Significance
During initiation of DNA replication in eukaryotes, the origin recognition complex, with Cdc6 and Cdt1, assembles an inactive Mcm2-7 double hexamer on the dsDNA. Later, the double hexamer recruits Cdc45 and GINS to form two active and separate DNA helicases. The active Cdc45–Mcm2-7–GINS helicase encircles the leading strand while excluding the lagging strand. One of the fundamental unanswered questions is how each Mcm2-7 hexamer converts from binding dsDNA to binding one of the single strands. The structure of the double hexamer on dsDNA reveals how DNA interacts with key elements inside the central channel, leading us to propose a lagging-strand extrusion mechanism. This work advances our understanding of eukaryotic replication initiation.
Keywords: DNA replication, helicase, DNA unwinding, mini chromosome maintenance, cryo-electron microscopy
Abstract
During replication initiation, the core component of the helicase—the Mcm2-7 hexamer—is loaded on origin DNA as a double hexamer (DH). The two ring-shaped hexamers are staggered, leading to a kinked axial channel. How the origin DNA interacts with the axial channel is not understood, but the interaction could provide key insights into Mcm2-7 function and regulation. Here, we report the cryo-EM structure of the Mcm2-7 DH on dsDNA and show that the DNA is zigzagged inside the central channel. Several of the Mcm subunit DNA-binding loops, such as the oligosaccharide–oligonucleotide loops, helix 2 insertion loops, and presensor 1 (PS1) loops, are well defined, and many of them interact extensively with the DNA. The PS1 loops of Mcm 3, 4, 6, and 7, but not 2 and 5, engage the lagging strand with an approximate step size of one base per subunit. Staggered coupling of the two opposing hexamers positions the DNA right in front of the two Mcm2–Mcm5 gates, with each strand being pressed against one gate. The architecture suggests that lagging-strand extrusion initiates in the middle of the DH that is composed of the zinc finger domains of both hexamers. To convert the Mcm2-7 DH structure into the Mcm2-7 hexamer structure found in the active helicase, the N-tier ring of the Mcm2-7 hexamer in the DH-dsDNA needs to tilt and shift laterally. We suggest that these N-tier ring movements cause the DNA strand separation and lagging-strand extrusion.
The initiation of DNA replication in eukaryotes is tightly regulated to ensure that duplication of DNA occurs only once within a single cell-division cycle (1–5). In Saccharomyces cerevisiae, the origin-recognition complex (ORC) is a stable six-subunit complex that binds replication origins throughout the cell-division cycle (6). After the M–G1 transition, Cdc6 binds to the ORC and then recruits a Cdt1-bound Mcm2-7 hexamer, resulting in Mcm2-7 loading onto DNA (7–15). Before loading, the Cdt1-bound Mcm2-7 hexamer adopts a spiral shape with a gap at the Mcm2/Mcm5 interface, which serves as the DNA entry gate for helicase loading (10, 15–18). In contrast, the final product of the multistep DNA loading process is an inactive head-to-head Mcm2-7 double hexamer encircling dsDNA. The double hexamer is characterized by a central channel, which is wide enough to allow dsDNA sliding in the presence of high salt in vitro (19, 20) or in a transcription-dependent manner in vivo (21). Loading the first Mcm2-7 hexamer involves an extensive interaction between the winged helix domains (WHDs) in ORC–Cdc6 and in Mcm2-7 (14, 22, 23). Different mechanisms have been proposed for the loading of the second MCM2-7 hexamer: (i) two face-to-face hexamers are formed by two ORC–Cdc6 ATPases at replication origins (24), or (ii) the first ORC–Mcm2-7 can directly recruit a second Cdc6 and another Cdt1-bound Mcm2-7 to form the double hexamer (3).
At the beginning of S phase, the inactive double hexamer is converted into two active Cdc45–Mcm2-7–GINS (Go, Ichi, Nii, San) (CMG) helicases by the actions of Cdc7-Dbf4–dependent protein kinase (DDK), cyclin-dependent protein kinase (CDK), and an array of protein factors (3, 25). Initially DDK-dependent phosphorylation of Mcm2-7 promotes the recruitment of Sld3 and Cdc45 (26–34) before CDK-dependent phosphorylation of Sld2 and Sld3 (35, 36) facilitates the binding of a preloading complex consisting of polymerase ε, GINS, Sld2, and Dpb11 (37). MCM10 binding is required to activate the CMG helicase for the production of ssDNA and splitting of the Mcm2-7 double hexamer (38–42). The process of helicase activation transforms the Mcm2-7 double hexamer that encircles dsDNA into two Mcm2-7 hexamers that have ssDNA in the central channel, but the mechanistic basis of this transition is not known (3, 43–45). The active CMG helicase, comprised of Cdc45, the GINS heterotetramer, and the Mcm2-7 hexamer, has coupled ATPase- and DNA-unwinding activities (14, 46–49). Cryo-EM structures of the S. cerevisiae and Drosophila CMG helicases show that Cdc45 and GINS seal the N-tier ring at the Mcm2/5 interface, while the C-tier AAA+ containing the motor ring is relatively flexible (22, 45, 50). Previous studies demonstrated that the CMG helicase surrounds the leading strand and excludes the lagging strand from the central channel (51, 52). This is consistent with a recent cryo-EM structure of the yeast CMG bound to a forked DNA in which the leading strand threads through the central Mcm2-7 channel (53). These studies suggested that the parental dsDNA enters the helicase from the N-tier ring, indicating that the C-tier motor ring pushes from behind the N-tier ring, and hence the N-tier ring faces the fork junction and is engaged in strand separation (54); however, alternative models have been proposed in which the C-terminal domain of the helicase contacts the parental dsDNA (50, 55–58).
The cryo-EM structure of the double hexamer in the absence of DNA revealed that the two hexamers are interlocked at the N-tier zinc finger (ZnF) domain regions and that the central channel is twisted due to the staggered fitting of the two hexamers (59). The double-stranded origin DNA passes through the central channel (19, 20). Multiple models have been proposed for the initial DNA strand-separation process that yields ssDNA templates for DNA synthesis (60). Since in the CMG the N-tier moves ahead of the C-tier motor ring (53, 54), one possibility is that, during activation, the two head-to-head hexamers of the double hexamer could provide torsional strain and push one strand of the DNA outside the double-hexamer central channel, essentially extruding the lagging strand from either the central N-tier (59, 61, 62) or the outer C-tier rings (53). A key missing piece of information that would help resolve how origin DNA is melted inside the double-hexamer chamber is the structure of the double hexamer bound to the dsDNA origin. We describe the cryo-EM structure of the S. cerevisiae double hexamer on dsDNA. The structure shows that the DNA is pressed against the Mcm2–Mcm5 gate at the N-terminal ZnF region and suggests that strand extrusion commences at the middle of the double hexamer.
Results and Discussion
Cryo-EM of the S. cerevisiae Double Hexamer–dsDNA Complex.
Mcm2-7 double-hexamer formation has been reconstituted in vitro using DNA containing a single budding yeast replication origin ARS1 and purified proteins (Fig. 1A) (19, 20). However, upon releasing the double hexamer from DNA coupled magnetic beads with DNase I, the DNA is lost from the double hexamer since the complex is able to diffuse on dsDNA over a short distance in the absence of ATP hydrolysis, resulting in the DNA being removed from the central channel, consistent with in vitro MCM double-hexamer sliding assays (20, 62, 63). Consequently, dsDNA is also absent from the double hexamer purified from an endogenous source (59). Moreover, the standard prereplicative complex (pre-RC) assay yields only small amounts of double hexamer, insufficient for cryo-EM. To overcome these limitations, we engineered a multiorigin DNA that allows the assembly of multiple double hexamers onto a single piece of DNA. Optimized pre-RC assay conditions allow efficient assembly of double hexamers at multiple replication origins with increased yields. Each origin is separated by AluI restriction sites, allowing restriction enzyme-mediated release of the double hexamer from the magnetic beads and the generation of ∼248-bp DNA fragments. Optimal AluI digestion conditions allow rapid digestion, minimizing the chance that the double hexamer can slide off from the DNA. In the raw electron micrograph recorded in a 300-kV Titan Krios microscope using a K2 Summit direct electron detector of the AluI-treated sample embedded in vitreous ice, a long stretch of DNA visibly protruded from many double-hexamer particles (Fig. 1B and SI Appendix, Fig. S1). 2D class averages confirmed the presence of DNA in virtually all double-hexamer particles (Fig. 1C). We reconstructed a 3D cryo-EM map at a resolution of 3.9 Å using RELION 2 software (Fig. 1D and SI Appendix, Figs. S2–S4) (64). The dsDNA density is continuous, running all the way through the axial channel of the double hexamer. We observe strong density for the nucleotide bases and weaker density for some of the backbone phosphate groups (Fig. 1D). The DNA density tapers off at the two ends of the DNA, indicating that the DNA is less ordered as it exits the C-tier rings of the double hexamer.
Fig. 1.
Cryo-EM of the yeast Mcm2-7 double hexamer loaded in vitro on dsDNA. (A) Main steps for the in vitro assembly of the double hexamer on 10× ARS1-containing DNA. (B) A raw electron micrograph of the AluI-treated sample embedded in vitreous ice. Red arrows point to the exposed DNA. (C) Selected 2D class averages. For scale, the length of the particle is ∼20 nm. (D) The central section of the surface-rendered cryo-EM 3D map at 3.9-Å resolution. The dsDNA passing through the central channel is segmented and showed in orange.
The Double Hexamer Binds to a 60-bp-Long and Zigzagged dsDNA.
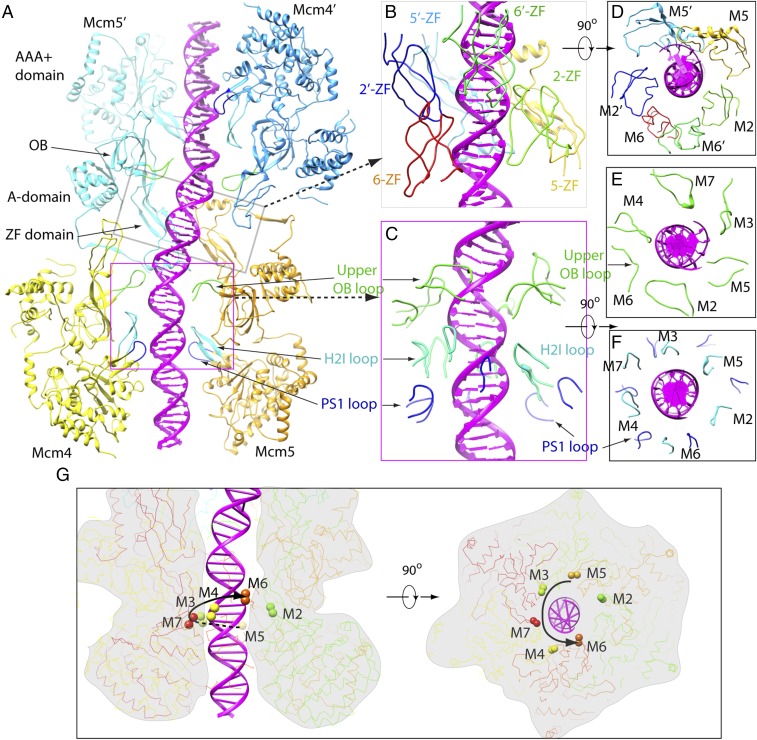
We built an atomic model into the cryo-EM map of the double-hexamer–dsDNA using the available apo–double-hexamer structure as the initial model (Fig. 2 and Movie S1) (59). As observed before, one hexamer in the double-hexamer–dsDNA structure is tilted off axis by ∼14° with respect to the other hexamer, and the two hexamers are laterally offset by ∼20 Å. Because the major and minor grooves and many individual nucleotide groups (bases, riboses, or phosphates) are clearly resolved in the 3D density map, we were able to model 60 bp into the continuous DNA density. Since the double hexamer can move on the DNA during sample preparation, the DNA sequence information is averaged out. Therefore, the nucleobase densities do not contain sequence identities. We modeled a random DNA sequence without using the same sequence in the two symmetry-related Mcm2-7 hexamers because origin DNAs are not palindromic. Overall, the DNA has a zigzag shape due to the presence of two ∼15° turns (Fig. 2). These DNA turns are a result of the off-axis tilt described above and the lateral offset of the two hexamers. The inflection points are inside the N-tier rings of the hexamers between the ZnF domains and the oligosaccharide–oligonucleotide (OB) domains. In the double-hexamer–DNA structure, the five WHDs from Mcm3, 4, 5, 6, and 7 were not resolved, similar to the apo structure suggesting these do not play a major role in DNA binding. In contrast, WHDs were found to play a prominent role in recruitment of the Mcm2-7 hexamer by ORC–Cdc6 (65). The double-hexamer diameter is ∼3% larger than the apo structure, allowing accommodation of the dsDNA in the axial channel (Movie S2).
Fig. 2.
The dsDNA follows a zigzag path in the axial channel. (A) Atomic model of double-hexamer–dsDNA shown in cartoon view. Visible Mcm subunits are individually labeled with a matching color. (B) A side view along the twofold axis showing the zigzag DNA structure (magenta) inside the axial channel. Subunits labeled with and without a prime, e.g., M3 and M3′, are twofold symmetry-related.
The Two Opposing Mcm3:Mcm5:Mcm7 Half Rings, Especially the N-Terminal Elements of Mcm5, Mediate Double-Hexamer Interactions.
The offset arrangement of the double hexamer is a result of differential interactions around the hexamer–hexamer interface (Figs. 2A and 3 A–C): Most hexamer–hexamer interactions are between the Mcm3:Mcm5:Mcm7 half rings of both hexamers, which bury an interface of 11,848 Å2. The two Mcm3:Mcm5:Mcm7 half rings lie on one side of the dsDNA. The other two half rings comprised of Mcm2:Mcm4:Mcm6 are separated from each other by a sizable gap and have only a few interactions that bury a small surface of 670 Å2. This feature is notable, because Mcm2, Mcm4, and Mcm6, but not Mcm3, Mcm5, and Mcm7, have an extra N-terminal serine/threonine-rich domain (NSD) that inhibits premature replication initiation (26, 27, 32, 66–69). The NSDs of Mcm2, Mcm4, and Mcm6 are not resolved in the cryo-EM structure and thus are disordered. However, their physical presence likely accounts for the apparent gap and the off-axis tilt between the two hexamers in the region. Therefore, the tilted arrangement of the double hexamer may expose the NSDs of Mcm2, Mcm4, and Mcm6 to access by DDK kinase to promote the eventual separation of the two hexamers. The interaction between the two Mcm3:Mcm5:Mcm7 half rings involves many important motifs: the N-terminal extension (NTE) of Mcm5 and the N-terminal insertion (NTI) of Mcm7 observed previously in the apo–double hexamer structure (59) and a previously unknown α-helix inserted into ZnF domain of Mcm5 which we have called the “Zn-finger insertion” (ZFI) motif (Fig. 3D). This motif is present only in Mcm5, not in any other yeast Mcm subunits, and is absent in Mcm5 proteins of higher eukaryotes (SI Appendix, Fig. S5). Computational analysis of the amino acid sequence suggests it is an unstructured loop. Therefore, the observed helical structure of ZFI was unexpected and may be induced upon double-hexamer assembly and be stabilized by interaction with dsDNA since the insertion is disordered in the apo–double hexamer structure as well as in the ORC–Cdc6–Cdt1–Mcm2-7 (OCCM) and CMG structures (22, 59, 65). Indeed, Mcm5 lysine residue 233 in the ZFI points toward the DNA. The Mcm5 ZFI helix interacts extensively with Mcm3 and Mcm7 of the opposing hexamer, indicating a key role in double-hexamer assembly and possibly separation (see below). The Mcm5 ZFI is also structurally connected to the Mcm5 α-helix bundle that harbors the Mcm5 P83L mutation that eliminates the requirement for DDK activity during the initiation of DNA replication (70).
Fig. 3.
The Mcm5:Mcm3:Mcm7 half rings of the top and bottom hexamers mediate double-hexamer assembly. (A) Side view of the cryo-EM density map segmented and colored by individual subunits. The two hexamers are stacked against each other with a tilt angle of 14°. (B) A 90° counterclockwise-rotated side view relative to A showing the gapped interface region formed by the top and the bottom half rings of Mcm2, Mcm4, and Mcm6. DNA is shown as a semitransparent magenta cartoon. The buried area at this interface is 670 Å2. (C) A 90° clockwise-rotated side view relative to A showing the main interaction interface formed by the two Mcm5:Mcm3:Mcm7 half rings. The buried area reaches 11,848 Å2. (D) Mcm5 has two unique structural elements involved in the double-hexamer assembly: the NTE and ZFI motifs. They form extensive interactions with Mcm3 and Mcm7 of the opposing hexamer. Subunits labeled with and without a prime, e.g., M3 and M3′, are twofold symmetry-related.
Four of the Six Nucleotide Sites Are Occupied by ADP.
Among the six available ATP sites in each Mcm2-7 hexamer, only four were occupied: at the interface between Mcm2:Mcm6, Mcm5:Mcm2, Mcm3:Mcm5, and Mcm7:Mcm3, respectively (SI Appendix, Fig. S6). The densities best fit with the ADP structure, indicating that ATP may have hydrolyzed during the preceding Mcm2-7 hexamer recruitment and double-hexamer assembly processes. Indeed, Mcm2-7 ATPase activity is required for double-hexamer assembly (14, 49). The nucleotide pockets between Mcm4:Mcm6 and between Mcm7:Mcm4 are empty. Such nucleotide occupancy in double-hexamer–dsDNA contrasts with the apo–double hexamer in which all six sites are occupied. It is possible that the loss of dsDNA in the apo structure may have allowed the empty sites at Mcm4:Mcm6 and Mcm7:Mcm4 interfaces to open up, leading to the binding of the nucleotides at the two sites, as the purification buffer used in that work contained 3 mM ATP (59). Consistent with this possibility, the nucleotide at the Mcm3:Mcm7 interface in the apo–double hexamer structure was noted to be an ATP rather than an ADP. Interestingly, in the active CMG helicase bound to either a ssDNA or a forked DNA, the three sites that are filled with nucleotides, Mcm5:Mcm3, Mcm2:Mcm5, and Mcm6:Mcm2, are the same as in the double hexamer–dsDNA, with the exception of Mcm3:Mcm7, which is empty in the CMG but is occupied in the double hexamer–dsDNA structure (53).
The Mcm2-7 Presensor 1 Loops Spiral Around the DNA.
We further analyzed how DNA interacts with the double hexamer and observed that the DNA was surrounded by and in direct contact with many structural elements in the axial channel, such as the ZnF domains, the channel-lining upper OB loops, the helix-2 insertion (H2I) loop, and the presensor 1 (PS1) loops (Fig. 4 and SI Appendix, Figs. S7 and S8) (45). These interactions could explain why the observed DNA sliding of the double hexamer in vitro is rather slow and why it is dependent on high-salt conditions, as the ions are probably necessary to destabilize the nucleic acid–protein contacts (19, 20).
Fig. 4.
Protein–DNA interactions in the double-hexamer–dsDNA structure. (A) A cut-open side view of the double-hexamer–dsDNA showing only Mcm4 and Mcm5 in each of the two hexamers. Mcm subunits and DNA are shown in cartoon. In the top Mcm5 structure, the N-terminal ZnF domain, the OB domain, the α-helical (A) domain, and the C-terminal AAA+ domains are labeled. The DNA-binding channel loops, i.e., upper OB loops, the H2i loops, and the PS1 loops, are highlighted in green, blue, and cyan, respectively. (B) An enlarged view of the middle interface region of the double hexamer in the gray box in A. Six ZnF domains of Mcm2, 2′, 5, 5′, 6, and 6′ surround the dsDNA. (C) A zoomed view of the N-tier region of the lower hexamer contained in the pink square in A. (D) A 90°-rotated view of B looking down the DNA axis. (E and F) Views of C at the upper OB loop region (E) and the H2I loop and PS1 loop region (F), rotated 90°, showing interactions with the DNA. (G) Side (Left) and top (Right) views of one Mcm2-7 hexamer showing the spiral arrangement of the PS1 loops. The top of each PS1 loop is marked by two spheres. The curved black arrow illustrates the helical rise of the PS1 loops of Mcm5, Mcm3, Mcm7, Mcm4, and Mcm6. The PS1 loop of Mcm2 does not contact DNA. Subunits labeled with and without a prime are twofold symmetry-related.
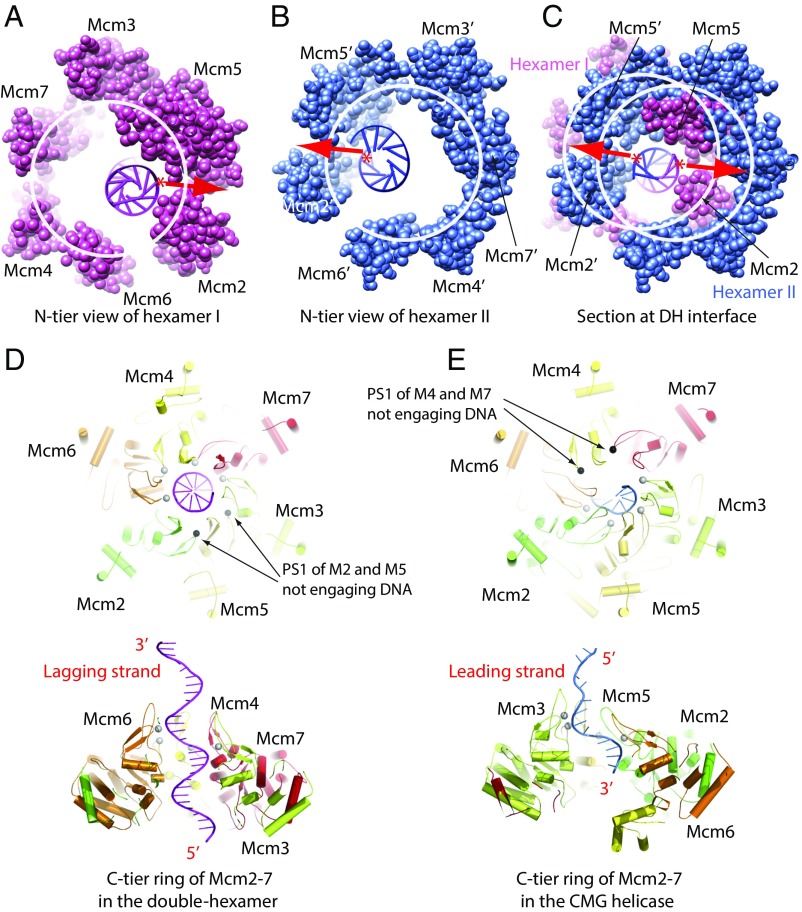
The two hexamers bound DNA in the same manner, with each hexamer covering approximately two full turns of dsDNA. This was possible only because the double-hexamer interface region covers exactly one full turn of the DNA, positioning each hexamer in the same, albeit opposing, orientation with respect to the bound dsDNA (Figs. 2 and 4). Importantly, among the 12 ZnF domains of the two hexamers, only six are close to the dsDNA: the two pairs of ZnF domains of Mcm2 and Mcm5 (with its ZFI helix) contact the dsDNA directly, with the two Mcm6 ZnF domains nearby but not in contact with the DNA (Fig. 4 B–D and SI Appendix, Fig. S7). Thus, one purpose of the staggered coupling of the two hexamers may be to position the dsDNA in the middle section next to the Mcm2–Mcm5 gates of both hexamers, a point that is discussed further below. Indeed, with the interface of Mcm2–Mcm5 serving as the DNA entry gate during helicase loading (10, 16, 17, 65), the proximity of the Mcm2 and Mcm5 ZnFs to the interface DNA suggests that initial DNA unwinding occurs here (see below).
There are two known DNA-binding loops in the C-tier AAA+ motor domain of each Mcm subunit, the H2I and PS1 loops. Inside each hexamer channel, the upper OB loop of Mcm4 inserts into the minor DNA groove, apparently responsible for DNA bending at each inflection point at the ends of the 14 base pairs of DNA that is surrounded by the double-hexamer interface (Fig. 4 A, C, and E). Notably, the same Mcm4 loop is at the DNA fork junction that splits the double strand in the cryo-EM structure of the CMG–forked DNA complex (53). Other upper OB loops make contact with either the phosphate backbone or DNA grooves, or both, but they generally do not follow a strict helical path (Fig. 4 C and E). However, five of the six PS1 loops (in Mcm5, Mcm3, Mcm7, Mcm4, and Mcm6) approximately follow a right-handed helical path, with each subunit rising approximately one base (Fig. 4 C, F, and G). The PS1 loop of Mcm2 does not follow the helical path, as it does not interact with DNA. Instead, the loop is localized between the lowest-positioned Mcm5 PS1 and the highest-position Mcm6 PS1. The six H2I loops also make contact with DNA, but these loops do not follow a helical path. Compared with the apo–double hexamer, these loops in the double-hexamer–dsDNA all move slightly outwards to enlarge the central channel to accommodate the dsDNA; however, the H2I loop of Mcm2 moved a significant distance of ∼6 Å (SI Appendix, Fig. S9 and Movie S3). Interestingly, we found that a section of one α-helix of the AAA+ fold, just below the PS1 loop, is close to and interacts with the dsDNA (SI Appendix, Fig. S10). The significance of the helix–DNA contact awaits further study.
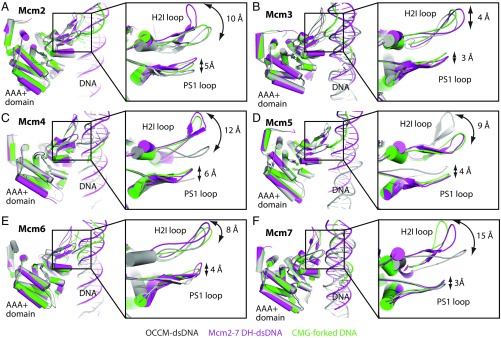
The Positions of the DNA-Binding H2I and PS1 Loops Vary to a Different Degree in Different Mcm2-7–Containing Structures.
The S. cerevisiae Mcm2-7 hexamer structure bound to DNA has been determined at least three times: in the current structure of double-hexamer–dsDNA, in the Mcm2-7 loading intermediate OCCM (65), and in the active CMG helicase bound to a forked DNA (53). We extracted the individual Mcm proteins from the three cryo-EM structures and superimposed them according to their respective AAA+ domains (Fig. 5). Such alignments reveal possible locations of the DNA-binding loops, but they may not reflect actual loop movements during DNA translocation. In general, the H2I loops were observed in very different positions, being 4 Å to 15 Å apart in the three structures. However, the PS1 loops are localized in a narrow range of 3–5 Å. Studies on an archaeal MCM hexamer suggested that the H2I loop functions to reduce the helicase affinity for DNA, as its deletion increases the affinity by nearly 100-fold (71). Therefore, the H2I loop could facilitate unwinding by destabilizing helicase–DNA interactions but may not directly drive DNA translocation. Indeed, this loop is present only in superfamily 6 MCM helicases and is absent in all five of the other super families of helicases, consistent with the idea that it does not participate in the direct unwinding process (45, 72, 73). Interestingly, loops that are involved in active DNA unwinding are usually associated with movements in a narrow range, as observed with the bovine papillomavirus E1 helicase (74, 75). The observation that H2I loops can adopt very different conformations supports the idea that this loop may not play an active role in DNA translocation. However, the 3- to 4-Å range of PS1 loop locations in the three different structures, together with the above-mentioned observation that five PS1 loops are arranged in a right-handed helical manner that rises by one base per subunit, is consistent with an active role in DNA translocation. The PS1 loop is conserved among the AAA+ family of hexameric replicative helicases (45, 62, 76). Mutations in PS1 loops of Mcm3, Mcm4, and Mcm5 are lethal or cause growth defects in vivo and decrease DNA unwinding activity in vitro (77–79). The facts that PS1 loops are arranged in spiral in the double hexamer and that their positions vary when the AAA+ domains are aligned suggest that the loops move during DNA translocation in Mcm2-7, as found in the virus DNA helicase (45). However, eukaryotic helicases differ fundamentally from the homo-hexameric viral helicases in that they have two additional factors, Cdc45 and GINS. The mechanism of DNA translocation by the eukaryotic replicative helicase needs further investigation (16, 22, 50, 53).
Fig. 5.
The range of movement of the DNA-binding H2I and PS1 loops of the AAA+ domains of Mcm proteins in three different contexts: in the loading-intermediate OCCM, in the double hexamer, and in the active CMG helicase. (A–F, Left) Superimposition of the AAA+ domains (based on secondary structure alignment) of each of the six Mcm proteins extracted from the current double-hexamer–dsDNA (magenta) with that from OCCM in complex with dsDNA (gray, PDB ID code 5UDB) and from CMG helicase in complex with a forked DNA (green, PDB ID code 5U8S). (Right) Zoomed views of the DNA-interacting H2I and PS1 loops.
Lagging-Strand Extrusion May Initiate from the N-Tier Regions of Mcm2-7 Double Hexamer.
It is widely accepted that the interface between Mcm2 and Mcm5 is the site where the dsDNA passes through during the Mcm2-7 hexamer loading and may also be the site where the ssDNA is extruded during helicase activation. However, how and from which part of the Mcm2–Mcm5 interface the lagging strand is extruded is not understood. In the double-hexamer–dsDNA structure, the two hexamers are staggered such that the dsDNA is pressed directly against Mcm2 and Mcm5 at the N-tier regions of both hexamers, with each strand facing one potential gate upon reopening, apparently poised for extrusion (Fig. 6 A–C and SI Appendix, Fig. S7). These two Mcm2–5 gates sit on opposite sides of the DNA double helix, with opposite DNA strands in the 5′–3′ orientation facing each gate (Fig. 6 A and B). This architecture is consistent with the knowledge that the strand to be extruded from the hexamer will become the lagging strand for that particular Mcm2-7 hexamer (Fig. 6C). Because of the topological constraint, the DNA segment must be pushed or pulled to be perpendicular to the axial channel during extrusion. This structural feature suggests that the initial strand separation and extrusion occur in this region between the two Mcm4 upper OB folds (Fig. 4A), the same region that contains the two Mcm4 NSDs that are the target for DDK regulation of initiation of DNA replication (26, 27). The proposed N-tier-first extrusion is supported by a previous crystallographic study of an archaeal MCM hexamer in which ssDNA was found to bind the N-tier upper OB loops perpendicular to the channel axis (80). Corresponding OB loops of Mcm2 and Mcm5 were shown by mutagenesis to be important for initiation in S. cerevisiae (79, 80). Mcm10 is a key replication factor that interacts with ssDNA, and it is essential during helicase activation to allow the CMG complex to gain processive DNA unwinding activity (38–40, 81, 82). It was recently shown that Mcm10 binds to an N-tier region of Mcm2 (the Mcm10-binding motif of Mcm2, referred to as “mbm”) to facilitate the replication initiation (83). The interaction between Mcm10 and the N-tier of Mcm2 is consistent with our proposed N-tier-first extrusion model.
Fig. 6.
A comparison of protein–DNA interactions in the double hexamer and in the CMG helicase. (A and B) N-tier views of hexamer I (A) and II (B) in the double hexamer, revealing that the DNA is off center and each strand of the dsDNA is pressed against the Mcm2–Mcm5 gate at the N-tier ZnF domain region of each hexamer. (C) A central section of the double hexamer showing that the two strands of DNA are aligned with the two Mcm2–Mcm5 gates of the two hexamers. The two hexamers are shown in all-atom mode. (D) A top view (Upper) and a side view (Lower) of the C-tier motor ring of the double hexamer. Only the PS1 loops of Mcm3, Mcm7, Mcm4, and Mcm6 (shown as gray spheres) engage the lagging-strand DNA. The PS1 loops of Mcm2 and Mcm5 (shown as black spheres) do not engage DNA. For clarity, the leading-strand DNA is not shown. (E) The C-tier motor ring of the Mcm2-7 of the CMG helicase. The PS1 loops of Mcm6, Mcm2, Mcm5, and Mcm3 (shown as white spheres) engage the leading-strand DNA. The PS1 loops of Mcm7 and Mcm4 (shown as black spheres) do not engage DNA. For clarity, the lagging-strand DNA outside the motor ring is not shown. Subunits labeled with and without a prime are twofold symmetry-related.
Remarkably, the four PS1 loops of Mcm3, Mcm7, Mcm4, and Mcm6 of the double hexamer interact directly with the future lagging-strand DNA but not with the future leading-strand DNA (Fig. 6D). This feature is consistent with the concept that the lagging strand is extruded by the double hexamer during helicase activation. In the active CMG helicase, there are also four PS1 loops that engage the DNA (Fig. 6E). However, the PS1 loops interact with the leading-strand DNA, as the lagging strand is excluded from the central channel (53, 84). Further, different PS1 loops interact with DNA: in the double hexamer, the PS1 loops of Mcm2 and Mcm5 do not interact with DNA, but in CMG, the PS1 loops of Mcm2 and Mcm5, along with Mcm3 and Mcm6, engage with DNA, and instead the PS1 loops of Mcm4 and Mcm7 do not contact DNA. Thus, the initiation of DNA replication must lead to a reorganization of the PS1 loops from the helicase-activation mode (Mcm 3/4/6/7 PS1 loops interacting with the lagging strand) to a DNA-unwinding mode (Mcm2/3/5/6 PS1 loops interacting with leading strand).
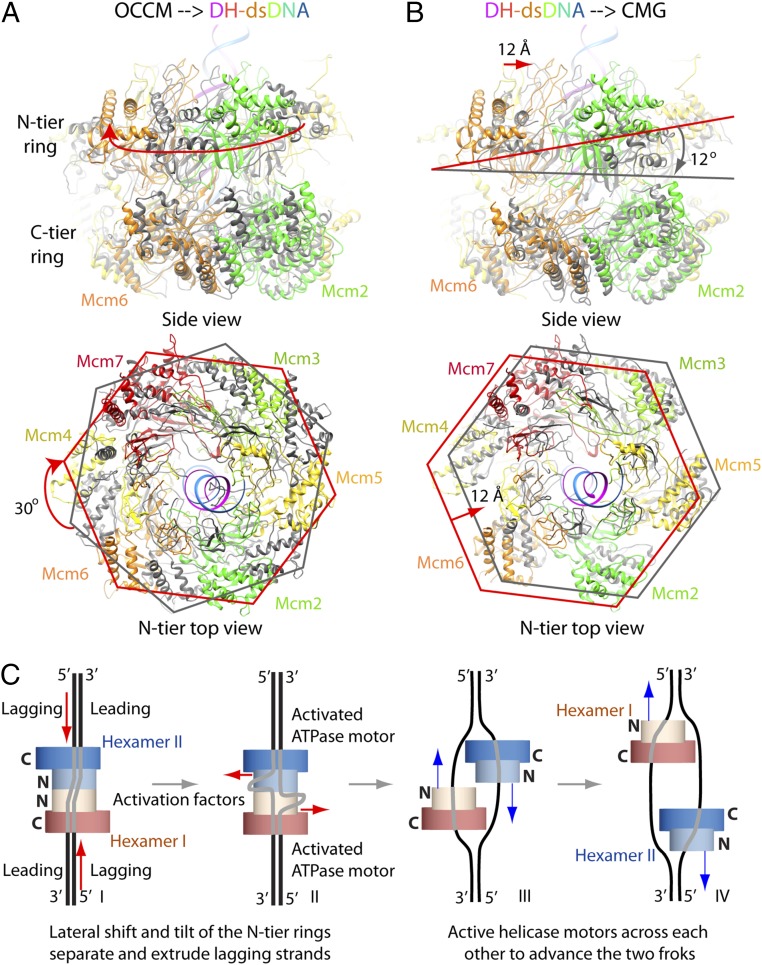
N-Tier Ring Movement of the Mcm2-7 Hexamer During Double-Hexamer Assembly and Subsequent Activation.
The yeast Mcm2-7 hexamer structures on DNA (i) as an intermediate preceding the double hexamer (the OCCM) and (ii) as a form following the double-hexamer activation (the CMG helicase) are both available (53, 65), providing an opportunity to analyze the conformational changes required for double-hexamer activation. We superimposed the Mcm2-7 hexamer of the OCCM with one hexamer of the double hexamer by aligning their respective C-tier motor ring. Such alignment reveals potential movement and conformational changes in the N-tier rings, because it is the untangling and dissociation of these rings that underlies double-hexamer assembly and subsequent disassembly, respectively. We found the two N-tier rings are rotated by nearly 30° during double-hexamer assembly (Fig. 7A). Because ATP hydrolysis by the Mcm2-7 subunits is required for double-hexamer loading (14, 49), it is possible that the N-tier ring rotation stores the energy derived from ATP hydrolysis during double-hexamer assembly for its subsequent disassembly during helicase activation (18, 85). Importantly, the 30° rotation apparently ensures that the DNA faces the Mcm2–Mcm5 gate at the N-tier region. We further compared the Mcm2-7 structure of the double-hexamer–dsDNA with that of CMG–fork DNA by aligning their respective C-tier rings and found that the N-tier ring translates laterally by ∼12 Å and tilts by ∼12° (Fig. 7B and Movie S4). These movements may underlie the DNA strand separation during the double-hexamer activation, as described below.
Fig. 7.
Conformational changes of the N-tier rings of Mcm2-7 hexamers during double-hexamer conversion to the CMG helicase may underlie the lagging-strand separation and extrusion mechanism. (A) The Mcm2-7 hexamers found in the OCCM (gray) and in the double hexamer (colored) are aligned based on all atoms in their respective C-tier rings. Note the ∼30° clockwise rotation of the N-tier ring during double-hexamer formation, which is an energy-consuming process (requiring ATP hydrolysis). Therefore, the double hexamer may be in a strained high-energy configuration. (B) The Mcm2-7 hexamers found in the double hexamer and in the CMG helicase, aligned based on all atoms of their respective C-tier rings. Note a ∼12° tilt and a ∼12-Å lateral translation of the N-tier ring. These movements may be driven by releasing the stored energy in the double hexamer. In both A and B, leading- and lagging-strand DNA in the double-hexamer–dsDNA structure are shown in blue and magenta, respectively. (C) A lagging-strand DNA extrusion model. Step I: The double hexamer may be in a strained high-energy state, as the two hexamers are offset, and the dsDNA is zig-zagged inside the central channel. The activation factors, such as DDK, Sld2, Sld3, Sld7, Mcm10, and Dpb11, may trigger conformational changes in the double hexamer, leading to the release of the stored energy to pump the lagging strands toward the inside. The two vertical red arrows indicate the direction of lagging-strand movement. Step II: The released energy may also drive the observed tilt and lateral shift of the N-tier rings. These N-tier ring movements may extrude the accumulating lagging strands through the Mcm2–Mcm5 gates at the N-tier regions of the double hexamer. The two horizontal red arrows indicate the N-tier ring shift and slight tilt. Step III: The two active helicases on the leading strand move past each other in the 3′–5′ direction with their respective N-tier rings ahead of the C-tier motor rings. Step IV: Bidirectional fork advancement by the two CMG helicases. The blue arrows indicate the helicase movement directions.
A Plausible Lagging-Strand Extrusion Model.
Based on the above-described features observed in the double-hexamer–dsDNA structure, we propose the following lagging-strand extrusion and helicase-activation model (Fig. 7C): First, the double hexamer may be in a strained state, storing energy derived from the ATP hydrolysis–powered Mcm2-7 hexamer loading and dimerization process. We note that the Mcm2-7 motor is inactive in this state (Fig. 7 A and C, step I) (63). During helicase activation, several protein factors, such as Sld3/7, Sld2, DDK, Dpb11, and Mcm10, and the helicase accessory components GINS and Cdc45 may interact with the N-tier rings, triggering the release of stored energy to drive Mcm2-7 movement and conformational changes (the observed N-tier ring tilt and lateral shift). Because the translocating PS1 loops are engaged with the lagging strand, a 3′–5′ translocation of the coupled (hence stationary) double hexamer would pump the lagging strand toward the inside (Fig. 7B). We also propose that the accumulation of the lagging strand inside the channel accompanied by the shift and tilt of the N-tier ring may separate the DNA double strand and extrude the lagging strand from the axial channel via the Mcm2–Mcm5 gate starting at the N-tier region (Fig. 7C, step II). This structural rearrangement would also lead to the activation of the Mcm2-7 ATPase motor, allowing the two helicases to move on leading strands past each other in 3′–5′ direction (Fig. 7C, step III) and go on to advance the replication forks using a strand-exclusion mechanism (Fig. 7C, step IV) (53).
DDK is required to activate the intertwined double hexamer. The DDK-docking site on each Mcm4 subunit on one hexamer (residues 174–333) sits immediately adjacent to the Mcm5-Mcm2 N-terminal domains on the opposite hexamer (26). Next to the DDK-docking domain is an unstructured region of Mcm4 that contains a domain (residues 74–174) that prevents premature initiation of DNA replication. When this domain is deleted, DDK is no longer required for cell viability and initiation of DNA replication (27). Likewise, a mutation in the N terminus of Mcm5 (P83L) that also eliminates the requirement for DDK is located in a region of the N terminus of Mcm5 that interacts with Mcm7 and Mcm3, and the mutation could alter the structure of the hexamer–hexamer interface, particularly near the Mcm5 ZnF with its ZFI helix (70). We suggest that DDK phosphorylation of Mcm4 inactivates the Mcm4-inhibitory domain, and this may be the major factor that triggers the ensuing series of conformational changes that lead to the separation of the two N-tier rings and the dissociation of two hexamers (27, 32). Furthermore, because the dsDNA–double hexamer appears to have ADP bound at four of the Mcm2-7 subunit–subunit interfaces and is in an ATPase-inactive state (63), the conformational changes in the Mcm2-7 during helicase activation may also provide a mechanism for ATP rejuvenation as well as ATPase activation.
There is a caveat to the proposed strand-extrusion model. The double-hexamer separation and helicase activation are likely multistep processes that eventually lead to CMG formation. Therefore, the observed tilt and lateral shift of the N-tier ring required for the double hexamer-to-CMG conversion may be a net effect of several steps and may not necessarily be solely responsible for the separation and extrusion of the DNA strand. An alternative strand-separation model would be to pump the DNA toward the inside. As the DNA accumulates at the middle N-tier region, it buckles and eventually is forced out at the middle of the double hexamer. Such a pumping-DNA-inwards model was previously proposed for origin melting in archaeal MCM (80) as well as for the large T antigen (86). However, the established 3′–5′ DNA translocation direction of the CMG helicase dictates that DNA should be pulled outwards rather than pushed inwards (87). It is unclear if the C-tier motor ring is able to operate in reverse to pump the DNA inwards. In yet another model, Mcm10 plays a major role in origin melting by binding to and stabilizing the melted and extruded lagging strand (42). Determining the structures of the activation intermediates of the double hexamer would be a key to clarify the important question in chromosome replication.
Experimental Procedures
10× ARS1 DNA Magnetic Bead Preparation.
To produce the 10× ARS1 DNA fragment, a 5-kb PCR fragment was amplified using pCS735 [pUC57 carrying a 10× cassette of ARS1 (248 bp), each flanked by AluI restriction sites], primer 1693 (GAAGATCAGTTGGGTGCACG), and primer 1568 (biotin-GCATCTTTTACTTTCACCAGCG). The resultant 5-kb DNA was bound to Dynabeads MyOne Streptavidin T1 beads (Invitrogen). Subsequently, a HindIII fragment containing the nonorigin DNA was released, resulting in ∼3 kb of 10× ARS1 DNA bound to the beads.
Double-Hexamer Preparation.
In vitro loading of double hexamer on an ARS1-containing linear DNA followed the standard protocol (20, 63) with the following modifications. Here, AluI instead of DNase I was used to release the double hexamer from the magnetic beads. Briefly, ORC (80 nM), Cdc6 (160 nM), Cdt1 (160 nM), and Mcm2-7 (160 nM) were incubated with the 10× ARS1 DNA-coupled beads (15 nM) in buffer A1 (50 mM Hepes⋅KOH at pH 7.5, 10 mM MgAc, 0.1 mM ZnAc, 3 mM ATP, 5 mM DTT, 0.1% Triton X-100, 10% glycerol, 0.05 mM EDTA) plus 300 mM KGlu and 0.8 mg/mL BSA for 60 min. The beads were washed with buffer A1 plus 300 mM NaCl and 100 mM KGlu before digestion with 2 U AluI (New England Biolabs) in buffer C (50 mM Hepes⋅KOH at pH 7.5, 100 mM KAc, 5 mM MgAc) with the addition of 5% buffer A1 plus 100 mM KGlu for 1 min at 24 °C.
Cryo-EM.
To prepare cryo-EM grids, we pooled 16 double-hexamer DNA assembly reactions together and concentrated the sample to a protein concentration of about 1.2 mg/mL with a Microcon centrifugal filter unit. Before EM grid preparation, we checked the sample for homogeneity by negative-stain EM. Then 3 μL of the double-hexamer DNA sample was applied to glow-discharged C-flat 1.2/1/3 holey carbon grids or lacey grids covered with a layer of continuous carbon film, incubated for 10 s at 6 °C and 95% humidity, blotted for 3 s, and then plunged into liquid ethane using an FEI Vitrobot IV. In C-flat R1.2/1.3 holey carbon film grids, the particles bound to holey carbon film or aggregated in the hole, even at high protein concentrations. In lacey grids with continuing carbon film, the double-hexamer particles distributed well with no aggregation problem. We loaded the grids into an FEI Titan Krios electron microscope operated at a high tension of 300 kV and collected images semiautomatically with EPU (FEI) under low-dose mode at a nominal magnification of 130,000× and a pixel size of 1.09 Å per pixel. A Gatan K2 Summit direct electron detector was used in super resolution mode for image recording with an under-focus range from 1.5–2.5 μm. A BioQuantum energy filter (Gatan) installed in front of the K2 detector was operated in zero-energy-loss mode with an energy slit width of 20 eV. The dose rate was 10 electrons⋅Å−2⋅s−1, and the total exposure time was 6 s. The total dose was divided into a 30-frame movie so each frame was exposed for 0.2 s.
Image Processing and 3D Reconstruction.
We collected ∼3,000 raw movie micrographs. The movie frames were first aligned and superimposed by the program MotionCorr 2.0 (88). Contrast transfer function parameters of each aligned micrograph were calculated using the program CTFFIND4 (89). All the remaining steps, including particle auto selection, 2D classification, 3D classification, 3D refinement, and density map postprocessing, were performed using RELION 2.0 (64). The template for automatic picking was generated from a 2D average of about ∼10,000 manually picked particles in different views. Automatic particle selection was performed for the entire dataset, and 312,403 particles were initially picked. We then carefully inspected the selected particles, removed “bad” ones that were broken or aggregated particles, repicked some initially missed “good” ones with expected size and shape, and sorted the remaining good particles by similarity to the 2D references; the bottom 10% of the particles with the lowest z-scores were removed from the particle pool. 2D classification of all good particles was performed and particles in the classes with features unrecognizable by visual inspection were removed. A total of 163,255 particles was used for further 3D classification. We derived five 3D models from the dataset and chose the two best models with strong DNA density for final refinement; the other three models were distorted or had weak DNA density, and those particles were discarded. The final dataset having 58,772 particles was used for further 3D refinement with application of the expected C2 symmetry, resulting in the 3.9-Å 3D density map. The resolution of the map was estimated by the gold-standard Fourier shell correlation at a correlation cutoff value of 0.143. The 3D density map was sharpened by applying a negative B-factor of −168 Å2. We also refined the 3D map without applying symmetry (C1) and obtained a map at 4.2-Å resolution. After atomic modeling and careful analysis of the two maps, we found the two hexamer structures and how they interact with the DNA are virtually the same. We therefore focused our analysis on the twofold symmetric 3.9-Å 3D map.
Structural Modeling, Refinement, and Validation.
The cryo-EM structure of the yeast Mcm2–7 double hexamer [Protein Data Bank (PDB) ID code 3JA8] was used as the initial model directly docking into the EM map. Each Mcm protein was then split into two parts, an N-terminal domain and a C-terminal domain, for subsequent rigid body-fitting into the 3D density map with COOT and Chimera. The dsDNA was then manually built into the long density that ran through the two Mcm2-7 hexameric structures in the program COOT. The DNA sequence used in preparation of the double hexamer is an origin DNA (ARS1) that had been multimerized. However, when the DNA is cut by the restriction enzyme to release the multimers of the double hexamer on DNA, it is possible that the DNA is not registered the same way relative to the origin in the double hexamers. For this reason we did not try to register the DNA sequence. We modeled the dsDNA in a random sequence, and the twofold symmetry was not enforced either in the DNA sequence during modeling or in refinement. The entire Mcm2–7 double-hexamer DNA model was first refined by rigid body refinement of individual chains in the PHENIX program and subsequently was adjusted manually in COOT. Finally, the atomic model was validated using MolProbity. Structural figures were prepared in Chimera and PyMOL (https://pymol.org/2/).
The final model was cross-validated using a method described previously (90). Briefly, we randomly added 0.1 Å noise to the coordinates of the final model using the PDB tools in Phenix and then refined the noise-added against the first half map (Half1) that was produced from one half of the particle dataset during refinement by RELION. We performed one round of coordinate refinement, followed by a B-factor refinement. The refined model was then correlated with the 3D maps of the two half maps (Half1 and Half2) in Fourier space to produce two forward scatter (FSC) curves: FSCwork (the model versus the Half1 map) and FSCfree (the model versus the Half2 map), respectively. A third FSC curve was calculated between the refined model and the final 3.9-Å resolution density map produced from all particles. The general agreement of these curves was taken as an indication that the model was not overfitted.
The map coordinates for the Mcm2-7 double hexamer on DNA were deposited as EMD-9400, PDB ID code 5BK4.
Supplementary Material
Acknowledgments
Cryo-EM data were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite in the Van Andel Research Institute. This work was funded by NIH Grants GM111472 (to H.L.) and GM45436 (to B.S.), UK Biotechnology and Biological Sciences Research Council Grant BB/N000323/1 (to C.S.), Wellcome Trust Investigator Award 107903/Z/15/Z (to C.S.), UK Medical Research Council Grant MC_U120085811 (to C.S.), and the Van Andel Research Institute (H.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryo-EM 3D map of double hexamer-dsDNA at 3.9 Å resolution has been deposited at the Electron Microscopy Data Bank (EMDB) database (accession no. EMD-9400). The corresponding atomic model was deposited at the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database (ID code 5BK4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712537114/-/DCSupplemental.
References
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Stillman B. Origin recognition and the chromosome cycle. FEBS Lett. 2005;579:877–884. doi: 10.1016/j.febslet.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Bell SP, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203:1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remus D, Diffley JF. Eukaryotic DNA replication control: Lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Bleichert F, Botchan MR, Berger JM. Mechanisms for initiating cellular DNA replication. Science. 2017;355:eaah6317. doi: 10.1126/science.aah6317. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Stillman B. The origin recognition complex: A biochemical and structural view. Subcell Biochem. 2012;62:37–58. doi: 10.1007/978-94-007-4572-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evrin C, et al. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization. Nucleic Acids Res. 2013;41:3162–3172. doi: 10.1093/nar/gkt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández-Cid A, et al. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell. 2013;50:577–588. doi: 10.1016/j.molcel.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Samel SA, et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28:1653–1666. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 2015;161:513–525. doi: 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 13.Frigola J, Remus D, Mehanna A, Diffley JF. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coster G, Frigola J, Beuron F, Morris EP, Diffley JF. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell. 2014;55:666–677. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ticau S, et al. Mechanism and timing of Mcm2-7 ring closure during DNA replication origin licensing. Nat Struct Mol Biol. 2017;24:309–315. doi: 10.1038/nsmb.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai Y, et al. Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nat Struct Mol Biol. 2017;24:300–308. doi: 10.1038/nsmb.3374. [DOI] [PubMed] [Google Scholar]
- 17.Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2-7 and the unequal role of active sites in ATP hydrolysis and viability. Mol Cell Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frigola J, et al. Cdt1 stabilizes an open MCM ring for helicase loading. Nat Commun. 2017;8:15720. doi: 10.1038/ncomms15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gros J, et al. Post-licensing specification of eukaryotic replication origins by facilitated Mcm2-7 sliding along DNA. Mol Cell. 2015;60:797–807. doi: 10.1016/j.molcel.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23:217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bleichert F, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519:321–326. doi: 10.1038/nature14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coster G, Diffley JFX. Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science. 2017;357:314–318. doi: 10.1126/science.aan0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araki H. Elucidating the DDK-dependent step in replication initiation. EMBO J. 2016;35:907–908. doi: 10.15252/embj.201694227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrera MC, et al. A reconstituted system reveals how activating and inhibitory interactions control DDK dependent assembly of the eukaryotic replicative helicase. Nucleic Acids Res. 2015;43:10238–10250. doi: 10.1093/nar/gkv881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519:431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang D, Cao Q, Lou H. Sld3-MCM interaction facilitated by Dbf4-dependent kinase defines an essential step in eukaryotic DNA replication initiation. Front Microbiol. 2016;7:885. doi: 10.3389/fmicb.2016.00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruck I, Dhingra N, Kaplan DL. A positive amplification mechanism involving a kinase and replication initiation factor helps assemble the replication fork helicase. J Biol Chem. 2017;292:3062–3073. doi: 10.1074/jbc.M116.772368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka S, Nakato R, Katou Y, Shirahige K, Araki H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr Biol. 2011;21:2055–2063. doi: 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 34.Itou H, Shirakihara Y, Araki H. The quaternary structure of the eukaryotic DNA replication proteins Sld7 and Sld3. Acta Crystallogr D Biol Crystallogr. 2015;71:1649–1656. doi: 10.1107/S1399004715010457. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka S, et al. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 36.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 37.Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31:2182–2194. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–2206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 41.Quan Y, et al. Cell-cycle-regulated interaction between Mcm10 and double hexameric Mcm2-7 is required for helicase splitting and activation during S phase. Cell Rep. 2015;13:2576–2586. doi: 10.1016/j.celrep.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Arnaiz P, Bruck I, Colbert MK, Kaplan DL. An intact Mcm10 coiled-coil interaction surface is important for origin melting, helicase assembly and the recruitment of Pol-α to Mcm2-7. Nucleic Acids Res. 2017;45:7261–7275. doi: 10.1093/nar/gkx438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tognetti S, Riera A, Speck C. Switch on the engine: How the eukaryotic replicative helicase MCM2-7 becomes activated. Chromosoma. 2015;124:13–26. doi: 10.1007/s00412-014-0489-2. [DOI] [PubMed] [Google Scholar]
- 44.Parker MW, Botchan MR, Berger JM. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol. 2017;52:107–144. doi: 10.1080/10409238.2016.1274717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller JM, Enemark EJ. Archaeal MCM proteins as an analog for the eukaryotic Mcm2-7 helicase to reveal essential features of structure and function. Archaea. 2015;2015:305497. doi: 10.1155/2015/305497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Botchan M, Berger J. DNA replication: Making two forks from one prereplication complex. Mol Cell. 2010;40:860–861. doi: 10.1016/j.molcel.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang S, Warner MD, Bell SP. Multiple functions for Mcm2-7 ATPase motifs during replication initiation. Mol Cell. 2014;55:655–665. doi: 10.1016/j.molcel.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou JC, et al. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. Proc Natl Acad Sci USA. 2017;114:4141–4146. doi: 10.1073/pnas.1700530114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Georgescu R, et al. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc Natl Acad Sci USA. 2017;114:E697–E706. doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langston L, O’Donnell M. Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. Elife. 2017;6:e23449. doi: 10.7554/eLife.23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pellegrini L, Costa A. New insights into the mechanism of DNA duplication by the eukaryotic replisome. Trends Biochem Sci. 2016;41:859–871. doi: 10.1016/j.tibs.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 56.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat Struct Mol Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 57.Costa A, et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. Elife. 2014;3:e03273. doi: 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J Biol Chem. 2007;282:34229–34234. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 59.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524:186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- 60.Vijayraghavan S, Schwacha A. The eukaryotic Mcm2-7 replicative helicase. Subcell Biochem. 2012;62:113–134. doi: 10.1007/978-94-007-4572-8_7. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: Mechanism of the MCM2-7 DNA helicase. Trends Biochem Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 62.Costa A, Onesti S. Structural biology of MCM helicases. Crit Rev Biochem Mol Biol. 2009;44:326–342. doi: 10.1080/10409230903186012. [DOI] [PubMed] [Google Scholar]
- 63.Sun J, et al. Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Genes Dev. 2014;28:2291–2303. doi: 10.1101/gad.242313.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan Z, et al. Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol. 2017;24:316–324. doi: 10.1038/nsmb.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Randell JC, et al. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stead BE, Brandl CJ, Davey MJ. Phosphorylation of Mcm2 modulates Mcm2-7 activity and affects the cell’s response to DNA damage. Nucleic Acids Res. 2011;39:6998–7008. doi: 10.1093/nar/gkr371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruck I, Kaplan D. Dbf4-Cdc7 phosphorylation of Mcm2 is required for cell growth. J Biol Chem. 2009;284:28823–28831. doi: 10.1074/jbc.M109.039123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lei M, et al. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jenkinson ER, Chong JP. Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc Natl Acad Sci USA. 2006;103:7613–7618. doi: 10.1073/pnas.0509297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Medagli B, Onesti S. Structure and mechanism of hexameric helicases. Adv Exp Med Biol. 2013;767:75–95. doi: 10.1007/978-1-4614-5037-5_4. [DOI] [PubMed] [Google Scholar]
- 73.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 74.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 75.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lam SK, et al. The PS1 hairpin of Mcm3 is essential for viability and for DNA unwinding in vitro. PLoS One. 2013;8:e82177. doi: 10.1371/journal.pone.0082177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramey CJ, Sclafani RA. Functional conservation of the pre-sensor one beta-finger hairpin (PS1-hp) structures in mini-chromosome maintenance proteins of Saccharomyces cerevisiae and archaea. G3 (Bethesda) 2014;4:1319–1326. doi: 10.1534/g3.114.011668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leon RP, Tecklenburg M, Sclafani RA. Functional conservation of beta-hairpin DNA binding domains in the Mcm protein of Methanobacterium thermoautotrophicum and the Mcm5 protein of Saccharomyces cerevisiae. Genetics. 2008;179:1757–1768. doi: 10.1534/genetics.108.088690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Froelich CA, Kang S, Epling LB, Bell SP, Enemark EJ. A conserved MCM single-stranded DNA binding element is essential for replication initiation. Elife. 2014;3:e01993. doi: 10.7554/eLife.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Du W, Stauffer ME, Eichman BF. Structural biology of replication initiation factor Mcm10. Subcell Biochem. 2012;62:197–216. doi: 10.1007/978-94-007-4572-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baxley RM, Bielinsky AK. Mcm10: A dynamic scaffold at eukaryotic replication forks. Genes (Basel) 2017;8:E73. doi: 10.3390/genes8020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lõoke M, Maloney MF, Bell SP. Mcm10 regulates DNA replication elongation by stimulating the CMG replicative helicase. Genes Dev. 2017;31:291–305. doi: 10.1101/gad.291336.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Riera A, et al. From structure to mechanism-understanding initiation of DNA replication. Genes Dev. 2017;31:1073–1088. doi: 10.1101/gad.298232.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhai Y, et al. Unique roles of the non-identical MCM subunits in DNA replication licensing. Mol Cell. 2017;67:168–179. doi: 10.1016/j.molcel.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 86.Gai D, Wang D, Li SX, Chen XS. The structure of SV40 large T hexameric helicase in complex with AT-rich origin DNA. Elife. 2016;5:e18129. doi: 10.7554/eLife.18129. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Kunkel TA, Burgers PMJ. Arranging eukaryotic nuclear DNA polymerases for replication: Specific interactions with accessory proteins arrange Pols α, δ, and ε in the replisome for leading-strand and lagging-strand DNA replication. Bioessays. 2017;39:1700070. doi: 10.1002/bies.201700070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.