Significance
To extract critical information from Alzheimer’s disease (AD) postmortem brains that may otherwise be lost, we chose to screen epigenetic signatures. Epigenome analysis is a robust methodology in terms of its cell type and gene specificity, suitability for high-throughput analysis, and resistance to postmortem degradation. Analysis of the neuron-specific methylome revealed a variety of differentially methylated genes, including BRCA1. We demonstrate the pathogenic relevance of compromised genomic integrity by analyzing the neuroprotective function of BRCA1 against amyloid β (Aβ)-induced DNA double-strand breaks. Furthermore, insolubility of BRCA1 under the presence of aggregated tau suggested the reason for its dysfunction despite enhanced expression. We provide insight into the pathomechanism of AD and demonstrate the potential of screening neuron-specific methylome to reveal new pathogenic contributors.
Keywords: Alzheimer’s disease, methylome, DNA repair, BRCA1
Abstract
Alzheimer’s disease (AD) is a chronic neurodegenerative disease characterized by pathology of accumulated amyloid β (Aβ) and phosphorylated tau proteins in the brain. Postmortem degradation and cellular complexity within the brain have limited approaches to molecularly define the causal relationship between pathological features and neuronal dysfunction in AD. To overcome these limitations, we analyzed the neuron-specific DNA methylome of postmortem brain samples from AD patients, which allowed differentially hypomethylated region of the BRCA1 promoter to be identified. Expression of BRCA1 was significantly up-regulated in AD brains, consistent with its hypomethylation. BRCA1 protein levels were also elevated in response to DNA damage induced by Aβ. BRCA1 became mislocalized to the cytoplasm and highly insoluble in a tau-dependent manner, resulting in DNA fragmentation in both in vitro cellular and in vivo mouse models. BRCA1 dysfunction under Aβ burden is consistent with concomitant deterioration of genomic integrity and synaptic plasticity. The Brca1 promoter region of AD model mice brain was similarly hypomethylated, indicating an epigenetic mechanism underlying BRCA1 regulation in AD. Our results suggest deterioration of DNA integrity as a central contributing factor in AD pathogenesis. Moreover, these data demonstrate the technical feasibility of using neuron-specific DNA methylome analysis to facilitate discovery of etiological candidates in sporadic neurodegenerative diseases.
Alzheimer’s disease (AD) is the most prevalent form of sporadic neurodegeneration. Although various studies have indicated that amyloid β (Aβ) and phosphorylated tau (pTau) are deeply involved in its pathogenesis, our current knowledge about these molecules is insufficient for the introduction of drugs that may ameliorate or cure the disease (1). There are several different strategies commonly used to investigate the pathogenesis of sporadic neurodegenerative diseases. The first approach has been to analyze genomic data of patients (i.e., SNPs) and compare with existing disease phenotypic signatures. This is a rather straightforward method since causal relation is often clear and also suitable when an underlying factor produces a particularly large effect. The discovery of the APOE ε4 allele as a risk factor is an important example of successful implementation of this approach in AD research (2). Despite this success, the methodology has not led to the discovery of additional factors largely involved in disease pathogenesis, since the effects of other genetic risk loci are often relatively small (3–5). A second approach has been to analyze postmortem brain samples from AD patients biochemically or histopathologically. While a more conventional method compared with genomic analysis, difficulties remain in extracting useful information from such samples. Unlike classic pathological analysis, high-throughput screening by transcriptome analysis have been proved to be powerful providing us with insights into the etiology of AD (6–9). However, many of these studies have analyzed bulk messenger RNA (mRNA) extracted from unprocessed brain samples, which are a mixture of mRNA originating from different cell types, including neurons, astrocytes, oligodendrocytes, microglia, and blood cells. Laser microdissection has been used to exclusively extract neuronal mRNA to overcome this drawback; however, the relatively small number of cells that can be analyzed (a maximum of ∼1 × 103 for each sample) and manually selected dissection areas can lead to problems of sampling bias. In addition, the vulnerability of mRNA molecules to postmortem degradation may also bias results (10, 11).
DNA methylation is an epigenetic gene regulation mark that occurs at the cytosine residue of CG dinucleotide sequences. In the genome, regions rich in CG doublet sequences, known as CpG islands, tend to be located at gene promoter regions. Methylation occurring at promoter regions inhibits downstream gene expression through interaction with methyl-CpG–binding proteins. In contrast, hypomethylated promoter CpG islands correspond to positive downstream gene expression (12). Compared with histone modification, another epigenetic mark that occurs in a more rapid manner, DNA methylation is thought to reflect constitutive regulation of gene expression (13). Although methylome does not provide direct information about gene expression, it serves as a rough representation of the entire transcriptome. Analysis of DNA methylation offers three significant benefits: (i) stability of DNA methylation in postmortem brains (14, 15), (ii) high-throughput isolation of up to 1 × 106–7 neuronal from nonneuronal nuclei by fluorescence-activated cell sorting (FACS) to eliminate any sampling bias (16), and (iii) implementation of high-throughput analysis using a DNA microarray system (17, 18).
Here, we present an analysis of the neuron-specific DNA methylome from human AD brains. Using this strategy, we identified three candidate molecules potentially related to AD but whose link to the disease has not been well studied. One of the molecules that emerged from the screen was BRCA1. We investigate the role of BRCA1 in human AD brains and model systems and demonstrate the importance of its interaction with the tau protein in the pathogenesis of AD.
Results
Genome-Wide Neuron-Specific DNA Methylome Analysis Reveals Dysregulated Genes in the AD Brain.
We examined inferior temporal gyrus samples from 30 age-matched normal control (NC) and 30 AD patients (Table 1). Neuronal and nonneuronal nuclei were separated by FACS using an anti-NeuN antibody (SI Appendix, Fig. S1 A and B). Starting material of 100–400 mg of brain tissue resulted in yields of ∼1 × 106–7 nuclei and 600–2,000 ng of genomic DNA. Extracted DNA was bisulfite converted and subjected to genome-wide DNA methylation analysis using an Infinium 450k methylation array. The data were analyzed using a filtering procedure described in Materials and Methods. The Infinium 450k contains 485,512 probes in total. Data with detection P values ≥ 0.01 were excluded for quality control purposes, along with probes on the sex chromosomes, to obtain 414,222 probes. Principal-component analysis of the top 1,000 most variable probes showed no evidence of separation between NC and AD, suggesting that there was little difference between the two methylomes (SI Appendix, Fig. S1C). Subsequently, probes that met the following criteria of statistical difference between the two groups were further analyzed: (i) mean β-value difference > 0.05 (ii), value of P < 0.05 by two one-sided t tests. Using this procedure, the number of probes of interest was reduced to 278 (SI Appendix, Fig. S1D). Since transcriptional regulation through DNA methylation requires regional modification rather than a single isolated change, we attempted to further refine the results by testing if the methylation occurred at a regional level. We defined differentially methylated regions (DMRs) as those including three or more consecutive differentially methylated probes. By analyzing the 278 probes in this manner, we found eight DMRs consisting of 36 statistically significant probes (SI Appendix, Fig. S1 E–G).
Table 1.
Clinical demographics of postmortem NC and AD brains used in neuron-specific methylome analysis
| Demographic variable | NC | AD | Statistics |
| No. of samples | 30 | 30 | |
| Male ratio, % | 50 | 50 | |
| Age at death | 76.7 ± 7.4 | 79.4 ± 7.4 | P = 0.16 |
| APOE ε4 ratio | 26.7 | 66.7 | P = 0.004 |
P values are calculated by t test or χ2 test.
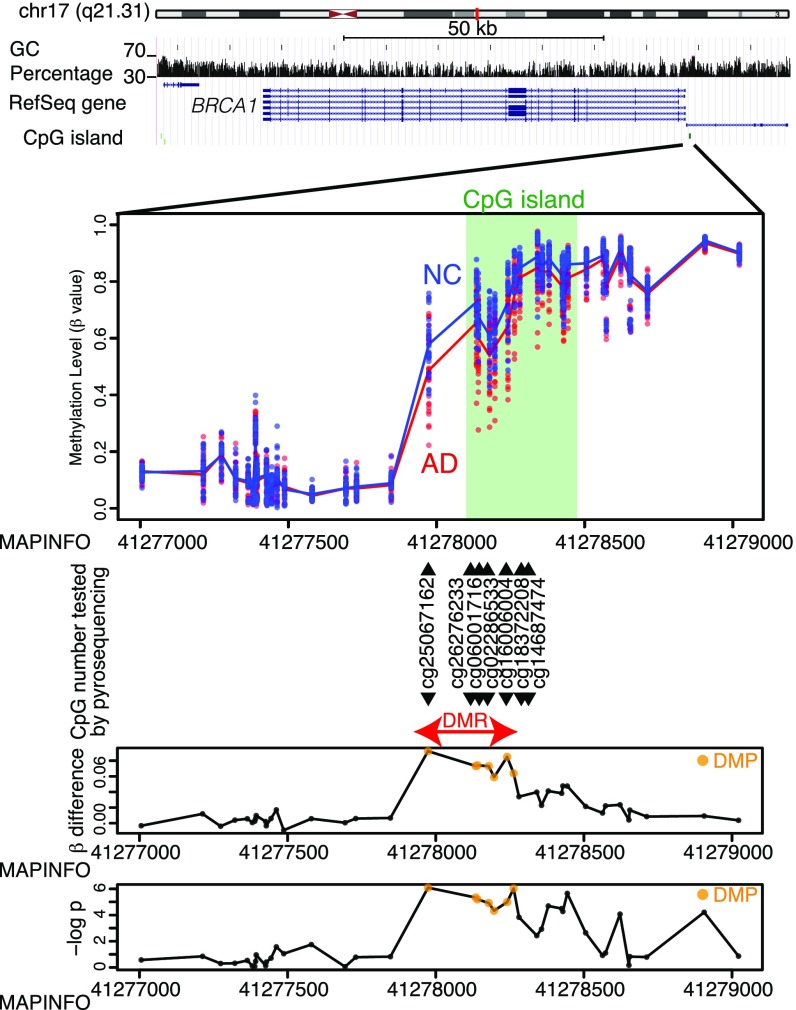
Three of the detected DMRs were located at the promoter CpG islands of the BRCA1, ZNF714, and AURKC genes. Another DMR was found at the promoter CpG island of LOC441666; however, this locus is documented as a pseudogene. The remaining four DMRs at DUSP5P1, PCDHB7, SSR1, and ERICH1 were located at intergenic regions or the gene body, where the biological significance was considered less significant and so were not pursued further. Our analysis focused on the DMRs at BRCA1, ZNF714, and AURKC. The DMRs at BRCA1 and AURKC were hypomethylated in AD samples, while that of ZNF714 was hypermethylated (Fig. 1 and SI Appendix, Fig. S1 H and I). Methylation levels of the DMRs at each of these three genes were revealed by correlation analysis and found to be tightly regulated, implying that methylation levels correlate with downstream gene regulation (SI Appendix, Fig. S2 A–C).
Fig. 1.
Details of DMR associated with BRCA1. The uppermost panel shows genomic structures from the University of California, Santa Cruz, genome browser including chromosomal ideogram, GC content (vertical bars), RefSeq gene map (blue lines and bars), and CpG islands (green bars). The second panel depicts β values of each sample at each CpG site. Dots are β values from each sample, and solid lines are mean β values of each group. Light green areas correspond to CpG islands. CG identifiers listed are the sites where methylation levels were validated using an additional pyrosequencing assay. The last two panels show β-value differences (Upper, βNC − βAD) and significance levels on a −log(P) scale at each CpG site. Orange dots represent differentially methylated probes (DMPs) discovered by Infinium screening.
Next, we analyzed the CpG methylation levels of the DMRs at BRCA1, AURKC, and ZNF714 by pyrosequencing to confirm the results of the aforementioned screening. Pyrosequencing and methylation array analysis were found to be consistent (SI Appendix, Fig. S3A). Methylation levels of the differentially methylated probes (DMPs) associated with these three genes did not correlate with age at death of the subjects (SI Appendix, Fig. S4A). The methylation levels of the ZNF714 and AURKC DMPs did demonstrate significant differences according to sex, whereas the BRCA1 DMPs showed no such difference (SI Appendix, Fig. S4B). Interestingly, methylation levels of the BRCA1 and AURKC DMRs were significantly correlated with the number of APOE ε4 alleles, but only in AD neuronal samples (SI Appendix, Fig. S4C). This suggests that the amount of Aβ deposition and subsequent neurodegeneration could be related to their altered methylation.
To test whether these results were specific to AD, we analyzed neuronal DNA from 16 inferior temporal gyrus samples of dementia with Lewy bodies (DLB) (SI Appendix, Table S2). For BRCA1 and AURKC, the differences in methylation levels between the NC and DLB groups were not statistically significant, while for ZNF714 the changes observed for AD were also seen in DLB (SI Appendix, Fig. S3B). Next, we monitored differences in methylation levels of nonneuronal samples from the inferior temporal gyrus or neuronal samples from the cerebellum of the same subjects. Surprisingly, the differences remained at several CpGs of nonneuronal DNA (SI Appendix, Fig. S3C), and even in DNA from cerebellar neuronal cells (predominantly granular cells as Purkinje cells are NeuN negative) (SI Appendix, Fig. S3D). Thus, the aberrant DNA methylation pattern appeared to be a widespread change within AD brains. Based on this, we decided to perform transcriptional analysis from postmortem brains using bulk mRNA. For this experiment, mRNA extracted from the temporal lobe in previously untested samples was used to validate the results of the methylome analysis (SI Appendix, Table S3). In AD brains, expression of BRCA1 and AURKC were clearly up-regulated; however, ZNF714 showed no statistically significant difference between the two groups (SI Appendix, Fig. S3E). This finding was mostly in agreement with the expression changes postulated from the methylation changes. Together, we hypothesized that the difference in promoter methylation levels of BRCA1 and AURKC would have a significant influence on expression of these genes, particularly in AD brains.
BRCA1 Is Mislocalized and Insoluble in the AD Brain.
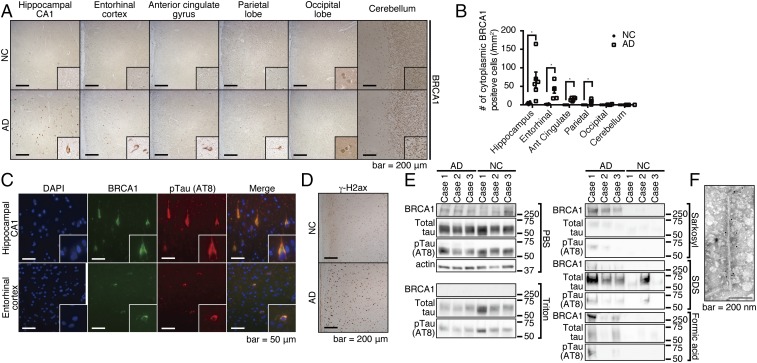
We next examined the protein expression levels of BRCA1 and AURKC in postmortem brains, starting with NC and advanced AD subjects at Braak stages 5–6 (SI Appendix, Fig. S5A). Immunohistochemical analysis showed remarkable staining of BRCA1 in the cytoplasm of neuronal cells in only the AD brains. Staining was prominent in the hippocampal CA1 region and entorhinal cortex, less so in the parietal lobe and anterior cingulate gyrus. Localization of BRCA1 in the occipital lobe or the cerebellum was nuclear, and no significant cytoplasmic signal was detected (Fig. 2 A and B). Furthermore, BRCA1 was localized to the cytoplasm of neuronal cells that well colocalized with pTau (Fig. 2C). The result was far less remarkable for AURKC, which showed a slightly increased staining in the hippocampal CA1 region and entorhinal cortex (SI Appendix, Fig. S5B). As changes in AURKC staining were less marked than those of BRCA1, further analysis was focused on BRCA1.
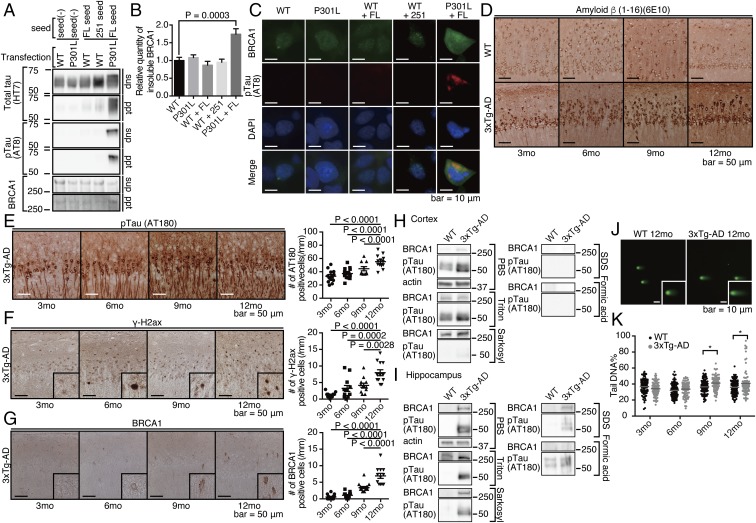
Fig. 2.
BRCA1 is mislocalized at the cytoplasm of AD brains, and it occurs at the insoluble fraction. (A) Immunohistochemical images of various regions from advanced-stage AD or NC by anti-BRCA1 antibody. Representative immunohistochemical images are shown from a total of n = 6 (hippocampus, entorhinal cortex, anterior cingulate gyrus, and parietal lobe) or n = 4 (occipital lobe and cerebellum) each. (B) Statistical analysis of A. Error bars represent means ± SEM. Significance was determined using one-way ANOVA followed by post hoc Holm–Sidak method. Mean ± SEM: NC: 3.72 ± 1.37/mm2 in the hippocampus, 1.35 ± 0.52/mm2 in the entorhinal cortex, 0/mm2 in the anterior cingulate gyrus, the parietal lobe, the occipital lobe, and the cerebellum. AD: 67.32 ± 21.27/mm2 in the hippocampus, 39.69 ± 8.05/mm2 in the entorhinal cortex, 15.33 ± 1.70/mm2 in the anterior cingulate gyrus, 8.46 ± 2.31/mm2 in the parietal lobe, 0.68 ± 0.48/mm2 in the occipital lobe, and 0/mm2 in the cerebellum. (C) Immunofluorescence images of advanced stage AD brains colabeled with DAPI, anti-BRCA1, and anti-pTau antibodies. Representative images from n = 4. (D) Immunohistochemical images of inferior temporal gyrus samples from NC or advanced-stage AD brains by anti–γ-H2ax antibody. Representative images are shown from a total of n = 3 samples of AD and NC. (E) Serial solubilization of postmortem brain samples using various detergents. The samples were treated with indicated detergents in a serial manner, each time saving the centrifuged supernatants as soluble fractions for each detergent. Three AD and NC samples were tested. (F) Immuno-EM of purified PHF by anti-BRCA1 antibody. *P < 0.05.
The distribution pattern of BRCA1 was similar to that of advanced AD pathology, consistent with a model where the AD pathology itself could be driving expression (Fig. 2 A and B). To test this hypothesis, the immunostaining pattern of BRCA1 was compared with those of pathological hallmarks of AD (e.g., deposition of Aβ or pTau) in subjects harboring earlier stage AD pathology. At Braak stage 3, Aβ- and pTau-positive pathology was already present in both the hippocampal CA1 region and the entorhinal cortex, while cytoplasmic BRCA1 was detected mainly in the hippocampal CA1 region (SI Appendix, Fig. S5 C–E), especially in the neuronal cells having prominent pTau deposition (SI Appendix, Fig. S5F). This indicates that accumulation of cytoplasmic BRCA1 may be a consequence of pTau deposition, which well explained the observation that the region without tau pathology, that is, the occipital lobe and the cerebellum, is mostly free from cytoplasmic accumulation of BRCA1 despite decreased methylation.
BRCA1 is a nuclear protein with a central role in DNA repair. Among various DNA repair related proteins, only γ-H2ax, a well-established marker for initiation of DNA double-strand break (DSB) repair, was prominent in AD brains (Fig. 2D and SI Appendix, Fig. S5G). This result suggested that Aβ induced DNA DSBs, of which BRCA1 specializes in their repair and possible failure of the downstream process. Considering that aberrant cytoplasmic localization of BRCA1 could be indicative of its dysfunction, we tested its solubility in AD brains. Extraction via a series of detergents revealed a substantial shift in BRCA1 solubility in AD brains, suggesting that the majority of BRCA1 in the AD brain is dysfunctional (Fig. 2E). For further assessment, we performed immuno-electron microscopy (EM) of the paired helical filaments (PHF) fraction to find that PHFs were positive with anti-BRCA1 antibodies (Fig. 2F). Immunoprecipitation of soluble tau from human AD brain lysates showed no association of BRCA1, indicating no direct binding of BRCA1 to soluble tau (SI Appendix, Fig. S5H). Taken together, we concluded that insolubilized tau as PHF sequesters BRCA1 by coaggregation.
Aβ Burden Induces DNA DSBs and Up-Regulation of BRCA1 Expression in Vitro and in Vivo.
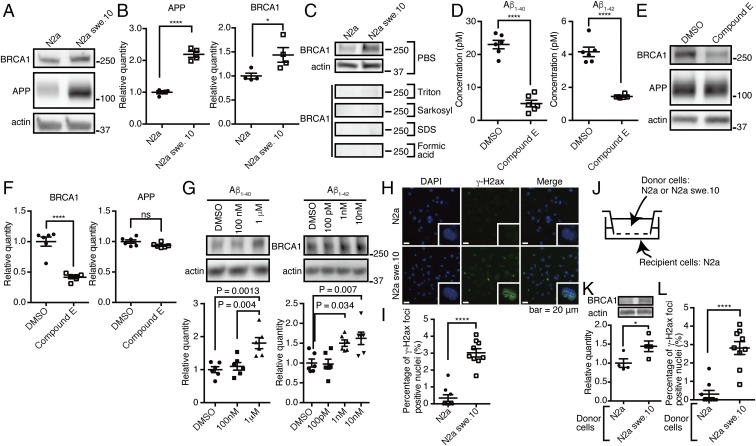
We next focused on the mechanism of BRCA1 up-regulation. Since the BRCA1 DMR was also hypomethylated in nonneuronal cells that are essentially free of tau pathology, we reasoned that its up-regulation might be more closely related to Aβ than tau. To test whether Aβ itself was sufficient to up-regulate BRCA1, we analyzed N2a swe.10 cell line that expresses human amyloid precursor protein (APP) (KM670/671NL). This cell line produces significant levels of intracellular APP (Fig. 3 A and B and SI Appendix, Fig. S6A) and secretes extracellular Aβ40 and Aβ42 into the medium (SI Appendix, Fig. S6B). N2a swe.10 cells expressed more soluble BRCA1 compared with control cells (mean ± SEM: 1.00 ± 0.06 in N2a cells vs. 1.44 ± 0.15 in N2a swe.10 cells) (Fig. 3 A and B). Full-length BRCA1 was found in the soluble fractions of both control and N2a swe.10 cells (Fig. 3C). To confirm that BRCA1 expression was caused by Aβ and not by APP overexpression, we treated N2a swe.10 cells with a potent γ-secretase inhibitor, compound E. The treatment significantly decreased BRCA1 expression as well as Aβ, without having any effect on APP expression in N2a swe.10 cells, while BRCA1 expression was not affected in N2a cells (Fig. 3 D–F and SI Appendix, Fig. S6 C and D). To eliminate any off-target effect of compound E, we treated N2a cells with recombinant Aβ40 or Aβ42 and found that both of them increased BRCA1 but Aβ40 required much higher concentration compared with Aβ42, suggesting that their toxicity is driving BRCA1 expression (Fig. 3G). Immunostaining revealed multiple positive nuclear γ-H2ax signals only in N2a swe.10 cells, consistent with the activation of DNA repair machinery (Fig. 3 H and I). To exclude the possibility that overexpressed APP may affect formation of nuclear γ-H2ax signals and BRCA1 up-regulation, N2a cells were cocultured with N2a swe.10 or N2a cells (Fig. 3J). Aβ secreted from donor N2a swe.10 cells was able to induce up-regulation of BRCA1 and multiple positive nuclear γ-H2ax signals were detected in recipient cells (Fig. 3 K and L). We analyzed the methylation level of Brca1 CpG island, but there was no difference between the two cell lines (SI Appendix, Fig. S6E). To evaluate the amount of DNA damage in N2a swe.10 cells, single-cell gel electrophoresis (comet assay) was performed. Control experiment using etoposide resulted in significant DNA fragmentation (Fig. 4 A and B). We found no significant DNA fragmentation in N2a swe.10 cells compared with controls (Fig. 4 C and D), indicating that sufficient DNA repair continued to occur despite activated DSBs in these cells. Knockdown of BRCA1 by lentiviral shRNA impaired DNA repair to increase DNA damage in N2a swe.10 cells (Fig. 4 E and F). These data indicated that the presence of Aβ triggers DNA DSBs, with the recruitment of BRCA1 sufficient to neutralize Aβ toxicity.
Fig. 3.
Aβ confers BRCA1 overexpression. (A) Western blot of N2a and swe.10 cells using anti-mouse BRCA1 and anti-APP antibodies. Actin was detected as a loading control. (B) Quantitative measurement of the relative amount of APP and BRCA1 from experiments in A. n = 4. Relative quantity against actin expression level was normalized to N2a = 1.0. Mean ± SEM: APP: 1.00 ± 0.05 in N2a cells vs. 2.19 ± 0.10 in N2a swe.10 cells; BRCA1: 1.00 ± 0.06 in N2a cells vs. 1.44 ± 0.15 in N2a swe.10 cells. (C) Serial fractionation of N2a and swe.10 cells using various detergents. (D) Concentrations of Aβ40 and Aβ42 in the culture media supernatant of N2a swe.10 cells treated with dimethyl sulfoxide (DMSO) or 25 nM compound E were measured by ELISA. n = 6. Mean ± SEM: Aβ40: 23.00 ± 1.28 pM in DMSO vs. 5.08 ± 1.00 pM in compound E; Aβ42: 4.16 ± 0.29 pM in DMSO vs. 1.45 ± 0.04 pM in compound E. (E) Western blot of N2a swe.10 cells treated with DMSO or compound E by anti-mouse BRCA1 and anti-APP antibodies. (F) Quantitative measurement of the relative amount of BRCA1 and APP from E. n = 6. Relative expression level was normalized to DMSO = 1.0. Mean ± SEM: BRCA1: 1.00 ± 0.08 in DMSO vs. 0.41 ± 0.02 in compound E; APP: 1.00 ± 0.03 in DMSO vs. 0.94 ± 0.02 in compound E. (G) Effect of recombinant Aβ40 and Aβ42 on BRCA1 expression. Western blot of N2a cells by anti-mouse BRCA1 and anti-actin antibodies. Quantitative measurement of the relative amount of BRCA1 is shown below (n = 6 independent wells). Each relative expression level of BRCA1 was normalized to DMSO = 1. One-way ANOVA [Aβ40: F(2,15) = 11.63, P = 0.0009; Aβ42: F(3,20) = 7.981, P = 0.0011] with post hoc Turkey method. Mean ± SEM: Aβ40: 1.00 ± 0.09 in DMSO, 1.10 ± 0.11 in 100 nM, 1.80 ± 0.17 in 1 μM; Aβ42: 1.00 ± 0.10 in DMSO, 0.97 ± 0.12 in 100 pM, 1.50 ± 0.08 in 1 nM, 1.62 ± 0.16 in 10 nM. (H) Immunofluorescence images of N2a and swe.10 cells stained with DAPI, and anti–γ-H2ax antibody. Insets show single nuclei at high magnification. (I) Quantitative analysis of the number of cells with nuclear γ-H2ax foci in H. n = 9 visual fields (3 visual fields from 3 experiments). Mean ± SEM: 0.34 ± 0.20% in N2a cells and 3.02 ± 0.25% in N2a swe.10 cells. (J) Diagram of coculture system. Recipient cells were used for biochemical and immunohistochemical analysis. (K) Western blot of recipient N2a cells by anti-mouse BRCA1 and anti-actin antibodies. Quantitative measurement of the relative amount of BRCA1 is shown below (n = 4 independent wells). Each relative expression level was normalized to N2a-donor culture. Mean ± SEM: 1.00 ± 0.11 in N2a cells and 1.44 ± 0.14 in N2a swe.10 cells. (L) Quantitative analysis of the number of cells with nuclear γ-H2ax foci in the recipient N2a cells. n = 9 visual fields (3 visual fields from 3 experiments). Mean ± SEM: 0.31 ± 0.20% in N2a cells and 2.81 ± 0.35% in N2a swe.10 cells. *P < 0.05 and ****P < 0.0001. ns, not significant.
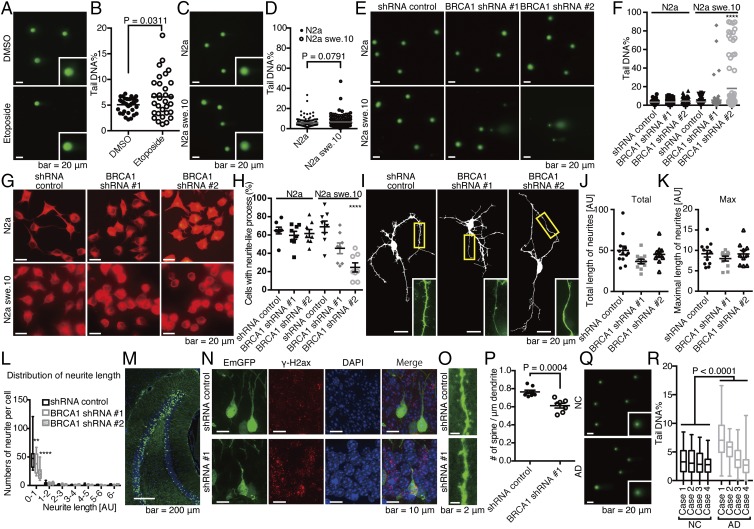
Fig. 4.
Functional relevance of BRCA1 dysfunction in an in vitro and in vivo neuronal model with Aβ burden. (A) Comet assay images of N2a treated with DMSO or 50 μM etoposide for 6 h. (B) Quantification of tail DNA%. Closed circles represent N2a cells treated with DMSO, and open circles, etoposide. n = 39 cells (DMSO) and 31 cells (etoposide). Mean ± SEM: 5.07 ± 0.21% in DMSO and 6.60 ± 0.73% in etoposide. (C) Comet assay images of N2a and N2a swe.10 cells. Representative images are shown. Insets are a high magnification of single nuclei. (D) Quantification of tail DNA%. Closed circles represent N2a cells, and open circles, swe.10. n = 150 cells. Gray bars represent means. Mean ± SEM: 5.72 ± 0.34% in N2a cells and 6.68 ± 0.42% in N2a swe.10 cells. Biological replicates are shown in SI Appendix, Fig. S10A. (E) Representative comet assay images are shown from a total of n = 80 N2a and swe.10 cells after lentiviral transduction of BRCA1 shRNA. (F) Statistical significance of BRCA1 knockdown on DNA fragmentation was determined using the Tukey method. Bars represent means. Mean ± SEM: N2a: 3.45 ± 0.24% in shRNA control, 3.59 ± 0.27% in shRNA #1, 4.40 ± 0.34% in shRNA #2; N2a swe.10: 3.67 ± 0.35% in shRNA control, 5.59 ± 1.37% in shRNA #1, 18.06 ± 3.10% in shRNA #2. Biological replicates are shown in SI Appendix, Fig. S10B. (G) Representative images of the differentiated N2a and swe.10 cells treated with shRNAs are shown (n = 8 visual fields). (H) Quantification of cells with neurite-like process. Statistical significance was calculated by the Tukey method. Mean ± SEM: N2a: 64.86 ± 3.65% in shRNA control, 59.64 ± 4.33% in shRNA #1, 61.62 ± 4.36% in shRNA #2; N2a swe.10: 68.98 ± 6.62% in shRNA control, 45.46 ± 5.81% in shRNA #1, 24.61 ± 4.98% in shRNA #2. (I) Representative images of primary neuronal cultures of cortical tissues from 3×Tg mice. Total length of neurites (J), maximal length of neurites (K), and neurite length distribution (L) were analyzed. Control (n = 12 neurons), shRNA #1 (n = 11 neurons), and shRNA #2 (n = 10 neurons). Statistical significance was determined by the Tukey method. Boxes extend from the 25th to 75th percentiles, and the lines in the boxes represent the median. The whiskers show the minimum and maximum values. (M) Knockdown of BRCA1 in vivo. Lentivirus expressing shRNA against BRCA1 was stereotactically injected into the dentate gyrus (DG) of APP/PS1 mice at 3 mo of age, and mice were killed 3 wk after the surgery. (N) Immunofluorescence images of the neuronal cell in DG stained with DAPI, and anti–γ-H2ax antibody. (O) Spine density of the neuronal cells in DG. n = 8 (control) and 7 (shRNA #1) dendrites. Representative images are shown. (P) Statistical analysis of O. Mean ± SEM: 0.76 ± 0.02/μm in shRNA control and 0.61 ± 0.03/μm in shRNA #1. (Q) Nucleus from inferior temporal gyrus of four NC and four AD brains were subjected to comet assay. Representative images are shown. (R) Statistical analysis of Q. Boxes extend from the 25th to 75th percentiles, and the lines in the boxes represent the median. The whiskers show the minimum and maximum values. n = 70–100 nuclei from each sample. Statistical significance was determined using two-way ANOVA [NC vs. AD: F(1,707) = 100.6, P < 0.0001; sample: F(3,707) = 52.30, P < 0.0001]. Mean ± SEM: NC: 3.54 ± 0.24%, 3.36 ± 0.23%, 3.13 ± 0.21%, and 2.58 ± 0.15%; AD: 7.45 ± 0.34%, 5.46 ± 0.19%, 3.64 ± 0.22%, and 2.80 ± 0.24%. **P < 0.01 and ****P < 0.0001.
To assess the functional relevance of BRCA1 function against Aβ in neuronal cells, we analyzed their morphological changes upon BRCA1 knockdown. BRCA1 knockdown suppressed formation of neurite-like processes in N2a swe.10 cells upon differentiation but not in N2a cells (Fig. 4 G and H). Furthermore, in primary cultures of neuronal cells from 3×Tg-AD mice harboring APP KM670/671NL and MAPT P301L transgenes, along with knocked-in PSEN1 M146V, which is a well-established model for both Aβ and pTau deposition (19), BRCA1 knockdown did not have significant effect on the global morphology (Fig. 4 I–K), but reduced the number of short neurites (mean ± SEM: for dendrites with 0–1 AU in length, 50.50 ± 7.16 in shRNA control, 39.18 ± 5.38 in shRNA #1, 22.60 ± 4.07 in shRNA #2) (Fig. 4L). We further assessed in vivo function of BRCA1 in APP/PS1 mice that expresses APP KM670/671NL and PSEN1dE9 (20, 21). Knocking down BRCA1 using stereotactic lentivirus injection into the dentate gyrus (DG) induced γ-H2ax foci and reduced spine density (Fig. 4 M–P).
We next used APP/PS1 mice to assess the effect of Aβ toxicity on BRCA1 expression and DNA damage in vivo. These mice exhibit Aβ pathology, but tau pathology is absent (SI Appendix, Fig. S7 A and B). Although γ-H2ax could be detected from the earliest stage (SI Appendix, Fig. S7 C and D), suggesting DNA DSBs were triggered in the hippocampal neurons, comet assays of hippocampal cells in the mice showed no significant fragmentation of genomic DNA (SI Appendix, Fig. S7 E and F). Immunohistochemistry showed no cytoplasmic BRCA1 in the hippocampal regions in these mice, and biochemical analysis demonstrated increased soluble BRCA1 [mean ± SEM: 1.00 ± 0.10 in wild type (WT) vs. 1.59 ± 0.09 in APP/PS1 in PBS fraction] (SI Appendix, Fig. S7 G–I). Data from these mice indicated that DNA DSBs were induced by Aβ but sufficiently repaired by BRCA1. To see whether DNA damage was actually happening in human AD brain, temporal lobe samples were subjected to comet assay, which showed significant DNA damage in AD brains (Fig. 4 Q and R).
If this is the case, what makes BRCA1 insoluble and dysfunctional in human AD brains? We hypothesized that aggregated tau could have a role in this phenomenon, since there were almost no glial cells positive for cytoplasmic BRCA1, despite a widespread methylation change in AD brains (Fig. 2 A and B and SI Appendix, Fig. S3 A, C, and D). To evaluate its role in BRCA1 inactivation, we first tested whether BRCA1 was insolubilized in in vitro seed-dependent tau aggregation model (22). Upon aggregation of P301L tau by adding in vitro-generated seed, endogenous BRCA1 was shifted to insoluble fraction (Fig. 5 A and B) and cytoplasmic tau aggregates colocalized with BRCA1, precluding its nuclear localization (Fig. 5C). To investigate whether this was also true in in vivo model, we histopathologically and biochemically analyzed 3×Tg-AD mice. As previously reported, Aβ and pTau accumulate in an age-dependent manner in hippocampal CA1 neurons (19, 23) (Fig. 5 D and E). In these mice, positive γ-H2ax signal preceded the appearance of cytoplasmic BRCA1 and the number of positive cells increased in an age-dependent manner (Fig. 5F). Cytoplasmic BRCA1 was also detected from the age of 6 mo, gradually increasing with age up to 12 mo (Fig. 5G). Together with hippocampal tau deposition in the 3×Tg-AD mice brain, we postulated that the cytoplasmic BRCA1 observed was the result of neuronal tau deposition. Consistent with this hypothesis, cortical regions of 3×Tg-AD mice show only Aβ pathology without tau deposition (23), similar to the APP/PS1 mice. These same cortical regions also exhibited increased levels of soluble BRCA1 (Fig. 5H). In contrast, the hippocampal region of 3×Tg-AD mice had increased levels of insoluble BRCA1 along with increased insoluble pTau at the age of 12 mo (Fig. 5I).
Fig. 5.
Cytoplasmic tau is important for mislocalization and insolubilization of BRCA1. (A) SH-SY5Y cells transfected with wild type (WT) or P301L mutant 4-repeat tau were treated with in vitro-aggregated tau seeds [either full length (FL) or C-terminal fragment (251)]. After fractionation, supernatant (sup) or pellet (ppt) was subjected to Western blot. (B) Quantification of BRCA1 in the pellet fraction. Mean ± SEM: 1.00 ± 0.09 in WT, 1.08 ± 0.08 in P301L, 0.87 ± 0.11 in WT + FL, 0.95 ± 0.09 in WT + 251, and 1.74 ± 0.15 in P301L + FL. (C) Immunocytochemical images of seed-dependent tau aggregation colabeled with DAPI, pTau, and BRCA1 antibodies. (D) Representative images of Aβ staining of 3×Tg-AD mice hippocampal region. (E–G) Representative immunohistochemical images of 3×Tg-AD mice hippocampus by anti-pTau (E), anti-γ-H2ax (F), and BRCA1 (G) staining. Samples are from 3×Tg-AD mice at 3, 6, 9, and 12 mo of age. n = 10–15 visual fields from six to seven mice for 3×Tg-AD mice. Quantitation of positive cells for each protein are shown at the Right side of the images. Statistical significance was determined by the Tukey method. Mean ± SEM: pTau: 33.26 ± 2.57/mm in 3 mo, 37.30 ± 1.99/mm in 6 mo, 44.56 ± 3.19/mm in 9 mo, and 55.12 ± 2.69/mm in 12 mo; γ-H2ax: 1.28 ± 0.22/mm in 3 mo, 3.23 ± 0.90/mm in 6 mo, 4.14 ± 0.66/mm in 9 mo, and 7.97 ± 0.90/mm in 12 mo; BRCA1: 0.56 ± 0.21/mm in 3 mo, 1.07 ± 0.24/mm in 6 mo, 3.41 ± 0.33/mm in 9 mo, and 6.94 ± 0.77/mm in 12 mo. (H and I) Expression of BRCA1 in detergent fractionated the cortical (H) and hippocampal (I) regions of 3×Tg-AD mice at 12 mo of age. n = 2 mice at 12 mo of age. Representative blot was shown. (J) Microscopic images of comet assays. Representative images are shown from a total of n = 136 cells from WT and 3×Tg-AD animals. Insets are magnified nuclei. (K) Quantification of tail DNA% in E. Solid circles are WT, and open circles, 3×Tg-AD mice. Gray bars represent means. Statistical significance was determined using the Holm–Sidak method. Mean ± SEM: WT: 36.71 ± 1.03% in 3 mo, 32.11 ± 0.92% in 6 mo, 36.55 ± 0.92% in 9 mo, and 36.32 ± 0.98% in 12 mo; 3×Tg-AD: 34.10 ± 0.88% in 3 mo, 33.67 ± 0.92% in 6 mo, 41.24 ± 0.90% in 9 mo, and 40.87 ± 1.65% in 12 mo. *P < 0.05.
Hippocampal cells from 3×Tg mice were monitored for DNA damage by comet assay. DNA fragmentation increased from 9 mo in an age-dependent manner, suggesting that the balance between DNA damage and repair was significantly compromised along with tau accumulation in 3×Tg-AD mice (mean ± SEM: for 9 mo, 36.55 ± 0.92% in WT, vs. 41.24 ± 0.90% in 3×Tg; for 12 mo, 36.32 ± 0.98% in WT, vs. 40.87 ± 1.65% in 3×Tg) (Fig. 5 J and K). These data indicated that the presence of aggregated tau induced mislocalization and insolubility of BRCA1, leading to accelerated DNA fragmentation.
BRCA1 expression was up-regulated by promoter demethylation in human AD brains. To determine whether this phenomenon could be reproduced in the brain of an AD model mouse, we measured the methylation level of the Brca1 gene in 3×Tg-AD mice. Pyrosequencing clearly showed that the Brca1 promoter in hippocampal DNA from 3×Tg-AD mice was significantly hypomethylated compared with that of WT mice (SI Appendix, Fig. S8), suggesting that Aβ-induced DNA damage leads to up-regulation of BRCA1 expression through demethylation of the promoter region.
Discussion
Genome-Wide Neuron-Specific DNA Methylation Analysis Reveals Previously Undiscovered Genes.
The methodology adopted in this study initially focused on genome-wide neuron-specific methylome analysis of the postmortem brain and the successful identification of biologically relevant molecules. Previous studies of AD epigenetics have used bulk DNA from postmortem brains and, in several instances, have demonstrated aberrant CpG methylation at genes related to familial AD and APP processing (24) or newly discovered genes (25, 26), which were not confirmed in our neuron-specific analysis (SI Appendix, Table S7). This discrepancy could be attributed to the level of resolution reached by the Infinium microarray [the exact CpGs detected in the previous study (24) are not on the array] or to the advantage of our neuron-specific analysis over a bulk DNA analysis made up of various cell types. Our FACS purification strategy demonstrated that the DNA amount of neuronal/nonneuronal cell ratios varied among samples from 1:0.19 to 1:2.8 [mean, 1:1.20 ± 0.65 (SD)], which could easily skew the methylome analysis, given that patterns of methylation differ among cell types (16, 27).
The sample numbers used in our study are relatively small, which does limit our capacity to perform precise statistical analyses such as multiple comparisons. These analyses were nonetheless performed; however, none of our probes was able to meet statistical criteria. Recent studies of site-directed methylation editing have revealed that methylation changes over broader ranges (rather than a single mark) are responsible for the expression level of mRNA (28). Thus, strict correction of multiple comparisons with statistical methods that have been useful in detecting single, differentially methylated spots may not be as supportive for detecting broader differential regions reflecting the transcriptome. The goal of our study was to identify a biologically relevant molecule for AD through methylome analysis. Therefore, rather than correcting multiple comparisons by statistically stringent method, we attached more importance to the discovery of methylation signature related to transcriptional regulation. Additionally, we chose to directly confirm results at the mRNA or protein level rather than simply replicating the methylome analysis in a different sample sets, since we considered it as a screening method for intraneuronal environment alteration in AD brains. We believe that neuron-specific analysis can reduce the intragroup variation of methylation level and enhance the specificity and sensitivity in detecting DMRs.
Methylation levels were validated independently in the DMR of interest. Moreover, of three candidate genes extracted from the methylome analysis, two corresponding proteins were found to be differentially expressed in AD brains, strongly supporting the validity of our method. Methylome analysis is not a method to directly analyze protein expression in the postmortem brain. However, given the limitations of previously reported methodologies, such as postmortem degradation and sampling bias, we believe that this methodology produced a more precise representation of molecular neuronal pathology compared with previous studies. In doing so, it has the potential to be used for further in-depth analysis of the pathomechanisms underlying AD and other neurodegenerative diseases.
Methylome Analysis Reveals a Unique Pathological Pathway.
Using methylome analysis, we found that BRCA1 and AURKC were up-regulated in AD brains. We also demonstrated that the methylation levels at the promoter regions of these genes were, to some extent, correlated with the number of APOE ε4 alleles, whose gene product is known to impair Aβ clearance in the brain compared with other alleles, such as ε2 or ε3 (29). Polymorphic variants of TREM2 gene are also the known risk factor influencing for developing AD on the same level as one copy of the APOE ε4 allele (30); however, there was no polymorphic variants detected in our cases. Together with our analyses using postmortem human and mouse brains, this supports our hypothesis that increased Aβ burden induces up-regulation of BRCA1 and AURKC to repair DSBs. However, BRCA1 protein was mislocalized to the cytoplasm and predominantly found in the detergent-insoluble fraction of both human AD and 3×Tg-AD mice brains that suggested it might be dysfunctional (31). Indeed, significant DNA fragmentation could be seen in both human AD and mouse model brains. Similar finding has also been reported in a previous study (32); however, our data have suggested an alternative role for BRCA1. In addition to the precision and relevance of our results, we provide three pieces of information that also support our interpretation. First, the methylome, transcriptome, proteome, and neuropathological analyses reported here consistently demonstrate BRCA1 up-regulation during the pathological process of AD. Second, we and others have shown that the molecular weight of BRCA1 is always >250 kDa (33, 34) (SI Appendix, Fig. S9 A–D), but the band indicated on immunoblots from a conflicting report (32) appears to be much smaller. In addition, knockdown of human and murine BRCA1 by multiple shRNAs confirmed that the band was BRCA1 (SI Appendix, Fig. S9 B–D). Third, our data corroborated the previous result demonstrating that large number of neurons with neurofibrillary tangles were positive for BRCA1 (35).
Differential extraction demonstrated the presence of Sarkosyl-insoluble BRCA1 in the hippocampus of 3×Tg AD mice at 12 mo without SDS-insoluble BRCA1. SDS-insoluble tau represents a later and more extensive state of aggregation than Sarkosyl-insoluble tau (36, 37). In human AD, the disease time course is quite long, usually 10–15 y from the beginning of tau aggregation to autopsy, whereas mice have a life expectancy of only 2 y. Considering the results of in vitro seed-dependent tau aggregation model and immuno-EM, BRCA1 aggregates in a tau-dependent manner. The amount of insoluble tau is very small in mice model compared with human AD brain (Figs. 2E and 5 H and I), even though in these mice mutant tau is more aggregate-prone and is also overexpressed, suggesting that the time factor is important in tau aggregation and its change in solubility. Thus, in the mouse model, tau is not insoluble enough compared with human disease as previously reported (38). Considering that coaggregation of BRCA1 with PHF was demonstrated by EM, we speculate that a relatively short time (e.g., 12 mo) is sufficient for endogenous BRCA1 to become Sarkosyl-insoluble and dysfunctional, but insufficient to become a highly aggregated state like SDS-insoluble.
BRCA1 is a DNA repair protein, whose mutation drastically increases the risk of breast cancer (39). It is well studied in the field of cancer research, but its role in neurodegeneration remains unknown. Recent reports have indicated the importance of DNA damage in AD pathogenesis (40), with evidence of activity-induced DNA breaks governing the expression of neuronal early-response genes (41). Despite this potential relevant activity, its role remains elusive (32, 35, 40, 42). Since differentiated neurons do not proliferate, impaired DNA cannot be repaired by the precise process of homologous recombination, which uses homologous sister chromatids as templates during mitosis. Thus, nonhomologous end joining, including microhomology-mediated end joining, is the only alternative for neurons. Preserving the integrity of neuronal DNA at the best possible level over its life span would be a key to maintaining the cellular functions of neurons (43–45). Therefore, the recruitment of DNA repair proteins, such as BRCA1, would appear to be important for neurons. Interestingly, several epidemiological studies reported that the risk for developing AD is increased by diabetes mellitus or atherosclerosis, which are known to induce DNA damage by producing oxidative stress (46–48). Furthermore, an inverse association between cancer and AD has been reported (49–52). These observations suggest that deterioration in the maintenance of DNA integrity could play an important role in AD pathogenesis. We demonstrated that knockdown of BRCA1 in neuronal cells in the presence of Aβ burden in vitro and in vivo resulted in a decrease in the number of short neurites and dendritic spines. Considering the decreased spine density in brains of human AD patients and AD model mice (53, 54), these results suggest a potential mechanism underlying neuronal dysfunction in the AD brain. That is, Aβ may impair synaptic plasticity in neuronal cells by causing deterioration of DNA integrity, thereby resulting in memory impairment.
BRCA1 Dysfunction Leads to Deterioration of DNA Integrity.
Our results clearly demonstrate that Aβ-induced DSBs were efficiently repaired by BRCA1 in the absence of aggregated tau, and that repair by this mechanism was impaired in its presence due to mislocalization and insolubilization of BRCA1. Among many DNA repair markers, only γ-H2ax was positive in AD brain. Considering γ-H2ax is a scaffold for recruitment of DNA repair proteins at an early stage of DNA repair (55–57). BRCA1 is one of the early-phase proteins recruited to the γ-H2ax foci (58) and, therefore, could possibly fail to form the foci of DNA repair proteins due to its mislocalization and insolubilization impeding the subsequent DNA repair process. A recent study showed that BRCA1 nuclear export was related to susceptibility to DNA damage (59), which suggests that the mislocalization of BRCA1 to the cytoplasm and insolubility once there, interfered with DNA damage repair. Our data with cellular and animal model only with Aβ burden suggested that BRCA1 is up-regulated for a protective purpose. However, in human AD brain, BRCA1 is coaggregated with tau that makes BRCA1 dysfunctional. Despite its increased expression, coaggregation leads to insufficient DNA repair. In other words, although BRCA1 is up-regulated, it does not reach to the sufficient level necessary to counteract Aβ-induced DNA damage leading to its accumulation.
BRCA1 is involved primarily in homologous recombination, but also, to some extent, in nonhomologous end joining (60). This raises the question of why BRCA1, rather than other DNA repair proteins, is recruited in response to DNA damage in nondividing neuronal cells. Several reports have shown the importance of BRCA1 function in the developing brain (61, 62) and the accumulation of somatic mutations in neuronal cells during the developmental process in a transcriptional activity-dependent manner (63). However, activity-induced DNA DSBs have previously been shown to govern the expression of neuronal early-response genes (41). These reports collectively suggest that DNA DSBs occur frequently in neurons of highly active regions, such as the hippocampus and entorhinal cortex, and are efficiently repaired by BRCA1 in normal brains. However, in AD brains, this maintenance system seems to be impaired by BRCA1 dysfunction, resulting in neuronal deterioration in these regions. Indeed, comet assay on postmortem brains showed increased DNA fragmentation in AD samples.
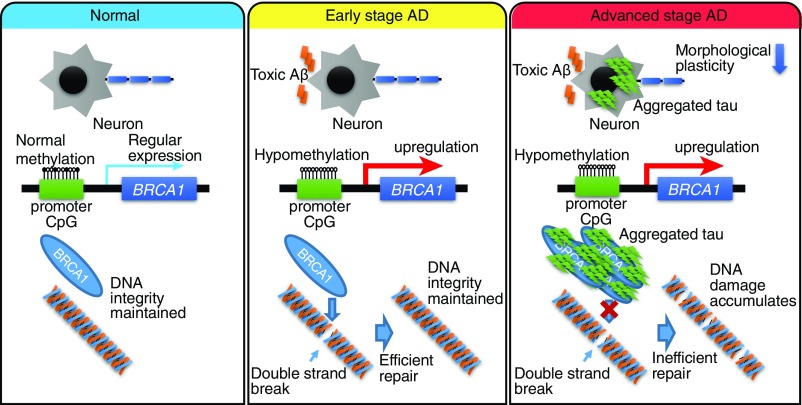
In this study, we present findings of neuron-specific DNA methylation alterations in AD brains. Taken together with expression analyses, these results suggest that BRCA1 plays an important role in the pathogenesis of AD, since methylome, transcriptome, proteome, and neuropathology results are all consistent with its up-regulation. Aβ induces DNA DSBs, which should be efficiently repaired by BRCA1 when aggregated tau is absent. The balance of DNA damage and repair is compromised by dysfunction of BRCA1, due to cytoplasmic mislocalization and consequential insolubility as a result of aggregated tau accumulation (Fig. 6). In this study, methylome analysis has directly facilitated identification of biologically relevant molecules for a neurodegenerative disease. Future analyses of neuron-specific methylome may also prove useful in understanding the mechanisms of pathogenesis for sporadic neurodegenerative disorders.
Fig. 6.
Schematic illustration summarizing our current hypothesis regarding Aβ-induced DNA damage, tau, and epigenetic regulation of BRCA1 in AD. In normal brain without Aβ or tau accumulation, there is no need for BRCA1 up-regulation (Left). At an early stage of AD with no accumulated tau, BRCA1 efficiently repairs DNA DSBs induced by toxic Aβ (Middle). However, at an advanced stage of AD, cytoplasmic aggregated tau sequestrates BRCA1 to an insoluble fraction, resulting in its dysfunction (Right). While neurons try to cope with this situation by up-regulating expression of the BRCA1 gene through epigenetic mechanisms, they are eventually overwhelmed by the accumulation of DNA damage.
Materials and Methods
Sample Preparation and DNA Extraction.
This research received approval from the ethics committee of the University of Tokyo (approval 2183-15). We collected postmortem brains with written consent from patients’ families and maintained them at −80 °C until use. According to criteria established by Braak and McKeith (64–67), trained neuropathologists made a diagnosis of AD, DLB, or NC, using hematoxylin–eosin, Nissl and silver staining, and immunostaining (68). We diagnosed samples fulfilling Braak stage ≥4 and amyloid stage ≥C as AD (64, 65), and samples fulfilling Lewy body score ≥4, Braak stage ≤3, and amyloid stage ≤B as DLB (66, 67). Brain samples of 18 AD, 15 NC, and 21 DLB subjects were obtained from Tokyo Metropolitan Geriatric Hospital brain bank; 11 AD and 3 NC subjects were from the University of Tsukuba; and 1 AD and 12 NC subjects were from the University of Tokyo. Detailed clinical demographics of the postmortem brains, including sex, age, and APOE ε4 and TREM2 variants, are shown in Table 1 and SI Appendix, Table S1. For the preparation of neuronal nuclei, we excised the gray matter from the inferior temporal gyrus and the cerebellum. Nuclear preparation and FACS nucleus sorting (FACSAria; BD Biosciences) were performed at the Support Unit for Bio-Material Analysis in RIKEN Brain Science Institute, according to a published protocol (16). Following the extraction of genomic DNA by the phenol–chloroform method, its concentration was measured using a Qubit dsDNA BR assay kit (Invitrogen).
Infinium 450k Methylation Microarray Analysis.
Genomic DNA (500 ng) was bisulfite-converted using an EZ DNA methylation kit (Zymo Research) and analyzed using an Illumina Infinium HumanMethylation450 BeadChip (Illumina) and iScan scanner (Illumina). We processed the methylation array data using the statistical computing package R 3.0.2 (www.r-project.org/) and the Bioconductor package ChAMP (69). Idat files were imported to calculate methylation levels (β values) for each probe. The β-mixture quantile (BMIQ) normalization method (70) was used to reduce probe-type bias, and batch correction was performed using the ComBat method (71). For each probe, we compared β values between AD and NC samples using two one-sided Student’s t tests: P (AD < NC) and P (AD > NC). We defined probes with P values < 0.05 and mean β differences between AD and NC > 0.05 as DMPs. Regions with no less than three consecutive DMPs were defined as DMRs.
Single-Cell Gel Electrophoresis (Comet Assay).
The comet assay was performed under neutral pH conditions, using a CometAssay kit (Trevigen), following the manufacturer’s instructions. DNA was visualized with SYBR Gold, and images were obtained using an Axioplan 2 fluorescent microscope (Carl Zeiss) and an Axiocam HRc CCD camera system (Carl Zeiss). The head and tail of each individual comet were detected semiautomatically to calculate Tail DNA% using CASPLab software (72).
Additional material and methods are included in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful for the technical support provided by Yuki Inukai and Fuyuko Suto. We are also grateful for editing by John C. Christianson, for scientific advice provided by Toshihiro Hayashi, for 3×Tg mice provided by Jun-ichi Kira, and for N2a swe.10 cells provided by Sam Sisodia. This study was supported by Japan Agency for Medical Research and Development Strategic Research Program for Brain Sciences (15656513), Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology (4216), Japan Society of Promotion of Science KAKENHI (16H05319 and 17H16113), the Cell Science Research Foundation, the Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care, the Takeda Science Foundation, Janssen Pharmaceutical, and Eisai Company.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All of the new data and analysis scripts used in this study have been deposited in the Github repository (https://github.com/TatsuoMano/AD-neuronal-methylome).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707151114/-/DCSupplemental.
References
- 1.Selkoe DJ. Resolving controversies on the path to Alzheimer’s therapeutics. Nat Med. 2011;17:1060–1065. doi: 10.1038/nm.2460. [DOI] [PubMed] [Google Scholar]
- 2.Strittmatter WJ, et al. Apolipoprotein E: High-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert JC, et al. European Alzheimer’s Disease Initiative (EADI) Genetic and Environmental Risk in Alzheimer’s Disease Alzheimer’s Disease Genetic Consortium Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerreiro R, Hardy J. Genetics of Alzheimer’s disease. Neurotherapeutics. 2014;11:732–737. doi: 10.1007/s13311-014-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jun G, et al. IGAP Consortium A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–117. doi: 10.1038/mp.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler JM, Fox HS. Transcriptome meta-analysis reveals a central role for sex steroids in the degeneration of hippocampal neurons in Alzheimer’s disease. BMC Syst Biol. 2013;7:51. doi: 10.1186/1752-0509-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan MG, et al. Genome wide profiling of altered gene expression in the neocortex of Alzheimer’s disease. J Neurosci Res. 2010;88:1157–1169. doi: 10.1002/jnr.22290. [DOI] [PubMed] [Google Scholar]
- 8.Podtelezhnikov AA, et al. Molecular insights into the pathogenesis of Alzheimer’s disease and its relationship to normal aging. PLoS One. 2011;6:e29610. doi: 10.1371/journal.pone.0029610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Twine NA, Janitz K, Wilkins MR, Janitz M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS One. 2011;6:e16266. doi: 10.1371/journal.pone.0016266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekar S, et al. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging. 2015;36:583–591. doi: 10.1016/j.neurobiolaging.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson JE, et al. MRC Cognitive Function and Ageing Neuropathology Study Group Microarray analysis of the astrocyte transcriptome in the aging brain: Relationship to Alzheimer’s pathology and APOE genotype. Neurobiol Aging. 2011;32:1795–1807. doi: 10.1016/j.neurobiolaging.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto L, et al. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson’s disease. PLoS One. 2010;5:e15522. doi: 10.1371/journal.pone.0015522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrachina M, Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J Neuropathol Exp Neurol. 2009;68:880–891. doi: 10.1097/NEN.0b013e3181af2e46. [DOI] [PubMed] [Google Scholar]
- 16.Iwamoto K, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bibikova M, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Dedeurwaerder S, et al. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 19.Oddo S, et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: Intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 20.Jankowsky JL, et al. Co-expression of multiple transgenes in mouse CNS: A comparison of strategies. Biomol Eng. 2001;17:157–165. doi: 10.1016/s1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- 21.Holcomb L, et al. Accelerated Alzheimer-type phenotype in transgenic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat Med. 1998;4:97–100. doi: 10.1038/nm0198-097. [DOI] [PubMed] [Google Scholar]
- 22.Nonaka T, Watanabe ST, Iwatsubo T, Hasegawa M. Seeded aggregation and toxicity of alpha-synuclein and tau: Cellular models of neurodegenerative diseases. J Biol Chem. 2010;285:34885–34898. doi: 10.1074/jbc.M110.148460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh KJ, et al. Staging of Alzheimer’s pathology in triple transgenic mice: A light and electron microscopic analysis. Int J Alzheimers Dis. 2010;2010:1–24. doi: 10.4061/2010/780102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwata A, et al. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum Mol Genet. 2014;23:648–656. doi: 10.1093/hmg/ddt451. [DOI] [PubMed] [Google Scholar]
- 25.De Jager PL, et al. Alzheimer’s disease: Early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci. 2014;17:1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lunnon K, et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat Neurosci. 2014;17:1164–1170. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vojta A, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verghese PB, et al. ApoE influences amyloid-β (Aβ) clearance despite minimal apoE/Aβ association in physiological conditions. Proc Natl Acad Sci USA. 2013;110:E1807–E1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerreiro R, et al. Alzheimer Genetic Analysis Group TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winklhofer KF, Tatzelt J, Haass C. The two faces of protein misfolding: Gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27:336–349. doi: 10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suberbielle E, et al. DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nat Commun. 2015;6:8897. doi: 10.1038/ncomms9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorrini C, et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med. 2013;210:1529–1544. doi: 10.1084/jem.20121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, et al. A cell cycle-dependent BRCA1-UHRF1 cascade regulates DNA double-strand break repair pathway choice. Nat Commun. 2016;7:10201. doi: 10.1038/ncomms10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans TA, et al. BRCA1 may modulate neuronal cell cycle re-entry in Alzheimer disease. Int J Med Sci. 2007;4:140–145. doi: 10.7150/ijms.4.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ksiezak-Reding H, Morgan K, Dickson DW. Tau immunoreactivity and SDS solubility of two populations of paired helical filaments that differ in morphology. Brain Res. 1994;649:185–196. doi: 10.1016/0006-8993(94)91063-4. [DOI] [PubMed] [Google Scholar]
- 37.Ksiezak-Reding H, Wall JS. Mass and physical dimensions of two distinct populations of paired helical filaments. Neurobiol Aging. 1994;15:11–19. doi: 10.1016/0197-4580(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 38.Hunter JM, et al. Biochemical and morphological characterization of the AbetaPP/PS/tau triple transgenic mouse model and its relevance to sporadic Alzheimer’s disease. J Alzheimers Dis. 2011;27:361–376. doi: 10.3233/JAD-2011-110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suberbielle E, et al. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madabhushi R, et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bucholtz N, Demuth I. DNA-repair in mild cognitive impairment and Alzheimer’s disease. DNA Repair (Amst) 2013;12:811–816. doi: 10.1016/j.dnarep.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 45.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 46.Tornovsky-Babeay S, et al. Type 2 diabetes and congenital hyperinsulinism cause DNA double-strand breaks and p53 activity in β cells. Cell Metab. 2014;19:109–121. doi: 10.1016/j.cmet.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Mercer JR, et al. DNA damage links mitochondrial dysfunction to atherosclerosis and the metabolic syndrome. Circ Res. 2010;107:1021–1031. doi: 10.1161/CIRCRESAHA.110.218966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garinis GA, van der Horst GT, Vijg J, Hoeijmakers JH. DNA damage and ageing: New-age ideas for an age-old problem. Nat Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Driver JA. Inverse association between cancer and neurodegenerative disease: Review of the epidemiologic and biological evidence. Biogerontology. 2014;15:547–557. doi: 10.1007/s10522-014-9523-2. [DOI] [PubMed] [Google Scholar]
- 50.Roe CM, Behrens MI, Xiong C, Miller JP, Morris JC. Alzheimer disease and cancer. Neurology. 2005;64:895–898. doi: 10.1212/01.WNL.0000152889.94785.51. [DOI] [PubMed] [Google Scholar]
- 51.Roe CM, et al. Cancer linked to Alzheimer disease but not vascular dementia. Neurology. 2010;74:106–112. doi: 10.1212/WNL.0b013e3181c91873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Musicco M, et al. Inverse occurrence of cancer and Alzheimer disease: A population-based incidence study. Neurology. 2013;81:322–328. doi: 10.1212/WNL.0b013e31829c5ec1. [DOI] [PubMed] [Google Scholar]
- 53.Sheng M, Sabatini BL, Südhof TC. Synapses and Alzheimer’s disease. Cold Spring Harb Perspect Biol. 2012;4:a005777. doi: 10.1101/cshperspect.a005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shankar GM, Walsh DM. Alzheimer’s disease: Synaptic dysfunction and Abeta. Mol Neurodegener. 2009;4:48. doi: 10.1186/1750-1326-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonner WM, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: To be or not to be. FEBS Lett. 2010;584:3717–3724. doi: 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Welcsh PL, Owens KN, King MC. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 2000;16:69–74. doi: 10.1016/s0168-9525(99)01930-7. [DOI] [PubMed] [Google Scholar]
- 59.Jiang J, et al. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer Res. 2011;71:5546–5557. doi: 10.1158/0008-5472.CAN-10-3423. [DOI] [PubMed] [Google Scholar]
- 60.Wu J, Lu LY, Yu X. The role of BRCA1 in DNA damage response. Protein Cell. 2010;1:117–123. doi: 10.1007/s13238-010-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pulvers JN, Huttner WB. Brca1 is required for embryonic development of the mouse cerebral cortex to normal size by preventing apoptosis of early neural progenitors. Development. 2009;136:1859–1868. doi: 10.1242/dev.033498. [DOI] [PubMed] [Google Scholar]
- 62.Pao GM, et al. Role of BRCA1 in brain development. Proc Natl Acad Sci USA. 2014;111:E1240–E1248. doi: 10.1073/pnas.1400783111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lodato MA, et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science. 2015;350:94–98. doi: 10.1126/science.aab1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 65.Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. doi: 10.1007/s00401-006-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKeith IG, et al. Consortium on DLB Diagnosis and management of dementia with Lewy bodies: Third report of the DLB Consortium. Neurology. 2005;65:1863–1872. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- 67.McKeith IG, et al. Consensus guidelines for the clinical and pathologic diagnosis of dementia with Lewy bodies (DLB): Report of the consortium on DLB international workshop. Neurology. 1996;47:1113–1124. doi: 10.1212/wnl.47.5.1113. [DOI] [PubMed] [Google Scholar]
- 68.Murayama S, Saito Y. Neuropathological diagnostic criteria for Alzheimer’s disease. Neuropathology. 2004;24:254–260. doi: 10.1111/j.1440-1789.2004.00571.x. [DOI] [PubMed] [Google Scholar]
- 69.Morris TJ, et al. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2014;30:428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 72.Końca K, et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutat Res. 2003;534:15–20. doi: 10.1016/s1383-5718(02)00251-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.