Abstract
The transcription factor zinc finger homeobox 3 (ZFHX3) plays a key role in coupling intracellular transcriptional-translational oscillations with intercellular synchrony in mouse suprachiasmatic nucleus (SCN). However, like many key players in central nervous system function, ZFHX3 serves an important role in neurulation and neuronal terminal differentiation while retaining discrete additional functions in the adult SCN. Recently, using a dominant missense mutation in mouse Zfhx3, we established that this gene can modify circadian period and sleep in adult animals. Nevertheless, we were still concerned that the neurodevelopmental consequences of ZFHX3 dysfunction in this mutant may interfere with, or confound, its critical adult-specific roles in SCN circadian function. To circumvent the developmental consequences of Zfhx3 deletion, we crossed a conditional null Zfhx3 mutant to an inducible, ubiquitously expressed Cre line (B6.Cg-Tg(UBC-cre/ERT2)1Ejb/J). This enabled us to assess circadian behavior in the same adult animals both before and after Cre-mediated excision of the critical Zfhx3 exons using tamoxifen treatment. Remarkably, we found a strong and significant alteration in circadian behavior in tamoxifen-treated homozygous animals with no phenotypic changes in heterozygous or control animals. Cre-mediated excision of Zfhx3 critical exons in adult animals resulted in shortening of the period of wheel-running in constant darkness by more than 1 h in the majority of homozygotes while, in 30% of animals, excision resulted in complete behavioral arrhythmicity. In addition, we found that homozygous animals reentrain almost immediately to 6-h phase advances in the light-dark cycle. No additional overt phenotypic changes were evident in treated homozygous animals. These findings confirm a sustained and significant role for ZFHX3 in maintaining rhythmicity in the adult mammalian circadian system.
Keywords: conditional mutagenesis, suprachiasmatic nucleus, intracellular coupling, period, neuropeptide
In mammals, circadian oscillations are evident in all tissues where these oscillations are believed to be directed by a well-characterized, cell autonomous transcription-translation feedback loop (TTFL). Briefly, this comprises the period (PER) and cryptochrome (CRY) proteins that inhibit their own transcription via inhibition of regulatory CLOCK and BMAL1 proteins. This TTFL also directly regulates the expression of numerous additional genes that are cell- or tissue-specific and mediates tissue-specific clock-relevant functions (Mohawk et al., 2012). The ability of the whole organism to synchronize all of these cell or tissue oscillations is not fully understood, but the master oscillator residing in the suprachiasmatic nucleus (SCN) of the hypothalamus is known to play a crucial role. The SCN maintains an intercellular synchrony of cellular oscillations through the coordinated activities of numerous neurotransmitters acting as coupling factors, including vasoactive intestinal peptide (VIP), arginine vasopressin (AVP), and γ-aminobutyric acid (GABA) (Herzog et al., 2017). Consequently, robust signals are relayed to other cells and tissues through orchestrated neural and humoral signals. These activities and signals are believed to be aligned to the SCN intracellular TTFL, and numerous candidate mechanisms have been proposed.
Recent studies (Parsons et al., 2015) have highlighted the key role of the transcription factor zinc finger homeobox 3 (Zfhx3) in coupling intracellular TTFL oscillations with intercellular synchrony in mouse SCN. Although Zfhx3 is not expressed at high levels throughout adult brain, Zfhx3 expression is highly enriched in adult SCN. Moreover, a dominant missense mutation in Zfhx3 (short circuit, Sci) shortens circadian wheel-running period by almost 1 h and SCN molecular oscillations by a similar margin. ZFHX3 regulates gene expression in SCN through its interaction with conserved AT motifs in promoters of target genes including clock-related genes and multiple SCN-enriched neuropeptides and neuropeptide receptors. In Zfhx3Sci/+ mutants, expression of many of these genes is affected. However, we wanted to establish further whether null mutations in Zfhx3 had a similar effect on circadian function in vivo and whether, by circumventing developmental effects of Zfhx3 function, we could still establish an effect on the circadian system.
In determining the significance of circadian phenotypes in adult mutant mice, it is important to consider whether phenotypes might be secondary consequences of primary developmental anomalies. ZFHX3 plays important roles in multiple systems in developing and adult tissues, and the pleiotropic nature of mutations in Zfhx3 testifies to this. The Sci mutation is homozygous perinatal lethal (Parsons et al., 2015), while a constitutive knockout of Zfhx3 can result in preweaning sublethality in heterozygotes (Sun et al., 2012). These observations confirm the nature of the Sci allele, already suggested (Parsons et al., 2015), as a gain of function, most probably dominant negative, allele. Moreover, the sublethality in heterozygous constitutive null mice suggests a haploinsufficiency in knockout mice. ZFHX3 plays vital roles in development and in other systems including mammary gland (Zhao et al., 2016) and the nervous system (Jung et al., 2005). Additionally, the transcription factor is known to play a role in tumorigenesis (Cho et al., 2007; Sun et al., 2015) and has been consistently associated with atrial fibrillation (Huang et al., 2015). Moreover, Zfhx3 is expressed in developing mouse SCN from at least E13.5 and may be important in the terminal differentiation of its peptidergic neurons (VanDunk et al., 2011). In comparison, it is notable that other transcription factors expressed in developing SCN have arguable adult circadian regulatory roles. Notably, deletion of the Lim homeodomain transcription factor (Lhx1) has contrasting effects on terminal differentiation and adult circadian function depending on when the gene is deleted in the maturing SCN (Bedont et al., 2014; Hatori et al., 2014).
To circumvent early developmental changes associated with Zfhx3 deletion and to assess its role specifically in adult mice, we used a tamoxifen-inducible transgenic line expressing Cre from the human ubiquitin C promoter (UBC CreERT2) (Ruzankina et al., 2007). Using this approach we demonstrate that ZFHX3 maintains an active role in setting the pace of the circadian clock in adult mice, thus sustaining a key role in the maintenance of stable circadian rhythms independent of any earlier developmental disruptions.
Materials and Methods
Mice
All animal studies were performed under the guidance issued by the Medical Research Council in Responsibility in the Use of Animals for Medical Research (July 1993) and Home Office Project License 30/3206, with local ethical approval. When not being tested, mice were group housed in individually ventilated cages under 12:12 h light-dark conditions with food and water available ad libitum. Floxed Zfhx3 mice, Zfhx3Tm1.1Jtd, were kindly provided by Dr. Jin-Tang Dong (Emory University) (Sun et al., 2012). These mice arrived on a mixed background of 129S6 and C57BL/6J and were subsequently backcrossed onto C57BL/6J to congenicity before being used for experimental crosses. An inducible Cre line expressing UBC-cre (B6.Cg-Tg(UBC-cre/ERT2)1Ejb/J) was obtained from the Jackson Laboratory (Ruzankina et al., 2007). Mice were genotyped for Zfhx3 using a quantitative PCR (qPCR) copy count Taqman assay. Primers were as follows: WT primers: AAGAAGCGATAAGCTAACACCAGG and ACGCCAAAGGTTGAGGAGAATG, probe sequence: TTAAAGGAATTCACGGGGTTAGGGC; mutant primers: GCCATAACTTCGTATAATGTATGCTATACG and ACGCCAAAGGTTGAGGAGAATG, probe sequence: TTATAAGCTTACGGGGTTAGGGCTGT. The mutant assay was designed to target the floxed region of the gene to distinguish between allele types while the copy count assay confirmed that the gene was not deleted prior to tamoxifen dosing. For the UBC-cre line, the same copy count assay was used with the following primers: GGGCTGCAGGTCGACTCT and TCGTTGCATCGACCGGTAATG, probe sequence: AGAGGATCCAGTTAACCTCGAGGGC.
Tamoxifen Dosing
Tamoxifen was administered via oral gavage for 5 days, and a 7-day recovery period was allowed before further testing. A stock concentration of 20 mg/mL tamoxifen solution was prepared by dissolving tamoxifen (Cambridge Bioscience, Cambridge, UK) in corn oil containing 1% ethanol (MP Biomedicals, Leicester, UK). At 8 weeks, mice were dosed daily according to their weights for 5 days; the total dose given over the 5 days was 1 g/kg.
Circadian Phenotyping
Wheel-running activity was performed as described previously (Banks and Nolan, 2011). Briefly, mice were singly housed in cages containing running wheels placed in light-controlled chambers, and wheel-running activity was monitored via ClockLab (Actimetrics, Wilmette, IL). Default settings were used for analysis in ClockLab. Animals were monitored for 5 days in a 12-h light-dark cycle (100-lux light intensity) followed by 12 days in constant darkness. For the jet lag protocol, mice were maintained for 3 days in standard light-dark conditions, after which light onset was abruptly advanced by 6 h, and the new LD schedule was maintained for 14 days. Light onset was then delayed by 6 h, and mice were again maintained for 14 days under these new conditions. Reentrainment to the altered LD cycle was assessed by measuring the difference between lights-off and activity onset. An onset of sustained activity within 30 min following lights-off was used as the cutoff for an individual mouse to be deemed entrained.
Statistics
All data were analyzed by repeated-measures, mixed-model parametric analysis with Bonferroni correction and Dunnett test for multiple testing corrections using InVivoStat (Clark et al., 2012).
Immunofluorescence
Mouse brains were dissected and immediately embedded in molds containing OCT compound (VWR International, Radnor, PA); molds were then floated in isopentane on dry ice to freeze the samples. Next, 12-µm sections were cut on a cryostat and collected onto slides. Slides were fixed in 4% paraformaldehyde for 4 h at 4 °C, washed in phosphate-buffered saline (PBS), and then incubated in 3% hydrogen peroxide in methanol for 30 min at 4 °C before washing with PBS again and then blocking in 5% normal goat serum made up in 0.5% PBS-Triton. Anti-ZFHX3 raised in rabbit (Parsons et al., 2015) was diluted 1:1000 in the blocking solution, and primary incubation was 48 h at 4 °C. Anti-rabbit 488 raised in goat (Abcam, Cambridge, UK) was diluted 1:200 in PBS, and secondary incubation was 2 h at room temperature, following a PBS wash. Slides were coverslipped using ProLong Gold Antifade mountant with DAPI (Life Technologies, Carlsbad, CA). Sections were imaged using an inverted confocal microscope running Zen software; settings were reused between images, and the “best fit” function was applied to all captured images.
Results
Adult Specific Knockout of Zfhx3 Using an Inducible Cre Line
To generate inducible knockout Zfhx3 animals, tamoxifen inducible cre mice (UBC-Cre) were crossed to Zfhx3 floxed (Zfhx3Flox) mice to produce an initial stock of Zfhx3Flox/+;UBC-Cre+ mice. These animals were subsequently crossed to Zfhx3Flox/+ mice to generate experimental cohorts (Sun et al., 2012). This resulted in progeny of 6 possible genotype combinations: animals that carried neither the Cre nor Zfhx3Flox alleles (Zfhx3+/+;UBC-Cre−), animals heterozygous or homozygous for the Zfhx3Flox allele but not carrying the Cre allele (Zfhx3Flox/+;UBC-Cre− or Zfhx3Flox/Flox;UBC-Cre−), animals that were hemizygous for the Cre allele but did not carry the Zfhx3 allele (Zfhx3+/+;UBC-Cre+), and animals that were hemizygous for the Cre allele and were either heterozygous or homozygous for the Zfhx3Flox allele (Zfhx3Flox/+;UBC-Cre+ or Zfhx3Flox/Flox;UBC-Cre+). The final group would result in heterozygous or homozygous deletion of critical floxed Zfhx3 exons following tamoxifen administration, thus creating a temporal null allele.
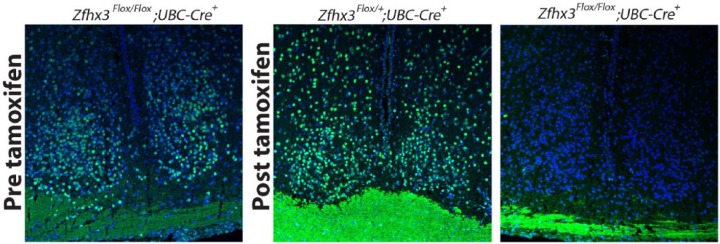
Immunofluorescence was undertaken on SCN sections from these animals to validate protein loss following tamoxifen treatment (Fig. 1). Prior to tamoxifen treatment, there was no obvious decrease in ZFHX3 staining in homozygous mutants compared with control SCN. Following treatment, however, ZFHX3 staining was effectively absent in SCN. SCN sections from control genotypes showed no evident decrease in ZFHX3 staining following tamoxifen treatment.
Figure 1.
ZFHX3 immunofluorescence in representative SCN sections (n = 3 for each genotype); although detectable prior to tamoxifen treatment, ZFHX3 immunopositive cells are undetectable following treatment in Zfhx3Flox/Flox;UBC-Cre+ mice, while staining remains robust in SCN of Zfhx3Flox/+;UBC-Cre+, used here as a representative control. Blue, DAPI; green, ZFHX3.
Tamoxifen-mediated Deletion of Zfhx3 in Adult Mice Shortens τDD
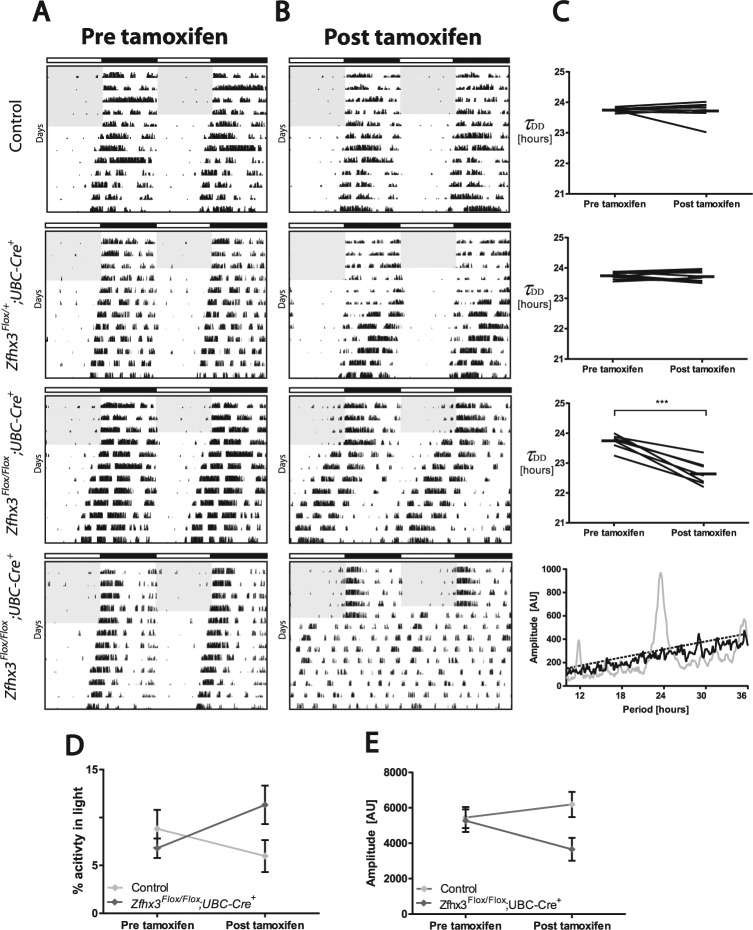
The effect of tamoxifen treatment on all groups was assessed using repeated-measures parametric analysis with tamoxifen treatment as a repeated-measures variable. All genotypes underwent circadian analysis (data shown in Table 1); however, Zfhx3Flox/Flox;UBC-Cre− mice have been used as the representative “control” group in figures for uniformity and clarity. Ten Zfhx3Flox/Flox;UBC-Cre+ mice and all controls were screened for changes in circadian parameters both before and after tamoxifen treatment. We found no significant group differences in any circadian parameters analyzed prior to tamoxifen treatment (Table 1). Following tamoxifen treatment, 7 out of 10 Zfhx3Flox/Flox;UBC-Cre+ mice displayed a significant shortening of τDD (F4,41 = 28.4, p < 0.0001). Multiple test correction of pairwise comparisons between the experimental groups confirmed a significant shortening of τDD in treated Zfhx3Flox/Flox;UBC-Cre+ mice compared with all other genotypes (p < 0.005 for Zfhx3Flox/Flox;UBC-Cre+ vs. all other genotypes). Furthermore, only Zfhx3Flox/Flox;UBC-Cre+ animals displayed a significant change in τDD following tamoxifen treatment (Zfhx3Flox/Flox;UBC-Cre+ circadian period: before vs. after tamoxifen treatment; p = 0.0045). Percentage activity in the light phase for Zfhx3Flox/Flox;UBC-Cre+ animals (Fig. 2D) showed an increase following tamoxifen treatment. Amplitude was also decreased in Zfhx3Flox/Flox;UBC-Cre+ animals (Fig. 2E); however, neither of these parameters remained statistically significant following multiple testing correction.
Table 1.
Circadian behavioral data for all genotypes tested.
| Circadian Parameter and Genotype | n | Before Tamoxifen | After Tamoxifen | p Value | Adjusted p Value |
|---|---|---|---|---|---|
| τDD (hours) | |||||
| Zfhx3Flox/Flox;UBC-Cre+ | 7 | 23.72 ± 0.09 | 22.66 ± 0.15 | 0.0001 | 0.0045 |
| Zfhx3Flox/+;UBC-Cre+ | 11 | 23.76 ± 0.03 | 23.74 ± 0.05 | 0.7968 | 1.0000 |
| Zfhx3Flox/Flox;UBC-Cre− | 8 | 23.75 ± 0.02 | 23.73 ± 0.11 | 0.8102 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre− | 10 | 23.76 ± 0.04 | 23.78 ± 0.04 | 0.8155 | 1.0000 |
| Zfhx3+/+;UBC-Cre+ | 10 | 23.69 ± 0.09 | 23.74 ± 0.04 | 0.4852 | 1.0000 |
| Amplitude (AU) | |||||
| Zfhx3Flox/Flox;UBC-Cre+ | 7 | 5272 ± 636 | 3658 ± 646 | 0.0034 | 0.1530 |
| Zfhx3Flox/+;UBC-Cre+ | 11 | 5284 ± 361 | 5568 ± 541 | 0.4964 | 1.0000 |
| Zfhx3Flox/Flox;UBC-Cre− | 8 | 5405 ± 517 | 6165 ± 614 | 0.1459 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre− | 10 | 4218 ± 320 | 4830 ± 320 | 0.1666 | 1.0000 |
| Zfhx3+/+;UBC-Cre+ | 10 | 4571 ± 423 | 5789 ± 656 | 0.0076 | 0.3420 |
| Activity in light phase (%) | |||||
| Zfhx3Flox/Flox;UBC-Cre+ | 10 | 6.79 ± 1.02 | 11.32 ± 2.01 | 0.0135 | 0.6075 |
| Zfhx3Flox/+;UBC-Cre+ | 11 | 4.51 ± 1.04 | 5.25 ± 1.50 | 0.8215 | 1.0000 |
| Zfhx3Flox/Flox;UBC-Cre− | 10 | 8.81 ± 1.99 | 5.98 ± 1.67 | 0.1141 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre− | 10 | 10.17 ± 1.99 | 6.10 ± 1.03 | 0.0253 | 1.0000 |
| Zfhx3+/+;UBC-Cre+ | 10 | 6.46 ± 1.32 | 3.69 ± 0.98 | 0.1236 | 1.0000 |
| ϕ (degrees) | |||||
| Zfhx3Flox/Flox;UBC-Cre+ | 10 | 179 ± 0.9 | 169 ± 7.1 | 0.0055 | 0.2475 |
| Zfhx3Flox/+;UBC-Cre+ | 11 | 175 ± 3.7 | 172 ± 7.5 | 0.3587 | 1.0000 |
| Zfhx3Flox/Flox;UBC-Cre− | 9 | 175 ± 5.1 | 176 ± 4.0 | 0.9868 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre− | 10 | 179 ± 0.7 | 181 ± 0.5 | 0.6193 | 1.0000 |
| Zfhx3+/+;UBC-Cre+ | 10 | 180 ± 0.7 | 181 ± 0.6 | 0.8466 | 1.0000 |
| αLD (hours) | |||||
| Zfhx3Flox/Flox;UBC-Cre+ | 10 | 12.09 ± 0.14 | 11.59 ± 0.52 | 0.0796 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre+ | 11 | 12.10 ± 0.16 | 11.85 ± 0.14 | 0.3470 | 1.0000 |
| Zfhx3Flox/Flox;UBC-Cre− | 9 | 12.22 ± 0.22 | 12.03 ± 0.13 | 0.4906 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre− | 10 | 12.11 ± 0.17 | 12.07 ± 0.09 | 0.8707 | 1.0000 |
| Zfhx3+/+;UBC-Cre+ | 10 | 11.93 ± 0.12 | 11.86 ± 0.10 | 0.8508 | 1.0000 |
| αDD (hours) | |||||
| Zfhx3Flox/Flox;UBC-Cre+ | 10 | 12.20 ± 0.82 | 10.73 ± 1.26 | 0.0229 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre+ | 11 | 12.84 ± 0.27 | 13.22 ± 0.23 | 0.5089 | 1.0000 |
| Zfhx3Flox/Flox;UBC-Cre− | 8 | 13.64 ± 0.39 | 13.08 ± 0.39 | 0.5556 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre− | 10 | 12.67 ± 0.24 | 12.77 ± 0.21 | 0.8686 | 1.0000 |
| Zfhx3+/+;UBC-Cre+ | 10 | 12.68 ± 0.19 | 12.27 ± 0.18 | 0.4978 | 1.0000 |
τDD = period in constant darkness; AU = arbitrary units; ϕ = phase angle of entrainment in light-dark (180° denotes a phase angle consistent with a ZT12 activity onset); αLD = activity duration in light-dark; αDD = activity duration in constant darkness. Both unadjusted and adjusted p values are shown for pairwise comparisons of group data before and after tamoxifen treatment; Bonferroni adjustment was used for correction of multiple comparisons. Units of measurement for parameters refer to measurements before and after tamoxifen administration, expressed as mean ± standard error of the mean.
Figure 2.
Representative double-plotted actograms for Zfhx3Flox/Flox;UBC-Cre+, Zfhx3Flox/+;UBC-Cre+, and control (Zfhx3Flox/Flox;UBC-Cre−) mice (A) before and (B) after tamoxifen treatment; shading denotes lights-on. Animals are in constant darkness from day 5. (C) Change in period in constant darkness following tamoxifen treatment for genotypes shown (control, n = 8; Zfhx3Flox/+;UBC-Cre+, n = 11; Zfhx3Flox/Flox;UBC-Cre+, n = 7) (***p < 0.001). A representative chi-square periodogram in constant darkness is shown for Zfhx3Flox/Flox;UBC-Cre+ mice that became arrhythmic following tamoxifen treatment (n = 3); gray line denotes periodogram before tamoxifen, black line denotes periodogram after tamoxifen treatment, and dashed line denotes chi-square line of significance. (D) Change in percentage activity in the light phase following tamoxifen treatment for control (n = 8) and Zfhx3Flox/Flox;UBC-Cre+ mice (n = 7). (E) Change in amplitude following tamoxifen treatment for control (n = 8) and Zfhx3Flox/Flox;UBC-Cre+ mice (n = 7).
In addition to the period shortening seen in the 7 homozygous mutants described, 3 Zfhx3Flox/Flox;UBC-Cre+ mice became arrhythmic in constant darkness (DD) following tamoxifen treatment (Fig. 2, B and C) and so could not be included in the statistical analysis of τDD. A comparison of chi-square periodograms in these animals before and after tamoxifen treatment showed a loss of significant peak amplitude after tamoxifen (Fig. 2C). Zfhx3Flox/+;UBC-Cre+ mice and all control genotypes screened showed no significant changes in other circadian parameters following tamoxifen treatment. The interaction between genotype and tamoxifen treatment showed no significant differences in the phase angle of entrainment (ϕ) (F4,45 = 1.9, p = 0.1266) or in alpha (activity duration) in LD (αLD) and DD (αDD) (LD: F4,45 = 0.43, p = 0.7878; DD: F4,41 = 1.39, p = 0.2558), suggesting that these parameters were unaffected by disruption of Zfhx3. To confirm this statistical analysis, a Dunnett test for multiple corrections was used to compare Zfhx3Flox/Flox;UBC-Cre+ mice against all other genotypes. This analysis found no additional significant differences.
Activity Pattern Relative to LD Is Altered in Zfhx3Flox/Flox;UBC-Cre+ Mutants
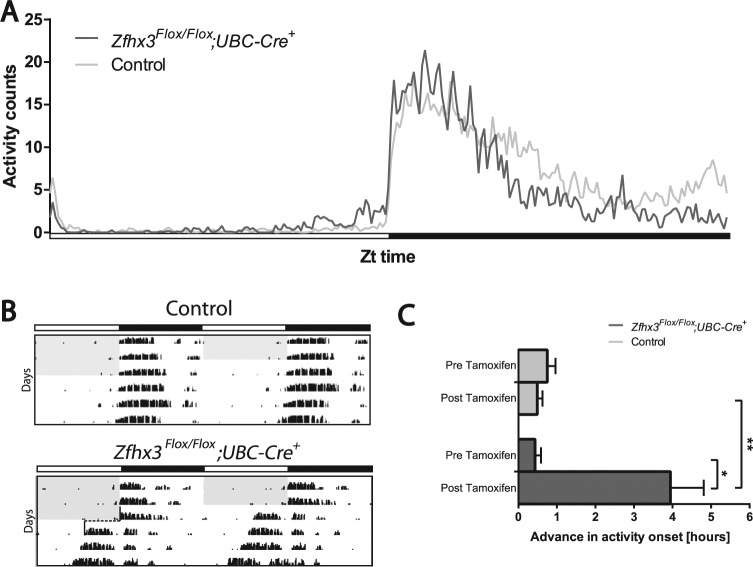
To analyze whether the daily pattern of activity over 24 h differed in tamoxifen-treated Zfhx3Flox/Flox;UBC-Cre+ animals compared with controls, average wheel-running activity was measured in 6-min time bins over the 24-h LD cycle. Following tamoxifen treatment, all Zfhx3Flox/Flox;UBC-Cre+ mice showed an increased proportion of activity in the light phase of a standard 12:12 LD cycle. The interaction between this and tamoxifen treatment was found to be significant (F4,446 = 3.91, p = 0.0081); however, significant changes did not survive multiple testing correction. Investigation of total activity over 24 h (Fig. 3A) revealed that tamoxifen-treated Zfhx3Flox/Flox;UBC-Cre+ animals showed a significantly different average pattern of activity compared with controls (F240,4079 = 2.03, p = 0.0001). This difference was clear at 2 stages of the dark phase: First, during the early period of the dark phase, Zfhx3Flox/Flox;UBC-Cre+ animals showed an elevated activity compared with controls; second, during the late period of the dark phase, Zfhx3Flox/Flox;UBC-Cre+ animals showed decreased activity compared with controls. Moreover, although not identified as a significant phase angle of entrainment effect, mutants appeared to show some increase in activity relative to the onset of the dark phase compared with controls.
Figure 3.
(A) Average activity profile for Zfhx3Flox/Flox;UBC-Cre+ mice (dark gray line) and control (Zfhx3Flox/Flox;UBC-Cre−) mice (light gray line) after tamoxifen treatment over 12:12 LD cycle (n = 10). (B) Representative actograms showing advance in activity onset upon release into DD for control and Zfhx3Flox/Flox;UBC-Cre mice after tamoxifen treatment. (C) Advance in activity onset upon release into DD conditions. Difference in activity onset from last day of LD before (upper bars) and after (lower bars) tamoxifen for Zfhx3Flox/Flox;UBC-Cre+ mice (dark gray) and control mice (light gray) (n = 6). *p < 0.05, **p < 0.01.
Zfhx3Flox/Flox;UBC-Cre+ Mutants Show Rapid Reentrainment to an Advance in the LD cycle
Upon release into DD, all tamoxifen-treated Zfhx3Flox/Flox;UBC-Cre+ mice that maintained stable rhythms showed an initial advance in activity onset compared with this measurement prior to tamoxifen treatment (p = 0.01) or in controls (p = 0.002). This advance in activity was, on average, around 4 h greater than the modest advances typically seen upon release into DD from LD in controls (Fig. 3, B and C). This apparent strong advance in activity onset prompted us to investigate the behavior of these mutants in jet lag protocols. To analyze reentrainment to a shift in the LD cycle, animals were subjected to an abrupt 6-h advance, and also delay, in lights-on. The number of days taken to reentrain to the new LD cycle was measured for all groups following tamoxifen treatment (data shown in Table 2). Since a defective clock may cause entrainment effects that are difficult to interpret, Zfhx3Flox/Flox;UBC-Cre+ mice that were arrhythmic in constant darkness were excluded from this analysis. However, it is notable that when tested, these arrhythmic animals did resynchronize to the new LD cycle in a similar manner to rhythmic animals (number of days to reentrain or resynchronize: rhythmic animals = 3.0 ± 0.52; arrhythmic animals = 2.0 ± 0.57; p = 0.28).
Table 2.
Days to reentrain in jet lag protocol for all genotypes tested.
| Protocol and Genotype | n | Days to Reentrain | p value |
|---|---|---|---|
| Advancing LD | |||
| Zfhx3Flox/Flox;UBC-Cre+ | 6 | 3.0 ± 0.5 | n/a |
| Zfhx3Flox/+;UBC-Cre+ | 10 | 5.6 ± 0.5 | 0.0471 |
| Zfhx3Flox/Flox;UBC-Cre− | 9 | 4.9 ± 0.7 | 0.097 |
| Zfhx3Flox/+;UBC-Cre− | 10 | 6.6 ± 0.5 | 0.0109 |
| Zfhx3+/+;UBC-Cre+ | 10 | 6.3 ± 0.7 | 0.0146 |
| Delaying LD | |||
| Zfhx3Flox/Flox;UBC-Cre+ | 5 | 3.0 ± 0.8 | n/a |
| Zfhx3Flox/+;UBC-Cre+ | 6 | 2.8 ± 0.2 | 1.0000 |
| Zfhx3Flox/Flox;UBC-Cre− | 4 | 3.8 ± 0.6 | 1.0000 |
| Zfhx3Flox/+;UBC-Cre− | 8 | 3.1 ± 0.3 | 1.0000 |
| Zfhx3+/+;UBC-Cre+ | 4 | 3.3 ± 0.5 | 1.0000 |
The table provides the number of days (mean ± standard error of the mean) taken to reentrain to a 6-h advance and a 6-h delay in the light-dark cycle for all genotypes tested in the jet lag protocol. All data shown are for animals after tamoxifen treatment. P values shown are for that genotype compared with Zfhx3Flox/Flox;UBC-Cre+ and Bonferroni corrected for multiple comparison.
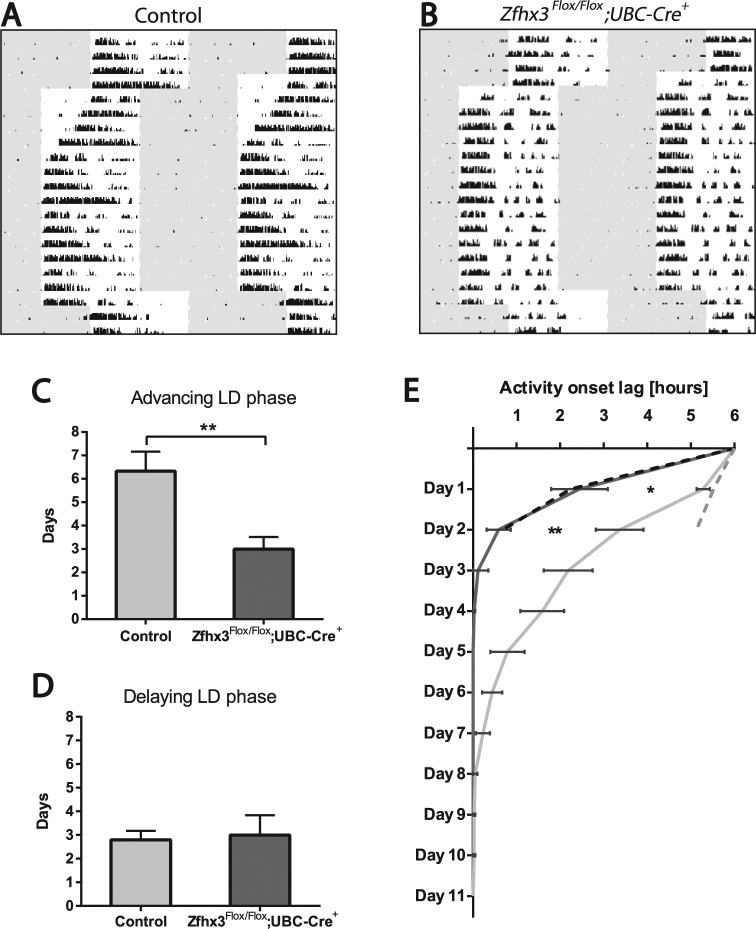
No significant difference was noted in response of activity onsets to a 6-h delay in the LD cycle between genotypes (F4,214 = 0.37, p = 0.825). Similarly, changes in activity offsets were not significantly different. However, there was a marked, significant difference between Zfhx3Flox/Flox;UBC-Cre+ mice and controls in response to a 6-h advance (F4,39 = 3.94, p = 0.0088), with mutants reentraining on average twice as fast as control mice (Fig. 4). Average activity onsets in these mice advanced by around 4 h on day 1 and then by almost a further 2 h on day 2. Pairwise comparisons following multiple test correction demonstrated that Zfhx3Flox/Flox;UBC-Cre+ animals had an earlier onset of activity for the first 2 days following the phase advance (p < 0.05 and p < 0.01 for Zfhx3Flox/Flox;UBC-Cre+ for days 1 and 2, respectively, following phase advance). There were no other significant differences between genotypes throughout the time course of the experiment. Since the reduction in reentrainment time demonstrated by Zfhx3Flox/Flox;UBC-Cre+ animals could be due to the combined effects of a faster running clock and of negative masking, we compared the advances in activity onsets that occurred on the first 2 days of release into DD with those that occurred on the first 2 days of the phase advance protocol. These advances were comparable in Zfhx3Flox/Flox;UBC-Cre+ animals, suggesting that the effects of negative masking upon release into constant darkness can fully account for differences in reentrainment in the phase advance protocol.
Figure 4.
Representative double-plotted actograms showing reentrainment of wheel-running activity in response to 6-h phase advances or phase delays for (A) control (Zfhx3Flox/Flox;UBC-Cre−) mice and (B) Zfhx3Flox/Flox;UBC-Cre+ mice. (C, D) Average number of days taken for control (light gray) and Zfhx3Flox/Flox;UBC-Cre+ (dark gray) mice to reentrain to (C) a 6-h advance in LD and (D) a 6-h delay in LD. (E) Average activity onset for control (light gray, n = 9) and Zfhx3Flox/Flox;UBC-Cre+ (dark gray, n = 6) mice following an advance in LD cycle of 6 h. All jet lag experiments were undertaken after tamoxifen treatment. For reference, dashed lines show respective advances in activity onset over the first 2 days following release into DD for control (gray) and mutant (black) mice. *p < 0.05, **p < 0.01.
Discussion
The transcription factor ZFHX3 has a range of crucial functions in cell cycle regulation, development, and cellular differentiation (Ishii et al., 2003; Jung et al., 2005; Sun et al., 2012). In addition, recent studies have demonstrated that the protein modulates circadian activity in mice by regulating the transcription of many key genes in the SCN, including neuropeptides and their receptors (Parsons et al., 2015). In instances where circadian gene mutations have pleiotropic effects, it is often difficult to establish which mutant phenotypes are consequences of developmental defects and which are specifically due to circadian rhythm disruption in the adult. By using a tamoxifen-inducible Cre line, we have confirmed that ZFHX3 has a sustained function in regulating adult circadian rhythms that is distinct from any of its developmental functions.
Genetic pleiotropy is evident in many clock components, affecting diverse functions from neural development and differentiation to metabolic homeostasis and cell cycle regulation. Inducible and conditional knockout tools enable one to study specific gene effects in the adult while still maintaining the gene’s developmental function. Surprisingly, such techniques are not routinely used in the circadian field. Constitutive mutations in one of the core circadian genes, Rora, for example, result in ataxia and perinatal lethality (Brown and Moore, 2012; Gold et al., 2007), while conditional mutagenesis has not been used to establish this gene’s role in adult rhythm maintenance. In a study contrasting developmental and adult functions of Bmal1 using inducible conditional knockout mice, many of the phenotypes previously attributed to circadian disruption of Bmal1 could be reassigned to developmental functions of the gene (Yang et al., 2016). Such studies demonstrate the usefulness of tamoxifen-inducible conditional knockout approaches in dissecting circadian function. Similar studies could be undertaken for other core clock genes such as Per, Cry, and Clock, whose roles in SCN development may have been underestimated or not distinguished from adult circadian rhythm maintenance. Moreover, with inducible Cre driver lines, the ability to use the same animal for data collection before and after tamoxifen treatment allows each animal to act as its own control, eliminating some of the natural variability in circadian parameters observed between animals.
The experimental data reported here reinforce a pivotal function for Zfhx3 in maintaining normal circadian rhythms and a robust circadian clock. It is particularly notable that 30% of the animals displayed behavioral arrhythmicity in DD conditions following adult knockout of Zfhx3. Single gene knockouts capable of inducing arrhythmia are rare (Baggs et al., 2009), with Bmal1 currently being the only known example sufficient to cause arrhythmia in constant conditions (Bunger et al., 2000). The remaining 70% of Zfhx3 mutants described all showed shortening of τDD by around an hour, which is comparable to the effect of the heterozygous Sci mutation in Zfhx3 previously reported (Parsons et al., 2015). These varied effects of gene knockout on behavioral rhythms reflect elements of those seen in both Lhx1 (Bedont et al., 2014; Hatori et al., 2014) and Vip mutants (Aton et al., 2005; Colwell et al., 2003) and may represent combined effects of the mutation on the robustness of SCN intracellular clocks and on maintenance of SCN intercellular synchrony. It is also notable that heterozygous knockout of Zfhx3 in adult mice in this study results in no detectable phenotype. This is in contrast to the reported effects of the Sci/+ mutation and of localized Zfhx3 gene knockdown in SCN (Parsons et al., 2015). Differences here could be attributable to the as-yet undefined, integrated effects of Zfhx3 knockout in other hypothalamic and thalamic brain nuclei, while they might also be related to differential effects of Zfhx3 knockout on dorsal AVP-expressing and ventral VIP-expressing SCN neurons. Similar inconsistencies in SCN-neural and whole-animal behavioral rhythms have been reported in circadian period aftereffects following the entrainment of animals to light-dark cycles that are longer or shorter than 24 h (Aton et al., 2004; Azzi et al., 2017; Molyneux et al., 2008). Further investigations are required to clarify these effects in Zfhx3 mutants. Furthermore, as the Cre driver used in this study has strong expression in all tissues, effects of gene deletion on peripheral clocks would be possible. Despite significant changes to period, other key circadian parameters (such as changes in alpha and phase angle of entrainment) were unaffected following the loss of Zfhx3. While we found no significant effect of genotype and tamoxifen treatment on the phase angle of entrainment, we did see an apparent increase in activity immediately prior to the onset of darkness in adult Zfhx3 knockout animals. Although noticeable, the changes show no significant effect using ClockLab algorithms to determine phase angle of entrainment.
In addition to changes in free-running period, rapid reentrainment to advancing LD cycles was observed in adult Zfhx3 knockout mice. Consistent with the apparent increases in activity in mutants in LD, this effect can be explained by the negative masking in adult Zfhx3 knockout mice. The effect is also reflected in the 4-h phase advances when mutants are released into constant darkness. Conversely, however, no differences were detected in time taken to reentrain to 6-h phase delays as reflected in changes to both activity onsets and offsets. Phase-resetting phenotypes have previously been shown in mice lacking the vasopressin receptors V1a and V1b (Yamaguchi et al., 2013). Our previous data demonstrating that Zfhx3 modulates the expression of a number of neuropeptides, including vasopressin (Parsons et al., 2015), suggested that similar disturbances would be observed in adult Zfhx3 knockout mice. Notably, however, in contrast to adult Zfhx3 knockout mice, V1a and V1b knockout animals showed no changes in circadian period (Yamaguchi et al., 2013). This indicates the complexity of the phenotype when Zfhx3 is deleted in the adult, indicating that its transcriptional effects are mediated through an integration of numerous downstream effector systems and are not restricted to neuropeptidergic effects. This complexity may also underlie why, in contrast with adult Zfhx3 knockout mice, V1a and V1b knockout animals also reentrain faster to a phase-delaying light-dark cycle. It remains to be determined whether combined effects on AVP and VIP peptidergic systems can explain the full effect of the Zfhx3 mutant deficits or whether additional downstream effectors might contribute to this. It is also notable that similar complex effects have been achieved by knocking out the transcription factor Eif4ebp1 (which also modulates neuropeptide expression). Knockout of this transcription factor can lead to a greater resistance to light-induced disruption (Cao et al., 2013).
This study confirms that ZFHX3 plays an active role in maintaining circadian period in adult mice in contrast to its well-characterized developmental functions. Based on aforementioned published data (Parsons et al., 2015) and the new data presented in this publication, it is likely that Zfhx3 is acting as a master regulator of coupling agents in the circadian system (Mohawk et al., 2012) to maintain synchrony in the SCN. Nevertheless, we are only part of the way in resolving ZFHX3’s multiple roles in developing and adult SCN. This is further confounded by the fact that the constitutive null mutant is subviable even in the heterozygous state. To investigate ZFHX3 function further, it will be necessary to investigate the consequences of its limited deletion using a number of spatially restricted and inducible Cre driver lines. Similar approaches have been used in further defining the roles of clock genes and SCN-enriched genes in circadian regulatory processes (Bedont et al., 2014; Hatori et al., 2014; Lee et al., 2015; Mieda et al., 2015; Smyllie et al., 2016). The use of hypothalamic Cre driver lines expressed in differentiating SCN should help to clarify how Zfhx3 disruption from early development might affect circadian behavior in the adult, while the use of inducible Cre lines that specify particular neuronal classes in adult SCN will help to establish how acute ablation of Zfhx3 in specific adult SCN cells can disrupt the clock. Such work will assist in defining whether the mechanisms of ZFHX3-mediated circadian disruption are similar in differentiating and adult SCN neurons.
Acknowledgments
The authors thank Greg Joynson, Tamzin Osbourne, and Adele Austin, MRC Harwell, for their work relating to this project. P.M.N. was supported by the MRC (grant code MC_U142684173) and by the 6th Framework Project EUCLOCK (No. 018741).
Footnotes
Conflict of Interest Statement: The author(s) have no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Aton SJ, Block GD, Tei H, Yamazaki S, Herzog ED. (2004) Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non-24-hour light cycles. J Biol Rhythm 19:198-207. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. (2005) Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 8:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A, Evans JA, Leise T, Myung J, Takumi T, Davidson AJ, Brown SA. (2017) Network dynamics mediate circadian clock plasticity. Neuron 93(2):441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. (2009) Network features of the mammalian circadian clock. PLoS Biol 7:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks GT, Nolan PM. (2011) Assessment of circadian and light-entrainable parameters in mice using wheel-running activity. Curr Protoc Mouse Biol 1:369-381. [DOI] [PubMed] [Google Scholar]
- Bedont JL, LeGates TA, Slat EA, Byerly MS, Wang H, Hu J, Rupp AC, Qian J, Wong GW, Herzog ED. (2014) Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep 7:609-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Moore MW. (2012) The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm Genome 23:632-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Robinson B, Xu H, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, et al. (2013) Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron 79:712-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YG, Song JH, Kim CJ, Lee YS, Kim SY, Nam SW, Lee JY, Park WS. (2007) Genetic alterations of the ATBF1 gene in gastric cancer. Clin Cancer Res 13:4355-4359. [DOI] [PubMed] [Google Scholar]
- Clark RA, Shoaib M, Hewitt KN, Stanford SC, Bate ST. (2012) A comparison of InVivoStat with other statistical software packages for analysis of data generated from animal experiments. J Psychopharmacol 26:1136-1142. [DOI] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. (2003) Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Reg I 285:R939-R949. [DOI] [PubMed] [Google Scholar]
- Gold DA, Gent PM, Hamilton BA. (2007) ROR alpha in genetic control of cerebellum development: 50 staggering years. Brain Res 1140:19-25. [DOI] [PubMed] [Google Scholar]
- Hatori M, Gill S, Mure LS, Goulding M, O’Leary DD, Panda S. (2014) Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. Elife 3:e03357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH. (2017) Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harbor Perspect Biol 9(1). doi: 10.1101/cshperspect.a027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang C, Yao Y, Zuo X, Chen S, Xu C, Zhang H, Lu Q, Chang L, Wang F, et al. (2015) Molecular basis of gene-gene interaction: cyclic cross-regulation of gene expression and post-GWAS gene-gene interaction involved in atrial fibrillation. PLoS Genet 11:e1005393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y, Kawaguchi M, Takagawa K, Oya T, Nogami S, Tamura A, Miura Y, Ido A, Sakata N, Hashimoto-Tamaoki T. (2003) ATBF1-A protein, but not ATBF1-B, is preferentially expressed in developing rat brain. J Comp Neurol 465:57-71. [DOI] [PubMed] [Google Scholar]
- Jung CG, Kim HJ, Kawaguchi M, Khanna KK, Hida H, Asai K, Nishino H, Miura Y. (2005) Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development 132:5137-5145. [DOI] [PubMed] [Google Scholar]
- Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, et al. (2015) Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85:1086-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, Sakurai T. (2015) Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85:1103-1116. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux PC, Dahgren MK, Harrington ME. (2008) Circadian entrainment aftereffects in suprachiasmatic nuclei and peripheral tissues in vitro. Brain Res 1228:127-134. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Brancaccio M, Sethi S, Maywood ES, Satija R, Edwards JK, Jagannath A, Couch Y, Finelli MJ, Smyllie NJ. (2015) The regulatory factor ZFHX3 modifies circadian function in SCN via an AT motif-driven axis. Cell 162:607-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. (2007) Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1:113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyllie NJ, Chesham JE, Hamnett R, Maywood ES, Hastings MH. (2016) Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc Natl Acad Sci U S A 113:3657-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Fu X, Li J, Xing C, Martin DW, Zhang HH, Chen Z, Dong JT. (2012) Heterozygous deletion of Atbf1 by the Cre-loxP system in mice causes preweaning mortality. Genesis 50:819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Xing C, Fu X, Li J, Zhang B, Frierson HF, Jr, Dong JT. (2015) Additive effect of Zfhx3/Atbf1 and Pten deletion on mouse prostatic tumorigenesis. J Genet Genom 42:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDunk C, Hunter LA, Gray PA. (2011) Development, maturation, and necessity of transcription factors in the mouse suprachiasmatic nucleus. J Neurosci 31:6457-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, et al. (2013) Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342:85-90. [DOI] [PubMed] [Google Scholar]
- Yang GR, Chen LH, Grant GR, Paschos G, Song WL, Musiek ES, Lee V, McLoughlin SC, Grosser T, Cotsarelis G, et al. (2016) Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 8(324):324ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Ma G, Zhang X, He Y, Li M, Han X, Fu L, Dong XY, Nagy T, Zhao Q, et al. (2016) Zinc finger homeodomain factor Zfhx3 is essential for mammary lactogenic differentiation by maintaining prolactin signaling activity. J Biol Chem 291:12809-12820. [DOI] [PMC free article] [PubMed] [Google Scholar]