Abstract
Background
Protein disulfide isomerase (PDI) and PDI-like proteins contain thioredoxin domains that catalyze protein disulfide bond, inhibit aggregation of misfolded proteins, and function in isomerization during protein folding in endoplasmic reticulum and responses during abiotic stresses.Chinese cabbage is widely recognized as an economically important, nutritious vegetable, but its yield is severely hampered by various biotic and abiotic stresses. Because of, it is prime need to identify those genes whose are responsible for biotic and abiotic stress tolerance. PDI family genes are among of them.
Results
We have identified 32 PDI genes from the Br135K microarray dataset, NCBI and BRAD database, and in silico characterized their sequences. Expression profiling of those genes was performed using cDNA of plant samples imposed to abiotic stresses; cold, salt, drought and ABA (Abscisic Acid) and biotic stress; Fusarium oxysporum f. sp. conglutinans infection. The Chinese cabbage PDI genes were clustered in eleven groups in phylogeny. Among them, 15 PDI genes were ubiquitously expressed in various organs, while 24 PDI genes were up-regulated under salt and drought stress. By contrast, cold and ABA stress responsive gene number were ten and nine, respectively. In case of F. oxysporum f. sp. conglutinans infection 14 BrPDI genes were highly up-regulated. Interestingly, BrPDI1–1 gene was identified as putative candidate against abiotic (salt and drought) and biotic stresses, BrPDI5–2 gene for ABA stress, and BrPDI1–4, 6–1 and 9–2 were putative candidate genes for both cold and chilling injury stresses.
Conclusions
Our findings help to elucidate the involvement of PDI genes in stress responses, and they lay the foundation for functional genomics in future studies and molecular breeding of Brassica rapa crops. The stress-responsive PDI genes could be potential resources for molecular breeding of Brassica crops resistant to biotic and abiotic stresses.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4277-2) contains supplementary material, which is available to authorized users.
Keywords: Pdi, Chinese cabbage, Gene expression, Gene evolution, Biotic and abiotic stresses
Background
Protein disulfide isomerases (PDIs) are enzymes found primarily in the endoplasmic reticulum (ER) in eukaryotes play a vital role in protein folding. Proteins, immediately after biosynthesis must be folded into correct three-dimensional shape for proper functioning. Misfolded proteins are often nonfunctional by producing aggregates and interferes cellular functions. PDIs can bind into misfolded or unfolded proteins preventing to produce such aggregates [1]. Venetianer and Straub, [2] firstly identified PDIs as protein-folding catalysts and demonstrated their catastrophic consequences of a defective protein folding process. Plants utilize multiple mechanisms to fold proteins correctly for proper functioning. PDIs are one of the mechanisms, which work through formation and breakage of connections between cysteine residues by producing disulfide bonds, which was described by Anfinsen [3]. These bonds stabilize the folded protein and provide correct structure to perform its particular function.
PDIs contain four thioredoxin (TRX)-like domains, two of which contain a catalytic site for disulfide bond formation. The reduced (dithiol) forms of PDIs catalyze the reduction of impaired thiol residues of a particular substrate, acting as an isomerase [4]. In human, PDIs usually contain four TRX-like domains (a, b, b’ and a’), a linker (x) and a C-terminal extension domain (c) [5]. The ‘a’ and ‘a’ domains are TRX domains, containing an active Cys-Gly-His-Cys motif joined to α-helices and β-strands (β-α-β-α-β-α-β-β-α) that is essential for polypeptide redox and isomerization [6]. Although the ‘b’ and ‘b’ domains share some structural identity with TRX domain, they do not possess specific active motif site [6]. The α-helices are particularly important for DNA binding motifs, including helix-turn-helix motifs, leucine zipper motifs and zinc finger motifs. The helix-turn-helix (HTH) motif is a major structural motif capable of DNA binding. This motif is composed of two α-helices joined by a short strand of amino acids found in many regulatory proteins for gene expression [7]. The C-terminal region contains a C-domain rich in acidic residues typical as calcium binding proteins and ends with an ER retention signal composed of four amino acids, generally KDEL/GKNF/VASS [8].
PDI genes have been identified in many higher plants, such as 21 PDI genes in Arabidopsis thaliana, 12 in Oryza sativa, 12 in Zea mays, 10 in Vitis vinifera, 9 in Triticum aestivum and so on [9]. In eukaryotes, PDI family proteins are divided into eleven groups based on phylogenetic analysis. Proteins in groups’ I–V contain two thioredoxin-like active domains, whereas those in groups VI–XI contain a single thioredoxin-like active domain. PDI proteins in higher plants are involved in signal transduction pathways and in transcriptional complexes that regulate the responses of genes to environmental stimuli. PDI proteins ERP57, PDIp, P5, ERP72, PDIR, and PDI-D act as redox catalysts and isomerases and exhibit differential functions, such as peptide binding, cell adhesion, and chaperone activities [10]. These proteins play key role as storage protein and plasma membrane maturation [9] GmPDIL-3a and GmPDIL-3b are highly expressed during seed maturation suggesting that they are involved in folding or accumulation of storage proteins in soybean [11]. PDIL2–1 of A. thaliana is directly involved in ovule structure and embryo sac development and determining proper direction of pollen tube growth [12]. The PDI-like protein RB60 plays important roles providing redox potential to regulate photosynthesis [13]. Most wheat PDI and PDIL genes are expressed during endosperm development, indicating their association with storage protein biosynthesis and deposition, which is directly related to gluten quality [14]. BdPDIL1–1 is significantly up-regulated under abiotic (drought, salt, ABA and H2O2) stress, suggesting their involvement in multiple stress responses [15]. All maize PDI genes are highly responsive to cold, salt, dehydration, and ABA stress [16]. During drought, heat and cold stress, TaPDI1, TaPDI2 and TaPDI3 are highly up-regulated in roots and other tissues [17]. There are many family genes involved in tolerance to abiotic and biotic stresses in plants; however we have selected PDI because of its abundance of ubiquitous sulfydryl oxidoreductase protein, which is an important cellular protein with multiple biological functions of all eukaryotic cells. This sulfydryl oxidoreductase protein of PDI displays versatile redox behavior, which also interact with other proteins and assumed its potential role against various diseases and abiotic stresses [18]. In addition, PDI is an important redox proteins regulating reactive oxygen species (ROS) production in the cells and alter redox status of cells to activate defense mechanism [19]. In this study, we make effort to identify the members of PDI gene family in Brassica rapa through genome wide exploration by using different bioinformatic tools. In addition, the putative candidates of PDI genes responsive to abiotic and biotic stresses are predicted through expression profiling using stress induced Chinese cabbage materials.
Results
Identification of PDI genes in B. rapa
A total of 32 BrPDI genes were identified using SWISSPROT of the B. rapa genomic database (http://brassicadb.org/brad/searchAll.php) (BRAD; [20]); using key word “PDI”, NCBI, and annotations of microarray Br135K dataset from cold-treated B. rapa (Chiifu & Kenshin), removing any duplicates. A BLAST search was performed using Arabidopsis PDI sequences as the query, and picked the encoded protein and genomic sequences of PDI genes from BRAD database for 32 BrPDI genes. A high degree of similarity of these 32 BrPDI protein sequences was also picked for other plant species. Isoelectric points, molecular weights and residual size (ranged from 144 to 596 aa) of putative 32 BrPDI proteins are presented in Table 1.
Table 1.
In silico analysis of PDI genes identified in Chinese cabbage with their Arabidopsis orthologs
| Gene name | Gene ID | Chromosome | Stand | Sub-genome | Iso electric point (Pi) | Molecular weight (Mw) | Protein length | Orthologous gene | ||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Start | End | ||||||||
| BrPDI1–1 | Bra016405 | A08 | 17,525,813 | 17,528,427 | – | MF1 | 4.66 | 55.78 | 501 | AT1G21750 |
| BrPDI1–2 | Bra012293 | A07 | 11,001,072 | 11,003,432 | – | MF2 | 4.92 | 55.04 | 496 | AT1G21750 |
| BrPDI1–3 | Bra017948 | A06 | 8,725,538 | 8,728,086 | + | LF | 5.21 | 55.52 | 498 | AT1G21750 |
| BrPDI1–4 | Bra008311 | A02 | 13,630,848 | 13,633,652 | + | MF1 | 4.79 | 56.24 | 511 | AT1G77510 |
| BrPDI1–5 | Bra015665 | A07 | 24,746,925 | 24,749,554 | + | LF | 4.85 | 55.8 | 509 | AT1G77510 |
| BrPDI2–1 | Bra007120 | A09 | 28,945,749 | 28,948,773 | – | LF | 4.70 | 64.28 | 579 | AT3G54960 |
| BrPDI2–2 | Bra002464 | A10 | 9,445,124 | 9,447,998 | – | LF | 4.59 | 66.16 | 596 | AT5G60640 |
| BrPDI2–3 | Bra020239 | A02 | 4,767,927 | 4,771,274 | + | MF2 | 4.58 | 65.06 | 588 | AT5G60640 |
| BrPDI3–1 | Bra014319 | A08 | 1,670,650 | 1,673,502 | – | MF1 | 4.74 | 46.36 | 413 | AT1G52260 |
| BrPDI3–2 | Bra018958 | A06 | 1,024,377 | 1,027,518 | – | LF | 4.98 | 59.04 | 529 | AT1G52260 |
| BrPDI4–1 | Bra000454 | A03 | 11,214,808 | 11,217,122 | + | MF2 | 5.80 | 39.48 | 362 | AT2G47470 |
| BrPDI4–2 | Bra004455 | A05 | 202,365 | 204,674 | – | LF | 6.05 | 39.45 | 361 | AT2G47470 |
| BrPDI5–1 | Bra015375 | A10 | 1,739,233 | 1,741,963 | + | LF | 6.43 | 46.64 | 432 | AT1G04980 |
| BrPDI5–2 | Bra005546 | A05 | 6,147,811 | 6,150,674 | + | LF | 5.71 | 47.85 | 443 | AT2G32920 |
| BrPDI6–1 | Bra018672 | A06 | 2,704,568 | 2,705,424 | – | LF | 4.93 | 16.43 | 144 | AT1G07960 |
| BrPDI7–1 | Bra034408 | A05 | 13,683,334 | 13,685,270 | – | MF2 | 5.28 | 49.08 | 435 | AT1G35620 |
| BrPDI8–1 | Bra001793 | A03 | 18,521,034 | 18,525,290 | + | MF2 | 6.85 | 54.29 | 484 | AT3G20560 |
| BrPDI8–2 | Bra035770 | A05 | 17,795,565 | 17,799,462 | – | LF | 6.96 | 54.31 | 483 | AT3G20560 |
| BrPDI8–3 | Bra019071 | A03 | 26,497,077 | 26,500,750 | + | MF1 | 6.82 | 53.75 | 478 | AT4G27080 |
| BrPDI8–4 | Bra010413 | A08 | 13,621,192 | 13,624,765 | – | MF2 | 6.72 | 53.89 | 480 | AT4G27080 |
| BrPDI8–5 | Bra030465 | A05 | 11,654,854 | 11,657,768 | – | MF2 | 7.73 | 54.20 | 483 | AT1G50950 |
| BrPDI8–6 | Bra018881 | A06 | 1,468,257 | 1,472,162 | – | LF | 6.26 | 53.57 | 477 | AT1G50950 |
| BrPDI9–1 | Bra026786 | A09 | 35,442,828 | 35,445,717 | + | MF2 | 6.85 | 60.45 | 532 | AT1G15020 |
| BrPDI9–2 | Bra014330 | A08 | 1,583,063 | 1,585,667 | + | MF1 | 6.65 | 58.34 | 523 | AT2G01270 |
| BrPDI10–1 | Bra001092 | A03 | 14,721,770 | 14,723,230 | + | MF2 | 6.75 | 33.86 | 303 | AT3G03860 |
| BrPDI10–2 | Bra031969 | A05 | 24,598,071 | 24,599,646 | – | MF1 | 7.09 | 34.22 | 302 | AT3G03860 |
| BrPDI10–3 | Bra036758 | A08 | 7,293,480 | 7,294,725 | + | MF1 | 8.57 | 33.54 | 298 | AT3G03860 |
| BrPDI10–4 | Bra036429 | A01 | 26,156,526 | 26,158,281 | _ | LF | 8.91 | 36.75 | 326 | AT1G34780 |
| BrPDI11–1 | Bra019406 | A03 | 24,393,210 | 24,394,793 | _ | MF1 | 6.02 | 51.69 | 468 | AT4G21990 |
| BrPDI11–2 | Bra013579 | A01 | 6,445,389 | 6,446,955 | _ | LF | 6.39 | 51.58 | 468 | AT4G21990 |
| BrPDI11–3 | Bra034466 | Scaffold000096 | 53,632 | 55,924 | _ | LF | 6.74 | 53.67 | 479 | AT1G62180 |
| BrPDI11–4 | Bra029505 | A09 | 17,850,361 | 17,852,000 | + | LF | 6.74 | 53.57 | 479 | AT4G04610 |
Phylogenetic analysis and domain location
A phylogenetic tree was constructed using 76 full-length protein sequences of PDI and PDI-like genes, which included 32 from Chinese cabbage, 11 from Brachypodium distachyon, 12 from maize, and 21 from Arabidopsis to investigate evolutionary relationship. Eleven phylogenetic groups were denoted among the considered PDI genes from different plant species (Fig. 1) with distinct 4 clades. Clade 1 consisted phylogenetic groups I, II, III and VII, whose members contain two active thioredoxin domains, whereas members of group VII encoded proteins with a single N-terminal active domain (Fig. 2). Clade 2 contained phylogenetic group IV, V and VI, among them members of group IV and V possessing two active thioredoxin domains in tandem at their N-termini. Group members of VI may have lost one of the two active thioredoxin domains due to shared structural features of a common progenitor. Clade 3 contained phylogenetic groups VIII, which included genes encoding proteins with a single active thioredoxin domain. The members of groups IX, X and XI formed clade 4; encoded proteins with an active thioredoxin domain. Phylogenetic and domain analyses revealed that these PDI family genes are divergent in plant. Most BrPDI proteins are contained an N-terminal signal peptide and a C-terminal KDEL signal responsible for translocation and ER retention, respectively. In the domain structures, ‘a’ and ‘a’ are homologous to thioredoxin and contain a -CXXC- active site for isomerase and redox activities. By contrast, the ‘b’ and ‘b’ domains have no homology to thioredoxin and lack of -CXXC- catalytic site. However, the secondary structures of four (a, a’, b, b’) domains are similar to thioredoxin rather than the active catalytic site. The thioredoxin-like domain comprises βαβαβαββα motifs forming four layers of β-sheets that are sandwiched three layers of α-helixes (Fig. 3).
Fig. 1.
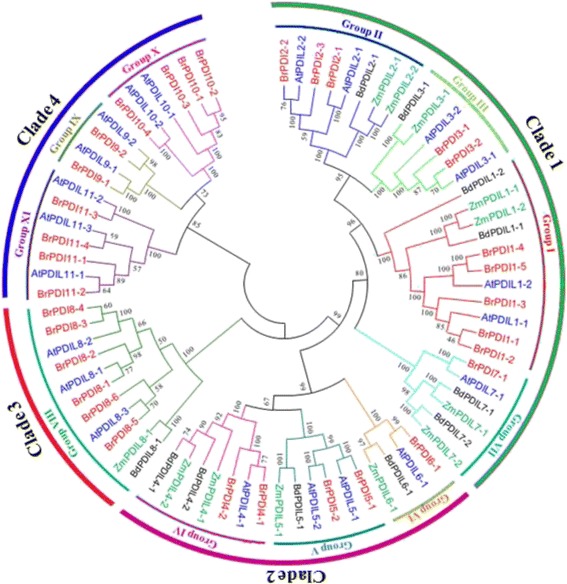
Phylogenetic analysis of PDI protein identified in B. rapa, Arabidopsis, Brachypodium distachyon and Zea mays. The phylogenetic tree was generated by the neighbor-joining method in MEGA6.0 with 1000 bootstrap replicates using full-length PDI protein sequences from B. rapa, Arabidopsis, B. distachyon and Z. mays. Blue, red, Green, and black letters indicate Arabidopsis, B. rapa, Z. mays and B. distachyon proteins, respectively
Fig. 2.

Domain structure of the deduced amino acid sequences of B. rapa PDI genes. The putative signal peptides (SP), the a and b type domains, the N-terminal calcium binding domain calsequestrin, the D domains (Erp29), the transmembrane domains (TM) and the C_ERV (COPII-coatedERV) domain are shown. The thioredoxin-like catalytic domains with two active sites (shown in detail in the boxes) are also shown. Numbers above indicate domain boundaries (aa), and numbers on the right indicate ORF (aa). Domains ao (light gray) and a’ (gray) are homologous to TRX and contain the catalytic CxxC motif (red). Domains b (yellow) and b’ (light yellow) also exhibit a TRX fold, but they do not share high sequence similarity with each other or with domains a or a’. The C-terminal extension (red) contains a (K/H)DEL retention signal for the ER
Fig. 3.
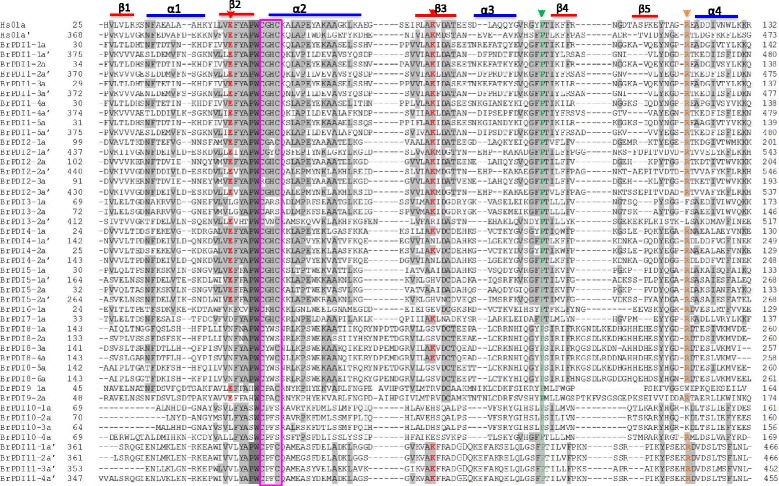
Multiple sequence alignment of the a-type domains of B. rapa PDI proteins and a typical human PDI. Residues highlighted in gray and light gray share 100% and >50% identity, respectively. Elements of the secondary structure are specified by blue colour bars (α-helices) and red colour bars (β-sheets). Red arrows indicate the two buried charged residues in the vicinity of the active site, orange arrow indicates the conserved arginine (R) and green arrow indicates the cis prolines (P) near each active site. Active-site residues within a domain are pink colour boxed
We performed multiple sequence alignment of ‘a’- and ‘a’-type domain sequences of BrPDI proteins to analyze the features of them. These domain sequences contained five β-sheets, four α-helices and the -CGHC- motif of the active site. They also contained conserved arginine, glutamic acid, proline, and lysine residues (Fig. 3). All BrPDI members ‘a’-type domain with conserved arginine residue, critical for their catalytic function, except BrPDI3–1 and BrPDI3–2 which contained phenylalanine or leucine residues instead of arginine residue. In addition, BrPDI9–1 contained tryptophan and BrPDI10–1, BrPDI10–2 and BrPDI10–3 contained lysine in the place of arginine residues. BrPDI2–3 accomplished with RGHC non-characteristic active sites; but BrPDI3–1 and BrPDI3–2 with CARS. Whereas, BrPDI8–1, BrPDI8–2, BrPDI8–5, and BrPDI8–6 comprised with CYWS; and BrPDI10–1, BrPDI10–2, BrPDI10–3, and BrPDI10–4 contained CPFS non-characteristic active sites (Fig. 2) which may affect their redox potential and lose their function.
Chromosomal distribution of BrPDI genes
Thirty two BrPDI genes are located on different chromosomes not less than one on each except chromosome 04. The highest number (six) of BrPDI genes was identified on chromosome A05 (19.35%), while chromosome A01, A02, A07, and A10 contained two BrPDI genes (6.45%) for each, and chromosome A04 has no BrPDI genes (0%). Only one gene (BrPDI11–3) is located in scaffold (Fig. 4a and b). None of the BrPDI gene clusters was detected in Chinese cabbage. The sequence alignment of BrPDI proteins showed higher similarity within groups. BrPDI proteins showed ≥68% similarity within groups, except group10 and group 2. By contrast, similarity between groups was ≤65%. Seventeen pairs of BrPDI proteins showed ˃80% similarity indicating duplications are predominant for those genes (Table 2).
Fig. 4.
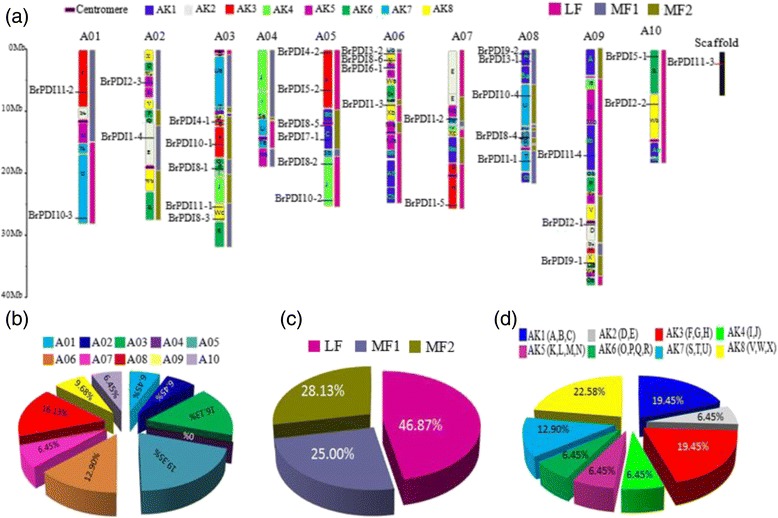
a Distribution of BrPDI genes on 10 chromosomes. The 24 (A to X) ancestral blocks and three sub-genomes are plotted, based on the report of Schranz et al., [46]. b The percentages of BrPDI genes on ancestral blocks are demonstrated in graph. c The percentages of BrPDI genes on each subgenomes are demonstrated in pie graph. Least fractionated (LF), medium fractionated (MF1) and most fractionated (MF2) genomes. d The percentages of BrPDI genes on each chromosome are also demonstrated in pie graph
Table 2.
For each pair wise alignment, the similarity (relative to the maximum similarity) and the number of identical amino acids (in %) is given
| Protein | BrPDI 1–2 |
BrPDI 1–3 |
BrPDI 1–4 |
BrPDI 1–5 |
BrPDI 2–1 |
BrPDI 2–2 |
BrPDI 2–3 |
BrPDI 3–1 |
BrPDI 3–2 |
BrPDI 4–1 |
BrPDI 4–2 |
BrPDI 5–1 |
BrPDI 5–2 |
BrPDI 6–1 |
BrPDI 7–1 |
BrPDI 8–1 |
BrPDI 8–2 |
BrPDI 8–3 |
BrPDI 8–4 |
BrPDI 8–5 |
BrPDI 8–6 |
BrPDI9–1 | BrPDI 9–2 |
BrPDI 10–1 |
BrPDI 10–2 |
BrPDI 10–3 |
BrPDI 10–4 |
BrPDI 11–1 |
BrPDI 11–2 |
BrPDI 11–3 |
BrPDI 11–4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BrPDI1–1 | 90 | 84 | 73 | 76 | 30 | 33 | 32 | 19 | 21 | 24 | 26 | 19 | 18 | 25 | 20 | 9 | 9 | 12 | 10 | 32 | 32 | 26 | 18 | 20 | 23 | 24 | 28 | 27 | 25 | 28 | 30 |
| BrPDI1–2 | 83 | 74 | 77 | 32 | 33 | 32 | 18 | 18 | 25 | 26 | 20 | 19 | 25 | 20 | 8 | 12 | 12 | 10 | 32 | 32 | 25 | 19 | 20 | 24 | 23 | 29 | 29 | 29 | 32 | 34 | |
| BrPDI1–3 | 72 | 74 | 28 | 31 | 32 | 19 | 19 | 24 | 26 | 19 | 19 | 24 | 21 | 9 | 9 | 10 | 10 | 27 | 32 | 26 | 22 | 21 | 24 | 24 | 29 | 27 | 25 | 27 | 29 | ||
| BrPDI1–4 | 82 | 28 | 32 | 33 | 19 | 17 | 28 | 26 | 21 | 20 | 21 | 16 | 12 | 12 | 12 | 11 | 33 | 31 | 28 | 20 | 20 | 20 | 24 | 27 | 23 | 23 | 24 | 25 | |||
| BrPDI1–5 | 29 | 29 | 31 | 17 | 21 | 24 | 25 | 21 | 19 | 25 | 19 | 11 | 12 | 12 | 13 | 34 | 32 | 29 | 20 | 20 | 24 | 24 | 27 | 31 | 25 | 24 | 27 | ||||
| BrPDI2–1 | 55 | 54 | 28 | 29 | 25 | 25 | 18 | 21 | 27 | 19 | 11 | 15 | 11 | 14 | 27 | 28 | 29 | 23 | 22 | 23 | 22 | 24 | 36 | 35 | 38 | 36 | |||||
| BrPDI2–2 | 86 | 30 | 30 | 27 | 28 | 20 | 18 | 22 | 17 | 11 | 13 | 12 | 13 | 33 | 31 | 25 | 23 | 23 | 20 | 21 | 26 | 34 | 33 | 37 | 41 | ||||||
| BrPDI2–3 | 29 | 28 | 27 | 28 | 19 | 18 | 27 | 20 | 12 | 11 | 14 | 14 | 31 | 30 | 24 | 22 | 23 | 17 | 20 | 25 | 37 | 32 | 37 | 34 | |||||||
| BrPDI3–1 | 80 | 18 | 14 | 15 | 15 | 19 | 16 | 13 | 11 | 11 | 11 | 26 | 25 | 25 | 21 | 25 | 26 | 29 | 25 | 35 | 32 | 34 | 30 | ||||||||
| BrPDI3–2 | 18 | 17 | 16 | 16 | 23 | 18 | 9 | 8 | 11 | 9 | 26 | 26 | 24 | 12 | 29 | 29 | 31 | 29 | 31 | 33 | 40 | 30 | |||||||||
| BrPDI4–1 | 93 | 28 | 23 | 31 | 18 | 13 | 13 | 14 | 15 | 31 | 35 | 27 | 15 | 27 | 26 | 27 | 31 | 29 | 29 | 28 | 29 | ||||||||||
| BrPDI4–2 | 29 | 25 | 34 | 15 | 13 | 13 | 16 | 16 | 33 | 37 | 32 | 14 | 27 | 25 | 27 | 31 | 30 | 28 | 28 | 28 | |||||||||||
| BrPDI5–1 | 81 | 27 | 15 | 11 | 13 | 15 | 13 | 29 | 32 | 29 | 16 | 25 | 26 | 27 | 27 | 34 | 33 | 32 | 29 | ||||||||||||
| BrPDI5–2 | 23 | 13 | 13 | 14 | 13 | 10 | 28 | 27 | 27 | 14 | 27 | 28 | 29 | 25 | 30 | 30 | 34 | 30 | |||||||||||||
| BrPDI6–1 | 26 | 27 | 27 | 28 | 29 | 31 | 34 | 27 | 21 | 27 | 32 | 22 | 23 | 25 | 25 | 29 | 32 | ||||||||||||||
| BrPDI7–1 | 12 | 11 | 11 | 11 | 21 | 23 | 23 | 16 | 26 | 19 | 20 | 25 | 29 | 27 | 29 | 26 | |||||||||||||||
| BrPDI8–1 | 86 | 70 | 70 | 74 | 69 | 31 | 18 | 28 | 25 | 23 | 23 | 25 | 28 | 23 | 30 | ||||||||||||||||
| BrPDI8–2 | 68 | 69 | 74 | 70 | 31 | 16 | 27 | 23 | 27 | 41 | 28 | 24 | 24 | 26 | |||||||||||||||||
| BrPDI8–3 | 89 | 68 | 65 | 32 | 11 | 65 | 22 | 22 | 41 | 24 | 23 | 23 | 27 | ||||||||||||||||||
| BrPDI8–4 | 67 | 63 | 32 | 18 | 65 | 23 | 25 | 41 | 26 | 26 | 26 | 25 | |||||||||||||||||||
| BrPDI8–5 | 82 | 31 | 14 | 24 | 25 | 26 | 47 | 23 | 27 | 23 | 26 | ||||||||||||||||||||
| BrPDI8–6 | 35 | 16 | 25 | 28 | 25 | 23 | 23 | 27 | 28 | 27 | |||||||||||||||||||||
| BrPDI9–1 | 69 | 21 | 33 | 27 | 30 | 22 | 24 | 22 | 35 | ||||||||||||||||||||||
| BrPDI9–2 | 32 | 23 | 24 | 27 | 21 | 22 | 18 | 31 | |||||||||||||||||||||||
| BrPDI10–1 | 83 | 83 | 34 | 29 | 40 | 44 | 27 | ||||||||||||||||||||||||
| BrPDI10–2 | 80 | 35 | 27 | 27 | 41 | 28 | |||||||||||||||||||||||||
| BrPDI10–3 | 36 | 29 | 29 | 23 | 25 | ||||||||||||||||||||||||||
| BrPDI10–4 | 26 | 30 | 30 | 27 | |||||||||||||||||||||||||||
| BrPDI11–1 | 92 | 75 | 84 | ||||||||||||||||||||||||||||
| BrPDI11–2 | 75 | 83 | |||||||||||||||||||||||||||||
| BrPDI11–3 | 75 |
Duplications are given bold
Three fractionated subgenomes, like least fractionated (LF), medium fractionated (MF1), and most fractionated (MF2) subgenomes are found in B. rapa genome. Notably, 32 BrPDI genes are distributed onto ten chromosomes with fractionation into three subgenomes, of which are 15 LF (46.87%), 8 MF1 (25%), and MF2 9 (28.13%; Fig. 4c and Table 1). In addition, 32 BrPDI genes are distributed in different block during evolution. Among them, 7 genes (22.58%) belong to AK8 block, followed by 19.45% of BrPDI genes into AK1, AK3 blocks; while only 6.45% of BrPDI genes were allocated to AK2, AK4, AK5 and AK6 (Fig. 4d). We have also found a total of 17 segmental duplicated PDI genes pairs in B. rapa genome (Fig. 5 and Table 3). Furthermore, the substitution rate of non-synonymous (Ka) and synonymous (Ks) ratios (Table 3) were calculated to assess the selection pressures among duplicated BrPDI gene pairs. In these analysis, Ka/Ks ratios were identified as lower than 1, 1 and above 1 indicating negative or purifying selection, neutral selection and positive selection, respectively. Nine out of seventeen BrPDI duplicated gene pairs, had Ka/Ks value below 1 (purifying selection) and rest of them had Ka/Ks value above 1(positive selection). Moreover, the estimated divergence time of BrPDI genes showed that duplication event started at 29.74 million years ago (MYA) and continued up to 1.62 MYA (Table 3).
Fig. 5.
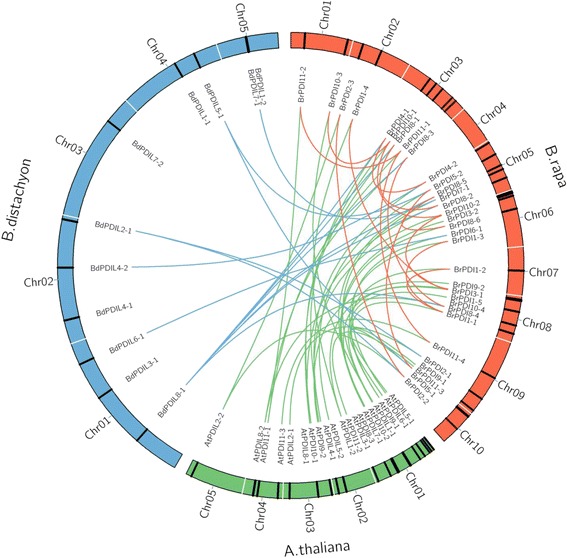
Microsynteny analysis of PDI genes among B. rapa, Brachypodium distachyon and A. thaliana. The chromosomes from the three species are indicated in different colors, orange, green and blue colors represent B. rapa, A. thaliana and B. distachyon chromosome respectively. Orange lines depiction of duplicated BrPDI genes on 10 B. rapa chromosomes
Table 3.
Estimated Ka/Ks ratios of the duplicated BrPDI genes with their divergence time in Chinese cabbage
| Duplicated gene pairs | Ks | Ka | Ka/Ks | Duplication type | Types of selection | Time (mya) | ||
|---|---|---|---|---|---|---|---|---|
| BrPDI1–1 | vs. | BrPDI1–2 | 0.0569 | 0.1138 | 2.0000 | Segmental | Positive | 1.89 |
| BrPDI1–1 | vs. | BrPDI1–3 | 0.1064 | 0.0726 | 0.6823 | Segmental | Purifying | 3.54 |
| BrPDI1–2 | vs. | BrPDI1–3 | 0.1113 | 0.0715 | 0.6424 | Segmental | Purifying | 3.71 |
| BrPDI1–4 | vs. | BrPDI1–5 | 0.2421 | 0.0349 | 1.4415 | Segmental | Positive | 8.07 |
| BrPDI2–2 | vs. | BrPDI2–3 | 0.0486 | 0.1554 | 3.1975 | Segmental | Positive | 1.62 |
| BrPDI3–1 | vs. | BrPDI3–2 | 0.1203 | 0.0698 | 0.5802 | Segmental | Purifying | 4.01 |
| BrPDI4–1 | vs. | BrPDI4–2 | 0.1093 | 0.1515 | 1.3860 | Segmental | Positive | 3.64 |
| BrPDI5–1 | vs. | BrPDI5–2 | 0.1010 | 0.4750 | 4.7029 | Segmental | Positive | 3.37 |
| BrPDI8–1 | vs. | BrPDI8–2 | 0.1773 | 0.0527 | 0.2972 | Segmental | Purifying | 5.91 |
| BrPDI8–3 | vs. | BrPDI8–4 | 0.1179 | 0.0343 | 0.2909 | Segmental | Purifying | 3.93 |
| BrPDI8–5 | vs. | BrPDI8–6 | 0.8922 | 0.3621 | 0.4058 | Segmental | Purifying | 29.74 |
| BrPDI10–1 | vs. | BrPDI10–2 | 0.2892 | 0.1337 | 0.4623 | Segmental | Purifying | 9.64 |
| BrPDI10–1 | vs. | BrPDI10–3 | 0.4183 | 0.2768 | 0.6613 | Segmental | Purifying | 13.94 |
| BrPDI10–2 | vs. | BrPDI10–3 | 0.7166 | 0.2026 | 0.2827 | Segmental | Purifying | 23.89 |
| BrPDI11–1 | vs. | BrPDI11–2 | 0.0895 | 0.1179 | 1.3173 | Segmental | Positive | 2.98 |
| BrPDI11–1 | vs. | BrPDI11–4 | 0.0969 | 0.2040 | 2.1052 | Segmental | Positive | 3.23 |
| BrPDI11–2 | vs. | BrPDI11–4 | 0.0985 | 0.3570 | 3.6243 | Segmental | Positive | 3.28 |
Ks the number of synonymous substitutions per synonymous site, Ka the number of nonsynonymous substitutions per nonsynonymous site, MYA million years ago
Motif composition and intron–exon analysis
The intron–exon structures were almost consistent among the groups of BrPDI genes. Group I contained nine to ten exons; whereas, members of group V contained nine exons (Additional file 1: Figure S1). All members of group II possessed 10 to 11 exons, while members of group IV and IX contained 11 exons, and the member of group VIII composed with the highest number (15) of exons. Member of groups X and XI possessed 3–4 exons. Finally, member of groups VI and VII contained four and five exons, respectively. Conserved motifs among BrPDI proteins were investigated using the MEME motif search tool. Motifs 1 and 2 contained-CXXC- catalytic sites, which are necessary for isomerase and redox activity. Motifs 3 was present in all groups, but motifs 2 was absent in groups VI, VII, VIII, IX and X. Motif 1 and 5 were absent in group IV and XI, respectively. Relatively less conserved motif 6 was found only in groups I, II, III and VII. Motifs 4, 7, 8 and 9 were unique to group VIII and motif 10 was present in group VIII and XI only (Additional file 1: Figure S2).
Microsyntenic analysis
A microsynteny map was constructed to find out orthologous gene pairs of PDI genes among B. rapa, B.distachyon and A. thaliana for exploration of evolutionary history and relationships among the genomes (Fig. 5). We found 22 orthologous gene pairs between B. rapa and A. thaliana; whereas, 13 orthologous gene pairs were identified between B. rapa and B. distachyon (Fig. 5). We found 17 duplicated PDI gene pairs in B. rapa genome. All of the segmental duplications are denoted with an orange line in fig. 5, no tandem duplications were found in PDI genes in B. rapa.
Microarray data analysis
We used our previously published microarray data set to analyze the expression patterns of 32 BrPDI genes using two contrasting inbred lines ‘Chiifu’ and ‘Kenshin’, treated with cold and freezing stress (4 °C, 0 °C, −2 °C and −4 °C) for 2 h [20]. All of the BrPDI genes were differentially expressed in response to cold or freezing stress in two lines (Fig. 6). In Chiifu, most of the BrPDI genes were highly expressed upon exposure to cold and freezing temperatures. While, BrPDI2–1, 4–1, 8–2, 8–6, 9–1 and 10–1 genes showed higher expression during cold stresses in ‘Kenshin’ compared to ‘Chiifu’.
Fig. 6.
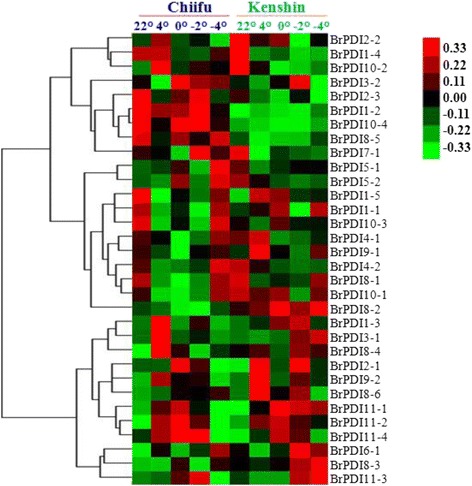
Heat map showing microarray expression of 32 B. rapa PDI genes against cold and freezing treatments in two inbred lines Chiifu and Kenshin
Promoter analysis of BrPDI genes
PDI genes of Chinese cabbage might contain biotic and abiotic stress-responsive cis-acting elements in their promoter regions. Most of the BrPDI genes bear cold-responsive LTR cis-acting element, drought inducible MBS cis-element, and salt- or environmental stresses cis-acting element (WBOX and TC-rich repeats; Additional file 2: Table S1). The genes BrPDI2–1, BrPDI2–2, BrPDI2–3, BrPDI3–2, BrPDI6–1, BrPDI7–1, BrPDI8–1, BrPDI8–2, BrPDI8–4, BrPDI10–1, BrPDI10–2, BrPDI10–3, BrPDI10–4, BrPDI11–1, BrPDI11–2, BrPDI11–3, and BrPDI11–4 have no ABA-responsive CE3 cis-acting element in their promoter regions. Majority of BrPDI genes contained more than one disease resistance or defense responsive cis-acting element (Box-W1, WBOX, and TC-rich repeats) except BrPDI8–5, BrPDI10–3, BrPDI11–3 and BrPDI11–4.
Organ-specific gene expression
We have used gene-specific primers of 32 BrPDIs for RT-PCR to check the expression patterns of BrPDIs in different organs (root, stem, leaf, flower buds, sepals, petals, stamens, and pistils) in Chinese cabbage line SUN-3061. Semi-quantitative RT-PCR analysis revealed that among the 32 genes, 15 were ubiquitously expressed in all organs (Fig. 7). Three genes (BrPDI 1–2, 9–1 and 11–4) were slightly expressed in all of the tested organs. BrPDI 7–1 was highly expressed in floral parts but slightly expressed in roots, stems, leaves and flower buds. On the other hands, BrPDI 1–2, 1–3, 1–4, 2–3 and 8–1 were totally absent in roots, stems and leaves but only expressed in flower buds and floral parts. In addition, BrPDI 2–1 was weakly expressed in roots, leaves and pistil; while BrPDI3–1 was expressed slightly in roots, stems, and leaves. BrPDI8–5 was expressed in leaves, flower buds, and pistils; the BrPDI 8–6 was expressed in stamens only. BrPDI10–4 was appeared with very low signal in leaves, sepals and pistils. BrPDI 11–1 and BrPDI1 11–2 were highly expressed in vegetative organs but weekly expressed in reproductive organs (Fig. 7).
Fig. 7.
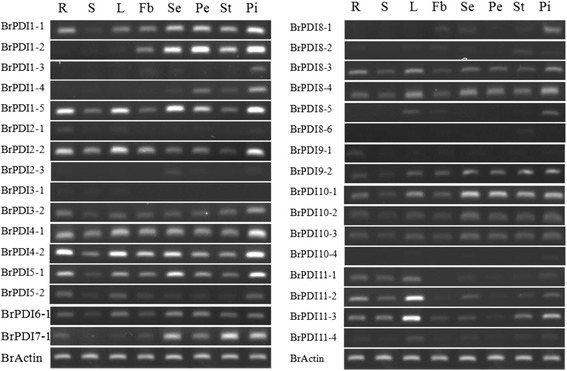
Expression profiles of BrPDI genes in various tissues, as determined by RT-PCR analysis. Eight amplified bands (from left to right) per gene represent amplified products from R- roots; S- stems, L- leaves; Fb- flower buds; Se- sepals; Pe- petals; St- stamens and Pi- pistils
Analysis of abiotic stress-responsive gene expression
Gene-specific primers of BrPDI genes were used to obtain expression profiles in response to various abiotic stresses. Expression patterns of BrPDI genes in response to cold was obtained by analyzing qPCR data from cold treated two contrasting Chinese cabbage lines, ‘Chiifu’ and ‘Kenshin’. In addition, we have calculated the expression patterns of different BrPDI genes during salt, drought and ABA stresses using qPCR data of Chinese cabbage line ‘kenshin’. Most of the BrPDI genes were significantly down regulated over the time course of cold stress, while six genes of ‘Chiifu’ (BrPDI 1–3, BrPDI 3–1, BrPDI 6–1, BrPDI 9–2, BrPDI 10–2 and BrPDI 11–1) were significantly up-regulated on the advancement of cold stress durations (Fig. 8a). By contarst, most of the BrPDI genes in ‘Kenshin’ were significantly down-regulated throughout the stress period, however, only four genes (BrPDI 1–1, BrPDI 4–1, BrPDI 4–2, and BrPDI 5–2) showed high expression in ‘Kenshin’ compared to ‘Chiifu’ (Fig. 8a).
Fig. 8.
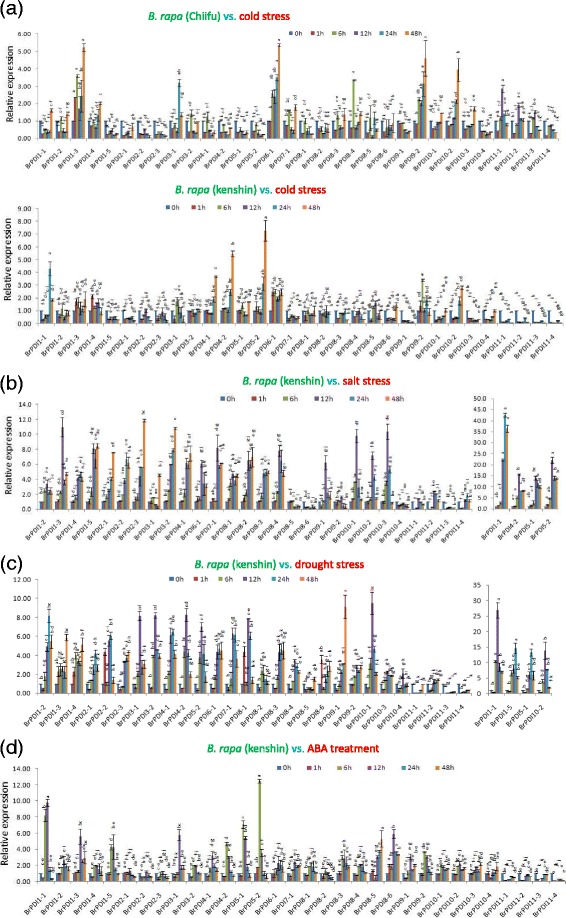
Real-time PCR expression analysis of BrPDI genes after treatment with (a) cold, (b) salinity, (c) drought, (d) ABA. The error bars represent the standard error of the means of three replications
High transcript levels were detected in 24 BrPDI genes over time courses and these genes were up-regulated in response to salt stress. BrPDI 1–1 gene was exhibited approximately 42- fold higher expression at 24 h, whereas BrPDI 4–2, BrPDI 5–1 and BrPDI 5–2 were exhibited 15-, 13-, and 19-fold higher expression, respectively than control at 12 h time point (Fig. 8b). In addition, BrPDI 1–3, BrPDI 6–1, BrPDI 7–1, BrPDI 8–4, BrPDI 9–1, BrPDI 10–1, BrPDI 10–2, and BrPDI 10–3 were significantly up-regulated up to 12 h time point (Fig. 8b). Besides, BrPDI 1–4, BrPDI 1–5, BrPDI 2–1, BrPDI 2–3, BrPDI 3–2, BrPDI 4–1 and BrPDI 8–3 were up-regulated approximately 5-, 8-, 7-, 11-, 11-, 7-, and 5-fold, respectively at 48 h.
In drought stress, most of the BrPDI genes were up-regulated at 12 h and 24 h; then gradually down-regulated with the advancement of time courses (Fig. 8c). Eight genes (BrPDI 1–1, BrPDI 3–1, BrPDI 3–2, BrPDI 4–2, BrPDI 5–2, BrPDI 8–1, BrPDI 10–1, and BrPDI 10–2) were significantly up-regulated at 12 h. Five genes (BrPDI 1–2, BrPDI 1–5, BrPDI 2–1, BrPDI 2–2, and BrPDI 5–1) were significantly up-regulated at 24 h drought stress, thereafter being down-regulated (Fig. 8c). BrPDI 1–1, BrPDI 1–5, BrPDI5–1 and BrPDI 10–2 exhibited about 27-, 15-, and 14- fold higher expression, respectively compared to control. Most of the BrPDI genes had low level of expression in response to ABA treatment (Fig. 8d). BrPDI 1–1, BrPDI 1–3, BrPDI 1–5, BrPDI 3–1, BrPDI 4–2, BrPDI 5–1, and BrPDI 5–2 were significantly up-regulated at 6 h and 12 h time point. However, those BrPDI genes were down-regulated at the beginning of ABA stress, but start up-regulation at 6 h and continued up to 12 h time point, thereafter gradually down-regulated over the remaining time courses. Four genes (BrPDI 11–1, BrPDI 11–2, BrPDI 11–3 and BrPDI 11–4) were down-regulated over the time courses. BrPDI 1–1 gene was 10-fold up-regulated at 12 h; while BrPDI 5–2 gene showed 13-fold up-regulation at 6 h time points compared to control (Fig. 8d).
Expression profiling of the BrPDI genes during chilling injury treatment
Chilling injury experiment with cold tolerance ‘Chiifu’ and cold sensitive ‘Kenshin’ was conducted to get the deep insight of the expression of the target cold tolerance gene as an alternative of functional analysis of the predicted genes. We exposed the ‘Chiifu’ and ‘Kenshin’ seedlings in cold injury treatment at 0 °C for 72 h until complete chilling injury of the susceptible (Kenshin). First remarkable chilling injury was recognized in ‘Kenshin’ seedlings at 24 h time point and gradually progress with the enhancement of time courses, ‘Kenshin’ seedlings were completely injured at 72 h time course. Whereas, no chilling injury was observed in ‘Chiifu’ seedlings up to 72 h time point, but plant growth become stunted (Fig. 9). In ‘Chiifu’, eight genes showed higher transcript level. BrPDI1–4 and BrPDI6–1 genes showed significant up-regulation about 12- and 9-fold higher expressions, respectively at 72 h time point in ‘Chiifu’ compared to ‘Kenshin’ (Fig. 10). Moreover, BrPDI9–2 and BrPDI11–1 showed about 6- and 8-fold higher expressions at 48 h and 24 h time point, respectively. In addition, BrPDI10–2 gene exhibited higher expression in both line at 72 h time point. In ‘kenshin’, four genes (BrPDI4–2, BrPDI5–1, BrPDI5–2, and BrPDI7–1) showed higher expression against chilling injury treatment (0 °C).
Fig. 9.

Comparative chilling injury symptoms in cold tolerant ‘Chiifu’ and ‘Kenshin’ plant at different time points (0 h, 24 h, 48 h, and 72 h) against chilling treatment (0 ° C). Chilling injury first appeared at 24 h in ‘Kenshin’ thereby gradually progress with the advancement of time points. The ‘kenshin’ plants are completely chill injured at 72 h time point but no chilling injury symptoms appeared in ‘Chiifu’ plants at any time points
Fig. 10.
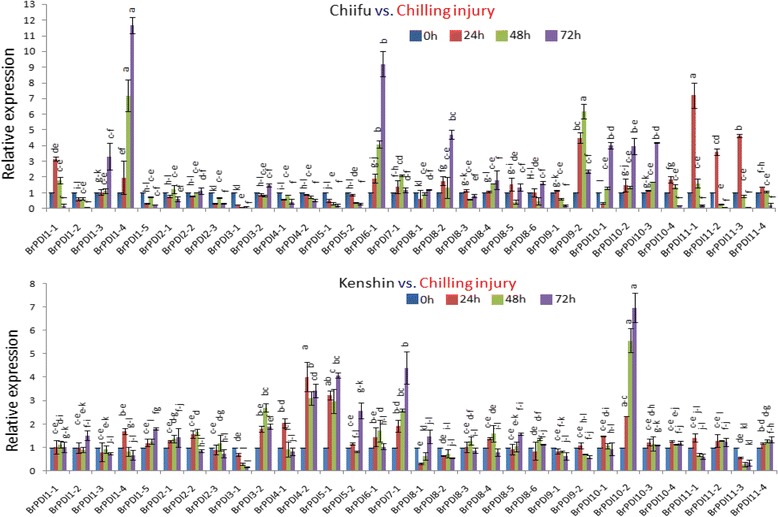
Real-time PCR expression analysis of BrPDI genes after treatment with Chilling injury. The error bars represent the standard error of the means of three replications
Analysis of biotic stress-related gene expression
To elucidate the expression patterns of BrPDI genes in response to Fusarium oxysporum f. sp. conglutinans infection, we have collected leaf samples from infected and mock-treated Chinese cabbage line ‘Chiifu’ for performing qPCR. This fungus causes wilt and root rot diseases in Brassica crops. The BrPDI genes exhibited distinct expression patterns in response to Fusarium infection. Out of 32 genes, 14 showed high expression at 6 h time point (Fig. 11). The expression levels of BrPDI 1–1, BrPDI 2–3, and BrPDI 5–1 were approximately 27-, 10-, and 11-fold higher, respectively compared to mock-treated plants. Fifteen genes (BrPDI 6–1, BrPDI 7–1, BrPDI 8–1, BrPDI 8–2, BrPDI 8–3, BrPDI 8–4, BrPDI 8–5, BrPDI 8–6, BrPDI 9–1, BrPDI 10–1, BrPDI 10–2, BrPDI 10–4, BrPDI 11–1, BrPDI 11–2, and BrPDI 11–4) had very little or insignificant expression compared to mock-treated (uninfected) plants. However, BrPDI10–3 and BrPDI11–3 showed no response against F. oxysporum infection.
Fig. 11.
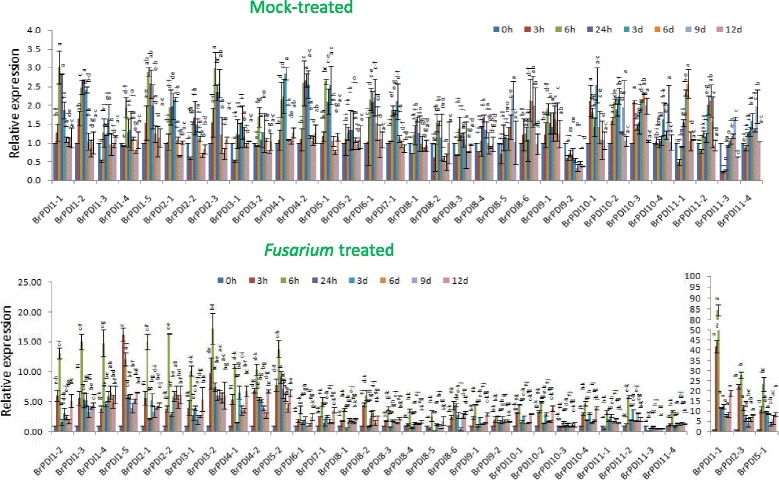
Real-time PCR expression analysis of BrPDI genes after treatment with Fusarium oxysporum f.sp. conglutinans. The error bars represent the standard error of the means of three replications
Discussion
In genome-wide exploration study of Chinese cabbage PDI genes; we have identified 32 PDI genes identical to 21 PDI genes of Arabidopsis. The Chinese cabbage genome has gone under two duplication events since its divergence from Arabidopsis [21]. The evolutionary relationship between Arabidopsis and Chinese cabbage is also supported by our results. We analyzed 32 BrPDI genes expression pattern based on the microarray data set in two contrasting Chinese cabbage inbred lines, ‘Chiifu’ and ‘Kenshin’ exposed to cold and freezing stress (4 °C, 0 °C, −2 °C, and −4 °C) [20]. From an evolutionary point of view, gene duplications, tandem and segmental duplications can increase the gene numbers of a particular gene family [22]. Distribution of BrPDI genes in the B. rapa genome are also affected by segmental duplication, tandem duplication, polyploidization etc. during evolution [23, 24]. In addition, genome triplication events of B. rapa may also be played important role in the expansion of PDI gene family. We found 17 pairs of segmental duplicated PDI genes in Chinese cabbage genome (Fig. 5 and Table 3), this result indicates that the expansion of PDI gene family in B. rapa genome depicted on segmental gene duplication.
We also investigated evolutionary history of BrPDI gene family and calculated the Ka (the number of synonymous substitutions per synonymous site), Ks (the number of nonsynonymous substitutions per nonsynonymous site) and Ka/Ks ratios of segmental duplicated gene pairs. Nine gene pairs had Ka/Ks < 1 (purifying selection) and eight gene pairs had Ka/Ks >1(positive selection; Table 3) indicating purifying selection and positive selection together accelerated evolution and functional divergence of BrPDI genes in B. rapa genome. Based on the synonymous substitution rate, we calculated divergence time of the BrPDI genes and found that duplication started on 29.74 MYA and stop at 1.62 MYA (Table 3) which indicate PDI genes divergence take place after the genome triplication events in B. rapa. Genome triplication event of B. rapa has occurred from the ancestor of A. thaliana between five to nine million years ago (MYA; [25]). Besides this, the exons and introns distribution of BrPDI genes are also consistent with the number of exons and introns of PDI genes in Arabidopsis, which indicate their close evolutionary relationship. In addition with, by drawing microsyntenic map, we conclude that BrPDI genes are strongly related to those of B. distachyon and A. thaliana.
The Chinese cabbage PDI genes appeared as functionally differentiated for short time, and keeps as functional genes in the genome and maintaining complex functions in organ development. The features and functions of PDI family genes have been extensively studied in Triticum aestivum [14] and Brachypodium distachyon [15]. Our identified and characterized PDI genes in Chinese cabbage help to increase the understanding of role of this family gene. Most of the BrPDI genes were predominantly expressed in all organs suggested their functions in regulation of plant growth and development. Based on our organ-specific expression analysis, all BrPDI genes are expressed at least in one organ at variable level. BrPDI 2–1 and BrPDI 9–1 were highly expressed in root, which is consistent with the previous findings [14, 15]. In addition to their involvement in plant growth and development, PDI genes are also responsive to adverse environmental conditions [15]. We have found BrPDI 1–2, BrPDI 1–3, BrPDI1–4, BrPDI 2–3 BrPDI 8–1 and BrPDI 8–6 were expressed in flower buds and floral parts, suggesting their possible role in flower development.
The expression patterns of 32 BrPDI genes were analyzed using a whole-genome microarray dataset of two inbred lines of B. rapa, ‘Chiifu’ and ‘Kenshin’ imposed to various temperature treatment [26]. Thereof, we have selected BrPDI family genes for analyzing their differential expression patterns compared to control by constructing heat-map (Fig. 6). Our results suggest that BrPDI genes play a vital role against abiotic stress responses in Chinese cabbage. In addition, our main objectives were to identify putative candidate PDI genes related to stress response during plant growth and development. It is established fact that environmental stresses like, cold, salt, and drought are severely affected crop production and make threaten in food security worldwide. Therefore, application of artificial stress treatment helps to know, how plants adapt to stresses via molecular, morphological, and physiological mechanisms. There is intense interest in identifying stress-responsive genes and elucidate them to develop stress-tolerant crop cultivars. Relative expression of PDI family genes were estimated from the sample of the plants subjected to abiotic stresses (cold, salt and drought), hormone (ABA), chilling injury, and biotic (F. oxysporum f.sp.conglutinans) stress treatment over different time periods (Fig. 8a–d, 9~11) using qPCR data. In plants, PDIs are localized to the ER and other cellular compartments, such as nucleus and chloroplasts [27, 28]. Proteins in the ER of plants can lead to unfolding or misfolding due to complex physiological processes during different environmental stresses [29]. Plants possess multiple mechanisms for ensuring correct folding of proteins. PDI was the first reported catalyst of protein folding (2). In the current study, BrPDI1–3, BrPDI3–1, BrPDI6–1, BrPDI9–2, BrPDI10–2, and BrPDI11–1 were highly expressed in cold-tolerant ‘Chiifu’ than in cold-susceptible ‘Kenshin’ during cold stress treatment. PDI genes that are highly expressed under cold stress may encode proteins catalyze for breakage of disulfide bonds in misfolded proteins, restoring their proper functioning through correction of folding patterns. Lu & David [30] reported six Arabidopsis genes; those were up-regulated due to chemicals such as dithiothreitol (DTT), β-mercaptoethanol and tunicamycin induction. In current study, we have found BrPDI1–1, BrPDI4–1, BrPDI4–2, and BrPDI5–2 were highly expressed under cold stress in cold-susceptible ‘Kenshin’ compared to cold-tolerant ‘Chiifu’, suggesting that these four genes might be related to the cold-susceptibility in ‘Kenshin’. However, BrPDI2–1, BrPDI2–2, BrPDI5–2, BrPDI8–1, BrPDI8–6, BrPDI9–1, BrPDI10–4, and BrPDI11–4 were not expressed or even down-regulated in ‘Chiifu’ under cold treatment confirmed that these genes contain putative cold-inducible LTR cis-acting elements. Notably, these genes also contain a heat-inducible HSE cis-acting element. Hence, both cold- and heat-inducible cis-acting elements are presented in the promoter regions of the same genes suggesting that the latter cis-acting element may down-regulate these genes under cold stress. Two contrasting cold tolerant Chinese cabbage lines were used in this study to predict the putative cold susceptible and cold tolerant BrPDI gene(s) of this crop. Four BrPDI genes (BrPDI1–4, BrPDI6–1, BrPDI9–2, and BrPDI11–1) exhibited higher expression in ‘Chiifu’ compared to ‘Kenshin’ at the level of chilling injury. Among these four genes BrPDI6–1, BrPDI9–2, and BrPDI11–1 genes were common in the higher expression in cold stress and chilling injury level treatments revealed their active involvement to overcome the cold and chilling injury at different time points of the cold tolerant line ‘Chiifu’. Therefore, from the in depth expression data, it is clearly evident that BrPDI6–1, and BrPDI9–2 genes are the putative candidate in Chinese cabbage for overcoming cold and even chilling injury temperature (0 ° C).
With few exceptions, BrPDI genes are highly expressed in Chinese cabbage under salt and drought stress, among them BrPDI1–1 to BrPDI5–2 genes encode the proteins which containing two active sites and a C-terminal ER retention signal composed of four amino acids (KDEL/ VASS/ MADD), might be involved in enhancing protein-folding in the ER. BrPDI1–1, BrPDI1–3, BrPDI1–5, BrPDI3–1, BrPDI4–2, BrPDI5–1, BrPDI5–2, BrPDI8–5, and BrPDI8–6 were highly expressed under ABA stress compared to control. Mittler et al. [31] and Sakamoto et al. [32] observed rapid accumulation of reactive oxygen species (ROS) in plant cells subjected to environmental stresses such as salt, drought, and ABA. Indeed, during environmental stress (e.g., drought, salt, ABA, UV, and heat exposure) ROS levels can increase dramatically, which may significantly damage cell structure. PDI proteins play a significant role in thioredoxin-based redox pathway, which comprises part of the antioxidative defense system [33]. Abiotic stress usually leads to protein unfolding, misfolding, and aggregation; which represent a common threat to living cell [34]. Efficient protein repair systems and/or protein folding stability helps plant to survive in adverse environmental conditions [35]. Most BrPDI genes were differentially expressed in response to salt, drought, and ABA treatment, which is consistent with the findings of Zhu et al. [15]. Those genes are highly expressed in response to salt, drought, and ABA might function in the repair of misfolded proteins, thereby playing a role in plant resistance to the stresses. The up-regulated genes contain the respective stress-responsive cis-elements in their promoter regions (Additional file 2: Table S1).
PDI genes have been previously shown to be contributed against powdery mildew resistance in common wheat [36]. In this study, we have analyzed expression profiles of these genes in response to Fusarium oxysporum f.sp. conglutinans infection. Moreover, 14 genes were up-regulated compare to mock treated one due to infection of fungal pathogen at 6 h time point, whose genes containing a disease resistance Box-W1 cis-acting element (Fig. 11, Additional file 2: Table S1). By contrast, BrPDI 6–1~ BrPDI 11–4 do not contain the Box-W1 element, those exhibited almost no expression due to fungal infection. Thus, the highly expressed BrPDI genes might play a vital role in response against fungal pathogen. Therefore, this might be the putative candidate genes for developing Fusarium oxysporum f.sp. conglutinans resistant Chinese cabbage cultivars through marker assisted back crossing (MAB) or gene technology. Besides, the up-regulated BrPDI genes might play differential roles in signal transduction pathways and/or cooperate with other genes to form networks for defending plants against adverse environmental conditions.
Conclusion
In conclusion, this is the first report of genome-wide characterization of PDIs in Chinese cabbage. We identified 32 BrPDI genes in the Chinese cabbage genome and characterized them based on motif distribution, protein structure, classification, number of introns -exons, sequence similarities, and expression patterns in response to abiotic (cold, salt, and drought) stresses, ABA treatment and biotic stress (F. oxysporum f.sp. conglutinans inoculation). We have also recognized BrPDI genes may play roles in biotic and abiotic stress responses. Several BrPDI genes (BrPDI1–1, BrPDI1–4, BrPDI1–5, BrPDI3–1, BrPDI3–2, BrPDI4–1, BrPDI4–2, BrPDI5–1, BrPDI5–2, BrPDI6–1, BrPDI7–1, BrPDI8–2, BrPDI9–1, BrPDI9–2, and BrPDI11–1) might function in responses to multiple stresses. The identified stress-induced BrPDI genes help to elucidate the complex regulatory network underlying stress resistance mechanisms. In addition, these genes represent a useful resource to molecular breeders for marker assisted back crossing (MAB) and/or engineering transgenic plants in future research with increased resistance to biotic and abiotic stresses.
Methods
Identification and sequence analysis of BrPDI genes
Chinese cabbage PDI family members were identified using the SWISSPROT tool of the B. rapa database (http://brassicadb.org/brad/searchAll.php; 20) and NCBI using the keyword “PDI”. We also searched these genes from a microarray Br135K dataset of two contrasting Chinese cabbage inbred lines, Chiifu and Kenshin exposed to low-temperature stress. The Arabidopsis PDI sequences were used as the query to perform a BLAST search setting a cutoff e-value of <10−10. The CDS (coding DNA sequences) and protein sequences of the identified PDIs were obtained from the B. rapa genomic database. The PDI protein sequences were further analyzed for the presence of a thioredoxin domain using the web tool “SMART” (http://smart.embl-heidelberg.de/ [37]) and NCBI (https://www.ncbi.nlm.nih.gov/cdd). Additionally, the primary structures of the genes (protein length, molecular weight and isoelectric point) were analyzed using ExPasy (http://web.expasy.org/compute_pi/). ORFs were identified using ORF finder at NCBI (https://www.ncbi.nlm.nih.gov/orffinder/). Sequence alignment of PDI proteins were carried out using CLUSTAL Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Sub-genome fractionation and positional information for all idetified BrPDI genes on 10 chromosomes of B. rapa were retrieved from the Brassica database, and draft maps of the locations of the BrPDI genes were constructed using Map Chart version 2.30 (http://www.wageningenur.nl/en/show/Mapchart.htm ). The Gene Structure Display Server (GSDS) web tool (http://gsds.cbi.pku.edu.cn/) was used to determine the number of introns and exons by aligning CDS and genomic sequences of the PDI genes [38]. Putative cis-acting regulatory elements in the BrPDI genes were predicted in regions of approximately1500-bp upstream of the translation initiation site [ATG] using the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Phylogenetic and conserved motif analysis of Chinese cabbage PDI genes
The predicted BrPDI protein sequences were aligned with those of Arabidopsis, Brachypodium distachyon and Zea mays and Chinese cabbage using Clustal Omega, and a phylogenetic trees was constructed based on the condensed alignment in MEGA6.06 using the Neighbor-Joining (NJ) algorithm [39], with the parameters set at 1000 replications for bootstrap values and complete deletion mode to analyze tree topology and reliability. The genes which were used in phylogenetic analysis, name and accession number provided in Additional file 2: Table S2. Motif analysis of proteins was performed using MEME (Expectation Maximization for Motif Elicitation v4.10.1) [40] with the following parameters: (1) number of motifs = 10, (2) Motif width ≥ 6 and ≤50.
Chromosome localization and gene duplications
The physical locations of BrPDI genes were collected from B. rapa genomic database and the positions of the BrPDI genes were drafted to ten B. rapa chromosomes by map chart program. Duplicated BrPDI genes were identified using BLAST searched against themselves, notably identity and query coverage were >80% of those particular genes [41]. Tandem duplicated genes were marked as an array of two or more homologous genes within a distance of 100 kb. The synonymous rate (Ks), nonsynonymous rate (Ka), and evolutionary constraint (Ka/Ks) were calculated between the duplicated PDI gene pairs (Table 3), using the method of Nei & Gojobori [42] by Mega 6.0 software. The value of Ka/Ks ratio like >1, <1 and =1 are depicted for positive selection, purifying selection and neutral selection, respectively [43]. The divergence time was calculated using the formula T = Ks/2r Mya (Millions of years) where, Ks being the synonymous substitutions per site and r is considered 1.5 × 10–8 substitutions rate per site per year for dicot plants [44]. We reconstructed 24 conserved chromosomal blocks (labelled A–X) of B. rapa genome. Colour coding of the blocks was assigned based on their positions in a proposed ancestral karyotype (AK1–8) following the procedure stated by Cheng et al. [45] and Schranz et al. [46].
Microarray and microsynteny of PDI gene family
Two contrasting inbred Chinese cabbage (B. rapa ssp. pekinensis) lines, ‘Chiifu’ and ‘Kenshin’ in respect to cold tolerance and cold susceptible, were used for microarray expression analysis Kayum et al. [47]. A heat map was generated based on transcript abundance value of PDI genes using Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm). The microsyntenic relationship of PDI genes among B. rapa, Brachypodium distachyon and A. thaliana were detected using Blast against whole genome of such crop species. Chromosomal positions of PDI genes were collected from respective databases and the relationship among the three crop species were plotted using circos software (http://circos.ca/) [48].
Plant growth conditions, treatments and sampling
Surface sterilize seeds of two Chinese cabbage inbred lines ‘Chiifu’ and ‘Kenshin’ were grown in incubation room maintaining 24 °C temperature with 14/10 h (light/dark) condition at the Department of Horticulture, Sunchon National University, Korea. Abiotic stress treatments were imposed to four-week-old seedlings according to the methods described by Ahmed et al. [49], three biological replicates were maintained for abiotic stress treatments. Two contrasting Chinese cabbage inbred lines, cold tolerance ‘Chiifu’ and cold susceptible ‘Kenshin’ were used for abiotic stress cold and chilling injury stress experiment. Whereas, only ‘Kenshin’ was used to predict the BrPDI genes against other stress treatments (salt, drought, and ABA) due to unavailability of any contrasting genotypes for other abiotic stress treatments (salt, drought, and ABA) in our hand. By contrast, we assumed ‘Kenshin’ would be suitable for molecular characterizing of BrPDI genes for abiotic stress responsiveness rather than cold, because these type of genotypes are widely grown in tropics and sub-tropics. Fresh roots and leaves (third and fourth leaves) were excised from stress-treated plants over time points (0 h, 1 h, 4 h, 12 h, 24 h, and 48 h). Samples were immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction. For chilling injury experiment, the Chinese cabbage inbred line ‘Chiifu’ and ‘Kenshin’ plants were grown in culture room under a 16 h light photoperiod maintaining 24 °C. The three week old seedlings were transferred to incubator (TOGA clean system; model: TOGA UGSR01) maintained 0 °C temperature and keep the seedling until clear remarkable cold injury symptoms appeared. Cold treated plants leaves were excised at 0 h, 24 h, 48 h and 72 h time points with the progression of cold injury in cold susceptible line ‘Kenshin’ (Fig. 9), thereby leaves were immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction. Total five biological replications were maintained for chilling injury treatment. However, inbred line ‘Chiifu’ was used for organ-specific expression profiling of BrPDI genes and also infection with F. oxysporum f. sp. conglutinans for biotic stress as described by Ahmed et al. [50]. Fusarium- and mock-infected leaves (4th and 5th leaves) were collected at 0 h, 3 h, 6 h, 24 h, 3d, 6d, 9d, 12d after inoculation and immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
RNA extraction and cDNA synthesis
Total RNA was extracted from the samples (roots and leaves) using an RNeasy mini kit (Qiagen, USA) following the manufacturer’s instructions. DNA contamination was removed using RNase-free DNase (Promega, USA) following manufacturer’s protocol. The extracted RNA was quantified by UV spectrophotometry at A260 using a NanoDropND-1000 and NanoDrop v3.7 software (Nano Drop Technologies, USA). Complementary DNA (cDNA) was synthesised from total RNA using a First-Strand cDNA synthesis kit (Invitrogen, Japan) following the manufacturer’s instructions.
Qualitative and quantitative PCR expression analysis
Qualitative expression analysis was conducted using one-step Emerald Amp GT PCR Master Mix (Takara, Japan) by RT-PCR. Gene-specific primers for BrPDIs were used for RT-PCR and the BrActin gene of Chinese cabbage was used as an internal control. RT-PCR was performed using 1 μL of 50 ng template cDNA using master mix contained 10 pmol of each primer (forward and reverse), 9 μL sterile water and 8 μL Emerald mix in a total volume of 20 μL. The PCR conditions were initial denaturation 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 30s, annealing at 58 °C for 30s and extension at 72 °C for 45 s, with a final extension at 72 °C for 5 min. The PCR products were visualized on 1.2% agarose gel.
Quantitative Real-time PCR (qRT-PCR) was conducted using 10 μL reaction volume consisted of 5 μL 2× Quanti speed SYBR mix, 1 μL (10 pmol) each of forward (F) and reverse (R) gene-specific primers (Additional file 2: Table S3), 1 μL template cDNA (50 ng) and 2 μL ultra-pure water (ddH2O). The conditions for real-time PCR was initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 58 °C for 10 s and extension at 72 °C for 15 s. The qRT-PCR reactions were normalized using the Chinese cabbage Actin gene as a reference for all comparisons [51]. Fluorescence was measured at last step of each cycle, and three replicates were used for each sample. Amplification detection and data were processed using the Light cycler® 96 SW 1.1 software and the cq value was calculated using the 2-ΔΔCT method to determine the relative expression. The relative expression data was statistically analyzed (Tukey HSD test) and lettering was done using Minitab 17 software (https://www. minitab . com/products/ minitab /).
Additional files
The genomic structures of BrPDI genes. Solid green boxes and red lines indicate exons and introns, respectively. The bottom scale indicates length of exons and introns. Figure S2. Schematic representation of motif compositions in the BrPDI protein sequences. Different motifs logo, numbered 1–10, motif are displayed in different colored boxes. The names of all members are displayed on the left-hand side. (PPT 932 kb)
Putative cis-elements, more than 6 bp, were identified in 32 BrPDI genes in Chinese cabbage. Table S2. A total of 76 PDI and PDIL genes name and accession numbers from 4 species used for constructing phylogenetic tree, including 32 from Brassica rapa,(Br) 21 from Arabidopsis thaliana (At),11 from Brachypodium distachyon (Bd) and 12 from Zea mays (Zm). Table S3. Primers for RT-PCR andreal-time PCR analysis of 32 BrPDI genes (DOC 472 kb)
Acknowledgments
Special thanks to Professor YoonKang Hur, Department of Biology, College of Biological Sciences and Biotechnology, Chungnam National University, Daejeon, Korea Republic for providing seeds of B. rapa inbred lines ‘Chiifu’ and ‘Kenshin’.
Funding
This research was supported by Golden Seed Project (Center for Horticultural Seed Development, No. 213007–05-01-SB510) Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Service (KFS), South Korea. The funding bodies were not involved in the design of the study or in any aspect of the collection, analysis and interpretation of the data or paper writing.
Availability of data and materials
We declare that the dataset(s) supporting the conclusions of this article are included within the article (and its additional file(s)).
The complete raw microarray data have been deposited in the Omics database of NABIC (http://nabic.rda.go.kr) as enrolled numbers NC-0024-000001 and NC-0024-000002. Phylogenetic data have been deposited in the TreeBASE as submission ID 21739.
Abbreviations
- ABA
Abscisic acid
- AK
Karyotype
- At
Arabidopsis thaliana
- Bd
Brachypodium distachyon
- Br
Brassica rapa
- BRAD
Brassica database
- C
Chiifu
- CDS
Coding DNA sequence
- Cl
Cluster
- dai
Days after infection
- Er
Endoplasmic reticulum
- F.oxysporum
Fusarium oxysporum
- f.sp.
Fungal species
- HTH
Helix-turn-helix
- K
Kenshin
- Ka
Nonsynonymous substitutions per nonsynonymous site
- kb
Kilo basepair
- Ks
Synonymous substitutions per synonymous site
- LF
Least fractionated
- MF1
Medium fractionated
- MF2
Most fractionated
- MYA
Million year
- PD
Protein disulfide isomerase
- qPCR
Quantitative polymerase chain reaction
- RT-PCR
Reverse transcription polymerase chain reaction
- TRX
Thioredoxin
- Zm
Zea mays
Authors’ contributions
The work presented here was carried out in collaboration among all authors. MAK carried out the computational analysis, plant culture and sample preparation, performed RT-PCR and real-time PCR, analyzed the data and drafted the manuscript. JIP and UKN collected primary data regarding genes and UKN revised the manuscript and made necessary corrections for giving its final shape. HTK and GS designed the stress experiments and cultured the plants and gave stress treatments to the inbred lines ‘Chiifu’ and ‘Kenshin’. MKB and JIP perform in silico analysis. ISN designed and participated in all the experiments and assisted in improving the technical sites of the project. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The seeds of B. rapa inbred lines ‘Chiifu’ and ‘Kenshin’ used in this study was provided by YoonKang Hur, Department of Biology, College of Biological Sciences and Biotechnology, Chungnam National University, Daejeon, South Korea. The corresponding author of this manuscript has bilateral collaboration at personal level. The plant materials are maintained following the institutional guidelines of department of Horticulture, Sunchon National University, South Korea.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4277-2) contains supplementary material, which is available to authorized users.
Contributor Information
Md. Abdul Kayum, Email: kayumagb@gmail.com.
Jong-In Park, Email: jipark@sunchon.ac.kr.
Ujjal Kumar Nath, Email: ujjalnath@gmail.com.
Gopal Saha, Email: saha.18@gmail.com.
Manosh Kumar Biswas, Email: manosh24@yahoo.com.
Hoy-Taek Kim, Email: htkim@sunchon.ac.kr.
Ill-Sup Nou, Phone: +82-61-750-3249, Email: nis@sunchon.ac.kr.
References
- 1.Hayano T, Hirose M, Kikuchi M. Protein disulfide isomerase mutant lacking its isomerase activity accelerates folding in the cell. FEBS Lett. 1995;377:505–511. doi: 10.1016/0014-5793(95)01410-1. [DOI] [PubMed] [Google Scholar]
- 2.Venetianer P, Straub FB. The enzymatic reactivation of reduced ribonuclease. Biochim Biophys Acta. 1963;67:166–168. doi: 10.1016/0926-6569(63)90223-2. [DOI] [PubMed] [Google Scholar]
- 3.Anfinsen C. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 4.Hatahet F, Ruddock LW. Substrate recognition by the protein disulfide isomerases. FEBS J. 2007;274(20):5223–5234. doi: 10.1111/j.1742-4658.2007.06058.x. [DOI] [PubMed] [Google Scholar]
- 5.Gruber CW, Cemažar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Kemmink J, Darby NJ, Dijkstra K, Nilges M, Creighton TE. The folding catalyst protein disulfide isomerase is constructed of active and inactive thioredoxin modules. Curr Biol. 1997;7:239–245. doi: 10.1016/S0960-9822(06)00119-9. [DOI] [PubMed] [Google Scholar]
- 7.Kevin S. Helix-Turn-Helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem Sci. 1989;14(4):137–140. doi: 10.1016/0968-0004(89)90145-X. [DOI] [PubMed] [Google Scholar]
- 8.Denecke J, DeRycke R, Botterman J. Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 1992;11:2345–2355. doi: 10.1002/j.1460-2075.1992.tb05294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houston NL, Fan CZ, Xiang QY, Schulze JM, Jung R, Boston RS. Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol. 2005;137:762–778. doi: 10.1104/pp.104.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari DM, Soling HD. The protein disulphide-isomerase family: unravelling a string of folds. J Biol Chem. 1999;339:1–10. [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki K, Kamauchi S, Wadahama H, Ishimoto M, Kawada T, Urade R. Molecular cloning and characterization of soybean protein disulfide isomerase family proteins with nonclassic active center motifs. FEBS J. 2009;276:4130–4141. doi: 10.1111/j.1742-4658.2009.07123.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang HZ, Leonor C, Boavida MR, McCormick S. Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis Thaliana. Plant Cell. 2008;20:3300–3311. doi: 10.1105/tpc.108.062919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trebitsh T, Levitan A, Sofer A, Danon A. Translation of chloroplast psbA mRNA is modulated in the light by counter acting oxidizing and reducing activities. Mol Cell Biol. 2000;20:1116–1123. doi: 10.1128/MCB.20.4.1116-1123.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d’Aloisio E, Paolacci AR, Dhanapal AP, Tanzarella OA, Porceddu E, Ciaffi M. The protein disulfide isomerase gene family in bread wheat (T. Aestivum L.) BMC Plant Biol. 2010;10:101. doi: 10.1186/1471-2229-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C, Luo N, He M, Chen G, Zhu J, Yin G, Li X, Hu Y, Li J, Yan Y. Molecular characterization and expression profiling of the protein disulfide Isomerase gene family in Brachypodiumdistachyon L. PLoS One. 2014;9(4):e94704. doi: 10.1371/journal.pone.0094704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YH, Wang XT, Shi YS, Huang YQ, Song YC, Wang TY, Li Y. Expression and characterization of protein disulfide isomerases in maize (Zea Mays L.) Chinese J Biochem Mol Biol. 2009;25:229–234. doi: 10.1016/j.cbpb.2008.12.017. [DOI] [Google Scholar]
- 17.Han HC, Khurana N, Tyagi AK, Khurana JP, Khurana P. Identification and characterization of high temperature stress responsive genes in bread wheat (Triticumaestivum L.) and their regulation at various stages of development. Plant Mol Biol. 2011;75:35–51. doi: 10.1007/s11103-010-9702-8. [DOI] [PubMed] [Google Scholar]
- 18.Hatahet F, Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 19.Stolf BS, Smyrnias I, Lopes LR, Vendramin A, Goto H, Laurindo FR, Shah AM, Santos CX. Protein disulfide isomerase and host-pathogen interaction. Sci World J. 2011;11:1749–1761. doi: 10.1100/2011/289182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11(1):136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song XM, Huang ZN, Duan WK, Ren J, Liu TK, Li Y, et al. Genome-wide analysis of the HLH transcription factor family in Chinese cabbage (Brassica Rapa Ssp. Pekinensis) Mol Genet Genomics. 2014;289(1):77–91. doi: 10.1007/s00438-013-0791-3. [DOI] [PubMed] [Google Scholar]
- 22.Bancroft I. Duplicate and diverge: the evolution of plant genome microstructure. Trends Genet Tig. 2001;17(2):89–93. doi: 10.1016/S0168-9525(00)02179-X. [DOI] [PubMed] [Google Scholar]
- 23.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 24.Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung HJ, Dong X, Park JI, Thamilarasan SK, Lee SS, Kim YK, Lim YP, Nou IS, Hur Y. Genome-wide transcriptome analysis of two contrasting Brassica Rapa doubled haploid lines under cold-stresses using Br135K oligomeric chip. PLoS One. 2014;9(8):e106069. doi: 10.1371/journal.pone.0106069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Mayfield PS. Protein disulfide Isomerase as a regulator of chloroplast translational activation. Science. 1997;278(5345):195421957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- 28.Trebitsh T, Meiri E, Ostersetzer O, Adam Z, Danon A. The protein disulfide isomerase2like RB60 is partitioned between stroma and thylakoids Chlamydomonas Reinhardtii chloroplasts. J Biol Chem. 2001;276(3):456424569. doi: 10.1074/jbc.M005950200. [DOI] [PubMed] [Google Scholar]
- 29.Liu JX, Howell SH. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell. 2010;22:2930–2942. doi: 10.1105/tpc.110.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu DP, David AC. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis Thaliana. Mol Genet Genomics. 2008;280:199–210. doi: 10.1007/s00438-008-0356-z. [DOI] [PubMed] [Google Scholar]
- 31.Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. The reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 32.Sakamoto H, Matsuda O, Iba K. ITN1, a novel gene encoding an ankyrinrepeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis Thaliana. Plant J. 2008;56:411–422. doi: 10.1111/j.1365-313X.2008.03614.x. [DOI] [PubMed] [Google Scholar]
- 33.Gilbert HF. Protein disulfide isomerase and assisted protein folding. J Biol Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9(5):244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Ortbauer M. Abiotic Stress Adaptation: Protein Folding Stability and Dynamics. INTECH 2013; 10.5772/53129.
- 36.Faheem M, Li Y, Arshad M, Jiangyue C, Jia Z, et al. A disulphide isomerase gene (PDI-V) from Haynaldia villosa contributes to powdery mildew resistance in common wheat. Sci Rep. 2016;6:24227. doi: 10.1038/srep24227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan. 2007;29:1023e1026. [PubMed] [Google Scholar]
- 39.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 40.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong X, Lv W, Jiang S, Zhang D, Cai G, Pan J, Li D. Genomewide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics. 2013;14:433. doi: 10.1186/1471-2164-14-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 43.Tang J, Wang F, Hou XL, Wang Z, Huang ZN. Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassica Rapa ssp.pekinensis ) reveals collinearity and their expression patterns under Abiotic and biotic stresses. Plant Mol Bio Rep. 2014;32(4):781–795. doi: 10.1007/s11105-013-0672-2. [DOI] [Google Scholar]
- 44.Koch MA, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae) Mol Biol Evol. 2000;17(10):1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- 45.Cheng F, Mandáková T, Wu J, Xie Q, Lysak MA, Wang X. Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell Online. 2013;25:1541–1554. doi: 10.1105/tpc.113.110486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schranz ME, Lysak MA, Mitchell-Olds T. The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant sci. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Kayum MA, Jung HJ, Park JI, Ahmed NU, Saha G, Yang TJ, Nou IS. Identification and expression analysis of WRKY family genes under biotic and abotic stresses in Brassica rapa. Mol Genet Genomics. 2014;290(1):79–95. doi: 10.1007/s00438-014-0898-1. [DOI] [PubMed] [Google Scholar]
- 48.Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;9(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed NU, Park JI, Jung HJ, Seo M, Kumar TS, Lee I, Nou IS. Identification and characterization of stress resistance related genes of Brassica rapa. Biotechnol Lett. 2012;34:979–987. doi: 10.1007/s10529-012-0860-4. [DOI] [PubMed] [Google Scholar]
- 50.Ahmed NU, Park JI, Jung HJ, Kang KK, Lim YP, Hur Y, Nou IS. Molecular characterization of thaumatin family genes related to stresses in Brassica rapa. Sci Hortic. 2013;152:26–34. doi: 10.1016/j.scienta.2013.01.007. [DOI] [Google Scholar]
- 51.Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, Korff MV, Varshney RK, Graner A, Valkoun J. Differentially expressed genes between drought-tolerant and drought sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot. 2009;60:3531–3544. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genomic structures of BrPDI genes. Solid green boxes and red lines indicate exons and introns, respectively. The bottom scale indicates length of exons and introns. Figure S2. Schematic representation of motif compositions in the BrPDI protein sequences. Different motifs logo, numbered 1–10, motif are displayed in different colored boxes. The names of all members are displayed on the left-hand side. (PPT 932 kb)
Putative cis-elements, more than 6 bp, were identified in 32 BrPDI genes in Chinese cabbage. Table S2. A total of 76 PDI and PDIL genes name and accession numbers from 4 species used for constructing phylogenetic tree, including 32 from Brassica rapa,(Br) 21 from Arabidopsis thaliana (At),11 from Brachypodium distachyon (Bd) and 12 from Zea mays (Zm). Table S3. Primers for RT-PCR andreal-time PCR analysis of 32 BrPDI genes (DOC 472 kb)
Data Availability Statement
We declare that the dataset(s) supporting the conclusions of this article are included within the article (and its additional file(s)).
The complete raw microarray data have been deposited in the Omics database of NABIC (http://nabic.rda.go.kr) as enrolled numbers NC-0024-000001 and NC-0024-000002. Phylogenetic data have been deposited in the TreeBASE as submission ID 21739.


