Significance
Fluorine is an element that has been exploited for its unique properties in the design of therapeutic small molecules. As such, there is significant interest in understanding the interaction of organofluorine compounds with living systems. Here, we explore the metabolism of one of the few known fluorinated natural products, fluorothreonine, in its native host Streptomyces cattleya. While this amino acid is efficiently incorporated into protein in place of threonine, mistranslation is avoided through the action of a trans-acting aminoacyl-transfer RNA deacylase with 670-fold selectivity for the fluorine substituent. This finding demonstrates how new enzyme activities can be recruited to biosynthetic gene clusters to enable the evolution of biosynthetic capacity and expands the range of known organofluorine biochemistry.
Keywords: tRNA editing, translation, secondary metabolism, organofluorine
Abstract
Fluorine is an element with unusual properties that has found significant utility in the design of synthetic small molecules, ranging from therapeutics to materials. In contrast, only a few fluorinated compounds made by living organisms have been found to date, most of which derive from the fluoroacetate/fluorothreonine biosynthetic pathway first discovered in Streptomyces cattleya. While fluoroacetate has long been known to act as an inhibitor of the tricarboxylic acid cycle, the fate of the amino acid fluorothreonine is still not well understood. Here, we show that fluorothreonine can be misincorporated into protein in place of the proteinogenic amino acid threonine. We have identified two conserved proteins from the organofluorine biosynthetic locus, FthB and FthC, that are involved in managing fluorothreonine toxicity. Using a combination of biochemical, genetic, physiological, and proteomic studies, we show that FthB is a trans-acting transfer RNA (tRNA) editing protein, which hydrolyzes fluorothreonyl-tRNA 670-fold more efficiently than threonyl-RNA, and assign a role to FthC in fluorothreonine transport. While trans-acting tRNA editing proteins have been found to counteract the misacylation of tRNA with commonly occurring near-cognate amino acids, their role has yet to be described in the context of secondary metabolism. In this regard, the recruitment of tRNA editing proteins to biosynthetic clusters may have enabled the evolution of pathways to produce specialized amino acids, thereby increasing the diversity of natural product structure while also attenuating the risk of mistranslation that would ensue.
In adapting to a variety of ecological niches, living systems have evolved a wide range of chemical phenotypes. One striking example is the soil bacterium, Streptomyces cattleya, which biosynthesizes organofluorine compounds, including the antibiotics fluoroacetate and 4-fluorothreonine (FThr) (1). While the unique elemental properties of fluorine have driven its widespread use in synthetic small molecules, ranging from pharmaceuticals and imaging agents to polymers and liquid crystals (2–7), only a handful of natural products contain fluorine (8, 9). Given that S. cattleya is one of the few known genetic hosts for fluorine biology, this organism serves as a platform for exploring how the enzymatic utilization of this unusual element can be achieved.
While the products of organofluorine metabolism in S. cattleya are simple, they pose a challenging enzymatic selectivity problem due to their close structural resemblance to key central metabolites. For example, fluoroacetate has been show to manifest its toxicity through potent mechanism-based inhibition of the tricarboxylic acid cycle (10, 11). To avoid toxicity due to tricarboxylic acid cycle shutdown, S. cattleya employs both regulatory and enzymatic detoxification strategies (12–14). In comparison, the fate of FThr remains relatively unknown, although it provides a possible substrate for both amino acid metabolism and translation based on its structural similarity to threonine (15, 16).
In this work, we examine the downstream metabolism of FThr in S. cattleya, with a focus on understanding its interaction with the translational machinery. The low error rate of protein synthesis (∼10−3–10−4) is controlled largely by the ability of aminoacyl-transfer RNA (tRNA) synthetases (ARS) to correctly charge tRNA substrates with their cognate amino acid and discriminate against other near-cognate amino acids found in the cell (17). In addition to displaying selectivity at the level of amino acid activation, many of the ARSs also contain an editing domain for hydrolysis of misacylated tRNAs to further reduce errors in translation (18, 19). While this “double sieve” approach is able to limit the mischarging of commonly occurring amino acids, some xenobiotic amino acids can bypass these filters, enabling the in vitro and in vivo translation of polypeptides that contain a variety of noncanonical amino acids (20–23). In particular, the threonyl aminoacyl-tRNA synthetase (ThrRS) has been observed to display permissiveness toward γ-substituted analogs similar to FThr (16, 20).
We have used bioinformatic analysis to identify a conserved gene, SCAT_p0564 (fthB), in the fluorothreonine biosynthetic gene cluster of S. cattleya. This protein shares homology with freestanding aminoacyl-tRNA deacylases of the INS superfamily, which along with members of the AlaX and D-Tyr deacylase families, have been shown to provide hydrolytic editing activities in trans (24–26). Biochemical characterization of FthB shows that this enzyme hydrolyzes fluorothreonyl-tRNA with 670-fold selectivity over threonyl-tRNA, consistent with a role in excluding FThr from the proteome. Indeed, the deletion of fthB from the S. cattleya genome leads to significant charging of tRNAs with FThr and consequent incorporation of fluorothreonine into the proteome. Although the knockout strain does not show any apparent growth defect, the deletion of a transporter (SCAT_p0565, fthC) in the same gene cluster increases both intracellular FThr levels and sensitivity toward exogenous FThr. In contrast, heterologous expression of FthB in a nonfluorothreonine-producing streptomycete host does mitigate FThr toxicity. Taken together, these results are consistent with a model where S. cattleya has evolved cellular machinery consisting of a fluorothreonyl-tRNA deacylase and FThr-selective transporter to handle its unique fluorometabolism.
Results and Discussion
Conserved Genomic Context of Fluorothreonine Biosynthesis.
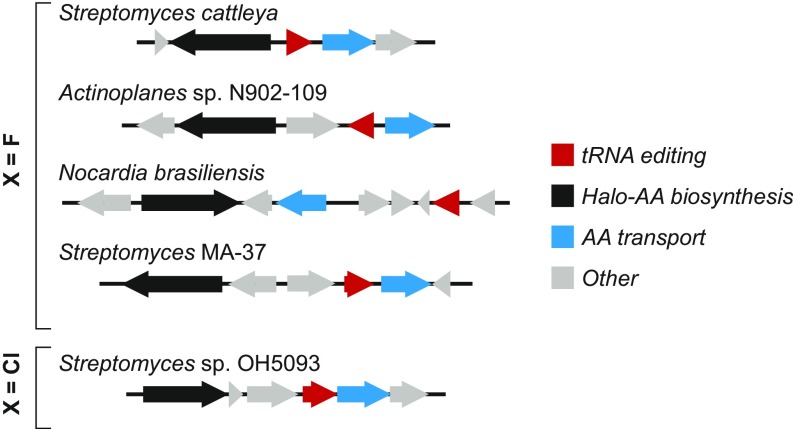
S. cattleya was the first documented genetic host for production of FThr, and remained the only known producer for many years (1). More recently, a number of bacterial genome sequences encoding organofluorine biosynthesis clusters have been published (27–30), enabling the use of comparative genomic approaches to identify new processes related to fluorometabolism. Using this approach to analyze genomes encoding the genes responsible for C–F bond formation (fluorinase) and FThr production (FThr transaldolase), we identified two previously uncharacterized conserved coding sequences (Fig. 1). These genes in S. cattleya, fthB and fthC, encode an aminoacyl-tRNA deacylating protein of the INS superfamily (NCBI CDD cl00022) and a putative amino acid exporter of the EamA-like superfamily (NCBI CDD cl23754), respectively.
Fig. 1.
Conservation analysis of fluorothreonine-associated proteins. A total of four gene clusters for fluorothreonine (X = F) biosynthesis were found within sequenced organisms. Organofluorine loci were identified by the presence of a fluorothreonine transaldolase CDS (black) and all were found to contain homologs of both the INS superfamily protein FthB (red) and the EamA-like protein FthC (blue). In addition, the biosynthetically unrelated chlorothreonine (X = Cl) gene cluster also contains similar organization with respect to these two functions.
Previous reports have described the role of INS superfamily proteins in maintaining translational fidelity (26, 31, 32). We were intrigued by the possibility that FthB could prevent the misincorporation of Fthr into proteins via hydrolysis of fluorothreonyl-tRNAThr arising from the acylation of FThr onto tRNAThr in place of Thr. The conservation of an amino acid transporter in the FThr biosynthetic cluster also suggested that it might serve a complementary role by exporting FThr from the cell. It is also interesting to note that the chlorothreonine biosynthesis cluster from Streptomyces sp. OH-5093, which shares no biosynthetic similarity with the fluorothreonine cluster, also contains an EamA-type transporter and an ORF with homology to the aminoacyl-tRNA deacylase AlaX (33) (Fig. 1). This functional conservation is consistent with both the tRNA-editing protein and transporter playing a role in adapting to the production of halogenated threonine analogs.
Phylogenetic analysis of close homologs (BLAST e-values < 1E-50) further supported the assignment of FthB as a fluorothreonine-associated protein. The sequences from organofluorine biosynthesis clusters group together to form a clade that contains just one protein that does not originate from a FThr transaldolase-containing gene cluster, suggesting that they may all descend from an ancestral FThr biosynthetic cluster (SI Appendix, Fig. S1). The majority of the remaining proteins are found in other Streptomyces species, and many are found within a set of five distinct conserved genomic contexts (SI Appendix, Fig. S2). Some of these contexts feature domains frequently associated with amino acid biosynthesis, and other editing proteins are clustered with nonribosomal peptide synthetase modules, which frequently employ specialized amino acid extender units to introduce structural diversity. These findings suggest the possibility that the recruitment of these freestanding tRNA editing proteins may be a common adaptation in natural product biosynthetic gene clusters that use or produce nonproteinogenic amino acids.
FthB Prevents Build-up of Fluorothreonyl-tRNA in Vitro.
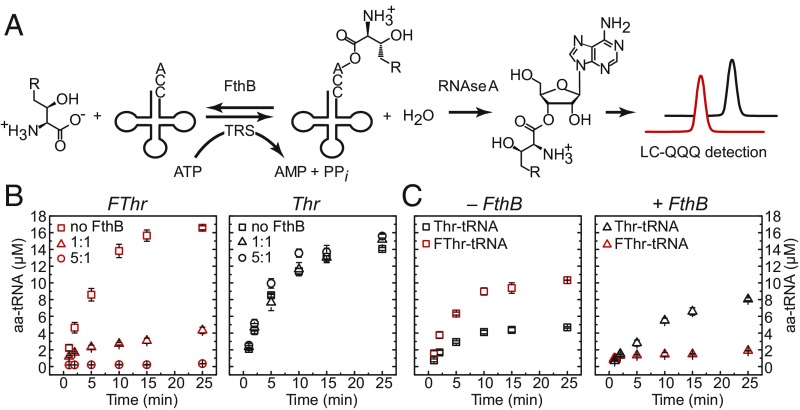
To characterize the role of FthB in fluorothreonine metabolism, we first examined its effects on aminoacylation in vitro. Both FthB and the ThrRS from S. cattleya were cloned, heterologously expressed in Escherichia coli, and purified to homogeneity (SI Appendix, Fig. S3). S. cattleya tRNAThr (GGU) was prepared by in vitro transcription from PCR-amplified template using the T7 polymerase, followed by urea-PAGE gel purification. To measure aminoacylation state, RNase A was used to cleave adenosine from the 3′ end of uncharged tRNA and 3′-O-threonyladenosine from threonyl-tRNA. These compounds were then analyzed by LC-MS/MS (34) (Fig. 2A and SI Appendix, Fig. S4). To enable quantification of the 3′-O-aminoacyladenosine esters, threonyl- and fluorothreonyl-tRNA standards were prepared and purified by anion exchange before measuring their concentration by amino acid analysis (SI Appendix, Fig. S5). Internal standards composed of aminoacyl tRNAs prepared with 13C-labeled ATP were also employed to extended the linear range of the assay and control for matrix-dependent and temporal variation in MS response (SI Appendix, Fig. S6).
Fig. 2.
Aminoacylation levels of tRNAThr with threonine (R = H) or fluorothreonine (R = F) as a function of FthB. (A) The amino acid was charged by 50 nM S. cattleya ThrRS and monitored by LC-MS/MS following enzymatic digestion with RNase A. In vitro transcribed tRNAThr was present at 24 μM in all reactions. (B) Aminoacylation of fluorothreonine (5 mM) and threonine (5 mM) in the presence of 0, 50, or 250 nM FthB. (C) Competitive assay containing equimolar fluorothreonine (5 mM) and threonine (5 mM) in the presence of 0 or 50 nM FthB. Each data point represents the mean ± SE (n ≥ 3).
This assay was applied to investigate the biochemical activity of FthB. In the absence of deacylating protein, FThr was observed to be an excellent substrate for the S. cattleya ThrRS, with 69 ± 1% charging achieved after 30 min in the presence of 50 nM ThrRS (Fig. 2B). When FthB was included in the aminoacylation reaction at a 1:1 molar ratio with respect to ThrRS, the time-dependent accumulation of fluorothreonyl-tRNA dropped dramatically to 18 ± 2%. Increase of FthB to a 5:1 molar ratio reduced the measured level of fluorothreonyl-tRNA below the calibrated range of the assay (<2%). In contrast, 58.6 ± 0.7% charging of threonine was achieved in the absence of FthB under the same conditions, and the tRNA aminoacylation state remained unaffected by even the highest level of FthB (Fig. 2B).
This experiment was also run under competitive conditions at equimolar concentrations of FThr and Thr (5 mM), because both aminoacyl-tRNA–derived species could be quantified simultaneously based on their difference in mass. While the total pool of charged tRNA reached approximately the same level as with the individual amino acids (62.6 ± 0.4% total charging), FThr (43.1 ± 0.1%) was able to outcompete Thr (19.0 ± 0.4%) for aminoacylation in the absence of FthB (Fig. 2C). When FthB was added at a 1:1 ratio with ThrRS, threonyl-tRNA (33.5 ± 0.5%) was then able to accumulate compared with fluorothreonyl-tRNA (7.9 ± 0.1%) (Fig. 2C). However, FThr remained inhibitory to Thr aminoacylation, which is consistent with effective competition by FThr for the charging reaction. Together, these results show that FthB can selectively hydrolyze fluorothreonyl- over threonyl-tRNA, and could potentially control the in vivo aminoacylation balance between these two amino acids.
The Fluorothreonyl-tRNA Selectivity of FthB Is Primarily Mediated by kcat.
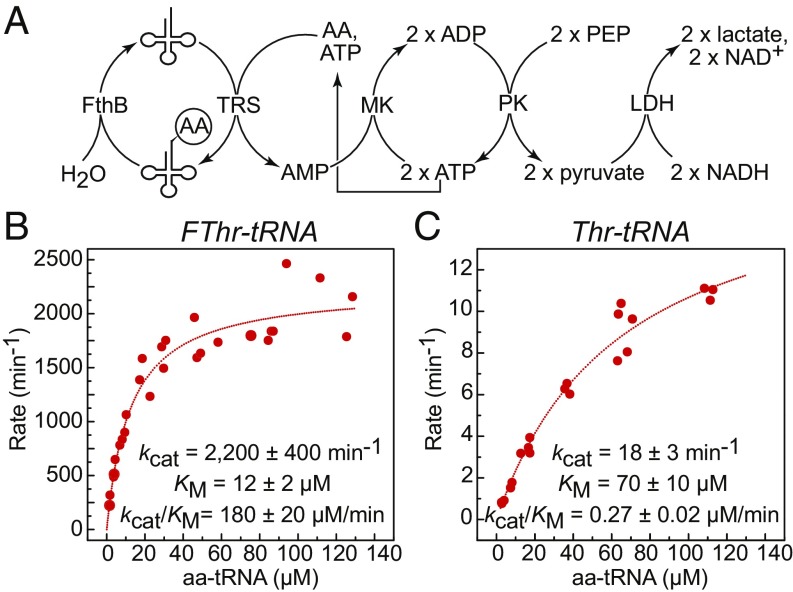
To further characterize the selectivity of FthB between fluorothreonyl- and threonyl-tRNA, a spectrophotometric assay employing a coupled aminoacylation/deacylation system was developed based on a similar assay reported for TyrRS (35). In this assay, the steady-state rate of deacylation was measured by coupling the hydrolysis of aminoacyl-tRNA to the aminoacylation reaction that occurs upon release of the free tRNA. The subsequent production of AMP results in NADH consumption by pyruvate kinase/lactate dehydrogenase, enabling a spectrophotometric readout of the reaction (Fig. 3A). In addition to monitoring the rate of the reaction, the concentration of the aminoacyl-tRNA substrate was measured directly by sampling from the cuvette, followed by quenching and LC-MS/MS analysis, as described above.
Fig. 3.
Steady-state kinetic analysis of the deacylation of fluorothreonyl- and threonyl-tRNA by FthB. (A) Coupled assay for monitoring aminoacyl-tRNA deacylation. Charged tRNA substrates were generated in situ by S. cattleya ThrRS. Enzymatic rates were measured spectrophotometrically by coupling AMP release to NADH consumption while substrate concentrations were measured by LCMS. (B) Hydrolysis of fluorothreonyl-tRNAThr. (C) Hydrolysis of threonyl-tRNAThr.
Using this assay, we discovered that FthB discriminates between fluorothreonyl- and threonyl tRNA with a 670-fold difference in kcat/KM (Fig. 3 B and C). The majority of selectivity is accounted for by a 130-fold increase in kcat with respect to the fluorinated substrate, with a lesser contribution from KM (Fig. 3 B and C). Interestingly, this behavior is similar to that observed for the fluoroacetyl-CoA thioesterase (FlK), an enzyme involved in fluoroacetate detoxification from S. cattleya that also displays high selectivity toward a single fluorine substitution for hydrogen (14, 36). The observed KM value for the fluorinated substrate (12 ± 2 μM) is slightly higher than the estimated in vivo concentration of threonyl-tRNA in bacterial cells, indicating that FthB is active at relevant substrate concentrations (37). Relative to other characterized proteins from the INS superfamily, the catalytic rate of FthB toward fluorothreonyl-tRNA (2,200 ± 400 min−1) appears to be high, although a direct comparison cannot be made. It has been noted that high enzyme concentrations must be employed to achieve appreciable rates of hydrolysis for many superfamily members (31). For example, 1 µM YbaK is required to achieve ∼50% hydrolysis of 0.2 μM Cys-tRNAPro in 2.5 min, corresponding to a rate of ∼0.04 min−1 (26). Under similar conditions, 5 nM FthB can hydrolyze >50% of 5 µM Fthr-tRNA in under 2 min. FthB is thus more comparable to the D-Tyr deacylase in catalytic activity, although these proteins are from different superfamilies (38). It is intriguing to speculate that this difference may be the result of selective pressure. The preferred substrate of YbaK, Cys-tRNAPro, is produced slowly by ProRS, so even a relatively slow hydrolytic enzyme is sufficient to prevent the accumulation of misacylated tRNA. In contrast, fluorothreonyl-tRNA must be hydrolyzed quickly because it is produced more quickly thanks to the competence of fluorothreonine as a substrate for ThrRS.
Elucidating the Contribution of FthB and FthC to S. cattleya Fluorothreonine Physiology.
With the capacity of FthB to preferentially hydrolyze fluorothreonyl-tRNA established in vitro, we sought to explore its function in vivo. We initiated these experiments by deleting the gene encoding FthB in the S. cattleya genome using the REDIRECT method (39) (SI Appendix, Fig. S7). In addition, we deleted the putative FThr transporter (fthC) to test its contribution to S. cattleya fluorothreonine physiology. The resulting ΔfthB::AmR and ΔfthC::AmR strains were grown alongside the WT strain for 6 d in GYM media supplemented with 2 mM sodium fluoride and all three strains displayed comparable growth and viability (SI Appendix, Figs. S8 and S9). Competitive growth experiments also confirmed that the WT and ΔfthB strains display similar growth patterns in the presence and absence of fluoride (SI Appendix, Fig. S10). After 6 d, cultures were harvested and the markers for FThr physiology were assessed.
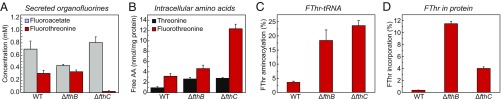
We analyzed these samples for intracellular and secreted FThr as well as its abundance in the aminoacyl-tRNA and protein pool. The concentrations of fluoroacetate and FThr in culture supernatant were monitored with 19F-NMR spectroscopy (Fig. 4A and SI Appendix, Fig. S11). The ΔfthB::AmR strain produced concentrations of FThr and fluoroacetate comparable to the WT strain, while the ΔfthC::AmR strain showed dramatically reduced secretion of FThr (P < 0.01). Intracellular levels of FThr and Thr were then quantified using the ratio of free amino acid to soluble protein to control for any differences in cell pellet recovery or lysis efficiency. These measurements showed that intracellular FThr content was comparable in WT and ΔfthB::AmR strains and significantly elevated (P < 0.001) in the ΔfthC::AmR strain, with a fourfold increase over WT (Fig. 4B). These data are consistent with the proposed role of FthC as a FThr exporter. Because the FThr secretion and accumulation phenotypes of the ΔfthB::AmR strain are more similar to those of the WT, it is likely that polar effects on the expression FthC are minimal. In contrast, intracellular Thr levels displayed a modest but significant increase (P < 0.01) in both knockout strains compared with the WT, which may be indicative of a broader regulatory response to FThr-related stress.
Fig. 4.
Analysis of fluorothreonine physiological markers in WT, ΔfthB::AmR, and ΔfthC::AmR S. cattleya strains after onset of organofluorine production (6 d). (A) Levels of secreted fluoroacetate and fluorothreonine in culture supernatant measured by 19F NMR. (B) Intracellular amino acid levels in cell pellet lysates measured by LC-MS/MS. (C) Fluorothreonyl-tRNA abundance compared with total fluorothreonyl- and threonyl-tRNA measured by RNase digestion followed by LC-MS/MS. (D) Incorporation of fluorothreonine into the total protein fraction measured by LC-MS/MS analysis of total protein hydrolysate. Data are mean ± SE (n = 4).
Whole RNA was prepared from cell pellets, digested with RNase A, and characterized by LC-MS to assess tRNA aminoacylation state. The baseline fluorothreonyl-tRNA level in WT was observed to be 4 ± 1% of the total FThr- and Thr-charged tRNA pool. This fraction increased five- to sevenfold upon deletion of the fluorothreonyl-tRNA deacylase (18 ± 7%) and the FThr transporter (23 ± 3%) (Fig. 4C). These results, like the in vitro aminoacylation experiments, support the hypothesis that FThr competes well for aminoacylation onto tRNA. Furthermore, it appears that that the elevated Fthr concentration resulting from knockout of the FThr transporter can overwhelm the trans-editing activity of FthB. The ability of fluorothreonyl-tRNA to be incorporated into protein was subsequently examined by amino acid analysis of total protein hydrolysate using LC-MS/MS, as described above (SI Appendix, Fig. S5). Hydrolyzed whole-protein samples contained very little fluorothreonine in WT S. cattleya (0.36 ± 0.02%) compared with the knockout strains (Fig. 4D). Strikingly, the ΔfthB::AmR strain exhibited the highest level of FThr incorporation (11.5 ± 0.2%) with respect to total Thr and FThr, whereas ΔfthC::AmR showed FThr to a lower extent (4.0 ± 0.3%). As the observed FThr content in protein does not scale linearly with the charging of FThr onto tRNA, the strains examined may exhibit time-dependent differences in FThr incorporation. From these studies, we conclude that FthB plays a role in preventing FThr mistranslation in vivo by reducing the intracellular levels of fluorothreonyl-tRNA.
Targeted and Shotgun Proteomic Experiments to Examine FThr Incorporation in the Proteome.
FThr incorporation into protein was confirmed by targeted proteomic experiments on the WT and ΔfthB::AmR strains. The most readily observed peptides from four extremely abundant proteins were selected for analysis (SI Appendix, Fig. S12 and Table S2). While the ability to make quantitative inferences about incorporation rates is limited in the absence of synthetic standards, the results were consistent with the data obtained from amino acid analysis of hydrolyzed protein. Peak areas for transitions corresponding to Thr- and FThr-containing peptides were tabulated and used to estimate FThr incorporation, which varied between 0.03% and 0.32% for WT and 2.0% and 12.9% for the fluorothreonyl-tRNA deacylase knockout strain. Higher FThr incorporation was observed in the ΔfthB::AmR strain across all peptides measured (Table 1).
Table 1.
Estimated fluorothreonine incorporation percentages in select proteins measured by targeted proteomics of WT and ΔfthB S. cattleya strains
| Protein/peptide | WT (% FThr) | ΔfthB (% FThr) |
| GapA | ||
| AAAENIIPTTTGAAK | 0.07 ± 0.02 | 3.4 ± 1.1 |
| LVDLTTFVGGR | 0.03 ± 0.01 | 2.0 ± 1.1 |
| ATALVIPELK | 0.10 ± 0.003 | 4.4 ± 0.6 |
| FadH | ||
| ILSYIDIGTAEGAK | 0.31 ± 0.02 | 10.9 ± 0.7 |
| VLTGGER | 0.31 ± 0.03 | 10.5 ± 0.5 |
| VDLGGSLSGGYYVAPTIFEGDNR | 0.36 ± 0.07 | 10.3 ± 0.7 |
| IFQEEIFGPVVSVTR | 0.3 ± 0.1 | 10.7 ± 0.7 |
| DLSTAYR | 0.49 ± 0.09 | 15.6 ± 1.8 |
| TufA | ||
| TTLTAAITK | 0.18 ± 0.03 | 6.5 ± 0.4 |
| LLGLMHTIDEAIPTPQR | 0.23 ± 0.03 | 8.2 ± 0.7 |
| VNETVDIIGIK | 0.22 ± 0.03 | 7.3 ± 0.6 |
| FlA | ||
| FFPEGTVFATTTYPATGTTTR | 0.42 ± 0.04 | 9.5 ± 0.2 |
| VIPEQPEPTFYSR | 0.32 ± 0.04 | 12.9 ± 0.7 |
Transitions corresponding to a set of FThr- and Thr-containing peptides from abundant proteins were monitored, and relative peak areas were used to estimate the rate of incorporation. All measurements were performed with n ≥ 3; values are reported as mean ± SE.
The global landscape of FThr incorporation into protein was probed in an unbiased fashion with shotgun proteomics. Analysis of tryptic peptides prepared from the ΔfthB::AmR strain yielded many high-quality peptide-spectrum matches (PSMs) corresponding to fluorothreonine-containing peptides, and manual examination of MS/MS spectra served to corroborate these assignments (Table 2 and SI Appendix, Fig. S13). As expected, FThr was found much less frequently in samples from the WT strain (Table 2). Data from the FThr-rich ΔfthB::AmR samples was further scrutinized to examine possible drivers of FThr incorporation. Given the extreme GC-rich bias exhibited at the wobble position of Streptomyces codons, differential incorporation by codon was considered as one possibility. Examination of the codon distribution of fluorothreonine- and threonine-containing PSMs in single-threonine coding peptides found quite similar codon distributions in both cases (SI Appendix, Table S3). Peptide- and protein-level incorporation was also estimated using precursor ion peak areas determined by the LFQ routine of MaxQuant (40). These results suggest that the integration of FThr varies across the proteome, with mean and median values consistent with the rate of incorporation measured by amino acid analysis (SI Appendix, Fig. S14). Despite this variation, very few proteins have significantly elevated FThr content, possibly due to low signal-to-noise ratios for peaks corresponding to FThr-containing peptides (SI Appendix, Table S4). Differential accumulation of FThr in proteins on the basis of protein function was also considered; to this end, Clusters of Orthologous Groups gene ontology assignments for proteins with threonine- and fluorothreonine-containing PSMs were tabulated. Again, a similar distribution was observed for both WT and ΔfthB::AmR strains (SI Appendix, Fig. S15). These results are consistent with an underlying random incorporation of FThr into the proteome, in the context of changing intracellular FThr concentrations.
Table 2.
Assessment of fluorothreonine incorporation across the proteome of S. cattleya measured by shotgun proteomics of WT and ΔfthB S. cattleya strains
| Strain | PSM or protein | FThr | Total |
| WT | PSMs | 20 ± 10 | 25,000 ± 3,000 |
| Proteins | 9 ± 2 | 1,630 ± 90 | |
| ΔfthB | PSMs | 1,800 ± 300 | 24,000 ± 3,000 |
| Proteins | 300 ± 20 | 1,600 ± 100 |
Database searches were performed with FThr for Thr substitution as a variable modification. The number of fluorothreonine-containing and total peptide spectrum matches and proteins IDs were then tabulated. All measurements were performed with n = 4; values are reported as mean ± SE.
FthB Expression Mitigates Fluorothreonine Toxicity in a Heterologous Streptomyces Host.
The role of the fluorothreonyl-tRNA deacylase in the context of FThr toxicity was probed via expression in a model Streptomyces species. Consistent with prior reports (1), FThr produces a zone of inhibition when applied to freshly plated spores of Strepomyces coelicolor M1152 on media with limited amino acid availability (Table 3 and SI Appendix, Fig. 16). This inhibition can be partially alleviated by the genomic integration of FthB under the control of the constitutive ermEp* promoter. In comparison, the negative control with a strain where only the ermEp* promoter is integrated shows no evidence of protection against FThr toxicity. The effect of FthB expression is even more pronounced in liquid culture, although quantification of growth is impeded by the clumpy phenotype characteristic of streptomycetes (SI Appendix, Fig. S17). FThr also inhibits the growth of the producing strain S. cattleya in disk diffusion assays, despite its ability to accumulate near-millimolar concentrations of this compound in fermentation media. While deletion of the fluorothreonyl-tRNA deacylase does not result in any change in FThr growth inhibition, deletion of the FThr transporter causes a marked increase in the zone of inhibition (ZOI) (Table 3 and SI Appendix, Fig. 16). Given the lower level of FThr incorporation into the proteome in the transporter knockout strain (Fig. 4D), it is possible that targets beyond translation may play a role in growth defects observed upon the addition of exogenous FThr. However, the ability of FthB to mitigate fluorothreonine toxicity in S. coelicolor M1152 suggests that it is mediated in part by the misacylation of FThr onto tRNAThr.
Table 3.
Growth inhibition of Streptomyces coelicolor M1152 and S. cattleya strains by fluorothreonine
| S. coelicolor | ZOI (mm) | S. cattleya | ZOI (mm) |
| WT | 50 ± 1 | WT | 49 ± 1 |
| ermEp*-empty | 53 ± 1 | ΔfthB::AmR | 51 ± 1 |
| ermEp*-FthB | 13 ± 2 | ΔfthC::AmR | 66 ± 1 |
Spores were plated on Hopwood media, and 200 μg fluorothreonine was applied via filter paper before incubation at 30 °C. Inhibition assays were performed in triplicate; results show mean ± SE.
Conclusion
The unusual ability of S. cattleya to utilize fluorine places a high demand on the cellular machinery to evolve new enzymes to handle the challenging selectivity problem of distinguishing a single substitution of fluorine for hydrogen. The well-characterized poison fluoroacete provides one such example, but the amino acid fluorothreonine also has potent antibiotic activity. In this work, we explore the mechanisms evolved by S. cattleya to alleviate this toxicity and report the discovery of a fluorine-selective aminoacyl-tRNA editing protein (SCAT_p0564, FthB) as well as a FThr transporter (SCAT_p0565, FthC) that are involved in this process.
FThr represents a natural candidate for mistranslation by ThrRS, and we find indeed that it is efficiently aminoacylated in vitro and in vivo. Biochemical characterization of FthB shows that it can hydrolyze fluorothreonyl-tRNA with 670-fold selectively in kcat/KM compared with threonyl-tRNA, suggesting that its physiological function is to protect against misacylation of FThr onto tRNAThr. The high selectivity of FthB with respective to the single γ-fluorine substitution is quite striking. In comparison, other naturally occurring fluorine-selective enzymes target the C–F bond itself in the case of the haloacetate dehalogenase, or hydrolyze a bond vicinal to the fluorine substituent in the case of the fluoroacetyl-CoA thioesterase, FlK (36, 41) In contrast, the scissile bond of fluorothreonyl-tRNA is located quite distantly from the fluorine atom yet still enables a 102-fold effect on kcat. While the catalytic basis for recognizing this subtle structural change remains unknown, it is possible that a difference in the pKa of the β-hydroxyl group between threonyl- and fluorothreonyl-tRNA could be exploited to direct substrate positioning or to directly facilitate hydrolysis. In addition, other features, such as the anticodon stem or the tRNA-synthetase complex, could tune recognition as found in other members of the INS superfamily (42, 43).
Genetic and physiological studies show that deletion of the gene encoding the fluorothreonyl-tRNA deacylase leads to an increase in the levels of tRNA charged with FThr, as well as FThr incorporation into the proteome. In the absence of this activity, these two markers rise to quite high levels of ∼18% and ∼11.5%, respectively. Further analysis demonstrates that FThr is indeed introduced at Thr sites across the proteome. At this time, we have not identified any significant bias in codon usage or protein identity that would indicate a physiological function for the introduction of FThr into ribosomally synthesized proteins and peptides. Despite this extensive mistranslation, the ΔfthB strain does not suffer a loss of fitness under standard growth conditions. Although initially surprising, previous studies of mistranslation-prone E. coli systems have indicated that additional stress may be required to reveal a growth defect (44, 45). In contrast, the overexpression of FthB in a heterologous host results in protection from toxicity due to exogenous FThr, although native levels of FthB are insufficient to provide a protective effect in S. cattleya. We have also assigned the function of FthC as a FThr transporter. Deletion of FthC leads to a significant increase in intracellular FThr concentration and increased toxicity associated with the addition of exogenous FThr. Taken together, the data suggest a model where the FThr transporter is required to avoid accumulation of critically toxic intracellular levels of FThr and that the fluorothreonyl-tRNA deacylase protects against the accumulation of FThr-tRNAThr.
These two functions of transport and tRNA editing are conserved across other FThr and chlorothreonine biosynthetic gene clusters, and thus appear to facilitate the management of halogenated amino acids that are closely related to a proteinogenic amino acid. Under normal conditions, the ARS are able to differentiate between cognate and noncognate substrates at the level of aminoacylation, or by hydrolytic editing domains encoded on the same polypeptide. However, trans-acting hydrolytic editing proteins have been found to resolve particularly difficult discrimination problems arising from the misacylation of near-cognate primary metabolites. For example, AlaX-like proteins can hydrolyze Ser-tRNAAla, while members of the INS superfamily have been shown to act on various substrates including misacylated cysteinyl-, threonyl-, seryl-, and aminobutyryl-tRNA (25, 26, 32, 46). In contrast to these housekeeping functions, FthB participates in mediating secondary metabolism, a finding that gives rise to the possibility that related proteins have been adapted to prevent mistranslation and enable the usage of specialized amino acids for natural product biosynthesis. Indeed, we find that homologous proteins are widespread in other streptomycetes, suggesting that there are other cases where tRNA editing proteins have been recruited to secondary metabolic clusters that induce novel translational stresses.
Experimental Procedures
Detailed procedures for cell culture, plasmid construction, gene deletion, protein purification, in vitro transcription, enzyme assays, organofluorine quantification, amino acid quantification, proteomics, and fluorothreonine toxicity can be found in SI Appendix.
Supplementary Material
Acknowledgments
This work was funded by the generous support of the National Institutes of Health (Grant R01 GM123181). J.L.M. also acknowledges the support of a National Institutes of Health National Research Service Award Training Grant 1 T32 GMO66698. The College of Chemistry NMR Facility at the University of California, Berkeley is supported in part by the National Institutes of Health (Grants 1S10RR023679-01 and S10 RR16634-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711482114/-/DCSupplemental.
References
- 1.Sanada M, et al. Biosynthesis of fluorothreonine and fluoroacetic acid by the thienamycin producer, Streptomyces cattleya. J Antibiot (Tokyo) 1986;39:259–265. doi: 10.7164/antibiotics.39.259. [DOI] [PubMed] [Google Scholar]
- 2.Müller K, Faeh C, Diederich F. Fluorine in pharmaceuticals: Looking beyond intuition. Science. 2007;317:1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 3.O’Hagan D. Understanding organofluorine chemistry. An introduction to the C-F bond. Chem Soc Rev. 2008;37:308–319. doi: 10.1039/b711844a. [DOI] [PubMed] [Google Scholar]
- 4.Purser S, Moore PR, Swallow S, Gouverneur V. Fluorine in medicinal chemistry. Chem Soc Rev. 2008;37:320–330. doi: 10.1039/b610213c. [DOI] [PubMed] [Google Scholar]
- 5.Furuya T, Kamlet AS, Ritter T. Catalysis for fluorination and trifluoromethylation. Nature. 2011;473:470–477. doi: 10.1038/nature10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger R, Resnati G, Metrangolo P, Weber E, Hulliger J. Organic fluorine compounds: A great opportunity for enhanced materials properties. Chem Soc Rev. 2011;40:3496–3508. doi: 10.1039/c0cs00221f. [DOI] [PubMed] [Google Scholar]
- 7.Le Bars D. Fluorine-18 and medical imaging: Radiopharmaceuticals for positron emission tomography. J Fluor Chem. 2006;127:1488–1493. [Google Scholar]
- 8.O’Hagan DB, Harper D. Fluorine-containing natural products. J Fluor Chem. 1999;100:127–133. [Google Scholar]
- 9.Gribble GW. The diversity of naturally produced organohalogens. Chemosphere. 2003;52:289–297. doi: 10.1016/S0045-6535(03)00207-8. [DOI] [PubMed] [Google Scholar]
- 10.Clarke DD. Fluoroacetate and fluorocitrate: Mechanism of action. Neurochem Res. 1991;16:1055–1058. doi: 10.1007/BF00965850. [DOI] [PubMed] [Google Scholar]
- 11.Lauble H, Kennedy MC, Emptage MH, Beinert H, Stout CD. The reaction of fluorocitrate with aconitase and the crystal structure of the enzyme-inhibitor complex. Proc Natl Acad Sci USA. 1996;93:13699–13703. doi: 10.1073/pnas.93.24.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker MC, Wen M, Weeks AM, Chang MCY. Temporal and fluoride control of secondary metabolism regulates cellular organofluorine biosynthesis. ACS Chem Biol. 2012;7:1576–1585. doi: 10.1021/cb3002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang F, et al. The gene cluster for fluorometabolite biosynthesis in Streptomyces cattleya: A thioesterase confers resistance to fluoroacetyl-coenzyme A. Chem Biol. 2006;13:475–484. doi: 10.1016/j.chembiol.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Weeks AM, Chang MCY. Catalytic control of enzymatic fluorine specificity. Proc Natl Acad Sci USA. 2012;109:19667–19672. doi: 10.1073/pnas.1212591109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb HK, Matthews RG. 4-Chlorothreonine is substrate, mechanistic probe, and mechanism-based inactivator of serine hydroxymethyltransferase. J Biol Chem. 1995;270:17204–17209. doi: 10.1074/jbc.270.29.17204. [DOI] [PubMed] [Google Scholar]
- 16.Minajigi A, Deng B, Francklyn CS. Fidelity escape by the unnatural amino acid β-hydroxynorvaline: An efficient substrate for Escherichia coli threonyl-tRNA synthetase with toxic effects on growth. Biochemistry. 2011;50:1101–1109. doi: 10.1021/bi101360a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 18.Fersht AR, Kaethner MM. Enzyme hyperspecificity. Rejection of threonine by the valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry. 1976;15:3342–3346. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 19.Bullwinkle T, Lazazzera B, Ibba M. Quality control and infiltration of translation by amino acids outside of the genetic code. Annu Rev Genet. 2014;48:149–166. doi: 10.1146/annurev-genet-120213-092101. [DOI] [PubMed] [Google Scholar]
- 20.Hartman MCT, Josephson K, Szostak JW. Enzymatic aminoacylation of tRNA with unnatural amino acids. Proc Natl Acad Sci USA. 2006;103:4356–4361. doi: 10.1073/pnas.0509219103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melo Czekster C, Robertson WE, Walker AS, Söll D, Schepartz A. In vivo biosynthesis of a β-amino acid-containing protein. J Am Chem Soc. 2016;138:5194–5197. doi: 10.1021/jacs.6b01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenthal GA, Dahlman DL, Janzen DH. A novel means for dealing with L-canavanine, a toxic metabolite. Science. 1976;192:256–258. doi: 10.1126/science.1257764. [DOI] [PubMed] [Google Scholar]
- 24.Calendar R, Berg P. D-Tyrosyl RNA: Formation, hydrolysis and utilization for protein synthesis. J Mol Biol. 1967;26:39–54. doi: 10.1016/0022-2836(67)90259-8. [DOI] [PubMed] [Google Scholar]
- 25.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 27.Vera-Cabrera L, Ortiz-Lopez R, Elizondo-Gonzalez R, Perez-Maya AA, Ocampo-Candiani J. Complete genome sequence of Nocardia brasiliensis HUJEG-1. J Bacteriol. 2012;194:2761–2762. doi: 10.1128/JB.00210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng H, et al. Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining. ChemBioChem. 2014;15:364–368. doi: 10.1002/cbic.201300732. [DOI] [PubMed] [Google Scholar]
- 29.Huang S, et al. Fluoroacetate biosynthesis from the marine-derived bacterium Streptomyces xinghaiensis NRRL B-24674. Org Biomol Chem. 2014;12:4828–4831. doi: 10.1039/c4ob00970c. [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Ren S-X, Yang S, Hu H-F. Comparative analysis of rapamycin biosynthesis clusters between Actinoplanes sp. N902-109 and Streptomyces hygroscopicus ATCC29253. Chin J Nat Med. 2015;13:90–98. doi: 10.1016/S1875-5364(15)60012-7. [DOI] [PubMed] [Google Scholar]
- 31.Ruan B, Söll D. The bacterial YbaK protein is a Cys-tRNAPro and Cys-tRNACys deacylase. J Biol Chem. 2005;280:25887–25891. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, et al. Homologous trans-editing factors with broad tRNA specificity prevent mistranslation caused by serine/threonine misactivation. Proc Natl Acad Sci USA. 2015;112:6027–6032. doi: 10.1073/pnas.1423664112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fullone MR, et al. Insight into the structure-function relationship of the nonheme iron halogenases involved in the biosynthesis of 4-chlorothreonine–Thr3 from Streptomyces sp. OH-5093 and SyrB2 from Pseudomonas syringae pv. syringae B301DR. FEBS J. 2012;279:4269–4282. doi: 10.1111/febs.12017. [DOI] [PubMed] [Google Scholar]
- 34.Chen YG, Kowtoniuk WE, Agarwal I, Shen Y, Liu DR. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat Chem Biol. 2009;5:879–881. doi: 10.1038/nchembio.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson CJ, First EA. A continuous tyrosyl-tRNA synthetase assay that regenerates the tRNA substrate. Anal Biochem. 2015;486:86–95. doi: 10.1016/j.ab.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Weeks AM, Coyle SM, Jinek M, Doudna JA, Chang MCY. Structural and biochemical studies of a fluoroacetyl-CoA-specific thioesterase reveal a molecular basis for fluorine selectivity. Biochemistry. 2010;49:9269–9279. doi: 10.1021/bi101102u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakubowski H, Goldman E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J Bacteriol. 1984;158:769–776. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad S, et al. Mechanism of chiral proofreading during translation of the genetic code. eLife. 2013;2:e01519. doi: 10.7554/eLife.01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 41.Liu J-Q, Kurihara T, Miyagi M, Esaki N, Soda K. Reaction mechanism of L-2-haloacid dehalogenase of Pseudomonas sp. YL. Identification of Asp10 as the active site nucleophile by 18O incorporation experiments. J Biol Chem. 1995;270:18309–18312. [PubMed] [Google Scholar]
- 42.Vargas-Rodriguez O, Musier-Forsyth K. Exclusive use of trans-editing domains prevents proline mistranslation. J Biol Chem. 2013;288:14391–14399. doi: 10.1074/jbc.M113.467795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An S, Musier-Forsyth K. Cys-tRNA(Pro) editing by Haemophilus influenzae YbaK via a novel synthetase. YbaK.tRNA ternary complex. J Biol Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 44.Bacher JM, de Crécy-Lagard V, Schimmel PR. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci USA. 2005;102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruan B, et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bacusmo JM, et al. Quality control by trans-editing factor prevents global mistranslation of non-protein amino acid α-aminobutyrate. RNA Biol. July 24, 2017 doi: 10.1080/15476286.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.