Significance
RNA–protein (RNP) granules contribute to spatiotemporal regulation of gene expression in eukaryotes. RNP granules have also been implicated in the pathology of neurodegenerative diseases. Insights into mechanisms of assembly and disassembly are fundamental to understanding the biology of RNP granules. In this manuscript, we provide evidence to support a model for P-body assembly which suggests that a summation of scaffolding interactions drives P-body assembly and scales the size of P body-related assemblies. Since the multivalent nature of factors and scaffolding interactions is a conserved feature of most RNP granules, our model provides an overarching theme for how other RNP granules in biology could assemble and function in vivo.
Keywords: P bodies, RNP granules, mRNA decay
Abstract
Eukaryotic cells contain multiple RNA–protein assemblies referred to as RNP granules, which are thought to form through multiple protein–protein interactions analogous to a liquid–liquid phase separation. One class of RNP granules consists of P bodies, which consist of nontranslating mRNAs and the general translation repression and mRNA degradation machinery. P bodies have been suggested to form predominantly through interactions of Edc3 and a prion-like domain on Lsm4. In this work, we provide evidence that P-body assembly can be driven by multiple different protein–protein and/or protein–RNA interactions, including interactions involving Dhh1, Psp2, and Pby1. Moreover, the relative importance of specific interactions can vary with different growth conditions. Based on these observations, we develop a summative model wherein the P-body assembly phenotype of a given mutant can be predicted from the number of currently known protein–protein interactions between P-body components.
Eukaryotic cells contain a wide diversity and number of RNP granules, which are nuclear or cytoplasmic assemblies of RNA and protein. RNP granules are broadly characterized by the lack of a limiting membrane, and thought to be held together by an extensive network of protein–protein, protein–RNA, and possibly RNA–RNA interactions (1, 2). One class of conserved RNP granules consists of P bodies, which are formed from populations of nontranslating mRNAs that are translationally repressed and/or targeted for interactions with the mRNA degradation machinery (3).
As non–membrane-bound organelles, it is of interest to understand how cells assemble large RNP granules and regulate their dynamics. The components of P bodies, and other RNP granules, are generally dynamic as assessed by fluorescence recovery after photobleaching (4). Moreover, some RNP granules exhibit liquid-like behavior, including a spherical shape, and liquid-like flow and viscosity (5, 6). Current models for messenger (m)RNP granule formation, often based on macroscopic behavior of RNP granules and the behavior of simplified model systems, suggest that proteins and RNA undergo extensive intermolecular interactions, which causes a liquid demixing event and drives liquid–liquid phase separation of these RNPs into liquid droplets (7, 8). A missing aspect in this model is identifying the full range of interactions that drive an RNP granule assembly in vivo and then relating those interactions to the larger properties of the assembly.
P bodies in Saccharomyces cerevisiae are a good model system for understanding RNP granule assembly since they are relatively discrete structures with a limited number of abundant components and are easily subjected to genetic analyses. The predominant components of yeast P bodies are Dcp1 and Dcp2, which constitute the decapping enzyme, Edc3, Pat1, Dhh1, and the Lsm1–7 complex, all of which are mRNA-binding proteins that stimulate the process of mRNA decapping (9–13). P-body assembly requires a pool of nontranslating mRNAs (14), and is thought to be predominantly driven by multivalent interactions of the Edc3 protein, which can serve as a scaffold for P-body assembly largely based on experiments done during glucose deprivation, when P-body formation is robust (15). P-body assembly is also enhanced by a prion-like domain of Lsm4, and by features of Pat1 (15, 16). Indeed, edc3∆ lsm4∆C yeast strains show a very strong defect in P-body formation during glucose depletion (15). In S. cerevisiae and Schizosaccharomyces pombe, Dcp2 can also enhance P-body assembly (7, 17), which is explained by the C terminus of Dcp2 containing six HLM motifs, which can interact with Edc3 and/or Pat1 to create a multivalent assembly between mRNPs (7). This role of Dcp2 illustrates the possibility of diverse interactions that can drive P-body formation.
During unrelated experiments, we observed that the edc3∆ lsm4∆C strain, deficient for P-body assembly in midlog growth, formed P bodies during the approach to stationary phase (see below), suggesting that there are additional interactions that promote P-body assembly. By genetic analyses, we provide evidence that the P-body components Psp2 and Pby1 and the DEAD-box helicase Dhh1 provide multivalent interactions that contribute to P-body formation. Although P bodies are increased with stress, we provide evidence that wild-type and even P-body assembly mutants form smaller constitutive P bodies. Based on these observations, we develop a summative model, wherein the P-body assembly phenotype of a given mutant can be predicted from the number of interactions contributing to P-body assembly. This highlights the redundant nature of RNP granule assembly, and underscores that many mutants that fail to make visibly detectable P bodies still form smaller related assemblies.
Results
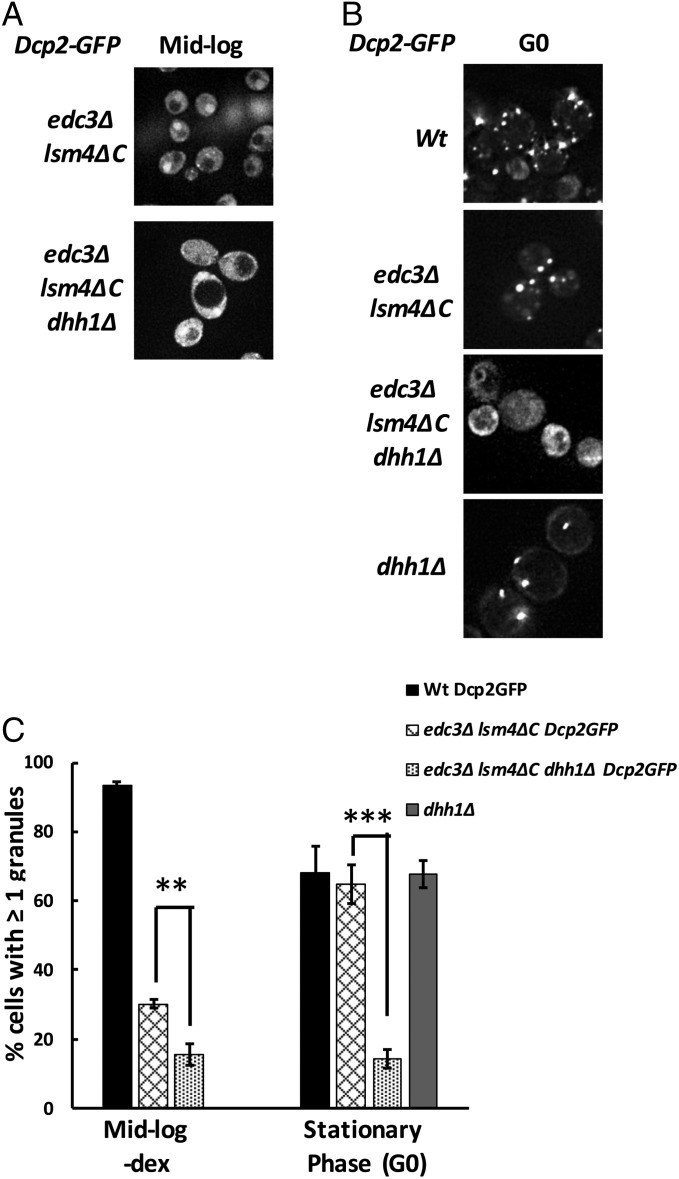
edc3Δ lsm4ΔC Yeast Assemble P Bodies in Stationary Phase.
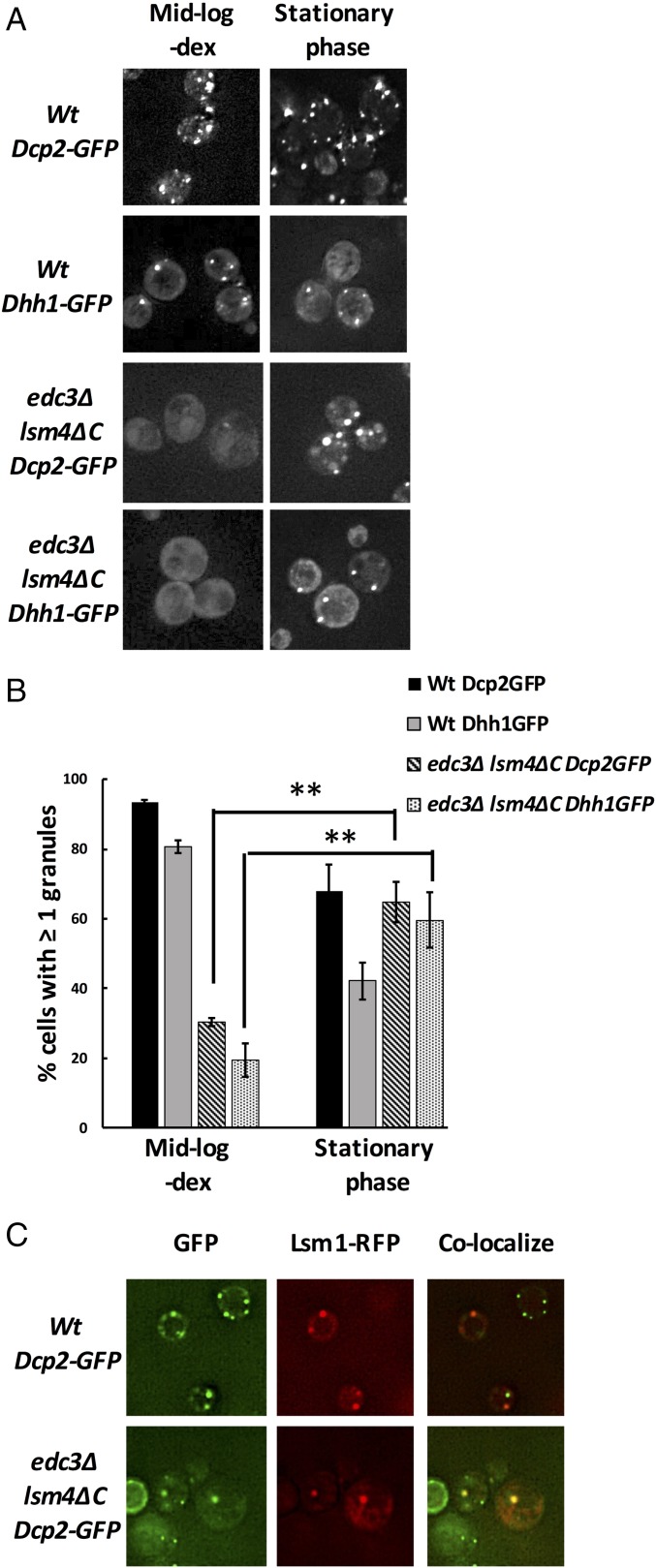
Previous work has indicated that Edc3 and the C-terminal prion-like domain of Lsm4 are important for the assembly of P bodies during glucose depletion (15). However, we observed that P-body assembly in edc3∆ lsm4∆C strains in stationary-phase cultures was markedly improved compared with glucose-starved midlog conditions as assessed by the presence of at least one Dcp2-GFP– or Dhh1-GFP–containing P body per cell (Fig. 1 A and B). This is in contrast to the edc3∆ lsm4∆C strain showing a strong reduction in P bodies during glucose deprivation of midlog cultures (Fig. 1). Additionally, the stationary-phase Dcp2-GFP foci exhibit consistent colocalization with Lsm1-RFP (red fluorescent protein), which is also a P-body marker, suggesting that the observed foci are P bodies (Fig. 1C).
Fig. 1.
edc3Δ lsm4ΔC yeast assemble P bodies in stationary phase. (A) Representative images of Dcp2-GFP– or Dhh1-GFP–containing wild-type and edc3Δ lsm4ΔC yeast grown in SComplete media containing 2% dextrose (dex). Images were taken with cells in midlog phase under glucose-depleted conditions and in stationary phase. (B) Quantification of P bodies in cells from the images obtained in A, in which images were similarly thresholded for fluorescence intensity and the percentage of cells with at least one Dcp2-GFP or Dhh1-GFP granule was determined. Error bars indicate the SD of mean values. Statistical significance for changes in cells with at least one P body was determined using a Student’s t test (**P ≤ 0.01). (C) Dcp2-GFP–containing wild-type and edc3Δ lsm4ΔC strains used in A and B were cotransformed with plasmid expressing Lsm1-RFP and grown to stationary phase to test colocalization with Dcp2-GFP granules. (Magnification: A and C, 100×.)
We note that a total number of P bodies per cell in edc3∆ lsm4∆C yeast in stationary phase are reduced compared with wild-type cells. However, the difference in P bodies in stationary vs. midlog edc3∆ lsm4∆C cultures demonstrates that P-body assembly is not restricted to interactions mediated by Edc3 and the C-terminal Q/N domain of Lsm4, and that alternative interactions can drive P-body formation.
Overexpression of Dhh1 Restores P-Body Assembly in Midlog edc3Δ lsm4ΔC Yeast.
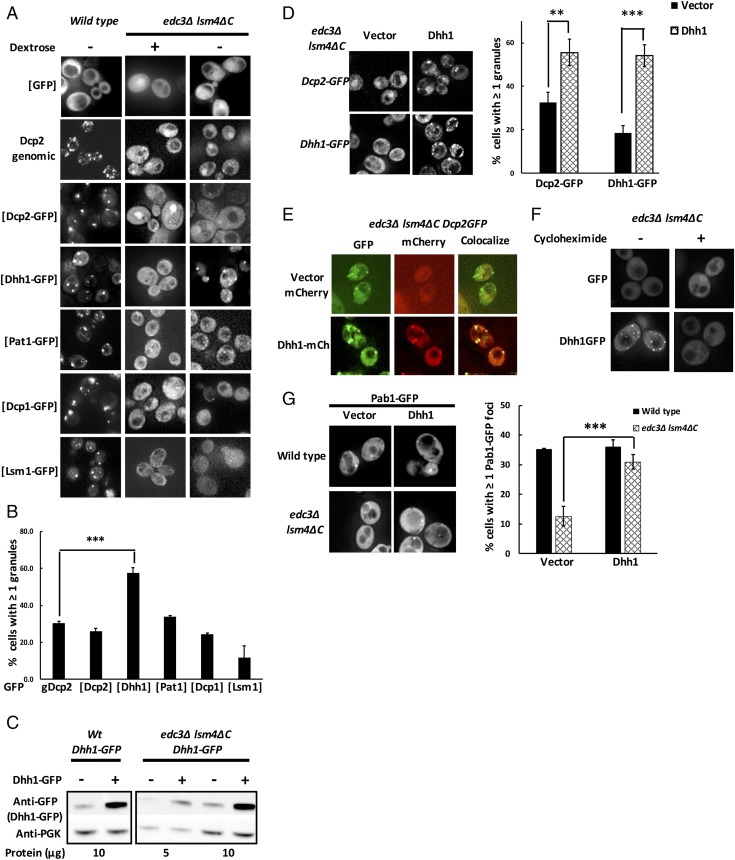
We hypothesized that other P-body components capable of driving P-body formation in a non–Edc3-, non–Lsm4-dependent manner in stationary phase would increase P-body assembly in the edc3∆ lsm4∆C strain when overexpressed during midlog growth. Hence, we introduced a GFP-tagged extra copy of several core P-body components under their native promoters, on a centromere plasmid into the edc3∆ lsm4∆C yeast strain, and assessed whether the formation of P bodies was increased relative to an edc3Δ lsm4ΔC strain with a genomic Dcp2-GFP.
We observed that during glucose deprivation only Dhh1-GFP overexpression rescued P-body formation in edc3Δ lsm4ΔC yeast cells compared with those that expressed the Dcp2-GFP fusion either as a genomic copy or ectopically (Fig. 2 A and B). Conversely, we observed that Dcp1-GFP and Lsm1-GFP failed to yield any increase in P bodies compared with the Dcp2 background. A slight increase in cells with green puncta was also observed in yeast expressing Pat1-GFP, but the difference compared with Dcp2-expressing control yeast was not statistically significant (Fig. 2 A and B). Since, on average, a centromere plasmid should provide only a single extra copy of the Dhh1 gene, this suggests that approximately a two- to threefold increase in Dhh1 levels, as seen by Western blots, is sufficient to rescue P-body assembly in the edc3Δ lsm4∆C strain (Fig. 2C).
Fig. 2.
Overexpression of Dhh1 in edc3Δ lsm4ΔC yeast led to partial recovery of P-body assembly under glucose-depleted conditions. (A) edc3Δ lsm4ΔC yeast were transformed with either GFP-only or GFP-tagged variants of Dcp2, Dhh1, Pat1, Dcp1, and Lsm1, and tested for P-body assembly in glucose starvation under midlog growth. Figures contain representative images of cells observed via fluorescence microscopy. For comparison, edc3Δ lsm4ΔC with a genomically GFP-tagged Dcp2 was similarly treated and imaged. (B) Quantification of cells with GFP-positive puncta from A. (C) Western blot indicating two- to threefold overexpression of Dhh1-GFP off a CEN vector. Wild-type and edc3Δ lsm4ΔC yeast containing genomic Dhh1-GFP were transformed with either vector (−) or an additional copy of Dhh1-GFP (+), and expression was tested under midlog growth. (D) Representative images and quantification of overexpression of untagged Dhh1 on a CEN vector in edc3Δ lsm4ΔC Dcp2-GFP or edc3Δ lsm4ΔC Dhh1-GFP. (E) Colocalization of overexpressed Dhh1-mCherry with Dcp2-GFP in edc3Δ lsm4ΔC Dcp2-GFP yeast under midlog glucose-depleted conditions. (F) Dhh1-GFP–overexpressing edc3Δ lsm4ΔC yeast were glucose-starved in the presence of cycloheximide (100 µg/mL) and tested for its effect on P-body formation. (G) Wild-type and edc3Δ lsm4ΔC yeast were cotransformed with an untagged Dhh1 and Pab1-GFP, and tested for stress granule formation (Pab1-GFP foci) after 30 min of glucose starvation. The number of yeast cells with at least one Pab1-GFP stress granule was quantified. Error bars indicate the SD of the mean. Statistical significance was determined using a Student’s t test (**P ≤ 0.01, ***P ≤ 0.001). (Magnification: A and D–G, 100×.)
Four observations suggest that the increase in Dhh1-GFP–positive foci represented an increase in P-body assembly in vivo. First, overexpression of untagged Dhh1 also increased P-body formation in edc3∆ lsm4∆C strains, as assessed by recruitment of either genomically expressed Dcp2-GFP or Dhh1-GFP to P bodies (Fig. 2D), which demonstrates that the increase in P bodies is not simply due to increased recruitment of the overexpressed Dhh1-GFP. Second, in strains with Dcp2-GFP, overexpression of Dhh1-mCherry led to the accumulation of both Dcp2-GFP and Dhh1-mCherry in the same foci, which is consistent with these being P bodies (Fig. 2E). Third, addition of cycloheximide to cultures during glucose deprivation blocked P-body assembly, consistent with earlier results that P bodies require a pool of nontranslating mRNA (14) (Fig. 2F). Fourth, we observed that overexpression of Dhh1 also rescued the formation of stress granules, as assessed by Pab1-GFP, during glucose deprivation (Fig. 2G), where stress granule formation is promoted by P bodies (16). This last observation demonstrates that the P bodies formed by Dhh1 overexpression are functionally similar to P bodies in wild-type cells. Thus, a modest increase in Dhh1 expression can rescue P-body formation in the edc3∆ lsm4∆C strain.
Overexpression of Psp2 and Pby1 Recovers P-Body Assembly in edc3∆ lsm4∆C Yeast.
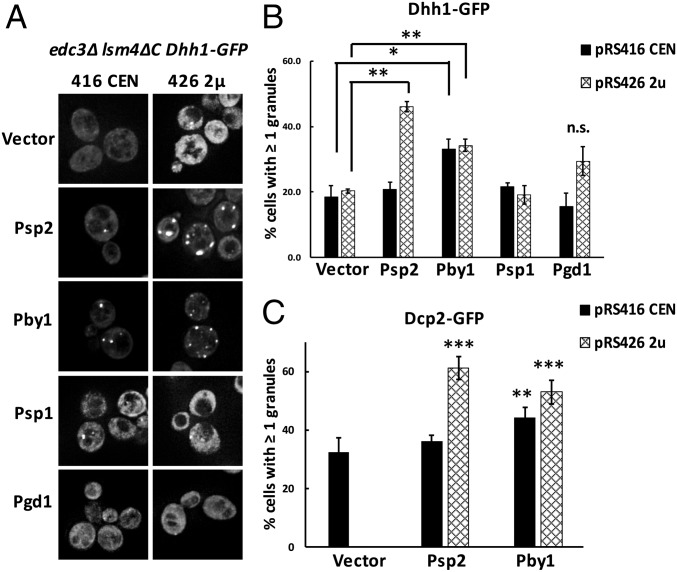
The rescue of P-body formation in edc3∆ lsm4∆C strains by a modest increase in Dhh1 demonstrates that there are redundant, and multiple, sets of interactions allowing efficient P-body formation. Several other proteins have been identified as accumulating in P bodies (18–20), including the RNA-binding proteins Psp1 and Psp2, both of which contain a Q/N-rich region analogous to the Lsm4 C-terminal extension, the Pgd1 protein, and Pby1, a tubulin tyrosine ligase. Since the function of Psp1, Psp2, Pgd1, and Pby1 in P-body assembly is unclear, we hypothesized that some of these proteins might contribute to P-body assembly but normally not be required due to redundant assembly mechanisms. To test this hypothesis, we determined if overexpressing Psp1, Psp2, Pby1, or Pgd1 could rescue P-body formation in the edc3∆ lsm4∆C strain. In these experiments, we overexpressed the Psp1, Psp2, Pby1, and Pgd1 proteins on CEN (pRS416) and 2-μ (pRS426) vectors to obtain different levels of expression in vivo. These experiments led to four observations.
First, we observed that overexpression of Psp1 or Pgd1 was unable to rescue P-body formation in the edc3∆ lsm4∆C strain. Second, we observed that Pby1 overexpression caused a clear increase in the number of Dhh1-GFP–positive P bodies compared with a vector-only control (Fig. 3 A and B). Interestingly, the effect of Pby1 overexpression on a centromere plasmid or a high-copy plasmid was similar, which implies there is a saturable role for Pby1 in promoting P-body assembly. Third, we observed that Psp2 rescued P-body formation in the edc3∆ lsm4∆C strain, but only when expressed from a 2-μ plasmid (Fig. 3 A and B). Fourth, overexpression of Psp2 and Pby1 also led to an increase in Dcp2-GFP–positive P bodies (Fig. 3C). Taken together, these observations identify Psp2 and Pby1 as two additional components that can promote P-body formation. We suggest that they are normally not required for P-body assembly due to the redundant mechanisms of P-body formation (Discussion). Thus, in addition to Dhh1, Psp2 and Pby1 can also rescue P-body formation in the edc3∆ lsm4∆C strain, demonstrating that multiple redundant interactions can enhance P-body assembly.
Fig. 3.
Psp2 and Pby1 overexpression recovers P-body assembly in edc3Δ lsm4ΔC yeast. (A) edc3Δ lsm4ΔC Dhh1-GFP yeast transformed with CEN and 2-μ (2u) vectors containing Psp2, Pby1, Psp1, and Pgd1 were tested for P-body assembly under midlog glucose-depleted conditions. (Magnification: 100×.) (B) The number of cells with at least one Dhh1-GFP–positive P body was quantified. (C) The number of cells with at least one Dcp2-GFP–positive P body in edc3Δ lsm4ΔC Dcp2-GFP yeast transformed with Psp2 and Pby1 on CEN and 2-μ vectors was quantified. Error bars indicate the SD of the mean. Statistical significance was determined using a Student’s t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; n.s., not significant). For C, the significance was determined by comparing with P bodies in a CEN vector-only control.
Mechanism of P-Body Rescue by Dhh1.
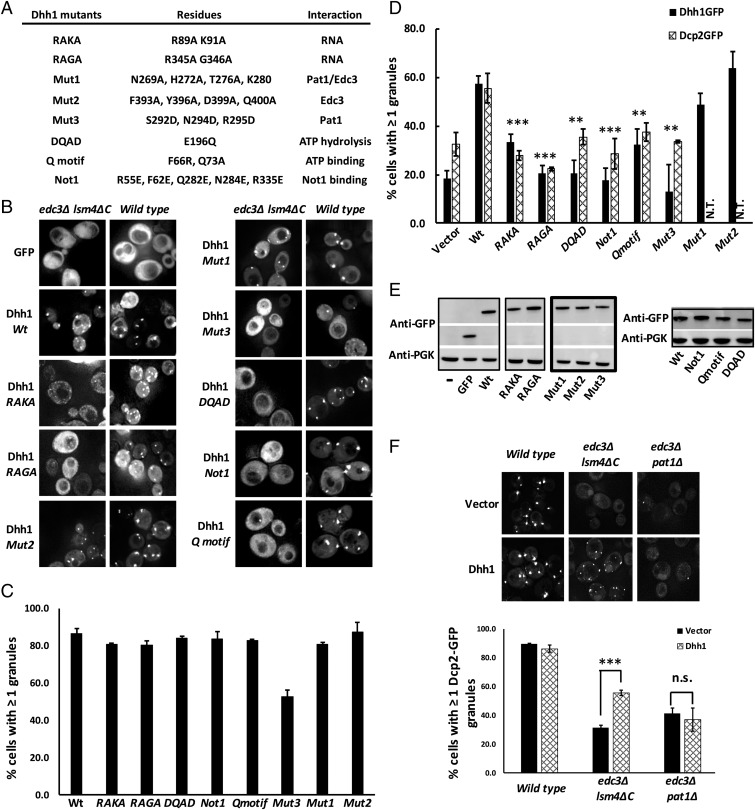
To understand how additional components could promote P-body assembly, we focused on determining the interactions that allow Dhh1 to rescue P-body formation. Dhh1 is a member of the DEAD-box family of ATP-dependent helicases, and is known to interact with Edc3, Pat1, Not1, and RNA, as well as possessing ATPase activity (10, 21–23). Dhh1 has been suggested to promote P-body assembly by binding ATP, interacting with RNA, and then forming a higher-order assembly (24). We tested whether each of these Dhh1 interactions was important for P-body assembly in the edc3∆ lsm4∆C strain. In each case, we made mutations in Dhh1 that disrupt a specific interaction (Fig. 4A) or activity, and then tested how they affected the restoration of P bodies in the edc3∆ lsm4∆C strain. The experiments led to the following conclusions.
Fig. 4.
Multivalent interactions of Dhh1 are essential for recovery of P-body assembly. (A) Table showing the mutations made in Dhh1. (B) GFP-tagged wild-type and mutant Dhh1 proteins were expressed in edc3Δ lsm4ΔC and its corresponding isogenic wild-type yeast, and the effect of the mutation on P-body recovery was tested. (C) Quantification of P-body assembly in wild-type yeast using Dhh1-GFP (wild type and mutants) to test recruitment of Dhh1-GFP variants to P bodies. (D) The number of cells with at least one Dhh1-GFP–positive (for tagged wild-type and mutant Dhh1 proteins transformed into edc3Δ lsm4ΔC yeast; black bars) or Dcp2-GFP–positive P body (for untagged wild-type and mutant proteins transformed into edc3Δ lsm4ΔC Dcp2-GFP yeast; patterned bars) was quantified. N.T., not tested. (E) Western blot showing expression of GFP-tagged Dhh1 mutants in edc3Δ lsm4ΔC yeast. (F) Dhh1 overexpression in edc3Δ pat1Δ yeast does not rescue P-body assembly. Wild type, edc3Δ lsm4ΔC, and edc3Δ pat1Δ yeast were cotransformed with plasmids encoding Dcp2-GFP and untagged Dhh1 (or vector-only), and P-body assembly was tested under glucose-depleted conditions. Error bars indicate the SD of the mean. Statistical significance was determined using a Student’s t test (**P ≤ 0.01, ***P ≤ 0.001; n.s., not significant). (Magnification: B and F, 100×.)
First, RNA binding is important for P-body rescue, since two GFP-tagged double mutants of Dhh1, R89A K91A (Dhh1-RAKA) and R345A G346A (Dhh1-RAGA), which reduce Dhh1 RNA binding (23), prevent the rescue of P bodies in the edc3∆ lsm4∆C strain (Fig. 4 A and B). Similar results were also seen with overexpressing an untagged Dhh1 protein with or without the RAKA and RAGA mutations in the edc3∆ lsm4∆C yeast strains expressing a genomically GFP-tagged Dcp2 (Fig. 4D). These Dhh1 RNA-binding mutants are expressed well by Western analysis (Fig. 4E), and are recruited normally to P bodies in a wild-type strain (Fig. 4 B and C), which suggests their overall folding and interactions are not drastically altered. Interestingly, the recruitment of Dhh1 variants defective in RNA binding to wild-type P bodies indicates that RNA binding per se is not required for recruitment into a P body. Since RNA binding is required for Dhh1 to serve as an assembly factor, or scaffold, for P bodies in edc3Δ lsm4ΔC yeast, we suggest that the RNA-binding mutant has changed the role of Dhh1 from a scaffold protein to a client protein in P bodies (8).
A second conclusion is that interaction with Pat1 is required for Dhh1 to rescue P-body assembly. We made mutations in the C-terminal RecA domain of Dhh1 that either disrupt interaction with Edc3, Pat1, or both (21) (Fig. 4A). The key observation was that the “Mut3” mutation (21), which specifically disrupts interaction with Pat1, prevents Dhh1 from rescuing P-body formation in the edc3∆ lsm4∆C strain (Fig. 4 B and D). In contrast, “Mut2” (21), which specifically disrupts Dhh1 interaction with Edc3, does not alter the ability of Dhh1 to rescue P-body assembly in the edc3∆ lsm4∆C strain, while “Mut1,” which disrupts both Edc3 and Pat1 interaction, albeit to a lesser extent, does not hinder Dhh1 rescue of P bodies (Fig. 4 B and D). The GFP-tagged Dhh1-Mut3 protein was expressed normally, but exhibited a slightly reduced recruitment to P bodies in the wild-type yeast strain (Fig. 4 B–E), suggesting it might be partially misfolded and/or is dominant-negative over the wild-type counterpart. Nevertheless, the inability of the Mut3 mutant to rescue P-body assembly tested using two P-body markers indicated that in the edc3∆ lsm4∆C strain, interaction of Dhh1 with Pat1 is essential for P-body assembly. Consistent with this interpretation, overexpression of Dhh1 in an edc3∆ pat1∆ strain fails to rescue P-body formation (Fig. 4F).
Third, and consistent with earlier work (24), we observed that a mutation that prevents Dhh1-binding ATP (Q motif) prevents its ability to promote P-body formation (Fig. 4).
Finally, we also observed that mutations in Dhh1 that inhibit its ATPase activity, either by mutation of the DEAD motif (DQAD mutant) or by limiting its interaction with Not1, which stimulates its ATPase activity (24) (Dhh1-Not1-R55E, F62E, Q282E, N284E, R335E), prevent Dhh1 from rescuing P bodies in the edc3∆ lsm4∆C strain (Fig. 4).
Taken together, these results argue that Dhh1 enhances P-body formation by binding ATP, interacting with RNA and Pat1, and thereby creating increased numbers of intermolecular interactions that give enhanced P-body formation.
Dhh1 Contributes to Residual P-Body Formation in edc3∆ lsm4∆C Yeast.
The above results indicate that overexpressed Dhh1 can contribute to P-body formation under conditions when P-body formation is limited (edc3∆ lsm4∆C), which implies that the normal levels of Dhh1 might promote P-body formation, although dhh1∆ cells show no, or minor, effects on P-body assembly (17, 24). We hypothesized that Dhh1’s role in P-body assembly is minimal in wild-type cells due to Edc3 and Lsm4 dominating P-body assembly. To test this prediction, we deleted Dhh1 from the edc3∆ lsm4∆C strain and examined its effect on P-body formation. We observed that the residual Dcp2-GFP–containing P bodies were significantly reduced (twofold) in the edc3∆ lsm4∆C dhh1∆ compared with the edc3∆ lsm4∆C strain under midlog conditions (Fig. 5 A and C). Finally, we observed that the edc3∆ lsm4ΔC dhh1∆ yeast exhibit a marked reduction in the number of Dcp2-GFP–containing P bodies in stationary phase (G0) compared with the edc3∆ lsm4∆C yeast, suggesting a role for Dhh1- and Dhh1-mediated interactions on P-body assembly in stationary phase (Fig. 5 B and C). Interestingly, the deletion of Dhh1 alone did not have a significant effect on P-body assembly in stationary phase, suggesting the observed reduction in P bodies is not a defect specifically due to Dhh1 but dependent on the simultaneous loss of three P-body components (Fig. 5C). Taken together, these observations argue that Dhh1 contributes to P-body assembly at endogenous levels but that Dhh1’s contribution is redundant with Edc3 and Lsm4.
Fig. 5.
Dhh1 contributes to residual P-body assembly in vivo. (A and B) The contribution of Dhh1-mediated interactions on residual P-body assembly in (A) midlog phase and in (B) stationary phase (G0) was tested. (C) Quantification of P-body assembly by visualizing Dcp2-GFP–positive foci in wild-type, edc3Δ lsm4ΔC, edc3Δ lsm4ΔC dhh1Δ, and dhh1Δ only (stationary phase only). Error bars indicate the SD of the mean. Statistical significance was determined using a Student’s t test (**P ≤ 0.01, ***P ≤ 0.001). (Magnification: A and B, 100×.)
Analysis of P-Body Assembly by Nanoparticle Tracking.
The above results suggested that a diversity of protein interactions could contribute to P-body assembly in a redundant manner. This redundancy would suggest that there should be smaller P body-like assemblies that form even when P bodies are not easily visible by light microscopy. This is consistent with theoretical models of polymer separations where assemblies form at smaller scales until they cross a system-spanning size that allows condensation into a larger mesoscopic phase separation (25). Indeed, examination of simple model systems shows that assembly size increases until the critical concentration wherein phase separation occurs (figure S10 in ref. 26). However, an examination of the size and number of P bodies in cells has been limited by quantitative techniques.
To analyze the size and number of P bodies in yeast cells and how they were affected by various mutations of P-body components, we examined if nanoparticle tracking analysis (NTA) could be used to analyze the size distribution and number of P bodies in yeast. We grew various strains, prepared lysates, and analyzed the P bodies within them following either GFP-tagged Dhh1 or Dcp2 by NTA (Materials and Methods).
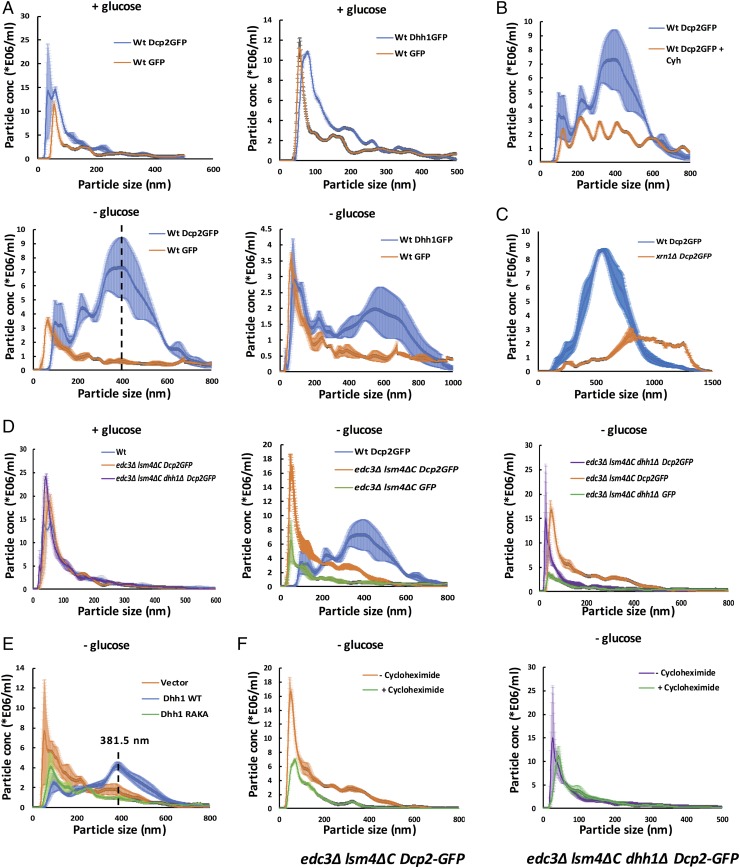
Several observations suggest NTA is a reliable approach to observing P bodies. First, in wild-type cells, we observed dramatic redistribution of Dcp2 or Dhh1 into larger assemblies when cells were exposed to glucose deprivation (Fig. 6A, + glucose vs. − glucose), which stimulates the formation of large P bodies (14). Consistent with these assemblies detected being P bodies, their formation was blocked by cycloheximide (Fig. 6B), which inhibits P-body formation (14). Moreover, an xrn1∆ strain, which has larger P bodies (17, 27), shows larger P bodies than its corresponding isogenic wild-type control (Fig. 6C). We interpret these results to indicate that NTA is a reliable method for analyzing yeast P bodies. The use of NTA to examine yeast P bodies revealed several observations.
Fig. 6.
Submicroscopic P body-related mRNP assemblies exist in visible P body-deficient yeast. (A) Traces derived from nanoparticle tracking analysis of Dcp2-GFP, Dhh1-GFP, and GFP-only particles obtained from wild-type (Wt) yeast cells. Total cell lysates derived from yeast grown under glucose-replete (Top; + glucose) and glucose-depleted (Bottom; − glucose) conditions were analyzed using NTA. The dashed line indicates the peak height (395.5 nm) of the largest Dcp2-GFP granule observed in wild type. (B) Cycloheximide (Cyh; 100 μg/mL) treatment reduces P-body assembly as assessed by visualizing Dcp2-GFP particles from wild-type yeast. (C) Deletion of xrn1Δ increases mean P-body size compared with wild type. These experiments were conducted in an alternate yeast genetic background strain. (D) Distinct reduction in the mean size of Dcp2-GFP particles obtained from glucose-starved edc3Δ lsm4ΔC or edc3Δ lsm4ΔC dhh1Δ yeast. The particles observed in edc3Δ lsm4ΔC Dcp2-GFP and edc3Δ lsm4ΔC dhh1Δ Dcp2-GFP yeast exhibit enrichment of Dcp2-GFP over GFP in the corresponding control experiment (green trace; − glucose). (E) Overexpression of wild-type Dhh1 (blue) but not the RNA-binding mutant, RAKA (green), partially restores P-body assembly in edc3Δ lsm4ΔC Dcp2-GFP yeast. (F) Cycloheximide inhibits assembly of Dcp2-GFP particles in edc3Δ lsm4ΔC but not edc3Δ lsm4ΔC dhh1Δ yeast. The shaded area around the trend line in all traces represents the SEM derived from replicates.
First, we observed that even in +glucose conditions, yeast strains had assemblies of Dcp2 and Dhh1 with a mean particle size of ∼100 nm (Fig. 6A). These assemblies were notably larger, and more abundant, than assemblies detected when GFP alone was analyzed (Fig. 6A). This suggests that in the absence of stress, there are smaller abundant P bodies present in yeast cells.
Second, we observed that the wild-type, edc3∆ lsm4∆C, and edc3∆ lsm4∆C dhh1Δ strains had similar levels and sizes of P-body assemblies in the presence of glucose (Fig. 6D, + glucose) but, compared with wild-type, edc3Δ lsm4ΔC and edc3∆ lsm4∆C dhh1Δ (see below) yeast formed smaller assemblies during glucose deprivation, which were more abundant than the corresponding GFP control (Fig. 6D, − glucose). This provides additional evidence for a defect in P-body assembly during glucose deprivation in the edc3∆ lsm4∆C and edc3∆ lsm4∆C dhh1Δ strains. Strikingly, these data also show that P bodies during glucose-replete conditions are not affected by the edc3∆, lsm4∆C, and dhh1Δ mutations, suggesting that formation of P bodies during glucose-replete and -depleted conditions can be dominated by different interactions.
Third, consistent with the microscopic analyses, NTA also shows that the dhh1∆ strain further reduces the size of P bodies that form during glucose deprivation (Fig. 6D), and that overexpression of wild-type Dhh1, but not the RNA-binding mutant RAKA, can rescue P-body formation in the edc3∆ lsm4∆C strain (Fig. 6E). In support of P-body rescue, we observed that the diameter of the largest Dcp2-GFP particle in the Dhh1 rescue trace (381.5 nm) is similar to the wild-type Dcp2-GFP–derived P body (395.5 nm) (Fig. 6 A and E, dashed lines). Additionally, the mean P-body size in edc3Δ lsm4ΔC yeast is similar to that in wild-type yeast when rescued with wild-type Dhh1 (371 ± 50 nm) as opposed to Dhh1-RAKA (313 ± 31 nm) (see Dhh1 interactions, Table 1; Table 2).
Table 1.
P-body proteins contribute to multivalent intermolecular RNA–protein and protein–protein interactions
| Protein | Known P-body interactions (no. of interactions) | Binding sites per mRNA | Protein–protein valency | Total valency | Refs. |
| Dcp1 | Dcp2 (1) | 0 | 1 | 1 | 37 |
| Dcp2 | Dcp1 (1), Edc3 (4), Pat1 (1)* | 1 | 6 | 7 | 9 and 38 |
| Pat1 | Edc3 (1), Pat1 (1), Dhh1 (1), Lsm1–7 (1) | 1 | 4 | 5 | 21, 39, and 40 |
| Edc3 | Pat1 (1), Dcp2 (1), Edc3 (1), Dhh1 (1) | ≥1† | 4 | 5 | 15 and 21 |
| Dhh1 | Dhh1 (1),‡ Edc3 (1), Pat1 (1) | ≥1‡ | ≥3‡ | 4 | 41 |
| Lsm1–7 complex | Pat1 (1), Lsm4 Q/N domain (valency unclear) | 1 + (1)§ | ≥2§ | 4 | 15 |
| Psp2 | Scd6 (1) | 1 | 1 | 2 | 28 |
| Scd6 | Edc3 (1), Dhh1 (1), Psp2 (1) | 0 | 3 | 3 | 13 |
| Pby1 | Pby1 (1), Edc3 (1) | 0 | 2 | 2 | 29 and 32 |
Summary of known P body-related interactions of conserved P-body components. The assumptions made in deriving the table are mentioned in the footnotes. Specifically, the data assume at least one mRNA binding site for Dhh1, Edc3, and Psp2 due to previous observations, but the total mRNA binding might exceed that number. Similarly, the number of interactions catalyzed by the intrinsically disordered domain (Q/N) of the Lsm4 C terminus is not known.
The interaction between Dcp2 and Pat1 is not demonstrated to be direct.
The RNA binding site and mRNA occupancy of Edc3 have not been demonstrated. Hence, Edc3 could bind at more than one location.
Dhh1 can self-interact and undergo oligomerization. However, the stoichiometry of the monomers in the oligomers is unknown. Additionally, UV cross-linking and immunoprecipitation (CLIP)-derived data suggest more than one binding site for Dhh1 on mRNA.
The number of RNA and protein interactions of the intrinsically disordered Q/N domain of Lsm4 is not known.
Table 2.
Interaction “score” for P-body assembly in yeast strains
| Yeast strain | Interactions contributed by P-body proteins | Total interactions | Visible P bodies (average diameter, nm) | ||||||||
| Dcp1 | Dcp2 | Pat1 | Edc3 | Dhh1 | Lsm1–7 | Psp2 | Scd6 | Pby1 | |||
| Wild type | 1 | 7 | 5 | 5 | 4 | 4 | 2 | 3 | 3 | 34 | ++++ (381 ± 15) |
| edc3Δ lsm4ΔC | 1 | 3 | 4 | 0 | 3 | 2 | 2 | 2 | 2 | 19 | ++ (210 ± 24) |
| edc3Δ lsm4ΔC + Dhh1 | 1 | 3 | 4 | 0 | ≥6 | 2 | 2 | 2 | 2 | ≥22 | +++ (371 ± 50) |
| edc3Δ lsm4ΔC + Dhh1 RAKA | 1 | 3 | 4 | 0 | ≥5 | 2 | 2 | 2 | 2 | ≥21 | ++ (313 ± 31) |
| edc3Δ lsm4ΔC dhh1Δ | 1 | 3 | 3 | 0 | 0 | 2 | 2 | 1 | 2 | 14 | + (125 ± 48) |
| edc3Δ pat1Δ | 1 | 2 | 0 | 0 | 2 | 3 | 2 | 2 | 2 | 14 | + (NA) |
| edc3Δ pat1Δ + Dhh1 | 1 | 2 | 0 | 0 | 4 | 3 | 2 | 3 | 2 | 17 | + (NA) |
Sum total of known interactions catalyzed by key P-body components. The summation is derived from the number of interactions listed in Table 1. The number of “+” is a measure of the degree to which visible P bodies assemble; “++++” indicates optimal P-body assembly while “+” indicates a strong P-body assembly deficiency. For Dhh1 overexpression, the increase in the number of interactions is calculated for a twofold increase in Dhh1 concentration. NA, not available.
We also observed that the smaller Dcp2-GFP particles observed in the edc3∆ lsm4∆C yeast were sensitive to cycloheximide treatment, indicating their dependence on nontranslating mRNA for assembly and that their assembly mechanism mirrors that of wild-type P bodies (Fig. 6F). Interestingly, the Dcp2-GFP particles obtained from edc3∆ lsm4∆C dhh1Δ yeast were not significantly affected by cycloheximide treatment, suggesting these particles either form on a stable pool of untranslating mRNAs that are not affected by cycloheximide (perhaps decapped mRNAs) or do not contain mRNA.
Taken together, we demonstrate that NTA can reproducibly detect Dcp2-GFP particles under various physiological conditions and from different yeast genetic backgrounds, which corroborate P-body assembly phenotypes observed microscopically. Consequently, these observations support the notion that submicroscopic Dcp2-GFP–related assemblies detected in lysates from edc3Δ lsm4ΔC and edc3Δ lsm4ΔC dhh1Δ yeast are physiologically relevant and derived via P-body assembly-dependent mechanisms. In conclusion, we provide evidence that P-body assembly proceeds via a continuum of smaller mRNP assemblies, which represent intermediates on the pathway to assembling larger visible P bodies and which are directly affected by the availability and summation of scaffolding interactions.
Discussion
Several observations in this manuscript argue that yeast P-body assembly can occur by multiple redundant mechanisms. This was first suggested by observations that no single P-body component was absolutely required for P-body formation (15, 17), although a strong defect was observed in the edc3∆ lsm4∆C strain (15). More recently, Dhh1 has been demonstrated to self-assemble and to have a small effect on P-body assembly (24). We now provide evidence for alternative assembly mechanisms, since the edc3∆ lsm4∆C strain efficiently forms P bodies during stationary phase (Fig. 1) and overexpression of Psp2, Pby1, or Dhh1 can rescue the defect in P-body assembly in edc3∆ lsm4∆C yeast seen in glucose depletion (Figs. 2 and 3). Thus, P-body assembly can occur by multiple interdependent assembly mechanisms.
The ability of Dhh1 to promote P-body assembly requires its ability to interact with RNA and bind Pat1, and can be affected by its ability to bind ATP and undergo ATP hydrolysis (Fig. 4). This is consistent with a model whereby Dhh1 promotes P-body assembly through multivalent interactions, most notably binding mRNAs and then interacting with Pat1. It is notable that while Dhh1 overexpression can rescue P-body formation in edc3∆ lsm4∆C, we do not observe a comparable rescue with overexpression of Pat1 (Fig. 2). A simple explanation for this difference comes from the demonstration that Pat1 predominantly binds to mRNAs at their very 3′ end, and only one binding site for Pat1 is identified in most mRNAs (19). Thus, overexpression of Pat1 would not be expected to increase P-body formation, since its binding sites are likely to be saturated. In contrast, Dhh1 can bind mRNAs in multiple regions, including the 5′ and 3′ UTRs as well as the coding region (19). Thus, overexpression of Dhh1 would be expected to increase the average number of Dhh1 molecules per mRNA and, by interacting with Pat1, increase the number of interactions between individual mRNPs, thereby enhancing P-body assembly.
The ability of Psp2 and Pby1 to increase P-body assembly when overexpressed suggests they will also have multivalent interactions between P-body components that allow them to enhance the interactions between individual mRNPs. Psp2 binds mRNA (19) and also interacts with Scd6, a known P-body component (28), suggesting it could enhance P-body formation through these interactions. Pby1 is known to coimmunoprecipitate and/or have two-hybrid interactions with Dcp1 and Edc3 as well as interact with itself (29–33), suggesting a model where Pby1 could cross-link individual mRNPs through these protein–protein interactions. Since increasing the gene dosage of Pby1 did not further increase the assembly of P bodies, it suggests that the binding sites (e.g., Dcp1) are limited and become saturated when Pby1 is overexpressed even twofold. Although the molecular details of how these proteins promote P-body assembly remain to be determined, the fact that overexpression of three different P-body components (Dhh1, Psp2, or Pby1) can rescue P-body assembly highlights the diversity of mechanisms by which P bodies can form.
The above observations highlight the redundant nature of P-body assembly in yeast. Given this, we developed a summative model for how individual proteins would affect P-body formation based on their expected or hypothesized binding sites per mRNA molecule and the number of identified protein–protein interactions for each component (Table 1). Although there are some assumptions made to build this model (detailed in comments for each protein), the model explains the behavior of how various mutations affect P-body assembly. For example, in wild-type cells, we predict that P bodies assemble through a summation of 34 interactions per mRNP (Table 2). Conversely, in the edc3∆ lsm4∆C strain, only 19 interactions are preserved, and P-body assembly is very poor.
Analysis of P-body assembly via the proposed summation model provides a molecular explanation for key assembly phenotypes reported in this manuscript. First, we predict that due to the ability of Dhh1 to oligomerize (34) and to presumably bind nontranslating RNA at multiple sites along the length of the molecule (19), Dhh1 overexpression restores, in part, the sum total of interactions required to assemble larger visible P bodies (Table 2). Mutations that affect RNA binding directly (RAKA and RAGA) or indirectly (ATP binding, hydrolysis, and Not1 interaction) reduce the net contribution of the overexpressed Dhh1 molecules, and thus reduce the impact of Dhh1 on P-body recovery. Second, we predict that the deletion of Dhh1 from the edc3Δ lsm4ΔC yeast strains leads to a further reduction in total interactions (Table 2), causing a stronger P-body assembly defect, which is corroborated by the drastic decrease in P-body assembly observed via fluorescence microscopy and NTA (Figs. 5 and 6).
Analysis of yeast deletion mutants suggests that the summation of interactions is the least in the edc3∆ pat1∆ yeast. Furthermore, our analysis predicts that due to the lack of Edc3 and Pat1, overexpression of Dhh1 would not significantly add to the sum total of interactions in the edc3Δ pat1Δ yeast, and hence fail to rescue the P-body assembly defect of the edc3Δ pat1Δ yeast. Consistent with this prediction, we did not observe any quantifiable increase in visible P bodies in edc3Δ pat1Δ yeast overexpressing Dhh1-GFP (Fig. 4F), further supporting the notion that P bodies assemble via a sum total of redundant interactions in vivo and require a threshold of interactions to assemble a minimalistic P body-related microscopically visible RNA–protein complex.
This work highlights four principles that we expect will be generalizable to other RNP granules. First, RNP granules can form by multiple redundant interactions. Second, because of redundant assembly mechanisms, mutations that inactivate a given assembly factor will often still give rise to the assembly of RNP granules, which in some cases may be below the detection limit of light microscopy (as we show for P bodies in the edc3∆ lsm4∆C strains; Fig. 6). Thus, one should be cautious about concluding that an RNP granule has no function when no assembly is detected by light microscopy. A third key principle is that different interactions can predominate under different conditions, which we illustrate by the importance of Edc3 and Lsm4 for P-body formation during glucose depletion but having a reduced effect on P-body formation during stationary phase (Fig. 1). Similar results are seen with mammalian stress granules wherein G3BP is important for stress granule formation in sodium arsenite stress but dispensable during sorbitol stress (35). Finally, this work highlights that some proteins (e.g., Pat1 and Pby1) will have saturable roles in granule assembly, presumably due to limiting binding sites on granule components, which illustrates two types of assembly factors for RNP granules that can have distinct effects on client recruitment to RNP granules.
Materials and Methods
Plasmid Construction.
PCR amplifications were conducted using the Phusion Hot Start II Kit (Thermo Fisher), unless stated otherwise. Dhh1-GFP with the Dhh1 promoter was amplified using genomic DNA from the Dhh1-GFP yeast strain from the Euroscarf yeast GFP library collection and the primers BSR_DhhGFP416NF and BSR_DhhGFP416NR. The Adh1 terminator was PCR-amplified using the BSR_Adh1SacF and BSR_Adh1SacR primers. The Dhh1-GFP and Adh1 terminator-containing PCR fragments were sequentially cloned into XhoI- and SacI-digested pRS416 vector, respectively, using the In-Fusion Cloning Kit (Takara). Specific mutants for Dhh1 were generated using the Phusion mutagenesis protocol (Thermo Fisher). Constructs lacking GFP were constructed using the primers BSR_416GFPdel and BSR_1577DelGFPR and the Phusion mutagenesis protocol.
Yeast Media and Growth.
The yeast strains and plasmids used in this study are listed in Tables S1 and S2, respectively. For P-body recovery assays, yeast strains transformed with the desired expression vectors were grown overnight in synthetic media with 2% dextrose and the appropriate amino acid dropout mix at 30 °C with aeration. The cultures were diluted to an OD600 of 0.15 in fresh synthetic media and grown to a midlog OD600 of 0.4 to 0.5. Cells were pelleted, washed, and resuspended in synthetic media lacking glucose, and incubated at 30 °C with aeration for 10 min to induce P-body assembly. Cells were repelleted and imaged using microscopy.
Table S1.
Yeast strains used in this manuscript
| Yeast strain | Genotype | Source |
| yRP1496 | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 | Lab stock |
| yRP1727 | MATa leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK 1pG/MFA2pG DCP2-GFP-NEO | 27 |
| yRP1923 | MATa leu2-3 trp1 ura3-52 his4-539 cup1::LEU2/PGK 1pG/MFA2pG xrn1::URA3 DCP2-GFP-NEO | 14 |
| yRP3205 | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 lsm4∆C::kanmx edc3∆::HygB | This study |
| yRP3206 | MATα leu2∆0 lys2∆0 ura3∆0 lsm4∆C::kanmx edc3∆::HygB DCP2-GFP-HIS3 | This study |
| yRP3207 | MATα leu2∆0 lys2∆0 ura3∆0 lsm4∆C::kanmx edc3∆::HygB DHH1-GFP-HIS3 | This study |
| yRP3208 | MATα lys2∆0 ura3∆0 lsm4∆C::kanmx edc3∆::HygB dhh1Δ::KLLeu2 DCP2-GFP-HIS3 | This study |
| yRP2313 | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 pat1Δ::kanmx edc3∆::HygB | This study |
| MATa his3∆1 leu2∆0 met15∆0 ura3∆0 DCP2-GFP-KanMX | Lab stock | |
| MATa his3∆1 leu2∆0 met15∆0 ura3∆0 DHH1-GFP-KanMX | Lab stock |
Table S2.
Plasmids used in this manuscript
| Plasmid no. | Construct | Source |
| pRP2690 | pRS416 + Dhh1 promoter-Dhh1-Adh1 terminator | This study |
| pRP2691 | pRS416 + Dhh1 promoter-Dhh1 (RAKA mutations)-Adh1 terminator | This study |
| pRP2692 | pRS416 + Dhh1 promoter-Dhh1 (RAGA mutations)-Adh1 terminator | This study |
| pRP2693 | pRS416 + Dhh1 promoter-Dhh1 (Mut1 mutations)-Adh1 terminator | This study |
| pRP2694 | pRS416 + Dhh1 promoter-Dhh1 (Mut2 mutations)-Adh1 terminator | This study |
| pRP2695 | pRS416 + Dhh1 promoter-Dhh1 (Mut3 mutations)-Adh1 terminator | This study |
| pRP2696 | pRS416 + Dhh1 promoter-Dhh1 (DQAD mutations)-Adh1 terminator | This study |
| pRP2697 | pRS416 + Dhh1 promoter-Dhh1 (Q motif mutations)-Adh1 terminator | This study |
| pRP2698 | pRS416 + Dhh1 promoter-Dhh1 (Not1 mutations)-Adh1 terminator | This study |
| pRP2699 | pRS416 + Dhh1 promoter-GFP-Adh1 terminator | This study |
| pRP2700 | pRS416 + Dhh1 promoter-Dhh1-GFP-Adh1 terminator | This study |
| pRP2701 | pRS416 + Dhh1 promoter-Dhh1 (RAKA mutations)-GFP-Adh1 terminator | This study |
| pRP2702 | pRS416 + Dhh1 promoter-Dhh1 (RAGA mutations)-GFP-Adh1 terminator | This study |
| pRP2703 | pRS416 + Dhh1 promoter-Dhh1 (Mut1 mutations)-GFP-Adh1 terminator | This study |
| pRP2704 | pRS416 + Dhh1 promoter-Dhh1 (Mut2 mutations)-GFP-Adh1 terminator | This study |
| pRP2705 | pRS416 + Dhh1 promoter-Dhh1 (Mut3 mutations)-GFP-Adh1 terminator | This study |
| pRP2706 | pRS416 + Dhh1 promoter-Dhh1 (DQAD mutations)-GFP-Adh1 terminator | This study |
| pRP2707 | pRS416 + Dhh1 promoter-Dhh1 (Q motif mutations)-GFP-Adh1 terminator | This study |
| pRP2708 | pRS416 + Dhh1 promoter-Dhh1 (Not1 mutations)-GFP-Adh1 terminator | This study |
| pRP2709 | pRS415 + Dhh1 promoter-Dhh1-mCherry-Adh1 terminator | This study |
| pRP2710 | pRS415 + Dhh1 promoter-mCherry-Adh1 terminator | This study |
| pRP2711 | pRS416 + Psp1 promoter-Psp1-Adh1 terminator | This study |
| pRP2712 | pRS416 + Psp2 promoter-Psp2-Adh1 terminator | This study |
| pRP2713 | pRS416 + Pby1 promoter-Pby1-Adh1 terminator | This study |
| pRP2714 | pRS416 + Pgd1 promoter-Pgd1-Adh1 terminator | This study |
| pRP2715 | pRS426 + Psp2 promoter-Psp2-Adh1 terminator | This study |
| pRP2716 | pRS426 + Pby1 promoter-Pby1-Adh1 terminator | This study |
| pRP1891 | pRS416 + Dcp2 promoter-Dcp2-GFP-Adh1 terminator | 38 |
| pRP1502 | yPlac33 + Pat1-GFP | 39 |
| pRP1176 | yPlacIII + Lsm1 promoter-Lsm1-GFP-Lsm1 terminator | Lab stock (J. Coller) |
| pRP1535 | pRS415 + Dcp1 promoter-Dcp1-GFPKanmx-Dcp1 terminator | Lab stock (C. J. Decker) |
| pRP2478 | pRS415 + Pab1-GFP | Lab stock (J. Ross Buchan) |
| pRS426 (2µ URA3 AmpR) | Lab stock | |
| pRS415 (CEN LEU2 AmpR) | Lab stock | |
| pRS416 (CEN URA3 AmpR) | Lab stock |
For nanoparticle tracking analysis, an overnight culture of the desired yeast culture grown in synthetic media was diluted to an OD600 of 0.1 in 150 mL synthetic media and grown to a midlog OD600 of 0.4 to 0.5. Cells were pelleted, washed, and resuspended in 100 mL synthetic media lacking dextrose, and incubated at 30 °C with aeration for 10 min. Cells were pelleted, washed twice with PBS, and fixed using 2% formaldehyde in PBS for 10 min at room temperature. The formaldehyde was quenched using 250 mM Tris (pH 8.0) for 10 min, followed by two PBS washes. Cells were flash-frozen using liquid nitrogen. For unstressed cells, formaldehyde was added directly to the 150 mL culture, followed by pelleting and washing.
Microscopy.
Yeast were analyzed via fluorescence microscopy on a DeltaVision Elite microscope with a 100× objective using a PCO Edge sCMOS camera; ≥2 images comprising nine Z sections were obtained for each replicate. Images were analyzed using ImageJ (NIH). Z projections derived from summation of the Z sections with constant thresholding were used to count the number of yeast cells with ≥1 GFP-positive granule, and the percentage of cells with at least one granule was calculated. Z projections with maximum intensity were used to make the figures.
Nanoparticle Tracking Analysis.
P body-enriched yeast cell lysate was prepared in a manner similar to the stress granule enrichment described previously (18, 36). Briefly, frozen yeast cell pellets resuspended in 1× lysis buffer were disrupted using four 10-min rounds of glass bead beating with intermittent cooling. The lysate was clarified by centrifugation at 850 × g. P bodies were enriched using two 18,000 × g centrifugation steps with an interval wash with lysis buffer. The final P body-enriched fraction was resuspended in 500 μL 1× lysis buffer. A 1:3 dilution of the enriched fraction was analyzed using the NanoSight NS300 (Malvern) and 488-nm laser. Three to five runs of five 60-s videos each (total 15 to 35 videos) were collected and analyzed by NTA 3.0 software (Malvern). Average particle concentrations and sizes were calculated and analyzed using standard statistical methods.
Western Analysis.
Total cell lysates were obtained from midlog yeast cultures. Protein concentrations were measured using BCA reagent (Thermo Fisher). Expression of Dhh1-GFP (wild type and mutants) was determined using anti-GFP (BioLegend) and anti-PGK1 (Novex) antibodies. The Western blots were imaged using the ImageQuant LAS 4000 Imaging System (GE Healthcare).
Supplementary Material
Acknowledgments
We thank Josh Wheeler, Erika Lasda, and Denise Muhlrad for their assistance with NTA and for their valuable insights that helped shape this manuscript. We also thank Dr. Olke Uhlenbeck, Mansi Arora, and other members of the R.P. laboratory for their helpful comments on the experiments and data interpretation. We thank Anne Webb for her assistance with figures. This work was supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712396114/-/DCSupplemental.
References
- 1.Buchan JR. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain S, Parker R. The discovery and analysis of P bodies. Adv Exp Med Biol. 2013;768:23–43. doi: 10.1007/978-1-4614-5107-5_3. [DOI] [PubMed] [Google Scholar]
- 4.Kedersha N, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol. 2013;203:875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fromm SA, et al. In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery. Angew Chem Int Ed Engl. 2014;53:7354–7359. doi: 10.1002/anie.201402885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banani SF, et al. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tharun S, et al. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 10.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1093/genetics/166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain S, et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell SF, Jain S, She M, Parker R. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sweet TJ, Boyer B, Hu W, Baker KE, Coller J. Microtubule disruption stimulates P-body formation. RNA. 2007;13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharif H, et al. Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res. 2013;41:8377–8390. doi: 10.1093/nar/gkt600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maillet L, Collart MA. Interaction between Not1p, a component of the Ccr4-Not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem. 2002;277:2835–2842. doi: 10.1074/jbc.M107979200. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mugler CF, et al. ATPase activity of the DEAD-box protein Dhh1 controls processing body formation. eLife. 2016;5:e18746. doi: 10.7554/eLife.18746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flory PJ, Krigbaum WR. Thermodynamics of high polymer solutions. Annu Rev Phys Chem. 1951;2:383–402. [Google Scholar]
- 26.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidner J, Wang C, Prescianotto-Baschong C, Estrada AF, Spang A. The polysome-associated proteins Scp160 and Bfr1 prevent P body formation under normal growth conditions. J Cell Sci. 2014;127:1992–2004. doi: 10.1242/jcs.142083. [DOI] [PubMed] [Google Scholar]
- 29.Babu M, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 32.Gavin A-C, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 33.Decourty L, et al. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci USA. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol Biol Cell. 2009;20:2464–2472. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kedersha N, et al. G3BP–Caprin1–USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler JR, Jain S, Khong A, Parker R. Isolation of yeast and mammalian stress granule cores. Methods. 2017;126:12–17. doi: 10.1016/j.ymeth.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol Cell Biol. 2010;30:1446–1456. doi: 10.1128/MCB.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol Cell Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]