Abstract
We have identified a nuclear-encoded Hb from plants (GLB3) that has a central domain similar to the “truncated” Hbs of bacteria, protozoa, and algae. The three-dimensional structure of these Hbs is a 2-on-2 arrangement of α-helices, distinct from the 3-on-3 arrangement of the standard globin fold [Pesce, A., Couture, M., Dewilde, S., Guertin, M., Yamauchi, K., Ascenzi, P., Moens, L. & Bolognesi, M. (2000) EMBO J. 19, 2424–2434]. GLB3-like genes are not found in animals or yeast, but our analysis reveals that they are present in a wide range of Angiosperms and a Bryophyte. Although cyanobacteria and Chlamydomonas have 2-on-2 Hbs (GLBN), GLB3 is more likely related to GLBO-type 2-on-2 Hbs from bacteria. Consequently, GLB3 is unlikely to have arisen from a horizontal transfer between the chloroplast and nuclear genomes. Arabidopsis thaliana GLB3 protein exhibits unusual concentration-independent binding of O2 and CO. The absorbance spectrum of deoxy-GLB3 is unique; the protein forms a transient six-coordinate structure after reduction and deoxygenation, which slowly converts to a five-coordinate structure. In A. thaliana, GLB3 is expressed throughout the plant but responds to none of the treatments that induce plant 3-on-3 Hbs. Our analysis of the sequence, ligand interactions, and expression profile of GLB3 indicates that this protein has unique biochemical properties, evolutionary history, and, most likely, a function distinct from those of other plant Hbs.
Hbs are ubiquitous in eukaryotes but are not found in some fully sequenced bacterial and archaeal genomes. Although Hb sequences are highly variable (some share <20% amino acid identity), they have a conserved structure, the globin fold, which consists of a 3-on-3 sandwich of α-helices. By identifying the cd1 Phe and F8 His residues that are always conserved and by considering classes of amino acid residues crucial for forming the globin fold, these Hbs can be readily distinguished from non-Hbs (1).
A second group of Hbs has been identified from prokaryotes, protozoa, and eukaryotic algae. These have shorter sequences and are known as “truncated” Hbs (2). High-resolution crystal structures of these Hbs from Paramecium caudatum and Chlamydomonas eugametos revealed an alternative folding pattern with a 2-on-2 sandwich of α-helices (2). It has been proposed that the 2-on-2 Hbs arose from 3-on-3 Hbs by deletion of the N-terminal A-helix and the D-helix and replacement of the proximal heme-binding F-helix with a nonhelical loop (2, 3). Alternatively, the 2-on-2 Hbs of unicellular organisms may have an origin distinct from 3-on-3 Hbs (2, 4). The 2-on-2 Hbs display high sequence conservation, and the presence of these genes in bacteria has been attributed to horizontal gene transfer from a common ancestor of protozoa and algae (3).
We show that plants also contain a 2-on-2 Hb-like gene (GLB3), with strong similarity to a subset of the bacterial 2-on-2 Hbs but with less similarity to 2-on-2 Hbs from P. caudatum and C. eugametos.
Materials and Methods
Database Searches.
Homology searches of the Arabidopsis thaliana genome were performed (5), and the expressed sequence tag (EST) database (www.ncbi.nlm.nih.gov) was searched by using the tblastn program. The barley and A. thaliana sequences were confirmed by resequencing. A full-length Medicago truncatula GLB3 sequence was present in EST database.
Three motifs conserved in GLB3 proteins were compared to the A. thaliana and SwissProt protein databases. A. thaliana GLB3 was the only eukaryotic protein matching any of the three motifs. The N- and C-terminal sequences from A. thaliana GLB3 were also compared with a database of conserved plant signal peptide sequences (6) and the entire sequence analyzed for predicted secondary structure (http://molmod.angis.org.au/predictprotein/).
Comparison of Sequences.
A similarity matrix of Hbs based on amino acid sequence was generated by using the olddistances program in the GCG (Ver. 10.0-UNIX) package. The BLOSUM62 matrix was used with a scoring matrix threshold of 1.0, and the denominator set as the length of the shortest sequence without gaps.
Phylogenetics.
Amino acid sequences of 2-on-2 and 3-on-3 Hbs were aligned by eye by using the crystal structures of sperm whale myoglobin, lupin leg Hb (LUPlu GLB2S;II), and C. eugametos and P. caudatum 2-on-2 Hbs as a guide [a list of the sequences (Table 4) and the alignment file (Table 5) are published as supplemental data on the PNAS web site, www.pnas.org].
Cladistic analyses of the amino acid alignments were carried out by using paup (GCG). The central portion of the data matrix used for phylogeny reconstruction was from residue 33 of the alignment (F in ARAth GLB1; absent from most 2-on-2 Hbs) to residue 215 (the end of the longest bacterial 2-on-2 Hb). Bootstrap (1,000 replicates) minimum evolution and maximum parsimony trees were generated. Outgroups were the globin domain of the O2-sensing protein HtB from Halobacterium salinarum (U75436), the Hb-like domain from Escherichia coli colicin A (AAA23592), and the Synechococcus C-phycocyanin α-protein (P03943). Relationships between the GLB3, GLBO, and GLBN groups were tested by outgroup substitution by using 19 different combinations of 3-on-3 Hbs or Hb-like proteins as outgroups (Table 6, which is published as supplemental data on the PNAS web site).
Hypoxic Stress Treatments.
Plants were grown to 3 weeks in Murashige-Skoog (MS) agar, then transferred to liquid MS and treated for 12 h in a hypoxic chamber filled with a 5% O2/95% N2 mix, as described by Ellis et al. (7).
Hormone Treatments.
After 2 weeks of growth on MS-agar, plants were transferred into liquid MS medium on a shaker under constant light. After 24 h, the plants were transferred to treatment medium containing either 5 μg⋅ml−1 6-(γ-γ-dimethylallylamino) purine (2iP), 10 μM cis-trans-abscisic acid (ABA) or 0.5 μg⋅ml−1 2,4-dichlorophenoxyacetic acid (2,4-D) and returned to the shaker for a 24 h treatment period.
Northern Analysis of Expression.
Total RNA was extracted by using the Qiagen (Chatsworth, CA) RNeasy extraction kit. RNA formaldehyde gel electrophoresis, transfer to nylon membranes, prehybridization, hybridization, and washing conditions were as described (8). The membranes were probed with 32P-labeled antisense riboprobes synthesized on an ARAth GLB3 cDNA clone template.
Recombinant GLB3 Expression and Purification.
GLB3 was overexpressed in E. coli as described for GLB1 and GLB2 (9), except that growth was at 28°C, and induction was with 300 μM isopropyl β-d-thiogalactoside for 16 h. Cells were harvested, lysed, and protein purified as described for GLB2 (9), except that the 30–90% NH4SO4 fraction was used, the dialysis medium was 20 mM Tris⋅Cl, pH 7.6, and the dialysate was reduced with Na2S2O4 in the presence of CO with subsequent purification at 4°C. Purified CO-GLB3 was concentrated and left at 4°C to reequilibrate to the oxy form. The quaternary structure of purified ferric GLB3 was measured at concentrations of 3 and 30 μM by analytical ultracentrifugation by using published methods (10).
Spectral and Kinetic Analysis.
Oxygenated samples were prepared by reducing the protein with Na2S2O4 and then desalting over a G25 column equilibrated in air and 0.1 M phosphate buffer at pH 7.0. Deoxy-GLB3 was generated by reducing either a ferric or oxy sample, and CO-GLB3 was generated from deoxy-GLB3. The spectral transition shown in Fig. 4B occurs when Na2S2O4 is added to either ferric or oxy-GLB3. Kinetics of O2 and CO binding were measured by laser flash photolysis (11) and stopped flow rapid mixing was measured by using the five-coordinate species (12). Time courses for CO rebinding after flash were measured between 10 ns and 0.1 s. These data were concatenated into a single time course, fitted to single exponential decays, and rate constants calculated by using published procedures (13). Detailed methods are provided as supplemental text on the PNAS web site.
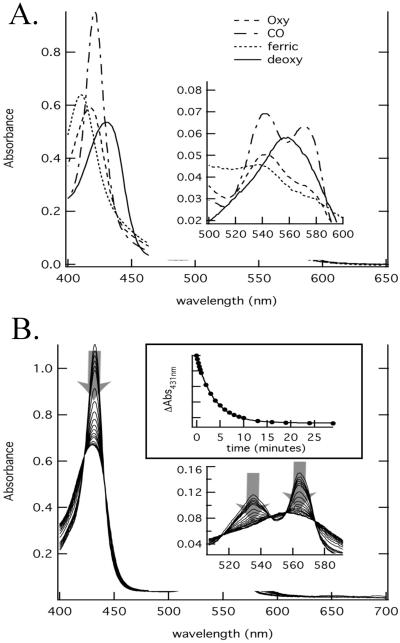
Figure 4.
The absorption spectra of A. thaliana GLB3 are shown (A). The absorption spectra of deoxyferrous GLB3 (solid line), O2-GLB3 (dashed), CO-GLB3 (dot-dash), and ferricGLB3 (dotted) are shown. The change in spectrum of deoxy-GLB3 over time is depicted (B), with the time dependence of the decay from six- to five-coordinate shown (Inset).
Results
A 2-on-2 Hb Gene from Plants.
The N-terminal region of the A. thaliana putative protein CAA18592.1 (At4 g32690) has up to 48% similarity to algal, bacterial, and protist 2-on-2 Hbs. Three Arabidopsis cDNA clones (R90000, N37398, and Z17742) encode part of the At4 g32690 ORF. Clone N37398 (14) was resequenced and found to contain a 528-bp ORF that includes the first 413 bp of the predicted At4 g32690 ORF; we named this gene GLB3.
The A. thaliana GLB3 gene contains three introns. Although the 5′ intron is positioned as in 3-on-3 Hbs at position B12.2, the central (ef9.1) and 3′ (phase 2 in H-helix) introns are in different positions from those of other plant Hb genes (E14.3, G6.3). None of the GLB3 introns correspond to intron positions in the truncated Hbs of Paramecium species or C. eugametos (15–17).
Expressed sequence tags with high similarity to A. thaliana GLB3 were identified from the dicots Populus hybrid (hybrid aspen), Glycine max (soybean), Solanum lycopersicon (tomato), M. truncatula (barrel medic), and Gossypium hirsutum (cotton), the monocots Oryza sativa (rice) and Hordeum vulgare (barley), and the moss Physcomitrella patens (Table 4). Those from barley, barrel medic, and cotton were found to encode proteins with between 50 and 65% identity to A. thaliana GLB3.
The plant GLB3 proteins contain a central region homologous to known 2-on-2 Hbs, flanked by ≈20-aa N- and C-terminal sequences (Fig. 1). The N-terminal flanking sequence is likely to be an α-helix, but the C-terminal sequence is not predicted to be an α-helical or β-sheet. The N-terminal sequence is highly conserved among the seven plant GLB3 sequences we have available but is not predicted to be a signal peptide, and the consensus sequence does not match any non-Hb proteins from A. thaliana or other organisms.
Figure 1.
The N- and C-terminal domains of some plant GLB3 proteins are shown in A, with a portion of the central region connecting them aligned with other Hbs in B. In A, the full C-terminal portion of cotton GLB3 is not shown, as it has not been determined. Shown in B is a structure-based sequence alignment of plant GLB3 proteins from A. thaliana, barley, barrel medic, and cotton, with 2-on-2 Hbs from microorganisms (2) and plant Hbs that have a myoglobin-like fold (11). Boxed regions denote α-helical regions in the structures of rice GLB1;1 and the 2-on-2 Hb of P. caudatum. Residues conserved between M. tuberculosis GLBO and GLB3 proteins are highlighted. The proximal (F8) and distal (E7) residues are marked with a star, and the helix designations for the beginning and end of the central domain are given.
Plant GLB3 Genes Are Homologous to the 2-on-2 Hbs from Bacteria.
A. thaliana GLB3 shares <25% amino acid identity with the GLB1 and GLB2 proteins from the same species. Using GLB3 sequences from H. vulgare, A. thaliana, M. truncatula, and G. hirsutum (partial), we have compared plant GLB3 sequences with groups of GLBO and GLBN type (2, 18) 2-on-2 Hb sequences (Table 1 and Table 7 and Fig. 6, which are published as supplemental data on the PNAS web site). Plant GLB3 proteins have more residues in common with GLBO than GLBN Hbs and are clearly more similar to either type of 2-on-2 Hb than to 3-on-3 Hbs from plants or animals. Most similarity is observed (40–45%) between GLB3 and GLBO 2-on-2 Hbs from Staphylococcus aureus, Bacillus subtilis, and Bacillus anthracis (Fig. 7, which is published as supplemental data on the PNAS web site). In addition, a number of amino acid residues and other signature sequences (19) (see Table 2 and Fig. 8, which is published as supplemental data on the PNAS web site) are conserved between GLB3 and GLBO proteins. GLB3 proteins have conserved B9 PHE, B10 TYR, F8 HIS, and GLY residues in the A-helix and ef-loop regions in common with other 2-on-2 Hbs. In contrast, plant 2-on-2 Hbs have ALA in the distal ligand E7 position, which is usually GLN or HIS in GLBN 2-on-2 Hbs and in 3-on-3 Hbs.
Table 1.
Amino acid similarity among groups of 2-on-2 HBs
The number of sequences used from each group.
Table 2.
Comparison of sequence compositions for GLB3, GLBO, and GLBN 2-on-2 HBs
| Residue or domain | GLB3 | GLBO | GLBN |
|---|---|---|---|
| F1 | H | H | Not conserved |
| F2 | P | P | Not conserved |
| F7 | R | R | Not conserved |
| fg6 | V | VI | Not conserved |
| bc loop | 7–8 residues | 5–9 residues | 5 residues |
| ef loop | 15 residues | 15 residues | 11 residues |
| G helix | 25 residues | 25 residues | 17–21 residues |
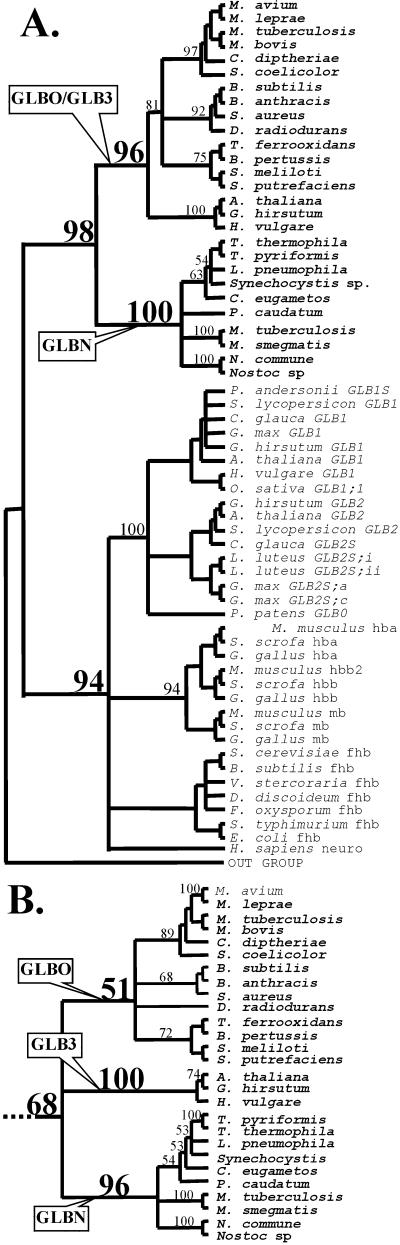
In our analyses (Fig. 2 and Fig. 9, which is published as supplemental data on the PNAS web site), the 3-on-3 Hbs form a clade with high bootstrap support [94% minimum evolution (ME), 92% maximum parsimony (MP)], separate from the 2-on-2 Hbs that may or may not form a monophyletic group (bootstrap 98% ME, 68% MP). GLB3 and GLBN form monophyletic groups, but the status of GLBO is unclear (Fig. 2B, low bootstrap score for GLBO under parsimony). On the basis of evolutionary distances, plant GLB3 proteins are more closely related to bacterial GLBO proteins, or some subset thereof, than to GLBN proteins. Parsimony yields the same result but with low bootstrap support (GLBO/GLB3 clade 49%); bootstrap support for the rival placement of GLB3 with GLBN was less than 1%. The results are stable to choice of outgroup.
Figure 2.
(A) Consensus tree showing the relationships among Hbs using minimum evolution. (B) An alternative arrangement of 2-on-2 Hb clades, found by using maximum parsimony. Bootstrap scores are indicated above the line for important clades; some scores have been omitted for clarity of presentation. See supplemental data for more details (www.pnas.org).
A. thaliana GLB3 mRNA Is Expressed Throughout the Plant.
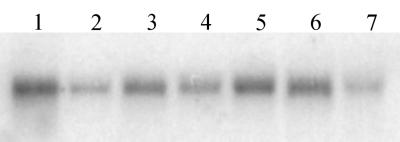
Northern analysis (Fig. 3) shows that A. thaliana GLB3 is expressed in both roots and aerial tissues. Expression in root tissue appears to be about four times higher than for shoot tissue, suggesting some tissue specificity of expression. GLB3 mRNA levels in whole plants are reduced by hypoxia and not markedly changed by 2,4-D, 2iP, or ABA treatment. GLB3 expression is up-regulated by auxin in some experiments listed in the microarray database (http://genome-www4.stanford.edu/).
Figure 3.
Northern blot analysis of ARAth GLB3 expression in A. thaliana. Lanes contain 25 μg of RNA from (Lane 1) roots, (Lane 2) leaves, and hypocotyl, (Lane 3) whole plants, (Lane 4) ABA, (Lane 5) 2,4-D, (Lane 6) 2iP, and (Lane 7) hypoxia-treated whole plants.
Spectral and Kinetic Characterization of the Arabidopsis GLB3 Protein.
A. thaliana GLB3 protein was expressed in E. coli, extracted from the soluble fraction of bacterial cell lysates, and purified to an A410/A280 ratio of 4 with a yield of 0.5 ml−1. Purified ferric GLB3 was determined to be dimeric over the concentration range 3–30 μM by analytical ultracentrifugation. Spectra of the recombinant protein in the CO, O2, and deoxy forms are shown in Fig. 4. The deoxy-GLB3 equilibrium absorbance spectra are characteristic of a five-coordinate Hb. The ferric form could be six-coordinate, as the Soret absorbance band at 411 nm is similar to six-coordinate plant Hbs (Fig. 4A). On addition of Na2S2O4 to either ferric or O2-GLB3, a six-coordinate deoxy absorbance spectrum is generated initially. The visible absorbance bands of this spectrum are similar to GLB1 or GLB2 nonsymbiotic plant Hbs with characteristic peaks at 538 and 565 nm. Over 20–30 min (with a rate constant of 0.3 s−1), the initial GLB3 six-coordinate deoxy spectrum converts to a typical five-coordinate spectrum with a single broad peak at ≈548 nm. This transition is accompanied by a decrease in the absorbance of the Soret band (Fig. 4B).
The O2-binding rate constant (k′O2) determined by stopped-flow methods was 0.2 μM−1⋅s−1, and the O2 dissociation rate constant (kO2) was 0.3 s−1 (Table 3, Fig. 5A). Equilibrium constants can be calculated for GLB3 by using these kinetic rate constants. The calculated O2 concentration at 50% saturation (P50 = 1.5 μM) is around 150-fold higher than for ARAth GLB1 (0.0097 μM) and 10 times than that of ARAth GLB2 (0.14 μM) (9) but only slightly higher than for GLBN type 2-on-2 Hbs studied from P. caudatum (20), C. eugametos (21), N. commune (22), Mycobacterium. tuberculosis GlbN (18), and Synechocystis PCC6803 (23). No kinetic data are available for comparison of GLB3 with bacterial GLBO Hbs. The affinity of GLB3 for O2 is higher than that of vertebrate Hbs and myoglobins (e.g., sperm whale myoglobin; see Table 3) but is lower than that reported for terminal cytochrome C-oxidase from soybean (24), so the protein could potentially participate in O2 transport within plant cells.
Table 3.
Comparison of ligand interaction constants for A. thaliana GLB3 and other HBs
| Protein | k′O2, μM−1⋅s−1 | kO2, s−1 | k′CO, μM−1⋅s−1 | kCO, s−1 |
|---|---|---|---|---|
| A. thaliana GLB3 | 0.2 | 0.3 | 0.014 | 0.001 |
| A. thaliana GLB2 | 86 | 0.14 | 22 | 0.0013 |
| A. thaliana GLB1 | 74 | 0.12 | 0.55 | 0.0012 |
| G. max GLB2S;a | 130 | 5.6 | 17 | 0.0078 |
| Sperm whale Mb | 15 | 18 | 0.53 | 0.019 |
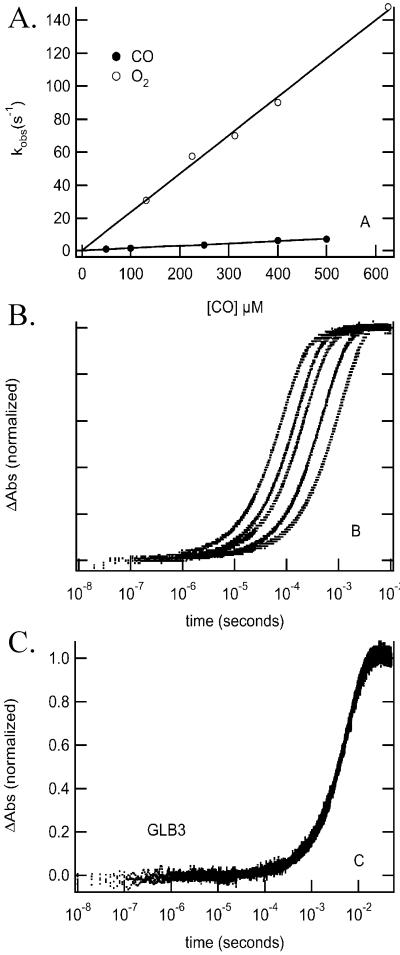
Figure 5.
Concentration dependence of CO and O2 binding to A. thaliana GLB3 after rapid mixing (A). Concentration dependence of CO binding to soybean GLB2S;a (B) and A. thaliana GLB3 (C), after flash photolysis.
The CO association and dissociation rate constants for GLB3 as measured by the stopped-flow technique are given in Table 3 (also see Fig. 5A). The calculated P50 for CO is 0.071 μM, approximately one-quarter of the P50 for O2. In contrast, GLB1 has approximately equal affinities for O2 and CO. As a result, GLB3 would be expected to bind CO preferentially when the O2:CO ratio is low, consequently any important cellular function performed by O2-GLB3 should be sensitive to CO treatment.
When laser-flash photolysis was used to analyze O2- and CO-binding kinetics for GLB3, some unusual results were obtained. For typical Hbs that are five-coordinate in the deoxyferrous state, the rate constants measured by flash photolysis and rapid mixing are identical and depend on ligand concentration. Fig. 5B shows CO rebinding to soybean GLB2S;a following flash photolysis is an example of a typical ligand-concentration-dependent set of data. The time course on the far left is 1,000 μM CO and moving from Left to Right are shown time courses for 700-, 500-, 200-, and 100-μM samples. In contrast, Fig. 5C shows time courses for CO rebinding to GLB3 at the same concentrations used in Fig. 5B. In this case, the rate constant for CO binding is 170 s−1 regardless of the concentration of CO in the buffer. The rate constant for binding of O2 to GLB3 after flash photolysis was also independent of ligand concentration with a value of 400 s−1. Fig. 5A shows the concentration dependence of CO binding after rapid mixing. Under these conditions, the observed rate constant is linearly dependent on CO concentration with a value of 0.014 μM−1⋅s⋅1.
Discussion
We have identified a gene from A. thaliana that encodes a protein similar to truncated Hbs of bacteria. The spectral properties of the protein show it contains a five-coordinate ferrous heme group when the protein has no bound ligand and that CO and O2 bind to the heme moiety in a reversible manner. These are properties of a functional Hb. A similar gene is active in a range of other vascular and nonvascular plants. We conclude that this is a type of Hb gene that occurs widely, perhaps universally, in plants.
It is likely that the plant GLB3 gene family has an evolutionary history distinct from other plant Hbs. Our analyses identify the 2-on-2 Hbs as a probable clade of Hbs that are separate from the 3-on-3 Hbs, supporting a previous analysis (3, 4). The plant 3-on-3 Hbs have a 3-intron/4-exon structure with intron positions being absolutely conserved. Although the A. thaliana GLB3 gene also has a 3-intron/4-exon structure, two of the introns are in different positions and phase supporting a separate origin of GLB3 genes. The first intron position of A. thaliana GLB3 is identical to that of 3-on-3 Hbs, but the gene structure of other GLB3 genes is needed to know whether this intron position is highly conserved.
The 2-on-2 Hbs may have arisen from other Hbs by deletion of the N-terminal α-helix and replacement of the heme-binding F-helix with a nonhelical loop. An alternative hypothesis, that 2-on-2 and 3-on-3 Hbs are separate gene families (3, 4), is supported by our observations. The 2-on-2 Hb gene is present in three of the five kingdoms, and both 2-on-2 and 3-on-3 Hbs are present in plants and Gram-positive bacteria. This occurrence implies that the origin of both gene types is extremely ancient, and the two types have evolved independently for a large portion of evolutionary history.
2-on-2 Hb genes are not found in the completed genomes of Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, or Saccharomyces cerevisiae and so are probably absent in the fungi/animal group. This absence implies either a loss of this gene in the fungi/animal lineage after the divergence of plants or an acquisition by plants of the gene by horizontal transfer. Our comparison of the plant and bacterial 2-on-2 Hbs (Tables 1 and 2, Fig. 2) indicates that the GLB3 proteins more closely resemble GLBO Hbs from Gram-positive bacteria. If the GLB3 proteins are homologous to the GLBO Hbs but not the GLBN type found in cyanobacteria, it is unlikely that a horizontal transfer event was coincident with acquisition of the endosymbiotic cyanobacteria, which later became chloroplasts. A horizontal transfer later in plant evolution from an alternative group of bacteria would seem likely and is perhaps indicative of an ancient close relationship between protoplants and nonphotosynthetic bacteria. The early history of the GLB3 gene will become clearer with the completion of more protozoal genomes. For example, if a GLBO Hb gene is present in C. eugametos, the origin of GLB3 might be before the advent of multicellularity in the plant kingdom.
Functional Properties of A. thaliana GLB3.
The A. thaliana GLB3 protein has a lower O2 affinity than other 2-on-2 Hbs and plant 3-on-3 Hbs. GLB3 might be an O2 transport protein, but our spectral characterization and laser photolysis results reveal that it is an unusual Hb. Deoxygenation of GLB3 leaves the protein in a six-coordinate state with an endogenous (i.e., amino acid side-chain) or uncharacterized exogenous ligand (i.e., not O2, CO, or NO) bound to the sixth bond of the heme iron molecule. This state is transitory, reverting to a five-coordinate form in ≈20 min. This extremely slow conversion has not been observed with any other Hb. The most direct explanation for the observed results would be rapid binding of a ligand to deoxy-GLB3 after Na2S2O4 treatment, followed by a slow conformational change, excluding the ligand from the distal pocket. The mechanism of the conformational change might involve changes in tertiary or quaternary structure or both.
Indeed, the unusual six- to five-coordinate deoxy-GLB3 kinetics and the unusual ligand concentration-independent rebinding in flash photolysis both might depend on quaternary structure. We have observed that GLB3 is dimeric. Possibly, the arrangement of the subunits obscures the distal pocket, slowing the exit of endogenous ligands and allowing molecules trapped between the two subunits to slowly rebind to the heme iron. Such a dimeric model might provide an explanation for the unusual laser-flash photolysis results obtained, where O2 and CO rebinding were found to be independent of ligand concentration in the external medium. The results are consistent with geminate rebinding of ligands, but at a rate much slower than has been observed previously (25, 26). Additionally, conformational changes after deoxygenation might be expected to attract and arrange water molecules around the subunit interface, a mechanism that might change tertiary structure or attract ligands from the distal pocket to the subunit interface and provide an explanation for the transitory six-coordinate nature of deoxy-GLB3. A water molecule might be a transitory ligand in deoxy-GLB3, which is removed from the distal pocket and attracted to the subunit interface leaving deoxy-GLB3 in a five-coordinate state.
Scapharca inaequivalvis dimeric Hb forms a tightly assembled dimer in the unliganded state, which becomes more open in the O2 and CO-Hb forms (27). The crystal structure of this Hb reveals that the interface between the two subunits overlies the distal pocket (28) and contains a highly ordered lattice of water molecules (29). Before the dimer binds to O2 or CO, structural changes displace the interface water molecules. Consequently, the dimer has a lower affinity for exogenous ligands when compared with the isolated subunits. The quaternary structure of GLB3 might impart similar biochemical properties to that of S. inaequivalvis Hb, but the structure itself would most likely be distinct. We predict that the GLB3 molecule will have a tertiary structure with core helices arranged similarly to P. caudatum and C. eugametos Hbs but with flanking N- and C-terminal domains. The longer ef-loop, F2 proline, and E7 alanine residues distinguish GLB3 proteins from GLBN Hbs and probably contribute to biochemical differences observed between GLB3 and other characterized 2-on-2 Hbs.
GLBN 2-on-2 Hbs are a diverse group; individual molecules exhibit a number of unusual properties, including pH-dependent conformational changes and cooperative ligand binding (2, 17, 22). GLB3 is distinct from most of these, but no GLBO 2-on-2 Hbs have been studied in detail.
Expression of the A. thaliana 2-on-2 Hb.
GLB3 is expressed widely in the plant and is likely to have a function that is different from GLB1 and GLB2, as it does not respond to treatments that induce those two genes. Expressed sequence tags for the other plant GLB3 genes were derived from cDNA libraries made from a wide variety of tissue types, including quiescent seeds, cotton fiber, root nodules, cambium, and inflorescence. The P. patens cDNA clone was obtained from ABA-treated protonema.
GLB3 expression is regulated differently from GLB1 or GLB2. Similar to GLB3, GLB1 is expressed predominantly in the roots, but GLB2 is expressed predominantly in shoots (9). The expression of GLB1 is increased by hypoxia (9), and GLB2 is induced by cytokinin treatment (30); however, GLB3 mRNA levels are decreased by hypoxia, as are many plant genes, and may be up-regulated slightly by 2,4-D (auxin), and 2iP (cytokinin) (Fig. 3).
Supplementary Material
Acknowledgments
John Trueman assisted us with interpretation of phylogenetic data. Preliminary sequence data were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org).
Abbreviations
- GLBO
2-on-2 Hbs similar to M. tuberculosis GlbO
- GLBN
2-on-2 Hbs similar to M. tuberculosis GlbN
- 2iP
6-(γ-γ-dimethylallylamino) purine
- ABA
cis-trans-abscisic acid
- 2,4-D
2,4-dichlorophenoxyacetic acid
Footnotes
References
- 1.Bashford D, Chothia C, Lesk A M. J Mol Biol. 1987;196:199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- 2.Pesce A, Couture M, Dewilde S, Guertin M, Yamauchi K, Ascenzi P, Moens L, Bolognesi M. EMBO J. 2000;19:2424–2434. doi: 10.1093/emboj/19.11.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moens L, Vanfleteren J, Van de Peer Y, Peeters K, Kapp O, Czeluzniak J, Goodman M, Blaxter M, Vinogradov S. Mol Biol Evol. 1996;13:324–333. doi: 10.1093/oxfordjournals.molbev.a025592. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki T, Imai K. Cell Mol Life Sci. 1998;54:979–1004. doi: 10.1007/s000180050227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 7.Ellis M H, Dennis E S, Peacock W J. Plant Physiol. 1999;119:57–64. doi: 10.1104/pp.119.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolferus R, de Bruxelles G L, Dennis E S, Peacock W J. Ann Bot. 1994;74:301–308. [Google Scholar]
- 9.Trevaskis B, Watts R A, Andersson C R, Llewellyn D J, Hargrove M S, Olson J S, Dennis E S, Peacock W J. Proc Natl Acad Sci USA. 1997;94:12230–12234. doi: 10.1073/pnas.94.22.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springer B A, Sligar S G. Proc Natl Acad Sci USA. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargrove M S. Biophys J. 2000;79:2733–2738. doi: 10.1016/S0006-3495(00)76512-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hargrove M S, Barry J K, Brucker E A, Berry M B, Phillips G N, Olson J S, Arredondo-Peter R, Dean J M, Klucas R V, Sarath G. J Mol Biol. 1997;266:1032–1042. doi: 10.1006/jmbi.1996.0833. [DOI] [PubMed] [Google Scholar]
- 13.Trent, J., Hvitved, A. N. & Hargrove, M. S. (2001) Biochemistry, in press. [DOI] [PubMed]
- 14.Newman T, Bruijn F J d, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamauchi K, Tada H, Usuki I. Biochim Biophys Acta. 1995;1264:53–62. doi: 10.1016/0167-4781(95)00114-v. [DOI] [PubMed] [Google Scholar]
- 16.Yamauchi K, Ochiai T, Usuki I. Biochim Biophys Acta. 1992;1171:81–87. doi: 10.1016/0167-4781(92)90142-m. [DOI] [PubMed] [Google Scholar]
- 17.Couture M, Chamberland H, St-Pierre B, Lafontaine J, Guertin M. Mol Gen Genet. 1994;243:185–197. doi: 10.1007/BF00280316. [DOI] [PubMed] [Google Scholar]
- 18.Couture M, Yeh S R, Wittenberg B A, Wittenberg J B, Ouellet Y, Rousseau D L, Guertin M. Proc Natl Acad Sci USA. 1999;96:11223–11228. doi: 10.1073/pnas.96.20.11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta R S. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das T K, Weber R E, Dewilde S, Wittenberg J B, Wittenberg B A, Yamauchi K, Van Hauwert M-L, Moens L, Rousseau D L. Biochemistry. 2000;39:14330–14340. doi: 10.1021/bi001681d. [DOI] [PubMed] [Google Scholar]
- 21.Couture M, Guertin M. Eur J Biochem. 1996;242:779–787. doi: 10.1111/j.1432-1033.1996.0779r.x. [DOI] [PubMed] [Google Scholar]
- 22.Thorsteinsson M V, Bevan D R, Potts M, Dou Y, Eich R F, Hargrove M S, Gibson Q H, Olson J S. Biochemistry. 1999;38:2117–2126. doi: 10.1021/bi9819172. [DOI] [PubMed] [Google Scholar]
- 23.Couture M, Das T K, Savard P Y, Ouellet Y, Wittenberg J B, Wittenberg B A, Rousseau D L, Guertin M. Eur J Biochem. 2000;267:4770–4780. doi: 10.1046/j.1432-1327.2000.01531.x. [DOI] [PubMed] [Google Scholar]
- 24.Millar A H, Bergersen F J, Day D A. Plant Physiol Biochem (Paris) 1994;32:847–852. [Google Scholar]
- 25.Henry E R, Sommer J H, Hofrichter J, Eaton W A. J Mol Biol. 1983;166:443–451. doi: 10.1016/s0022-2836(83)80094-1. [DOI] [PubMed] [Google Scholar]
- 26.Rohlfs R J, Olson J S, Gibson Q H. J Biol Chem. 1988;263:1803–1813. [PubMed] [Google Scholar]
- 27.Royer W E, Fox R A, Smith F R, Zhu D, Braswell E H. J Biol Chem. 1997;272:5689–5694. doi: 10.1074/jbc.272.9.5689. [DOI] [PubMed] [Google Scholar]
- 28.Chiancone E, Verzili D, Boffi A, Royer W E, Hendrickson W A. Biophys Chem. 1990;37:287–292. doi: 10.1016/0301-4622(90)88028-q. [DOI] [PubMed] [Google Scholar]
- 29.Royer W E, Pardanani A, Gibson Q H, Peterson E S, Friedman J M. Proc Natl Acad Sci USA. 1996;93:14526–14531. doi: 10.1073/pnas.93.25.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt, P. W., Watts, R. A., Trevaskis, B., Llewellyn, D. J., Burnell, J., Dennis, E. S. & Peacock, W. J. (2001) Plant Mol. Biol., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.