Significance
Wheat provides a substantial proportion of the calories and proteins consumed by humans, but further production increases are necessary to feed a growing human population. Reducing yield losses caused by pathogens can contribute to these increases. In this study, we report the identification of Sr13, a gene from pasta wheat that confers resistance to the new virulent races of the stem rust pathogen that appeared in Africa at the beginning of this century. We identified three different resistance forms of Sr13 and developed a diagnostic marker to accelerate their deployment in wheat breeding programs. In addition, Sr13 can be a useful component of transgenic cassettes including multiple resistance genes.
Keywords: durum wheat, stem rust, Ug99, resistance genes, CC-NBS-LRR
Abstract
The Puccinia graminis f. sp. tritici (Pgt) Ug99 race group is virulent to most stem rust resistance genes currently deployed in wheat and poses a threat to global wheat production. The durum wheat (Triticum turgidum ssp. durum) gene Sr13 confers resistance to Ug99 and other virulent races, and is more effective at high temperatures. Using map-based cloning, we delimited a candidate region including two linked genes encoding coiled-coil nucleotide-binding leucine-rich repeat proteins designated CNL3 and CNL13. Three independent truncation mutations identified in each of these genes demonstrated that only CNL13 was required for Ug99 resistance. Transformation of an 8-kb genomic sequence including CNL13 into the susceptible wheat variety Fielder was sufficient to confer resistance to Ug99, confirming that CNL13 is Sr13. CNL13 transcripts were slightly down-regulated 2–6 days after Pgt inoculation and were not affected by temperature. By contrast, six pathogenesis-related (PR) genes were up-regulated at high temperatures only when both Sr13 and Pgt were present, suggesting that they may contribute to the high temperature resistance mechanism. We identified three Sr13-resistant haplotypes, which were present in one-third of cultivated emmer and durum wheats but absent in most tested common wheats (Triticum aestivum). These results suggest that Sr13 can be used to improve Ug99 resistance in a large proportion of modern wheat cultivars. To accelerate its deployment, we developed a diagnostic marker for Sr13. The identification of Sr13 expands the number of Pgt-resistance genes that can be incorporated into multigene transgenic cassettes to control this devastating disease.
Stem rust of wheat, caused by the fungus Puccinia graminis f. sp. tritici (Pgt), is a serious wheat disease that has historically caused large yield losses worldwide. An isolate of Pgt identified in Uganda in 1998 (known as Ug99) is virulent to multiple stem rust resistance genes including the widely deployed Sr31 (1). This race was later designated TTKSK using the North American stem rust nomenclature system (2). As TTKSK spread through Africa and the Middle East, it gained new virulences to wheat cultivars carrying resistance genes Sr24 (2), Sr36 (3), Sr9h (4, 5), and SrTmp (6). The expansion of the Ug99 race group to 10 countries in Africa, Egypt, Yemen, and Iran (7, 8) has prompted the search for new sources of resistance. This search has been broadened to other virulent races outside the Ug99 race group, such as TKTTF that caused severe stem rust epidemics in southern Ethiopia since 2013 (9). The recent experience in Ethiopia emphasizes the need for resistance genes with broader resistance or for combinations of resistance genes that can confer a broader and more durable resistance.
Among more than 70 Sr resistance loci that have been cataloged and assigned official or temporary designations, roughly half of them are effective against the Ug99 race group (7). However, only two slow-rusting adult plant resistance genes and five all-stage resistance genes effective against Ug99 have been cloned so far, likely reflecting the difficulties imposed by the large and complex wheat genomes. The two slow-rusting genes include Sr57/Lr34 and Sr55/Lr67, which encode a putative ABC transporter (10) and a hexose transporter (11), respectively. The five all-stage resistance genes identified so far include Sr35 (12) and Sr22 (13) from diploid wheat Triticum monococcum L.; Sr33 (14) and Sr45 (13) from diploid Aegilops tauschii Coss.; and Sr50 from diploid rye (Secale cereale L.) (15). These five genes encode coiled-coil nucleotide-binding leucine-rich repeat (CC-NBS-LRR) proteins, hereafter designated as CC-NBS-LRR genes (NLRs). Chromosomes of T. monococcum and rye carrying these resistance genes recombine poorly with wheat chromosomes (16, 17), so the Pairing homeologous 1 mutation (ph1b) is required to induce homeologous recombination to reduce the size of the introgressed chromosome segments. The chromosomes of A. tauschii recombine well with the polyploid wheat D genome chromosomes, although the frequencies of recombination are sometimes reduced in the most divergent accessions (18).
The transfer of resistance genes directly from the wheat primary gene pool minimizes the need for additional rounds of homeologous recombination in the ph1b background. One of these resistance genes, Sr13, was identified first in durum wheat [Triticum turgidum ssp. durum (Desf.) Husn.] and was then transferred to hexaploid wheat (Khapstein/9*LMPG) (19). The domesticated emmer wheat [T. turgidum ssp. dicoccon (Schrank ex Schüebler) Thell.] Khapli (20) and the tetraploid Ethiopian accession ST464 (21) were used as sources of Sr13 in durum wheat. Sr13 confers resistance to all races in the Ug99 group (typical infection types range from 2 to 2+) (22), but the resistant reaction is influenced by temperature and genetic background (23, 24).
Sr13 has been mapped on the distal region of chromosome arm 6AL (25), but the development of diagnostic molecular markers has been delayed likely due to the different germplasm sources used to transfer Sr13 into modern cultivated wheats (20, 21, 25). The map-based cloning and characterization of the stem rust resistant gene Sr13 reported in this study revealed the existence of three different resistant haplotypes with some differences in resistance profiles. All three haplotypes confer resistance to the Ug99 race group and to the virulent races TKTTF and TRTTF outside the Ug99 race group.
Results
Construction of Genetic and Physical Maps of the Sr13 Region.
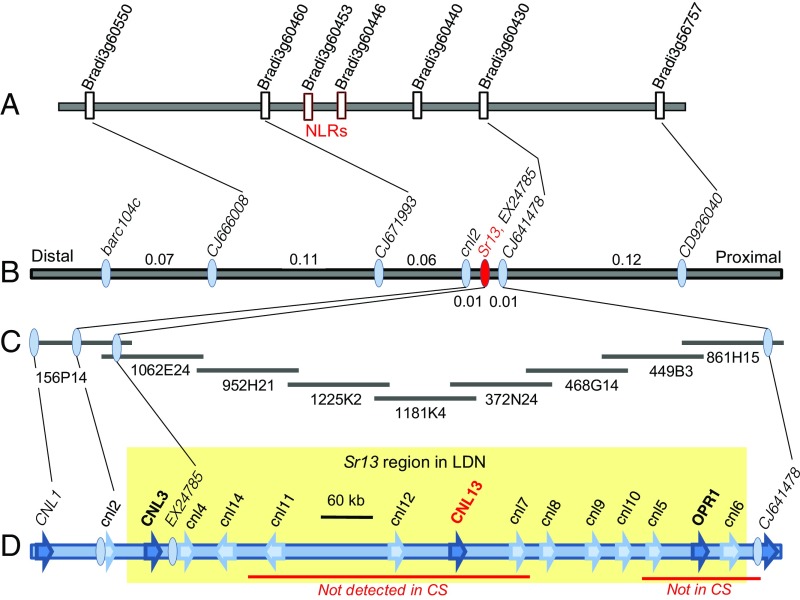
Sr13 was previously mapped on the distal region of the long arm of chromosome 6A of durum wheat within a ∼1.2-cM interval flanked by markers CD926040 and barc104c (25). In this study, we generated a larger mapping population (12,162 gametes) from the cross Kronos × Rusty and identified 45 recombination events between these two flanking markers, providing a better estimate of their genetic distance (0.4 cM). We generated additional markers in the region (SI Appendix, Table S1 and Method S1) and mapped Sr13 completely linked to EX24785, within a 0.08-cM interval delimited by markers CJ641478 and CJ671993 (Fig. 1 A and B).
Fig. 1.
Genetic and physical maps of Sr13. (A) B. distachyon chromosome 3 region colinear with the wheat Sr13-candidate region. Rectangles in red indicate NLR genes (likely nonfunctional since they carry frame-shift mutations and premature stop codons). (B) High-density genetic map of Sr13 on chromosome arm 6AL. (C) Physical map of the Sr13 region constructed with overlapping BACs from the Sr13-resistant durum wheat variety Langdon. (D) Diagrammatic representation of the annotated sequence of the Sr13 region (GenBank accession no. KY924305). Genes are indicated by arrows (uppercase names indicate complete genes and lowercase names 5′ or 3′ truncated genes). Red lines below this graph indicate regions where no CS orthologs were found.
We screened the bacterial artificial chromosome (BAC) library of the Sr13-resistant durum wheat variety Langdon (26) with marker EX24785 and identified BAC 156P14. Sequencing of this BAC revealed two NLR genes (CNL1 and CNL3) and two 5′ or 3′ truncated NLR genes (cnl2 and cnl4). We developed markers for these complete and truncated NLR genes and mapped Sr13 within a 0.02-cM interval delimited by cnl2 and CJ641478 (Fig. 1B). We developed a physical map of this region including nine overlapping BACs (Fig. 1C, 955 kb) that were sequenced, annotated, and deposited in GenBank as accession no. KY924305.
The 858-kb region between flanking markers cnl2 and CJ641478 (Fig. 1D) included two complete NLR genes (CNL3 and CNL13), one gene annotated as 12-oxophytodienoate reductase 1 (OPR1), and 10 5′- or 3′-truncated NLR genes (Fig. 1D). Among the three complete genes, we prioritized the two NLR genes, which encode proteins frequently associated with pathogen recognition and initiation of resistance responses in plants. The linked gene OPR1, which encodes an enzyme involved in the jasmonic acid biosynthesis pathway, was considered a less likely candidate.
Validation of Sr13 Candidate Genes Using Ethyl Methanesulfonate Mutants.
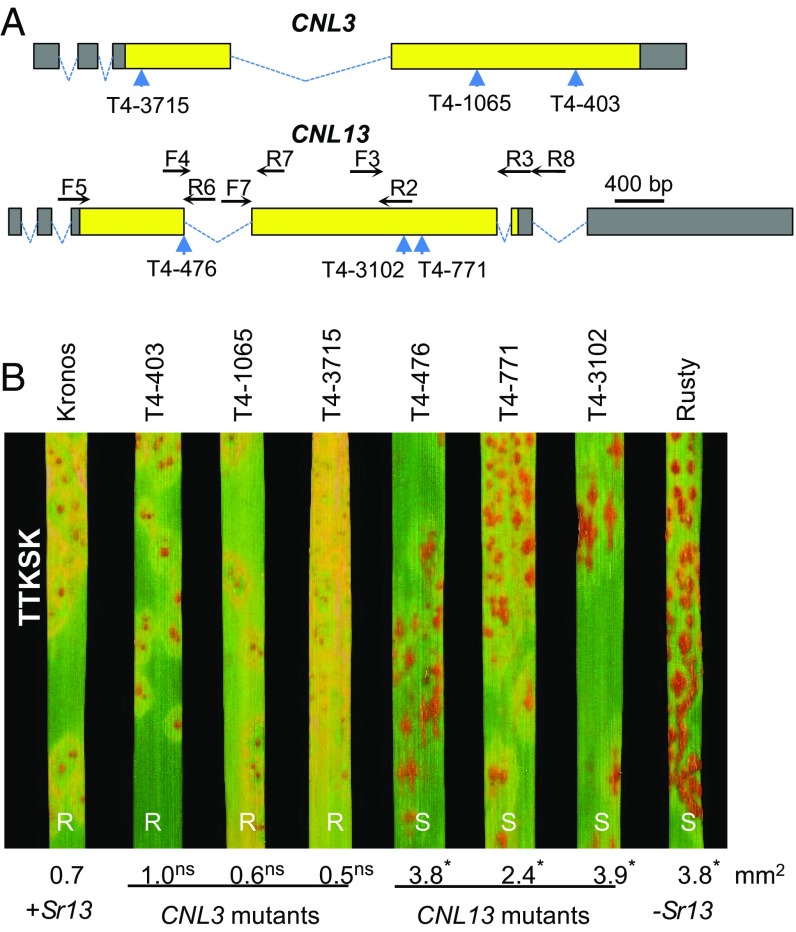
To determine if CNL3 or CNL13 were required for resistance to Ug99 (TTKSK), we selected three truncation mutations for each gene from the recently published database of sequenced ethyl methanesulfonate mutations in Kronos (27). The three mutations identified in CNL3 (mutant lines T4-3715, T4-1065, and T4-403) were all predicted to generate premature stop codons (Fig. 2A) and to encode nonfunctional truncated proteins. Plants homozygous for each of the three CNL3 mutations showed levels of resistance to TTKSK similar to those observed in the resistant Kronos control (Fig. 2B). Similar results were observed when the three CNL3 mutants were inoculated with races TKTTF, JRCQC, and TRTTF (SI Appendix, Fig. S1), which suggests that CNL3 is not Sr13.
Fig. 2.
Sr13 mutants. (A) Schematic representation of NLR candidate genes CNL3 and CNL13. Dotted lines indicate introns, yellow rectangles coding exons, and gray rectangles 5′ and 3′ UTRs. The positions of the selected truncation mutations are indicated by blue arrows. Mutations in Kronos mutant lines T4-403, T4-1065, T4-3715, T4-771, and T4-3102 are expected to result in premature stop codons, whereas the mutation in mutant line T4-476 is expected to eliminate the donor splice site of the third intron. (B) CNL3 and CNL13 mutant lines inoculated with race TTKSK. Kronos is the Sr13-resistant control and Rusty is the susceptible control. CNL3 mutants are all resistant, whereas CNL13 mutants are all susceptible to TTKSK (Ug99). Numbers below the leaves are average pustule sizes (n = 3). Superscripts indicate significance of the differences with Kronos using Dunnett’s test (ns, not significantly larger than Kronos, *P < 0.05). Results for races TKTTF, JRCQC, and TRTTF are presented in SI Appendix, Fig. S1.
By contrast, plants homozygous for CNL13 mutations encoding premature stop codons (mutant lines T4-3102 and T4-771) or an altered donor splice site (mutant line T4-476; Fig. 2A) were highly susceptible to TTKSK, and showed average pustule sizes significantly larger (P < 0.05) than those in Kronos (Fig. 2B). Similar results were obtained when the homozygous CNL13 mutants were inoculated with races TKTTF and JRCQC (SI Appendix, Fig. S1). However, the same mutants inoculated with race TRTTF were all resistant, suggesting that Kronos carries another resistance gene effective against this race. To test this hypothesis, we selected F2 plants from the cross Kronos × Rusty (25) that were either homozygous for the resistant (three plants) or for the susceptible (nine plants) CNL13 alleles. Inoculation of the F3 progeny of these plants with races TTKSK and JRCQC showed a perfect cosegregation between the CNL13 allele and the phenotype (SI Appendix, Fig. S2). However, when the same plants were inoculated with race TRTTF, we observed resistant plants among the progeny of F2 plants homozygous for the susceptible CNL13 allele (SI Appendix, Fig. S2). This result confirmed the presence of a second TRTTF resistance gene in Kronos.
To characterize better the role of the CNL13 LRR domain, we inoculated four additional lines carrying mutations in this region with Pgt race TTKSK. Mutations L757F (T4-3913), S730F (T4-4581), and D704N (T4-2561) had no effect on resistance to TTKSK, whereas mutation A717V (T4-4367) resulted in fully susceptible plants. This suggests that this LRR amino acid is important for TTKSK resistance (SI Appendix, Fig. S3). Taken together, these results demonstrated that CNL13 is required for the Sr13-mediated resistance to races TTKSK, TKTTF, and JRCQC.
Validation of Sr13 Using Transgenic Complementation.
To test if CNL13 was also sufficient to confer resistance to these races, we performed a transgenic complementation experiment using a complete genomic copy of CNL13. First, we identified the transcriptional start of the CNL13 gene using 5′ rapid amplification of cDNA end (RACE, see Materials and Methods). The end of the transcript was determined from a previous Kronos transcriptome (27). Comparing Kronos CNL13 transcripts with the corresponding genomic sequences revealed the presence of five introns, two of them in the 5′ untranslated region (UTR) and one in the 3′ UTR (Fig. 2A). The coding region, which starts in the third exon and ends in the fifth exon, extends for 2,847 bp (including the stop codon and excluding introns) and encodes a protein of 948 aa. The 538-bp 5′ untranslated region (UTR) included two introns, whereas the 2,187-bp 3′ UTR included a single intron (Fig. 2A, GenBank accession no. KY924305).
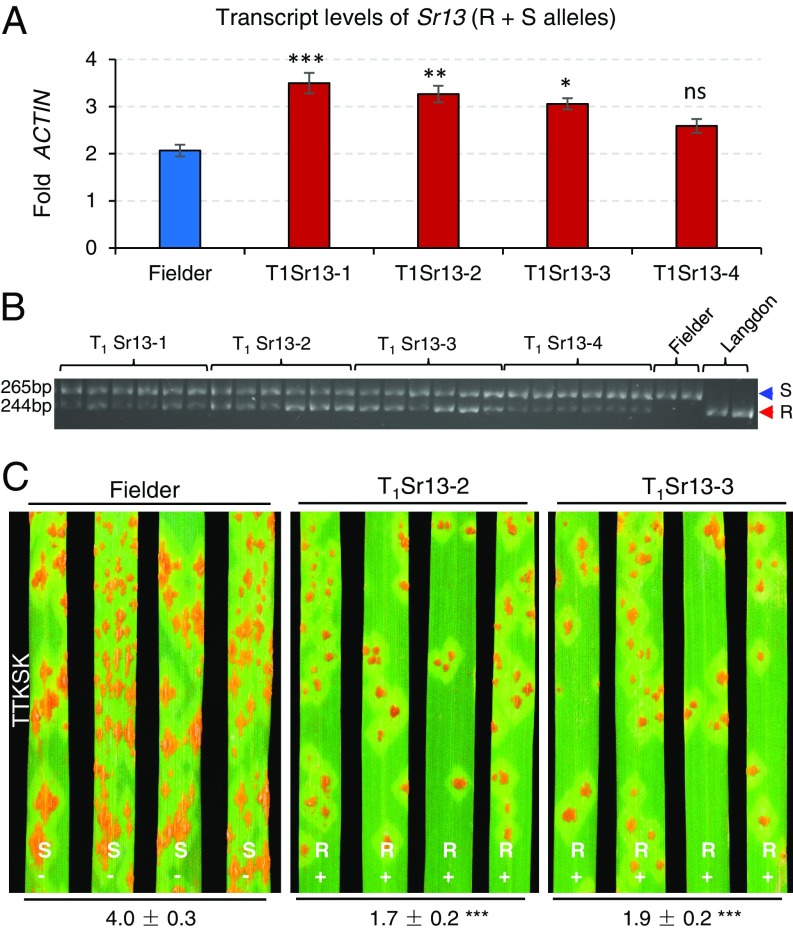
We cloned an 8,055-bp genomic fragment encompassing the complete transcribed region and transformed this construct into the Ug99-susceptible hexaploid common wheat variety Fielder. We obtained four independent transgenic events (T0), which were confirmed with two independent sets of primers (Materials and Methods). Transgenic events T1Sr13-1, T1Sr13-3, T1Sr13-4 showed a 3:1 transgene segregation ratio, suggesting the presence of a single copy of CNL13. This result was confirmed using a TaqMan Copy Number Assay (SI Appendix, Table S2). By contrast, we detected the presence of the transgene in all 80 T1 plants from family T1Sr13-2, suggesting the presence of at least three CNL13 copies. This estimate was also supported by the TaqMan Copy Number Assay (SI Appendix, Table S2).
The differences in the number of inserted CNL13 copies in the different transgenic events were not reflected in the expression levels. CNL13 transcript levels in T1Sr13-1, T1Sr13-2, and T1Sr13-3 were similar to each other and significantly higher than in the susceptible control Fielder (Fig. 3A). Those differences were not significant for plants from the T1Sr13-4 family (Fig. 3A). In addition to the transgenic CNL13 transcripts, the qRT-PCR primers used in this study also amplified the susceptible CNL13 allele present in Fielder (see Sr13 Allelic Variation). To separate both alleles, we performed a semiquantitative PCR assay using the restriction enzyme HhaI, which generated a 265-bp fragment for the susceptible allele and a 244-bp fragment for the transgenic resistant allele (Fig. 3B). The semiquantitative PCR study confirmed that the transcript levels of the CNL13-resistant allele were higher in T1Sr13-1, T1Sr13-2, and T1Sr13-3 than in T1Sr13-4 (Fig. 3B).
Fig. 3.
Sr13 transgenic plants. (A) Transcript levels of CNL13 in the susceptible control Fielder and in transgenic plants from four independent events (Fielder background). qRT-PCR primers amplify both the susceptible CNL13 allele in Fielder (S1) and the resistant allele (R3) in the transgenic plants. Transcript levels are expressed as fold-ACTIN using the 2−ΔCT method. *P < 0.05, **P < 0.01, ***P < 0.001, NS, not significant (Dunnett’s test). (B) Semiquantitative PCR products from dCAPS marker Sr13F/R digested with restriction enzyme HhaI. The lower band corresponds to the transcript from the resistant allele (R = LDN) and the upper band to the transcript from the susceptible allele (S = Fielder). Six independent T1 plants from every transgenic event were evaluated. (C) Reaction to Pgt race TTKSK (Ug99) in Fielder and transgenic families T1Sr13-2 and T1Sr13-3. R, Resistant; S, Susceptible; −, no resistant CNL13 allele; +, resistant CNL13 allele present. Numbers below leaves are average pustule sizes (n = 8). Superscripts indicate significance of the differences between Fielder and each of the transgenic families using Dunnett’s test (***P < 0.0001). Results for the four transgenic families for races TTKSK, TKTTF, and TRTTF are presented in SI Appendix, Fig. S4.
T1 plants from transgenic families T1Sr13-2 and T1Sr13-3 were more resistant to TTKSK than the untransformed control Fielder and showed significantly smaller pustules (P < 0.001; Fig. 3C). T2 plants from selected T1 plants from the four transgenic events showed resistance to races TTKSK, TKTTF, and TRTTF, and all exhibited average pustule sizes significantly smaller than those in Fielder (P < 0.01; SI Appendix, Fig. S4). Inoculation of Fielder and transgenic plants with race JRCQC provided no useful information because Fielder was highly resistant to this race. These transgenic experiments demonstrated that CNL13 is sufficient to confer resistance to races TTKSK, TKTTF, and TRTTF.
Taken together, the complementation results, the mutant analyses, and the high-density genetic and physical maps demonstrated that CNL13 is Sr13.
Effect of Temperature on Pathogen Growth and Gene Expression.
Effect of temperature on Pgt resistance.
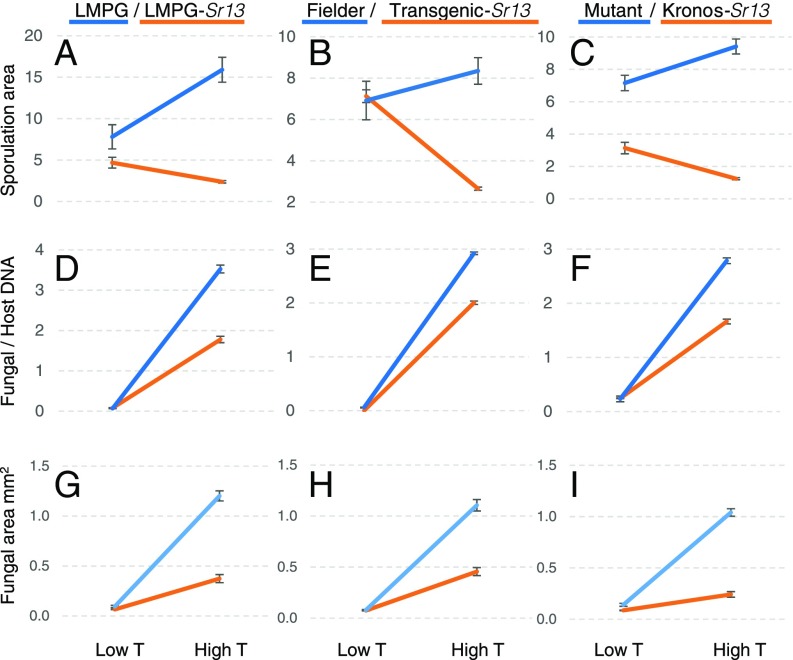
Sr13 showed higher levels of resistance to TTKSK when plants were grown at high temperature (25 °C day/22 °C night) than when grown at lower temperature (18 °C day/15 °C night). The tetraploid variety Kronos (Sr13) was more resistant than Rusty (no Sr13) at both high and low temperature regimes, but the hexaploid isogenic lines LMPG (no Sr13) and Khapstein/9*LMPG (=LMPG-Sr13) were both susceptible at low temperature and only differed at high temperature (SI Appendix, Fig. S5).
These observations were confirmed in a three-way ANOVA for average pustule size determined 13 d after inoculation (dpi) in eight independent plants. This analysis showed highly significant (P < 0.0001) effects for genotype, temperature, ploidy level, and for the three possible two-way interactions (SI Appendix, Table S3). The interactions between genotype and temperature can be visualized in Fig. 4 A–C. These interactions were also significant for Fielder and its derived Sr13 transgenic lines and for Kronos and its respective sr13-mutants (SI Appendix, Table S4).
Fig. 4.
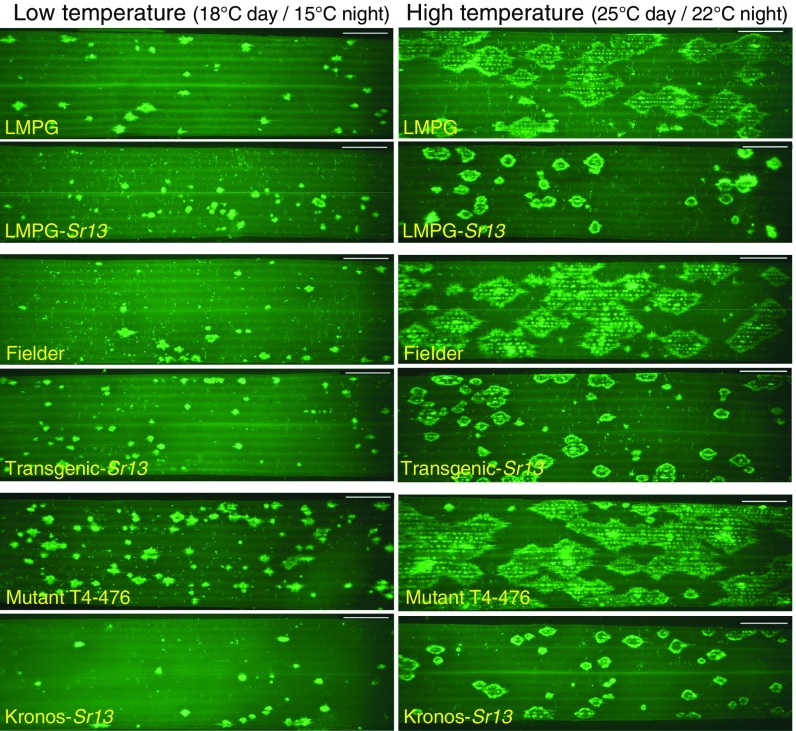
Pathogen growth in different genotypes at different temperatures. Interaction graphs for pathogen growth at two temperatures (low = 18 °C day/15 °C night and high = 25 °C day/22 °C night) and in two genotypes (Sr13 gene present or absent). Resistant genotypes are indicated in orange (LMPG-Sr13, transgenic-Sr13, and Kronos) and susceptible genotypes in blue (LMPG, Fielder, and Kronos sr13-mutant). Pathogen growth was estimated using average sporulation area 13 dpi (A–C), ratio between fungal and host DNA 5 dpi (D–F), and average size of individual fungal infection areas estimated by fluorescence microscopy 5 dpi (G–I). All plants were inoculated with TTKSK. Statistical analyses (including replication numbers) are detailed in SI Appendix, Table S4.
We also quantified the differences in TTKSK growth at 5 dpi by measuring the ratio of fungal DNA relative to host DNA (Fig. 4 D–F) and average infection areas using microscopy and a fluorescent dye that stains the pathogen (Figs. 4 G–I and 5). Both methods showed highly significant (P < 0.0001) differences between genotypes and between temperatures. We also detected highly significant interactions between genotype and temperature (SI Appendix, Table S4) that reflect the larger differences between genotypes at high than at low temperature (Fig. 4). The differences in fungal infection area between LMPG and LMPG-Sr13 were also validated with Pgt race BCCBC (isolate 09CA115-2; SI Appendix, Fig. S6).
Fig. 5.
Infection areas visualized by fluorescent staining. Visualization of TTKSK growth at different temperatures in resistant genotypes carrying Sr13 (LMPG-Sr13, transgenic-Sr13, and Kronos) and in susceptible genotypes without Sr13 (LMPG, Fielder, and Kronos sr13-mutant T4-476) 5 dpi. Infected leaves were cleared with KOH and stained with WGA-FITC.
Effect of temperature on Sr13 transcription profiles.
Transcript levels of Sr13 were similar at low and high temperature but were consistently lower in TTKSK inoculated than in mock-inoculated plants at the four sampling times (SI Appendix, Fig. S7). A three-way factorial ANOVA confirmed the absence of differences between temperatures (P = 0.84), and the significant differences between TTKSK inoculated and mock-inoculated plants (P < 0.0001) and among sampling times (P < 0.0001) (SI Appendix, Table S5). The consistency of the differences between inoculations across days and temperatures was reflected in nonsignificant interactions among these factors (SI Appendix, Table S5). Plants inoculated with TTKSK showed an 8.2% decrease in Sr13 transcript levels relative to the mock-inoculated control. No significant differences were detected between the first and second dpi or between the fourth and sixth dpi. However, a 15% decrease in CNL13 transcript levels was detected in a statistical contrast between the first two versus the last two time points (P < 0.0001). This decrease cannot be completely attributed to the pathogen, because an 11.5% decrease (P = 0.006) was also observed between the same time points for the mock-inoculated plants. However, the decrease was larger (18.7%) and more significant for the plants inoculated with TTKSK.
Effect of temperature on the transcript levels of pathogenesis-related genes.
We first compared the effect of TTKSK inoculation with mock inoculation at the high temperature regime on the transcript levels of pathogenesis-related genes (PR genes) PR1, PR2, PR3, PR4, PR5, and PR9 in different genotypes. These comparisons included resistant tetraploid Kronos versus susceptible sr13-mutant line T4-476 (Fig. 6), susceptible hexaploid LMPG versus its isogenic resistant line LMPG-Sr13 (SI Appendix, Fig. S8) and susceptible hexaploid Fielder versus the Sr13 transgenic line T1Sr13-2 (SI Appendix, Fig. S9).
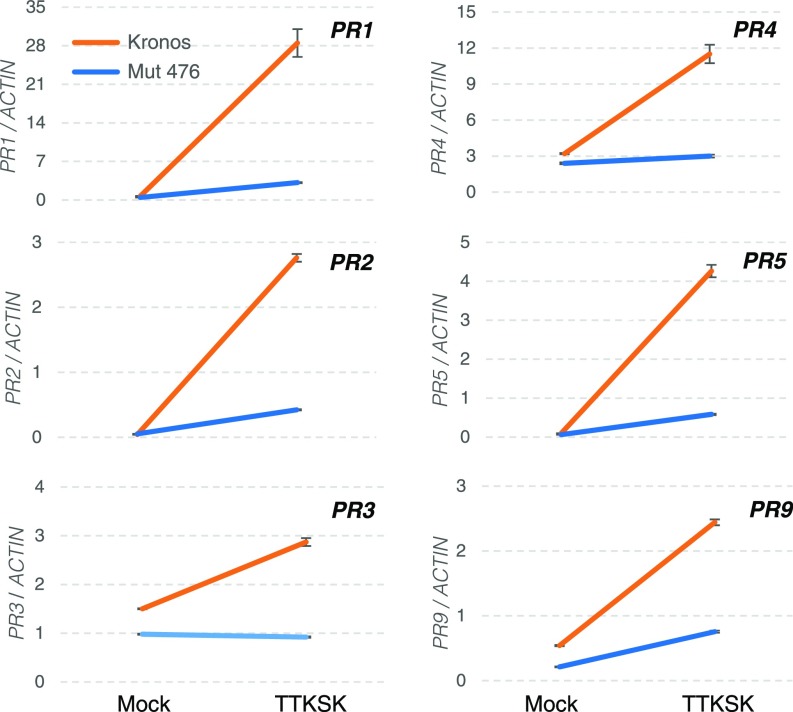
Fig. 6.
Transcript levels of PR genes. Transcript levels of PR genes PR1, PR2, PR3, PR4, PR5, and PR9 are presented in interaction graphs including two genotypes (Kronos vs. sr13-mutant T4-476) and two inoculation treatments (TTKSK vs. mock). Values relative to ACTIN endogenous control calculated using the ΔCT method. Error bars indicate SEMs. All interactions between genotype and inoculation were highly significant (P < 0.001). n = 6. Samples were collected 5 dpi. Statistical analyses are presented in SI Appendix, Table S6.
Two-way ANOVAs showed highly significant differences between resistant and susceptible genotypes and between inoculations (SI Appendix, Table S6). We also detected highly significant interactions between genotype and inoculation that reflect the different PR profiles observed in resistant and susceptible genotypes. In the genotypes carrying Sr13, we observed a strong up-regulation of all six PR genes after inoculation with TTKSK but not after mock inoculation (Fig. 6 and SI Appendix, Figs. S8 and S9). In the susceptible genotypes, we observed weaker responses that differed between genotypes. In the tetraploid sr13-mutants, all PR genes except PR3 showed a slight but significant up-regulation after inoculation with TTKSK relative to mock (Fig. 6). By contrast, in the susceptible hexaploid genotypes, most PR genes (except PR5 and PR9) showed a significant down-regulation after inoculation with TTKSK relative to mock (SI Appendix, Figs. S8 and S9 and Table S6). The opposite responses of susceptible and resistant hexaploid genotypes contributed to nonsignificant or less significant differences between inoculations for some PR genes (SI Appendix, Table S6).
We then studied the effect of different temperatures on the transcript levels of the PR genes (SI Appendix, Fig. S10). In Kronos plants inoculated with TTKSK or mock-inoculated at low and high temperatures, we observed highly significant (P < 0.0001) effects for temperature and inoculation (SI Appendix, Table S7). We also detected a highly significant interaction between temperature and inoculation because of the larger differences between TTKSK and mock-inoculated plants at high than at low temperatures (SI Appendix, Fig. S10). Similar results were observed for both sample collection times (4 and 6 dpi), although the overall transcript levels were higher (P < 0.001) 6 dpi than 4 dpi (except for PR2). In the mock-inoculated plants, we observed a small but significant increase (P < 0.05) in the transcript levels of all PR genes except PR2 at low relative to high temperatures (SI Appendix, Fig. S10).
Sr13 Allelic Variation.
Based on the sequence of Langdon BAC clone 1181K4 (Fig. 1; GenBank accession no. KY924305), we designed five pairs of gene-specific primers that amplify the complete coding and intron sequences of CNL13 (SI Appendix, Table S1). Using these primers, we sequenced cultivars previously used to map Sr13, including four cultivars resistant to TTKSK (Kronos, Kofa, Medora, and Sceptre) and three cultivars susceptible against the same race (Rusty, UC1113, and Mindum) (25). We also sequenced the complete coding region of CNL13 from the Ethiopian tetraploid landrace ST464 that carries Sr13 (21) and from the hexaploid genetic stock Khapstein/9*LMPG that defines the Sr13 gene name. As negative controls, we included the susceptible hexaploid lines LMPG, Fielder, and McNeal.
Among these lines, we found 10 DNA polymorphisms in the LRR region and two outside this region (SI Appendix, Fig. S11). We found three different haplotypes associated with resistance to TTKSK, which were designated R1 (Kronos and Khapstein/9*LMPG, KY825225), R2 (Kofa, Medora, and Sceptre, KY825226), and R3 (Langdon and ST464, KY924305). R2 and R3 differed from each other by two single nucleotide polymorphisms (SNPs), and both differed from R1 by three SNPs (SI Appendix, Fig. S11). We also identified four different haplotypes associated with susceptibility to TTKSK, which were designated S1 (Fielder, KY825227), S2 (UC1113, KY825228), S3 (LMPG, KY825229), and S4 (Rusty, Mindum, and McNeal). In the last three accessions, we failed to amplify any PCR product, which suggests that CNL13 is deleted in these lines (SI Appendix, Fig. S11).
Since most CNL13 polymorphisms were found in the LRR domain, we characterized the natural variation in this region in multiple accessions of T. urartu (74), T. turgidum ssp. dicoccoides (Koern. ex Asch. & Graebn.) Thell (50), T. turgidum ssp. dicoccon (90), T. turgidum ssp. durum (54), and T. aestivum (48, including three in which Sr13 was introgressed from T. turgidum ssp. dicoccon accession Khapli). No PCR product was obtained for 70 accessions of T. urartu (94.6%), 47 of T. turgidum ssp. dicoccoides (94%), 37 of T. turgidum ssp. dicoccon (41.1%), 10 of T. turgidum ssp. durum (18.5%), and 18 of T. aestivum (37.5%), which suggests that CNL13 is deleted in these accessions (SI Appendix, Table S8). The same DNAs were tested with primers for the positive control gene RAR1 (SI Appendix, Table S1) to confirm that the lack of amplification was not due to degraded DNA. We evaluated these 182 lines with five pairs of primers (Fig. 2 and SI Appendix, Table S9). We were able to amplify a complete NLR gene in 20 accessions (98.2% identical to Sr13, with a premature stop codon at the end of exon 2 and a frame shift mutation in exon 3). For the rest, we amplified partial fragments from 41 accessions and failed to amplify any product from 121 accessions. Some of the amplified PCR fragments, including those from T. urartu, are likely from paralogs or nonfunctional copies based on their low identity to Sr13 (SI Appendix, Table S9).
Among the 134 accessions for which we obtained PCR amplification products for the LRR region, 51 showed the 734R amino acid associated with resistance to TTKSK. Among these, we found 24 with the R1 haplotype, 21 with the R2 haplotype, and 6 with the R3 haplotype (SI Appendix, Table S8). To determine their resistance profiles, we inoculated accessions carrying each of these three haplotypes with races TTKSK, TKTTF, TRTTF, and JRCQC. Accessions carrying the R1 or R3 haplotypes showed resistance to all four races, but several of those carrying the R2 haplotype were susceptible to race JRCQC (SI Appendix, Fig. S12A). We confirmed that these differences were associated to Sr13 by inoculating two Sr13 homozygous resistant and two homozygous susceptible lines from the cross Kofa (R2) × UC1113 (S2) segregating for Sr13. Plants carrying the CNL13 R2 allele were resistant to TTKSK and TRTTF but susceptible to JRCQC (SI Appendix, Fig. S12B). Based on this result, we designated the resistance allele associated with the R1 and R3 haplotypes as Sr13a and the one associated with the R2 haplotype as Sr13b.
Haplotypes R1, R2, and R3 were restricted in this survey to T. turgidum ssp. dicoccon and T. turgidum ssp. durum accessions (except for three T. aestivum genetic stocks in which Sr13 was introgressed from Khapli). The only geographic region where we found all three resistant haplotypes was Ethiopia (in T. turgidum ssp. dicoccon accessions). Outside this region, we found two T. turgidum ssp. dicoccon accession carrying R2 (one in Spain and the other one in Saudi Arabia) and 11 carrying R1 [eight in India, one in Georgia, one in Russia, and one in Hungary (SI Appendix, Fig. S13 and Table S10)].
We obtained PCR products for the LRR region in T. urartu only for four (PI 428225, PI 428226, PI 428227, and PI 428251) of the 74 accessions tested. Sequencing of the PCR products revealed identical sequences among these four accessions that encoded a protein carrying the 734W amino acid associated with the susceptible alleles, but that were more divergent (93.8% identity) from the other 12 haplotypes, than those haplotypes were from each other (>98.6% identical, SI Appendix, Fig. S11). This result suggested that the T. urartu sequences correspond to a related paralog rather than to a CNL13 ortholog. Previous studies have shown that these and other accessions of T. urartu are highly susceptible to TRTTF and, therefore, do not have Sr13 (28). An identical sequence was found in a T. turgidum ssp. dicoccoides (PI 470955) collected close to the border between Syria and Turkey. The other two accessions of T. turgidum ssp. dicoccoides (collected in Israel) showed the CNL13 S5 haplotype (SI Appendix, Table S10).
Among the tetraploid and hexaploid accessions for which we obtained CNL13 LRR amplification products, we found the 734W amino acid associated with susceptibility in 78 accessions. These accessions included eight additional DNA polymorphisms, which defined six additional haplotypes. For each of these new haplotypes, we sequenced the complete gene for one accession (S5–S10, GenBank accession nos. KY825230–KY825235, SI Appendix, Fig. S11). In addition to these SNPs, haplotypes S8, S9, and S10 had insertions in the coding regions of 162, 17, and 3 bp, respectively. The 17-bp insertion in S9 altered the reading frame, modifying or eliminating the last 16 aa. A 48-bp insertion was found in the first intron of haplotypes S1 and S8. Inoculation of representative accessions for each of these new haplotypes with TTKSK revealed that at least one accessions per haplotype was susceptible to TTKSK (SI Appendix, Fig. S14), confirming that they were susceptible CNL13 alleles. The geographic distribution of the different CNL13 haplotypes is described in SI Appendix, Fig. S13 and Table S10.
The phylogenetic relationships among CNL1, CNL3, and CNL13 and their closest proteins in T. aestivum, T. urartu, S. cereale, Hordeum vulgare, and Brachypodium distachyon are described in SI Appendix, Fig. S15. These results suggest that the duplication of the three complete CNL proteins in this cluster originated before the wheat-rye divergence. Two of the three closest B. distachyon proteins (reconstructed by correcting frame shift mutations) were present in the CNL13 colinear region in the B. distachyon genome (Fig. 1).
An SNP Associated with CNL13-Resistant Haplotypes.
A comparison between the Sr13 susceptible and resistant haplotypes showed only one DNA polymorphism (T2200C, and its corresponding amino acid change W734R) that perfectly matched the differences between Sr13 haplotypes susceptible and resistant to TTKSK (SI Appendix, Fig. S11). The amino acid change W to R is associated with a negative BLOSUM62 score (−3), which is indicative of a disruptive change for protein structure and function. To visualize the effect of this change on the LRR structure, we modeled the LRR domain of the R3 haplotype with both the 734R and 734W amino acids using Phyre 2 (29). The two structures revealed an α/β horseshoe fold typical of many LRR domains, but displayed large differences in the region flanking the W734R polymorphisms and in the two proximal β-sheets facing the interior side of the horseshoe fold (SI Appendix, Fig. S16).
The complete association detected so far between the T2200C SNP and resistance to TTKSK (SI Appendix, Table S8) suggests that a marker for this polymorphism would be a useful tool for selection of Sr13 in wheat breeding programs. We designed a derived cleaved amplified polymorphic sequence (dCAPS) marker (SI Appendix, Table S1, primers Sr13F/R). Digestion of the amplification product with restriction enzyme HhaI yields a 244-bp band in accessions carrying the resistant haplotypes and a 265-bp band in accessions carrying the susceptible haplotypes (Fig. 3B). Lack of PCR amplification with these primers is indicative of the susceptible S4 allele.
We used this marker to clarify the relationship between Sr13 and Srdp2, a gene first identified in T. turgidum subsp. durum accession PI 94701 (30). Srdp2 was first proposed to be allelic to Sr13 (23) and later suggested to be distinct from Sr13 based on pathogenic variability studies (31). We found no PCR product for Sr13 in the Srdp2 donor lines PI 94701 and Golden Ball-derived W3504, supporting the hypothesis that Srdp2 and Sr13 are different genes.
Discussion
The Sr13 Region.
We mapped the Sr13 gene to a ∼858-kb region including two complete NLR genes and 10 truncated NLR genes. NLR genes in both mammal and plant species tend to be organized in clusters of variable size, which provide a reservoir of genetic variation (32). The Sr13 NLR cluster in Langdon (LDN, R3 haplotype) and Kronos (R1 haplotype) are almost identical. The two CNL13 resistance alleles are 99.9% identical at the DNA level, and the sequences of all other complete and truncated NLR genes described in Fig. 1D are 100% identical between the two haplotypes. These results suggest a close evolutionary relationship between the regions, including the CNL13 R1 and R3 haplotypes.
By contrast, the comparison of the 955-kb LDN sequence with the available genomic sequence of Chinese Spring (CS; haplotype S4) revealed limited conservation in this region. Only 8 of the 14 complete and truncated NLR genes were detected in CS (>98.5% DNA identity; Fig. 1D). The missing genes in the CS genome (including CNL13) define two large deleted regions of 368 kb and 50 kb, respectively (Fig. 1, red lines). Similar deletions seem to be present in the tetraploid Rusty (S4 haplotype), since primers for markers within these deletions did not yield PCR amplification products in this durum variety. We also identified four NLR-related sequences in CS and Rusty that were completely linked genetically to CNL13, which were not present in Kronos or Langdon. The extensive polymorphisms detected between the S4 and R3 haplotype in this region suggest rapid evolutionary changes within this NLR cluster in polyploid wheat, a characteristic that has been described before for other NLR clusters (33). The presence of large indels that differentiate Kronos and Rusty in this region explained the lack of recombination observed in the large region between cnl2 and CJ641478 (∼858 kb; Fig. 1).
In the colinear region of Brachypodium chromosome 3 between Bradi3g60460 (ortholog of wheat CJ671993) and Bradi3g60430 (ortholog of wheat CJ641478), we identified two NLR annotated genes (Bradi3g60446 and Bradi3g60453; Fig. 1), which encode proteins 75% and 66% similar (excluding indels larger than one amino acid) to wheat CNL13, respectively. This result suggests that this NLR cluster likely has a long evolutionary history that extends beyond the divergence between the Triticum and Brachypodium lineages (estimated 30–40 Mya; ref. 34). This conclusion is also consistent with the presence of large NLR clusters between CJ671993 and CJ641478 orthologs in CS chromosomes 6B (555 kb) and 6D (465 kb).
Identification and Initial Characterization of CNL13.
The susceptibility to Ug99 of the four independent CNL13 Kronos mutants and the resistance of the CNL13 transgenic Fielder lines confirmed that this gene was both necessary and sufficient to confer resistance to Ug99 and, therefore, that CNL13 is Sr13. We detected a single isoform of CNL13 in our expression studies, which compared with the genomic sequence revealed the presence of complex 5′ and 3′ UTR including two and one intron, respectively. A complex UTR region with multiple introns was also described for the NLR gene Sr35, which also confers resistance to Ug99 (12). However, the role of these complex UTR regions is currently unknown. CNL13 is expressed at relatively high levels in the leaves of Kronos (1- to 1.6-fold ACTIN) and is slightly down-regulated in the presence of the stem rust pathogen (8.2% decrease), suggesting that Pgt is not very effective in suppressing this gene.
Effect of Temperature on Sr13-Mediated Resistance and Gene Expression.
Temperature is known to affect plant resistance to diseases, but the mechanisms are only partially understood (35). Known mechanisms include reduced steady-state resistance protein levels at high temperatures (36), temperature-sensing NB-LRR proteins (37), and salicylic acid (SA) regulation involving EDS1 and PAD4 (38, 39). In these studies, elevated growth temperature was associated with inhibition of the resistance response (35). Similar results have been observed for wheat Pgt resistance genes Sr6, Sr10, Sr15, and Sr17, which become more susceptible at increasing temperatures.
By contrast, Sr13 was shown in previous studies (23, 24) and here to be more effective at high than at low temperature (Fig. 4 A–C). This observation was also supported by highly significant interactions between temperature and genotype in the three pairs of genotypes included in this study and in the three different methods used to estimate TTKSK growth (SI Appendix, Table S4). These consistent interactions indicate that the Sr13-mediated resistance is modulated by temperature. As wheat stem rust is often most problematic at relatively high temperatures (40), the temperature-sensitive Sr13 resistance response could have been selected to optimize the balance between disease responsiveness and fitness.
At the early stages of the infection (5 dpi), the growth of the pathogen at low temperatures was slower for both the susceptible and resistant genotypes (Fig. 4 D–I). However, at the sporulation stage (13 dpi), the resistant genotypes showed reduced pathogen growth at high temperatures (Fig. 4 A–C), suggesting an active resistance mechanism of Sr13. This cannot be explained by differences in Sr13 transcript levels because no significant differences were observed in its transcript levels between high and low temperatures.
By contrast, six PR genes showed a strong up-regulation at high but not at low temperature. We concluded that this up-regulation was dependent on the presence of the pathogen because we did not observe it in the mock-inoculated plants (SI Appendix, Fig. S10). This may reduce the fitness cost associated with Sr13 in the absence of the pathogen. We also concluded that the up-regulation of the PR genes at high temperature was dependent on the presence of Sr13 because it was not observed in the sr13-mutant (Fig. 6), or the LMPG (SI Appendix, Fig. S8) or Fielder (SI Appendix, Fig. S9) susceptible controls. Based on these results, we hypothesize that the strong and coordinated up-regulation of PR genes at high temperatures contributed to the higher levels of resistance to TTKSK conferred by Sr13 under these conditions. This is consistent with the type of resistance conferred by Sr13. The Sr13-mediated resistance is not associated with the rapid cell death characteristic of a typical hypersensitive response, but rather with a slower growth of the pathogen at high temperatures in the presence of the resistance gene (Fig. 5). A chlorotic halo is observed around the Pgt pustules in the resistant reactions, which can eventually lead to cell death.
Finally, we also observed Sr13-independent mechanisms of PR gene regulation in wheat. The slower growth of the pathogen at low temperatures, together with the slight up-regulation of most PR genes at low temperatures in the susceptible genotype, can explain the reduced sporulation areas in the susceptible genotypes at low temperatures. In addition, in the susceptible hexaploid backgrounds, most of the PR genes were down-regulated by inoculation with TTKSK relative to mock. This observation suggests that Pgt has developed some mechanisms to down-regulate this defensive mechanism in hexaploid wheat. It would be interesting to investigate why this response was not observed in tetraploid Kronos.
Identification of Different CNL13 Haplotypes.
Thirteen different haplotypes were detected in the CNL13 LRR domain from 311 diploid, tetraploid, and hexaploid Triticum species (including S4 where the gene appears to be deleted). Evaluation of these different haplotypes with Ug99 revealed three resistant haplotypes (R1–R3) and 10 susceptible haplotypes (S1–S10). The additional haplotype found in T. urartu is likely from a close paralogous gene (SI Appendix, Fig. S15) and is not included in the haplotype analysis.
These haplotypes differed by 27 SNPs at the DNA level (18 in the LRR) and 25 aa at the protein level (16 in the LRR, SI Appendix, Fig. S11). The large ratio of nonsynonymous (25) to synonymous (2) substitutions suggests that the CNL13 gene has been under positive selection, a hypothesis also supported by the Z test of positive selection as implemented in MEGA 6 (41). The number of nonsynonymous substitutions per nonsynonymous site (Ka) was significantly higher (P < 0.05) than the number of synonymous substitutions per synonymous site (Ks) for 47 of the 66 possible pairwise haplotype comparisons (SI Appendix, Table S11). Nonsynonymous substitutions were observed for 38 of the 66 pairwise comparisons (Ka/Ks = infinite), and for the other 28, the average Ka/Ks was 2.58 ± 0.17. This ratio is larger than one, which is the expected ratio under neutral selection. Positive selection has been reported in the LRR domains of multiple NLR genes (33). For CNL13, both the LRR and non-LRR region appear to be under positive selection. However, the number of polymorphisms available in the non-LRR region is too low to make a confident conclusion.
The only amino acid change that was consistent with the susceptible and resistant haplotypes was a W to R change at position 734 within the LRR domain (SI Appendix, Fig. S11). The W734R mutation is associated with large changes in the predicted LRR structure (SI Appendix, Fig. S16). Based on this result, we hypothesize that the W734R polymorphism may play an important role in the detection of the pathogen. An additional induced mutation in the LRR domain (A717V) located only 17 amino acids apart from W734R was also associated with susceptibility, further supporting the hypothesis that this region of the LRR domain is critical for the Sr13 resistance to TTKSK.
Evolution of Sr13.
To trace the origin of the Ug99-resistant haplotypes, we analyzed the sequence variation in the CNL13 LRR domain and the geographic distributions of the different haplotypes. A lack of amplification of the CNL13 LRR domain (haplotype S4) was detected in 95% of the T. urartu accessions and 94% of the T. turgidum subsp. dicoccoides accessions included in this study. This result suggests that the S4 haplotype was likely transferred from T. urartu to the wild tetraploid wheats, although we cannot rule out independent deletion events.
The only CNL13 allele amplified so far from T. turgidum subsp. dicoccoides was S5, detected in two accessions from Israel. The same allele was detected in five T. turgidum subsp. dicoccon accessions from Ethiopia, Iran, and Russia (SI Appendix, Fig. S13 and Table S10), suggesting a wide distribution. These results, together with the absence of differential SNPs in S5 (SI Appendix, Fig. S11, red) suggests that S5 may be similar to the ancestral CNL13 haplotype.
Three indirect observations point to Ethiopia and neighboring countries as a likely region for the origin of the CNL13-resistant haplotypes. First, this is the only region where all three Ug99-resistant haplotypes were found, and that has the largest diversity of CNL13 susceptible haplotypes (SI Appendix, Table S10). This diversity in CNL13 may reflect a higher diversity in the pathogen population in this region. Preliminary results suggest that Berberis holstii, which is endemic to the highlands of East Africa, is a functional alternate host where Pgt can complete its sexual cycle (42). This may have contributed to the origin of the new virulences in the Pgt population (including the development of Ug99) and promoted the increased diversity in the CNL13 gene in this region. Since a single SNP change can be used to diagnose the CNL13-resistant alleles, DNA extractions from ancient archeological samples of T. turgidum subsp. dicoccon from Ethiopia and India may provide interesting information on the origin and dispersion of the CNL13-resistant alleles. The presence of multiple T. turgidum subsp. dicoccon accessions from India carrying the R1-resistant haplotype are not surprising, given that connections between Ethiopia and India go back more than 2,000 y of recorded history.
So far, no CNL13-resistant haplotype has been detected in T. turgidum subsp. dicoccoides, suggesting that the haplotypes resistant to Ug99 may have originated in T. turgidum subsp. dicoccon. Selection likely contributed to the rapid increase in the frequency of the resistant alleles, found in ∼31% of the accessions from this subspecies (SI Appendix, Table S8). Sr13 was then incorporated into modern durum cultivars derived from T. turgidum subsp. dicoccon accessions from Ethiopia (ST464) and India (Khapli) (20, 21). Among the 54 T. turgidum ssp. durum accessions analyzed so far, only ∼37% carried the CNL13-resistant alleles (SI Appendix, Table S8), which suggests that there is still a large proportion of durum accessions that can benefit from the incorporation of Sr13.
At the hexaploid level, the most frequent CNL13 haplotypes were S1 (26 accessions) and S4 (18 accessions). The S3 haplotype was identified only in LMPG (SI Appendix, Table S8). The CNL13 allele R1 was the only resistant allele found in hexaploid wheat, where it was transferred from T. turgidum subsp. dicoccon Khapli (SI Appendix, Table S8). However, since Sr13 is present in several durum varieties, it is also expected to be present in synthetic wheat generated from T. turgidum subsp. durum × A. tauschii hybrids or in their derivatives (43). Indeed, we found Sr13-resistant alleles in three of the nine synthetic wheats we tested. The synthetics “ITMI Synthetic” and “CROC_1/A. tauschii (205)” showed the R3 haplotype of Sr13, and the synthetic “CETA/A. tauschii (327)” showed the R1 haplotype.
One additional explanation for the low frequency of the Sr13 in common wheat is the lower level of resistance conferred by this gene in hexaploid compared with tetraploid genetic backgrounds (23, 24). The reasons for this reduction and the opportunities to improve the impact of this gene in common wheat are discussed in more detail below.
Utilization of Sr13 in Breeding.
The lower level of Pgt resistance conferred by Sr13 when it is transferred from tetraploid to hexaploid wheat has been reported for other resistance genes transferred to hexaploid wheat from relatives of lower ploidy (44). This has been attributed to the presence of modifiers or inhibitors of the transferred resistance gene in hexaploid wheat. In few cases, such as the Pm8 powdery mildew resistance gene, the reduced resistance of the transferred gene was found to vary among hexaploid backgrounds. For Pm8, it was possible to identify the chromosome region responsible for this reduction and to use this knowledge to enhance the effect of Pm8 in hexaploid wheat (44). Similar studies may be useful to enhance the effectiveness of Sr13 resistance to TTKSK in common wheat-breeding programs.
Sr13 was previously reported to be ineffective against races TRTTF and JRCQC, based on high infection reactions observed on hexaploid varieties Khapstein/9*LMPG and Combination VII (R1 haplotype) in greenhouse and field nurseries in Ethiopia (45). However, our results show that, under controlled environmental conditions, Sr13a is effective against races JRCQC, TRTTF, and TKTTF in both tetraploid and hexaploid wheat (SI Appendix, Figs. S2 and S4). This result is important, because TKTTF has caused severe yield losses in southern Ethiopia in the Ug99-resistant variety Digalu in 2013 and 2014 (9). For hexaploid wheat, a test of the effectiveness of Sr13-resistant haplotypes against JRCQC, TRTTF, and TKTTF in the field is still pending.
The resistance conferred by Sr13 to different Pgt races suggests that this gene will be useful in tetraploid wheat breeding programs. For this purpose, the Sr13a allele is a better option than the Sr13b allele because of the additional resistance to race JRCQC. However, because of the rapid breakdown of Sr genes deployed singly, it would be better to combine Sr13 with additional TTKSK resistance genes. The diagnostic marker Sr13F/R developed in this study can be used to accelerate the generation of these resistance gene pyramids in durum wheat breeding programs.
Once transgenic approaches gain a wider public acceptance, the cloning of Sr13 will provide an additional resistance gene to include in transgenic cassettes combining multiple resistance genes. As more Pgt resistance genes become available, it would be important to combine genes that use different resistance mechanisms. The delayed pathogen growth and PR gene up-regulation associated with Sr13 can complement the hypersensitive resistance mechanism associated with Sr35 and other resistance gene (12). Transgenic cassettes of resistance genes have the advantage of introducing multiple resistance genes in a single step and in a single chromosome location. These multiple resistance loci can be traced with a single molecular marker and have a reduced risk of being disrupted by recombination, a problem frequently encountered when resistance gene pyramids are used by breeding programs based only on phenotypic selection.
In addition to the practical applications described above, the cloning of Sr13 provides a tool to identify the Pgt effectors recognized by this gene and the proteins targeted by these effectors. A study of the downstream components of this resistance response can provide useful knowledge to develop more durable resistance strategies against this devastating pathogen.
Materials and Methods
Plant Materials.
F2 plants (6,081) from the cross of Kronos × Rusty were used to generate the Sr13 high-density genetic map. A collection of 74 accessions of T. urartu, 50 of T. turgidum ssp. dicoccoides, 90 of T. turgidum ssp. dicoccon, 54 of T. turgidum ssp. durum, and 48 of T. aestivum was used to study the taxonomic and geographic distribution of the Sr13 alleles. These accessions included the original source of Sr13 (T. turgidum ssp. dicoccon Khapli), seven T. turgidum ssp. durum lines used as parents in Sr13 mapping populations (Kronos, Medora, Sceptre, Kofa, Rusty, Mindum, UC1113) (25), and three hexaploid accessions with Sr13 introgressions from Khapli (Khapstein/9*LMPG, W2691Sr13, and Combination VII). Seeds of the mutants used for validation were obtained from the Kronos sequenced EMS induced mutant population (27).
Stem Rust Assays.
We tested the efficacy of Sr13 resistance against Pgt races that caused epidemics in Kenya and/or Ethiopia [TTKSK (Ug99), TKTTF] in addition to other virulent races that were previously reported as virulent to Sr13 (TRTTF, JRCQC). Race TKTTF, originally detected in Turkey in the 1990s, is currently distributed throughout Europe, West Asia, North Africa, and East Africa (8, 10). TRTTF and the closely related RRTTF race were first detected in Iran in 1997 and have been found later in Ethiopia, Yemen, and Pakistan (8, 39). JRCQC has been detected in Ethiopia recently (39) and as early as 1987 (31). Seedling resistance evaluations were performed at the US Department of Agriculture-Agricultural Research Service (USDA-ARS) Cereal Disease Laboratory using multiple Puccinia graminis f. sp. tritici races [TTKSK (Ug99) isolate 04KEN156/04, TRTTF isolate 06YEM34-1, JRCQC isolate 09ETH08-3, and TKTTF isolate 13ETH18-1].
Procedures for inoculation, incubation, and scoring disease reactions were reported previously (28) except that seedling assays were incubated in growth chambers maintained at 25 °C day/22 °C night after inoculation unless otherwise described. The effectiveness of Sr13 in tetraploid and hexaploid wheat under two different temperature regimes (18 °C day/15 °C night and 25 °C day/22 °C night) was also evaluated at the USDA-ARS Cereal Disease Laboratory.
Marker Development and BAC Library Screening and Sequencing.
The initial markers flanking Sr13 were obtained from previous genetic maps (25). New genome-specific primers were developed from wheat genes orthologous to those located in the colinear region in Brachypodium. Additional markers were developed from NLR genes in the CS 6AL arm sequence and, as they became available, from the BACs of T. turgidum subsp. durum cultivar Langdon (26) used in the construction of the physical map. The physical map was assembled by chromosome walking, starting from the closest proximal marker CJ641478 and from the completely linked marker EX24785 in both directions.
DNAs were extracted from selected BACs using a large-construct kit (Qiagen) and sequenced with Illumina at the University of California (UC) Davis Genomic Center. Sequences were assembled using Galaxy (46), and gaps were filled by Sanger sequencing. Based on the BAC sequence, we designed additional genomic specific primers to rescreen the BAC library for new clones. The process was repeated until markers flanking Sr13 in both directions were identified.
Sequencing Annotation.
First, we identified and annotated repetitive elements using the Triticeae Repeat Sequence Database (https://wheat.pw.usda.gov/ITMI/Repeats/blastrepeats3.html). Host duplications flanking the repetitive elements were manually curated. The nonrepetitive sequences were then annotated using a combination of BLASTN searches against wheat EST collections and the TIGR Wheat Genome Database (blast.jcvi.org/euk-blast/index.cgi?project=tae1). We also used gene models generated by comparison of our genomic region with diploid and tetraploid wheat transcriptomes (wheat.pw.usda.gov/GG2/WheatTranscriptome/). Additional BLASTX searches were performed against the nonredundant GenBank database of plant proteins. The annotated sequences of the 955-kb physical map (including three gaps) and of the different CNL13 haplotypes were submitted to GenBank using Sequin.
5′ RACE.
To determine the transcriptional start of the CNL13 gene, we used 5′ RACE. High quality total RNA was extracted from the resistant parent Kronos, and the FirstChoice RLM-RACE Kit (Invitrogen) was used to perform the RACE reactions as described before (12). Briefly, primers developed at the 5′ coding region of CNL13 were used as the gene-specific primers for nested PCRs (SI Appendix, Table S1). The TA cloning kit (Invitrogen) was used to clone the PCR products from the 5′ RACE reactions, and selected positive clones were sequenced using the Sanger method.
Effect of Temperature and Genotype on Pathogen Growth.
Seedlings of different genotypes with and without Sr13 were grown in a greenhouse and at the three-leaf stage were transferred to growth chambers at two different temperature regimes (low temperature:18 °C day/15 °C night and high temperature: 25 °C day/22 °C night). Half of the plants were inoculated with Pgt race TTKSK, and the other half were mock-inoculated. All plants were grown under a long day photoperiod (16 h light and 8 h dark) with a light intensity of ∼500 μM·m−2·s−1 and a humidity of 95%.
Pathogen growth was estimated by three different methods. The amount of fungal DNA relative to host DNA and the average fungal infection area (visualized by fluorescence) were determined 5 dpi, whereas the average pustule size was determined 13 dpi. Relative DNA amount was determined as described before (12). Average infection area was determined in infected leaves cleared with KOH (37 °C, 12 h) and stained with WGA-FITC (L4895-10MG; Sigma-Aldrich). Images were obtained with a Zeiss Discovery V20 fluorescent dissecting scopes, and average fluorescent area per leaf was estimated from 20 individual infection sites. Average sporulation area was determined from images of infected leaves using ASSES (version 2) image analysis software for plant disease quantification from the American Phytopathology Society (47). In each leaf, all sporulation areas were measured in two nonoverlapping regions of 60 mm2.
Effect of Temperature and Pathogen Inoculation on Sr13 and PR Genes Transcript Levels.
Total RNA was extracted using Spectrum Plant Total RNA Kit (Sigma-Aldrich). First-strand cDNA was synthesized from 1 µg of total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was carried out on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) using Fast SYBR GREEN Master Mix. Transcript levels were expressed as fold-ACTIN levels (the number of molecules in the target/the number of ACTIN molecules) using the 2−ΔCT method as described before (48). We calculated the significance of the differences in expression levels using factorial ANOVAs and the SAS program version 9.4.
To evaluate the effect of temperature and Pgt inoculation on Sr13 transcript levels, we developed qRT-PCR primers Sr13RTF1R1 (SI Appendix, Table S1). The forward primer was designed in the third exon and the reverse primer in the junction between exons three and four to avoid amplification from genomic DNA. Primer efficiencies were estimated using five fourfold cDNA dilutions (1:1, 1:4, 1:16, 1:64, and 1:256), with each reaction carried out in triplicate. The qRT-PCR primers used in this study showed a single product in dissociation curves and its efficiency was higher than 95% (SI Appendix, Table S1). For each treatment, three samples from three different Kronos plants were collected at four time points: 1, 2, 4, and 6 dpi with TTKSK, always at the same time of the day.
We also characterized the expression of six pathogenesis-related (PR) genes (PR1-AJ007348, PR2-Y18212, PR3-AB029934, PR4-AJ006098, PR5-AF442967, and PR9-X56011) in two experiments. In the first experiment, we compared the expression of these genes in Kronos and the sr13-mutant T4-476 inoculated with TTKSK or mock-inoculated (both at high temperature). In the second experiment, we used only the variety Kronos and evaluated the plants 4 and 6 dpi with TTKSK or mock-inoculated under high and low temperature regimes. Data were transformed to restore normality of residuals in the factorial ANOVAs and was analyzed using SAS v. 9.4.
Transformation.
We cloned a genomic fragment of 8,055 bp from Langdon BAC 1181K4 into a HindIII-HF/SpeI linearized binary vector pLC41Hm (49). This fragment included the complete coding region and introns (3,513 bp) plus 2,108 bp upstream of the start codon (including the 538 bp 5′ UTR) and 2,434 bp downstream of the stop codon (including the 2,187 bp 3′ UTR). DNA was extracted using the QIAGEN Large-Construct Kit (QIAGEN), and PCR amplifications were performed using Phusion High-Fidelity DNA Polymerase (New England BioLabs). The construct was introduced into the Ug99-susceptible hexaploid wheat variety Fielder via Agrobacterium tumefaciens-mediated transformation. To validate the presence of the transgene, we used two independent pairs of PCR primers. Primers HptmikiF/R amplify the hygromycin resistance gene present in the pLC41Hm binary vector and primers 6ACNL13F3/R3 (digested with enzyme FauI) amplify a region of the CNL13 LRR domain (SI Appendix, Table S1). To estimate the number of copies inserted in each of the transgenic events, we used a TaqMan Copy Number Assay that includes CONSTANS2 as the single copy control gene (50). These results were further validated using progeny tests of the T1 plants.
Transcript levels in the transgenic plants were estimated with qRT-PCR primers Sr13RTF1R1 (SI Appendix, Table S1) using the 2−ΔCT method and ACTIN as an endogenous control (48). The significance of the differences in transcript levels between Fielder and the transgenic plants was estimated using the Dunnett’s test as implemented in SAS 9.4. Because Fielder has a susceptible CNL13 allele (S1) that is also amplified by the qRT-PCR primers, we complemented this test with a semiquantitative PCR test using primers Sr13F/R and restriction enzyme HhaI to distinguish the two alleles (SI Appendix, Table S1).
Supplementary Material
Acknowledgments
We thank M. Padilla for excellent technical support; L. Epstein (UC Davis) for her help in the quantification of the sporulation size; S. Chao (USDA-ARS Fargo), J. Dvorak (UC Davis), and Y. Jin (USDA Cereal Disease Laboratory) for providing critical materials; the UC Davis genome center for the BAC sequencing; and UC Davis plant transformation facility for the wheat transformation. Work at J.D.’s laboratory was supported by the Howard Hughes Medical Institute and USDA Grant 2017-67007-25939 from the USDA National Institute of Food and Agriculture (NIFA). Work at M.N.R. laboratory was supported by USDA-ARS appropriated project 5062-21220-021-00-D, the USDA-ARS National Plant Disease Recovery System, and National Research Initiative Competitive Grant 2017-67007-25939 from the USDA NIFA.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY825225–KY825235 and KY924305).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706277114/-/DCSupplemental.
References
- 1.Pretorius ZA, Singh RP, Wagoire WW, Payne TS. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 2000;84:203. doi: 10.1094/PDIS.2000.84.2.203B. [DOI] [PubMed] [Google Scholar]
- 2.Jin Y, et al. Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp tritici. Plant Dis. 2008;92:923–926. doi: 10.1094/PDIS-92-6-0923. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, et al. Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp tritici. Plant Dis. 2009;93:367–370. doi: 10.1094/PDIS-93-4-0367. [DOI] [PubMed] [Google Scholar]
- 4.Pretorius ZA, Szabo G, Boshoff WHP, Herselman L, Visser B. First report of a new TTKSF race of wheat stem rust (Puccinia graminis f. sp. tritici) in South Africa and Zimbabwe. A. Pretorius. Plant Dis. 2012;96:590. doi: 10.1094/PDIS-12-11-1027-PDN. [DOI] [PubMed] [Google Scholar]
- 5.Rouse MN, et al. Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor Appl Genet. 2014;127:1681–1688. doi: 10.1007/s00122-014-2330-y. [DOI] [PubMed] [Google Scholar]
- 6.Patpour M, et al. First report of the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Egypt in 2014. Plant Dis. 2016;100:863. [Google Scholar]
- 7.Singh RP, et al. Emergence and spread of new races of wheat stem rust fungus: Continued threat to food security and prospects of genetic control. Phytopathology. 2015;105:872–884. doi: 10.1094/PHYTO-01-15-0030-FI. [DOI] [PubMed] [Google Scholar]
- 8.Fetch T, Zegeye T, Park RF, Hodson D, Wanyera R. Detection of wheat stem rust races TTHSK and PTKTK in the Ug99 race group in Kenya in 2014. Plant Dis. 2016;100:1495–1495. [Google Scholar]
- 9.Olivera P, et al. Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013-14. Phytopathology. 2015;105:917–928. doi: 10.1094/PHYTO-11-14-0302-FI. [DOI] [PubMed] [Google Scholar]
- 10.Krattinger SG, et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- 11.Moore JW, et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat Genet. 2015;47:1494–1498. doi: 10.1038/ng.3439. [DOI] [PubMed] [Google Scholar]
- 12.Saintenac C, et al. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science. 2013;341:783–786. doi: 10.1126/science.1239022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steuernagel B, et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol. 2016;34:652–655. doi: 10.1038/nbt.3543. [DOI] [PubMed] [Google Scholar]
- 14.Periyannan S, et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science. 2013;341:786–788. doi: 10.1126/science.1239028. [DOI] [PubMed] [Google Scholar]
- 15.Mago R, et al. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat Plants. 2015;1:15186. doi: 10.1038/nplants.2015.186. [DOI] [PubMed] [Google Scholar]
- 16.Dubcovsky J, Luo M, Dvorak J. Differentiation between homoeologous chromosomes 1A of wheat and 1Am of Triticum monococcum and its recognition by the wheat Ph1 locus. Proc Natl Acad Sci USA. 1995;92:6645–6649. doi: 10.1073/pnas.92.14.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukaszewski AJ. Physical distribution of translocation breakpoints in homoeologous recombinants induced by the absence of the Ph1 gene in wheat and triticale. Theor Appl Genet. 1995;90:714–719. doi: 10.1007/BF00222138. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T, et al. Vrn-D4 is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theor Appl Genet. 2010;120:543–552. doi: 10.1007/s00122-009-1174-3. [DOI] [PubMed] [Google Scholar]
- 19.Knott DR. Near-isogenic lines of wheat carrying genes for stem rust resistance. Crop Sci. 1990;30:901–905. [Google Scholar]
- 20.Knott DR. The inheritance of rust resistance: IX. The inheritance of resistance to races 15B and 56 of stem rust in the wheat variety Khapstein. Can J Plant Sci. 1962;42:415–419. [Google Scholar]
- 21.Klindworth DL, Miller JD, Jin Y, Xu SS. Chromosomal locations of genes for stem rust resistance in monogenic lines derived from tetraploid wheat accession ST464. Crop Sci. 2007;47:1441–1450. [Google Scholar]
- 22.Jin Y, et al. Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp tritici. Plant Dis. 2007;91:1096–1099. doi: 10.1094/PDIS-91-9-1096. [DOI] [PubMed] [Google Scholar]
- 23.McIntosh RA, Wellings CR, Park RF. 1995 Wheat Rusts, an Atlas of Resistance Genes (CSIRO, Melbourne). Available at https://www.globalrust.org/sites/default/files/wheat_rust_atlas_full.pdf. Accessed October 11, 2017.
- 24.Roelfs AP, Mcvey DV. Low infection types produced by Puccinia graminis f. sp. tritici and wheat lines with designated genes for resistance. Phytopathology. 1979;69:722–730. [Google Scholar]
- 25.Simons K, et al. Genetic mapping of stem rust resistance gene Sr13 in tetraploid wheat (Triticum turgidum ssp. durum L.) Theor Appl Genet. 2011;122:649–658. doi: 10.1007/s00122-010-1444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cenci A, et al. Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum) Theor Appl Genet. 2003;107:931–939. doi: 10.1007/s00122-003-1331-z. [DOI] [PubMed] [Google Scholar]
- 27.Krasileva KV, et al. Uncovering hidden variation in polyploid wheat. Proc Natl Acad Sci USA. 2017;114:E913–E921. doi: 10.1073/pnas.1619268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouse MN, Jin Y. Stem rust resistance in A-genome diploid relatives of wheat. Plant Dis. 2011;95:941–944. doi: 10.1094/PDIS-04-10-0260. [DOI] [PubMed] [Google Scholar]
- 29.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rondon MR, Gough FJ, Williams ND. Inheritance of stem rust resistance in Triticum aestivum ssp. vulgare Reliance and Pi 94701 of Triticum durum. Crop Sci. 1966;6:177–179. [Google Scholar]
- 31.Huerta-Espino J. 1992. Analysis of wheat leaf and stem rust virulence on a worldwide basis. PhD thesis (University of Minnesota, St. Paul)
- 32.Jacob F, Vernaldi S, Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol. 2013;4:297. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelmore RW, Christopoulou M, Caldwell KS. Impacts of resistance gene genetics, function, and evolution on a durable future. Annu Rev Phytopathol. 2013;51:291–319. doi: 10.1146/annurev-phyto-082712-102334. [DOI] [PubMed] [Google Scholar]
- 34.Bossolini E, Wicker T, Knobel PA, Keller B. Comparison of orthologous loci from small grass genomes Brachypodium and rice: Implications for wheat genomics and grass genome annotation. Plant J. 2007;49:704–717. doi: 10.1111/j.1365-313X.2006.02991.x. [DOI] [PubMed] [Google Scholar]
- 35.Alcázar R, Parker JE. The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 2011;16:666–675. doi: 10.1016/j.tplants.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Bieri S, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y, Qian WQ, Hua J. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Path. 2010;6:e1000844. doi: 10.1371/journal.ppat.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S, Hua J. A haplotype-specific resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004;16:1060–1071. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Bao Z, Zhu Y, Hua J. Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact. 2009;22:498–506. doi: 10.1094/MPMI-22-5-0498. [DOI] [PubMed] [Google Scholar]
- 40.Roelfs AP, Singh RP, Saari EE. Rust Diseases of Wheat: Concepts and Methods of Disease Management. CIMMYT; Mexico: 1992. [Google Scholar]
- 41.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim M, et al. 2015 Berberis holstii is functional as an alternate host of Puccinia graminis in Ethiopia. 2015 BGRI Workshop (BGRI, Sydney). Available at https://www.globalrust.org/content/berberis-holstii-functional-alternate-host-puccinia-graminis-ethiopia. Accessed October 11, 2017.
- 43.Warburton ML, et al. Bringing wild relatives back into the family: Recovering genetic diversity in CIMMYT improved wheat germplasm. Euphytica. 2006;149:289–301. [Google Scholar]
- 44.McIntosh RA, et al. Rye-derived powdery mildew resistance gene Pm8 in wheat is suppressed by the Pm3 locus. Theor Appl Genet. 2011;123:359–367. doi: 10.1007/s00122-011-1589-5. [DOI] [PubMed] [Google Scholar]
- 45.Olivera PD, et al. Races of Puccinia graminis f. sp. tritici with combined virulence to Sr13 and Sr9e in a field stem rust screening nursery in Ethiopia. Plant Dis. 2012;96:623–628. doi: 10.1094/PDIS-09-11-0793. [DOI] [PubMed] [Google Scholar]
- 46.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamari L. ASSESS 2.0 Image Analysis Software for Plant Disease Quantification. Am Phytopatholog Soc; St. Paul: 2008. [Google Scholar]
- 48.Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013;163:1433–1445. doi: 10.1104/pp.113.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishida Y, Hiei Y, Komari T. High efficiency wheat transformation mediated by Agrobacterium tumefaciens. In: Ogihara Y, Takumi S, Handa H, editors. Proceedings of the 12th International Wheat Genetics Symposium. Springer Japan KK; Tokyo: 2015. pp. 167–173. [Google Scholar]
- 50.Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum) PLoS One. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.