Significance
Two well-characterized carboxylate residues catalyze vectorial proton transport in rhodopsin proton pumps, such as bacteriorhodopsin. We report the mechanism of a type of rhodopsin, a channelrhodopsin unique in that it exploits the same residues to accomplish a completely different process: opening and closing of a cation channel. This finding promises leaps in our understanding of how evolution modifies shared protein scaffolds to create new protein chemistry in homologous membrane proteins. Moreover, algal channelrhodopsins have become widespread molecular tools to manipulate membrane potential in cells with light (optogenetics). The structurally distinct, independently evolved channelrhodopsins studied here open the possibility for discovery of optogenetic tools with new properties. Our results provide insights into the conductance mechanisms of these channelrhodopsins.
Keywords: proton transfers, ion transport, photocycle, channelrhodopsins, optogenetics
Abstract
The recently discovered cation-conducting channelrhodopsins in cryptophyte algae are far more homologous to haloarchaeal rhodopsins, in particular the proton pump bacteriorhodopsin (BR), than to earlier known channelrhodopsins. They uniquely retain the two carboxylate residues that define the vectorial proton path in BR in which Asp-85 and Asp-96 serve as acceptor and donor, respectively, of the photoactive site Schiff base (SB) proton. Here we analyze laser flash-induced photocurrents and photochemical conversions in Guillardia theta cation channelrhodopsin 2 (GtCCR2) and its mutants. Our results reveal a model in which the GtCCR2 retinylidene SB chromophore rapidly deprotonates to the Asp-85 homolog, as in BR. Opening of the cytoplasmic channel to cations in GtCCR2 requires the Asp-96 homolog to be unprotonated, as has been proposed for the BR cytoplasmic channel for protons. However, reprotonation of the GtCCR2 SB occurs not from the Asp-96 homolog, but by proton return from the earlier protonated acceptor, preventing vectorial proton translocation across the membrane. In GtCCR2, deprotonation of the Asp-96 homolog is required for cation channel opening and occurs >10-fold faster than reprotonation of the SB, which temporally correlates with channel closing. Hence in GtCCR2, cation channel gating is tightly coupled to intramolecular proton transfers involving the same residues that define the vectorial proton path in BR.
Microbial, or type 1, rhodopsins use variations of a shared seven-transmembrane helix design and photochemistry to carry out distinctly different light-driven energy and sensory transduction processes in diverse prokaryotic and lower eukaryotic organisms (1, 2). Phototaxis by green (chlorophyte) algae such as the model microorganism Chlamydomonas reinhardtii is mediated by sensory rhodopsins of the microbial type that depolarize the algal plasma membrane when photoactivated (3). These proteins, named “channelrhodopsins,” have attracted much attention because of their light-gated passive cation transport upon heterologous expression of their genes in animal cells (4, 5). Today chlorophyte channelrhodopsins are widely used for depolarization of the cell membrane and neuronal excitation by light (optogenetics) (6). Molecular mechanisms of photoactivation and cation conductance in these proteins have been extensively studied by a combination of biochemical and biophysical methods (7, 8). A high-resolution X-ray crystal structure of a hybrid channelrhodopsin has been obtained (9).
Cryptophyte flagellates, a group of microorganisms phylogenetically distant from green algae, also exhibit phototaxis (10), the photoreceptors for which have not yet been identified. The genome of one cryptophyte species, the marine alga Guillardia theta, has been completely sequenced and is predicted to encode at least 53 microbial (type 1) rhodopsins (11). Among them, the closest homologs to chlorophyte channelrhodopsins conduct anions rather than cations and therefore have been named “anion channelrhodopsins” (ACRs) to distinguish them from cation channelrhodopsins (CCRs) of green algae (11). Many ACR homologs have also been found in other cryptophyte species, indicating that these proteins are not unique to G. theta (12–14). Not only ion selectivity, but also the conduction mechanism of ACRs, some of which exhibit up to 50-fold larger currents than CCRs (11), is very different from the mechanism of chlorophyte CCRs (15, 16). Several ACRs are >1,000-fold more efficient for membrane hyperpolarization than previously known inhibitory optogenetic tools and have been used for a specific and rapid inhibition of spiking in cultured neurons (11, 14, 17) and brain slices (18), and of a wide range of behaviors in live Drosophila (19).
Another cluster of rhodopsins predicted by the G. theta genome shows close sequence similarity to haloarchaeal proton-pumping rhodopsins (20), including conservation of the residues involved in the intramolecular transfer of the Schiff base (SB) proton, namely, those corresponding to Asp-85 (the proton acceptor) and Asp-96 (the proton donor) in bacteriorhodopsin (BR). A homologous protein has also been found in the related organism, Proteomonas sulcata, and shown to generate photocurrents when the encoding construct was expressed in neurons (21). Overall conservation patterns clearly distinguish these cryptophyte proteins from both chlorophyte CCRs, in which the acceptor position is most frequently occupied by Glu (or more rarely a neutral residue) and the donor position mostly by His, and from cryptophyte ACRs, in which Asp-85 is universally replaced with a noncarboxylate residue and Asp-96 usually with Gln (Fig. 1). Surprisingly, when these “BR-like” cryptophyte rhodopsins were tested by expression in cultured animal cells and patch clamp analysis, they demonstrated nonspecific cation channel activity similar to that of chlorophyte CCRs (20, 22). However, electrophysiological data obtained under continuous light stimulation, as in the above cited studies, do not allow precise deconvolution of photocurrent components and may lead to incorrect interpretation of their nature. There are two reasons why continuous light data give quantitatively unreliable results. First, under continuous light a complex mixture of different conductive and nonconductive intermediates is accumulated, and their ratio depends on the intensity and spectral composition of the light. Second, under such continuous light conditions photocurrents related to different possible processes (e.g., channel activity, active charge transfer, secondary changes in membrane due to the current flow) are integrated. In contrast, under single-turnover conditions (6-ns laser excitation) those processes can be kinetically separated and analyzed.
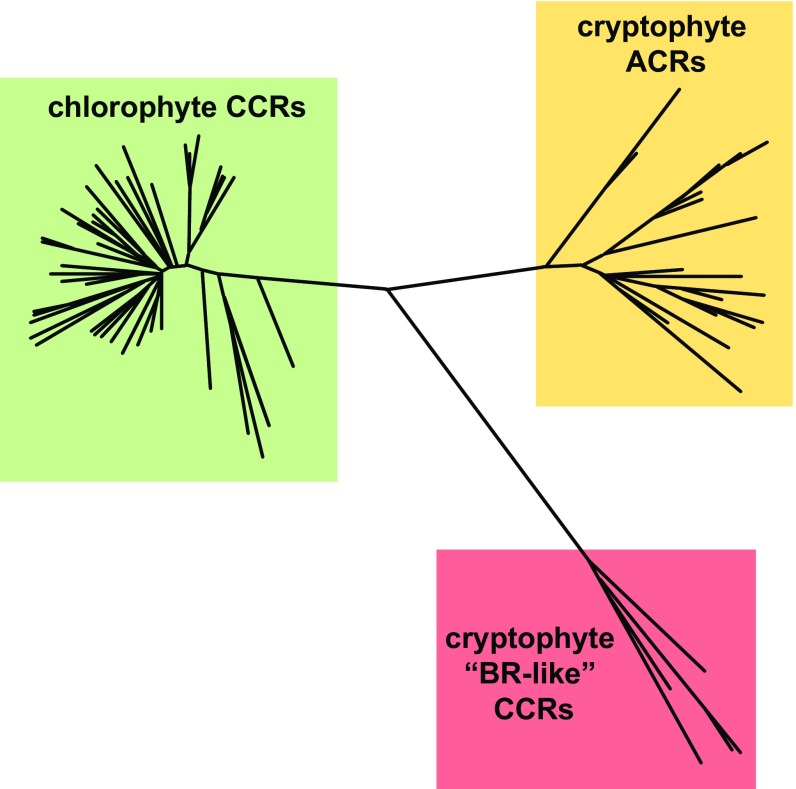
Fig. 1.
A phylogenetic tree of three families of channelrhodopsins constructed by the neighbor-joining method. Approximately ∼60 chlorophyte CCRs and ∼30 cryptophyte АCRs have been identified to date. Only 5 cryptophyte CCRs have been characterized by heterologous expression, but the fully sequenced G. theta genome harbors several other homologous genes, and more homologs are likely to be found in other cryptophyte species.
Among cryptophyte CCRs that we have previously tested, G. theta cation channelrhodopsin 2 (GtCCR2) generated the largest photocurrents under continuous light (20) and therefore lent itself to more detailed investigation. Here we report analysis of GtCCR2 photocurrents under single-turnover conditions. Under such conditions it is not only possible to separate quantitatively kinetically different electrogenic processes, but also to correlate them with 6-ns-flash-induced absorbance changes in purified pigments.
In BR and most other microbial rhodopsins, retinal isomerization is followed by a rapid transfer of the SB proton to an outwardly located acceptor(s) (23). We have previously demonstrated that the corresponding intramolecular charge displacements upon laser flash excitation can be detected by patch clamp recording in microbial rhodopsins expressed in cultured animal cells (24). Here we show that in GtCCR2 the SB rapidly deprotonates to the outer located acceptor Asp-87, as in BR. However, in contrast to BR, reprotonation of the SB occurs in GtCCR2 not from the Asp-96 homolog, but rather by eventual return of the proton from the protonated acceptor, thus preventing vectorial proton translocation across the membrane. Most importantly, the homolog of the BR proton donor (Asp-98) must be unprotonated to open the cation channel in GtCCR2, which implies that deprotonation of this residue takes place orders of magnitude faster than reprotonation of the SB. Channel closing correlates with reprotonation of the SB from the earlier protonated outwardly located acceptor Asp-87. These results show that cryptophyte CCRs not only structurally, but also functionally differ from their counterparts from green algae, forming a third family of channelrhodopsins, which can be described as “BR-like cation channelrhodopsins” (BCCRs).
Results
GtCCR2 Exhibits Both Intramolecular Proton Transfers and Cation Channel Activity.
Photoelectric activity of GtCCR2 was studied upon expression in human embryonic kidney 293 (HEK293) cells by whole-cell patch clamp. A contribution of multiple processes to recorded photocurrents was evident even under continuous light excitation. At negative holding potentials with 150 mM NaCl in the bath and 126 mM KCl in the pipette, both pH 7.4 (standard solutions; for their other components, see Materials and Methods), switching the light off after 1-s illumination caused a fast transient negative current followed by slower relaxation of channel current (Fig. S1, red line). No such fast transient was observed in current traces recorded from chlorophyte CCRs (Fig. S1, black line). The appearance of this transient could be explained either by cessation of a sustained positive current, or by dissipation of an accumulated electrical dipole within the protein molecule. As noted in the introduction, single-turnover measurements were required for a definitive discrimination between these two possibilities.
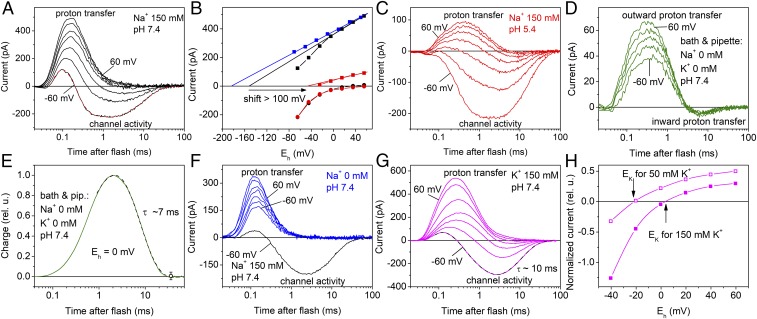
Under single-turnover conditions using 6-ns laser flashes for photoexcitation, two processes involved in generation of photocurrent could be kinetically resolved. At negative holding potentials with standard solutions, the fast current with a positive peak at ∼150 µs was followed by a slower oppositely directed current (Fig. 2A). The amplitude of the fast peak showed a shallow dependence on voltage [current (I)–voltage (E) relationship, IE curve] (Fig. 2B, black squares). When only the linear part of this dependence at the positive holding potentials was taken into account, its approximation crossed the x axis at approximately −150 mV (Fig. 2B, black line). After replacement of Na+ in the bath with nonpermeable N-methyl-d-glucamine (NMG+) the IE curve of the fast positive current became linear (Fig. 2B, blue squares), which showed that the deviation from linearity observed at 150 mM Na+ (Fig. 2B, black squares) was due to a contribution of Na+ channel current. Linear extrapolation of the IE curve measured at 0 mM Na+ crossed the x axis at −180 ± 6 mV (mean ± SEM, n = 3 cells), which was close to the value measured for the currents generated by BR expressed in oocytes (25). Acidification of the external medium from pH 7.4 to 5.4 decreased the amplitude of the fast positive current (Fig. 2C) and led to a >100-mV shift of the IE curve to more positive potentials (Fig. 2B, red squares and line), which was close to the shift in the Nernst equilibrium potential for protons. These results and optical data (below) strongly suggest that the fast current reflects outwardly directed transfer of the SB proton.
Fig. 2.
Two components of GtCCR2 photocurrents under single-turnover conditions and their ionic dependencies. (A, C, D, F, and G) Representative series of current traces recorded from wild-type GtCCR2 in response to a 6-ns laser flash at different ionic conditions. The main ions in the bath solution are indicated in the panels (for other components see Materials and Methods); the pipette solution was standard except in D and E, in which it contained no Na+, K+, or Ca2+. The holding voltage at the amplifier output was changed in 20-mV (A, C, F, and G) or 30-mV (D) steps from −60 mV. In A and G, multiexponential fits of experimental data recorded at −60 mV are shown as the dashed lines. In F, photocurrent recorded from the same cell at −60 mV with 150 mM Na+ in the bath is shown for comparison as the black line. (B) The voltage dependence of the fast (squares) and channel currents corrected for inward proton transfer (circles) at pH 7.4 (black) and 5.4 (red) of the bath medium in the presence of 150 mM Na+. For 0 mM Na+ (blue), only the scaled values of the fast current are shown, because no channel currents were detected under these conditions. The straight lines show linear fits to the data. The arrow shows a shift in Erev measured at 150 mM Na+ upon acidification of the medium from pH 7.4 to 5.4. (E) Kinetics of charge transfer calculated as the area under the current curve measured in the absence of any channel activity. (H) The voltage dependence of the channel current at 50 (empty symbols) or 150 (filled symbols) mM K+ in the bath, corrected for inward proton transfer. To calculate the error values, data for E and H obtained on different cells were normalized and plotted as relative units (rel. u.).
A neutralizing mutation of the homolog of the BR proton acceptor Asp-85 (GtCCR2_D87N) led to more than an order of magnitude reduction of the fast positive photocurrent (Fig. S2, red lines) at approximately the same expression level judged by the tag fluorescence, indicating that this residue is the primary acceptor of the SB proton. The residual positive current from the GtCCR2_D87N mutant may reflect proton transfer to an alternative acceptor, most likely, Asp-219, the homolog of Asp-212 in BR, as we observed earlier in cation-conducting channelrhodopsin from Chlamydomonas augustae (CaChR1) (24).
When all metal cations that may contribute to channel current (Na+, K+, and Ca2+) were replaced with NMG+ in both pipette and bath solutions adjusted to the same pH 7.4, the fast positive current was followed by a slower negative current (Fig. 2D), attributable to return of the proton from the outer acceptor to the SB. At zero holding voltage, when passive proton current was also excluded, the amount of charge (the area under the current curve) transferred outwardly was equal to that later transferred inwardly (Fig. 2E). Therefore, all protons transferred from the SB to the outer acceptor return back to the SB, and no active proton transport as in rhodopsin pumps takes place in GtCCR2.
At 150 mM Na+ in the bath, the amount of charge transported during the negative current was more than an order of magnitude larger than that carried by protons transported from the SB to the acceptor, which argued against active Na+ transport. Furthermore, replacement of Na+ in the bath with NMG+ led to a strong suppression of slow current (Fig. 2F), indicating passive transport of primarily Na+. The kinetics of this transport were very similar to that of inward intramolecular proton transfer, which therefore contributed to the amplitude of slow negative current. The IE curve of the main negative peak, observed in the wild-type GtCCR2 1–2 ms after the flash, revealed strong inward rectification (Fig. 2B, circles). Erev values for channel currents from most chlorophyte CCRs measured with the same standard solutions were close to zero, indicating high relative permeability for protons (24). In contrast, Erev of GtCCR2 channel current, corrected for contribution of intramolecular proton transfers, was 33 ± 4 mV (mean ± SEM, n = 4 cells; Fig. 2B, black circles), which further confirmed that this current reflected primarily passive Na+ transport.
Replacement of Na+ in the bath with K+ did not change the waveform of the currents (Fig. 2G), which indicated that this cation was also transported by the GtCCR2 channel. When the IE dependence of the negative current measured in the K+ bath and pipette was corrected for contribution of inward intramolecular proton transfers, its reversal potential was equal to the Nernst equilibrium potential for K+ for both tested gradients (Fig. 2H), which confirmed GtCCR2 passive permeability for this ion. In the presence of Na+, acidification of the bath solution by two pH units caused only a minor shift of the Erev of channel current (Fig. 2B, red circles), which indicated a very small passive conductance for protons. Replacement of Cl− in the bath with Asp− changed neither the current kinetics nor the IE dependence (Fig. S3), which showed that GtCCR2 had no Cl− permeability.
The Channel Open State Is Controlled by the Protonation State of Asp-98.
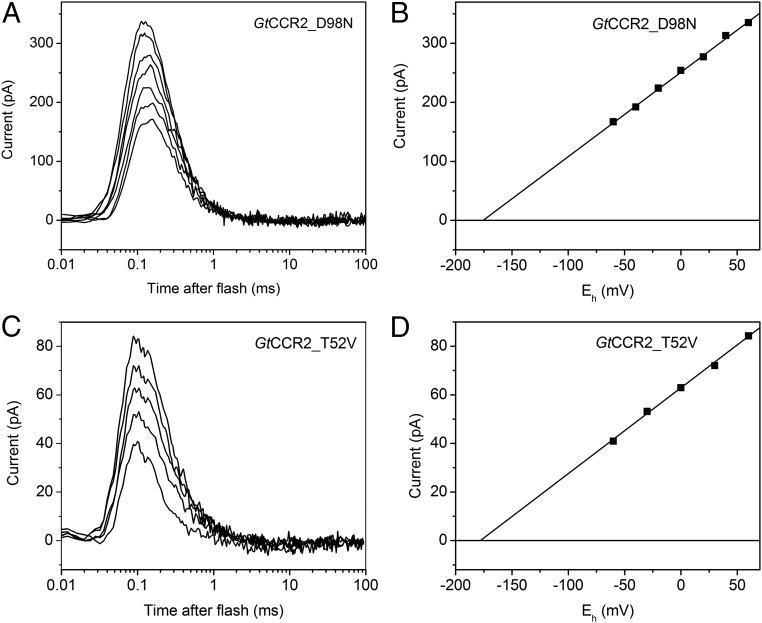
Neutralization of the BR Asp-96 homolog by mutagenesis (GtCCR2_D98N) led to complete elimination of cation channel activity, whereas intramolecular proton transfer was not impaired (Fig. 3 A and B). One possible explanation is that the channel in this mutant is already open in the dark, as has been found in the GtACR1_E68R mutant (15), so that only the fast proton transfer can be detected. This explanation, however, could be ruled out, as the average membrane conductance of the cells expressing the D98N mutant was not different from that of the wild type (3.0 ± 0.6 and 3.1 ± 0.3 nS, respectively; mean ± SEM, n = 15 cells for each variant), whereas opening of the wild-type channels caused by illumination corresponded to 6.7 ± 0.2 nS increase in the conductance.
Fig. 3.
Role of the homologs of the proton donor in BR and its hydrogen-bonded threonine in channel activity of GtCCR2. (A and C) Series of current traces recorded in response to a 6-ns laser flash at standard conditions from GtCCR2_D98N (A) and GtCCR2_T52V (C). No channel current was detected. (B and D) Corresponding voltage dependences of the peak amplitudes of proton transfer currents.
We conclude that unprotonated Asp-98 is required for opening of the channel in GtCCR2. Asp-96 in BR is protonated with a high pKa in the dark (26), in part because the protonated Asp-96 is stabilized by a hydrogen bond with Thr-46, as discussed below. Its deprotonation has been suggested to “unlatch” the cytoplasmic half channel (27–30). A role of Asp-96 as a “latch” in the BR cytoplasmic channel may be relevant to our finding that an unprotonated Asp-98 is necessary for opening the larger cation channel of GtCCR2. Comparison with the behavior of Asp-96 in BR suggests a model in which Asp-98 in GtCCR2 is protonated in the dark and its deprotonation during the photocycle is required for opening the passive cation channel, as proposed for BR. Nevertheless, there are also substantial differences in individual steps of intramolecular proton transfer between GtCCR2 and BR, as we show in the next section.
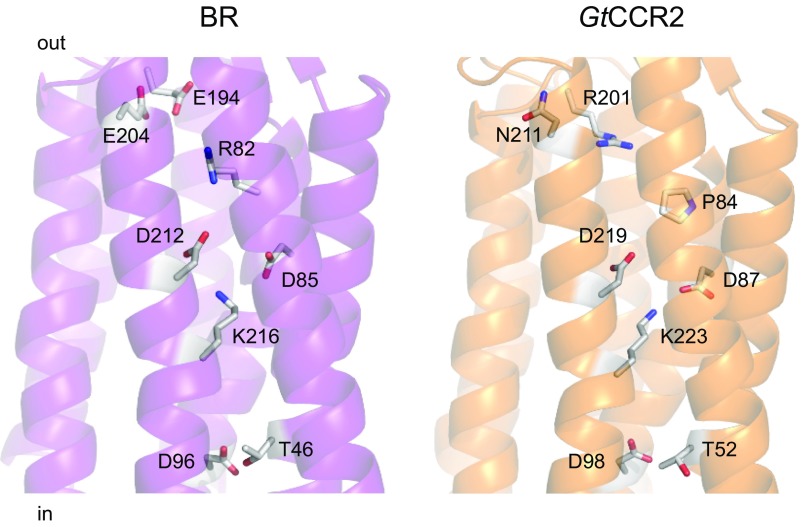
According to our homology model of GtCCR2, Thr-52 corresponds to Thr-46 in BR (Fig. 4). In BR, protonated Asp-96 forms a hydrogen bond with Thr-46, which was suggested to be involved in modulation of the pKa of Asp-96, because mutation of Thr-46 to Val affected the rates of proton transfer from and to Asp-96 (31) and formation and stabilization of water clusters in the cytoplasmic domain between Asp-96 and the SB (32, 33). An FTIR study and molecular dynamic simulations show that the structural rearrangements associated with the disruption of the D96–T46 hydrogen bond upon Asp-96 deprotonation are sufficient to reorganize internal water molecules between Asp-96 and the SB to facilitate proton transfer from Asp-96 to the deprotonated SB (29). We made the GtCCR2_T52V mutant and tested it upon laser excitation. Channel activity in the T52V mutant was suppressed, whereas the fast proton transfer signal was not affected (Fig. 3 C and D), as in the D98N mutant. This observation suggests that Thr-52 is involved in Asp-98 deprotonation upon photoexcitation.
Fig. 4.
A crystal structure of BR (1c3w; Left) and a homology model of GtCCR2 built by the Robetta server using a structure of a haloarchaeal sensory rhodopsin II (2ksy) as a template (Right). The side chains of the key residues involved in intramolecular proton transfers are shown as sticks.
Photocycle of Purified GtCCR2 and the Sequence of Intramolecular Proton Transfer Steps.
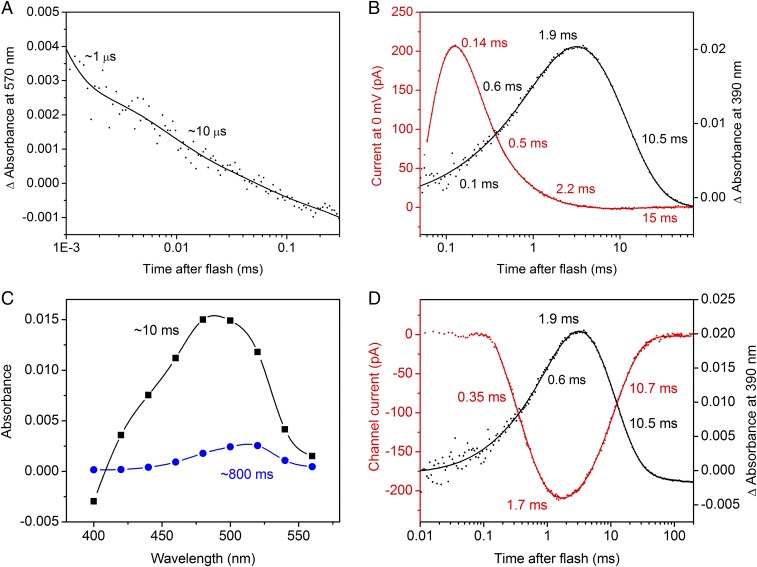
We produced photoactive GtCCR2 by heterologous expression of the encoding construct in Pichia followed by detergent extraction and studied the photocycle of the purified pigment by flash photolysis. The time course of the initial transition from the unphotolyzed state to a red-shifted (K) photoproduct with the maximum of the difference spectrum near 540 nm could not be resolved with our experimental setup. The K state decayed to a blue-shifted (L) intermediate in a biphasic manner with time constants (τ) of 0.8 ± 0.1 and 8.9 ± 1.9 µs (n = 7 measurements) (Fig. 5A).
Fig. 5.
Laser flash-induced absorbance changes in purified of GtCCR2 and their correlation with photocurrents. (A) Biphasic decay of the K intermediate. The dots show experimental data, and the solid line, a multiexponential fit. (B) The photocurrent trace recorded in the absence of Na+ at 0 mV holding potential is shown as red dots, and kinetics of the M intermediate, as black dots. The results of multiexponential fitting are shown as solid lines; the numbers are the τ-values derived from the fit. (C) The spectral dependence of the amplitudes of the two slowest components derived by global fit. (D) Correlation of the opening and closing of the channel (red) with the appearance and decay of the M intermediate (black). The channel current was calculated by subtraction of the properly scaled proton transfer current (obtained at the positive holding potentials) from the photoelectric signal measured at −60 mV in the standard bath.
Deprotonation of the SB could be followed by the formation of an M intermediate with absorption maximum <400 nm. Fitting of the M rise yielded three kinetic components with the τ-values 0.13 ± 0.02, 0.5 ± 0.2 and 2 ± 0.3 ms (Fig. 5B, black line). To compare intramolecular proton transfers with absorption changes, we recorded photocurrents at zero holding potential in the absence of Na+, thereby minimizing the contribution of channel current. Changes in the concentrations of intermediates should be reflected by the current decay. Indeed, three kinetic components similar to those in the M rise were found in the current decay, which confirmed that the fast positive current reflected proton transfer from the SB to the acceptor Asp-87 (Fig. 5B, red line). Reprotonation of the SB occurred with τ 8.9 ± 0.6 ms and led to the appearance of a blue-shifted N intermediate (Fig. 5C, black line). The longest-lived intermediate of the photocycle had a life time of ∼800 ms and red-shifted absorption, and corresponded probably to an O intermediate (Fig. 5C, red line). No significant contribution of the N-to-O transition to the current signal could be detected. The kinetics of pure channel current could be reconstructed by subtraction of the properly scaled proton transfer current (measured at the positive holding potential when the contribution of channel current is minimal) from the current trace obtained at strongly negative holding potentials recorded in the medium containing Na+. These kinetics are shown in Fig. 5D (red line). The kinetics of channel opening and closing closely matched that of de- and reprotonation of the SB, respectively (Fig. 5D, black line).
The large electrical currents associated with intramolecular proton transfer and the high sequence homology of cryptophyte CCRs with rhodopsin proton pumps suggest a simple analogy of ion transport mechanisms in these two rhodopsin families. Indeed, proton transfer from the SB to the Asp-85 homolog, and a reversible deprotonation of the Asp-96 homolog, were observed in GtCCR2, as in BR. Moreover, similar to BR (30), in GtCCR2 the open state of the cytoplasmic channel depends on the unprotonated state of the Asp-96 homolog (Fig. 3 A and B). However, the sequence of proton transfer steps appears to be fundamentally different in these two proteins.
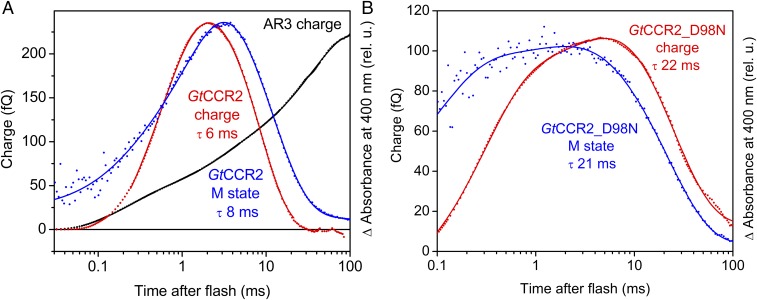
Photocurrent measurements reflect the rate of charge movements and therefore better characterize fast electrogenic processes. As shown in Fig. 5 B and D (black lines), reprotonation of the SB monitored by measuring absorbance changes at 380 nm was two orders of magnitude slower than its deprotonation. Therefore, intramolecular proton transfers corresponding to SB reprotonation could not be resolved in the current traces on the background of residual channel currents even in the absence of Na+. To analyze slow electrogenic processes, the charge displacement, i.e., the integral of the current curve needs to be calculated (23), the results of which are shown in Fig. 6. In rhodopsin proton pumps the SB proton is transferred to the outwardly located acceptor, and then another proton is received from the inwardly located donor. This is reflected by multicomponent outwardly directed charge displacement. For example, the charge displacement curve for the proton pump archaeorhodopsin-3 (AR3, or Arch) showed three outwardly directed components with τ-values 0.2, 2.3, and 38 ms (Fig. 6A, black trace), which closely match the τ-values of M rise (>0.1 ms) and biphasic M decay (2.2 and 46 ms), reported for this pigment (34).
Fig. 6.
Temporal correlation of the charge transfer and M formation in wild-type GtCCR2 and its D98N mutant. (A) Charge transfer in GtCCR2 (red trace) expressed in HEK293 cells recorded in response to a 6-ns laser flash, and the kinetics of the M intermediate in purified GtCCR2 (blue trace). Charge transfer in the proton pump Arch (black trace) is shown for comparison. (B) Charge transfer (red trace) recorded in response to a 6-ns laser flash and the M intermediate kinetics (blue trace) in the GtCCR2_D98N mutant. Absorption changes in panels are plotted in relative units (rel. u.) to compensate for different expression levels and normalized to the amplitudes of charge transfer plotted as femtocoulombs (fQ).
We compared the kinetics of charge displacement in GtCCR2 in HEK293 cells with that of de- and reprotonation of the SB in purified pigment. The charge signal was calculated from a photocurrent trace recorded in the absence of metal cations in the bath and pipette solutions and absence of a proton electrochemical gradient across the membrane, to eliminate the contribution of passive currents. As shown in Fig. 6A, in GtCCR2 an inwardly directed charge transfer (red line) took place during reprotonation of the SB (blue line), in contrast to proton pumps, in which an outwardly directed charge transfer was observed (black line). We conclude that in GtCCR2, reprotonation of the SB occurs from an outwardly located protonated residue, likely Asp-87, rather than from the inwardly located Asp-98. This would mean that proton shuttles between the SB and the acceptor during the GtCCR2 photocycle. Interestingly, a similar reverse transfer of proton from the counterion to the Schiff base was suggested for the M-to-N transition in the photocycle of a rhodopsin sodium pump in the presence of Na+ (35).
This interpretation of the wild-type data is consistent with the results obtained in the GtCCR2_D98N mutant. In BR, mutagenetic replacement of the donor Asp-96 with a noncarboxylic residue dramatically slowed SB reprotonation (36). In contrast, in GtCCR2, the corresponding mutation (D98N) decreased the rate of deprotonation not more than threefold (Fig. 6B, blue trace). An inwardly directed charge transfer slowed to the same degree in the mutant (Fig. 6B, red trace).
In BR, reprotonation of the SB from the acceptor is prevented, probably, by a very high pKa of Asp-85 in the M state (37) and fast deprotonation of the proton release complex (38). Residues contributing to this complex in BR (Arg-82, Glu-194, and Glu-204) are not conserved in GtCCR2 (20). We introduced these residues at the corresponding positions in GtCCR2 (Pro-84, Arg-201, and Asn-211, respectively) and tested the obtained mutant by patch clamp. No outwardly directed current in the time domain of SB reprotonation was found in the triple mutant; therefore, BR-like behavior was not restored by these three mutations (Fig. S4).
Discussion
Recently discovered cryptophyte CCRs are unique among channelrhodopsins, because they show higher sequence homology to haloarchaeal rhodopsin proton pumps rather than to chlorophyte CCRs or cryptophyte ACRs. By analysis of photocurrents recorded upon single-turnover excitation, we show that GtCCR2 and at least one other homolog (GtCCR1, Fig. S5) represent a type of retinal protein in which complex intramolecular proton transfers, involving the same two Asp residues that are responsible for vectorial proton transport in BR, also occur, but, in stark contrast to BR, do not result in proton pumping, but instead gate a channel for monovalent metal cations. Therefore, these cryptophyte proteins can be considered as BCCRs. However, molecular details of proton transfers and channel gating in BCCRs differ from those in BR and chlorophyte CCRs, respectively.
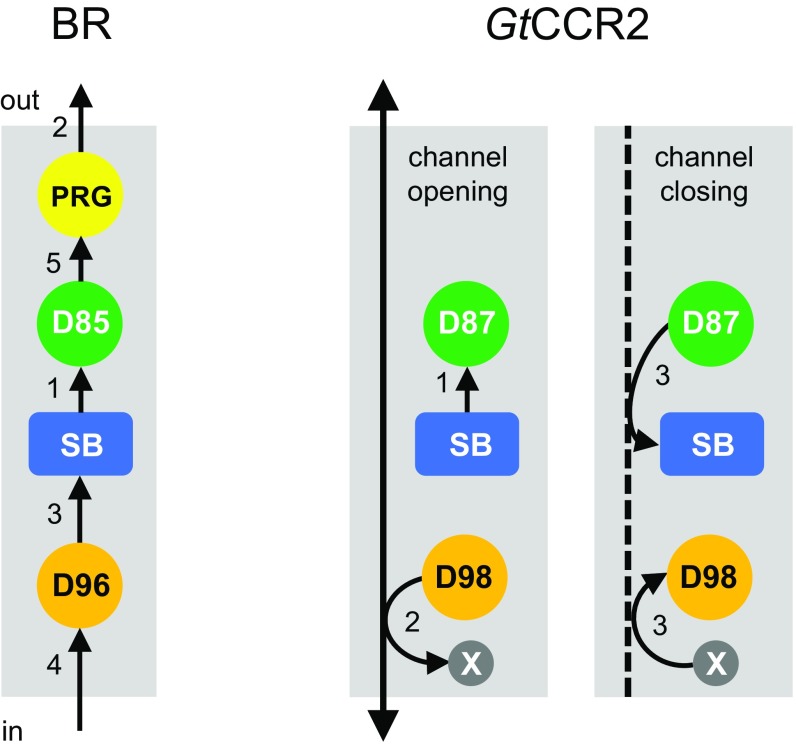
Two major events necessary for proton pumping in BR (transfer of the SB proton to the Asp-85 homolog and deprotonation of the Asp-96 homolog) also occur in GtCCR2. However, in GtCCR2, the proton accepted by the Asp-85 homolog returns back to the SB, and therefore, in contrast to pumps, no actual proton translocation across the membrane is achieved under physiological conditions. Also in contrast to proton pumps, in GtCCR2, deprotonation of the Asp-96 homolog (reflected by cation channel opening, see below) takes place >10-fold faster than reprotonation of the Schiff base, which indicates proton transfer from this residue to an unknown residue or water molecule rather than to the SB. A hypothetical scheme of key intramolecular proton transfer steps in GtCCR2, compared with those in BR, is shown in Fig. 7.
Fig. 7.
Schematic presentation of intramolecular proton transfers in GtCCR2 compared with BR. As reviewed in refs. 48 and 49, in BR deprotonation of the SB to Asp-85 via a water molecule (1) is followed by a fast release of another proton to the extracellular medium (2). The SB is then reprotonated from Asp-96 whose drop in pKa is facilitated by breakage of its hydrogen bond with Thr-46 (3), and which in turn takes up a proton from the cytoplasm (4). Finally, Asp-85 donates a proton to the proton release group (5). In GtCCR2, the SB is deprotonated to Asp-87 (1), whereas deprotonation of Asp-98 to an unidentified residue or water molecule is coupled to channel opening (2). Return of the proton from Asp-87 to the SB and a presumably synchronous reprotonation of Asp-98 are coupled to channel closing (3). PRG, proton release group; SB, Schiff base; X, an unidentified residue or water molecule.
In BR, disruption of the hydrogen bond between the proton donor Asp-96 and Thr-46 modulates deprotonation of the former by lowering its pKa (31). The homolog of Thr-46 in GtCCR2 (Thr-52) also plays an important role in deprotonation of Asp-98, the homolog of the proton donor in BR. However, in GtCCR2 the mutation of Thr-52 prevents Asp-98 deprotonation rather than lowers its pKa, as in BR. This difference may be related to a different orientation of the Thr-52 side chain indicated by our homology model of GtCCR2, which prevents formation of a hydrogen bond with the side chain of Asp-98.
Mutagenic neutralization of Asp-98 by substitution with Asn eliminates channel current, strongly indicating that in GtCCR2, opening of the cytoplasmic half-channel for monovalent cations requires Asp-98 to be unprotonated (Fig. 3). We propose that deprotonation of Asp-98 during the photocycle drives channel opening, which temporarily correlates with deprotonation of the SB, whereas channel closing correlates with reprotonation of the SB (Fig. 6A). In chlorophyte CCRs the homolog of Asp-98 is not conserved, and channel closing occurs nearly an order of magnitude slower than reprotonation of the SB, whereas its deprotonation precedes channel opening (39–42). Asp-156 (a homolog of Asp-115 in BR) has been identified as the proton donor to the SB in channelrhodopsin 2 from Chlamydomonas reinhardtii (CrChR2), the best-studied chlorophyte CCR (43). In BCCRs, an aspartate in this position is replaced with a noncarboxylate residue, although its hydrogen-bonding partner, Cys-128 in CrChR2, is conserved (20), which suggests that the latter residue is required for channel function in both CCR families. The difference in channel gating mechanisms between BCCRs and chlorophyte CCRs was also revealed by accurate calculation of the voltage dependence of GtCCR2 currents recorded upon single turnover excitation (Fig. 2 B and H), which showed much stronger cation channel rectification than in chlorophyte CCRs. From an evolutionary perspective, our results suggest that cryptophyte CCRs (BCCRs) can be considered as an early step in the branching of rhodopsin channels and proton pumps.
Although so-far identified BCCRs do not generate particularly large photocurrents, their small permeability for protons can be advantageous for optogenetic applications to avoid undesirable acidification of the cytoplasm, as results from expression of chlorophyte CCRs (44). Also, the strong voltage dependence of the SB protonation in BCCRs and the possibility to generate easily their nonconducting forms by a single point mutation suggest that such mutants might find use as fluorescent voltage indicators, either through direct retinal fluorescence, as was successfully done with derivatives of the proton pump archaerhodopsin (45), or by converting voltage-induced changes in retinal absorption spectrum into changes in quenching of an appended fluorescent protein tag.
Materials and Methods
All protocols used in this study have been approved by the University of Texas Health Biosafety Committee. The opsin domains of the wild-type GtCCR2 or its mutants were cloned into the mammalian expression vector pcDNA3.1 (Life Technologies) in frame with an EYFP tag. HEK293 cells were transfected using the ScreenFectA reagent (Waco Chemicals) and supplemented with 4 µM all-trans retinal (Sigma-Aldrich). Photocurrents were recorded by whole-cell patch clamp using an Axopatch 200B amplifier (Molecular Devices) and digitized with a Digidata 1440A using pClamp 10 software (both from Molecular Devices) at the sampling rate of 250 kHz. Continuous light pulses were provided by a Polychrome V light source (T.I.L.L. Photonics GmbH) in combination with a mechanical shutter (Uniblitz model LS6, Vincent Associates; half-opening time 0.5 ms). Laser excitation was provided by a Minilite Nd:YAG laser (532 nm, pulsewidth 6 ns, energy 12 mJ; Continuum). For more accurate quantitative deconvolution of overlapping current components, a sum of exponential functions was fit to experimental data using Origin 7.0 software. The data points recoded at the time <0.1 ms after the flash were excluded from fitting because they were not fully resolved. The IE curves were corrected for liquid junction potentials using the built-in pClamp calculator (46).
For expression in Pichia pastoris SMD1168 (his4, pep4) the constructs encoding the opsin domains of GtCCR2 or its mutants were fused with an eight-His tag and transferred to the pPIC9K vector (Invitrogen). The proteins were extracted with dodecyl maltoside and purified using nickel-nitrilotriacetic acid agarose beads (Qiagen). Light-induced absorption changes were measured with a laboratory-constructed crossbeam apparatus. Excitation flashes (532 nm, 6 ns, up to 40 mJ) were provided by a Surelite I Nd:YAG laser (Continuum). Signals were amplified by a low noise current amplifier (model SR445A; Stanford Research Systems) and digitized with a GaGe Octopus digitizer board (model CS8327, DynamicSignals), maximal sampling rate 50 MHz. Data analysis was performed with pClamp 10 (Molecular Devices) and OriginPro 7 (OriginLab) software. Logarithmic filtration of the data was performed using the GageCon program. Global fit of spectral transitions was performed using the FITEXP program (47). A detailed description of the acquisition, processing, and analysis of the data is provided in SI Materials and Methods.
SI Materials and Methods
Photocurrent Measurements.
The opsin domains of the wild-type GtCCR2 or its mutants were cloned into the mammalian expression vector pcDNA3.1 (Life Technologies) in frame with an EYFP tag. Human embryonic kidney 293 (HEK293) cells were transfected using the ScreenFectA transfection reagent (Waco Chemicals). All-trans retinal (Sigma-Aldrich) was added as a stock solution in ethanol at a final concentration of 4 µM. Measurements were performed 48–72 h after transfection with an Axopatch 200B amplifier (Molecular Devices). The signals were digitized with a Digidata 1440A using pClamp 10 software (both from Molecular Devices) at the sampling rate of 250 kHz. Patch pipettes with resistances of 2–5 MΩ were fabricated from borosilicate glass and filled with the following standard solution (in millimoles): KCl 126, MgCl2 2, CaCl2 0.5, EGTA 5, Hepes 25, pH 7.4. The standard bath solution contained (in millimoles): NaCl 150, CaCl2 1.8, MgCl2 1, glucose 5, Hepes 10, pH was adjusted with N-methyl-d-glucamine (NMG+) to the desired value. For analysis of intramolecular proton transfers, channel currents were eliminated by substituting NMG+ for all permeable metal cations in the pipette and bath solutions, the pH of which was adjusted to the same value of 7.4. To correct IE dependencies of channel current for slow inwardly directed proton movement, its amplitude was subtracted from the amplitude of channel current. To correct for different expression levels, the amplitude of slow inwardly directed current was scaled according to the amplitude of fast outwardly directed proton transfer. A 4 M KCl bridge was used in all experiments. All measurements were carried out at room temperature (25 °C). Liquid junction potentials were calculated using the built-in tool of pClamp 10. Continuous light pulses were provided by a Polychrome V light source (T.I.L.L. Photonics GmbH) in combination with a mechanical shutter (Uniblitz model LS6, Vincent Associates; half-opening time 0.5 ms). Maximal quantum density at the focal plane of the 40× objective lens was 7.7 mW/mm2 at 520 nm. Laser excitation was provided by a Minilite Nd:YAG laser (532 nm, pulsewidth 6 ns, energy 12 mJ; Continuum). A laser artifact was measured with a blocked optical path and digitally subtracted from the recorded traces. The signals were logarithmically averaged with a custom-created computer algorithm for further analysis. Curve fitting was performed using Origin 7 software (OriginLab).
Expression and Purification of GtCCR2 from P. pastoris.
The human-codon adapted opsin domain of GtCCR2 (amino acid residues 1–300) was fused in frame with a C-terminal eight-His tag and subcloned into the pPIC9K vector (Invitrogen) between EcoRI and NotI sites. Mutations were introduced using a QuikChange XL site-directed mutagenesis kit (Agilent Technologies) and verified by DNA sequencing. P. pastoris strain SMD1168 (his4, pep4) was transformed by electroporation with the resultant plasmids linearized by SalI. The transformants were screened according to the company protocol for their ability to grow on histidine-deficient medium and geneticin resistance. For each construct, a single colony that grew on 4 mg/mL geneticin and yielded the brightest color in a small-scale test was used for large-scale protein production, which was carried out as follows: A starter culture was inoculated into buffered complex glycerol medium and grown until A600 reached two to five. The cells were harvested by centrifugation at 2,975 × g and transferred to buffered complex methanol medium supplemented with 5 μM all-trans retinal (Sigma-Aldrich). Expression was induced by the addition of 0.5% methanol. The cells were harvested after 24 h and disrupted in a bead beater (BioSpec Products) or French press in buffer A (20 mM sodium phosphate, pH 7.4, 100 mM NaCl, 1 mM EDTA, 5% glycerol). Cell debris and unbroken cells were removed by centrifugation at 2,975 × g for 20 min. Membrane fragments were collected by ultracentrifugation at 40,000 rpm for 1 h in a Ti45 rotor (Beckman Coulter), resuspended in buffer B (20 mM Hepes, pH 7.0, 300 mM NaCl, 5% glycerol) and solubilized by incubation with 1% dodecyl maltoside (DDM) for 1 h at 4 °C. After removing nonsolubilized material by ultracentrifugation at 50,000 rpm for 1 h in a TLA-100 rotor (Beckman Coulter), the supernatant was mixed with nickel-nitrilotriacetic acid agarose beads (Qiagen) for 1 h and loaded on a column. The column was thoroughly washed with buffer C (20 mM Hepes, pH 7.0, 300 mM NaCl, 5% glycerol, 0.02% DDM) containing 40 mM imidazole, after which the protein was eluted with buffer C containing 300 mM imidazole. The pigment was concentrated and imidazole removed by repetitive washing with imidazole-free buffer C using YM-10 centrifugal filters (Amicon).
Flash-Induced Absorption Changes.
Laser flash-induced absorption changes were measured with a laboratory-constructed crossbeam apparatus. Excitation flashes (532 nm, 6 ns, up to 40 mJ) were provided by a Surelite I Nd-YAG laser (Continuum). Measuring light was from a 250-W incandescent tungsten lamp combined with a McPherson monochromator (model 272). A photomultiplier tube model R928 from Hamamatsu Photonics was protected from excitation laser flashes by a second monochromator of the same type and additionally with interference filters (12-nm bandwidth; Oriel Instruments). Signals were amplified by a low noise current amplifier (model SR445A, Stanford Research Systems) and digitized with a GaGe Octopus digitizer board (model CS8327, DynamicSignals), maximum sampling rate 50 MHz. The time interval between excitation flashes was 20 s, and up to 100 sweeps were averaged for each wavelength. Data analysis was performed with pClamp 10 (Molecular Devices) and OriginPro 7 (OriginLab) software. Logarithmic filtration of the data was performed using the GageCon program. Global fit of spectral transitions was performed using the FITEXP program provided by A. K. Dioumaev, University of California, Irvine, CA.
Supplementary Material
Acknowledgments
We thank Xin (Faith) Wang for technical assistance and Dr. Sergei P. Balashov (University of California, Irvine) for helpful discussions. This work was supported by NIH Grant R01GM027750 and Endowed Chair AU-0009 from the Robert A. Welch Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710702114/-/DCSupplemental.
References
- 1.Ernst OP, et al. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Govorunova EG, Sineshchekov OA, Li H, Spudich JL. Microbial rhodopsins: Diversity, mechanisms, and optogenetic applications. Annu Rev Biochem. 2017;86:845–872. doi: 10.1146/annurev-biochem-101910-144233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sineshchekov OA, Jung K-H, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagel G, et al. Channelrhodopsin-1: A light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 5.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–1225. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lórenz-Fonfría VA, Heberle J. Channelrhodopsin unchained: Structure and mechanism of a light-gated cation channel. Biochim Biophys Acta. 2014;1837:626–642. doi: 10.1016/j.bbabio.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Schneider F, Grimm C, Hegemann P. Biophysics of channelrhodopsin. Annu Rev Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 9.Kato HE, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erata M, Kubota M, Takahashi T, Inouye I, Watanabe M. Ultrastructure and phototactic action spectra of two genera of cryptophyte flagellate algae, Cryptomonas and Chroomonas. Protoplasma. 1995;188:258–266. [Google Scholar]
- 11.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. NEUROSCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govorunova EG, Sineshchekov OA, Spudich JL. Proteomonas sulcata ACR1: A fast anion channelrhodopsin. Photochem Photobiol. 2016;92:257–263. doi: 10.1111/php.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wietek J, Broser M, Krause BS, Hegemann P. Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. J Biol Chem. 2016;291:4121–4127. doi: 10.1074/jbc.M115.699637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govorunova EG, et al. The expanding family of natural anion channelrhodopsins reveals large variations in kinetics, conductance, and spectral sensitivity. Sci Rep. 2017;7:43358. doi: 10.1038/srep43358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sineshchekov OA, Govorunova EG, Li H, Spudich JL. Gating mechanisms of a natural anion channelrhodopsin. Proc Natl Acad Sci USA. 2015;112:14236–14241. doi: 10.1073/pnas.1513602112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sineshchekov OA, Li H, Govorunova EG, Spudich JL. Photochemical reaction cycle transitions during anion channelrhodopsin gating. Proc Natl Acad Sci USA. 2016;113:E1993–E2000. doi: 10.1073/pnas.1525269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahn M, Prigge M, Ron S, Levy R, Yizhar O. Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat Neurosci. 2016;19:554–556. doi: 10.1038/nn.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malyshev AY, et al. Chloride conducting light activated channel GtACR2 can produce both cessation of firing and generation of action potentials in cortical neurons in response to light. Neurosci Lett. 2017;640:76–80. doi: 10.1016/j.neulet.2017.01.026. [DOI] [PubMed] [Google Scholar]
- 19.Mohammad F, et al. Optogenetic inhibition of behavior with anion channelrhodopsins. Nat Methods. 2017;14:271–274. doi: 10.1038/nmeth.4148. [DOI] [PubMed] [Google Scholar]
- 20.Govorunova EG, Sineshchekov OA, Spudich JL. Structurally distinct cation channelrhodopsins from cryptophyte algae. Biophys J. 2016;110:2302–2304. doi: 10.1016/j.bpj.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamauchi Y, et al. Molecular properties of a DTD channelrhodopsin from Guillardia theta. Biophys Physicobiol. 2017;14:57–66. doi: 10.2142/biophysico.14.0_57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dér A, Keszthelyi L. Charge motion during the photocycle of bacteriorhodopsin. Biochemistry (Mosc) 2001;66:1234–1248. doi: 10.1023/a:1013179101782. [DOI] [PubMed] [Google Scholar]
- 24.Sineshchekov OA, Govorunova EG, Wang J, Li H, Spudich JL. Intramolecular proton transfer in channelrhodopsins. Biophys J. 2013;104:807–817. doi: 10.1016/j.bpj.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagel G, Möckel B, Büldt G, Bamberg E. Functional expression of bacteriorhodopsin in oocytes allows direct measurement of voltage dependence of light induced H+ pumping. FEBS Lett. 1995;377:263–266. doi: 10.1016/0014-5793(95)01356-3. [DOI] [PubMed] [Google Scholar]
- 26.Száraz S, Oesterhelt D, Ormos P. pH-induced structural changes in bacteriorhodopsin studied by Fourier transform infrared spectroscopy. Biophys J. 1994;67:1706–1712. doi: 10.1016/S0006-3495(94)80644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999;286:255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 28.Luecke H. Atomic resolution structures of bacteriorhodopsin photocycle intermediates: The role of discrete water molecules in the function of this light-driven ion pump. Biochim Biophys Acta Bioenerg. 2000;1460:133–156. doi: 10.1016/s0005-2728(00)00135-3. [DOI] [PubMed] [Google Scholar]
- 29.Freier E, Wolf S, Gerwert K. Proton transfer via a transient linear water-molecule chain in a membrane protein. Proc Natl Acad Sci USA. 2011;108:11435–11439. doi: 10.1073/pnas.1104735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, et al. Deprotonation of D96 in bacteriorhodopsin opens the proton uptake pathway. Structure. 2013;21:290–297. doi: 10.1016/j.str.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown LS. Reconciling crystallography and mutagenesis: A synthetic approach to the creation of a comprehensive model for proton pumping by bacteriorhodopsin. Biochim Biophys Acta Bioenerg. 2000;1460:49–59. doi: 10.1016/s0005-2728(00)00129-8. [DOI] [PubMed] [Google Scholar]
- 32.Maeda A, Gennis RB, Balashov SP, Ebrey TG. Relocation of water molecules between the Schiff base and the Thr46-Asp96 region during light-driven unidirectional proton transport by bacteriorhodopsin: An FTIR study of the N intermediate. Biochemistry. 2005;44:5960–5968. doi: 10.1021/bi047469h. [DOI] [PubMed] [Google Scholar]
- 33.Schobert B, Brown LS, Lanyi JK. Crystallographic structures of the M and N intermediates of bacteriorhodopsin: Assembly of a hydrogen-bonded chain of water molecules between Asp-96 and the retinal Schiff base. J Mol Biol. 2003;330:553–570. doi: 10.1016/s0022-2836(03)00576-x. [DOI] [PubMed] [Google Scholar]
- 34.Inoue K, et al. Converting a light-driven proton pump into a light-gated proton channel. J Am Chem Soc. 2015;137:3291–3299. doi: 10.1021/ja511788f. [DOI] [PubMed] [Google Scholar]
- 35.Balashov SP, et al. Light-driven Na(+) pump from Gillisia limnaea: A high-affinity Na(+) binding site is formed transiently in the photocycle. Biochemistry. 2014;53:7549–7561. doi: 10.1021/bi501064n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otto H, et al. Aspartic acid-96 is the internal proton donor in the reprotonation of the Schiff base of bacteriorhodopsin. Proc Natl Acad Sci USA. 1989;86:9228–9232. doi: 10.1073/pnas.86.23.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braiman MS, Dioumaev AK, Lewis JR. A large photolysis-induced pKa increase of the chromophore counterion in bacteriorhodopsin: Implications for ion transport mechanisms of retinal proteins. Biophys J. 1996;70:939–947. doi: 10.1016/S0006-3495(96)79637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balashov SP, et al. Glutamate-194 to cysteine mutation inhibits fast light-induced proton release in bacteriorhodopsin. Biochemistry. 1997;36:8671–8676. doi: 10.1021/bi970744y. [DOI] [PubMed] [Google Scholar]
- 39.Bamann C, Kirsch T, Nagel G, Bamberg E. Spectral characteristics of the photocycle of channelrhodopsin-2 and its implication for channel function. J Mol Biol. 2008;375:686–694. doi: 10.1016/j.jmb.2007.10.072. [DOI] [PubMed] [Google Scholar]
- 40.Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283:35033–35041. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 42.Govorunova EG, Sineshchekov OA, Li H, Janz R, Spudich JL. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga Platymonas subcordiformis. J Biol Chem. 2013;288:29911–29922. doi: 10.1074/jbc.M113.505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lórenz-Fonfría VA, et al. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc Natl Acad Sci USA. 2013;110:E1273–E1281. doi: 10.1073/pnas.1219502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barry PH. JPCalc, a software package for calculating liquid junction potential corrections in patch-clamp, intracellular, epithelial and bilayer measurements and for correcting junction potential measurements. J Neurosci Methods. 1994;51:107–116. doi: 10.1016/0165-0270(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 47.Dioumaev AK. Evaluation of intrinsic chemical kinetics and transient product spectra from time-resolved spectroscopic data. Biophys Chem. 1997;67:1–25. doi: 10.1016/s0301-4622(96)02268-5. [DOI] [PubMed] [Google Scholar]
- 48.Balashov SP. Protonation reactions and their coupling in bacteriorhodopsin. Biochim Biophys Acta Bioenerg. 2000;1460:75–94. doi: 10.1016/s0005-2728(00)00131-6. [DOI] [PubMed] [Google Scholar]
- 49.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757:1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.