Significance
Control of mRNA synthesis and decay is crucial for cells to adapt to their environment and for proper development. The 5′ end of eukaryotic mRNAs is modified by a structure called cap that protects them from rapid and uncontrolled decay. During mRNA decay, this cap is removed by a specialized and finely regulated multiprotein factory called decapping complex. Our results support a model in which the two major enzymes responsible for mRNA decapping (Dcp2) and decay (Xrn1) are sequentially recruited to mRNAs by the same surface from Pat1, a scaffolding protein central for decapping. As this Pat1 region is important for growth and specific to fungi, this is a potential target for the development of drugs against pathogenic yeasts.
Keywords: mRNA decapping, eukaryotic mRNA decay, protein–protein interaction, yeast
Abstract
The Pat1 protein is a central player of eukaryotic mRNA decay that has also been implicated in translational control. It is commonly considered a central platform responsible for the recruitment of several RNA decay factors. We demonstrate here that a yeast-specific C-terminal region from Pat1 interacts with several short motifs, named helical leucine-rich motifs (HLMs), spread in the long C-terminal region of yeast Dcp2 decapping enzyme. Structures of Pat1–HLM complexes reveal the basis for HLM recognition by Pat1. We also identify a HLM present in yeast Xrn1, the main 5′–3′ exonuclease involved in mRNA decay. We show further that the ability of yeast Pat1 to bind HLMs is required for efficient growth and normal mRNA decay. Overall, our analyses indicate that yeast Pat1 uses a single binding surface to successively recruit several mRNA decay factors and show that interaction between those factors is highly polymorphic between species.
Messenger RNA (mRNA) decay is a highly regulated process that finely tunes protein production, contributing thereby to numerous processes including cell cycle control, cellular responses to environmental cues, and development. In eukaryotes, mature cytoplasmic mRNAs are protected from rapid and uncontrolled degradation by two main cis-acting stability determinants: a 7-methylguanosine cap (m7GpppN, where N is any nucleotide) and a poly(A) tail present at their 5′ and 3′ extremities, respectively. The decay of functional mRNAs is initiated by 3′ poly(A) tail shortening followed by degradation via either the 5′-to-3′ or 3′-to-5′ mRNA decay mechanisms, the former being the principal mode of degradation (1). In the 5′-to-3′ pathway, the 5′ cap is eliminated by an essentially irreversible process known as decapping, which consists of the severing of the m7GDP moiety of the cap. This reaction is mediated by the decapping holoenzyme composed of the catalytic subunit Dcp2 and its cofactor Dcp1 (2–5). The resulting 5′ monophosphorylated RNA molecule can then be rapidly degraded by the 5′-to-3′ exonuclease Xrn1 (6). Because decapping definitively removes mRNAs from the pool of translatable templates present in cells, it is a critical step that is tightly regulated.
Hydrolytic cleavage of the cap is mediated by the Dcp2 NUDIX domain (3–5). However, Dcp2 and the decapping holoenzyme have a low intrinsic catalytic activity and requires accessory factors, such as Lsm1–7, Pat1, or Edc1–4 proteins, to be fully efficient (5, 7, 8). Among these cofactors, Pat1 (Pat1b in mammals, HPat in fruitfly) appears as a central and critical component of the decapping machinery. First, PAT1 gene deletion results in a strong inhibition of decapping in vivo in Saccharomyces cerevisiae (9, 10). Second, beyond DCP1 and DCP2, which are essential genes, the deletion of PAT1 gene exhibits the strongest phenotype among accessory decapping factors with slow growth at 30 °C and lethality at 37 °C (9, 11). Pat1 is considered a central platform, recruiting numerous mRNA decay factors and uses different regions to interact with these partners. Hence, the Pat1 N-terminal region contacts the Dhh1 helicase (RCK/p54/DDX6 in mammals, Me31B in fruitfly) and this interaction was proposed to modulate Dhh1 interaction with RNA (12–15). The central domain (M for middle) is involved in Lsm1–7 recruitment (8, 12, 13, 16) but also interacts with DCP2 as well as subunits of the CCR4–NOT RNA decay complex in metazoan (13, 17). Finally, Pat1 harbors an α-helical C-terminal domain (hereafter Pat1C). This region displays a strongly conserved surface responsible for the direct interaction with Lsm2 and Lsm3 proteins from the Lsm1–7 complex, which binds to the 3′ end of oligoadenylated mRNAs (9, 10, 18–21). Pat1C is also important for Dcp2 and Xrn1 binding in yeast and human (8, 17, 22) as well as for yeast Edc3 (21) and human EDC4 recruitment (12). While the Dcp2, Xrn1, and EDC4 binding sites on Pat1C remain to be identified, the region responsible for yeast Edc3 binding has been mapped using the two-hybrid assay to a conserved region, which is only present at the C-terminal extremity of fungal Pat1 proteins (21). However, whether this Pat1–Edc3 interaction is direct or bridged by other factors remains unclear.
In this paper, we show that the fungal-specific C-terminal Pat1 extension binds to several helical leucine-rich motifs (HLMs) located within S. cerevisiae Dcp2 C-terminal region, thereby bridging Pat1 and Edc3 proteins. We also identify a HLM signature present in fungal Xrn1 proteins and demonstrate that this motif is important for Xrn1 recruitment by the same Pat1 site. Specific disruption of the Pat1 surface interacting with HLMs results in a thermosensitive phenotype and impairs mRNA decay, demonstrating its functional significance. Altogether, our results support a model for efficacious mRNA decay through a Pat1-mediated coordinated recruitment of Dcp1–Dcp2 complex to capped mRNAs followed by recruitment of Xrn1 to the uncapped mRNAs for further degradation. Our data provide another example of the role of short linear motifs (SLiMs) in the formation of multiprotein assemblies involved in decapping and support further the plasticity of these interaction networks (23).
Results
The Pat1C Domain Binds Dcp2 That Bridges Interaction with Edc3.
We have previously shown using a yeast two-hybrid assay that the fungal-specific and conserved region located at the C-terminal part of Pat1C was important for interaction with Edc3 (21). Analysis of a deletion mutant demonstrated that the same region was required as well for growth at 37 °C (21) revealing its functional importance.
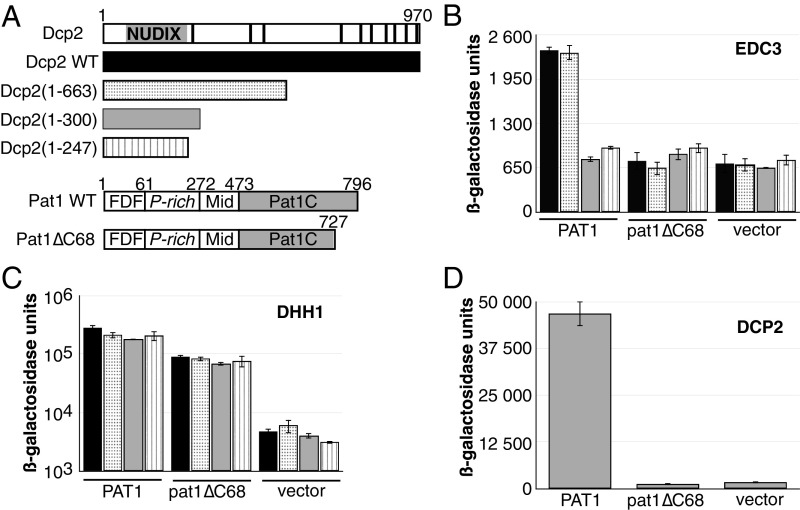
As both Edc3 and Pat1C are known to bind Dcp2 (24), we investigated whether the observed Edc3–Pat1 interaction was direct or whether it could be bridged by Dcp2. For this purpose, we tested the ability of Pat1 to interact with Edc3 using the two-hybrid assay in specifically built yeast host strains expressing endogenous Dcp2 versions with various truncations of their C-terminal domains (Fig. 1A). Indeed, our own data (see below) as well as data published while this work was in progress (22), supported the idea that the dispensable long C-terminal region of Dcp2 located downstream of its NUDIX domain mediates interaction with Pat1. As previously reported (21), a two-hybrid signal significantly above background was observed between full-length Pat1 and Edc3 in the wild-type (WT) host strain (Fig. 1B). A similar signal was detected in a two-hybrid host strain expressing a shorter Dcp2 protein encompassing residues 1–663, while it was reduced to background level in strains for which endogenous Dcp2 was essentially reduced to the critical NUDIX domain (residues 1–300 or 1–247, Fig. 1B). These observations do not result from a reduced ability of the mutant yeast strains to report interaction, as the Dhh1–Pat1 interaction, which is known to be direct (15), is similarly detected in the four yeast strains (Fig. 1C). No interaction of Edc3 with a Pat1 protein lacking its C-terminal conserved extension [i.e., residues 728–796 (Pat1ΔC68)] was detected, whichever form of endogenous Dcp2 was present (Fig. 1B). In contrast, Dhh1, which is known to bind to the N terminus of Pat1, interacted efficiently with the Pat1ΔC68 two-hybrid fusion, indicating that this construct is well expressed in all four yeast backgrounds. Overall, these data support the conclusion that the Dcp2 C-terminal extension bridges Pat1 and Edc3 in S. cerevisiae. They also indicate that residues 300–663 from Dcp2 are important for bridging Pat1 and Edc3. To confirm that Dcp2 interacts with the Pat1 C-terminal extension, we assayed the interaction of Dcp2 with Pat1 or Pat1ΔC68 using the two-hybrid assay in the wild-type yeast strain. This confirmed that, while Dcp2 interacts with full-length Pat1, it is unable to interact with the Pat1 construct lacking the C-terminal 68 residues (Fig. 1D).
Fig. 1.
Pat1C and Dcp2 interact directly. (A) Schematic representation of S. cerevisiae Dcp2 and Pat1 proteins. The Dcp2 catalytic NUDIX domain is depicted in gray. HLM motifs are depicted by black bars. The four different Dcp2 versions with various truncations of their C-terminal domains that are endogenously expressed in the yeast host strains used in this study are shown with a different color code, which is used in all of the panels of this figure. Pat1 domains are defined as follow: FDF, N-terminal domain containing the FDF signature involved in Dhh1 binding; P-rich, proline-rich domain; Mid, middle domain; and Pat1C, Pat1 C-terminal domain. The Pat1∆C68 construct expressed in the yeast host strains used in this study is also shown. (B) Effect of Dcp2 truncations on the Edc3–Pat1 interaction as monitored by yeast two-hybrid assay. Edc3 and Pat1 were fused to the DBD and AD domains, respectively. The interaction between these two factors was scored by assaying the β-galactosidase activity in yeast two-hybrid strains expressing the various forms of Dcp2. (C) Effect of Dcp2 truncations on the Dhh1–Pat1 interaction as monitored by yeast two-hybrid assay. Dhh1 and Pat1 were fused to the DBD and AD domains, respectively. The interaction between these two factors was scored by assaying the β-galactosidase activity as for B. (D) The fungal-specific C-terminal extension from Pat1 is required for Pat1 interaction with Dcp2. Dcp2 and Pat1 were fused to the DBD and AD domains, respectively. The interaction between these two factors was scored by assaying the β-galactosidase activity in the strain expressing the wild-type Dcp2 from its chromosomal locus.
Pat1C Domain Interacts Directly with Repeated Regions from Dcp2 C-Terminal Extension.
An in vitro pull-down experiment using proteins expressed in Escherichia coli demonstrated that the Pat1C–Dcp2 interaction is direct and that, for the Dcp2 [1–460] construct, the region encompassing residues 316–460 is necessary for binding (Fig. 2A). To delineate more precisely the region(s) of Dcp2 interacting with Pat1, various Dcp2 truncations were tested. Yeast two-hybrid assays demonstrated the C-terminal region located downstream of its NUDIX domain was important for binding and, in particular, that constructs encompassing Dcp2 residues 368–460, 450–663, and 436–663 interacted efficiently with Pat1 (Fig. 2B). Moreover, these constructs did not interact with Pat1ΔC68 (Fig. 2B). Interestingly, these Dcp2 regions share in common a linear motif (HLM). In S. cerevisiae Dcp2, 10 HLMs harboring a LLXΦL (Φ designing a hydrophobic residue) consensus motif have been previously identified (25) (Fig. 2 B and C) and such motifs were reported to mediate the interaction of Dcp2 with Edc3 and Scd6 Lsm domains in fission yeast (26). More recently, while this work was in progress, 8 of these HLMs were proposed to mediate Dcp2 binding to Pat1 in S. cerevisiae from yeast two-hybrid experiments (22).
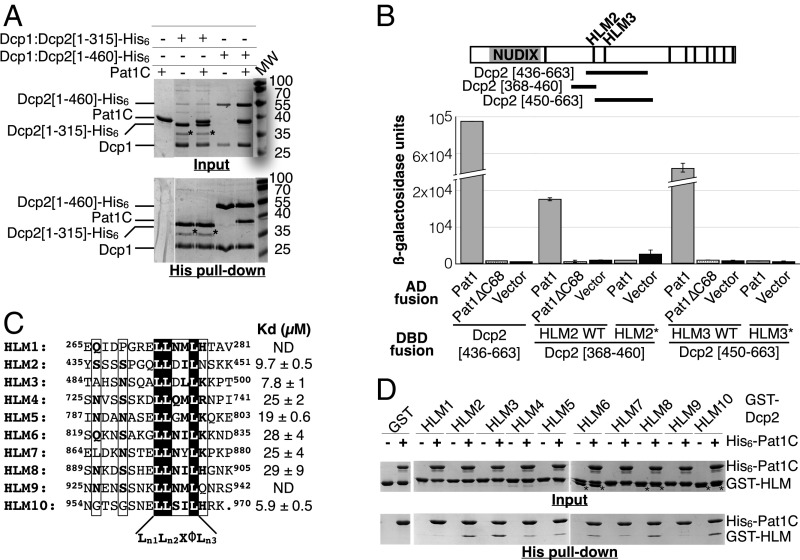
Fig. 2.
Pat1C binds to repeated linear motifs from the Dcp2 C-terminal region. (A) Pull-down experiments of untagged Pat1C with various Dcp1:Dcp2–His6 fragments. Input and eluted (His pull-down) samples were analyzed on 15% SDS/PAGE and Coomassie Blue staining. Asterisks denote Dcp2 degradation product or contaminant from E. coli. (B) Interaction of Pat1 with various Dcp2 regions located downstream of the NUDIX domain was assayed by monitoring β-galactosidase production in the two-hybrid assay. The location of the regions tested relative to the NUDIX domain and HLM motifs is indicated in the Upper schematic representation. Results obtained with the Pat1∆C68 and Dcp2 HLM mutants (HLM2* = L443A/L444A; HLM3* = L492A/L493A) from different constructs demonstrate that the in vivo interaction involves both of those and that different HLMs may bind Pat1 independently. (C) Sequence alignment of the 10 HLMs from S. cerevisiae Dcp2. Superscript numbers correspond to the numbering of amino acids located at the N- and C-terminal extremities of each HLM peptide from S. cerevisiae Dcp2. Strictly conserved residues are in white on a black background. Partially conserved residues are boxed. This panel was generated using the ESPript server (52). The consensus sequence is shown below the alignment and ɸ denotes hydrophobic residues. Kd values determined by fluorescence anisotropy for each FITC-labeled HLM peptide are indicated on the Right. ND, not determined. (D) Pull-down experiments of various GST–HLM fusions by His6–Pat1C. Input and eluate (His pull-down) samples were analyzed on 15% SDS/PAGE and Coomassie Blue staining. Asterisks denote GST proteins resulting from degradation of some GST–HLM fusions.
To investigate whether these Dcp2 HLMs are indeed directly responsible and sufficient for Pat1 binding, we produced and purified each of these 10 HLMs as GST fusions (GST–HLMs) and performed pull-down assays on NiNTA beads in the presence of His6-tagged Pat1C (Fig. 2D). With the exception of two GST–HLM fusions, which were not (GST–HLM1) or only slightly (GST–HLM9) retained on NiNTA beads when incubated with His6-tagged Pat1C, the remaining (GST–HLM2, 3, 4, 5, 6, 7, 8, and GST–HLM10) were efficiently pulled-down by His6–Pat1C, but with different apparent affinities. To quantify precisely the affinity between Pat1C and these 8 HLM peptides, we chemically synthesized short peptides corresponding to each of these 8 different HLMs N-terminally fused to FITC, as well as their unmarked counterpart, and performed fluorescence polarization experiments. In agreement with our pull-down experiments, all these peptides bind Pat1C with different affinities (Fig. 2C and SI Appendix, Fig. S1A). The HLM2, HLM3, and HLM10 peptides were found to interact with Pat1C with dissociation constants in the 5- to 10-µM range, while HLM4 to HLM8 exhibited dissociation constants higher than 19 µM. Chase experiments of the HLM2 fluorescent peptide by increasing concentrations of the corresponding unlabeled peptide led to a similar Kd value (3.8 µM), ruling out any major effect from FITC on binding of HLM peptides to Pat1C (SI Appendix, Fig. S1B).
Altogether, these results indicate that multiple short linear HLM motifs located within the Dcp2 C-terminal long extension are sufficient to directly bind the conserved fungal specific region of Pat1.
Structural Bases of Pat1C–Dcp2 Interaction.
To elucidate the molecular bases underlying the interaction between Pat1C and Dcp2, we crystallized Pat1C with the HLM peptides exhibiting the highest affinity for Pat1C (i.e., HLM2, HLM3, and HLM10). As wild-type Pat1C failed to crystallize in the presence of these peptides, we performed cocrystallization using the Pat1C mutant Q706A/L713A, which is not affected in HLM binding (SI Appendix, Fig. S1C). Crystals were obtained for this Pat1C mutant in the presence of a twofold molar excess of HLM3 peptide and were further used to obtain crystals of complexes with either HLM2 or HLM10 peptides by streak seeding. These crystals allowed us to determine the structures of Pat1C:HLM2, Pat1C:HLM3, and Pat1C:HLM10 complexes by molecular replacement and refining these at 2.6-Å, 2.15-Å, and 1.9-Å resolution, respectively (SI Appendix, Table S4). For each complex, two copies of Pat1C:HLM complex are present in the asymmetric unit and are arranged in an intricate head-to-tail organization characterized by a large interacting surface area (more than 1,800 Å2; SI Appendix, Fig. S2A). This arrangement is similar to that observed for our previously solved crystal structure of apo Pat1C WT (21). When comparing all Pat1C structures either in the apo form (WT) (21) or bound to various HLMs (this work), the rmsd values range from 0.25 to 0.65 Å, indicating that all these structures are virtually identical.
All three Dcp2 HLM peptides fold as an amphipathic α-helix and bind to the same Pat1C cavity (Fig. 3A and SI Appendix, Fig. S2B), forming a mean interface area of ∼570 Å2 (as determined by the PISA server) (27), a classical value for protein–peptide complexes (28). Chase experiments further confirmed that unlabeled HLM3 peptide competes with FITC-labeled HLM2 peptide to bind to Pat1C domain (SI Appendix, Fig. S2C). In addition to this main surface area, the HLM peptides are interacting with neighboring Pat1C molecules in the crystal packing. However, the contacts between the HLM peptides and lattice mates exhibit small interface areas and differ with respect to the three peptides used and between the two Pat1–HLM complexes present in the asymmetric units (SI Appendix, Fig. S3). This further supports that the Pat1–HLM interfaces with the largest surface is the biologically relevant one.
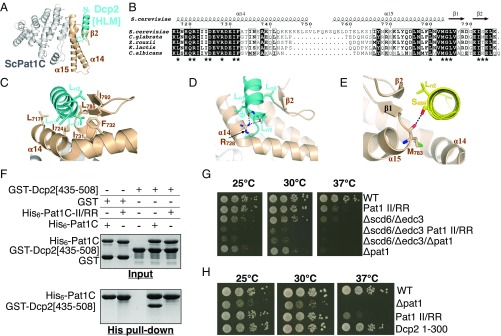
Fig. 3.
Structural basis of Pat1C/Dcp2 interaction. (A) Ribbon representation of the crystal structure of Pat1C in complex with a Dcp2 HLM peptide. The Dcp2 HLM is in cyan, Pat1C is in gray, and Pat1C fungi-specific C-terminal extension is in beige. (B) Sequence alignment of the fungal Pat1C. For the sake of clarity, only the C-terminal residues are shown. Strictly conserved residues are in white on a black background. Partially conserved residues are boxed. Residues involved in the interaction with HLM3 and HLM10 peptides are indicated by black stars below the alignment. This panel was generated using the ESPript server (52). (C) Detailed representation of hydrophobic interactions common to the Pat1C:Dcp2 [HLM] interface. Same color code as A is used. (D) Detailed representation of an electrostatic interaction common to all Pat1C:Dcp2 [HLM] interfaces (same color code as A). The black dashed lines indicate hydrogen bonds. (E) Detailed representation of an electrostatic interaction found in Pat1C:Dcp2 [HLM3] and Pat1C:Dcp2 [HLM10] complexes. The Pat1C:Dcp2 [HLM3] complex was shown to illustrate this interaction. The black dashed line indicates a hydrogen bond. (F) Pull-down experiment of GST–Dcp2 [435-508] (encompassing HLM2 and HLM3) by His6–Pat1C and His6–Pat1C-II/RR. Input and eluted (His pull-down) samples were analyzed on 15% SDS/PAGE and Coomassie Blue staining. (G) Growth analysis of Pat1C-II/RR and pat1Δ in an edc3Δ/scd6Δ background. Serial dilutions of strains with the indicated genotypes were spotted on YPDA plates and incubated at the indicated temperatures for 2 d. (H) Comparative growth analysis of isogenic wild type, pat1Δ, Pat1-II/RR, and Dcp2 [1–300] strains. All mutations were chromosomally integrated. Serial dilutions of the different strains were spotted on YPDA plates and incubated at the indicated temperatures for 2 d.
The Pat1C region involved in HLM binding is formed by the N-terminal half from helix α14, the C-terminal end from helix α15, and the short C-terminal two-stranded β-sheet. It then mostly corresponds to the highly conserved and fungi-specific Pat1C C-terminal extension that we have identified previously as functionally important (Fig. 3 A and B). Pat1C residues interacting with HLM peptides are strongly conserved within yeasts and form a large and central hydrophobic patch surrounded by charged residues (Fig. 3 B and C). The details of the interface between Pat1C and HLM peptides discussed here are solely based on the structures with HLM3 and HLM10 peptides, because the crystals of Pat1C–HLM2 complex suffered from high anisotropy diffraction and the quality of the 2Fo–Fc electron density map (particularly for the side chains) is not sufficient for detailed analysis. The hydrophobic face of the HLM amphipathic helix, composed of the three leucines from the Ln1Ln2XΦLn3 motif, is oriented toward the Pat1C hydrophobic patch. Indeed, HLM Ln1 is clamped between L717, I724, and L785 from Pat1C, HLM Ln2 interacts with L785 and I792, and HLM Ln3 contacts I731, F732, L785, and I792 (Fig. 3 B and C). The importance of these conserved leucine residues for the binding of Dcp2 HLM to Pat1 is confirmed by the disruption of the Pat1–Dcp2 interaction upon mutations of L443 (Ln1) and L444 (Ln2) from HLM2 into alanine as observed by fluorescence polarization experiments (SI Appendix, Fig. S4A). Similarly, the Dcp2 [368–460] fragment containing these Leu-to-Ala substitutions within HLM2 is affected in Pat1 interaction according to yeast two-hybrid assays (Fig. 2B). A similar observation was made when both L492 and L493 from HLM3 were mutated into alanines {Dcp2 [450–663]} (Fig. 2B). Because constructs carrying only a single HLM (either HLM2 or HLM3) interact with Pat1, we conclude that these motifs are independently able to interact with Pat1 in vivo. Moreover, a higher β-galactosidase is detected with the construct carrying both HLM2 and HLM3 {Dcp2 [436–663]}, suggesting that several Pat1 molecules may bind simultaneously to a single Dcp2 C-terminal tail (Fig. 2B).
Although the Pat1–Dcp2 HLM interaction is largely hydrophobic, some electrostatic contacts are observed in these Pat1C–HLM complexes. The side chain from the highly conserved R728 from Pat1C interacts with the carbonyl group of the Φ residue from the HLM α-helix (Leu495 from HLM3 and Ile966 from HLM10) and hence is well positioned to form an electrostatic interaction with the C-terminal negatively charged end of HLM helix (Fig. 3D). Such an interaction is very likely to be involved in the interaction of Pat1 with all its HLM partners and might rule the C-terminal end of Dcp2 HLM helices. Another hydrogen bond observed in both crystal structures is formed between Pat1 M783 carbonyl group and the hydroxyl groups from either HLM3 S489 (Fig. 3E) or HLM10 S960, which correspond to position −3 relative to Ln1. However, such an interaction would probably be restricted to HLM3, HLM4, and HLM7–10 as the residues present at the corresponding position on the other HLMs cannot form hydrogen bonds through their side chains (Fig. 2C). Additional electrostatic interactions are also observed in only one of the two complexes. For instance, H486 from HLM3 forms a hydrogen bond with Pat1 E794, while E962 from HLM10 forms a salt bridge with Pat1 R721 (SI Appendix, Fig. S4 B and C). According to HLM sequences, K821 from HLM6 and K891 from HLM8, which correspond to HLM3 H486, could also interact with Pat1 E794 side chain and form salt bridges (Fig. 2C). Similarly, residues from HLM5, HLM7, and HLM8 corresponding to HLM10 E962 are also glutamic acid, indicating that they are very likely to form a salt bridge with R721 from Pat1 (Fig. 2C). These HLM-specific interactions could explain the differences in the Kd values measured for the Dcp2 HLM peptides. Mutation of conserved polar residues in the periphery of the hydrophobic pocket of Pat1C (mutants Pat1C-Q720A/R721A/D725A/R728A and Pat1C-R721A/R728A/F732A/E794A) resulted in significantly reduced affinities for HLM3 and HLM10 peptides as determined by fluorescence polarization (SI Appendix, Fig. S4 D and E). Notably, both mutants displayed a more important reduction in affinity for HLM10 than for HLM3, which correlates with R721 from Pat1C being engaged in a salt bridge with E962 (HLM10) while not contacting any HLM3 residue (SI Appendix, Fig. S4 D and E). Interaction between Pat1C-R721A/R728A/F732A/E794A and HLM3 or HLM10 appeared to be weaker than the one observed with Pat1C-Q720A/R721A/D725A/R728A. This later observation can be explained first, by the disruption of hydrophobic contacts through the substitution of F732 by alanine (Fig. 3C) and, second, by the impairment of the hydrogen bonds between E794 and H486 (HLM3; SI Appendix, Fig. S4B).
An additional mutant was generated to disrupt the Pat1–Dcp2 interaction for functional studies. Two conserved isoleucines (I724 and I731) from Pat1, which contact Ln1 and Ln3 of the HLMs (Fig. 3C), respectively, and hence are at the center of the Pat1 hydrophobic patch, were substituted by arginine to generate the Pat1C-I724R/I731R mutant (hereafter named Pat1C-II/RR). This mutant protein displayed a gel filtration behavior similar to the one observed for the wild-type Pat1C domain, supporting a correct overall folding. These mutations were sufficient to abolish the interaction between Pat1C and Dcp2 [435–508] in vitro (Fig. 3F). The same mutation introduced in a full-length Pat1 prevented its interaction with Dcp2 in a two-hybrid assay (SI Appendix, Fig. S4F). This further indicates that this Pat1 region is essential for in vivo interaction with Dcp2. To assess the functional significance of HLM binding by Pat1C, we next examined the phenotype of the Pat1 II/RR mutant in combination with inactivation of DHH1 or with a double deletion of EDC3 and SCD6. We previously showed that pat1Δ/dhh1Δ and pat1Δ/edc3Δ/scd6Δ strains exhibit a growth defect phenotype, which cannot be rescued by expression of a Pat1 protein lacking either its entire C-terminal domain (Pat1ΔC) or the last 68 C-terminal residues (Pat1ΔC68) (21). Interestingly, the Pat1 II/RR introduced on a plasmid in the pat1Δ/dhh1Δ and pat1Δ/edc3Δ/scd6Δ strains behaves identically to Pat1∆C68 in being unable to rescue its growth defect, in contrast to wild-type Pat1 (SI Appendix, Fig. S4G). To confirm these observations without potential variability resulting from plasmid copy number heterogeneity, we constructed a yeast strain carrying the II/RR allele at the chromosomal PAT1 locus using the CRISPR/Cas strategy. This mutation was introduced in the scd6∆/edc3∆ context by crossing and growth of the resulting strains compared with isogenic derivatives carrying a pat1∆ allele instead. Combination of the Pat1 II/RR mutation with scd6∆/edc3∆ resulted in synthetic growth phenotype indistinguishable from the one observed for the association of pat1∆ with scd6∆/edc3∆ (Fig. 3G). Western blot analyses confirmed that wild-type Pat1 and the II/RR mutant were present at similar levels (SI Appendix, Fig. S5) in these strains, thus ruling out that instability of the mutant protein could be responsible for the synthetic phenotype. The Pat1 II/RR strain also displayed a thermosensitive growth phenotype (Fig. 3 G and H). Altogether, these results demonstrate that the Pat1 region involved in HLM binding is functionally important. The thermosensitive phenotype was nevertheless not as strong as the one observed in a ∆pat1 strain, indicating that the mutant protein harbors residual activity (Fig. 3 G and H). However, a yeast strain expressing a truncated version of Dcp2, which does not interact with Pat1, e.g., Dcp2 [1–300] containing only HLM1 does not phenocopy the thermosensitive phenotype of Pat1 II/RR at 37 °C (Fig. 3H). This observation indicates that the disruption of the Pat1–Dcp2 interaction in the II/RR mutant is not entirely responsible for the observed phenotype. In turn, this suggests that this Pat1 region might be responsible for the interaction with at least another partner.
Dcp2-Binding Site On Pat1C also Recruits Xrn1 Exonuclease and Is Important for RNA Decay.
To identify an additional putative Pat1 interacting factor, we searched for a potential HLM signature in two other Pat1 partners that were previously reported to interact with Pat1C, namely, Dcp1 and Xrn1 (9). Close inspection of the S. cerevisiae Xrn1 sequence revealed the presence of a putative HLM (with core sequence 1287LLNFI1291), reminiscent of Dcp2 HLMs, near its C terminus in a region that is predicted to be unstructured. Interestingly, similar motifs are conserved in Xrn1 C termini from different yeast species in which Pat1C also displays the yeast-specific C-terminal extension (Fig. 4A). To test whether this putative HLM can mediate interaction between Xrn1 and Pat1, we fused the Xrn1 region [1,277–1,301] to GST and performed an in vitro His pull-down assay in the presence of a His6-tagged version of either wild-type Pat1C or Pat1C-II/RR. This GST-Xrn1[HLM] was specifically pulled down by wild-type Pat1C but not by the Pat1C-II/RR mutant (Fig. 4B). This indicates, first, that this HLM is involved in Xrn1 binding to Pat1C and, second, that it binds to the same region as Dcp2 HLMs on Pat1C. A fluorescence anisotropy experiment on a FITC-labeled Xrn1[HLM] peptide revealed a dissociation constant (Kd) of 50 µM (SI Appendix, Fig. S6). These results suggest that Pat1/Xrn1 interaction could be, at least in part, mediated by this HLM. The interaction of Pat1 with Xrn1 was then monitored in vivo using a two-hybrid system (Fig. 4C). We detected a two-hybrid interaction between Pat1 and a truncated Xrn1 construct encompassing residues 1–1,456. This construct contains the Xrn1 HLM region and when Leu1287 and Leu1288 from this motif were simultaneously substituted by Ala, the two-hybrid interaction was lost (Fig. 4C). The Xrn1–Pat1 interaction was also obliterated by the Pat1-II/RR mutation (Fig. 4C). Altogether, the two-hybrid results confirm that the in vivo association of Xrn1 with Pat1 occurs through a HLM–Pat1C interaction.
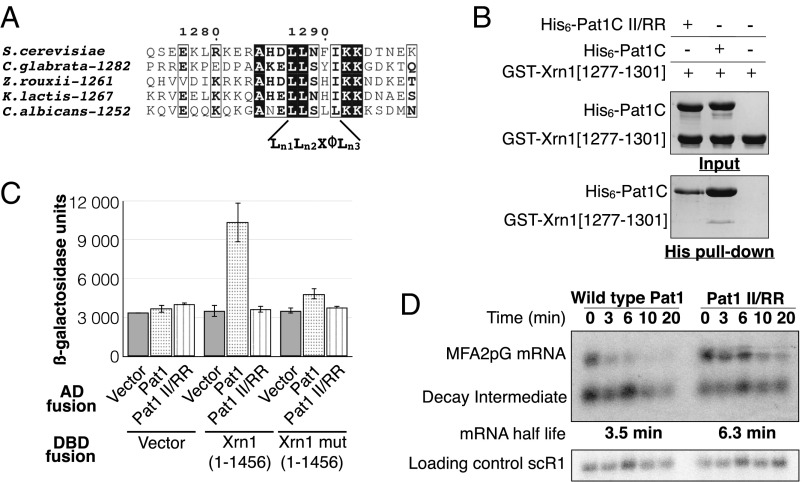
Fig. 4.
Pat1C C-terminal extension is responsible for Xrn1 recruitment. (A) Sequence alignment of fungal Xrn1 proteins focusing on the identified HLM motif. Strictly conserved residues are in white on a black background. Partially conserved residues are boxed. The numbers following the organism names correspond to the numbering of amino acids located at the N-terminal extremity of the HLM in Xrn1 from the corresponding organisms. (B) Pull-down experiment of GST–Xrn1 [1,277–1,301] by His6–Pat1C and His6–Pat1C-II/RR. Input and eluted (His pull-down) samples were analyzed on 15% SDS/PAGE and Coomassie Blue staining. (C) Interaction of Pat1 with Xrn1 monitored through β-galactosidase production in the two-hybrid assay. Results obtained with the Pat1-II/RR and Xrn1 [1–1,456] HLM mutant (L1287A/L1288A) demonstrate that the in vivo interaction depends on both of those structural elements. (D) Analysis of the decay of the MFA2pG reporter mRNA in the wild type and Pat1-II/RR mutant. The rate of disappearance of the MFA2pG mRNA following transcription shutdown was determined by monitoring the level of MFA2pG mRNA remaining at the time points indicated by Northern blot analysis. The scR1 RNA serves as a loading control. The MFA2pG mRNA half-life in the two strains was determined following quantification of the signal by fitting with exponential decay.
To analyze the functional impact of Pat1C HLM-binding surface, we compared the decay of the MFA2pG reporter mRNA in the wild-type strain and the isogenic strain carrying the Pat1-II/RR chromosomal mutation. The plasmid encoding the GAL-promoter driven MFA2pG reporter was introduced in both strains and the expression of the reporter mRNA was induced by growth in galactose. RNA samples were collected at various time points after glucose had been added to repress the GAL promoter and the levels of the MFA2pG reporter mRNA in each sample were determined by Northern blotting. While in the wild-type strain, the MAF2pG mRNA half-life is 3 min (Fig. 4D), in the Pat1-II/RR strain, the MAF2pG mRNA half-life is twice as long (i.e., half-life of 6.3 min). This demonstrates that the surface of Pat1 that recruits Dcp2 and Xrn1 is functionally important for mRNA decay in yeast.
Discussion
Eukaryotic mRNA decay is a highly regulated and concerted process involving several proteins that are mostly part of multiprotein assemblies. It typically initiates with the shortening of the 3′ poly(A) tail by deadenylases (29). It can then continue from the 3′ end through the action of the 3′-to-5′ exonucleolytic activity of the cytoplasmic exosome or alternatively through 5′-cap removal by decapping followed by 5′-to-3′ decay (1). The latter is considered the major mRNA decay pathway in yeast.
In this paper, we have analyzed the C-terminal domain from S. cerevisiae Pat1 protein, a central scaffolding protein inhibiting translation initiation and stimulating the Dcp2 decapping enzyme (8, 30). Our results have implications for the coordination of events leading to the degradation of mRNAs via the 5′-to-3′ pathway and for the evolution of the protein interaction networks involved in eukaryotic mRNA decay. Finally, the Pat1 region, which is important for the recruitment of mRNA decay enzymes and for growth, is specific to fungi and is therefore of potential interest for the development of future antifungal drugs.
Pat1-Coordinated Recruitment of Dcp2 and Xrn1 to the 5′ End of Deadenylated mRNAs.
Pat1 is a key player in mRNA degradation by serving as a platform that recruits decapping factors and their activators. Its C-terminal domain is particularly important as testified by the thermosensitive phenotype resulting from its deletion (21). A conserved surface located at the N-terminal edge of the Pat1C domain is functionally critical as it binds to the Lsm1–7 heteroheptameric complex to form the Lsm1–7/Pat1 complex, which interacts preferentially with the 3′ tail of oligoadenylated mRNAs (12, 18–21, 31, 32). Here, we identify another functionally important region located at the opposite edge of the Pat1C domain, i.e., the C-terminal extremity, which is conserved in fungal homologs (21). First, we show that this Pat1 region interacts with at least eight HLMs from S. cerevisiae Dcp2, in agreement with recent two-hybrid results (22). This would allow Dcp2 to recruit several mRNPs each containing a Lsm1–7/Pat1 complex associated with one mRNA targeted for degradation. This could result in a more efficient mRNA degradation offering an evolutionary advantage (Fig. 5A). In Schizosaccharomyces pombe, several HLMs from Dcp2 were described to be involved in Edc3 and Scd6 binding and this multivalency was shown to be involved in P-body formation (26). This does not seem to be the case for the Pat1–Dcp2 interaction, as the expression of the Dcp2 [1–300] fragment lacking all HLMs involved in the recruitment of Pat1 supports P-body formation in S. cerevisiae (24). Finally, as the yeast Dcp2 C-terminal extension was also shown to contain a cis-acting inhibitory element (residues 350–375) (22), the concomitant recruitment of Pat1 proteins to several HLMs on a single Dcp2 molecule could induce either conformational changes of this Dcp2 C-terminal tail or steric hindrance, thereby preventing Dcp2 inhibition by this cis-acting element. This could also rationalize the Pat1 role as a decapping activator.
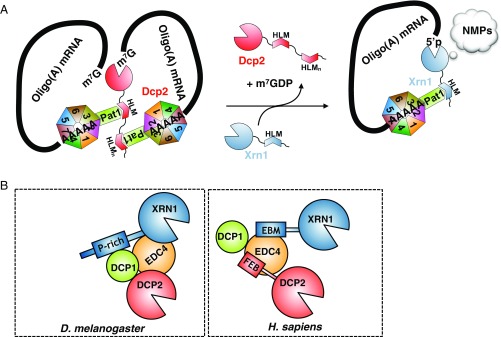
Fig. 5.
Comparison of mechanisms for the recruitment of Dcp2 and Xrn1 to deadenylated eukaryotic mRNAs. (A) Model for the Pat1-coordinated 5′-to-3′ mRNA decay mechanism in yeast S. cerevisiae. (B) Differences in 5′-to-3′ degradation pathway networks in fruitfly and human compared with budding yeast. EBM, EDC4-binding motif; FEB, phenylalanine-rich EDC4 binding; NMP, nucleoside monophosphate; P-rich, proline-rich sequence.
Furthermore, we show that this conserved Pat1C region specific to yeasts, is also responsible for direct recruitment of Xrn1 through binding to a HLM peptide. Thus, this Pat1C region can directly interact with the two main enzymes (Dcp2 and Xrn1) involved in the 5′-to-3′ mRNA decay pathway. Hence, Pat1C bridges 5′ and 3′ ends from mRNAs undergoing degradation and then is ideally positioned to coordinate the action of both enzymes on oligoadenylated mRNAs (Fig. 5A). The central role of this Dcp2- and Xrn1-binding surface on Pat1C in mRNA decay is perfectly illustrated both by the strong thermosensitive phenotype (Fig. 3H) and the stabilization of the MFA2pG mRNA reporter (Fig. 4D) associated with the Pat1-II/RR mutant. As the HLM peptides that we have identified in S. cerevisiae Dcp2 and Xrn1 compete to interact on the same surface of the Pat1C domain, we propose that in yeasts, the Lsm1–7/Pat1 complex recruits in a stepwise manner Dcp2 to deadenylated mRNA substrates to remove the 5′ cap and then Xrn1 to perform rapid 5′-to-3′ exonucleolytic digestion of decapped mRNAs (Fig. 5A). Such a Pat1-mediated stepwise recruitment of Dcp2 and Xrn1 would ensure an efficacious mechanism for rapid mRNA degradation by Xrn1 following decapping by Dcp2. Our model is supported by several observations: (i) the Lsm1–7/Pat1 complex enhances the interaction of Dcp2 with mRNAs (31); (ii) Pat1 has been described as an activator of Dcp2 (8–10, 16); (iii) Pat1 recognizes Dcp2 in one of its most active forms, i.e., bound to its two most potent activators Dcp1 (Fig. 2A) and Edc3 (Fig. 1B); (iv) capped oligoadenylated mRNAs accumulate in pat1Δ mutant in yeast (9); (v) the Lsm1–7/Pat1 complex still binds to deadenylated mRNAs after decapping (31); (vi) the Lsm1–7/Pat1 complex copurifies with Xrn1 (9); and (vii) xrn1Δ cells accumulate deadenylated mRNAs lacking cap structure (6). The exchange of the partner of the Lsm1–7/Pat1 complex, i.e., Dcp2 replacement by Xrn1, could be triggered by the removal of the 5′ cap. Although the 5′ cap does not enhance affinity of Dcp2 for capped mRNAs (33), it should prevent Xrn1 binding to these mRNAs according to the crystal structure of an Xrn1–substrate complex (34). Furthermore, although several HLMs from Dcp2 exhibit higher affinities for Pat1C than the Xrn1 HLM motif in vitro (Fig. 2C and SI Appendix, Fig. S6), Xrn1 but not Dcp2 copurifies with the Lsm1–7/Pat1 complex in vivo (9). This preferential association might result from the abundance of Xrn1, which is about two to seven times more abundant than Dcp2 in S. cerevisiae (35–37). It could also be due to additional partners or Xrn1 regions, which could strengthen the interaction between Xrn1 and the Lsm1–7/Pat1 complex in vivo. In summary, we propose an optimized model in which the Lsm1–7/Pat1 complex sequentially recruits Dcp2 and then Xrn1 for degradation of deadenylated mRNAs. A coordinated recruitment of decay factors to mRNAs undergoing decay was previously suggested for metazoan (12, 23, 38) but further experiments to investigate such mechanisms are definitely needed.
Recruitment of Xrn1 by the Lsm1–7/Pat1 Complex Perfectly Illustrates the Plasticity in the Eukaryotic 5′-to-3′ mRNA Decay Interaction Networks.
With the exception of the metazoan-specific decapping activator EDC4 (also called Ge-1), most decapping factors are well conserved between eukaryotic organisms. Decapping of mRNAs requires a tight interplay between several proteins that assemble dynamically as multiprotein complexes. Most of these factors are modular with defined structural domains flanked by less conserved regions. These later are mostly disordered regions containing SLiMs, which most of the time fold upon partner binding. These SLiMs actively participate in the rewiring of interacting networks made by these conserved decapping factors in S. cerevisiae, fruitfly, and human cells (23). For instance, S. cerevisiae Scd6, Edc3 (as well as their human and fruitfly orthologs), and Pat1 harbor conserved FDF motifs that compete to bind to the same region of Dhh1/DDX6 helicases (15, 39). A SLiM found in Edc3 proteins from Saccharomycetaceae phylum has also been shown to mediate Edc3 interaction with Rps28 ribosomal protein and then to participate in an autoregulatory feedback loop controlling RPS28B mRNA levels (40). The Lsm domains from metazoan EDC3 and LSm14A (yeast Scd6) bind to a HLM region present in DCP1 (26). Interestingly, the yeast Edc3 Lsm domain also interacts with a HLM-like peptide, which is located in Dcp2 (5, 22, 24, 26). Here, we demonstrate that several HLMs present within the long C-terminal region from S. cerevisiae Dcp2 directly interact with Pat1. These HLMs are located in a region that does not mediate the interaction with yeast Edc3. We also identify a HLM in yeast Xrn1, which is responsible for its interaction with Pat1 (8, 9). This differs from Drosophila melanogaster XRN1, which interacts directly with the DCP1 EVH1 domain via a proline-rich sequence (38) (Fig. 5B). To our knowledge, although a proline-rich sequence is also present in yeast Xrn1, there is no clear evidence in S. cerevisiae for a direct interaction between Xrn1 and the Dcp1 EVH1 domain, the latter being involved in the interaction with Edc1 and Edc2 (41, 42). In human, the DCP1 EVH1 domain interacts with PNRC2, which was recently proposed to be orthologous to yeast Edc1–2 factors (42) but not XRN1. The latter uses a conserved C-terminal extension (EBM for EDC4-binding motif) to interact with EDC4 (38, 43, 44). Finally, although some weak contact may exist between metazoan DCP1 and DCP2, their interaction requires EDC4 (44, 45). Hence, human DCP2 and XRN1 interact with EDC4 via SLiMs and can coexist on the same complex together with EDC4 and DCP1 (44) (Fig. 5B). Our study shows that rewiring of the interaction network between decapping factors is not only restricted to SLiMs that have emerged during evolution to create a new anchoring point between decapping factors or to recruit new factors such as EDC4 (23). Indeed, the fungal Pat1 C-terminal extension, which acts as binding platform for Dcp2 and Xrn1, is integrated within the fungal Pat1C domain architecture, thereby adding one layer of complexity in the evolution of decapping interaction networks. This study further reveals a mutually exclusive mechanism for the recruitment of Dcp2 and Xrn1 to mRNAs undergoing decay, which differs completely from those previously described for human and fruitfly (38, 44).
Conclusion
We have previously shown that a S. cerevisiae Pat1 protein lacking the fungal-specific C-terminal extension (Pat1ΔC68) did not rescue the thermosensitive phenotype resulting from the deletion of the PAT1 gene (21). Our current study reveals that point mutants targeting this region and precluding Dcp2 and Xrn1 interactions with Pat1 have the same effect, highlighting the importance of this region for Pat1 function, mRNA decay, and yeast growth at 37 °C. This Pat1 surface further appears as a potential target for the development of drugs against pathogenic yeasts such as Candida and Aspergillus species, which infect patients with compromised immune systems and can cause allergic diseases. Interestingly, LxxLL HLMs are also involved in the binding of coactivators to nuclear receptors (25). Such a motif is also crucial for the cellular ubiquitin ligase E6AP to interact with the viral oncoprotein E6 from human papilloma virus and then to trigger p53 degradation (46, 47). As proapoptotic peptides and small molecules targeting LxxLL pocket of E6 oncoprotein have been recently tested (48–50), future studies aimed at identifying molecules precluding Dcp2 or Xrn1 HLMs to bind to this fungal-specific Pat1 region could be of great medical interest to develop antifungal drugs.
Materials and Methods
Details on other experimental procedures are available as SI Materials and Methods.
Yeast Strains.
All yeast strains are listed in SI Appendix, Table S1. Yeast strains used for phenotypic and functional assays were derived from the W303 background or the related BMA64 derivative. Strains for two-hybrid assays were derived from MAV203 (Invitrogen). The lithium acetate transformation protocol was used to introduce in cells exogenous plasmids, oligonucleotides (listed in SI Appendix, Tables S2 and S3, respectively), or linear DNA fragments. Deletions were constructed using standard PCR-based methods while point mutations were introduced using the CRISPR/Cas system described by Ryan et al. (51). Briefly, plasmids encoding the Cas9 nuclease and a guide RNA targeting the site of interest in the desired gene were first constructed by site-directed mutagenesis of plasmid pRNR2-Cas9::8His-tRNA(Pro)::sgRNA (51) using specifically designed oligonucleotides (SI Appendix, Table S3). The parental yeast strain was cotransformed with one of these plasmids (SI Appendix, Table S2) and two repair oligonucleotides overlapping the target site and containing the mutation of interest (SI Appendix, Table S3). Transformants were recovered on YPDA-G418 plates. Screening for the presence of the desired mutation was performed by PCR and restriction digest, thanks to the polymorphic restriction site introduced in the repair oligonucleotide(s). The sequence of the mutation region was determined by sequencing the PCR product. A derivative, lacking the Cas9 gene and G418 marker following the random loss of the corresponding plasmid, was recovered and used for further studies.
Yeast growth assays.
Cells carrying chromosomal mutation were grown at the permissive temperature in YPDA media. Cultures were diluted to an optical density of 0.1 at 600 nm (OD600) with sterile water. Three microliters of this dilution as well as 10-fold serial dilutions were plated on YPDA plates. Cell growth was monitored after 48 h at the indicated temperature. For plasmid-encoded Pat1 mutation, the cognate plasmid and control vectors (SI Appendix, Table S2) were first introduced in Δpat1∆dhh1 and Δpat1∆scd6∆edc3 cells (YFW168 and BSY2601, SI Appendix, Table S1) and transformants selected on −TRP plates. Transformants were grown in liquid synthetic complete (SC) medium lacking tryptophan and containing 2% glucose. Growth assays were then performed as described above, except that −TRP selective plates incubated for 3 d were used.
Supplementary Material
Acknowledgments
We thank N. Cougot for participation in preliminary experiments, Pascal Eberling for peptide synthesis, and SOLEIL for provision of synchrotron radiation facilities and particularly staff from Proxima-1 and Proxima-2 beamlines. This work was supported by Ecole Polytechnique (M.G.), the Centre National pour la Recherche Scientifique (B.S. and M.G.), including specific support by the Action Thématique et Incitative sur Programme-AVENIR program (M.G.), the Agence Nationale pour la Recherche [Grant ANR-11-BSV800902 (to B.S. and M.G.), ANR-10-LABX-0030-INRT performed under the programme Investissements d’Avenir ANR-10-IDEX-0002-02 (to B.S.)], the Ligue Contre le Cancer (Equipe Labellisée 2017) (B.S.), and the Centre Européen de Recherche en Biologie et en Médecine–Institut de Génétique et de Biologie Moléculaire et Cellulaire and INSERM (B.S.). Z.F. was supported by the Fondation pour la Recherche Médicale. C.C. holds a PhD fellowship from the French Ministère de l’Enseignement Supérieur et de la Recherche and Ecole Normale Supérieure Cachan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5LM5, 5LMF, and 5LMG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711680114/-/DCSupplemental.
References
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dijk E, et al. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.She M, et al. Structural basis of dcp2 recognition and activation by dcp1. Mol Cell. 2008;29:337–349. doi: 10.1016/j.molcel.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charenton C, et al. Structure of the active form of Dcp1-Dcp2 decapping enzyme bound to m(7)GDP and its Edc3 activator. Nat Struct Mol Biol. 2016;23:982–986. doi: 10.1038/nsmb.3300. [DOI] [PubMed] [Google Scholar]
- 6.Hsu CL, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1:253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 8.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tharun S, et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 11.Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun JE, et al. The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa. EMBO J. 2010;29:2368–2380. doi: 10.1038/emboj.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas G, et al. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozgur S, Stoecklin G. Role of Rck-Pat1b binding in assembly of processing-bodies. RNA Biol. 2013;10:528–539. doi: 10.4161/rna.24086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharif H, et al. Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res. 2013;41:8377–8390. doi: 10.1093/nar/gkt600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol Cell Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharif H, Conti E. Architecture of the Lsm1-7-Pat1 complex: A conserved assembly in eukaryotic mRNA turnover. Cell Rep. 2013;5:283–291. doi: 10.1016/j.celrep.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Wu D, et al. Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation. Cell Res. 2014;24:233–246. doi: 10.1038/cr.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fourati Z, et al. The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment. PLoS One. 2014;9:e96828. doi: 10.1371/journal.pone.0096828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He F, Jacobson A. Control of mRNA decapping by positive and negative regulatory elements in the Dcp2 C-terminal domain. RNA. 2015;21:1633–1647. doi: 10.1261/rna.052449.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonas S, Izaurralde E. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 2013;27:2628–2641. doi: 10.1101/gad.227843.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol Cell Biol. 2010;30:1446–1456. doi: 10.1128/MCB.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaudon C, Chambon P, Losson R. Role of the essential yeast protein PSU1 in p6anscriptional enhancement by the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1999;18:2229–2240. doi: 10.1093/emboj/18.8.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fromm SA, et al. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 2012;31:279–290. doi: 10.1038/emboj.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 28.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Yan YB. Deadenylation: Enzymes, regulation, and functional implications. Wiley Interdiscip Rev RNA. 2014;5:421–443. doi: 10.1002/wrna.1221. [DOI] [PubMed] [Google Scholar]
- 30.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tharun S, Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 32.Chowdhury A, Kalurupalle S, Tharun S. Pat1 contributes to the RNA binding activity of the Lsm1-7-Pat1 complex. RNA. 2014;20:1465–1475. doi: 10.1261/rna.045252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshmukh MV, et al. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol Cell. 2008;29:324–336. doi: 10.1016/j.molcel.2007.11.027. [DOI] [PubMed] [Google Scholar]
- 34.Jinek M, Coyle SM, Doudna JA. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 36.Newman JR, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 37.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
- 38.Braun JE, et al. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat Struct Mol Biol. 2012;19:1324–1331. doi: 10.1038/nsmb.2413. [DOI] [PubMed] [Google Scholar]
- 39.Tritschler F, et al. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Kolesnikova O, Back R, Graille M, Séraphin B. Identification of the Rps28 binding motif from yeast Edc3 involved in the autoregulatory feedback loop controlling RPS28B mRNA decay. Nucleic Acids Res. 2013;41:9514–9523. doi: 10.1093/nar/gkt607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borja MS, Piotukh K, Freund C, Gross JD. Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA. 2011;17:278–290. doi: 10.1261/rna.2382011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valkov E, et al. Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation. Nat Struct Mol Biol. 2016;23:574–579. doi: 10.1038/nsmb.3232. [DOI] [PubMed] [Google Scholar]
- 43.Lai T, et al. Structural basis of the PNRC2-mediated link between mrna surveillance and decapping. Structure. 2012;20:2025–2037. doi: 10.1016/j.str.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Chang CT, Bercovich N, Loh B, Jonas S, Izaurralde E. The activation of the decapping enzyme DCP2 by DCP1 occurs on the EDC4 scaffold and involves a conserved loop in DCP1. Nucleic Acids Res. 2014;42:5217–5233. doi: 10.1093/nar/gku129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 46.Zanier K, et al. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science. 2013;339:694–698. doi: 10.1126/science.1229934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Zapien D, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–545. doi: 10.1038/nature16481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cherry JJ, et al. Structure based identification and characterization of flavonoids that disrupt human papillomavirus-16 E6 function. PLoS One. 2013;8:e84506. doi: 10.1371/journal.pone.0084506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malecka KA, et al. Identification and characterization of small molecule human papillomavirus E6 inhibitors. ACS Chem Biol. 2014;9:1603–1612. doi: 10.1021/cb500229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanier K, et al. The E6AP binding pocket of the HPV16 E6 oncoprotein provides a docking site for a small inhibitory peptide unrelated to E6AP, indicating druggability of E6. PLoS One. 2014;9:e112514. doi: 10.1371/journal.pone.0112514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryan OW, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3:e03703. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.