Abstract
In many higher plants, cellulose synthesis is inhibited by isoxaben and thiazolidinone herbicides such as 5-tert-butyl-carbamoyloxy-3-(3-trifluromethyl) phenyl-4-thiazolidinone. Semidominant mutations at the IXR1 and IXR2 loci of Arabidopsis confer isoxaben and thiazolidinone resistance. Isolation of the IXR1 gene by map-based cloning revealed that it encodes the AtCESA3 isoform of cellulose synthase. The two known mutant alleles contain point mutations that replace glycine 998 with aspartic acid, and threonine 942 with isoleucine, respectively. The mutations occur in a highly conserved region of the enzyme near the carboxyl terminus that is well separated from the proposed active site. Although the IXR1 gene is expressed in the same cells as the structurally related RSW1 (AtCESA1) cellulose synthase gene, these two CESA genes are not functionally redundant.
Isoxaben (N-3[1-ethyl-1-methylpropyl]-5-isoxazolyl-2,6, dimethoxybenzamide, EL-107, Flexidor, Gallery) is a preemergence broad-leaf herbicide used primarily on small grains, turf, and ornamentals. This compound is extremely toxic, with an I50 for Brassica napus of 20 nM (1). Isoxaben inhibits the incorporation of glucose into the cellulose-rich acid-insoluble fraction of isolated cell walls and has been proposed to be a specific inhibitor of cellulose biosynthesis (2, 3). Treated cells of sensitive species fail to elongate normally and consequently grow isodiametrically (1). Analysis of the effects of the compound on the polysaccharide composition of cell walls and other aspects of plant physiology has led to the proposal that the herbicidal action of isoxaben can be explained entirely by its effect on cellulose biosynthesis (4). Other cellular processes, such as seed germination, mitosis, respiration, photosynthesis, and lipid and RNA synthesis, are unaffected by isoxaben (1, 5).
Mutations at two genetic loci in Arabidopsis thaliana, originally termed ixrA and ixrB but since renamed ixr1 and ixr2, respectively, confer resistance to isoxaben (6, 7). Homozygous ixr1–1 and ixr1–2 mutant plants are 300 and 90 times, respectively, more resistant to isoxaben than wild-type plants (6). The ixr mutations appear to directly affect the herbicide target, because resistant cell lines show no alterations in uptake or detoxification of the herbicide (4).
Thiazolidinones such as 5-tert-butyl-carbamoyloxy-3-(3-trifluromethyl) phenyl-4-thiazolidnone (TZ) are a new class of N-phenyl-lactam-carbamate herbicides (8). TZ has a similar syndrome of effects on plants to isoxaben. The ixr1–1 mutant of Arabidopsis exhibits resistance to both isoxaben and TZ, indicating that isoxaben and TZ share a common mode of action (8). At 12 μM, TZ kills wild-type Arabidopsis but reduces growth of the ixr1–1 mutant by only 50% (8).
Here, we describe the isolation and characterization of the IXR1 gene and the identity of the amino acids changes associated with herbicide resistance in two ixr1 alleles. Resistance is caused by changes in the amino acid sequence of the presumed catalytic subunit of a cellulose synthase gene designated AtCESA3 (9, 10). Identification of the site of action of isoxaben and TZ enhances the potential utility of these compounds in mechanistic studies of cellulose synthase structure and function.
Materials and Methods
Nomenclature.
The 10 cellulose synthase genes in Arabidopsis have been designated CESA1 to CESA10 (9, 10). Some of these genes also have clone or genetic locus designations as follows: RSW1 = CESA1 (11), Ath-A = CESA2 (11), Ath-B = IXR1 = CESA3 (11), PROCUSTE = IXR2 = CESA6 (H. Höfte, personal communication), IRX3 = CESA7 (12). Because the results presented here are primarily related to the characterization of several mutants, we have used the genetic locus symbols throughout.
Genetic Materials.
Seeds from homozygous isoxaben-resistant mutants ixr1–1 (strain DH47; stock no. CS6201) and ixr1–2 (strain DH48; stock no. CS6202) (6) were obtained from the Arabidopsis Biological Resource Center, Columbus, OH. Plants were grown in a commercial peat–vermiculite–perlite mix (Premier Horticulture, Red Hill, PA) in a glasshouse under natural light conditions at a temperature of ≈25°C (day) to ≈20°C (night).
Transgenic plants containing RSW1 and IXR1 promoters fused to the Escherichia coli β-glucuronidase (GUS) coding sequence were obtained as follows. The promoters were amplified from genomic DNA with Expand High Fidelity polymerase (Roche Molecular Biochemicals) by using primer pairs CTGGTCGACGAGAAGAGATGGTAAAGAGAG and GGACTGCAGCCATGGCGCAGCCACCGACACACAGAG for RSW1 and GACGTCGACCTGGTGATACGAGGTGATGG and GACGTCGACCTGGTGATACGAGGTGATGG for IXR1. The 2,170-bp amplified fragment for the IRX1 promoter was cloned into pRITA1 as a SalI/PstI fragment, and then this construct was cut with NcoI and a snapback ligation was done to remove the small section of linker between the 3′ end of the promoter and the ATG of GUS. The 1,111-bp promoter fragment of RSW1 was cloned directly as a SalI/NcoI fragment into pRITA1. The clones were sequenced and then subcloned as NotI promoter-GUS fragments into Ti plasmid pMLBART, introduced into Agrobacterium tumefaciens, and used to transform Col-0 Arabidopsis (13). Plasmids pRITA1 and pMLBART were obtained from Bart Janssen and Kim Richardson at the Horticultural and Food Research Institute of New Zealand in Auckland.
Scoring Isoxaben Resistance.
Isoxaben (95%), supplied by DowElanco (Indianapolis, IN), was solubilized in DMSO. Beginning 2 days after germination, seedlings were sprayed 4 times at 3-day intervals with 1 ml/6 cm2 soil surface of 2.0 μM isoxaben in an aqueous solution of 0.025% DMSO. The growth of wild-type plants was completely inhibited by isoxaben. No effect of 0.025% DMSO on plant growth was noted. Homozygous mutants grew normally, although ixr1–1 grew better than ixr1–2. All heterozygous plants had clearly reduced growth as compared with the homozygous mutants.
Genetic Mapping of the ixr1 Locus.
Approximately 5,000 F2. seeds from an ixr1–2 × Landsberg erecta ecotype cross were planted at a density of ≈1 plant per 2.3 cm2. The seedlings were treated with 2.0 μM isoxaben, leaving only F2-plants homozygous for the ixr1 locus. DNA was prepared from tissue samples of 1,056 plants by alkaline lysis, as described (14).
Four percent agarose gels were used to resolve SSLP markers for mapping. Conditions for PCR amplification of SSLP markers were: 50 mM KCl/10 mM Tris⋅HCl (pH 9.0 at 25°C)/0.1% Triton X-100/200 μM each of dATP, dGTP, dTTP, dCTP/5 pmols of each primer/2.0–2.5 mM MgCl2/1.0 unit Taq polymerase (Promega)/10–50 ng of genomic DNA, final volume 20 μl. PCR conditions: 1 min 94°C; 40 cycles (20 sec 94°C, 20 sec 48–55°C, 40 sec 72°C), 3 min 72°C. The simple sequence length polymorphism and clearable amplified polymorphic sequence markers used for mapping were identified from the information provided by The Arabidopsis Information Resource (http://www.arabidopsis.org).
DNA Sequencing.
Overlapping PCR fragments, 2,034, 2,064, and 2,395 bp in length and spanning the entire coding sequence of the IXR1 gene on TAC-clone K2A11 (AB018111 bases 10620–15972), were amplified from genomic wild-type Columbia, ixr1–1 and ixr1–2 DNA with three oligonucleotide primer pairs (TTAGCCATCCCAAGATTCT, CTTCAAGGGGTCAACAGTA; TACCGAGCGTTTTTCCTAT, CCAGCACCTAAGTTTCACA; GTTCAGTTCCCACAAAGATT, TCATTCCGACCAAAAGTT). Genomic DNA for each genotype was prepared from a mixture of young growing leaves and inflorescence tissue by using a cetyltrimethylammonium bromide-detergent extraction method (14). The fragments from each genotype were PCR-amplified by using a mixture of Taq and proofreading Pfu polymerases (Promega) and sequenced with a set of 20 primers by cycle sequencing by using Big-Dye dideoxy-terminator reaction-mix (Perkin–Elmer Applied Biosystems) and resolved on an ABI310 sequenator. The fragments that carried the ixr1–1 and ixr1–2 point mutations were independently amplified and sequenced three times.
Cosmid Library Screen and Complementation.
Cosmids carrying IXR1 were isolated from a Landsberg erecta ecotype library constructed in the binary Ti cosmid vector pBIC20 (15).
Mutant ixr1 plants were transformed by A. tumefaciens (GV3101) carrying the various cosmid clones according to Bent and Clough (13), and T1 transformants were selected on MS plates containing kanamycin (50 μg ml−1). To score for isoxaben resistance, surface-sterilized seeds of the transgenic T1 plants were germinated on 0.8% agar-solidified medium containing Murashige and Skoog mineral salts (Sigma) and 600 nM isoxaben. The plates were incubated vertically at 25°C under continuous fluorescent illumination (≈50 μmols photons m−2⋅s−1) so that the roots grew on the surface of the agar. Isoxaben resistance was scored after 7 days of incubation.
RNA Gel Blot Analysis.
Total RNA was isolated from various tissues of 6-week-old flowering plants grown on soil or from roots and rosettes of 3-week-old plants grown in a hydroponic system, by using TRIzol reagent (Life Technologies, Rockville, MD). Fifteen micrograms of total RNA was electrophoresed in 1.2% formaldehyde agarose gels, transferred to a nylon membrane, immobilized by UV irradiation, and probed with random-primed DNA fragments encoding stretches of variable region 1 of the IXR1, RSW1, AtCESA2, and IRX3 cellulose synthase genes. The probes, 188–274 bp in length, were amplified by PCR from wild-type Columbia seedling cDNA with the following primers: IXR1 [AAATTTCAGAGCGGATGC, CAAGCTACATTCCCGAGTC], AtCESA2 [CGTGGTGGATTGGATTCAG, GCTTTTCGCCTTGTCGTC], RSW1 [GAGCTAACAAGGCGAGAC, ACATTACCAAGCCCATAAG], IRX3 [GGAGATTCCCACCTGTTAT, TTCTGGCCCAAGATTTC].
Microscopy.
Seeds were germinated and grown at 25°C for 5 days in continuous illumination (≈50 μmols photons m−2⋅s−1) on MS agar plus 1% sucrose. Whole seedlings were infiltrated under mild vacuum for 10 min with 50 μM ImaGene Green C12FDGlcU (Molecular Probes) in 0.1% Silwet L-77 detergent (Lehle Seeds, Round Rock, TX) and incubated for 4–6 h at room temperature before examination. Tissue was counterstained with 0.1 μg ml−1 propidium iodide just before visualization. Cleavage of C12FDGlcU by GUS generates 5-dodecanoylaminofluorescein, which was visualized by using a Bio-Rad 1024 confocal microscope (excitation 488 nm; emission 522/535 nm). Wild-type controls incubated with C12FDGlcU showed no evidence of background staining (16).
Results
Positional Cloning of the IXR1 Locus.
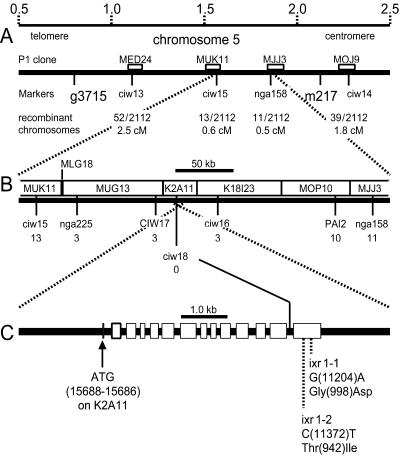
The IXR1 locus maps to the top arm of chromosome 5 within 3 cM of the visible marker lutescens. To obtain a high-resolution map position, 1,056 homozygous mutant plants were selected from the F2 progeny of a cross of the ixr1–2 mutant (Col background) to the Landsberg erecta line. DNA from each isoxaben-resistant F2 plant was examined with the two codominant PCR-based SSLP-markers, CIW13, and CIW14. CIW13 is ≈310 kb distal to g3715, and CIW14 is ≈150 kb distal to m217 (Fig. 1A). Initial mapping resulted in the identification of 52 lines with a recombination event between CIW13 and ixr1 and 39 lines with a recombination event between ixr1 and CIW14. By using six additional SSLP or CAPS markers, the genomic interval containing the IXR1 locus was subsequently narrowed down to a fully sequenced ≈50-kb genomic fragment between markers CIW16 and CIW17 (Fig. 1B).
Figure 1.
Map-based cloning of the IXR1 gene. (A) Representation of a region of Arabidopsis chromosome 5 showing a megabase-scale (Upper), the position of RFLP markers g3715 and m217, SSLP markers CIW13, 14, 15, and SSLP marker nga158. Chromosome 5 P1 clones (http://www.kazusa.or.jp/arabi/) containing the SSLP markers are represented by white boxes. The number of recombinant chromosomes (meiotic breakpoints) in a total of 2,112 examined chromosomes, found for each marker and their calculated genetic distance to the IXR1 locus [in centimorgans (cM)], is given. (B) The region containing the flanking SSLP-markers CIW15 and nga158, showing the position and the number of recombinants found for CAPS (CIW17, PAI2) and SSLP (CIW16, 18) markers and the nonoverlapping parts of P1 and TAC clones spanning the region. (C) An 8-kb segment of TAC clone K2A11 (nucleotides 17, 500–9, 500) showing the intron/exon structure of the IXR1 gene (note crossover of the dotted lines). The start codon and the point mutations in the ixr1–1 and ixr1–2 alleles in the last exon and the predicted amino acid exchanges in the mutant gene products are indicated.
Inspection of the region of chromosome V between CIW16 and CIW17 revealed the presence of a gene for cellulose synthase previously designated as AtCESA3 (9, 10). An additional SSLP marker, CIW18 (Fig. 1 B and C), located within this gene, did not yield any recombinant chromosomes, indicating that the IXR1 locus was in very close proximity. A cDNA clone corresponding to this same cellulose synthase gene was previously described by Arioli et al. (11). Comparison of the sequence of the AtCESA3 cDNA (GenBank accession no. AF027174) to the genomic sequence present on TAC-clone K2A11 (GenBank accession no. AB018111) indicated that the cDNA clone had 87 nucleotides at the 5′ end that are not present in the genomic sequence. This extra sequence corresponds to a 59-nt multiple cloning site (G GACTC GCGCGC CTGCAG GTCGAC ACTAGT GGATCC AAA GAATTC G CGGCCG C GTCGAC, restriction enzyme sites are shown in italics) that was introduced during cloning of the cDNA and an additional 28-nt fragment of DNA (TACGGCTGCGAGAAGACGACAGAAGGGG) that was also introduced at some stage during the cloning of the cDNA. A search of GenBank indicated that this 28-bp sequence is also found at the 5′ ends of other cDNA clones; thus, it is a common artifact in some libraries.
The first nucleotide of the AtCESA3 mRNA that corresponds to the genomic clone is nucleotide 15966 of TAC-clone K2A11. The ORF of the gene begins at nucleotide 15688 (start codon) of K2A11 and ends at nucleotide 10999 (stop codon). The genomic clone contains an intron in the 5′ nontranslated leader sequence that corresponds to nucleotides 15736–15845 of K2A11 and another 13 introns that divide the coding sequence into 14 exons (Fig. 1C), as predicted by grail (http://grail.sourceforge.net), genscan 1.0 (http://genes.mit.edu/GENSCAN.html), and netplantgene (http://www.cbs.dtu.dk/services/netpgene).
In view of the evidence indicating that the mechanism of action of the isoxaben and thiazolidinone herbicides is to inhibit cellulose synthesis, this observation suggested that AtCESA3 corresponds to the IXR1 gene. To test this hypothesis, the gene was cloned and sequenced from Columbia wild type and from the ixr1–1 and ixr1–2 mutants. Sequencing revealed that the AtCESA3 genes from the two ixr1 mutants were identical to the wild type except for a G to A change at nucleotide 11204 in ixr1–1 and a C to T change at nucleotide 11372 in ixr1–2 on the coding strand of TAC-clone K2A11. The point mutation in ixr1–1 leads to replacement of a conserved glycine at position 998 in the AtCESA3 protein with an aspartic acid residue, and the point mutation in ixr1–2 results in replacement of a conserved threonine residue at position 942 in the AtCESA3 protein with an isoleucine residue (Fig. 1C). Thus, both of the ixr1 mutations occurred in exon 14 of the AtCESA3 cellulose synthase gene.
Transformation of ixr1 Mutants with Wild-Type IXR1 Results in Isoxaben Sensitivity.
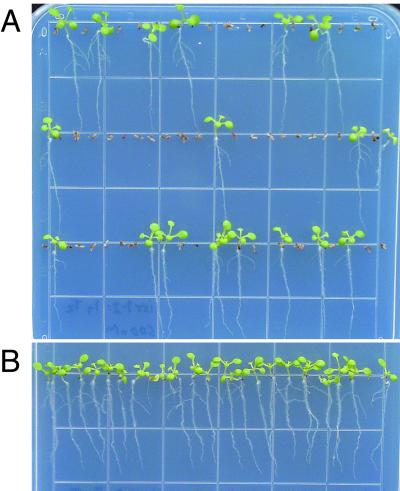
To confirm that the mutations found in the ixr1 mutants are responsible for the isoxaben-resistant phenotype, a series of overlapping clones was isolated from a genomic cosmid library of the Columbia wild type (15) and transformed into the ixr1–1 and ixr1-2 mutants. Cosmids C, H, I, K, and S (see Fig. 2) all contained the full IXR1 coding sequence along with variable amounts of 5′ upstream sequence. T2-progeny of ixr1 mutants transformed with these cosmid clones segregated for isoxaben-resistance (Fig. 3A), whereas the T2-progeny of ixr1 mutants transformed with cosmid clones F and G, which lack a complete IXR1 gene, were completely isoxaben-resistant (Fig. 3B). Because the only complete gene carried in common by the cosmids that produced isoxaben sensitivity was the IXR1 gene, we conclude that this gene is responsible for isoxaben sensitivity.
Figure 2.
Representation of the positions and sizes of various cosmid clones (C-S) tested for ability to complement an ixr1 mutation. The genomic DNA contained in TAC clones K2A11 and K18I23 (Fig. 1B) is represented as bold black and gray lines, respectively. Predicted and known genes (white boxes) were annotated by Kaneko et al. (23). HinDIII restriction sites (vertical thin lines) and the position of hybridization probes X, Y, and Z (small black boxes) used for cosmid library screening are shown.
Figure 3.
Transformation of the ixr1–2 mutant with a wild-type IXR1 gene results in isoxaben sensitive progeny. (A) Segregating T2 progeny of an ixr1–2 mutant transformed with cosmid I (Fig. 2). Approximately 75% of the progeny, representing heterozygous and homozygous ixr1–2 transformants, are herbicide-sensitive when grown on vertical MS plates containing 600 nM isoxaben. (B) Progeny of an ixr1–2 mutant transformed with cosmid F were 100% isoxaben-resistant.
The IXR1 Gene Is Expressed in Both Roots and Shoots.
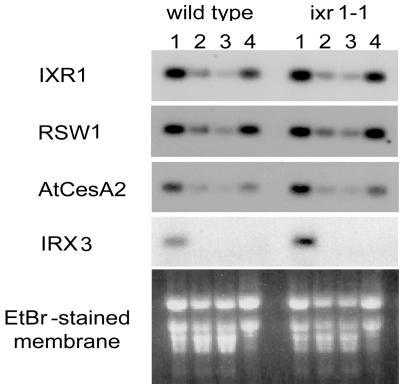
Total RNA was extracted from stems, rosette, and cauline leaves and flowers (Fig. 4), and from shoots and roots of Arabidopsis plants that were treated with or without isoxaben (Fig. 5). Northern blots of these RNA samples were probed with PCR-amplified DNA fragments encoding variable region 1 (9) of the IXR1 gene as well as the related CesA genes RSW1, AtCESA2 (11), and IRX3 (12). IXR1, RSW1, and AtCESA2 were highly expressed in stems, flowers, roots, and shoots, whereas IRX3 was expressed only in stems (Fig. 4). IXR1 appeared to be expressed more highly in roots than shoots.
Figure 4.
Northern analysis of cellulose synthase genes (IXR1, RSW1, AtCESA2, and IRX3) in (1) stems, (2) rosette leaves, (3) cauline leaves, and (4) flowers of wild-type and ixr1–1 mutant.
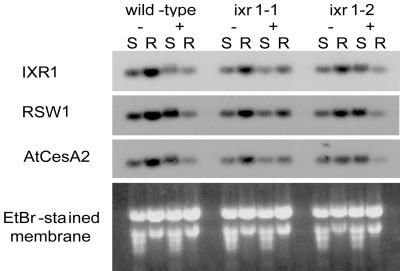
Figure 5.
Northern analysis of cellulose synthase genes (IXR1, RSW1, and AtCESA2) in shoots (S) and roots (R) of wild-type, ixr1–1, and ixr1–2 mutants grown in hydroponic culture and treated with (+) or without (−) 0.5 μM isoxaben for 2 days. No signal was detectable for IRX3.
To examine the possibility that one or more of these CESA genes might be induced by a deficiency in cellulose, wild-type and ixr1 plants were treated with isoxaben for 2 days, and a steady-state amount of mRNA was assessed by Northern blots (Fig. 5). The results of this experiment indicate that there was no detectable increase in mRNA for any of the CESA genes as a result of the isoxaben treatment.
Because we noted that the mRNA expression patterns for IXR1 and RSW1 were almost identical in the Northern blot experiments, we further explored the possibility that these two genes might be expressed in the same cell types and at the same time during development. The expression pattern of the GUS reporter gene was examined in transgenic plants containing the IXR1 or the RSW1 promoter fused to GUS. Five-day-old seedlings stained for GUS activity with 5-bromo-4-chloro-3-indoyl-β-d-glucuronide showed intense blue staining in most cell types (see Fig. 8, which is published as supplemental data on the PNAS web site, www.pnas.org). Staining of the root meristem and the newly emerging true leaves was absent or substantially reduced by comparison with staining in other tissues.
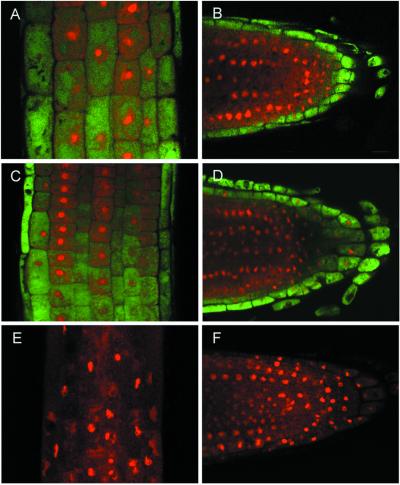
To obtain high-resolution images, GUS activity was visualized by confocal microscopy by using ImaGene Green as a substrate and propidium iodide as a nonspecific fluorescent indicator for substrate penetration into the tissue (Fig. 6). Longitudinal medial sections of root tips showed that the root cap and the epidermal layer of the transgenics had high levels of GUS activity, but activity was not detected in the internal cells of the meristem region of the root (Fig. 6 B and D). However, all of the cells of the root above the meristematic region exhibited strong GUS activity (Fig. 6 A and C). Activity could not be detected in the wild type (Fig. 6 E and F). Thus, IXR1 and RSW1 have indistinguishable patterns of expression with respect to cell type and stage of root development.
Figure 6.
Medial longitudinal sections of roots from transgenic plants containing the RSW1:GUS promoter fusion (A, B), IXR1:GUS promoter fusion (C, D) or nontransformed (E, F). A, C, and E show sections made just above the boundary of the root meristem and the elongation zone. Green fluorescence indicates GUS activity.
Discussion
Results presented here show that resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants is because of amino acid changes in the CESA3 cellulose synthase gene. The mutations corresponding to the ixr1–1 and ixr1-2 mutations cause amino acid substitutions in a 56-aa region near the carboxyl terminus of the protein. In the ixr1–1 mutation Gly-998 becomes Asp-998 and in the ixr1–2 Thr-942 becomes Ile-942. In the absence of a structural model for the protein, it is not currently possible to understand why these mutations render the enzyme resistant to two structurally dissimilar inhibitors. The simplest assumption is that these compounds bind to a site on the enzyme that includes, or is affected by, the indicated amino acids. However, the amino acids are located at a site on the enzyme quite distant from the highly conserved aspartic acid residues that have been proposed to be a component of the active site (17). Although it is formally possible that the identified residues fold into the proposed active site, this seems unlikely because the residues are near the center of a predicted transmembrane helix (ixr1-1) and in a short extracellular loop that connects two proposed transmembrane helices (ixr1–2) (9). If the herbicides are acting directly on the active site of the enzyme, this observation could suggest the unlikely possibility that the conserved aspartic acid residues are not in the active site but have another role in enzyme action. Another possibility for the mode of action of the herbicides is that these regions are necessary for binding of regulatory molecules and that the ixr1 mutations may alter these sites rather than directly affecting the catalytic site. According to this model, the herbicides may act by binding directly to the Ixr1 protein or to a regulatory subunit that binds near the site of the ixr1-1 and ixr1-2 mutations. Yet a third possibility is that these regions, being in or around proposed transmembrane helices, are critical for formation of a proposed pore (9) for glucan chain secretion, and that the hydrophobic herbicides bind to these regions and disrupt pore formation. Conceivably, mutations that prevent herbicide binding might allow proper pore formation in the presence of the herbicides.
Previous genetic studies of the mode of inheritance of the ixr1 mutations had shown that the mutations conferred a semidominant resistance phenotype in which heterozygous (ixr1/IXR1) plants had a level of herbicide resistance that was intermediate between that of mutant and wild type (6). Consistent with previous studies, transgenic ixr1 mutant plants that had been transformed with the wild-type IXR1 gene were much less resistant to isoxaben than the mutant. Our interpretation of this phenotype is based on a model in which cellulose synthase forms a multisubunit complex in the plasma membrane (9, 17–19). According to this hypothesis, the ≈36 individual β-1,4-glucan chains that comprise cellulose microfibrils in higher plants are produced by a corresponding number of catalytic centers in a multisubunit complex in such a way that the individual molecules are coordinately synthesized and hydrogen bond to form the microfibrils as they emerge from the complex. In a heterozygous ixr1IXR1 plant the putative cellulose synthase complexes would be composed of a mixture of resistant and sensitive subunits. Presumably in a complex that contained some sensitive subunits, those subunits would not be able to synthesize β-1,4-glucan chains in the presence of the herbicide. This could, in principle, lead to two outcomes: the resistant subunits might synthesize aberrant cellulose fibrils with fewer than the normal number of β-1,4-glucan chains. Alternatively, the presence of a certain number of sensitive subunits might cause the entire complex to stall, thereby preventing synthesis of a microfibril. Either possibility may cause weakening or disruption of the cell wall leading to cessation of growth.
The IXR1 gene is a member of the CesA multigene family (9, 10) that contains at least 10 closely related members in Arabidopsis (http://cellwall.stanford.edu/). At least two of the other CESA genes (RSW1, AtCESA2) appear to be expressed at levels similar to IXR1 on Northern blots of mRNA from roots, rosette, and cauline leaves, stems, and flowers. Thus, IXR1 is just one of several CesA genes expressed in the major tissues. Indeed, histochemical analyses presented here show that IXR1 and RSW1 are expressed in the same cells. These two CESA genes are also by far the most highly represented by Arabidopsis expressed sequence tags (http://cellwall.stanford.edu/cesa/species/arabidopsis_atcesa/). In view of the fact that rsw1 mutants are not viable at the nonpermissive temperature (11), it is apparent that RSW1 and IXR1 do not have redundant functions. Similarly, the observation that isoxaben and TZ inhibit the growth of wild-type plants implies that a functional Ixr1 protein is also required for cellulose synthesis, even though Rsw1 protein is present. Thus, we infer that at least two closely related cellulose synthase genes that are expressed in the same tissues at similar levels of steady-state mRNA are required for cellulose synthesis. The implication seems to be that IXR1 and RSW1 are not functionally redundant. This conclusion is similar to that of Taylor et al. (20), who found that both the IRX1 and IRX3 genes are required for synthesis of cellulose in secondary cell walls of xylem tissue. In addition, Taylor et al. (20) obtained direct biochemical evidence that the Irx1 and Irx3 proteins bound each other in detergent solubilized extracts, suggesting that they are normally present in the same complex.
Several possible explanations have been proposed for the requirement for expression of at least two nonidentical CESA genes in the same cells. One possibility is that two nonidentical subunits are required to catalyze β-1,4-glucan chain elongation (20, 21). However, an equally compelling hypothesis arises from consideration of the geometry of a self-assembling planar multisubunit complex such as cellulose synthase (21). Because there are estimated to be approximately 36 β-1,4-glucan chains in a cellulose microfibril (9, 19), it seems likely that the cellulose synthase complex has approximately that many CesA subunits arranged in a planar configuration in the membrane. One conceivable way of packing 36 structurally similar proteins into a regular structure is shown in Fig. 7. This arrangement resembles the six-subunit rosette structures observed by electron microscopy (17–19). At least two types of CesA subunits, designated α and β, would be required to allow such a structure to spontaneously assemble. Each α subunit must be able to interact with three other subunits (i.e., one α subunit and two β subunits). By contrast, each β subunit needs only to interact with two other subunits (i.e., two α subunits). More complex variants of this model are also possible, but less complex models seem unlikely, based on simple geometric considerations.
Figure 7.
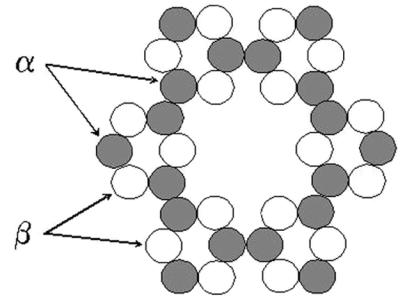
A heteromeric model for the structure of cellulose synthase. In this model, the complex is composed of 36 CesA subunits that are distinguished by the number of subunit binding sites. The α subunits have binding sites for three CesA subunits (one α and two β), whereas the β subunits have only two binding sites.
In addition to the ixr1 mutations, Heim et al. (7) isolated mutations at another locus, now designated ixr2, that also conferred isoxaben resistance. The gene corresponding to this mutation has recently been identified as CESA6 (H. Höfte, personal communication). Mutations at the PROCUSTE locus lead to loss of CESA6 function and result in a phenotype that is similar to the rsw1 mutants (22). This observation led Fagard et al. (22) to suggest that PROCUSTE and RSW1 also have at least partially nonredundant functions. By contrast, that either an ixr1 or an ixr2 mutation can give rise to herbicide resistance indicates that these two genes are functionally redundant with regard to each other. This would be consistent with the observation that ixr1 ixr2 double mutants do not show a higher level of isoxaben resistance than the single mutations (7). Because ixr mutations are sufficient to confer herbicide resistance, we infer that the Rsw1 enzyme and possibly other CesA proteins are resistant to isoxaben and TZ. None of the wild-type CESA genes, including RSW1, has either of the amino acids that confer herbicide resistance in the ixr1 mutants. However, all of the CesA proteins do exhibit other amino acid differences with Ixr1 in the carboxyl terminal region of the Ixr proteins, and it is possible that these differences may render them insensitive to the herbicide. Within the context of the model proposed here (Fig. 7), it is also possible that the herbicides act only on a function or a structural feature that is unique to either the α or β subunits.
We anticipate that, with further progress in understanding the structure and function of the cellulose synthase complex, it will eventually become possible to carry out detailed in vitro biochemical studies of catalysis by this enigmatic enzyme. Because it is now clear that isoxaben and TZ act either directly on cellulose synthase or on some other factor that interacts directly with the enzyme, these compounds may prove useful in future mechanistic studies of enzyme action.
Supplementary Material
Acknowledgments
We acknowledge the contributions of the other members of the Delmer laboratory, particularly Pat Hogan and Alison Roberts, who also contributed, along with R.E., to create the transgenic lines expressing the promoter/GUS constructs for the CESA1 and CESA3 genes. We thank Wolfgang Lukowitz and Stewart Gillmor for helpful discussions, Herman Höfte for communicating unpublished results, and the Arabidopsis Biological Resource Center for providing seeds of ixr1–1 and ixr1–2 mutants. This work was supported in part by a grant to C.R.S. from the U.S. Department of Energy (DOE-FG02–00ER20133) and to D.D. from the U.S. National Science Foundation (DBI 9872627). W.R.S. was the recipient of a fellowship (Sche 548 1/1) from the Deutsche Forschungsgemeinschaft.
Abbreviations
- TZ
5-tert-butyl-carbamoyloxy-3-(3-trifluromethyl) phenyl-4-thiazolidinone
- GUS
β-glucuronidase
References
- 1.Lefebvre A, Maizonnier D, Gaudry J C, Clair D, Scalla R. Weed Res. 1987;27:125–134. [Google Scholar]
- 2.Heim D R, Skomp F R, Tschabold E D, Larrinua I M. Plant Physiol. 1990;93:695–700. doi: 10.1104/pp.93.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corio-Costet M-F, Agnese M D, Scalla R. Pestic Biochem Physiol. 1991;40:246–254. [Google Scholar]
- 4.Heim D R, Skomp J R, Waldron C, Larrinua I M. Pestic Biochem Physiol. 1991;39:93–99. [Google Scholar]
- 5.Corio-Costet M-F, Lherminier J, Scalla R. Pestic Biochem Physiol. 1991;40:255–265. [Google Scholar]
- 6.Heim D R, Roberts J L, Pike P D, Larrinua I M. Plant Physiol. 1989;90:146–150. doi: 10.1104/pp.90.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heim D R, Roberts J L, Pike P D, Larrinua I M. Plant Physiol. 1990;92:858–861. doi: 10.1104/pp.92.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharples K R, Hawkes T R, Mitchell G, Edwards L S, Langford M P, Langton D W, Rogers K M, Townson J K, Wang Y. Pest Sci. 1998;54:368–376. [Google Scholar]
- 9.Delmer D P. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- 10.Richmond T, Somerville C R. Plant Physiol. 2000;124:495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arioli T, Peng L, Betzner A S, Burn J, Wittke W, Herth W, Camilleri C, Höfte H, Plazinski J, Birch R, et al. Science. 1998;279:717–720. doi: 10.1126/science.279.5351.717. [DOI] [PubMed] [Google Scholar]
- 12.Taylor N G, Scheible W-R, Cutler S, Somerville C R, Turner S R. Plant Cell. 1999;11:769–779. doi: 10.1105/tpc.11.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bent A F, Clough S J. In: Plant Molecular Biology Manual. 2nd Ed. Gelvin S B, Schilperoot R A, editors. Dordrecht, The Netherlands: Kluwer; 1998. , Section B7, pp. 1–14. [Google Scholar]
- 14.Lukowitz W, Gillmor C S, Scheible W R. Plant Physiol. 2000;123:795–805. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer K, Leube M P, Grill E. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 16.Fleming A J, Manzara T, Gruissem W, Kuhlemeier C. Plant J. 1996;10:745–754. doi: 10.1046/j.1365-313x.1996.10040745.x. [DOI] [PubMed] [Google Scholar]
- 17.Brown R M, Saxena I M, Kudlicka K. Trends Plant Sci. 1996;1:149–156. [Google Scholar]
- 18.Mueller S C, Brown R M., Jr J Cell Biol. 1980;84:315–326. doi: 10.1083/jcb.84.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura S, Laosinchai W, Itoh T, Cui X, Linder R, Brown R M., Jr Plant Cell. 1999;11:2075–2085. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor N G, Laurie S, Turner S R. Plant Cell. 2000;12:2529–2539. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perrin R M. Curr Biol. 2001;11:R213–R216. doi: 10.1016/s0960-9822(01)00108-7. [DOI] [PubMed] [Google Scholar]
- 22.Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Höfte H. Plant Cell. 2000;12:2409–2423. doi: 10.1105/tpc.12.12.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko T, Katoh T, Sato S, Nakamura Y, Asamizu E, Kotani H, Miyajima N, Tabata S. DNA Res. 1999;6:183–195. doi: 10.1093/dnares/6.3.183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.