Significance
Receptor tyrosine kinases (RTKs) and their stimulatory ligands regulate a variety of cellular processes. Many diseases, including cancer, are driven by mutations in or dysregulation of RTKs and their signaling pathways. The physiological ligands responsible for activating the RTKs, ALK and LTK, remained elusive. Recently, the ligands of each were identified and named “ALKAL1” and “ALKAL2” (“FAM150” or “augmentors”). Here, we demonstrate that zebrafish contain three distinct ligand molecules and describe their role in controlling pigment development in the embryo and adult zebrafish eye and body. These experiments show that these hormone-like molecules are critical factors influencing neural crest cell differentiation and progenitor/stem cell fates in zebrafish, suggesting a role for augmentors in control of similar processes in higher organisms.
Keywords: phosphorylation, cell signaling, cytokines, surface receptors, pigment development
Abstract
Anaplastic lymphoma kinase (Alk) and leucocyte tyrosine kinase (Ltk) were identified as “orphan” receptor tyrosine kinases (RTKs) with oncogenic potential. Recently ALKAL1 and ALKAL2 (also named “augmentor-β” and “augmentor-α” or “FAM150A” and “FAM150B,” respectively) were discovered as physiological ligands of Alk and Ltk. Here, we employ zebrafish as a model system to explore the physiological function and to characterize in vivo links between Alk and Ltk with their ligands. Unlike the two ligands encoded by mammalian genomes, the zebrafish genome contains three genes: aug-α1, aug-α2, and aug-β. Our experiments demonstrate that these ligands play an important role in zebrafish pigment development. Deficiency in aug-α1, aug-α2, and aug-β results in strong impairment in iridophore patterning of embryonic and adult zebrafish that is phenocopied in zebrafish deficient in Ltk. We show that aug-α1 and aug-α2 are essential for embryonic iridophore development and adult body coloration. In contrast, aug-α2 and aug-β are essential for iridophore formation in the adult eye. Importantly, these processes are entirely mediated by Ltk and not by Alk. These experiments establish a physiological link between augmentor ligands and Ltk and demonstrate that particular augmentors activate Ltk in a tissue-specific context to induce iridophore differentiation from neural crest-derived cells and pigment progenitor cells.
Receptor tyrosine kinases (RTK) represent a class of cell-surface receptors that convert an extracellular signal, in the form of a specific stimulatory ligand, into an intracellular response (1–3). Ligand binding to the extracellular domain induces tyrosine kinase activation, resulting in the stimulation of multiple intracellular signaling pathways that promote cell proliferation and differentiation among other processes necessary for normal cellular homeostasis (1–5).
As RTKs are key regulators of important cellular events, their dysregulation results in a variety of pathological conditions resulting in many diseases, including several cancers (4–6). One family of RTKs includes the homologous receptors designated “anaplastic lymphoma kinase” (ALK) and “leukocyte tyrosine kinase” (LTK) (1–3) that were initially discovered as RTKs with potential oncogenic properties (7). ALK has received significant attention during the past decade because of its important role as an oncogenic driver of several cancers. A subpopulation of non–small-cell lung carcinoma is driven by the EML4-ALK gene translocation, which encodes for an oncogenic protein composed of the cytoplasmic domain of ALK fused to a gene product, EML4, that is capable of dimerizing and stimulating ALK tyrosine kinase activity (8). Moreover, gain-of-function mutations of full-length ALK were identified in subpopulations of pediatric neuroblastoma (9, 10) and melanoma patients (11, 12).
The normal, physiological roles of LTK and ALK are not well understood. Mice deficient in Ltk or Alk do not display pronounced phenotypes. Alk-deficient mice display a decrease in newborn neurons (13) and defects in brain function (14). Ltk-knockout mice do not display a significant reduction in newborn neurons (13). However, Alk– and Ltk–double-knockout mice exhibit an 80% reduction in newborn neurons, suggesting a compensatory function of the two RTKs (13). It was also shown that in zebrafish (Danio rerio), morpholino-mediated knockdown and pharmacological inhibition of Alk result in reduced neuronal differentiation and survival in the central nervous system (15). Zebrafish deficient in ltk, first identified as “shady,” is a well-established model for the study of vertebrate pigmentation (16, 17). Shady larva and adult mutants lack iridophores, the blue-tinted cells that reflect light to give fish their metallic shine and which, together with yellow xanthophore and black melanophores, generate the zebrafish’s stripes (18–20).
Only recently did Alk and Ltk lose their “orphan” RTKs designation when their physiological activating ligands were identified (21–23). One ligand originally designated “FAM150A,” also named “augmentor-β” (AUG-β) and recently named “ALK and LTK-ligand 1” (ALKAL1), functions as a high-affinity ligand of LTK. A second ligand, designated “FAM150B”, also named “augmentor-α” (AUG-α) or “ALK and LTK-ligand 2” (ALKAL2), functions as an activating ligand of both ALK and LTK (22). AUG-α and AUG-β are basic proteins with a predicted mass of 14.5 and 11.4 kDa, respectively (22). They consist of a variable N-terminal region and a conserved C-terminal domain, termed the “augmentor domain,” with 65.9% sequence identity across the two molecules (22). It is noteworthy that genetic studies have shown that jelly belly (Jeb) and hesitation behavior-1 (Hen-1) function as ligands for Alk in Drosophila melanogaster and Caenorhabditis elegans, respectively (24–27). However, there is no evidence that Jeb or Hen-1 can directly bind to and activate Alk of Drosophila or nematode cells. Moreover, vertebrate homologs of Jeb and Hen-1 have not been identified, and, likewise, no fly or worm homologs of AUG-α and AUG-β are known. Additionally, we demonstrated that heparin can function as activating ligand for ALK in NB-1 cells (28), tumor cells overexpressing ALK.
Here, we use zebrafish pigment development to characterize the physiological function of the augmentors as ligands for Alk and Ltk. Our data show that zebrafish possess two aug-α gene homologs, aug-α1 and aug-α2, and a single aug-β gene. Iridophore patterning of embryonic and adult zebrafish deficient in all three augmentors phenocopy the ltk mutant but not the alk mutant. Genetic analysis experiments indicate that aug-α1 and aug-α2 are sufficient to allow the differentiation of neural crest-derived cells (NCCs) to embryonic iridophores. During adult pigmentation, aug-α1 and aug-α2 remain the favorable ltk ligands for iridophore development of the body. However, aug-α2 and aug-β are required for iridophore development in the adult eye. These results indicate that particular augmentors may allow the activation of Ltk in a tissue-specific context to induce iridophore differentiation from NCCs and adult pigment progenitor cells.
Results and Discussion
We use zebrafish as a model system to explore the physiological roles of Alk and Ltk ligands by analyzing the phenotypes of ligand-deficient fish. Unlike the human and other mammalian genomes that contain two augmentor genes, zebrafish contain three augmentor orthologs designated “aug-α1,” “aug-α2,” and “aug-β,” encoding for small, basic proteins with masses of 16.2, 22.5, and 17.6 kDa, respectively (Fig. S1A). We noticed that two highly conserved cysteine residues in the C terminus are missing from the sequence of aug-α1 deposited in the Zfin database (Zfin ID: ZDB-TSCRIPT-100915-712). We therefore cloned and determined the sequence of aug-α1 as well as the sequences of the two other augmentors. This analysis confirmed the sequences of aug-α2 and aug-β and corrected the flawed sequence of aug-α1 deposited in the Zfin database. Indeed, comparison of their primary structures showed that, like their human counterparts, zebrafish Alk and Ltk ligands are composed of two distinct regions: an N-terminal variable region and a C-terminal augmentor domain that contains four cysteines critical to maintaining augmentors’ structural integrity via disulfide bonds (Fig. 1A and Fig. S1A).
Fig. 1.
Generation of zebrafish ltk, alk, and augmentor loss-of-function mutants. (A) Schematic representation of augmentor ligands and sequence alignment of the augmentor domain (AugD) of human and zebrafish augmentors. “Zfin” indicates the sequence as given in the Zfin database, and “Exp” indicates the experimental and observed sequence of aug-α1. In all other zebrafish augmentors, the observed sequence matches the Zfin database. Color coding depicts the conservation of different protein domains in zebrafish and human: red, signal peptide (SP); orange, N-terminal variable region; brown, augmentor domain. Red arrowheads indicate CRISPR/Cas9 cut sites within the respective DNA-coding sequences. “AUG” indicates human orthologs, and “Aug” indicates zebrafish orthologs, as per convention. (B) Schematic representation of receptor and ligand protein domains in human and zebrafish. Color coding depicts the conservation of different protein domains in zebrafish and human: cyan, N-terminal region; green, MAM domain; yellow, LDLa repeat; black, EGF-like motif; purple, tyrosine kinase domain (KD). Proteins are not to scale. Red arrows indicate CRISPR/Cas9 cut sites within the respective DNA coding sequences. ALK and LTK indicate human orthologs; Alk and Ltk indicate zebrafish orthologs, as per convention (C) Lateral view (magnification: 27×) of whole-mount zebrafish embryos genotyped as indicated. (D) qPCR analysis of the indicated transcripts for the respective genotypes (n = 3–5; error bars indicate SEM).
Zebrafish orthologs of ALK and LTK differ from their human counterparts in the composition of their extracellular domains (ECDs) (8). In humans, the ALK ECD is composed of an N-terminal heparin-binding region (NTR) (28), one low-density lipoprotein receptor class A (LDLa) repeat flanked by two meprin/A5-protein/PTPμ (MAM) domains, an augmentor-binding glycine-rich region (GlyR), and an EGF-like motif (Fig. 1B) (22). Human LTK ECD is composed of only a GlyR and an EGF-like motif (8, 17). The presence of MAM domains is a distinctive feature between the two receptors in mammals. In contrast, the ECD of zebrafish Alk contains an LDLa repeat followed by a MAM domain, a GlyR, and an EGF-like motif (15). The ECD of zebrafish Ltk contains an LDLa domain flanked by two MAM domains followed by a GlyR and an EGF-like motif (Fig. 1B) (16, 17). The annotation of zebrafish Alk and Ltk was originally proposed based on a comparison of the sequences of the tyrosine kinase domains of human and zebrafish receptors. It is noteworthy that the zebrafish Ltk ECD sequence contains striking hallmarks of human ALK. The presence of MAM domains distinguishes ALK from LTK in mammals, but MAM domains are present in both RTKs in zebrafish. Unlike human LTK, the ECD of zebrafish Ltk contains an LDLa repeat flanked by two MAM domains with topology identical to the topology of corresponding domains in ECD of human ALK. Moreover, the primary structure of the GlyR of zebrafish Ltk is more similar to human ALK (64.0% identity) than to human LTK (59.7% identity) (Fig. S1B). Phylogenetic analysis suggests that vertebrate alk and ltk arose by a gene-duplication event in evolution (16). On the basis of these considerations, we propose that the zebrafish genome contains two orthologs of ALK-like RTKs, alk-1 and alk-2, rather than the so-called zebrafish “Ltk” and “Alk,” respectively.
Generation of Loss-of-Function Mutations of Alk, Ltk, and Their Three Ligands in Zebrafish.
We used the CRISPR/cas9 system (29, 30) to generate zebrafish mutants deficient in the aug-α1, aug-α2, and aug-β genes individually and combination and evaluated the phenotype of all mutants. Likewise, the CRISPR/cas9 system was used to generate zebrafish mutants deficient in alk or ltk and in both alk and ltk. Single-guide RNAs (sgRNA) were engineered to introduce insertion-deletion (indel) mutations in critical regions detrimental for ligand binding or receptor activity (Fig. 1 A and B, arrowheads, and Fig. S1C). To minimize any potential off-target affects, F0 fish were outcrossed to wild-type zebrafish (AB strain) to ultimately give rise to the F2 generation. Furthermore, potential off-target sites of each sgRNA were identified using a computational analysis as previously described (31). By using the endonuclease T7 (T7E1) assay, we observed that off-target genome sequences were found not to be mutated compared with the respective on-target loci (Fig. S2).
F1 aug-α1, aug-α2, and aug-β mutants carried a premature stop codon (Fig. 1A and Fig. S1C, arrowhead), which resulted in the incomplete translation of the variable region in addition to the augmentor domain, regions essential for receptor binding (22). Mutations in the primary sequence of Alk and Ltk are likely to prevent the expression of functional and active receptors (Fig. 1B and Fig. S1C, arrowhead).
F2 alk−/−;ltk−/− double mutants and aug-α1−/−;aug-α2−/−;aug-β−/− triple mutants displayed no gross morphological differences relative to wild-type embryos at 72 h postfertilization (hpf) (Fig. 1C). qPCR analysis conducted on alk−/− and ltk−/− single mutants and alk−/−;ltk−/− double mutants confirmed the loss of expression of each respective gene receptor with no genetic compensation detected (Fig. 1D). A decrease in the transcript level was also detected for each augmentor in the respective single-homozygous mutants and in the aug-α1−/−;aug-α2−/−;aug-β−/− triple mutant (Fig. 1D). qPCR analysis in all augmentor genotypes did not show any genetic compensation among this family of genes, not even in aug-α1 and aug-α2, duplicated gene paralogues of AUG-α (Fig. 1D).
Together, these results indicate that we successfully identified augmentor genes in zebrafish and generated five loss-of-function mutant fish to study the requirement for Alk, Ltk, and their ligands in development. Additionally, we report that such signaling is not required for major early developmental decisions, including survival of the neural system.
Aug-α1 and Aug-α2 but Not Aug-β Are Required for Iridophore Formation During Embryonic Development.
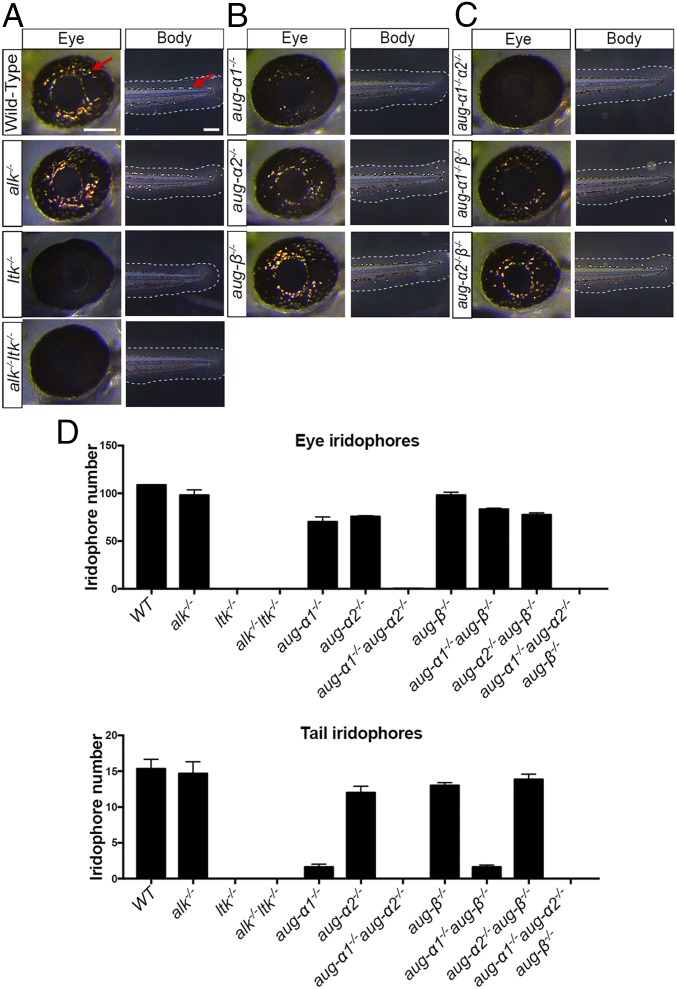
During the preliminary analysis of F2 mutants, we observed that while wild-type embryos developed iridophores in the eye, neither alk−/−;ltk−/− double-mutant nor aug-α1−/−;aug-α2−/−;aug-β−/− triple-mutant embryos developed these light-reflecting pigments (Fig. 1C, arrows). To further understand the individual contributions of Alk, Ltk, and the augmentors to this phenotype, we counted the number of iridophores present in the eye and tail in all genotypes. We first observed normal iridophore development in alk−/− embryos at 52 hpf, indicating that this receptor, by itself, was not able to contribute to eye pigmentation (Fig. 2 A and D). In contrast, ltk−/− embryos showed no iridophore development, in agreement with other Ltk loss-of-function mutants in which the role of Ltk in iridophore specification, proliferation, and survival is well-characterized (Fig. 2 A and D) (16, 17, 20). Interestingly, alk−/−ltk−/− embryos did not possess a more severe defect in any additional pigment cells than the ltk−/− mutant alone (Fig. 2 A and D). Therefore, Alk does not genetically interact with or contribute to Ltk function in the pigmentation of the developing embryo.
Fig. 2.
Iridophore phenotypes associated with ltk, alk, and augmentor loss-of-function embryos. (A–C) Lateral view of the eye and tail (Body) region of 52-hpf embryos. Red arrows indicate representative iridophore cells. (Scale bars: eye, 100 μm; tail, 200 μm.) (D) The number of iridophores in the eye and tail were counted as the number of cells located within the eye region or the number of cells located on the dorsal stripe between the yolk extension and the tail tip, respectively; n = 5; error bars indicate SEM.
Upon analysis of augmentor mutants, we discovered a 25% reduction in the number of iridophores in the eye of aug-α1−/− and aug-α2−/− single-mutant embryos (Fig. 2 B and D). In contrast, an 80% reduction in the number of iridophores was observed in the tail of aug-α1−/− mutants but not in aug-α2−/− mutants (Fig. 2 B and D). Interestingly, a synergistic effect, a 100% reduction in iridophore numbers of the eye and tail, was observed in the aug-α1−/−;aug-α2−/− double-mutant embryos, suggesting that Aug-α1 and Aug-α2 are the primary Ltk ligands driving iridophore development in embryogenesis (Fig. 2 C and D). In support of this hypothesis, no significant reduction of this cell type was detected in aug-β−/− single-mutant embryos, nor was an additive effect observed in aug-α1−/−;aug-β−/− or aug-α2−/−;aug-β−/− double-mutant embryos (Fig. 2 C and D). Therefore, aug-α1 and aug-α2 are the main contributors to the lack of iridophores observed in the aug-α1−/−;aug-α2−/−;aug-β−/− triple mutants (Fig. 2 C and D).
Our findings demonstrate that expression of Aug-α1 and Aug-α2 is required for iridophore formation of the developing embryo and that loss-of-function mutations of both genes uniquely phenocopy mutants deficient in Ltk but not in Alk.
Ltk, Aug-α1, and Aug-α2 Are Expressed in NCCs During Embryonic Iridophore Development.
Previous studies reported ltk expression in differentiated iridophores and in NCCs (16), the iridophore precursor expressing the transcription factor Sox10 during development (18, 19). Because our results suggest that Aug-α1 and Aug-α2 are the primary ligands of Ltk responsible for eye pigmentation, we investigated their expression in NCCs in this particular region.
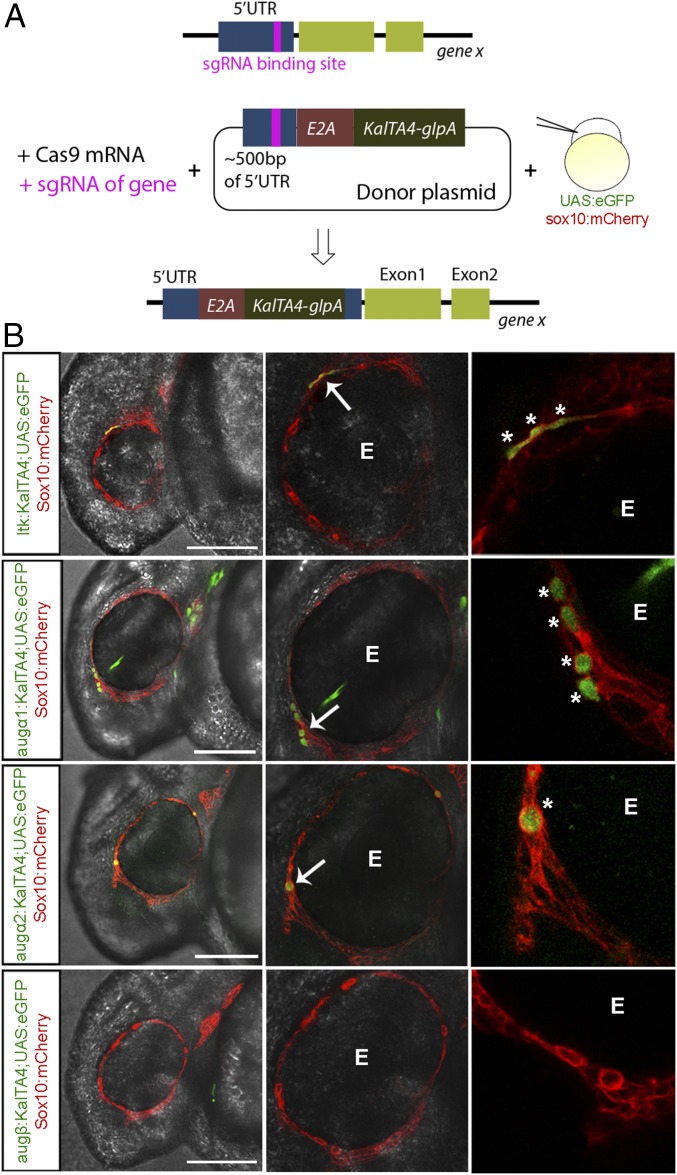
To this end, we took advantage of the Gal4 and upstream activation sequence (UAS) system in combination with CRISPR/Cas9 technology (32, 33). This strategy allows us to visualize the respective endogenous expression of Ltk and augmentors via an eGFP reporter in a transgenic sox10:mCherry;UAS:eGFP zebrafish line (Fig. 3A). After 26 h, the activation of the KalTA4-UAS-eGFP signal was analyzed using confocal microscopy. In accordance with previous ltk expression analysis, eGFP driven by KalTA4 within the ltk 5′ UTR was detected in Sox10+ NCCs in the peripheral regions of the eye (Fig. 3B, arrows). Interestingly, both the aug-α1:KalTA4 and aug-α2:KalTA4 transgenes activated UAS:eGFP in Sox10+ cells of the eye region (Fig. 3B, arrows). In contrast, aug-β:KalTA4;UAS:eGFP embryos showed no eGFP expression at this particular anatomical position (Fig. 3B). Therefore, aug-α1– and aug-α-2–expressing cells were visible in a region similar to that in which ltk and sox10 double-positive NCCs were visible, supporting the notion that these ligands could activate Ltk in a paracrine and/or autocrine fashion (Fig. 3B, asterisks). Accordingly, the mRNA expression of Ltk, Aug-α1, Aug-α2, and Aug-β observed in embryos at different stages, together with the expression in FACS-sorted Sox10+ cells, further confirmed the imaging results from our transgenic GFP lines (Fig. S3).
Fig. 3.
Ltk, Aug-α1, and Aug-α2 are expressed in NCCs during iridophore development. (A) Schematic representing the experimental strategy used to generate ltk and augmentor KalTA4 fish. (B) Lateral view of a confocal cross-section of 24-hpf zebrafish embryos injected as in A (the head region is to the left). UAS expression is driven by the KalTA4 ORF in the 5′ UTR region of each gene, thus activating UAS:eGFP+ cells. mCherry expression is driven by the sox10 promotor to indicate Sox10+ NCCs. eGFP and mCherry colocalization indicate ltk, aug-α1, and aug-α2 expression (green) in Sox10+ NCCs (red) within the peripheral area of the eye (E). White arrows indicate cells coexpressing eGFP and mCherry. Asterisks indicate single NCCs expressing eGFP. (Scale bars: 120 μm.)
The analysis of these embryos together with the genetic evidence provided above suggests that NCCs are committed to differentiating into eye iridophores by Ltk activation through interactions with Aug-α1 and Aug-α2.
Aug-α1, Aug-α2, and Aug-β Are Differentially Required for Iridophore Patterning in Adult Zebrafish.
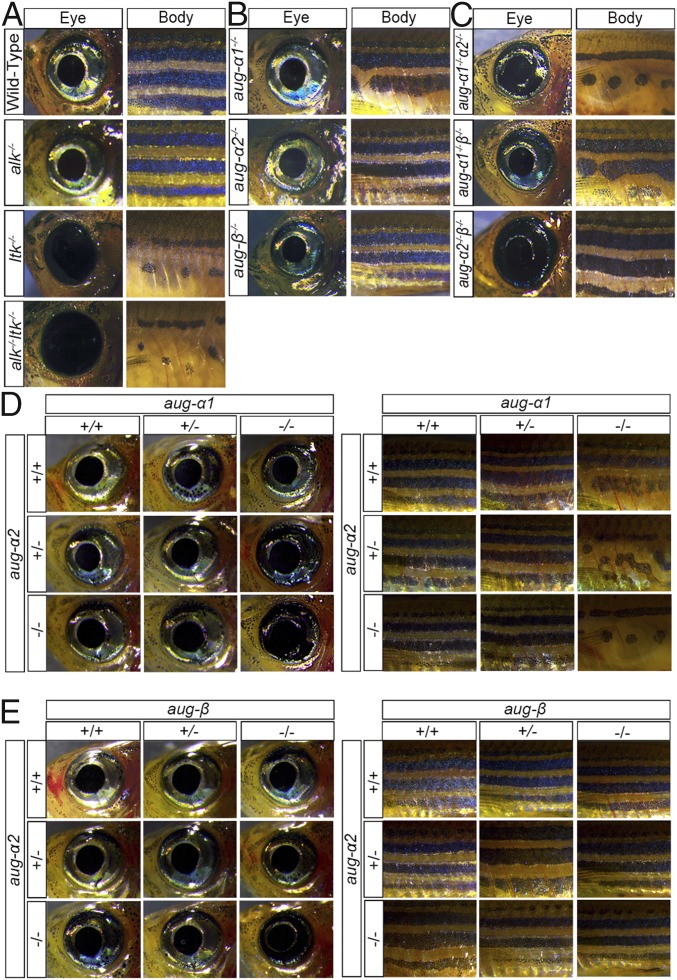
Previous studies have shown that Ltk activity is required for iridophore development of the adult eye and body (16, 17, 20). Specifically, Ltk is critical to committing adult pigment progenitor cells to iridophore specification (19). We aimed to understand the role of ltk–augmentor interactions in this biological process. First, we found that adult ltk−/− single mutants and alk−/−;ltk−/− double mutants do not develop any iridophores in the eye or body and display abnormal stripe patterning, but iridophore development of alk−/− single mutants was similar to that of wild-type zebrafish (Fig. 4A). Thus, our results recapitulate previously described phenotypes of adult Ltk mutants and exclude Alk participation in pigment cell development. Additionally, we observed that ltk−/− mutants comprise ∼25% of all offspring from a heterozygous cross but only 3% of adults, suggesting that Ltk may have another role in survival or in competing with wild-type peers. Importantly, we observed that iridophores develop normally in aug-α1−/−, aug-α2−/−, and aug-β−/− single mutants as well as in the aug-α1−/−;aug-β−/− double mutant (Fig. 4 B and C).
Fig. 4.
Ltk and its augmentor ligands are required for the development of iridophores in the adult zebrafish. (A–E) Lateral views of whole-mount zebrafish adults aged 1–3 mo. Eye and body regions were imaged at 1.6× magnification (the head region is to the left). Iridophore and stripe pigmentation were observed in the indicated genotypes.
The detailed analysis of the body coloration revealed an unexpected finding. The aug-α1−/−;aug-α2−/− double mutants displayed severe iridophore developmental defects of the body and a partial iridophore defect in the eye (Fig. 4C). Because iridophores influence melanophore differentiation and migration (34), these fish subsequently displayed abnormal stripe patterning similar to that in ltk−/− mutants (Fig. 4 A and C). In contrast, aug-α2−/−;aug-β−/− double mutants displayed a severe and specific loss of eye iridophores with no significant stripe defects (Fig. 4C). Therefore, ltk-expressing pigment progenitor cells could differentiate to iridophores in different and distinct anatomical regions based on augmentor availability.
In support of this, we observed a genetic dosing effect in combined augmentor homozygous and heterozygous genotypes. For example, in the case of eye and body pigmentation and patterning, fish genotypes such as aug-α1−/−;aug-α2−/+ displayed partial disruption of eye iridophore development and stripe patterning compared with wild-type fish (Fig. 4D). In this genotype, the aug-α2 wild-type allele partially contributes to iridophore development. Interestingly, only modest defects in eye iridophore development and normal stripe patterning are observed in aug-α1−/+;aug-α2−/− fish, thus suggesting that one aug-α1 allele is sufficient to allow normal body coloration (Fig. 4D).
The aug-α2−/−;aug-β−/+ and aug-α2−/+;aug-β−/− mutants displayed modest defects in eye iridophore development (Fig. 4E). In this case, one allele of aug-α2 or aug-β contributes equally to the eye, while neither is required for body iridophore development and stripe patterning (Fig. 4E). Thus, the α augmentors are the predominant ligands necessary and sufficient to drive stripe formation.
Together, these genetic experiments support the diverse requirement of augmentors in the development of adult pigmentation.
Conclusions
We demonstrate that Alk and Ltk ligands are required for iridophore development in embryonic and adult zebrafish. Our experiments show that aug-α1 and aug-α2 are essential to drive embryonic iridophore development and adult stripe development, while aug-β and aug-α2 together are critical for iridophore formation in the adult eye. Moreover, these processes are mediated entirely by Ltk and not by Alk (Fig. 5).
Fig. 5.
Schematic model of Ltk and augmentor function in iridophore differentiation from embryonic NCCs or from adult pigment progenitor cells. Different combinations of augmentors are required for Ltk-driven iridophore development at different times and locations.
This study fills a gap in the current literature describing iridophore development. In premetamorphic zebrafish (younger than 3 wk), iridophores arise from NCCs (20), which can also give rise to glia, chondrocytes, neurons, and other cell types (19). In postmetamorphosis zebrafish (older than 3 wk), neural crest-derived pigment cell progenitors retain some multipotency and can differentiate into neural cells as well as iridophores (18, 19). Both stem and progenitor cells express Ltk, even if they have not yet committed to pigment cell lineage (18), and it was proposed that determination of neural crest and progenitor cell fates occurs via specific Ltk–ligand interactions (18). Here, we provide genetic evidence that augmentors indeed stimulate this process.
Although all pigment cells of the adult emerge from a common origin, the striped patterning can vary within the same fish. For example, stripe development can differ in the fins and the flank region (35). While we do not yet understand what factors dictate this limited region-specific stripe formation, genetic evidence suggests that different mechanisms of development could be involved (36). One possibility is that particular ligand–receptor interactions activate a signaling cascade to determine the differentiation and proliferation of each pigment cell type at different sites (35).
We show that augmentors act as key factors influencing temporal and spatial differentiation of Ltk-expressing NCCs or adult progenitor cells. It should be noted that triple-augmentor–null mutants were not detected, suggesting that additional defects could be responsible for their lack of survival to adulthood. Our current data indicate that a combination of aug-α1, aug-α2, and aug-β is required for specific iridophore differentiation from Ltk+ stem/progenitor cells. Further analysis is required to determine the role played by aug-α1, aug-α2, and aug-β stimulation of Alk+ stem/progenitor cells.
The genomes of D. melanogaster and C. elegans contain a single ALK-like RTK designated “Alk” (26) or “suppressor of constitutive dauer 2” (SCD-2) (25), respectively. In zebrafish, mice, and humans, LTK is also recognized as part of this receptor family. We propose that the zebrafish genome contains two orthologs of ALK-like RTKs, alk-1 and alk-2, rather than Ltk and Alk, respectively. Direct binding and cellular experiments with human AUG-α and AUG-β have shown that AUG-α functions as a high-affinity ligand for ALK and LTK, while AUG-β functions as a high-affinity ligand of LTK (22). It is not yet clear whether zebrafish Alk and Ltk ligands follow a similar hierarchy and specificity toward their zebrafish receptors. Further study will be necessary to deepen our understanding of these receptor-specific functions in biology. This could be critical in light of a potential role proposed for ALK in human melanoma (12). The analysis of Alk-1 and Alk-2 and their ligands therefore could reveal ancestral signaling pathways that might be reactivated in human pigment cell malignancy. Our paper represents one step toward this fascinating perspective.
Materials and Methods
Zebrafish Strains and Husbandry.
D. rerio embryos and adults were maintained at 28.5 °C on a 10-h dark/14-h light cycle and were handled using standard methods and according to the provisions of The Yale University Institutional Animal Care and Use Committee (no. 2015-11473). Zebrafish lines include the AB wild-type strain, Tg(5xUAS:EGFP)nkuasgfp1a, and Tg(KDR:GFP; Sox10:mCherry) (37).
sgRNA Preparation.
The sgRNAs were prepared using a cloning-free procedure as previously published (31). sgRNA sequences were determined for each gene using CRISPRscan (Table S1) (29).
In Vitro-Transcribed mRNA Preparation.
In the case of Cas9 mRNA, the plasmid pT3TS-nCASn (Addgene no. 46757) was linearized with XbaI and purified for processing with an mMESSAGE mMACHINE T3 Transcription Kit (Ambion).
Zebrafish Injection.
For the generation of zebrafish mutants, embryos were injected at the single-cell stage with 2 nL of solution containing 100 ng/µL Cas9 mRNA, 30 ng/µL sgRNA, and 10% phenol red. To generate the KalTA4/UAS:eGFP fish, the donor plasmid was injected into zebrafish embryos at a concentration of 30 ng/µL in addition to 30 ng/µL sgRNA, 100 ng/µL Cas9 mRNA, and 10% phenol red. Donor plasmid preparation consisted of cloning 300–500 bp corresponding to the sequence flanking the sgRNA target-site primers specific to the 5′ UTR of each gene (Table S1) into eGFPbait-E2A-KalTA4-pA donor plasmid via BamHI and EcoRV (32). Injected embryos were subjected to image acquisition (see below).
Genotyping.
Genotyping was performed using 6-FAM PCR fragment analysis as previously described (38). Primers used are listed in Table S2.
Total RNA Preparation and Real-Time qPCR.
Isolation of total RNA from whole embryos was performed as previously described (38, 39) using primers listed in Table S2. The relative expression was normalized to β-actin and was calculated using the 2−ΔCt or 2−ΔΔCt method (40).
Image Acquisition.
Confocal microscopy was performed using the Leica Microsystem SP5 confocal microscope as previously described (38). For iridophores analysis, 52-hpf embryos were briefly anesthetized with Tricaine (Western Chemical), and pictures were captured with the Leica Application Suite (Leica), using the Leica MacroFluo system and Leica LED5000RL illumination system (Leica). The number of iridophores was counted by ImageJ (NIH). For eye iridophore quantification, individual cells within the eye were counted. For tail iridophore quantification, individual cells located on the dorsal stripe between the yolk extension and the tail tip were counted.
Supplementary Material
Acknowledgments
We thank Philip B. Murray and Gulhan Ercan-Sencicek for contributing to the initiation of this study, Meredith S. Cavanaugh for fish husbandry at the zebrafish facility of Yale’s Cardiovascular Research Center, and Albertomaria Moro for helping with the computational search of the off-target sequence.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710254114/-/DCSupplemental.
References
- 1.Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 2.Heldin CH, Lu B, Evans R, Gutkind JS. Signals and receptors. Cold Spring Harb Perspect Biol. 2016;8:a005900. doi: 10.1101/cshperspect.a005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 5.Hynes NE, Lane HA. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 6.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 7.Morris SW, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 8.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13:685–700. doi: 10.1038/nrc3580. [DOI] [PubMed] [Google Scholar]
- 9.Osajima-Hakomori Y, et al. Biological role of anaplastic lymphoma kinase in neuroblastoma. Am J Pathol. 2005;167:213–222. doi: 10.1016/S0002-9440(10)62966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George RE, et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS One. 2007;2:e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busam KJ, et al. Primary and metastatic cutaneous melanomas express ALK through alternative transcriptional initiation. Am J Surg Pathol. 2016;40:786–795. doi: 10.1097/PAS.0000000000000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiesner T, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature. 2015;526:453–457. doi: 10.1038/nature15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss JB, et al. Anaplastic lymphoma kinase and leukocyte tyrosine kinase: Functions and genetic interactions in learning, memory and adult neurogenesis. Pharmacol Biochem Behav. 2012;100:566–574. doi: 10.1016/j.pbb.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 14.Bilsland JG, et al. Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacology. 2008;33:685–700. doi: 10.1038/sj.npp.1301446. [DOI] [PubMed] [Google Scholar]
- 15.Yao S, et al. Anaplastic lymphoma kinase is required for neurogenesis in the developing central nervous system of zebrafish. PLoS One. 2013;8:e63757. doi: 10.1371/journal.pone.0063757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopes SS, et al. Leukocyte tyrosine kinase functions in pigment cell development. PLoS Genet. 2008;4:e1000026. doi: 10.1371/journal.pgen.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fadeev A, Krauss J, Singh AP, Nüsslein-Volhard C. Zebrafish leucocyte tyrosine kinase controls iridophore establishment, proliferation and survival. Pigment Cell Melanoma Res. 2016;29:284–296. doi: 10.1111/pcmr.12454. [DOI] [PubMed] [Google Scholar]
- 18.Singh AP, et al. Pigment cell progenitors in zebrafish remain multipotent through metamorphosis. Dev Cell. 2016;38:316–330. doi: 10.1016/j.devcel.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Singh AP, Schach U, Nüsslein-Volhard C. Proliferation, dispersal and patterned aggregation of iridophores in the skin prefigure striped colouration of zebrafish. Nat Cell Biol. 2014;16:607–614. doi: 10.1038/ncb2955. [DOI] [PubMed] [Google Scholar]
- 20.Kelsh RN, et al. Zebrafish pigmentation mutations and the processes of neural crest development. Development. 1996;123:369–389. doi: 10.1242/dev.123.1.369. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, et al. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proc Natl Acad Sci USA. 2014;111:15741–15745. doi: 10.1073/pnas.1412009111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reshetnyak AV, et al. Augmentor α and β (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc Natl Acad Sci USA. 2015;112:15862–15867. doi: 10.1073/pnas.1520099112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan J, et al. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. Elife. 2015;4:e09811. doi: 10.7554/eLife.09811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430:345–350. doi: 10.1038/nature02647. [DOI] [PubMed] [Google Scholar]
- 25.Reiner DJ, Ailion M, Thomas JH, Meyer BJ. C. elegans anaplastic lymphoma kinase ortholog SCD-2 controls dauer formation by modulating TGF-beta signaling. Curr Biol. 2008;18:1101–1109. doi: 10.1016/j.cub.2008.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorén CE, et al. A crucial role for the anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep. 2003;4:781–786. doi: 10.1038/sj.embor.embor897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss JB, Suyama KL, Lee HH, Scott MP. Jelly belly: A Drosophila LDL receptor repeat-containing signal required for mesoderm migration and differentiation. Cell. 2001;107:387–398. doi: 10.1016/s0092-8674(01)00540-2. [DOI] [PubMed] [Google Scholar]
- 28.Murray PB, et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal. 2015;8:ra6. doi: 10.1126/scisignal.2005916. [DOI] [PubMed] [Google Scholar]
- 29.Moreno-Mateos MA, et al. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narayanan A, et al. In vivo mutagenesis of miRNA gene families using a scalable multiplexed CRISPR/Cas9 nuclease system. Sci Rep. 2016;6:32386. doi: 10.1038/srep32386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auer TO, Duroure K, Concordet JP, Del Bene F. CRISPR/Cas9-mediated conversion of eGFP- into Gal4-transgenic lines in zebrafish. Nat Protoc. 2014;9:2823–2840. doi: 10.1038/nprot.2014.187. [DOI] [PubMed] [Google Scholar]
- 33.Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frohnhöfer HG, Krauss J, Maischein HM, Nüsslein-Volhard C. Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development. 2013;140:2997–3007. doi: 10.1242/dev.096719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nüsslein-Volhard C, Singh AP. How fish color their skin: A paradigm for development and evolution of adult patterns: Multipotency, plasticity, and cell competition regulate proliferation and spreading of pigment cells in zebrafish coloration. BioEssays. 2017;39:1600231. doi: 10.1002/bies.201600231. [DOI] [PubMed] [Google Scholar]
- 36.Singh AP, Frohnhöfer HG, Irion U, Nüsslein-Volhard C. Fish pigmentation. Response to comment on “Local reorganization of xanthophores fine-tunes and colors the striped pattern of zebrafish”. Science. 2015;348:297. doi: 10.1126/science.aaa2804. [DOI] [PubMed] [Google Scholar]
- 37.Asakawa K, Kawakami K. Targeted gene expression by the Gal4-UAS system in zebrafish. Dev Growth Differ. 2008;50:391–399. doi: 10.1111/j.1440-169X.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 38.Kasper DM, et al. MicroRNAs establish uniform traits during the architecture of vertebrate embryos. Dev Cell. 2017;40:552–565.e5. doi: 10.1016/j.devcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Ristori E, Nicoli S. miRNAs expression profile in zebrafish developing vessels. Methods Mol Biol. 2015;1214:129–150. doi: 10.1007/978-1-4939-1462-3_7. [DOI] [PubMed] [Google Scholar]
- 42.Ristori E, et al. A dicer-miR-107 interaction regulates biogenesis of specific miRNAs crucial for neurogenesis. Dev Cell. 2015;32:546–560. doi: 10.1016/j.devcel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bae S, Park J, Kim JS. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.