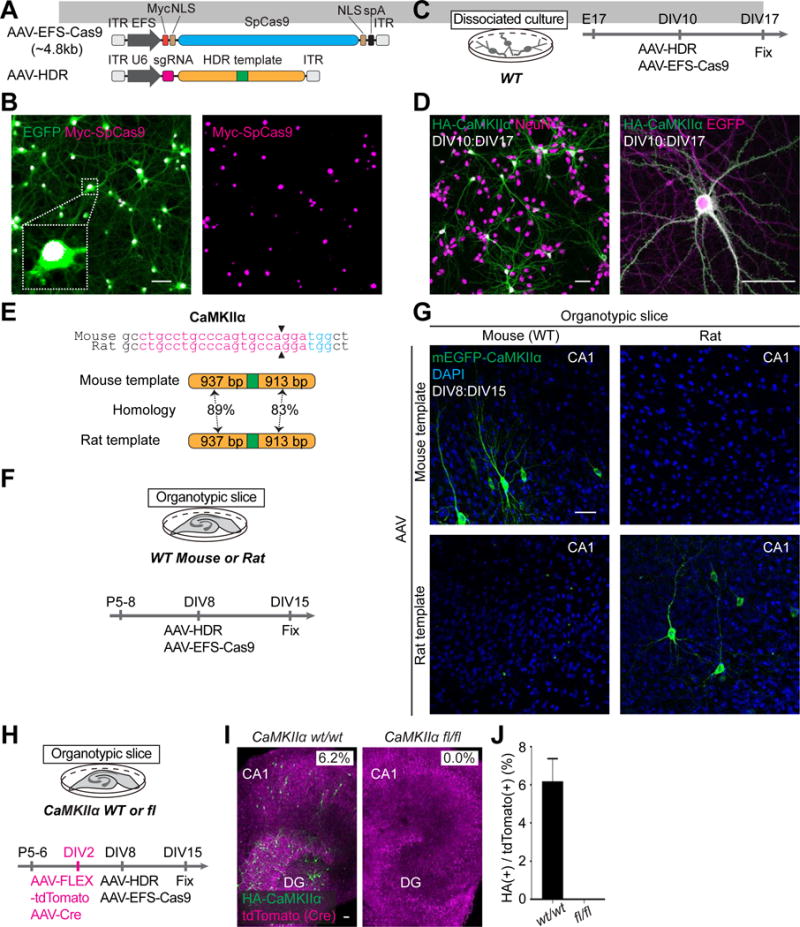
(A) Schematic of AAV-EFS-Cas9 and AAV-HDR. EFS, elongation factor-1 short promoter; Myc; c-Myc tag; NLS, nuclear localization signal; spA, synthetic polyadenylation signal.
(B) Confocal microscopic images of hippocampal dissociated neurons showing the EGFP fluorescence (green) and immunoreactivity for the Myc tag (magenta) fused to the N-terminus of SpCas9.
(C, F, and H) Schematic illustrations of experiments.
(E) Genomic sequence comparisons of mouse and rat CaMKIIα. The sgRNA targeting sequences (magenta), PAM sequences (blue), and Cas9 cleavage sites (black arrowheads) are shown (top). Schematics of mouse and rat HDR template for HA-CaMKIIα (bottom) showing homology arms (orange) and HA (green).
(G) Confocal microscopic images of organotypic hippocampal cultures prepared from wild type mice (left) and rats (right) showing DAPI signal (blue) and mEGFP fluorescence (green) fused to the N-terminus of endogenous CaMKIIα.
(I) Confocal microscopic images of organotypic hippocampal slice cultures prepared from wild type mice (left) or CaMKIIα conditional knockout mice (right) showing the tdTomato fluorescence (magenta) and immunoreactivity for the HA tag (green) fused to the N-terminus of endogenous CaMKIIα.
(J) The fraction of HA-CaMKIIα-positive cells in wild type and CaMKIIα deleted cells (the ratio of the number of HA-CaMKIIα-positive cells to that of tdTomato-positive cells). Wild-type, n = 375 cells, 3 slices; floxed, n = 374 cells, 3 slices.
Data are represented as mean ± SEM. Scale bars, 50 μm (B, D, G, and I).
See also Figures S5F–S5H.
Taking advantage of the flexibility of the dual AAV system which can be used for animals other than mice, we next assessed the specificity of AAV-HDR mediated genome editing using rats and mice. Because mouse and rat share sgRNA targeting sequences of CaMKIIα (100% match) (Figure 4E, top), AAV-HDR for mouse CaMKIIα can make DSBs at specific loci of rat CaMKIIα. However, because the mouse and rat genome has similar but not identical sequences to those of homology arm of AAV-HDR for CaMKIIα (~80–90% similarity) (Figure 4E; bottom, Table S2), HDR should occur only in targeted animals. To test if this is the case, we transduced AAV-HDR for mEGFP-CaMKIIα with AAV-EFS-Cas9 into organotypic slice cultures from wild-type mice or rats (Figure 4F). Consistent with our hypothesis, mEGFP signal was observed in mouse slice cultures, but not in rat slice cultures co-transduced with AAV-HDR for mEGFP-mouse CaMKIIα and AAV-EFS-Cas9 (Figure 4G, top). In contrast, when we transduced AAV-HDR for mEGFP-rat CaMKIIα and AAV-EFS-Cas9, mEGFP signal was observed in rat slice cultures, but not in mouse slice cultures (Figure 4G, bottom). These results demonstrate that AAV-mediated HDR in postmitotic neurons occurs in a homology-sequence dependent manner.
To confirm that the immunostained signal was specifically from the HA tag fused to the N-terminus of CaMKIIα as intended, we utilized a conditional knockout mouse, in which exon 2 of CaMKIIα can be deleted by the expression of Cre recombinase (Hinds et al., 2003). We prepared hippocampal organotypic slice cultures from wild type or conditional CaMKIIα knockout mice and transduced AAV encoding Cre recombinase (AAV-Cre) together with AAV which conditionally expressed tdTomato as a Cre reporter (AAV-FLEX-tdTomato) at DIV2. We then applied AAV-EFS-Cas9 and AAV-HDR for HA-CaMKIIα onto cultures at DIV8 and performed immunohistochemistry using anti-HA antibody at DIV15 (Figures 4H and S5F). The strong HA signal was observed in a significant number of wild type CA1 and DG neurons (CA1, HA/tdTomato: 6.2% ± 1.2%, n = 375 cells/3 slices), but not in tdTomato-positive, CaMKIIα deleted neurons (CA1, n = 374 cells/3 slices) (Figures 4I, 4J, S5G, and S5H). These results indicated that the observed HA signal was specifically from HA-CaMKIIα as intended.
HDR-Mediated Genome Editing in the Adult Brain In Vivo
Next, we asked if vSLENDR can be applied to neurons in the adult brain in vivo. We first applied the single AAV system to the adult mice brain and stereotaxically injected AAV-HDR for HA-CaMKIIα or HA-β-Actin together with AAV-EGFP into the cerebral cortex or hippocampus of adult Cas9 mice at P48–58 (Figures 5A and S6A). The mice were fixed around 3 weeks after injection and their brain sections were subjected to immunohistochemistry using anti-HA or NeuN antibodies. We observed strong HA-CaMKIIα staining in a large number of neurons, but not in glial cells, in the cerebral cortex and hippocampal DG and CA3 of Cas9 mice (Figures 5B–5F). The number of HA-CaMKIIα positive neurons reached at ~30% among infected cells ([cerebral cortex] HA/EGFP: 14.8%, n = 461 cells, HA/NeuN: 5.2%, n = 1310 cells; [DG] HA/EGFP: 28.2%, n = 529 cells, HA/NeuN: 11.3%, n = 1322 cells; [CA3] HA/EGFP: 20.4%, n = 260 cells, HA/NeuN: 14.9%, n = 356 cells). No HA-positive neurons were observed when we injected AAV-HDR for HA-CaMKIIα into the cortex of the wild type mouse (Figure 5C, right). In contrast, HA-β-Actin positive signals were observed both in neurons and glia (Figures S6B–S6D). The number of HA-β-Actin positive neurons was 5–7 % among mEGFP positive cells ([DG] HA/EGFP: 7.0%, n = 469 cells, HA/NeuN: 4.4%, n = 758 cells; [CA1] HA/EGFP: 5.2%, n = 346 cells, HA/NeuN: 4.1%, n = 436 cells).
Figure 5. Single and Dual AAV Systems for HDR-Mediated Genome Editing In Vivo.
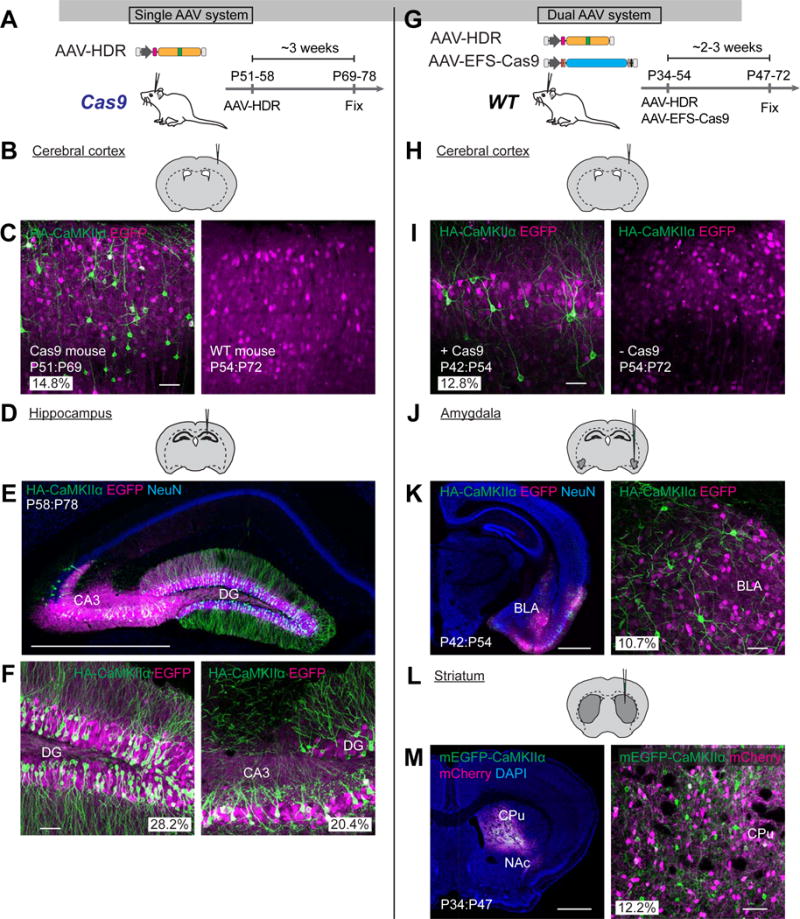
(A and G) Schematic illustrations of experiments.
(B, D, H, J, and L) Schematic illustrations of AAV injection sites.
(C, E, F, I, K, and M) Confocal microscopic images of coronal brain sections of the cerebral cortex (C and I), hippocampus (E and F), amygdala (K), and striatum (M) of Cas9 (C; left, E, and F) or wild type mice (C; right, I, K, and M) showing DAPI signal (blue), the EGFP fluorescence (magenta), and immunoreactivities for NeuN (blue) and the HA tag (green) fused to the N-terminus of endogenous CaMKIIα. Images of the cerebral cortex transduced with AAV-HDR and AAV-EFS-Cas9 (I, left) and AAV-HDR only (I, right) are shown. BLA, basolateral amygdala; CPu, caudate putamen; NAc, nucleus accumbens.
Scale bars, 50 μ,m (C, F, I, K; right, M: right); 1 mm (E, K; left, and M; left).
See also Figure S6.
We next applied the dual AAV system to various brain regions in vivo in 1–2 months old wild type mice (Figure 5G and S6E). We observed HA-CaMKIIα staining in neurons, but not in glia, in injected brain regions, i.e. the cerebral cortex, amygdala, and striatum (Figures 5H–5M). In the cerebral cortex, the knockin efficiency in the dual AAV system was similar to that achieved by the single AAV system in Cas9 mice (dual AAV system, HA/EGFP: 12.8%, n = 344 cells; HA/NeuN: 5.2%, n = 854 cells) (Figures 5C and 5I). No obvious HA signal was observed in a control experiment where we injected only AAV-HDR for HA-CaMKIIα into the cerebral cortex of wild type mice (Figure 5I, right). The efficiency of HDR was similar among different brain regions ([striatum] HA/EGFP: 15.0%, n = 748 cells, HA/NeuN: 7.3%, n = 1524 cells; [amygdala] HA/EGFP: 10.7%, n = 419 cells, HA/NeuN: 5.6%, n = 803 cells) (Figures 5H–5M). Moreover, the efficiency of mEGFP-CaMKIIα knockin was comparable to that of HA-CaMKIIα knockin in the striatum (Figures S6F and S6G, mEGFP/mCherry: 12.2%, n = 368 cells), suggesting that HDR efficiency is relatively independent of the length of the insertion. Taken together, the data demonstrate that the single and dual AAV systems allow for precise sequence insertion with comparable efficiency in both mitotic cells and postmitotic neurons in the adult brain in vivo.
HDR-Mediated Genome Editing in Aged, Disease-Model Mice
Given that the dual AAV system can be used for the brain of adult animals other than Cas9 knockin mice, the dual AAV system would greatly accelerate experiments using the disease mouse models, particularly models of age-related human neurological disorders, by skipping the process of crossing the animals with Cas9 knockin mice. To demonstrate this, we applied the dual AAV system for mEGFP-β-Actin to a mouse model of Alzheimer’s disease expressing a mutant form of the human amyloid protein precursor (J20 mice) (Mucke et al., 2000). Because the dendritic spine pathology has been implicated in Alzheimer’s disease, we aimed to image the structure and number of dendritic spines by the fluorescence of mEGFP fused to endogenous β-Actin (Figure S2H) (Mikuni et al., 2016).
We injected AAV-EFS-Cas9, AAV-HDR for mEGFP-β-Actin, and AAV-mCherry into the hippocampus of J20 mice and their littermate controls at ~1 year old (P354). Two weeks after injection, mice were fixed and subjected to immunohistochemistry using anti-NeuN antibody (Figure 6A). mEGFP-β-Actin fluorescence was observed in the injected side of the CA1 hippocampus in both J20 mice and littermate control (Figures 6B and 6C). The mEGFP-β-Actin fluorescence was highly accumulated in dendritic spines, and thus served as a convenient marker for highlighting spines (Figures 6D and 6E). Consistent with previous studies, we found that spine number was significantly reduced in J20 mice (Figure 6F; p = 0.01, Student’s t test) (Pozueta et al., 2013). These results showed that the dual AAV system can be applied to brains of aged mice with various genetic backgrounds.
Figure 6. HDR-Mediated Genome Editing in Aged, Disease-Model Mice.
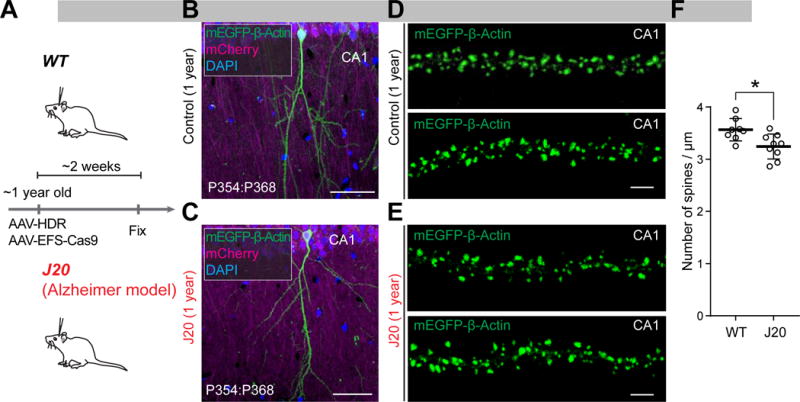
(A) Schematic illustration of experiments.
(B–E) Confocal microscopic images of coronal brain sections of the CA1 hippocampus of wild type (B and D) and littermate J20 (C and E) mice at 1 year old showing the DAPI signal (blue), mCherry fluorescence (magenta), and mEGFP fluorescence (green) fused to the N-terminus of endogenous β-Actin. Representative images of the apical secondary dendrites of CA1 pyramidal neurons of wild type (D) and J20 (E) mice are shown.
(F) The averaged density of spines on apical secondary dendrites of wild type (n = 946 spines/8 neurons) and J20 (n = 1001/9) mice. *p < 0.05, Student’s t test.
Data are represented as mean ± SEM. Scale bars, 50 μm (B and C); 5 μm (D and E).
DISCUSSION
In this study, we have developed vSLENDR, which allows HDR-mediated precise genome editing both in mitotic and postmitotic cells in the brain through the use of an AAV-mediated delivery system. Through this approach, we achieved highly efficient sequence insertion in primary dissociated and organotypic slice cultures in vitro and in various brain areas and cell-types in developing and adult brains in vivo. Previously, precise genome editing has been primarily limited to dividing cells, due to low HDR activity in non-dividing cells (Chapman et al., 2012; Cox et al., 2015; Heidenreich and Zhang, 2016; Orthwein et al., 2015; Panier and Durocher, 2013; Rothkamm et al., 2003). This poses a significant challenge especially for the in vivo application of HDR, since most adult tissue is composed of non-dividing cells. In the current study, we overcome this problem by using AAV to effectively deliver genome editing machinery in target cells.
The exact mechanism by which AAV enables highly efficient HDR remains elusive. AAV has been reported to allow gene targeting by conventional homologous recombination mechanisms (Deyle and Russell, 2009). Importantly, AAV-mediated gene targeting frequencies are much higher than conventional transfection or electroporation approaches, presumably due to the efficient nuclear delivery of vector genomes, the single-strand nature of the genome and the effect of ITRs (Barzel et al., 2015; Gaj et al., 2016; Russell and Hirata, 1998). Although the efficiency of AAV-mediated gene targeting by itself is too low for most of somatic tissue in vivo, especially for postmitotic cells, the induction of DSBs by endonuclease can increase the frequency of AAV-mediated gene targeting by several orders of magnitudes (Dever et al., 2016; Gaj et al., 2016; Porteus et al., 2003). This increased recombination efficiency by AAV likely enabled efficient HDR-mediated genome editing in postmitotic neurons in the adult brain in our study. Since HDR in postmitotic neurons requires much higher concentration of AAV-HDR than what is required for mitotic cells, it appears that a large copy number of template DNA is necessary for the induction of HDR in postmitotic neurons. Indeed, we achieved efficient HDR-mediated genome editing in dividing neuronal progenitors by intraventricular administration, thereby perhaps lower concentration than that of local injection in the adult brain, of AAV-HDR.
The unprecedentedly high HDR efficiency (up to ~15–30%) in postmitotic cells in vitro and in vivo for multiple genes (CaMKIIα, β-Actin, and ERK2) strongly suggests the broad applicability of vSLENDR for many other genes/loci. However, it should be noted that the HDR efficiency could be different from target to target. Indeed, recent studies have shown that the HDR efficiency is affected by many factors such as sequences in and around the target, sgRNA design, the distance between the insertion site and DSBs, the size of the insert, the length of the homology arms, and the activity of the endogenous repair systems (Lin et al., 2014; Moreno-Mateos et al., 2015; Paquet et al., 2016; Xu et al., 2015).
Because the efficiency of HDR is substantially lower than that of NHEJ in many systems, alternative strategies have been developed for targeted sequence insertion via NHEJ using engineered nucleases, where the exogenous DNA fragment is directly ligated at DSBs (Auer et al., 2014; Maresca et al., 2013; Suzuki et al., 2016). In the current study, we achieved ~15% in vivo knockin efficiency in the cerebral cortex both by the single AAV system in Cas9 mice and by the dual AAV system in wild type mice. These numbers are comparable to the reported efficiency of sequence insertion in postmitotic neurons using the NHEJ-mediated strategy (Suzuki et al., 2016). In addition, HDR-based genome editing is advantageous over NHEJ-based techniques as it enables precise introduction of any desired changes, such as point mutations and deletions, incorporated in the template DNA at the specific genomic locus homologous to the template. Furthermore, HDR allows for simultaneous genome editing at two different target genomic loci (Mikuni et al., 2016).
We show that the gene-editing efficiency achieved by the dual AAV system is comparable to that achieved by the single AAV system in the cerebral cortex in vivo (12.8% vs 14.8%, Figures 5C and 5I). Analogously, the editing efficiency was similar between these delivery systems in organotypic hippocampal slice cultures in vitro (CA1, 7.6% vs 6.2%; Figures 2C and 4J). On the other hand, in the embryonic brain, HDR occurred more efficiently in the single AAV system than in the dual AAV system (Figure S3). Thus, our results indicate that the dual AAV system allows us to perform highly efficient HDR-based genome editing in postmitotic cells (but not in embryonic cells) as well as providing us with much higher flexibility. The flexibility of the dual AAV system has been highlighted by our results showing that efficient HDR-based genome-editing is possible in organotypic slices in rats (Figure 4G) and in a mouse model of Alzheimer’s disease at ~1 year old (Figure 6). Cas9-knockin rats have not been developed yet and it would take a long time to breed the disease model crossed with Cas9 knock-in mice. In both cases, the dual AAV system would be the favorable solution.
We confirmed HA or mEGFP sequences were precisely inserted as intended at the N-terminus of β-Actin, CaMKIIα or ERK2 by DNA sequencing following PCR amplification of the targeted locus. To minimize the effects of on-target indel formation, we designed all constructs so that the DSBs are made in the non-coding region. Previously, using the similar strategy, we showed that the target protein was not deleted in 98.7% of the transfected neurons and the expression level was not changed (Mikuni et al., 2016). In this study, we showed that endogenous CaMKIIα was not deleted in 98.1% of the neurons infected with AAV-HDR for HA-CaMKIIα (Figure S5E). However, potential effects of NHEJ have to be carefully evaluated depending on the purpose of experiments.
Another potential caveat of vSLENDR is that CRISPR-Cas9 may potentially generate DSBs at unwanted off-target sites. However, sequence insertion is unlikely to occur at those sites since HDR is highly specific to the sequence of homology arms of the template DNA. Indeed, no HA-labeled cells were observed by immunohistochemistry when we knocked out the targeted gene. In addition, the subcellular localization pattern of tagged β-Actin, CaMKIIα, and ERK2 was always consistent with that reported previously. Furthermore, consistent with the endogenous gene regulation of β-Actin, CaMKIIα, and ERK2, tagged β-Actin or ERK2 was ubiquitously observed in both neurons and glia in the entire brain, while tagged CaMKIIα was specifically found in neurons in limited brain regions. The specificity of this technique could be further enhanced by using novel Cas9 variants with minimum or no off-target effects while retaining comparable on-target cleavage activity (Kleinstiver et al., 2016; Slaymaker et al., 2016).
We have demonstrated vSLENDR is applicable to virtually any brain area, cell-type, and age of interest. Recently, we and other groups reported techniques for site-specific sequence insertion for single-cell labeling of endogenous proteins via CRISPR-Cas9 mediated HDR by introducing HDR machinery to dividing neuronal progenitors in the embryonic brain with IUE (Mikuni et al., 2016; Uemura et al., 2016). However, because IUE-based techniques can be applied to limited area and age of the embryonic brain, it is difficult to target deep or broad areas in the brain. When applied to the embryo, AAV-based HDR can edit the genome in widespread regions of the brain including deep brain structures. Thus, systematic analysis of protein localization may be possible by combining this approach with whole-brain clearing and imaging method (Chung et al., 2013; Hama et al., 2011; Ke et al., 2016; Susaki et al., 2014). Precise genome editing in specific brain region is possible by injecting AAV encoding HDR machinery to specific brain regions in adult in vivo. Therefore, the production of Cre dependent AAV expressing Cas9 would enable cell type specific genome editing by combining with the hundreds of different Cre-driver lines (Gerfen et al., 2013; Taniguchi et al., 2011). Furthermore, the dual AAV system described here might be directly adapted to other mammalian species, opening the possibility of precise genome engineering in a variety of organisms, including non-human primates, in which genetic tools are limited. Thus, virus-mediated precise genome editing in the brain will provide a versatile tool for both basic and translational neuroscience.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-HA (C29F4) | Cell Signaling Technology | Cat# 3724S; RRID:AB 1549585 |
| Mouse anti-HA (16B12) | BioLegend | Cat# 901514; RRID:AB 2565336 |
| Chicken anti-GFP | EMD Millipore | Cat# AB16901; RRID:AB 90890 |
| Guinea pig anti-NeuN | EMD Millipore | Cat# ABN90P; RRID:AB_2341095 |
| Rabbit anti-Calbindin | EMD Millipore | Cat# AB1778; RRID:AB 2068336 |
| Chicken anti-Tyrosine hydroxylase | Aves labs | Cat# TYH; RRID:AB 10013440 |
| Guinea pig anti-Parvalbumin | Frontier Institute | Cat# PV-GP; RRID:AB 2571615 |
| Rabbit anti-Myc (71D10) | Cell Signaling Technology | Cat# 2278; RRID:AB 490778 |
| Mouse anti-CaMKIIα (6G9) | Cell Signaling Technology | Cat# 50049 |
| Bacterial and Virus Strains | ||
| AAV-HDR (HA-β-Actin, AAV1) | Penn Vector Core at the University of Pennsylvania | N/A |
| AAV-HDR (HA-CaMKIIα, AAV1) | Penn Vector Core at the University of Pennsylvania | N/A |
| AAV-HDR (HA-CaMKIIα, AAV9) | Vigene Biosciences | N/A |
| AAV-HDR (HA-rat-CaMKIIα, AAV1) | Vigene Biosciences | N/A |
| AAV-HDR (mEGFP-β-Actin, AAV1) | Penn Vector Core at the University of Pennsylvania | N/A |
| AAV-HDR (mEGFP-CaMKIIα, AAV1) | Penn Vector Core at the University of Pennsylvania | N/A |
| AAV-HDR (HA-ERK2, AAV1) | Vigene Biosciences | N/A |
| AAV-EFS-Cas9 (AAV1) | Penn Vector Core at the University of Pennsylvania | N/A |
| AAV1.CB7.CI.eGFP.WPRE.rBG | Penn Vector Core at the University of Pennsylvania | AV-1-PV1963 |
| AAV1.CB7.CI.mCherry.WPRE.rBG | Penn Vector Core at the University of Pennsylvania | AV-1-PV1969 |
| AAV1.CAG.Flex.tdTomato.WPRE.bGH | Penn Vector Core at the University of Pennsylvania | AV-1-ALL864 |
| AAV9.CamKII0.4.Cre.SV40 | Penn Vector Core at the University of Pennsylvania | AV-9-PV2396 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trypsin | Lonza | Cat# 17-160E |
| DNase I | Roche Applied Science | Cat# 10104159001 |
| Poly-L-lysine hydrobromide | Sigma Aldrich | Cat# P9155 |
| B-27 supplement | GIBCO | Cat# 17504044 |
| GlutaMAX supplement | GIBCO | Cat# 35050061 |
| Fast Green FCF | Sigma Aldrich | Cat# F7252 |
| Bicuculline | Tocris | Cat# 2503 |
| 20% Mannitol | Neogen | Cat# 09061 |
| Critical Commercial Assays | ||
| Phusion Hot Start II DNA Polymerase | Thermo Fisher | Cat# F549S |
| DNeasy Blood & Tissue Kit | QIAGEN | Cat# 69504 |
| QiaQuick Gel Extraction Kit | QIAGEN | Cat# 28704 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Swiss Webster | Charles River Laboratories | Strain Code: 024 |
| Mouse: C57BL/6J | Charles River Laboratories | Strain Code: 027 |
| Mouse: Igs2tm1.1(CAG-cas9*)Mmw/J (also known as H11Cas9 mice) | The Jackson Laboratory | JAX: 027650 |
| Mouse: C57BL/6-Camk2atm1Vyb/J | The Jackson Laboratory | JAX: 006575 |
| Mouse: B6.Cg-Tg(PDGFB-APPSwInd)20Lms/2J (also known as J20 line) | The Jackson Laboratory | JAX: 006293 |
| CD IGS Rat | Charles River Laboratories | Strain Code: 001 |
| Oligonucleotides | ||
| HDR template DNA sequences, see Table S2 | This study | N/A |
| sgRNA target sequences, see Table S3 | This study or Mikuni et al., 2016 | N/A |
| Genomic PCR primers, see Table S3 | This study or Mikuni et al., 2016 | N/A |
| DNA sequencing primers, see Table S3 | This study or Mikuni et al., 2016 | N/A |
| Recombinant DNA | ||
| p×551 | Swiech et al., 2015 | Addgene Plasmid #60957 |
| lentiCRISPR v2 | Sanjana et al., 2014 | Addgene Plasmid #52961 |
| p×330-β-Actin | Mikuni et al., 2016 | N/A |
| p×330-CaMKIIα | Mikuni et al., 2016 | N/A |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| ImageJ | NIH | https://imagei.nih.gov/ij/ |
| Adobe Photoshop CS6 | Adobe | https://www.adobe.com/products/photoshop.html?promoid=KLXLS |
| Zen software | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| Other | ||
| Millicell Cell Culture Insert, 30 mm, hydrophilic PTFE, 0.4 μm | EMD Millipore | Cat# PICM0RG50 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ryohei Yasuda (Ryohei.Yasuda@mpfi.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice and Rats
All the experiments were approved by the Max Planck Florida Institute for Neuroscience Institutional Animal Care and Use Committee and were performed in accordance with guidelines from the US National Institutes of Health. The mice (Swiss Webster, C57BL/6J, H11Cas9, C57BL/6-Camk2atm1Vyb/J, and J20 line) and rats (CD IGS) were purchased from Charles River Laboratories or The Jackson Laboratory and bred in the animal facility of the Max Planck Florida Institute for Neuroscience. All mice used in experiments were maintained under a 12 hr light/dark cycle. The day on which the vaginal plug was detected was designated as embryonic day 0 (E0). The first 24 h after birth was referred to as postnatal day 0 (P0). Fetal mice of Swiss Webster female or homozygous H11Cas9 female mated with Swiss Webster male at E10–16 were used for experiments of in utero intraventricular administration of AAV. Fetal mice of Swiss Webster female and homozygous H11Cas9 female mated with Swiss Webster male at E17 were used for primary dissociated cultures. P5–P6 pups of C57BL/6J, heterozygous H11Cas9, and homozygous C57BL/6-Camk2atm1Vyb/J mice and P7 pups of CD IGS rat were used for organotypic slice cultures. P48–58 heterozygous H11Cas9, P42–54 Swiss Webster mice, and P354 J20 line and their littermates were used for in vivo stereotaxic injection of AAV. Both of male and female mice were used and randomly assigned to experimental groups. For experiments using J20 line and their littermates, only male mice were used.
Primary dissociated cultures
Hippocampal primary dissociated cultures were prepared according to the standard procedures. Briefly, hippocampi dissected from both male and female mice at E17 were treated with 0.25% trypsin (Lonza) and 0.05 mg/ml DNasel (Roche Applied Science) in Hanks’ Balanced Salt Solution (GIBCO) at 37 °C for 20 min. The dissociated hippocampal neurons were plated on poly-L-lysine (Sigma Aldrich)-coated 12 mm glass coverslips in 24-well dish and maintained in neurobasal medium (GIBCO) supplemented with B-27 (GIBCO), GlutaMAX (GIBCO), and penicillin/streptomycin (GIBCO) at 37°C in a 5% CO2 humidified atmosphere. The half of the culture medium was exchanged with fresh medium every week.
Organotypic slice cultures
Organotypic cortical slice cultures were prepared according to the standard procedures (Stoppini et al., 1991). In brief, coronal hippocampal slices of 350 μm thickness were dissected from both male and female mice at P5–6 or rats at P7 using a McIlwain tissue chopper (Ted Pella, Inc). The slices were plated on a plated on hydrophilic PTFE membranes (Millicell, Millipore) and maintained in culture medium containing MEM medium (GIBCO), 20% horse serum (GIBCO), 1 mM L-glutamine, 1 mM CaCl2, 2 mM MgSO4, 12.9 mM D-Glucose, 5.2 mM NaHCO3, 30 mM Hepes, 0.075% Ascorbic acid, 1 μg/ml Insulin at 37 °C in a 5% CO2 humidified atmosphere. The culture medium was exchanged with fresh medium every other day.
METHOD DETAILS
DNA constructs
To generate AAV-HDR for HA or mEGFP-β-Actin/CaMKIIα/ERK2, we first constructed targeting vectors containing HDR donor template DNAs. The ~2 kb homology sequence flanking the each side of the target site for β-Actin/CaMKIIα/ERK2 was synthesized and subcloned into the pUC57 vector (Genewiz). HA or mEGFP sequence was then inserted into the target site of the homology sequence of β-Actin/CaMKIIα/ERK2. The resulting HDR template DNA was cloned into the pAAV backbone from PX551 gifted from F. Zhang (Addgene Plasmid #60957) (Swiech et al., 2015), yielding the HDR donor template for HA and/or mEGFP-β-Actin/CaMKIIα/ERK2 (Table S2). To incorporate the sgRNA expression cassette into the targeting vectors, pX330-β-Actin/CaMKIIα/ERK2 which contains human U6 promoter and sgRNA specific for β-Actin/CaMKIIα/ERK2 (sgRNA target sequence: β-Actin, tgtgtcttgatagttcgcca; CaMKIIα, ctgcctgcccagtgccagga, ERK2: cggcggctgtgcagccaaca) was used as a PCR template. The sgRNA expression cassettes were amplified with Phusion DNA polymerase (Thermo Scientific) and cloned into the targeting vectors for HA- and/or mEGFP-β-Actin/CaMKIIα/ERK2, yielding AAV-HDR for HA-β-Actin/CaMKIIα/ERK2 and mEGFP-β-Actin/CaMKIIα. To generate an AAV vector to express the human codon-optimized SpCas9 under the control of a ubiquitous promoter, we utilized the EFS promoter and synthetic polyadenylation signal (Gray et al., 2011; Kostic et al., 2003; Swiech et al., 2015). lentiCRISPR v2 construct was a gift from F. Zhang (Addgene Plasmid #52961) (Sanjana et al., 2014) and used as a PCR template for EFS promoter. Myc tag was added by PCR to the N-terminus of SpCas9 using pX551 as a template. PCR amplified Myc-SpCas9 fragment was then joined with EFS promoter by overlap extension PCR. The resulting EFS-Myc-SpCas9 was cloned into the pAAV backbone from pX551, yielding AAV-EFS-Cas9. All obtained constructs were sequenced and verified.
Genomic PCR and DNA sequencing
To isolate genomic DNA from the AAV-transduced brains or organotypic slices, DNeasy Blood & Tissue Kit (Qiagen) was used according to the manufacturer’s instruction. Using the extracted DNA as a template, we performed genomic PCR using the following primers: HA-F, 5′-cccatacgatgttccagatt-3′; HA-R, 5′-gcgtaatctggaacatcgtatg-3′; GFP-F, 5′-catggtcctgctggagttcgtg-3′; GFP-R, 5′-gctgaacttgtggccgtttac-3′; β-Actin-F, 5′-ttcacctgccctgagtgtttc-3′; β-Actin-R, 3′-gggagagcatagccctcgta-5′; CaMKIIα-F, 5′-agcacaagcagaaactggga-3′; CaMKIIα-R, 5′-tgaatcgggtgcaggtgatg-3′; ERK2-F, 5′-tgtgttgtcctccttcctcg-3′; ERK2-R, 5′-tcgccctgtaaaaggacttc -3′. The PCR product was purified by QiaQuick gel extraction kit (Qiagen) and then proceeded to DNA sequencing using the following primers: β-Actin-S, 5′-gaacagccttcttagcaccg-3′; CaMKIIα-S, 5′-agccctagttcccagcctaa-3′; ERK2-S, 5′-caggagtgaaggccaagaag-3′.
AAV vector production
All custom AAV vectors were produced by the Penn Vector Core at the University of Pennsylvania or Vigene Biosciences. The titers of AAV-HDR pseudotyped with serotype 1 were 1.8×1013 (HA-β-Actin), 1.1×1013 (HA-mouse-CaMKIIα), 1.3×1013 (HA-rat-CaMKIIα), 1.6×1013 (HA-ERK2), 1.1×1013 (mEGFP-β-Actin), 2.1×1013 (mEGFP-CaMKIIα), and 3.5×1012 (AAV-EFS-Cas9) GC/ml. The titer of AAV-HDR pseudotyped with serotype 9 was 6.5×1013 (HA-CaMKIIα) GC/ml. All premade AAV vectors were purchased from the Penn Vector Core at the University of Pennsylvania. The viral vectors used, with their abbreviated names as used in the text, are as follows: AAV1.CB7.CI.eGFP.WPRE.rBG (AAV-EGFP), AAV1.CB7.CI.mCherry.WPRE.rBG (AAV-mCherry), AAV1.CAG.Flex.tdTomato.WPRE.bGH (AAV-FLEX-tdTomato), and AAV9.CamKII0.4.Cre.SV40 (AAV-Cre).
Viral transduction into cultures in vitro
For viral transduction into hippocampal organotypic slice cultures, AAV-HDR and/or AAV-EFS-Cas9 and/or AAV-EGFP were directly added onto cultures at DIV5–8. The applied amount of AAV-HDR and AAV-EFS were as follows (per slice): AAV-HDR for HA-CaMKIIα, 5.4×109 GC; AAV-HDR for mEGFP-CaMKIIα, 1.0×1010 GC; AAV-HDR for mEGFP-β-Actin, 5.6×109 GC. AAV-ESF-Cas9, 0.9×109 GC. AAV-HDR for HA-ERK2, 8.0×109 GC; In some experiments, smaller amount of AAV-HDR was used. Cultures transduced with AAV were analyzed at indicated time points.
For viral transduction into hippocampal dissociated cultures, AAV-HDR and/or AAV-EFS-Cas9 together with AAV-mCherry or AAV-EGFP was directly added to culture media at DIV8-10. The applied amount of AAV-HDR and AAV-EFS were as follows (per well in 24-well plate): AAV-HDR for mEGFP-CaMKIIα, 2.1×1010 GC; AAV-HDR for mEGFP-β-Actin, 1.1×1010 GC; AAV-HDR for HA-CaMKIIα, 1.1×1010 GC; AAV-EFS-Cas9, 3.5×109 GC. In some experiments, smaller amount of AAV-HDR was used.
Intraventricular viral injection
For viral transduction of the embryonic brain in vivo, pregnant mice were deeply anesthetized with isoflurane (Piramal Healthcare; 5% for induction, 2–2.5% for maintenance in O2 at 1.5 L/min). Buprenorphine-SR (0.1 mg/mouse, ZooPharm) was subcutaneously administered for analgesia and laparotomy was performed to expose the uterus. For single AAV system, a mixture of AAV-HDR for HA-β-Actin (or HA-CaMKIIα or HA-ERK2) and AAV-EGFP or AAV-HDR for mEGFP-β-Actin (or mEGFP-CaMKIIα) alone or/and AAV-mCherry (3:1 ratio) was prepared. For dual AAV system, a mixture of AAV-ESF-Cas9, AAV-HDR for HA-β-Actin (or HA-CaMKIIα), and AAV-EGFP (3:3:1 ratio) was prepared. AAV was mixed with Fast Green (0.1 mg/ml, Sigma-Aldrich) to visualize the injection site. An aliquot of 1 μl of AAV mixture was unilaterally administrated into the lateral ventricle of the embryonic brain using a glass micropipette. After the injection, the uterus was repositioned in the abdominal cavity, the abdominal wall and skin were closed with sutures, and the mice were monitored during recovery in a warm cage. For viral transduction of neonatal brain in vivo, P0 or P1 pups were anaesthetized on ice for 2–4 min. An aliquot of 1–2 μl of AAV mixture was unilaterally administrated into the lateral ventricle of the neonatal brain using a glass micropipette. The pups were placed on a heat blanket for several minutes and then returned to their mother.
Stereotaxic viral injection
Adult mice were anesthetized with isoflurane (5% for induction, 2–2.5% for maintenance in O2 at 1.5 L/min). The mice were given an intraperitoneal injection of 500 μl of 20% mannitol (Neogen) for brain decompression and subcutaneous injection of 0.05 mg of buprenorphine-SR for analgesia. Body temperature was maintained at 37°C using a homeothermic blanket. The mice were placed in a stereotaxic frame (David Kopf Instruments). The skull was then exposed and the head holding angles were adjusted to achieve the flat skull position for stereotaxic alignment. A ~1 mm in diameter hole in the skull was made in the targeted area with dental drill. The following stereotaxic coordinates were used: Hippocampus, anteroposterior (AP), −2.0 mm from bregma; mediolateral (ML), +1.5 mm from midline; dorsoventral (DV), −2.0 to −1.5 mm from dura. Cerebral cortex, AP −1.1 mm from bregma; ML +3.3 mm from midline; DV −0.3 mm from dura. Amygdala, AP −1.6 mm from bregma; ML +3.0 mm from midline; DV −4.5 mm from dura. Striatum, AP +0.9 mm from bregma; ML +1.5 mm from midline; DV −3.0 mm from dura. For single AAV system, a mixture of AAV-HDR for HA-β-Actin (or HA-CaMKIIα) and AAV-GFP (10:1 ratio) was prepared. For dual AAV system, a mixture of AAV-ESF-Cas9, AAV-HDR for HA-CaMKIIα, and AAV-EGFP (5:5:1 ratio) was prepared. AAV was mixed with Fast Green (0.1 mg/ml, Sigma-Aldrich) to visualize the injection volume. An aliquot of 1 μl of AAV mixture was slowly injected into the targeted area over 10 min at a frequency of 0.3Hz using a glass micropipette connected to the Picospritzer III (Parker) controlled by a sweep function generator (Model 4045, B&K Precision). The needle was left in place for an additional 3–5 min and slowly withdrawn. The skull hole was covered with biocompatible silicone adhesive (Kwik-Sil, WorldPrecision Instruments) and the skin was closed with sutures. After surgery, the mice were monitored during recovery in a warm cage.
Histology
Under deep ketamine-xylazine anesthesia (100 μg of ketamine – 10 μg of xylazine per g of body weight, i.p.), mice were fixed by cardiac perfusion with 4% paraformaldehyde in 0.1 M phosphate buffer (4% PFA/PB), pH 7.4. The brain was then removed and soaked in 4% PFA/PB for 4–12 h. After rinsing with PBS, coronal or sagittal brain sections (50 or 100 μm in thickness) were prepared using a vibratome (VT1200, Leica). For immunohistochemistry, sections were permeabilized with 0.3–0.4% Triton X-100 in PBS, blocked with 5% normal goat serum and 2% BSA, and incubated overnight with the following primary antibodies: rabbit anti-HA (1:1000, Cell Signaling Technology), mouse anti-HA (1:1000, Biolegend), chicken anti-GFP (1:1000, EMD Millipore), guinea pig anti-NeuN (1:1000, EMD Millipore), chicken anti-Tyrosine hydroxylase (1:1000, Aves labs), guinea pig anti-Parvalbumin (1:200, Frontier Institute), rabbit-anti-Myc (1:1000, Cell Signaling Technology), rabbit anti-calbindin antibodies (1:1000, EMD Millipore), mouse anti-CaMKIIα (1:1000, Cell Signaling Technology). After 1–3 h incubation with Alexa Fluor-conjugated secondary antibodies (Invitrogen) followed by DAPI staining (0.1 μg/ml, Life technologies), the stained slices were imaged using a confocal laser-scanning microscope (LSM880 with Airyscan, Zeiss). The acquired images were processed using the Zen software (Zeiss) or Adobe Photoshop CS6 software (Adobe Systems) and analyzed with the ImageJ software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data collection and analyses were not performed blind. Data from all animals used in the experiments were included in the statistical analysis and data plots. Sample sizes were not predetermined with statistical methods. Quantitative data are presented as mean ± SEM. The details of sample number (n) including exact values are indicated in the main text and/or figure legends. The Student’s t test was used when two independent samples were compared. Statistical analyses were conducted using Prism 7 software (GraphPad Software). Differences between data sets were judged to be significant at p < 0.05.
Supplementary Material
Acknowledgments
The authors thank L. Colgan for critical reading of the manuscript; the R.Y. lab members for discussion; T. Laviv and Y. Sugaya for advice on in vivo virus injection; E. Szatmari for sharing aged J20 mice; K. Liu and J. Richards for technical assistance; and D. Kloetzer for laboratory management. This work was supported by the National Institute of Health (R01MH080047 and DP1NS096787 to R.Y.), the Human Frontier Science Program (long-term fellowship to T.M.) and JST/PRESTO, Japan (JPMJPR16F9 to T.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.N. and T.M. conceived the technique. J.N. and T.M. designed and performed all the experiments and data analysis. R.Y. supervised the study. J.N., T.M., and R.Y. wrote the paper.
The authors declare no competing financial interests.
References
- Aschauer DF, Kreuz S, Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PloS one. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzel A, Paulk NK, Shi Y, Huang Y, Chu K, Zhang F, Valdmanis PN, Spector LP, Porteus MH, Gaensler KM, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chiou SH, Winters IP, Wang J, Naranjo S, Dudgeon C, Tamburini FB, Brady JJ, Yang D, Gruner BM, Chuang CH, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29:1576–1585. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nature medicine. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyle DR, Russell DW. Adeno-associated virus vector integration. Curr Opin Mol Ther. 2009;11:442–447. [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Gaj T, Epstein BE, Schaffer DV. Genome Engineering Using Adeno-associated Virus: Basic and Clinical Research Applications. Mol Ther. 2016;24:458–464. doi: 10.1038/mt.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY. Local generation of glia is a major astrocyte source in postnatal cortex. Nature. 2012;484:376–380. doi: 10.1038/nature10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SJ, Foti SB, Schwartz JW, Bachaboina L, Taylor-Blake B, Coleman J, Ehlers MD, Zylka MJ, McCown TJ, Samulski RJ. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum Gene Ther. 2011;22:1143–1153. doi: 10.1089/hum.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci. 2016;17:36–44. doi: 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds HL, Goussakov I, Nakazawa K, Tonegawa S, Bolshakov VY. Essential function of alpha-calcium/calmodulin-dependent protein kinase II in neurotransmitter release at a glutamatergic central synapse. Proc Natl Acad Sci U S A. 2003;100:4275–4280. doi: 10.1073/pnas.0530202100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke MT, Nakai Y, Fujimoto S, Takayama R, Yoshida S, Kitajima TS, Sato M, Imai T. Super-Resolution Mapping of Neuronal Circuitry With an Index-Optimized Clearing Agent. Cell reports. 2016;14:2718–2732. doi: 10.1016/j.celrep.2016.02.057. [DOI] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic C, Chiodini F, Salmon P, Wiznerowicz M, Deglon N, Hornfeld D, Trono D, Aebischer P, Schorderet DF, Munier FL, et al. Activity analysis of housekeeping promoters using self-inactivating lentiviral vector delivery into the mouse retina. Gene Ther. 2003;10:818–821. doi: 10.1038/sj.gt.3301948. [DOI] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M, Lin VG, Guo N, Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013;23:539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-throughput, high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell. 2016;165:1803–1817. doi: 10.1016/j.cell.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1–42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol. 2013;14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- Passini MA, Watson DJ, Vite CH, Landsburg DJ, Feigenbaum AL, Wolfe JH. Intraventricular brain injection of adeno-associated virus type 1 (AAV1) in neonatal mice results in complementary patterns of neuronal transduction to AAV2 and total long-term correction of storage lesions in the brains of beta-glucuronidase-deficient mice. J Virol. 2003;77:7034–7040. doi: 10.1128/JVI.77.12.7034-7040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Cathomen T, Weitzman MD, Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozueta J, Lefort R, Ribe EM, Troy CM, Arancio O, Shelanski M. Caspase-2 is required for dendritic spine and behavioural alterations in J20 APP transgenic mice. Nat Communi. 2013;4:1939. doi: 10.1038/ncomms2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahim AA, Wong AM, Ahmadi S, Hoefer K, Buckley SM, Hughes DA, Nathwani AN, Baker AH, McVey JH, Cooper JD, et al. In utero administration of Ad5 and AAV pseudotypes to the fetal brain leads to efficient, widespread and long-term gene expression. Gene Ther. 2012;19:936–946. doi: 10.1038/gt.2011.157. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Hirata RK. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageot CM, Stiles CD. Molecular mechanisms controlling cortical gliogenesis. Curr Opin Neurobiol. 2002;12:244–249. doi: 10.1016/s0959-4388(02)00322-7. [DOI] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Mori T, Kurihara T, Kawase S, Koike R, Satoga M, Cao X, Li X, Yanagawa T, Sakurai T, et al. Fluorescent protein tagging of endogenous protein in brain neurons using CRISPR/Cas9-mediated knock-in and in utero electroporation techniques. Scientific reports. 2016;6:35861. doi: 10.1038/srep35861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, Wu D, Cong L, Zhang F, Liu JS, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai S, Ark ED, Parra-Bueno P, Yasuda R. Long-distance integration of nuclear ERK signaling triggered by activation of a few dendritic spines. Science. 2013;342:1107–1111. doi: 10.1126/science.1245622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


