Significance
Functionally diverse enzyme superfamilies are sets of homologs that conserve a structural fold and mechanistic details but perform various distinct chemical reactions. What are the evolutionary routes by which ancestral proteins diverge to produce extant enzymes? We present an approach that combines experimental data with computational tools to trace these sequence–structure–function transitions in a model system, the functionally diverse flavin mononucleotide-dependent nitroreductases (NTRs). Our results suggest an evolutionary model in which contemporary NTR classes have diverged in a radial manner from a minimal flavin-binding scaffold via insertions at key positions and fixation of functional residues, yielding the reaction versatility of contemporary enzymes. These principles will facilitate rational design of NTRs and advance general approaches for delineating the emergence of functional diversity in enzyme superfamilies.
Keywords: enzyme superfamilies, evolution, flavoenzyme, sequence similarity network, nitroreductase
Abstract
Insight regarding how diverse enzymatic functions and reactions have evolved from ancestral scaffolds is fundamental to understanding chemical and evolutionary biology, and for the exploitation of enzymes for biotechnology. We undertook an extensive computational analysis using a unique and comprehensive combination of tools that include large-scale phylogenetic reconstruction to determine the sequence, structural, and functional relationships of the functionally diverse flavin mononucleotide-dependent nitroreductase (NTR) superfamily (>24,000 sequences from all domains of life, 54 structures, and >10 enzymatic functions). Our results suggest an evolutionary model in which contemporary subgroups of the superfamily have diverged in a radial manner from a minimal flavin-binding scaffold. We identified the structural design principle for this divergence: Insertions at key positions in the minimal scaffold that, combined with the fixation of key residues, have led to functional specialization. These results will aid future efforts to delineate the emergence of functional diversity in enzyme superfamilies, provide clues for functional inference for superfamily members of unknown function, and facilitate rational redesign of the NTR scaffold.
Understanding functional divergence within enzyme superfamilies is a profound question for fundamental biological sciences (1–4). Enzyme superfamilies comprise homologous enzymes that share a structural fold, select active site traits, and a subset of mechanistic features but exhibit various functions; investigation of the sequence and structural transitions that accompany their divergence from a common ancestor can provide a framework to understand the molecular foundations of functional divergence. Do enzyme functions evolve in a sequential manner, driven by the fitness needs of the metabolic pathways in which they function (5, 6)? Or do the functions of contemporary enzymes emerge in a multitude of different ways that each maintain the key structural and catalytic capabilities of the ancestral scaffold (1, 7–9)? Elucidating the mechanisms of functional divergence in enzyme superfamilies, however, is extremely challenging, as the underlying processes occurred over billions of years of evolutionary history. An enzyme superfamily typically contains many distinct functional families, sequence divergence between functional families is often vast (pairwise sequence identity can be less than 10%), and existing sequence information is widely dispersed (ancestral sequences, whose features could link extant functional families, may be lost over evolutionary timescales). As a consequence, sequence signatures that differentiate distinct families are often ambiguous. In addition, the vast majority of enzymes within a superfamily remain uncharacterized; superfamilies often contain well over 20,000 sequences (10), and the investigation of such large datasets, which harbor significant diversity, is technically demanding.
Here, we have addressed these issues for the functionally diverse flavin mononucleotide (FMN)-dependent nitroreductase (NTR) superfamily by using a combination of in-depth bioinformatic analyses. The NTR superfamily is ancient, with a calculated evolutionary age of ∼2.5 billion years (11), and large, comprising more than 20,000 sequences (12). It was named after the nitroreduction reaction that was first characterized several decades ago (13, 14). In addition to nitroreduction, however, a diverse range of reactions can be catalyzed by the NTR superfamily, including dehydrogenation (15, 16), flavin fragmentation (17), and dehalogenation (18) activities that act upon a broad range of substrates including nitroaromatic (19), flavin (20), metal ion (21), enone (22), and quinone (23) compounds (Fig. 1). NTRs form an α+β fold and, like the majority of flavoproteins, noncovalently bind the flavin moiety (24). NTRs are typically homodimers that are composed of two monomeric subunits that form two FMN-binding active sites at the dimeric interface, that is, both monomers contribute to each active site (Fig. 1A). Dimerization is essential for FMN binding and enzymatic function in the NTR superfamily, in contrast to other prevalent flavin-binding proteins such as TIM barrels and Rossmann fold proteins. NTRs generally use a ping-pong bi-bi redox reaction mechanism (25), employing a nicotinamide cofactor to supply electrons to the bound FMN in an oxidative half reaction, which are subsequently transferred to a downstream electron acceptor in a reductive half reaction (Fig. 1 B and C).
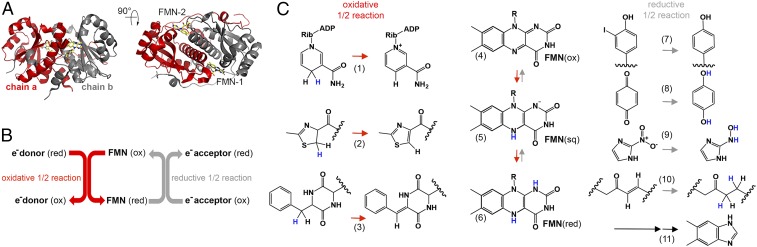
Fig. 1.
An overview of NTR superfamily structure and reaction diversity. (A) A representative NTR structure (PDB ID code 3E39) is depicted in cartoon display in two orientations, with individual monomers colored in gray and red and FMN depicted as a stick model with carbons in yellow (Dataset S1 includes a detailed list of NTR superfamily structures). (B) Diagram showing the ping-pong bi-bi reaction scheme. (C) Representative NTR superfamily reactions: Electron donor (oxidative) reactions, e.g., (1) nicotinamide oxidation, (2) thiazoline oxidation, (3) diketopiperazine oxidation; FMN reduction from (4) oxidized FMN, (5) FMN semiquinone to (6) reduced FMN; electron acceptor (reductive) reactions, e.g., (7) deiodination, (8) quinone reduction, (9) nitroimidazole reduction, (10) ene reduction, and (11) the fragmentation of reduced FMN to dimethylbenzamide.
The diversity of NTR reactions is partly facilitated by the variety of chemical states in which the bound FMN can exist (26). However, the chemical malleability of the flavin alone does not explain the extent of functional diversity observed. NTR enzymes have been used for various biotechnological applications that exploit their broad substrate specificity, including gene therapy for cancer treatment (27), developmental studies (28), bioremediation (21, 29), and biocatalysis (30). However, despite the biochemical and biotechnological importance of these enzymes, most investigations to date have focused on a limited set of NTRs, namely two Escherichia coli enzymes that catalyze nitroreduction reactions, NfsA and NfsB (31, 32), and a small number of their homologs. The bias inevitably resulting from these early focused studies has limited a broader exploration of NTR function and resulted in a vague classification system that is prone to misannotation; NTR sequences have been historically categorized by their similarity to NfsA or NfsB enzymes (19) or simply as outliers (33–35).
In this work, we elucidate the mechanisms of the functional divergence within the NTR superfamily by comprehensively characterizing sequence–structure–function relationships via a unique combination of sequence similarity networks (SSNs), multiple sequence alignments (MSAs), sequence profiles, structural comparisons, and phylogenetic reconstruction. Subsequent incorporation of literature-documented knowledge facilitated the identification of sequence and structural traits that are associated with known NTR superfamily functions. The integration of phylogeny-based reconstructions enabled the extrapolation of our findings to develop a theoretical evolutionary model that reflects the structural transitions that have led to the functional diversity of contemporary NTR superfamily enzymes. Interactive similarity networks and other data from this study are available from the University of California, San Francisco (UCSF), Structure-Function Linkage Database (SFLD; sfld.rbvi.ucsf.edu/django/superfamily/122/).
Results
A Global View of Sequence Diversity Within the NTR Superfamily.
To investigate the sequence diversity within the NTR superfamily, we collected from public databases a nonredundant set of all available sequences and structures that can be associated with this superfamily (Materials and Methods). This data set contains 24,270 nonredundant NTR sequences that range between 150 and 1,580 aa in length. The similarities among all these sequences were calculated by using “all-vs.-all” BLAST pairwise comparisons, and the resulting information was visualized by using SSNs (36–38). The SSN presented in Fig. 2 displays nodes (circles) that represent sets of sequences that share >60% pairwise sequence identity; this level of similarity ensures that the sequences within a single representative node can be aligned with statistical significance (SI Appendix, Fig. S1) and enables the entire superfamily to be visualized (as less abstraction will generate networks that are too computationally demanding). However, a 60% identity level may also condense enzymes that harbor distinct, different functions within a single representative node. Nodes are connected by an edge if the mean pairwise BLAST E-value between all sequences in each node is more significant than 1 × 10−18 (corresponding to an average sequence identity of 28.5%).
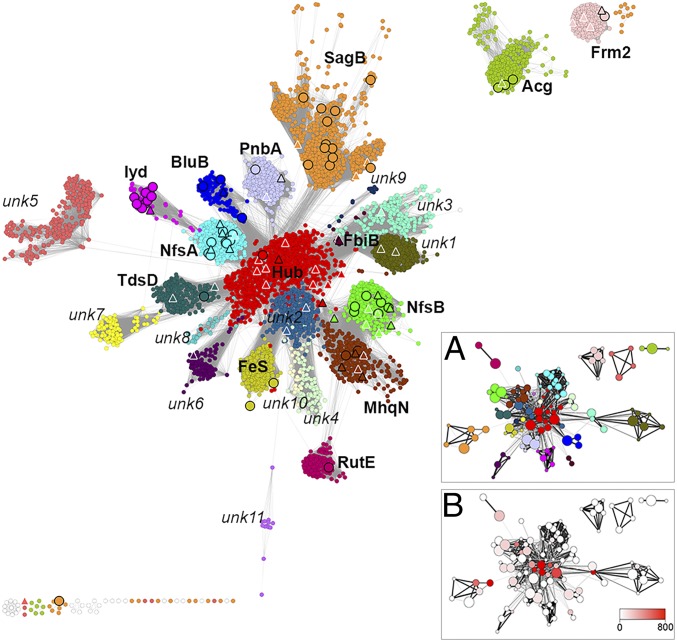
Fig. 2.
A representative SSN of the NTR superfamily: 24,270 protein sequences are depicted by 5,337 nodes (circles), which represent proteins sharing >60% sequence identity. Edges between nodes indicate an average pairwise BLAST E-value of at least 1 × 10−18. Node coloring represents subgroup classification. White nodes with light gray borders indicate remainder sequences that do not belong in any of the categorized subgroups. Large triangle nodes include at least one solved crystal structure; black borders indicate that a biochemical activity was also experimentally characterized. Large circular nodes with black borders include at least one protein associated with experimental evidence (but without structural information). Names in bold indicate subgroups that contain at least one protein with literature-documented functional information. The network is visualized by Cytoscape (74) using the organic layout algorithm (36). (Inset) HMM networks of the NTR superfamily. Nodes represent SSGs (Materials and Methods), and node size correlates with SSG size, from smallest (<100 proteins) to largest (>300 proteins). Edges represent pairwise HMM alignment between SSGs, and similarities with HHALIGN scores >154 (corresponding with an HMM alignment score more significant than 1 × 10−24) are shown. Edge color and width correspond with the HHalign score: <160 indicated by thin and light edges, >300 indicated by thick and dark edges. Nodes are colored based on (A) subgroup and (B) betweenness centrality.
Proposed Classification System for the NTR Superfamily.
To facilitate a more detailed study of this large sequence set, we used a “divide-and-conquer” strategy to parse the superfamily SSN into subgroups. We clustered sequences based on similarity, and thus defined “subgroups” as subsets of sequences in which members of one subgroup share more similarity among themselves than with members of other subgroups. We used specific criteria to quantify differences in similarity, for example, unique sequence profiles [hidden Markov model (HMM)] (39) and the persistence of subgroup boundaries across a wide range of similarity scores. Next, to validate subgroup delineation, we integrated available functional knowledge. Although such information is extremely sparse in the NTR superfamily, we found that it tracks broadly with the subgroup boundaries identified from sequence comparison (SI Appendix, Text S1). Of note, as few NTR members have been experimentally characterized (Table 1) and subgroups show significant sequence diversity (SI Appendix, Fig. S1), multiple functions may occur within a subgroup, as observed in other large and functionally diverse enzyme superfamilies (e.g., refs. 1, 10). This approach was developed to identify broad features, for example, structural modifications and/or active site motifs that may be associated with function(s), which are conserved within the emergent subgroups.
Table 1.
NTR subgroup summary and taxonomic distribution
| Subgroup | Sequences/ investigated enzymes* | EC number(s) | Activity (function)† | Taxonomic profiling, % representation‡ | ||||||||
| Bacteria | ||||||||||||
| ND | Ar | Eu | Bdt | Str | Pro | Frm | Act | Oth | ||||
| NfsB | 2,632/18 | 1.3.1.x, 1.5.1.x, 1.6.5.x, 1.6.99.x | Diverse (32, 84, 85) | 2 | 1 | — | 20 | — | 54 | 18 | 1 | 4 |
| Hub | 2,540/3 | 1.3.3.x§, 1.6.99.x | Diverse (15, 48, 49) | 3 | 9 | — | 17 | 1 | 7 | 50 | 3 | 10 |
| NfsA | 2,299/20 | 1.5.1.x, 1.6.3.x, 1.6.5.x, 1.6.99.x | Diverse (20, 21, 23, 31, 85) | 4 | 1 | — | 7 | — | 35 | 41 | 8 | 4 |
| SagB | 1,936/5 | 1.3.1.x, 3.4.21.x§ | Azole oxidation (TOMM biosynthesis) (16, 41, 86) | 5 | 7 | 1 | 5 | 7 | 26 | 24 | 13 | 12 |
| unk1 | 1,769/3 | — | Unknown (85, 87, 88) | 6 | — | — | — | 1 | 90 | — | 3 | — |
| MhqN | 1,688/5 | 1.6.5.x, 1.6.99.x, | Diverse (22, 89, 90) | 3 | 2 | 3 | 11 | — | 27 | 44 | 3 | 7 |
| Frm2 | 1,568/2 | 1.6.5.x, | Quinoline reduction (redox stress) (33, 91, 92) | 4 | 1 | 13 | 6 | — | 20 | 53 | 2 | 1 |
| PnbA | 1,455/7 | 1.6.5.x, 1.6.99.x | Diverse (93, 94) | 4 | — | 2 | — | 2 | 66 | 7 | 17 | 2 |
| TdsD | 943/1 | 1.5.1.x | FMN reduction (95) | 5 | 5 | — | 4 | 1 | 50 | 5 | 21 | 9 |
| RutE | 861/1 | 1.1.1.x | Malonate semialdehyde reduction (pyrimidine catabolism) (96) | 4 | — | — | — | 6 | 80 | — | 10 | — |
| BluB | 859/4 | 1.13.11.x, 1.16.8.x§ | Unknown (FMN fragmentation) (17) | 7 | 5 | — | 1 | 5 | 61 | 3 | 14 | 4 |
| unk2 | 827 | — | Unknown | 3 | 5 | 1 | 18 | — | 6 | 57 | 2 | 8 |
| unk3 | 789 | — | Unknown | 6 | — | 3 | 17 | — | 2 | 67 | 1 | 4 |
| Acg | 773/5 | — | Unknown (virulence) (35, 97) | 8 | 1 | — | 12 | 13 | 20 | — | 44 | 2 |
| Iyd | 625/12 | 1.21.x | Dehalogenation (iodine salvage) (18) | 13 | 3 | 25 | 5 | 14 | 29 | — | 9 | 2 |
| unk4 | 623 | — | Unknown | 10 | — | — | — | 15 | 1 | — | 73 | 1 |
| unk5 | 533 | — | Unknown | 3 | 2 | 1 | 4 | — | 1 | 70 | 14 | 5 |
| FeS | 529/2 | 1.6.99.x | Nitroaromatic reduction (98) | 2 | 7 | 3 | 7 | — | 28 | 47 | 2 | 4 |
| unk6 | 287 | — | Unknown | 4 | — | — | 14 | — | 20 | 36 | 5 | 21 |
| FbiB | 242/2 | 6.3.2.x§ | Unknown (F420 biosynthesis) (45) | 5 | — | — | — | 30 | — | — | 65 | — |
| unk7 | 135 | — | Unknown | 3 | 1 | — | 21 | — | 35 | 9 | 24 | 7 |
| unk8 | 129 | — | Unknown | 1 | — | — | — | 5 | — | — | 93 | 1 |
| unk9 | 71 | — | Unknown | 7 | — | — | — | — | 1 | 88 | 3 | 1 |
| unk10 | 59 | — | Unknown | 3 | — | 2 | 48 | — | 10 | 28 | 2 | 7 |
| unk11 | 14 | — | Unknown | — | — | 100 | — | — | — | — | — | — |
| Remainder | 84 | — | Unknown | 7 | 11 | 1 | — | 1 | 6 | 11 | 32 | 31 |
| Superfamily¶ | 24,270 | — | Diverse | 4.9 | 2.6 | 2.5 | 8 | 3 | 35 | 26 | 13 | 5 |
Act, Actinobacteria; Acg, acr coregulated gene; Ar, Archaea; Bdt, Bacteroidetes; BluB, Blush B; Eu, Eukaryota; FbiB, F420 biosynthetic pathway B; Frm, Firmicutes; Frm2, fatty acid repression mutant 2; Iyd, Iodotyrosine dehalogenase; MhqN, 2-methylhydroquinone reductase N; ND, sequences typically originating from metagenomic surveys; NfsA, nitrofurazone sensitivity A; NfsB, nitrofurazone sensitivity B; Oth, other; PnbA, p-nitrobenzoate reductase A; Pro, Proteobacteria; RutE, pyrimidine utilization E; SagB, SLS-associated gene B; Str, Streptomycetales; TdsD, Thermophilic desulfurization D.
To the best of our knowledge.
Subgroup activity and function were assigned based on literature associated with canonical members.
Taxonomical frequencies are based on UniProtKB/National Center for Biotechnology Information data retrieved for each subgroup member.
Multidomain enzymes.
Taxonomic profiling numbers represent percentages from all NTR enzymes.
Our analysis resulted in 22 major subgroups, each containing >100 unique sequences, as indicated by various colors in Fig. 2. Fourteen of these subgroups could be named by biochemically characterized representatives, for example, the NfsA subgroup includes E. coli NfsA and close homologs (31), and the BluB subgroup is exemplified by the BluB enzyme that catalyzes the fragmentation of reduced FMN (17) (Table 1). Among members of each subgroup, the average pairwise percent identities vary from >42% sequence identity for IyD, BluB, RutE, and Frm2 to <35% sequence identity for subgroups such as NfsA and NfsB (SI Appendix, Fig. S1).
Eight additional subgroups have, at present, no members with known biological roles or documented activity: These subgroups were named as “unknown (unk) subgroups,” for example, unk1 and unk2 (Fig. 2 and Table 1). In addition to the major 22 subgroups, there are four small subgroups that contain <100 sequences, including a “remainder” subgroup of outlier sequences, which share only an average of 27% sequence identity with any of the other superfamily subgroup members. The robustness of our classification system is further evidenced by visualizing a representative SSN that uses a higher similarity threshold that eliminates connections between subgroups but generally maintains subgroup clusters (SI Appendix, Fig. S2A). Finally, to eliminate the potential of bias arising from our visually based subgrouping and/or the SSN layout, we validated our method using the Markov cluster algorithm (40), which displayed significant agreement (98%) with our approach (SI Appendix, Text S1 and Table S1).
The Functional Diversity of the NTR Superfamily Remains Unknown.
Although 14 NTR subgroups can be associated with at least one experimentally validated function, multiple reactions may be represented in addition to their namesake function, especially for the large subgroups that contain >1,500 members. Furthermore, there are no experimentally characterized enzymes associated with the remaining eight major subgroups of the superfamily. Thus, the functional diversity of the NTR superfamily remains unknown. Although there are numerous experimental studies that are devoted to in depth biochemical and structural characterization of a select few members of the superfamily, our comprehensive analysis reveals that the vast majority of the enzymes in the NTR superfamily (∼99%) have not been experimentally characterized. In addition, as indicated in Table 1, the proportion of sequences with functional and/or structural information across the different subgroups is uneven. For example, very few sequences have been characterized from the SagB subgroup (16, 41), which is large and diverse (less than 32% average sequence identity; SI Appendix, Fig. S1) and likely to contain smaller “sub-subgroups” (SSGs) that may individually possess different substrate and catalytic specificities (Fig. 2). Similarly, less than 40% average sequence identity is observed within each of the well-studied NfsA and NfsB subgroups, and less than 1% of sequences have been characterized (i.e., 18 of 2,632 NfsB subgroup sequences and 20 of 2,299 NfsA subgroup sequences). In addition, many NTR superfamily enzymes have been shown to be promiscuous for multiple substrates and reactions, for example, NfsA, NfsB, and MhqN (Table 1), complicating the inference of their functional properties.
Taxonomic Representation Across the Biosphere.
Most NTRs are bacterial, but NTRs are also found in all forms of life: 2.6% of the sequences are from Eukaryotes and 2.5% are archaeal. Eukaryotic sequences are found within nine of the 22 major subgroups, and archaeal sequences are found within 14 subgroups (Table 1 and SI Appendix, Fig. S3). The distinctive nature of some subgroups is further evidenced by unique taxonomic distributions, for example, 25% of the Iyd subgroup are from Eukaryotes, and 90% of unk1 sequences are from Proteobacteria. Of note, the Actinobacteria phylum harbors the most diverse and redundant set of NTRs, as actinobacterial representatives are found in each of the 22 NTR subgroups and, for example, Mycobacterium sp. JLS encodes seven Acg subgroup paralogs. Organisms that reside in variable environments, such as those from the Actinobacteria phylum, may have evolved to rely upon the metabolic versatility conferred by flavoenzymes (24).
The SSN Topology Reveals Similarity Relationships Organized Around a Central “Hub” Subgroup.
SSN topology has been previously used to study the evolutionary and functional relationships between members of a superfamily (42–44), as the examination of subgroup connectivity can serve as a platform for knowledge-based inference of function. Perhaps the most striking feature of the NTR superfamily is a distinct and robust “hub topology,” which was revealed by the SSN and is consistently observed across a wide range of edge-inclusion E-value thresholds (1 × 10−12 to 1 × 10−20; Fig. 2 and SI Appendix, Fig. S2B); that is, most subgroups directly connect to a central “hub” subgroup and almost all subgroups show more significant sequence similarity to the hub sequences than to any other subgroup. There are two exceptions to this trend: The NfsB and MhqN subgroups connect most closely with each other, and a similar scenario is observed for the unk1 and unk3 subgroups.
To validate the robustness of the SSN hub topology, we investigated similarity relationships with respect to protein domain architecture, insertions in NTR sequences and alternative similarity calculations. We examined whether NTR sequences that harbor not only the NTR domain but also another domain associated with a different fold, for example, FbiB (45), or N- and C-terminal sequence extensions that flank the NTR domain or segments that reside within it, contribute to subgroup separation and SSN topology. We found that 94% of the superfamily are single-domain NTRs (SI Appendix, SI Methods). We also generated an SSN by using trimmed sequences that represent the minimal α + β homodimeric fold shared by all NTR superfamily members (Fig. 3 and SI Appendix, Fig. S4A). The results show that subgroup divisions are maintained, demonstrating that the pairwise similarity signal that underpins the topology is consistent and is not skewed by alterations of the minimal scaffold.
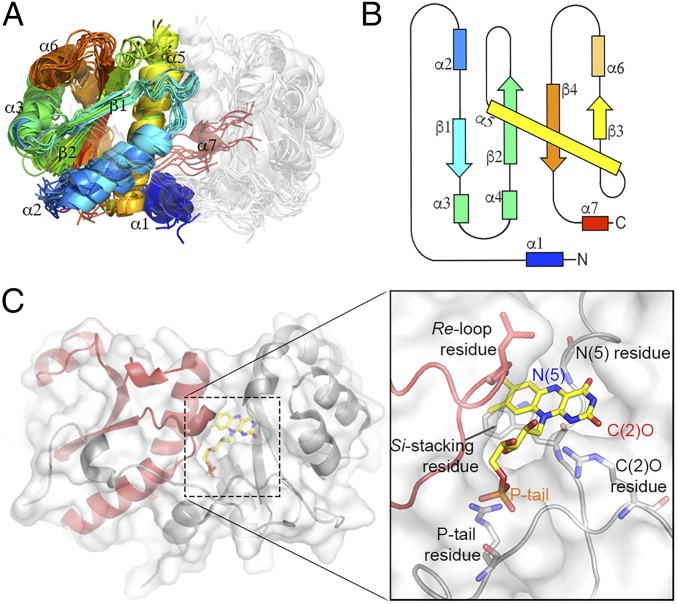
Fig. 3.
The NTR superfamily scaffold. (A) The NTR superfamily domain: An overlay of 17 representative NTR structures showing the conserved α+β FMN binding fold that was generated using MUSTANG-MR (76) at a sieving level of 2.0 Å. (B) A 2D topology map of the minimal NTR scaffold colored from blue (N terminus) to red (C terminus) with numbered α-helices and β-strands. (C) A ribbon representation of the hub subgroup structure PDB ID code 3E39 with monomers colored in gray and red, respectively. FMN is depicted in stick form with carbons in yellow. (Inset) Key FMN interacting residues: The FMN moiety and interacting active site residues are displayed in stick form and labeled.
To further substantiate the presence of a hub subgroup, we calculated the all-vs.-all similarities between NTR subgroups by using a distance metric derived from sequence profiles [HMMs (46); Fig. 2A and SI Appendix, SI Methods]. Although the underlying similarity measure is different from those used to compute the SSN (pairwise sequence similarity vs. multiple sequence similarity), this analysis also produced a network in which a hub subgroup can be visualized. The connectivity of the HMM network was analyzed by calculating the “betweenness centrality” of each node (47), that is, each node is ranked by the number of shortest paths that connect between any possible pair of nodes in the network and traverses that ranked node (Fig. 2B). These results show that the nodes representing hub sequences display the highest centrality scores, providing complementary evidence for the hub topology.
The Hub Subgroup May Represent “Ancestral-Like” NTRs.
These analyses of the hub subgroup allow us to hypothesize that the functional divergence observed within the NTR superfamily may originate from ancestral sequences that are most similar to those of the contemporary hub subgroup. The taxonomic distribution of NTR proteins lends support to this conjecture: Enzymes in the hub subgroup are primarily from bacterial organisms (88%) but contain a significantly higher proportion of proteins from archaeal organisms compared with the overall taxonomic distribution of the NTR superfamily. Within the hub subgroup, archaeal sequences are significantly enriched: 9% of hub subgroup sequences are archaeal, compared with 2.4% of the superfamily (P = 2.2 × 10−16, binomial test; Table 1). Taxonomically diverse subgroups may indicate a more ancient origin than taxonomically narrow subgroups, as they are more likely to have appeared before phyla branching.
Little is currently known about members of the Hub, making it difficult to investigate the structure–function relationships within this subgroup. To date, only three hub enzymes [AlbA (15), NitB (48), and Nox (49)] have been biochemically characterized. These three enzymes, however, display considerable substrate and catalytic diversity: AlbA is a cyclic dipeptide oxidase, forming α/β-unsaturated residues from a cyclized precursor. In contrast, NitB and NOX display NAD(P)H oxidase and nitroaromatic reductase activities. This suggests that, albeit with a limited sample number, the hub subgroup may consist of diverse enzymes with distinct functions (i.e., multiple functional families).
The “Hub of the Hub”.
To further examine the hub subgroup, we subdivided it into 15 second-level SSGs, each displaying an average pairwise sequence identity within the SSG of 35%. A representative SSN of the hub subgroup is shown in SI Appendix, Fig. S5. The three characterized hub enzymes are found in hub SSG-2 (Nox), hub SSG-3 (AlbA) and hub SSG-6 (NitB). Similar to the overall network topology of the NTR superfamily, hub SSGs display a hub-like arrangement, and hub SSG-5 appears to be the “Hub of the Hub.” As with the hub subgroup, hub SSG-5 shows an increased enrichment of archaeal sequences (22%, compared with 9% for the hub subgroup), indicating that proteins similar to hub SSG-5 likely appeared very early in the ancestry of the NTR superfamily.
The Hub Subgroup Represents a Minimal Scaffold.
Global studies of functionally diverse enzyme superfamilies suggest a common theme: Conservation of a core structural domain and active site architecture that can be associated with conserved chemical capabilities (50–52). Overlaid on this foundation, nature has diversified other structural features in ways that can be associated with functional differences [see, e.g., Burroughs et al. (51)]. The NTR superfamily also appears to follow this general theme, that is, comparison of available NTR structures reveals a conserved minimal scaffold that harbors key FMN interacting residues (Fig. 3); of note, these residues originate from both chains of the homodimer. It is especially intriguing that the majority of contemporary hub structures exhibit architectures that mimic the minimal NTR scaffold with little or no decorating features, for example, Protein Data Bank (PDB) ID code 3E39. Thus, the minimal structural architectures found in the hub subgroup and the consistency of the hub subgroup observed in the sequence and HMM similarity networks provide additional support for the notion that hub sequences may display “ancestral-like” features.
Extensions to the Minimal Scaffold.
To investigate how nature may have evolved functional variations from a minimal NTR scaffold ancestor, we manually compared all NTR structures to probe the structural basis of superfamily divergence: 54 NTR proteins are associated with crystal structures (Fig. 2), with 73% of superfamily subgroups (16 of 22) containing at least one crystal-solved structure. This set includes 22 structures associated with biochemically characterized enzymes and 32 structures without an associated function (Dataset S1). The diversity of NTR architecture is apparent in this set of structures, which includes fused monomeric proteins, that is, fusion of two NTR domains to create a protein that mimics an NTR dimer (e.g., PDB ID codes 2YMV and 3EO7), and domain fusions that link an NTR domain with a domain from a different superfamily (e.g., PDB ID codes 4EO3 and 4XOO). To further delineate structural diversity, we used the TM-align algorithm to compute pairwise structural similarity of a nonredundant set of NTR structures to generate a structure similarity network (Materials and Methods and Fig. 4A). The resulting structural network is in general agreement with the sequence-based networks (Fig. 2), with the hub subgroup structures observed as the central and most connected nodes. To show that the central positioning of the hub subgroup is statistically robust (regardless of the similarity method used), we used Infomap (53) to show that hub subgroup members are significantly more central in the sequence-based, HMM-based, and structure-based networks (SI Appendix, SI Methods and Fig. S4).
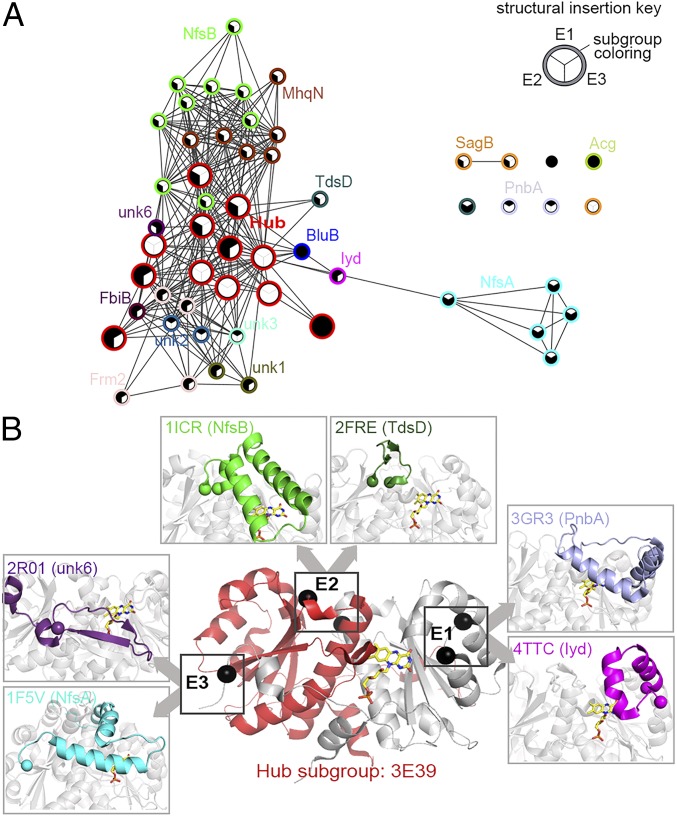
Fig. 4.
Structural analysis of the NTR superfamily. (A) A structure similarity network of the NTR superfamily. Each node represents a crystal structure, colored by subgroup as per Fig. 2 (red nodes represent hub subgroup members). Nodes are filled according to the presence or absence of the structural extensions inserted in any of the three hot spot sites, as depicted by the key (Inset). Edges represent pairwise structural similarity scored <0.746, as measured by TM-align. (B) A diagram of the structural diversity observed at the E1, E2, and E3 insertion sites relative to one FMN binding active site of the enzyme. A cartoon representation of a hub protein structure (PDB ID code 3E39) is shown with monomers depicted in gray and red. The locations of the E1, E2, and E3 structural insertion points are indicated by spheres that depict the bordering residues of each insertion (E3 has only one bordering residue, as it extends the C terminus). The FMN molecule is shown in a stick model with carbons colored in yellow. (Inset) Boxes display examples of subgroup specific diversity at each extension site labeled by PDB ID code and subgroup. Extensions are colored by subgroup as per Fig. 2.
Guided by length variations and alignment gaps among the NTR structures, manual examination revealed three “hot spots” of structural divergence, each associated with a structural extension to the minimal NTR scaffold that occurs proximal to the active site. Extension 1 (E1) represents an insertion of amino acids between α-helices 3 and 4, extension 2 (E2) is located between β-strand 2 and α-helix 5, and extension 3 (E3) is located at the C terminus of the enzyme (Fig. 4B). Of note, relative to one FMN active site, E1 and E3 arise from the same chain and E2 extends from the alternative chain of the homodimer. The structural similarity network, presented in Fig. 4A, shows that almost all NTR subgroups, excluding the hub, contain at least one extension (Fig. 4B and SI Appendix, Fig. S6A). Despite conservation of the relative position of each insert, the length and secondary structure elements of each varies, and this variation is more significant between subgroups than within subgroups. Extensions are often absent from hub subgroup structures and, if present, they are typically very short, for example, hub subgroup enzymes display average extension lengths of 9 aa (E1) and 12 aa (E2), in contrast to the average extension lengths of 15 aa (E1) and 29 aa (E2) across the superfamily. The extensions are, on average, >8 Å from the isoalloxazine ring of the bound flavin, and are therefore more likely to be involved in substrate interactions rather than in FMN binding (SI Appendix, Fig. S6B). Furthermore, extensions have been crystallized in multiple conformations within a single enzyme, likely indicating dynamic roles in enzyme function (22, 54). We generated an MSA that includes 47 representative structures to demonstrate the overall sequence conservation of minimal scaffold and the conservation of the insertion sites of the structural extensions (SI Appendix, Fig. S7).
FMN Interacting Residues Display Distinct Conservation Patterns.
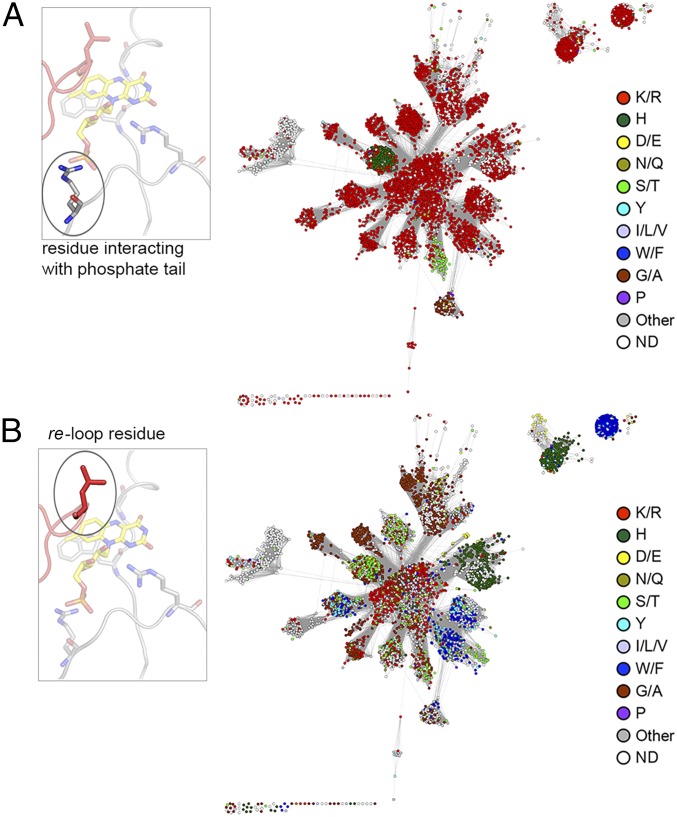
In contrast to the extensions, the conserved minimal NTR scaffold contains key residues that interact with the FMN isoalloxazine ring to modulate redox potential and influence catalysis. For example, a positively charged residue at the C(2)O locus increases redox potential by stabilizing the reduced form of the flavin, and the N(5) locus is typically within 3.5 Å of a hydrogen-bond donor, which is essential for dehydrogenation (55) and dehalogenation (56) (Fig. 3C). We calculated the superfamily-wide residue conservation of FMN interacting residues by manually assigning these residue positions in all available structures and subsequently inferring the location and the identity of the relevant amino acid within all sequences in the superfamily via structure-based pairwise alignment. Clear conservation patterns are seen for key FMN interacting residues throughout the NTR superfamily (Fig. 5 and SI Appendix, Fig. S8). For example, the FMN phosphate tail is almost ubiquitously interacting with a basic amino acid throughout the superfamily, that is, arginine (81% conservation), and the C(2)O interacting residue is typically a basic amino acid, for example, arginine/lysine (76% conservation; Fig. 5 and SI Appendix, Fig. S8). In contrast, both the re-loop residue (located ∼5 Å from the re side of the flavin) and the si-stacking residue (located on the si side of the flavin) show considerable, and subgroup-specific, diversity, potentially indicating their involvement in reaction specificity (Fig. 5 and SI Appendix, Fig. S8). Of note, and in contrast to the other interacting residues discussed earlier, the re-loop residue arises from the alternative chain of the homodimer and is depicted in red in the structural representations of Figs. 3 and 5. Additionally, the hub subgroup displays a diverse range of re-loop residues, in contrast to the conservation of subgroup specific residues in the rest of the superfamily (Fig. 5B).
Fig. 5.
Conservation of FMN-interacting positions across the NTR superfamily. (A) A representative SSN of the NTR superfamily is shown with nodes colored by the most frequent residue type found in the FMN phosphate moiety interacting position. (Inset) Ribbon representation of the active site of hub subgroup structure PDB ID code 3E39 is shown with FMN depicted in stick form with carbons in yellow. The residue (arginine) interacting with the phosphate moiety is circled. (B) A representative SSN of the NTR superfamily is shown with nodes colored by the most frequent residue type found in the re-loop position. (Inset) Ribbon representation of PDB ID code 3E39 (as per A) with the re-loop residue (leucine) circled. Note that the re-loop residue originates from the alternative chain of the homodimer (shown in red).
Structural Extensions Harbor Residues with Distinct Conservation Patterns and Important Roles for Substrate Specificity and Catalysis.
The enzymatic function(s) of NTRs are governed by the first shell of residues near the FMN isoalloxazine ring and also proximal amino acids that are likely to be involved in substrate recognition. To determine the extent to which the NTR scaffold extensions are associated with diverse functionality, we selected and analyzed eight subgroups that have structural and/or experimental data to support the identification of “functional amino acids,” for example, residues that may have key substrate binding or other functional roles. These subgroups, Iyd, BluB, Frm2, FbiB, PnbA, NfsA, NfsB, and MhqN, represent a diverse range of chemical and biological activities (Table 1). We individually aligned every sequence of the eight selected subgroups to the manually generated subgroup-specific MSA (Materials and Methods). The results identified the residues in each sequence that are most likely to be relevant to function based on their alignment to experimentally confirmed residues. The percentage conservation of functional residues within each subgroup of interest was then determined (SI Appendix, Text S2); location of the key residues, their conservation levels, and catalytic reaction(s) are detailed in Table 2. In all of the eight subgroups analyzed, the majority of the key substrate binding residues are found within the E1, E2, and E3 extensions (Dataset S2), and a wide range of subgroup-specific residue conservation was observed (SI Appendix, Text S2 and Fig. S9). In addition, we verified that our observed patterns of residue conservation do not stem from the surrounding context, that is, high sequence conservation is not an inherent feature of the sequence segments that include the functional residue (Dataset S2). The conservation of key residues was considerably higher (>80%) in subgroups that are hypothesized to target the same or very similar substrate(s), that is, Iyd, BluB, Frm2, and FbiB subgroups. In comparison, much lower conservation levels were observed in subgroups that are known to have a more diverse substrate range, that is, NfsA, NfsB, PnbA, and MhqN.
Table 2.
Conservation and location of functional residues within extensions
| Subgroup | Reaction or function | Catalytic residues* | % cons† |
| NfsB | Reduction of a diverse substrate range (32, 85, 65, 66, 99) | E1 & E2 | 34–55 |
| NfsA | Reduction of a diverse substrate range (20, 21, 23, 31, 67, 85) | E3 | 19–60 |
| MhqN | Diverse catalysis (22, 89, 90) | E1 & E2 | 5–51 |
| Frm2 | Reduction of 4NQO; oxidative stress (33, 92) | E1 & E2 | 81 |
| PnbA | Reduction of a diverse substrate range (93, 94) | E1 | 10–72 |
| BluB | FMN fragmentation (17, 100) | E2 & E3 | 99–100 |
| IyD | Dehalogenation of aromatic compounds (18) | E1 | 92–100 |
| FbiB | Biosynthesis of the F420 flavin cofactor (45) | E1 & E2 | 98 |
Location of catalytic residues.
Percentage conservation; Dataset S2 includes further details.
Large-Scale Phylogenetic Reconstruction Supports a Radial Model of NTR Functional Divergence.
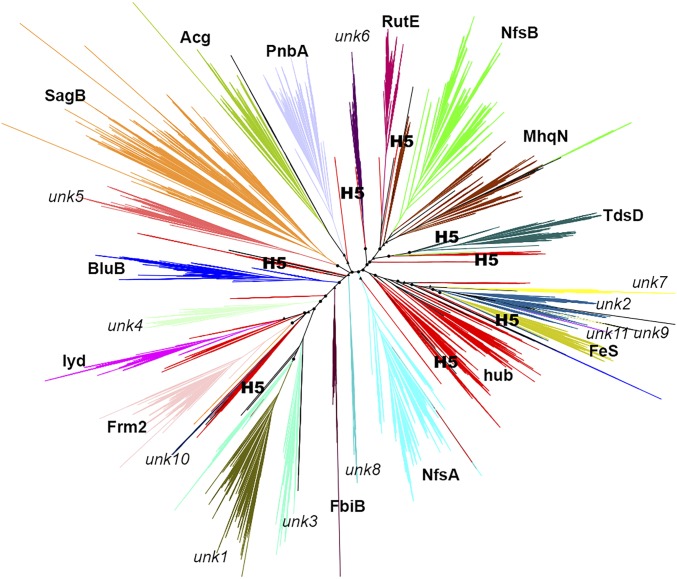
Although the SSNs and other investigations described in the present work provide clues about how evolutionary divergence may have produced the contemporary structures and functions of the NTR superfamily from an ancestral scaffold, they do not explicitly incorporate evolutionary information (e.g., SSNs are based on pairwise sequence similarity, thereby limiting inferences about divergence that are based on transitivity). We therefore constructed a maximum-likelihood phylogenic model of the NTR superfamily (24,270 sequences) to directly assess their evolutionary relationships. The resulting phylogenetic tree, shown in Fig. 6, is characterized by highly significant branching probabilities for the major branch points. Moreover, the branching supports the subgroupings independently identified from the SSN analyses (Fig. 2). The complementarity of the tree and SSN is illustrated by the different hypotheses that can be derived from each approach: For example, the tree reveals the existence of neither a discrete hub subgroup nor its key topological position in relation to other subgroups, and the SSN does not allow the classification of enzymes beyond the subgroup level (“metasubgroups,” described later), which can be identified in the tree.
Fig. 6.
A phylogenetic reconstruction of the NTR superfamily. Branches are colored and labeled by subgroup; dispersed red branches represent hub subgroup sequence sets, and black branches represent members of the remainder subgroup. The eight hub SSG-5 branches are labeled (H5). Circles represent branching points with probabilities >0.9; triangles represent probabilities >0.8.
Examination of the tree shows three metasubgroups with common descendance according to the phylogenetic model: MhqN-NfsB-RutE, FeS-unk2, and unk1-unk3. These subgroups also show high interconnectivity in the SSN, but so do others; the large-scale phylogenetic tree uniquely reveals their relatedness. Interestingly, additional factors suggest catalytic similarities between metasubgroup members: MhqN-NfsB-RutE enzymes display similar extension lengths and distances from extension atoms to N5 of the FMN, they cluster together in the structural similarity network, and they also share common si-stacking and re-loop residues. Unk1 and unk3 members display similar distances from extension atoms to the FMN N5 and cluster together in the structural similarity network, and FeS-unk2 enzymes share common re-loop, and N5 interacting, residues. These features may indicate shared aspects of catalysis for metasubgroup members (Figs. 4 and 5 and SI Appendix, Figs. S6–S8).
Of particular note, although the hub subgroup has a robust central location in the SSN, it does not have a singular position in the phylogenetic model (Fig. 6). Hub sequences are dispersed throughout the tree; most of the hub SSGs colocalize in one principal branch, but some hub SSGs appear in branch points that diverge before many of the other individual subgroups. This is most notable in hub SSG-5 sequences (the Hub of the Hub, indicated by “H5” in Fig. 6); interestingly, it is unlikely that the dispersion of hub SSG-5 would have been noticed from the tree if the topology of the SSN had not strongly suggested its existence. The observation that hub SSG-5 members are found in dispersed “presubgroup” branch points in the tree and at the center of the hub in the SSN, together with their phylogenetically diverse nature and minimal scaffold architecture, adds support to the conjecture that they represent ancestral-like sequences: Hub SSG-5 members may be modern-day sequence fossils that exemplify the evolutionary transitions between an ancient hub-like subgroup and the diverse structure/function subgroups of extant NTRs. These notions fit a scenario of a radial burst of functionalization that occurred early in the ancestry of the NTR superfamily (Figs. 2, 4, and 6, Table 1, and SI Appendix, Fig. S5).
Discussion
Deciphering the evolution of a large superfamily that has taken place over billions of years and has generated diverse contemporary functions is a challenging task. To achieve global views of the superfamily, comprehensive and exhaustive bioinformatics are essential. Similarity networks establish a global context for interpreting sequence, structural, and functional relationships, and facilitate hypotheses and observations that are not easily accessible from smaller scale approaches. In this work, we present a combination of integrated methodologies, utilizing large sets of sequences and structures, alignments, phylogenetic reconstructions, and biochemical data, to reveal sequence–structure–function associations and evolutionary relationships within the NTR superfamily. Our results illustrate the power of large-scale comparisons to provide new insights regarding the evolution of contemporary reaction types within enzyme superfamilies.
Our observations significantly revise the historical ad hoc NfsA/NfsB NTR grouping system and enable a new and robust classification system to be established. Our analysis guided the separation of the NTR superfamily into 22 distinct subgroups, which will facilitate the accurate assignment of NTRs to functions and pathways for future studies, and the correlation of active site profiles with assigned functions. More generally, these results guide the exploration and discovery of functions within uncharacterized subgroups and suggest important active site transitions that are necessary for functional divergence.
Together, the complementary analyses applied in the present study indicate that hub subgroup sequences represent ancestral-like proteins and suggest that functional divergence of the NTR superfamily has largely occurred in a radial manner from ancestral sequences that resemble extant hub subgroup enzymes. The SSN topologies observed for other enzyme superfamilies do not show hub topologies, and instead largely indicate a sequential manner of functional divergence (10, 57–61). Our results suggest that the functional expansion of enzyme superfamilies, and, by inference, their respective network topologies, may exhibit unique patterns that are specific to the evolutionary process by which variation has occurred (3). The molecular and evolutionary causes of radial and sequential divergence patterns, however, are unclear. Detailed and large-scale characterization of additional superfamilies is needed to reveal the trends by which molecular and structural features have diverged across the universe of enzyme superfamilies.
Our analyses have revealed potential molecular determinants that distinguish subgroup functions that are located in extensions to the minimal NTR scaffold as well as the FMN binding pocket. These findings let us hypothesize an evolutionary scenario for the NTR superfamily: Enzymes composed of a minimal scaffold existed in the early stages of NTR evolution. This scaffold, which we speculate in this work to share structural features similar to those of the contemporary hub subgroup proteins, may have provided an “evolvable platform” for diverse function, while at the same time exploiting a conserved structural fold and active site architecture for FMN-based chemistry. Substitutions within the scaffold as well as structural insertion events in three hot spots supported the innovation of new function, producing the contemporary array of NTR superfamily subgroups. Acquisition of extensions and the associated functional specificity, however, may have been achieved at the expense of “evolvability” (62), and therefore the contemporary specialized subgroups may now be less primed for functional divergence. The strong distinction between “scaffold,” which provides the majority of critical protein folding features and catalytic residues, and “loops” that determine catalytic and substrate specificity, is proposed to be one of the signatures of an “innovable” functionally diverse superfamily (63, 64). Further experimental characterization, which includes large-scale activity profiling and engineering experiments, ancestral reconstruction, and characterization of evolutionary pathways between distinct functional families, will be required to address the question.
The distinct scaffold and loops structure of the NTR superfamily may serve as an attractive enzyme engineering target for the generation of novel and efficient enzymes. As noted in the Introduction, NTRs have been exploited for various biotechnological applications such as cancer gene therapy, developmental studies, bioremediation, and biocatalysis. Only a handful of studies, however, have successfully engineered NTR enzymes for biotechnological applications, and these studies typically result in only small improvements in catalytic activity (65, 66, 67). Targeted mutagenesis and modification of NTR scaffold extensions that have been identified in the present study might offer a more effective starting point to enhance, diversify, and switch NTR specificity.
Historically, protein characterization efforts have been strongly skewed toward certain classes of proteins, protein families, and superfamilies, leaving the vast majority of superfamilies unexplored (68). Exhaustive bioinformatic approaches, such as demonstrated in the present study, can dramatically enhance our understanding of each superfamily and aid in the rational selection of protein targets (69). In particular, integrated approaches, such as those detailed here, will be applicable to other superfamilies that display broad sequence, structure, and function divergence, and thus will support the development of classification methods for functionally diverse protein superfamilies. Ultimately, the ability to decipher, understand, and predict the molecular mechanisms of functional diversity in such other superfamilies will not only aid our understanding of fundamental questions in evolutionary biology, but also enable the accurate, efficient, and evolutionary-informed design of new protein catalysts for biotechnology.
Materials and Methods
Gathering NTR Sequences.
The criteria used for gathering NTR sequences incorporated computational and experimental evidence, for example, sequence profiles, structural fold, and relevant enzyme commission (EC) numbers (70) (SI Appendix, SI Methods and Table S2). This work focuses on “canonical” NTRs that share a common unique fold, are capable of FMN binding, and belong to a homologous superfamily [NADH oxidase, CATH 3.40.109.10 (4)]. By using HMMSCAN (46) and Pfam sequence signatures, we verified that 94% of our superfamily members are single NTR domain sequences (SI Appendix, SI Methods). The resulting sequences were uploaded to the UCSF SFLD (10).
Generating an NTR-Representative SSN.
A representative SSN was created by using SFLD database tools (10, 71). Briefly, pairwise BLAST (72) E-values were calculated between all possible pairs of available sequences (omitting E-values less significant than 1 × 10−2). Pairwise similarities were used to generate a network in which a node represents a protein sequence and an edge represents a pairwise BLAST E-value [with E-values used as scores (36)]. We used “representative networks” to circumvent the computational limitations of visualizing large networks: Each node represents a set of proteins that share 60% sequence identity as measured by CD-HIT (73), and edges represent a mean E-value more significant than 1 × 10−18 between all E-value scores that connect the representative nodes (SI Appendix, SI Methods). This threshold was set via manual sampling of several edge inclusion cutoffs until a reasonable reconciliation was achieved between distinct similarity clusters and representation of remote homologies between them (SI Appendix, Text S1). Networks were visualized by Cytoscape (74) by using the organic layout (36).
Obtaining Sequence Profiles for NTR Subgroups.
Briefly, subgroup member sequences were selected to ensure appropriate coverage of the sequence space, generating a set of sequences that were subsequently aligned by using structural and functional information. After manual refinement, HMM models (46) were created, and a safe detection threshold was determined by minimizing cross-HMM detection (SI Appendix, SI Methods and Fig. S10).
Generating an NTR HMM Similarity Network.
To create the HMM similarity network, each subgroup was subdivided into SSGs in a similar manner to subgroup classification: Edge inclusion thresholds were sampled, and a specific cutoff was determined so that the grouping agreed with specific criteria, for example, sets of enzymes documented by the literature to belong to the same class, phylogenetic branches, or domain architectures (SI Appendix, Text S1). MSAs were generated and manually refined for each group, and an HMM was calculated by using HHblits (75). HMM–HMM alignments were calculated by HHalign (from the HHblits package), and scores were used to create the HMM similarity network.
Identification of the Core FMN Scaffold.
MUSTANG-MR (76) was used at an rmsd threshold of 2.0 Å to generate a multiple structural alignment of representative NTR structures. A structure-based MSA of the core sequences (after removal of structural extensions and N/C termini) was then generated by using UCSF Chimera (77), with extensive manual refinement to integrate information from literature (SI Appendix, Fig. S7).
Structural Similarity Network of the NTR Superfamily.
NTR structures were obtained from the RCSB database (Dataset S1) and manually examined to minimize redundancy, yielding a list of 54 representative structures. All pairs of structures were compared by using TM-align (78), and a TM-score of 0.746 was used as an edge-inclusion threshold, which was determined by sampling different thresholds while maintaining connections between clusters of similar structures. Note that a TM-score above 0.5 is considered to indicate the same fold (78).
Superfamily-Wide Profiling of Extension Lengths.
NTR structures were structurally aligned and inspected by using Chimera (77) to determine the start and end positions of each insertion hot spot. Each superfamily member was then paired with the most relevant structure (i.e., best BLAST hit), and 3D-Coffee (79) was used to generate a pairwise structure-based alignment from which the extension lengths were calculated.
Residue Profiling Across the Superfamily and Within Subgroups.
Relevant residues (as deduced from structural information) were assigned to specific column positions in the MSA of each subgroup. Each subgroup member was then individually added to the MSA by using a specific module of MAFFT (80); this allowed extraction of the amino acid identity of relevant positions.
Large-Scale Phylogenetic Analysis.
The ultra-large alignments using phylogeny-aware profiles (UPP) algorithm (81) was used to create a superfamily-wide alignment. A backbone MSA containing representative sequences was generated by using PASTA (82), which was then used to compute a backbone tree by using FastTree (83). The backbone MSA, backbone tree, and all 24,270 NTR sequences were then used as input to UPP. The resulting MSA was used for generating a phylogenetic model using FastTree and the tree was visualized via FigTree (SI Appendix, SI Methods).
Data Dissemination.
Data produced by the present study are available from the UCSF SFLD (10) (sfld.rbvi.ucsf.edu/django/superfamily/122/).
Supplementary Material
Acknowledgments
We gratefully acknowledge the UCSF Resource for Biocomputing, Visualization, and Informatics Laboratory for their support of the SFLD, and Dr. Ben Polacco for SFLD data management and the assistance with statistical analysis. This work was funded by National Institutes of Health Grant R01 GM60595 (to P.C.B.); Natural Sciences and Engineering Research Council of Canada Discovery Grants RGPIN 418262-12 and RGPIN 2017-04909 (to N.T.). N.T. is a Canadian Institute of Health Research new investigator and a Michael Smith Foundation of Health Research career investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706849114/-/DCSupplemental.
References
- 1.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 2.Almonacid DE, Babbitt PC. Toward mechanistic classification of enzyme functions. Curr Opin Chem Biol. 2011;15:435–442. doi: 10.1016/j.cbpa.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown SD, Babbitt PC. New insights about enzyme evolution from large scale studies of sequence and structure relationships. J Biol Chem. 2014;289:30221–30228. doi: 10.1074/jbc.R114.569350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sillitoe I, et al. CATH: Comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 2015;43:D376–D381. doi: 10.1093/nar/gku947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horowitz NH. On the evolution of biochemical syntheses. Proc Natl Acad Sci USA. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horowitz NH. The Evolution of Biochemical Syntheses–Retrospect and Prospect. Evolving Genes and Proteins. Elsevier; Amsterdam: 1965. pp. 15–23. [Google Scholar]
- 7.Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 8.Petsko GA, Kenyon GL, Gerlt JA, Ringe D, Kozarich JW. On the origin of enzymatic species. Trends Biochem Sci. 1993;18:372–376. doi: 10.1016/0968-0004(93)90091-z. [DOI] [PubMed] [Google Scholar]
- 9.Babbitt PC, Gerlt JA. Understanding enzyme superfamilies. Chemistry as the fundamental determinant in the evolution of new catalytic activities. J Biol Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- 10.Akiva E, et al. The Structure-Function Linkage Database. Nucleic Acids Res. 2014;42:D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, et al. A universal molecular clock of protein folds and its power in tracing the early history of aerobic metabolism and planet oxygenation. Mol Biol Evol. 2011;28:567–582. doi: 10.1093/molbev/msq232. [DOI] [PubMed] [Google Scholar]
- 12.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson FJ, Mason RP, Hovsepian J, Holtzman JL. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J Biol Chem. 1979;254:4009–4014. [PubMed] [Google Scholar]
- 14.Bryant DW, McCalla DR, Leeksma M, Laneuville P. Type I nitroreductases of Escherichia coli. Can J Microbiol. 1981;27:81–86. doi: 10.1139/m81-013. [DOI] [PubMed] [Google Scholar]
- 15.Gondry M, et al. Cyclic dipeptide oxidase from Streptomyces noursei. Isolation, purification and partial characterization of a novel, amino acyl alpha,beta-dehydrogenase. Eur J Biochem. 2001;268:1712–1721. doi: 10.1046/j.1432-1033.2001.02038.x. [DOI] [PubMed] [Google Scholar]
- 16.Melby JO, Li X, Mitchell DA. Orchestration of enzymatic processing by thiazole/oxazole-modified microcin dehydrogenases. Biochemistry. 2014;53:413–422. doi: 10.1021/bi401529y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taga ME, Larsen NA, Howard-Jones AR, Walsh CT, Walker GC. BluB cannibalizes flavin to form the lower ligand of vitamin B12. Nature. 2007;446:449–453. doi: 10.1038/nature05611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas SR, McTamney PM, Adler JM, Laronde-Leblanc N, Rokita SE. Crystal structure of iodotyrosine deiodinase, a novel flavoprotein responsible for iodide salvage in thyroid glands. J Biol Chem. 2009;284:19659–19667. doi: 10.1074/jbc.M109.013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roldán MD, Pérez-Reinado E, Castillo F, Moreno-Vivián C. Reduction of polynitroaromatic compounds: The bacterial nitroreductases. FEMS Microbiol Rev. 2008;32:474–500. doi: 10.1111/j.1574-6976.2008.00107.x. [DOI] [PubMed] [Google Scholar]
- 20.Chung HW, Tu SC. Structure-function relationship of Vibrio harveyi NADPH-flavin oxidoreductase FRP: Essential residues Lys167 and Arg15 for NADPH binding. Biochemistry. 2012;51:4880–4887. doi: 10.1021/bi3002314. [DOI] [PubMed] [Google Scholar]
- 21.Ackerley DF, Gonzalez CF, Keyhan M, Blake R, 2nd, Matin A. Mechanism of chromate reduction by the Escherichia coli protein, NfsA, and the role of different chromate reductases in minimizing oxidative stress during chromate reduction. Environ Microbiol. 2004;6:851–860. doi: 10.1111/j.1462-2920.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 22.Hou F, et al. Structure and reaction mechanism of a novel enone reductase. FEBS J. 2015;282:1526–1537. doi: 10.1111/febs.13239. [DOI] [PubMed] [Google Scholar]
- 23.Liochev SI, Hausladen A, Fridovich I. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:3537–3539. doi: 10.1073/pnas.96.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macheroux P, Kappes B, Ealick SE. Flavogenomics–A genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 25.Pitsawong W, Hoben JP, Miller AF. Understanding the broad substrate repertoire of nitroreductase based on its kinetic mechanism. J Biol Chem. 2014;289:15203–15214. doi: 10.1074/jbc.M113.547117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Colibus L, Mattevi A. New frontiers in structural flavoenzymology. Curr Opin Struct Biol. 2006;16:722–728. doi: 10.1016/j.sbi.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Williams EM, et al. Nitroreductase gene-directed enzyme prodrug therapy: Insights and advances toward clinical utility. Biochem J. 2015;471:131–153. doi: 10.1042/BJ20150650. [DOI] [PubMed] [Google Scholar]
- 28.Curado S, et al. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 29.Van Aken B. Transgenic plants for enhanced phytoremediation of toxic explosives. Curr Opin Biotechnol. 2009;20:231–236. doi: 10.1016/j.copbio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Yanto Y, et al. Asymmetric bioreduction of alkenes using ene-reductases YersER and KYE1 and effects of organic solvents. Org Lett. 2011;13:2540–2543. doi: 10.1021/ol200394p. [DOI] [PubMed] [Google Scholar]
- 31.Zenno S, et al. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zenno S, Koike H, Tanokura M, Saigo K. Gene cloning, purification, and characterization of NfsB, a minor oxygen-insensitive nitroreductase from Escherichia coli, similar in biochemical properties to FRase I, the major flavin reductase in Vibrio fischeri. J Biochem. 1996;120:736–744. doi: 10.1093/oxfordjournals.jbchem.a021473. [DOI] [PubMed] [Google Scholar]
- 33.Song HN, et al. Crystal structure of the fungal nitroreductase Frm2 from Saccharomyces cerevisiae. Protein Sci. 2015;24:1158–1163. doi: 10.1002/pro.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin Y, et al. Characterization of catabolic meta-nitrophenol nitroreductase from Cupriavidus necator JMP134. Appl Microbiol Biotechnol. 2010;87:2077–2085. doi: 10.1007/s00253-010-2666-4. [DOI] [PubMed] [Google Scholar]
- 35.Chauviac FX, et al. Crystal structure of reduced MsAcg, a putative nitroreductase from Mycobacterium smegmatis and a close homologue of Mycobacterium tuberculosis Acg. J Biol Chem. 2012;287:44372–44383. doi: 10.1074/jbc.M112.406264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enright AJ, Ouzounis CA. BioLayout–An automatic graph layout algorithm for similarity visualization. Bioinformatics. 2001;17:853–854. doi: 10.1093/bioinformatics/17.9.853. [DOI] [PubMed] [Google Scholar]
- 38.Brown SD, Babbitt PC. Inference of functional properties from large-scale analysis of enzyme superfamilies. J Biol Chem. 2012;287:35–42. doi: 10.1074/jbc.R111.283408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 40.van Dongen S, Abreu-Goodger C. Using MCL to extract clusters from networks. Methods Mol Biol. 2012;804:281–295. doi: 10.1007/978-1-61779-361-5_15. [DOI] [PubMed] [Google Scholar]
- 41.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: Microcin B17 synthase. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 42.Song N, Joseph JM, Davis GB, Durand D. Sequence similarity network reveals common ancestry of multidomain proteins. PLOS Comput Biol. 2008;4:e1000063. doi: 10.1371/journal.pcbi.1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin AJM, Walsh I, Domenico TD, Mičetić I, Tosatto SCE. PANADA: Protein association network annotation, determination and analysis. PLoS One. 2013;8:e78383. doi: 10.1371/journal.pone.0078383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corel E, Lopez P, Méheust R, Bapteste E. Network-thinking: Graphs to analyze microbial complexity and evolution. Trends Microbiol. 2016;24:224–237. doi: 10.1016/j.tim.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bashiri G, et al. Elongation of the poly-γ-glutamate tail of F420 requires both domains of the F420:γ-glutamyl ligase (FbiB) of Mycobacterium tuberculosis. J Biol Chem. 2016;291:6882–6894. doi: 10.1074/jbc.M115.689026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy SR. Accelerated profile HMM searches. PLOS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Kutty R, Bennett GN. Biochemical characterization of trinitrotoluene transforming oxygen-insensitive nitroreductases from Clostridium acetobutylicum ATCC 824. Arch Microbiol. 2005;184:158–167. doi: 10.1007/s00203-005-0036-x. [DOI] [PubMed] [Google Scholar]
- 49.Park HJ, et al. Purification and characterization of a NADH oxidase from the thermophile Thermus thermophilus HB8. Eur J Biochem. 1992;205:881–885. doi: 10.1111/j.1432-1033.1992.tb16853.x. [DOI] [PubMed] [Google Scholar]
- 50.Babbitt PC, et al. The enolase superfamily: A general strategy for enzyme-catalyzed abstraction of the alpha-protons of carboxylic acids. Biochemistry. 1996;35:16489–16501. doi: 10.1021/bi9616413. [DOI] [PubMed] [Google Scholar]
- 51.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 52.Ojha S, Meng EC, Babbitt PC. Evolution of function in the “two dinucleotide binding domains” flavoproteins. PLOS Comput Biol. 2007;3:e121. doi: 10.1371/journal.pcbi.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci USA. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang B, et al. Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole. Nitric Oxide. 2016;60:32–39. doi: 10.1016/j.niox.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraaije MW, Mattevi A. Flavoenzymes: Diverse catalysts with recurrent features. Trends Biochem Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 56.Mukherjee A, Rokita SE. Single amino acid switch between a flavin-dependent dehalogenase and nitroreductase. J Am Chem Soc. 2015;137:15342–15345. doi: 10.1021/jacs.5b07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mashiyama ST, et al. Large-scale determination of sequence, structure, and function relationships in cytosolic glutathione transferases across the biosphere. PLoS Biol. 2014;12:e1001843. doi: 10.1371/journal.pbio.1001843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hicks MA, et al. The evolution of function in strictosidine synthase-like proteins. Proteins. 2011;79:3082–3098. doi: 10.1002/prot.23135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lukk T, et al. Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily. Proc Natl Acad Sci USA. 2012;109:4122–4127. doi: 10.1073/pnas.1112081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baier F, Tokuriki N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J Mol Biol. 2014;426:2442–2456. doi: 10.1016/j.jmb.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 61.Ahmed FH, et al. Sequence-structure-function classification of a catalytically diverse oxidoreductase superfamily in mycobacteria. J Mol Biol. 2015;427:3554–3571. doi: 10.1016/j.jmb.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 62.Aharoni A, et al. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 63.Tóth-Petróczy A, Tawfik DS. The robustness and innovability of protein folds. Curr Opin Struct Biol. 2014;26:131–138. doi: 10.1016/j.sbi.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 64.Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 2013;425:2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 65.Race PR, et al. Kinetic and structural characterisation of Escherichia coli nitroreductase mutants showing improved efficacy for the prodrug substrate CB1954. J Mol Biol. 2007;368:481–492. doi: 10.1016/j.jmb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 66.Swe PM, et al. Targeted mutagenesis of the Vibrio fischeri flavin reductase FRase I to improve activation of the anticancer prodrug CB1954. Biochem Pharmacol. 2012;84:775–783. doi: 10.1016/j.bcp.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 67.Copp JN, et al. Engineering a multifunctional nitroreductase for improved activation of prodrugs and PET probes for cancer gene therapy. Cell Chem Biol. 2017;24:391–403. doi: 10.1016/j.chembiol.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 68.Schnoes AM, Ream DC, Thorman AW, Babbitt PC, Friedberg I. Biases in the experimental annotations of protein function and their effect on our understanding of protein function space. PLOS Comput Biol. 2013;9:e1003063. doi: 10.1371/journal.pcbi.1003063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pieper U, et al. Target selection and annotation for the structural genomics of the amidohydrolase and enolase superfamilies. J Struct Funct Genomics. 2009;10:107–125. doi: 10.1007/s10969-008-9056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Webb EC. Enzyme Nomenclature 1992. Academic; San Diego: 1992. [Google Scholar]
- 71.Barber AE, 2nd, Babbitt PC. Pythoscape: A framework for generation of large protein similarity networks. Bioinformatics. 2012;28:2845–2846. doi: 10.1093/bioinformatics/bts532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 73.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 74.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Remmert M, Biegert A, Hauser A, Söding J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 76.Konagurthu AS, et al. MUSTANG-MR structural sieving server: Applications in protein structural analysis and crystallography. PLoS One. 2010;5:e10048. doi: 10.1371/journal.pone.0010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Armougom F, et al. Expresso: Automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Katoh K, Frith MC. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics. 2012;28:3144–3146. doi: 10.1093/bioinformatics/bts578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen NP, Mirarab S, Kumar K, Warnow T. Ultra-large alignments using phylogeny-aware profiles. Genome Biol. 2015;16:124. doi: 10.1186/s13059-015-0688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mirarab S, et al. PASTA: Ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol. 2015;22:377–386. doi: 10.1089/cmb.2014.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Price MN, Dehal PS, Arkin AP. FastTree 2–Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koike H, et al. 1.8 A crystal structure of the major NAD(P)H:FMN oxidoreductase of a bioluminescent bacterium, Vibrio fischeri: Overall structure, cofactor and substrate-analog binding, and comparison with related flavoproteins. J Mol Biol. 1998;280:259–273. doi: 10.1006/jmbi.1998.1871. [DOI] [PubMed] [Google Scholar]
- 85.Prosser GA, et al. Creation and screening of a multi-family bacterial oxidoreductase library to discover novel nitroreductases that efficiently activate the bioreductive prodrugs CB1954 and PR-104A. Biochem Pharmacol. 2013;85:1091–1103. doi: 10.1016/j.bcp.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 86.Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: Complex natural products from ribosomal templates. Curr Opin Chem Biol. 2011;15:369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi JW, et al. Crystal structure of a minimal nitroreductase, ydjA, from Escherichia coli K12 with and without FMN cofactor. J Mol Biol. 2008;377:258–267. doi: 10.1016/j.jmb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 88.Copp JN, et al. Toward a high-throughput screening platform for directed evolution of enzymes that activate genotoxic prodrugs. Protein Eng Des Sel. 2014;27:399–403. doi: 10.1093/protein/gzu025. [DOI] [PubMed] [Google Scholar]
- 89.Takeda K, et al. Synechocystis DrgA protein functioning as nitroreductase and ferric reductase is capable of catalyzing the Fenton reaction. FEBS J. 2007;274:1318–1327. doi: 10.1111/j.1742-4658.2007.05680.x. [DOI] [PubMed] [Google Scholar]
- 90.Nguyen VD, et al. Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis. Proteomics. 2007;7:1391–1408. doi: 10.1002/pmic.200700008. [DOI] [PubMed] [Google Scholar]
- 91.Bang SY, et al. Confirmation of Frm2 as a novel nitroreductase in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2012;423:638–641. doi: 10.1016/j.bbrc.2012.05.156. [DOI] [PubMed] [Google Scholar]
- 92.Mermod M, et al. Structure and function of CinD (YtjD) of Lactococcus lactis, a copper-induced nitroreductase involved in defense against oxidative stress. J Bacteriol. 2010;192:4172–4180. doi: 10.1128/JB.00372-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guillén H, Curiel JA, Landete JM, Muñoz R, Herraiz T. Characterization of a nitroreductase with selective nitroreduction properties in the food and intestinal lactic acid bacterium Lactobacillus plantarum WCFS1. J Agric Food Chem. 2009;57:10457–10465. doi: 10.1021/jf9024135. [DOI] [PubMed] [Google Scholar]
- 94.Manina G, et al. Biological and structural characterization of the Mycobacterium smegmatis nitroreductase NfnB, and its role in benzothiazinone resistance. Mol Microbiol. 2010;77:1172–1185. doi: 10.1111/j.1365-2958.2010.07277.x. [DOI] [PubMed] [Google Scholar]
- 95.Takahashi S, Furuya T, Ishii Y, Kino K, Kirimura K. Characterization of a flavin reductase from a thermophilic dibenzothiophene-desulfurizing bacterium, Bacillus subtilis WU-S2B. J Biosci Bioeng. 2009;107:38–41. doi: 10.1016/j.jbiosc.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 96.Kim KS, et al. The Rut pathway for pyrimidine degradation: Novel chemistry and toxicity problems. J Bacteriol. 2010;192:4089–4102. doi: 10.1128/JB.00201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu Y, Coates AR. Mycobacterium tuberculosis acg gene is required for growth and virulence in vivo. PLoS One. 2011;6:e20958. doi: 10.1371/journal.pone.0020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Müller J, et al. Comparative characterisation of two nitroreductases from Giardia lamblia as potential activators of nitro compounds. Int J Parasitol Drugs Drug Resist. 2015;5:37–43. doi: 10.1016/j.ijpddr.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bai J, Zhou Y, Chen Q, Yang Q, Yang J. Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering. Chem BioChem. 2015;16:1219–1225. doi: 10.1002/cbic.201500070. [DOI] [PubMed] [Google Scholar]
- 100.Yu TY, et al. Active site residues critical for flavin binding and 5,6-dimethylbenzimidazole biosynthesis in the flavin destructase enzyme BluB. Protein Sci. 2012;21:839–849. doi: 10.1002/pro.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.