Significance
Tetraploid complementation, through which an entire organism is produced from pluripotent donor cells, is taken as the most stringent test for pluripotency. However, it remains unclear whether embryonic stem cells (ESCs) of other species besides mice can pass this test. Our results demonstrated the capacity of rat ESCs to produce live rats via tetraploid complementation and how the capacity is lost during in vitro culture. This report demonstrates that ESCs of other species besides mice can pass the tetraploid complementation test for pluripotency. We believe this original work will facilitate the understanding of evolution and regulation of pluripotency across mammalian species.
Keywords: pluripotency, embryonic stem cells, tetraploid complementation, rat, imprinted gene
Abstract
Pluripotency of embryonic stem cells (ESCs) can be functionally assessed according to the developmental potency. Tetraploid complementation, through which an entire organism is produced from the pluripotent donor cells, is taken as the most stringent test for pluripotency. It remains unclear whether ESCs of other species besides mice can pass this test. Here we show that the rat ESCs derived under 2i (two small molecule inhibitors) conditions at very early passages are able to produce fertile offspring by tetraploid complementation. However, they lose this capacity rapidly during culture due to a nearly complete loss of genomic imprinting. Our findings support that the naïve ground state pluripotency can be captured in rat ESCs but also point to the species-specific differences in its regulation and maintenance, which have implications for the derivation and application of naïve pluripotent stem cells in other species including human.
Pluripotency, the capacity to generate any type of cells of the body, can be captured from early embryonic tissues in the form of different types of pluripotent stem cells depending on the developmental stage of origin and culture conditions, such as the embryonic stem cells (ESCs) and epiblast stem cells (EpiSCs) derived from pre- and postimplantation epiblasts, respectively (1, 2). Pluripotent stem cells have broad applications in developmental studies, genome engineering, and regenerative medicine. Intriguingly, recent studies in mice have shown that pluripotency can be classified into two distinct states, termed the naïve (ground) state pluripotency and primed pluripotency (3). The mouse preimplantation epiblast cells and ESCs represent the naïve pluripotent and developmental ground state without any lineage commitment and self-renewal occurs autonomously under culture conditions shielding from the differentiation signals by two small molecule inhibitors (2i) (4). The mouse postimplantation epiblast cells and EpiSCs are developmentally more advanced and primed for lineage commitment and represent the primed pluripotent state (5, 6).
The pluripotent state also can be defined according to the developmental potency as measured by functional assessment assays, such as teratoma formation, chimera formation, germ-line transmission, and tetraploid complementation (7). The naïve pluripotent ESCs, such as mouse ESCs, can colonize the blastocyst to form chimera and transmit them into the germ line. However, the primed mouse EpiSCs can only colonize the postimplantation epiblast, not the blastocyst (5, 6). The authentic rat ESCs are derived in ground-state culture conditions with 2i and germ-line competence and thus are considered to be naïve pluripotent (8, 9). And the conventional human ESCs, although being derived from preimplantation blastocysts, have been shown to colonize the post- but not preimplantation epiblasts and thus are supposed to represent the primed pluripotent state (10–12).
Tetraploid complementation is an experimental approach of using the tetraploid host blastocyst to support the embryonic development of donor pluripotent cells. It is taken as the most stringent test for developmental potency, as by this approach the donor cells can mimic the early epiblasts to produce an entire organism (7). Previous studies showed that the mouse naïve pluripotent stem cells could pass this test, however the rat ESCs failed to do so (13–16). These results raise questions about whether naïve pluripotent stem cells from different species have the same developmental potency and, if not, how the naïve pluripotency is differentially regulated between species. Here we report the capacity of rat ESCs to pass the tetraploid complementation test and its rapid loss during cultivation due to loss of genomic imprinting.
Results
Derivation and Characterization of Rat ESCs.
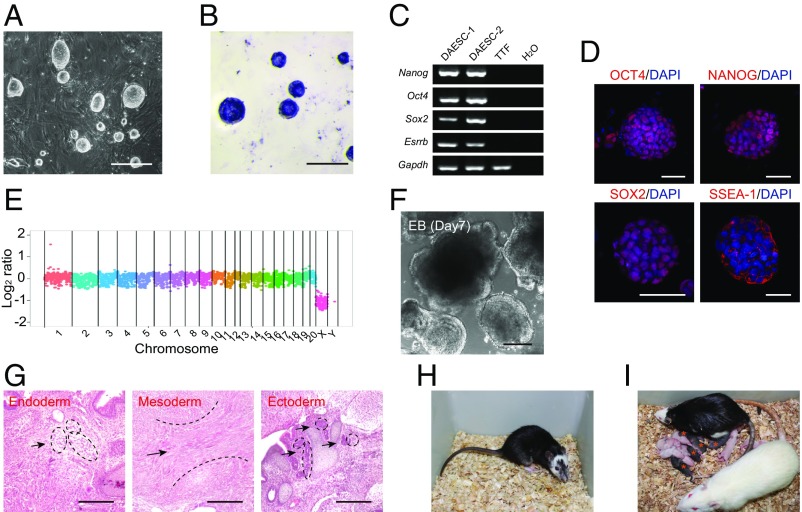
To derive rat ESC lines, we adopted the titrated 2i (1 μM MEK inhibitor and 1 μM GSK3 inhibitor) culture condition to cultivate the inner cell mass (ICM) isolated from embryonic day (E) 4.5 rat blastocysts from two genetic backgrounds [Dark Agouti (DA); Brown Norway × Sprague–Dawley, BN × SD] (17). All established ESC lines had typically dome-shaped colony morphology, exhibited positive staining for alkaline phosphatase, and expressed pluripotency markers Oct4, Nanog, Sox2, Esrrb, and SSEA-1 (Fig. 1 A–D). Whole genome sequencing revealed no gross deletions, insertions, or duplications in the repeatedly passaged rat ESC lines, indicating the maintenance of normal karyotype and intact genome in rat ESCs (Fig. 1E and Fig. S1).
Fig. 1.
Generation of germ line-competent rat ESCs. (A) The compact colony morphology of rat ESCs at passage 16. (Scale bar, 100 μm.) (B) Alkaline phosphatase staining of rat ESCs at passage 16. (Scale bar, 100 μm.) (C) RT-PCR analysis of pluripotent markers expressed in two rat ESC lines (DAESC-1 and DAESC-2). Rat tail-tip fibroblast cells and H2O served as negative controls. (D) Immunostaining of OCT4, NANOG, SOX2, and SSEA-1 in rat ESCs. (Scale bar, 100 μm.) (E) Whole-genome sequencing analysis showed the genome integrity of DAESC-1 at passage 13. Relative copy number is plotted using log2 scale. Each point represents the average coverage of 500 kb. (F) Morphology of EBs formed from rat ESCs. (Scale bar, 100 μm.) (G) Teratoma formation of rat ESCs. Shown are histological sections of teratoma including adipose tissue for endoderm (black arrows indicated, Left), muscle for mesoderm (black arrow indicated, Middle), and neural epithelium rosettes for ectoderm (black arrows indicated, Right). (Scale bar, 500 μm.) (H) Coat chimera generated from rat ESCs by blastocyst injection. Black indicates the contribution of donor rat ESCs, and white indicates the host diploid F344 embryos. (I) Germ-line offsprings produced by the chimeric female rat mated with a wild-type SD male rat. Five germ-line offsprings were obtained (black pups as indicated by the red stars) among 13 F1 pups.
We further examined the in vitro and in vivo differentiation potentials of the rat ESCs. After random differentiation for 7 d, the rat ESCs could form embryoid bodies (EBs) (Fig. 1F). They could also form teratomas containing all three germ layers after being s.c. injected into severe combined immunodeficient (SCID) mice (Fig. 1G). The essential feature of authentic ESCs is their capacity to colonize host embryos and contribute to the germ line of chimeric animals. Hence, we injected rat ESCs from three independent lines of DA background and one line of BN × SD background (passages ≥ 10) into a total of 329 E4.5 F344 blastocysts for chimera production. All of the four lines could produce chimeric rats, and 31 out of 64 live born pups exhibited coat color chimerism (Fig. 1H and Table S1). The chimeric rats from all three DA lines, but not the BN × SD line, produced DA pups after mating with SD rats, indicating the germ-line transmission of injected rat ESCs (Fig. 1I and Table S1). These results collectively demonstrated that the rat ESCs we derived fulfilled the criterion for naïve pluripotent ESCs.
Assessing Developmental Potency of Rat ICM Cells and ESCs by Tetraploid Complementation.
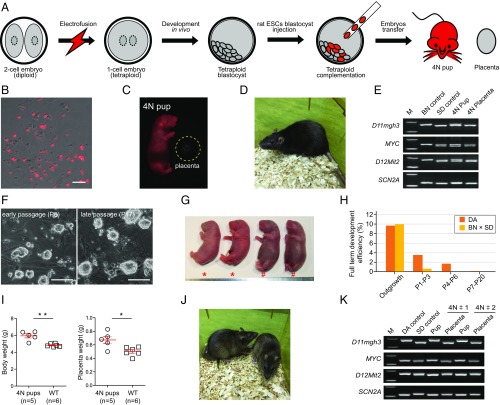
Next, we examined whether the rat ESCs could produce live offspring by tetraploid complementation, the most stringent test for pluripotency (Fig. 2A). Since experimental issues such as embryo manipulation may affect embryonic development, we first used the ICM cells as a proof-of-principle validation. Rat tetraploid host embryos were prepared by electrofusion of blastomeres of the two-cell embryos (SD strain, white coat color) into a one-cell embryo followed by transplantation into a pseudopregnant rat for in vivo development (Fig. S2 A–C). After 3-d in vivo development, ∼70% of the transplanted embryos developed into good-morphology blastocysts and were harvested for tetraploid complementation (Fig. S2D). ICM cells isolated from E4.5 blastocysts (BN × SD background) carrying a red fluorescent protein (RFP) reporter gene were trypsinized into single cells as donors for tetraploid blastocyst injection (Fig. 2B). From a total of 42 embryos injected, five live “all-ESC” rats were produced, and four grew into fertile adults (Fig. 2 C and D and Table S2). The presence of RFP expression in embryos but not the placenta as well as simple sequence length polymorphism (SSLP) genetic analysis confirmed that these rats were generated from injected ICM cells, while the placentas were generated from tetraploid host embryos (Fig. 2E). These results demonstrated that our experimental platform for tetraploid complementation assay for rats was reliable.
Fig. 2.
In vivo developmental ability of rat ESCs via tetraploid blastocyst complementation assay. (A) Schematic overview of tetraploid blastocyst complementation assay showing the steps of derivation of 4N pups with rat ESCs. (B) Individual RFP-positive ICM cells acquired from RFP-positive blastocysts by trypsinization with 0.05% trypsin. (Scale bar, 100 μm.) (C) Living pup was produced by RFP-positive rat ICM cells by tetraploid blastocyst complementation, and RFP was detected in the pup and absent in the placenta (the area marked by yellow dashed line). (D) Adult rat generated via tetraploid blastocyst complementation with rat ICM cells. (E) SSLP analysis of the pup and placenta. The wild-type SD and BN rat genome served as the control. (F) Morphology of rat ESC line DAESC-3 at early passage (P5) and late passage (P17). (Scale bar, 100 μm.) (G) Surviving and dead rat pups derived from rat ESCs at early passage via tetraploid complementation. *, surviving full-term pups; #, pups that died hours after being born. (H) The ratio of full-term pups that were produced with rat ESCs at different passages (from outgrowth to P20) via tetraploid blastocyst complementation. (I) The weight of the body and placenta of full-term developed 4N pups. *P ≤ 0.01, **P ≤ 0.001. (J) Two adult rats from rat ESCs at early passages via tetraploid complementation. (K) SSLP analysis of the body and placenta of 4N pups, with the pups derived from the DA strain background and the placentas derived from the SD strain background. The wild-type DA and SD rat served as controls.
We next injected tetraploid SD blastocysts with rat ESCs from two independent cell lines derived from DA backgrounds (DAESC-1 and DAESC-2) at more than 10 passages (Table S1). Of 255 tetraploid embryos injected, none developed beyond E11.5 due to severe growth retardation (Fig. S2E and Table S1). Since ICM cells could produce all-ESC rats, we reasoned that the derivation or cultivation of rat ESCs compromised the capacity of rat ESCs to produce all-ESC rats. Hence, we established another five DA background (DAESC-4, DAESC-5, DAESC-6, DAESC-7, and DAESC-8) and two BN × SD background (BSESC-2 and BSESC-3) cell lines and tested their developmental potency from early passages by the tetraploid complementation assay (Table 1). During the passage of rat ESCs, these cells maintained the typical doomed colony morphology as well as chimera formation capacity (Fig. 2F). Of 102 tetraploid embryos injected with cells isolated from the outgrowth of rat blastocysts, 10 full-term pups were produced, including five males and five females, and five pups grew into adulthood, indicating that the cells at the initial derivation stage maintained the capacity to generate all-ESC rats (Table 1). Three male rat ESC lines of DA background produced 18 full-term all-ESC pups from a total of 813 injected tetraploid embryos before passage 7 (∼2.21%) (Fig. 2G and Table 1). However, cells after passage 10 failed to produce any full-term pup from 687 injected tetraploid embryos (Table 1). For the BN × SD background male rat ESCs, only two full-term pups were produced from 317 injected tetraploid embryos before passage 3 (∼0.63%), and no pups were produced from 471 tetraploid embryos injected with cells after passage 3. Hence, the developmental potency of rat ESCs to produce 4N pups was rapidly lost during in vitro culture (Fig. 2H). Notably, both early and late passage female rat ESCs failed to generate any full-term pups from 609 injected tetraploid embryos (Table 1). The full-term all-ESC pups had significantly heavier bodies and placentas than the wild-type ones, consistent with the “large offspring syndrome” observed in tetraploid complementation-produced all-ESC mice (Fig. 2I). Six out of the 20 full-term pups survived into adulthood, and the rest died within 2 h after birth due to respiratory failure (Fig. 2 G and J and Table 1). The SSLP assay confirmed that the all-ESC rats were derived from the same background as the donor rat ESCs rather than the host tetraploid embryos (Fig. 2K).
Table 1.
Developmental efficiencies of rat ESCs at different passages for tetraploid complementation
| Background | Rat ESC lines | Sex | Passage | No. injected | No. recipient | No. development in vivo (%) | |
| Full-term | Live adult | ||||||
| DA | Outgrowth | Male/female | — | 72 | 5 | 4F/3M (9.7) | 1F/2M (4.2) |
| DAESC-4 | Male | P2 | 44 | 1 | 1 (2.3) | 1 (2.3) | |
| P3 | 75 | 3 | 4 (5.3) | 1 (1.3) | |||
| P4 | 152 | 5 | 2 (1.3) | 1 (0.7) | |||
| P6 | 47 | 1 | 0 | 0 | |||
| P7 | 37 | 1 | 1 (2.7) | 0 | |||
| P10 | 121 | 3 | 0 | 0 | |||
| P13 | 40 | 1 | 0 | 0 | |||
| P17 | 40 | 1 | 0 | 0 | |||
| DAESC-5 | Male | P3 | 110 | 4 | 3 (2.7) | 1 (0.9) | |
| P4 | 58 | 2 | 0 | 0 | |||
| P6 | 120 | 5 | 3 (2.5) | 1 (0.8) | |||
| P7 | 40 | 1 | 0 | 0 | |||
| P11 | 65 | 2 | 0 | 0 | |||
| P15 | 105 | 4 | 0 | 0 | |||
| P20 | 85 | 2 | 0 | 0 | |||
| DAESC-6 | Male | P4 | 130 | 4 | 4 (3.1) | 1 (0.8) | |
| P11–P13 | 231 | 4 | 0 | 0 | |||
| DAESC-7 | Female | P2 | 45 | 2 | 0 | 0 | |
| P3 | 62 | 4 | 0 | 0 | |||
| P10 | 123 | 6 | 0 | 0 | |||
| DAESC-8 | Female | P10–P12 | 87 | 5 | 0 | 0 | |
| BN × SD | Outgrowth | Male/female | — | 30 | 1 | 1F/2M (10.0) | 2M (6.7) |
| BSESC-2 | Male | P1 | 105 | 3 | 1 (0.9) | 0 | |
| P2 | 109 | 3 | 0 | 0 | |||
| P3 | 103 | 3 | 1 (1.0) | 0 | |||
| P5 | 92 | 2 | 0 | 0 | |||
| P6–P10 | 379 | 12 | 0 | 0 | |||
| BSESC-3 | Female | P2–P12 | 292 | 14 | 0 | 0 | |
BN × SD, Brown Norway strain mated with Sprague–Dawley strain; DA, Dark Agouti strain; F, female; M, male.
Global DNA Methylation and Genomic Imprinting Analysis of Rat ESCs During Cultivation.
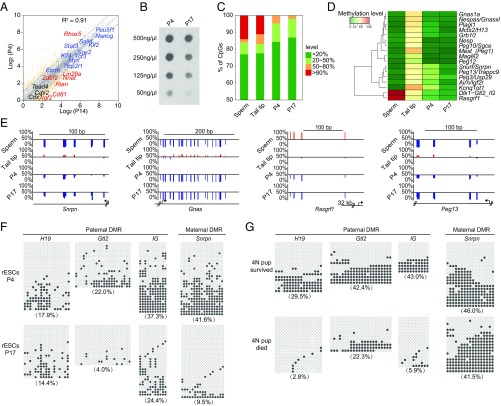
To decipher the underlying mechanisms that affect the developmental potency of rat ESCs during cultivation, we compared the transcriptomes of rat ESCs at early (passage 4) and late (passage 14) passages. The whole-genome expression patterns were highly similar (Pearson correlation, R2 = 0.91), including the similar expression levels of pluripotency genes (Fig. 3A). The differentially expressed genes were enriched in the cell differentiation- and developmental-related processes (Fig. S3A). Notably, several imprinted genes were differentially expressed in the rat ESCs between the early and late passages (Fig. S3B). Meanwhile, we also analyzed the genome methylation pattern of rat ESCs at early (passage 4) and late (passage 17) passages. The genome methylation level was largely reduced in ESCs at the late passage according to the immuno-dot blotting assay (Fig. 3B). The genome-wide methylation pattern at single-base resolution was detected by reduced representation bisulfite sequencing (RRBS). The RRBS revealed that the full methylated sites (methylation level > 80%) were almost lost in rat ESCs at both early and late passages. The moderate methylation sites (methylation level at 50% ∼ 80%) were further demethylated during culture in late-passage rat ESCs (Fig. 3C). The same trends were also observed in the imprinting control regions (ICRs), also known as differentially methylated regions (DMRs), of which both the paternal and maternal ICRs were hypomethylated compared with the somatic tissue, suggesting the loss of genomic imprinting (Fig. 3D). A single-base resolution examination for several ICRs, including Snrpn-, Gnas-, Rasgrf1-, and Peg13-ICR in late-passage rat ESCs, also exhibited a hypomethylation status (Fig. 3E). To further confirm the imprinting status, we performed bisulfite sequencing of four ICRs: the H19-, Gtl2-, IG-, and Snrpn-ICR. All imprinting regions showed severe demethylation in late-passage ESCs, indicating the loss of genomic imprinting in these regions (Fig. 3F). To explore whether the loss of ability to produce all-ESC rats was equal to all of the ESCs upon in vitro culture, we established 12 subclones from rat ESC line DAESC-6 at passage 15, and all of the subclones showed nearly complete loss of genomic imprinting in the Snrpn and H19 ICRs (Fig. S3C). Three subclones were used for the tetraploid complementation assay by injection into a total of 232 tetraploid embryos, and all of them failed to generate any full-term all-ESC pups, suggesting that the loss of ability to produce all-ESC rats equally affected all of the rat ESCs in culture.
Fig. 3.
Global DNA methylation and genomic imprinting analysis of rat ESCs during cultivation. (A) Scatter plot comparison of transcriptomes of rat ESCs at passages 4 and 14. The raw FPKM for each gene was transformed to a log2 value, and genes with more than one FPKM were shown. The yellow points represent the differentially expressed genes. The pluripotent genes, extraembryonic lineage genes, and imprinted genes were labeled with blue, black, and red text, respectively. Dashed lines depict twofold changes. The R2 was determined by Pearson’s correlation. (B) Dot blot of genomic DNA of rat ESCs at passages 4 and 17 with 5mC-specific antibody, showing reduced global 5mC levels at the later passage of rat ESCs. (C) Distribution of genome-wide CpG methylation levels in different rat cell types or different passages. (D) Heat map of the methylation level of ICRs in different rat cell types or different passages. (E) Representative CpG methylation profiles of imprinted genes (Snrpn, Gnas, Rasgrf1, Peg13) in ICR regions at different passages of rat ESCs. The red line shows the CpG site with the methylation level more than 50%, while the blue line represents the site with the methylation level less than 50%. (F) Bisulfite sequencing analysis for rat ESCs at different passages in paternal and maternal ICRs of the H19-, Gtl2-, IG-, and Snrpn-ICR. (G) Bisulfite sequencing analysis of tissues of surviving and dead 4N pups generated with rat ESCs via tetraploid complementation in paternal and maternal ICRs, respectively.
Finally, we analyzed the imprinting status in the all-ESC pups that died at birth. In contrast to the normal methylation level (∼50%) in survived all-ESC pups, the methylation levels of ICRs in dead pups were largely reduced (Fig. 3G). Collectively, these results revealed that the rat ESCs were disposed to genome-wide demethylation during culture, including demethylation of the ICRs that caused the thorough loss of genomic imprinting.
Discussion
In summary, we reported here that the rat ESCs at very early passages (usually before passage 5) were capable of producing fertile offspring by tetraploid complementation, suggesting that this most stringent test for developmental potency can also be used for assessing the pluripotency in other species besides mice. It was also demonstrated that the pluripotency of rat early epiblasts could be captured in vitro in rat ESCs using the 2i culture condition, the so-called ground-state culture condition, which supported the proposition of the naïve ground-state pluripotency (1, 3). This finding points out that the regulations of naïve pluripotency are generally conserved between mouse and rat.
Previous studies found that the naïve pluripotency of mouse ESCs was associated with global DNA hypomethylation induced by the 2i ground-state culture condition, though the methylation of imprinting regions in male ESCs was protected from erasure (18–20). During revision, two papers were published that showed that mouse ESCs upon prolonged culture in the 2i/LIF condition were also disposed to global demethylation and loss of imprinting and finally lost the ability to produce all-ESC mice (21, 22). Hence, both mouse and rat ESCs exhibited impaired developmental potential after prolonged culture in the 2i/LIF condition. However, the progression of the impairment of developmental capacity happened faster in rats than in mice, as the mouse studies showed that mouse ESC lines at passage 20 could still generate all-ESC mice (22). This difference between mouse and rat ESCs was consistent with another difference found in previous studies, showing that the canonical Wnt signaling was intrinsically higher in rat ESCs; thus, the 2i concentrations optimal for mouse ESC maintenance caused sporadic differentiation of rat ESCs during culture (23, 24).
It will be interesting to elucidate these species-specific differences in pluripotency maintenance in the future, which will shed light on the derivation and application of naïve pluripotent stem cells in other species, including humans (25). Our findings also suggest that the epigenetic state, especially the genomic imprinting status, needs to be considered or analyzed before the application of naïve pluripotent stem cells for developmental studies and regenerative medicine.
Experimental Procedures
Rat.
DA and BN strain rats were used to establish ESCs, strain rats were used to produce pregnant recipients, Fisher 344 (F344) was used to obtain diploid blastocysts, and CF-1 strain mice were used to produce feeder cells to the ESC culture. All experiments involving animals were conducted according to the guidelines for the care and use of laboratory animals established by the Beijing Association for Laboratory Animal Science and were approved under the Animal Ethics Committee of Institute of Zoology, Chinese Academy of Sciences.
Rat ESC Derivation and Cell Culture.
Two types of rat blastocysts of different genetic background were used to derive rat ESCs, and blastocysts were obtained from the DA/DA rat strain and the BN/SD rat strain that carried the RFP transgene. The blastocysts were flushed from the uteri of 4.5 d post coitus (dpc) pregnant rats, and then 4∼5 blastocysts were planted in a 4-well dish coated with feeder layer cells and the culture medium composed of N2B27 medium (Gibco), 1 mM l-glutamine, 0.1 mM β-mercaptoethanol, 50 U/mL penicillin, 50 μg/mL streptomycin, 1 μM CHIR99021 (Stemgent), and 1 μM PD0325901 (Stemgent) and rLIF (ESGRO). The embryos were incubated at 37 °C in a 5% CO2 incubator for 4∼5 d, and then the formed outgrowth (named passage 0) was picked separately by a glass pipette and dissociated with rat digest solution (0.025% trypsin–100 μM EDTA + 1% chicken serum). The cells that replanted and survived the the first trypsinization of the outgrowth were counted as passage 1 (P1). For the routine passage, the passage ratio was about 1:4, and rat ESCs were passaged around every 3 d.
Tetraploid Blastocyst Aggregation.
SD strain female rats aged 6∼8 wk were superovulated by i.p. injections of 150 IU/kg pregnant mare’s serum gonadotropin (PMSG) and 75 IU/kg human CG (hCG) at an interval of 48 h. The female rats were mated with fertile SD males after the hCG injection, and two-cell embryos were recovered from the oviducts at 46 h post-hCG injection with M2 medium (Sigma). To produce tetraploid rat embryos, an electrofusion apparatus (Multiporator, Eppendorf) was used to electrofuse the blastomeres of two-cell stage embryos within fusion medium consisting of 0.3 M mannitol, 0.1 mM MgSO4, 3 mg/mL BSA (Sigma), and 0.1 mg/mL polyvinylalcohol (PVA; Sigma). After pre-equilibrium treatment with fusion medium in a microfusion chamber (gap width 0.2 mm), embryos were exposed to a fusion DC pulse (25 V, 30 μs, 2 cycles) followed by application of AC pulses (2.5 V, 15 s) in a microfusion chamber (Eppendorf) with a gap width of 0.2 mm, and then embryos were transferred to mR1ECM medium (310 mOsm) cultured in 5% CO2 at 37 °C. For most embryos (about 90%), the blastomeres were successfully fused after electrofusion treatment in a half hour. Further, these one-cell tetraploid embryos were transferred into the oviduct of pseudopregnant SD females at 0.5 dpc by standard methods. The tetraploid blastocyst embryos were flushed with mR1ECM medium in addition to HEPAS (270 mOsm) from the uteri of SD recipient females at 3.5 dpc.
The introduction of rat ESCs into tetraploid blastocysts was performed as described with some modifications. The harvested blastocysts were transferred into mR1ECM medium (270 mOsm) cultured in 5% CO2 at 37 °C for a half hour, while the blastocoele of blastocysts was apparent. Embryo injections were performed with a micromanipulation system equipped with an inverted microscope with differential interference contrast (DIC) optical components (DMI-3000B, Lecia), manual manipulator (MMO-202, Narishige), and piezo drive system (PMM-150, Prime Tech). The rat ESC colonies, including different passages, were briefly dissociated to single cells with 0.05% trypsin for 3∼4 min. About 15∼20 cells were injected into one tetraploid blastocyst, and after being cultured in mR1ECM medium for a half hour, the ESC aggregated tetraploid blastocysts were transferred into the uteri of 3.5 dpc pseudopregnant SD recipient rats.
To assess the developmental ability of rat ESCs, the pregnant rats were killed at different days after embryo transfer to obtain full-term offspring. The ICM cell and primary outgrowth (passage 0) tetraploid blastocyst aggregation was performed with a method similar to ESC aggregation. ICM cells were first generated from rat blastocysts by immunosurgery, and the outgrowth was picked up from the bottom of the culture disk by a glass tube. Then those cells were dissociated to single cells by trypsinization for tetraploid blasstocyst aggregation.
RNA-Seq Data Processing.
Sequencing was performed on an Illumina HiSeq 2500 sequencer with 125 bp paired-end sequencing reactions. For the bulk cell RNA-seq data, the clean reads were mapped to a rn6 genome assembly by HISAT (version 0.1.6-beta) (26). Due to the incomplete gene annotation for rats, the transcripts were assembled by Cufflinks (version 2.0.2) with the known annotation as a guide (27). Then the assembled transcripts without gene function annotation were aligned to the mouse Refseq transcripts with BLAST, and the transcripts with more than 200 nt and 30% identity sequences similar to the mouse transcripts were annotated as the mouse homolog transcripts. The merged transcripts from known and annotated homolog transcripts were used for the gene expression-level calculation.
Quantification and Statistical Analysis.
Statistical parameters including statistical analyses, statistical significance, and n values are reported in the figure legends. Statistical analyses were performed using Prism Software (GraphPad). For statistical comparison, one-way ANOVA was employed. A value of P < 0.05 was considered significant.
Data and Software Availability.
The accession numbers for the RRBS and RNA-seq expression values reported in this paper are GSE97968 and GSE97966, respectively.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China Grants 31422038, 31471395, and 31501188; National High Technology Research and Development Program Grants 2015AA020307 and 2014BAI02B01; and National Basic Research Program of China Grants 2014CB964900 and 2014CB964800.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The reduced representation bisulfite sequencing (RRBS) and RNA-sequencing expression values reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE97968 and GSE97966, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708710114/-/DCSupplemental.
References
- 1.Nichols J, Smith A. Pluripotency in the embryo and in culture. Cold Spring Harb Perspect Biol. 2012;4:a008128. doi: 10.1101/cshperspect.a008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 4.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 6.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 7.De Los Angeles A, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- 8.Li P, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buehr M, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 11.Tam PP. Human stem cells can differentiate in post-implantation mouse embryos. Cell Stem Cell. 2016;18:3–4. doi: 10.1016/j.stem.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Mascetti VL, Pedersen RA. Human-mouse chimerism validates human stem cell pluripotency. Cell Stem Cell. 2016;18:67–72. doi: 10.1016/j.stem.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirabayashi M, et al. Ability of tetraploid rat blastocysts to support fetal development after complementation with embryonic stem cells. Mol Reprod Dev. 2012;79:402–412. doi: 10.1002/mrd.22043. [DOI] [PubMed] [Google Scholar]
- 14.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy A, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 16.Zhao XY, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Blair K, Smith A. Robust self-renewal of rat embryonic stem cells requires fine-tuning of glycogen synthase kinase-3 inhibition. Stem Cell Rep. 2013;1:209–217. doi: 10.1016/j.stemcr.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leitch HG, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackett JA, et al. Synergistic mechanisms of DNA demethylation during transition to ground-state pluripotency. Stem Cell Rep. 2013;1:518–531. doi: 10.1016/j.stemcr.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ficz G, et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yagi M, et al. Derivation of ground-state female ES cells maintaining gamete-derived DNA methylation. Nature. 2017;548:224–227. doi: 10.1038/nature23286. [DOI] [PubMed] [Google Scholar]
- 22.Choi J, et al. Prolonged Mek1/2 suppression impairs the developmental potential of embryonic stem cells. Nature. 2017;548:219–223. doi: 10.1038/nature23274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, et al. Generation and application of mouse-rat allodiploid embryonic stem cells. Cell. 2016;164:279–292. doi: 10.1016/j.cell.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 24.Meek S, et al. Tuning of β-catenin activity is required to stabilize self-renewal of rat embryonic stem cells. Stem Cells. 2013;31:2104–2115. doi: 10.1002/stem.1466. [DOI] [PubMed] [Google Scholar]
- 25.Martello G, Smith A. The nature of embryonic stem cells. Annu Rev Cell Dev Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- 26.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, et al. Derivation of germline competent rat embryonic stem cells from DA rats. J Genet Genomics. 2012;39:603–606. doi: 10.1016/j.jgg.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.