Significance
Inflammation is a major risk factor for many cancers and the role of metabolic reprogramming in the inflammatory progression of cancer is not clear. We used a quantitative proteomic approach to identify mitochondrial proteins that are altered early in intestinal inflammation. We show that mitochondrial iron dysregulation is an early event that initiates mitochondrial dysfunction. Through the proteomic analysis, we identified a mitochondrial iron reductase, six-transmembrane epithelial antigen of prostate 4 (STEAP4), as being highly elevated during inflammation. Using intestinal epithelial-specific STEAP4 mice, we show that an increase in STEAP4 is sufficient to alter mitochondrial iron homeostasis. Chronic increase in mitochondrial iron leads to tissue injury and potentiates colon cancer, whereas mitochondrial iron chelation is protective in colitis and colitis-associated colon cancer models.
Keywords: STEAP4, hypoxia, inflammatory bowel disease, colorectal cancer, mitochondrial iron
Abstract
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder and is a major risk factor for colorectal cancer (CRC). Hypoxia is a feature of IBD and modulates cellular and mitochondrial metabolism. However, the role of hypoxic metabolism in IBD is unclear. Because mitochondrial dysfunction is an early hallmark of hypoxia and inflammation, an unbiased proteomics approach was used to assess the mitochondria in a mouse model of colitis. Through this analysis, we identified a ferrireductase: six-transmembrane epithelial antigen of prostate 4 (STEAP4) was highly induced in mouse models of colitis and in IBD patients. STEAP4 was regulated in a hypoxia-dependent manner that led to a dysregulation in mitochondrial iron balance, enhanced reactive oxygen species production, and increased susceptibility to mouse models of colitis. Mitochondrial iron chelation therapy improved colitis and demonstrated an essential role of mitochondrial iron dysregulation in the pathogenesis of IBD. To address if mitochondrial iron dysregulation is a key mechanism by which inflammation impacts colon tumorigenesis, STEAP4 expression, function, and mitochondrial iron chelation were assessed in a colitis-associated colon cancer model (CAC). STEAP4 was increased in human CRC and predicted poor prognosis. STEAP4 and mitochondrial iron increased tumor number and burden in a CAC model. These studies demonstrate the importance of mitochondrial iron homeostasis in IBD and CRC.
Inflammatory bowel disease (IBD) is an idiopathic chronic inflammatory disease of the intestine that manifests as ulcerative colitis (UC) or Crohn’s disease (CD) (1, 2). In UC, inflammation is restricted to the colon and affects the superficial inner lining of the epithelium. CD can appear throughout the gastrointestinal tract and affects all layers of the intestinal mucosa. Moreover, there are several other differences with respect to the immune responses, susceptibility genes, and environmental factors that may alter risk. However, both UC and CD exhibit an increase in the metabolic demands due to increased injury, regenerative proliferation, and influx of inflammatory cells. In IBD patients, intestinal mitochondrial function is dysregulated, causing less energy and more reactive oxygen species (ROS) to be produced by the mitochondria (3–6). This leads to a focal hypoxic environment requiring altered cell metabolism to meet the demands for ATP and macromolecule anabolism for efficient repair (7, 8). Hypoxia-inducible factor (HIF) regulates many of these adaptive pathways, including genes controlling glycolytic metabolism, inflammatory responses, and tumor development (9, 10). Due to the hypoxic nature of both inflammation and cancer, many similar metabolic reprogramming pathways are altered in inflammation and cancer. Currently, it is not clear what initiates changes in mitochondrial metabolism during inflammation, if changes in epithelial metabolism during inflammation can exacerbate tissue injury, and if alteration of inflammatory metabolism is a major mechanism enhancing tumorigenesis.
To further identify the initial mitochondrial changes that take place during intestinal inflammation, mitochondrial proteomics analysis of colonic tissues from an experimental mouse model of colitis was performed. Several iron metabolism-related proteins were highly increased in the mitochondria in inflamed colons compared with controls. Six-transmembrane epithelial antigen of prostate 4 [STEAP4, also called six-transmembrane protein of prostate 2 (STAMP2) or TNF-α–induced protein 9 (TNFIAP9)] was one of several iron-metabolism–related mitochondrial proteins induced in experimental colitis. STEAP4 belongs to a family of oxidoreductases that can function as a metalloreductase (11). Previous work demonstrated that STEAP4 is involved in responses to nutrients, inflammatory and oxidative stress, fatty acid metabolism, and glucose metabolism (12–15). The present work demonstrates that STEAP4 is localized to the mitochondria and regulated by hypoxia in mouse models of colitis. Increased STEAP4 expression in inflammatory foci leads to mitochondrial iron accumulation and elevated oxidative stress and increases susceptibility to experimental colitis and colitis-associated colon cancer (CAC). Chelating mitochondrial iron is protective in mouse models of colitis and colorectal cancer (CRC). Collectively, these data provide compelling evidence that mitochondrial iron dysfunction via STEAP4 is an integral link between inflammatory tissue injury and CAC.
Results
Mitochondrial Dysfunction Is an Early Event in Colitis.
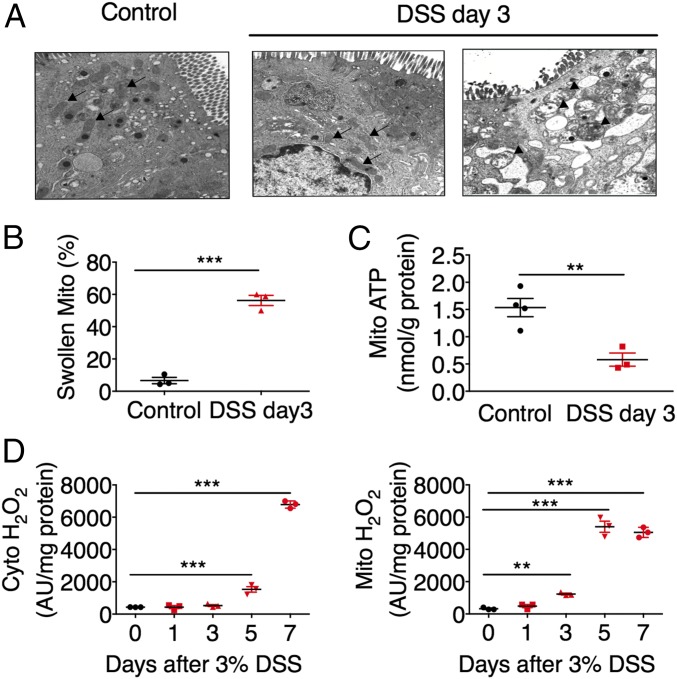
To investigate the role of mitochondria in the pathogenesis of IBD, wild-type mice were treated with 3% dextran sulfate sodium (DSS) or Salmonella Typhimurium SL1344 (S. Typhimurium) to induce colitis. Massive tissue damage and significant cytokine induction were observed at day 7 but not at day 3 after DSS treatment (Fig. S1 A and B). However, swollen mitochondria (Fig. 1 A and B), reduced mitochondrial ATP production (Fig. 1C), increased mitochondrial ROS generation (Fig. 1D), and increased expression of antioxidant proteins NRF2 and NQO1 (Fig. S1C) in colonic epithelial cells were observed as early as day 3 after DSS administration. In addition, S. Typhimurium treatment significantly increased the mitochondrial size of colonic epithelial cells (Fig. S1D). These results indicate that mitochondrial dysfunction is an early marker for colitis.
Fig. 1.
Mitochondrial alterations during colitis. (A) Electron microscopy (Magnification: A, 30,600×; arrows indicate normal mitochondria, arrowheads indicate swollen mitochondria) and (B) quantification of swollen mitochondria. (C) Mitochondrial (Mito) ATP levels and (D) cytosolic (cyto) and Mito H2O2 levels in colon tissues from mice treated with 3% DSS for indicated times (n = 3–5). **P < 0.01 and ***P < 0.001. One-way ANOVA followed by Dunnett’s multiple comparisons test or Student’s t test.
STEAP4 Is a Mitochondrial Protein That Is Increased in Colitis.
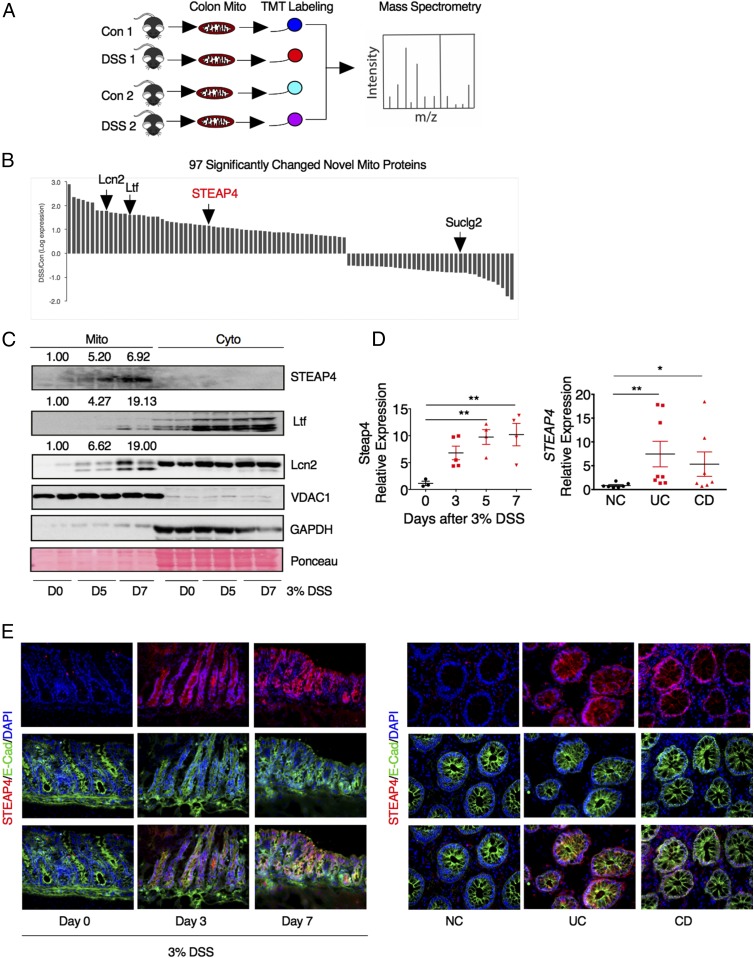
Mitochondrial proteomics analysis was assessed in colons from mice treated with 3 d of 3% DSS or regular drinking water using tandem mass spectrometry following isobaric peptide tagging to identify mitochondrial proteins altered following inflammation (Fig. 2A). Marginal changes were identified for 415 known mitochondrial proteins (Fig. S1A and Dataset S1). Among the 95 novel mitochondrial proteins significantly changed by DSS treatment, several iron metabolic proteins, including lipocalin 2 (Lcn2), lactoferrin (Ltf), and STEAP4 were identified (Fig. 2B). Western blot analysis confirmed that STEAP4 was selectively increased in the mitochondria following DSS treatment, whereas Ltf and Lcn2 were induced in both the mitochondrial and cytoplasmic fraction (Fig. 2C). STEAP4 was increased as early as day 3 in inflamed mouse colon tissues compared with normal control tissues by qPCR (Fig. 2D). Immunofluorescent costaining with the epithelial cell marker E-cadherin demonstrated that STEAP4 is predominantly induced in epithelial cells (Fig. 2E). The expression of STEAP4 was also assessed in IBD specimens. qPCR and immunofluorescent staining showed that STEAP4 was significantly increased compared with normal colon tissues (Fig. 2 D and E), but the expression levels of other STEAP family genes were not changed (Fig. S2B). Taken together, these data suggest that STEAP4 may play a role in the pathogenesis of colitis.
Fig. 2.
Characterization of STEAP4 as a mitochondrial protein increased in colitis. (A) Schematic diagram of the colonic mitochondrial proteomics analysis in mouse model of colitis. Mice were treated with 3% DSS for 3 d, and then colonic mitochondria were extracted and labeled with a tandem mass tag (TMT) labeling kit for mass spectrometry identification. (B) Histogram for 97 significantly changed novel mitochondrial proteins. Lcn2, lipocalin2; Ltf, lactoferrin; STEAP4, six-transmembrane epithelial antigen of prostate 4; Suclg2, succinate-CoA ligase, GDP-forming, β-subunit. (C) Western blot analysis of STEAP4, mitochondrial marker VDAC1, and cytosolic protein GAPDH after 3% DSS treatment. Ponceau staining was used to examine protein loading levels. Numbers above the blots indicate the mean value normalized with VDAC1. (D) qPCR analysis and (E) representative images of immunofluorescent staining for STEAP4 and epithelial cell marker E-cadherin (E-Cad) in colon tissues from DSS-treated mice (n = 3), UC (n = 8), CD (n = 7), and normal control (NC, n = 8) patients. (Magnification: E, 40×.) *P < 0.05 and **P < 0.01. One-way ANOVA followed by Dunnett’s multiple comparisons test.
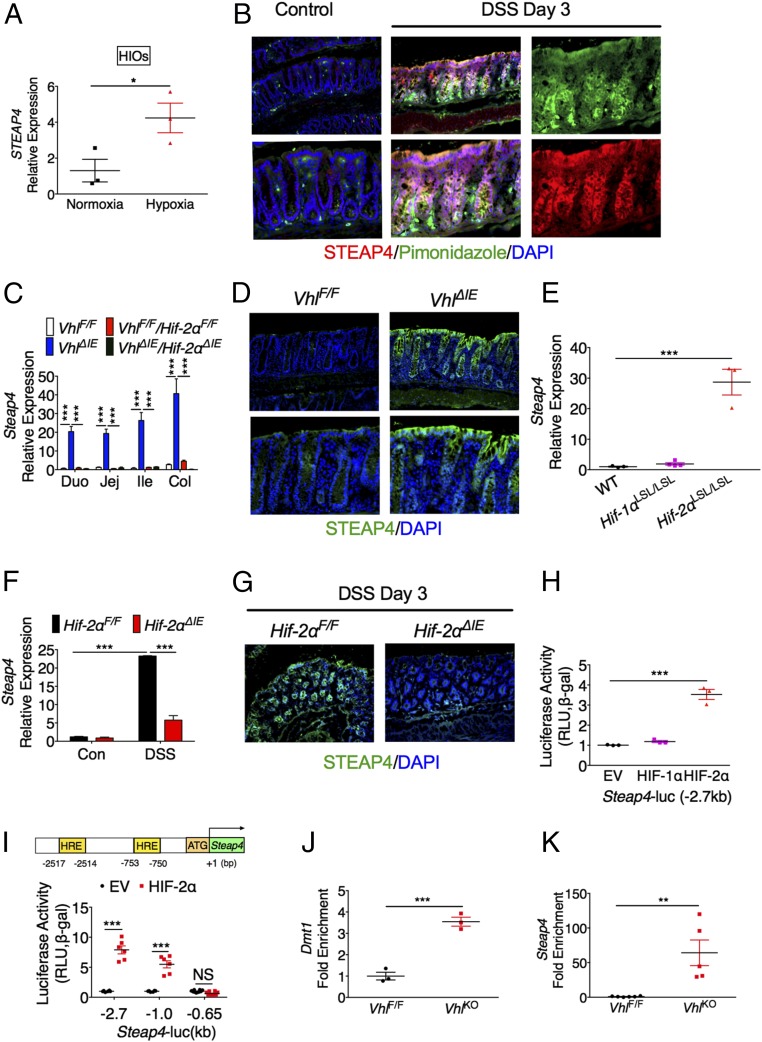
Steap4 Is a Direct Target Gene of HIF-2α.
STEAP4 is increased by the inflammatory cytokine TNF-α in adipocytes (13). In human intestinal organoids (HIOs), TNF-α or IL-6 did not induce STEAP4 (Fig. S3A), whereas TNF-α and IL-6 significantly increased the expression of their target genes IL-8 and SOCS3, respectively (Fig. S3 B and C). Interestingly, STEAP4 was significantly induced by hypoxia in HIOs (Fig. 3A). Pimonidazole staining demonstrated that hypoxic areas colocalized with STEAP4 expression after 3 d of DSS treatment (Fig. 3B). Hypoxia signaling is mainly mediated by HIF-1α and HIF-2α (16). Intestinal epithelial-specific disruption of von Hippel-Lindau protein (VhlΔIE) activates HIF signaling (16). By qPCR analysis, the expression of Steap4 was highly induced in the intestines from VhlΔIE mice compared with VhlF/F mice. This induction was dependent on HIF-2α, as assessed in mice with an intestine specific disruption of both Vhl and Hif-2α (VhlΔIE/Hif-2αΔIE) (Fig. 3C). Furthermore, immunofluorescent staining confirmed that STEAP4 is induced in the intestinal epithelial cells of VhlΔIE mice (Fig. 3D). To directly understand the role of HIF in the induction of Steap4, mouse models with intestinal epithelial-specific overexpression of HIF-1α (Hif-1αLSL/LSL) or HIF-2α (Hif-2αLSL/LSL) were assessed. Overexpression of HIF-2α but not HIF-1α increased Steap4 expression (Fig. 3E). To demonstrate that HIF-2α is essential in STEAP4 regulation in colitis, wild-type and HIF-2α intestine-specific knockout (Hif-2αΔIE) mice were treated with DSS to induce colitis. DSS treatment induced Steap4 expression in intestines from wild-type mice, but this induction was reduced by disruption of HIF-2α (Fig. 3 F and G). To understand if HIF-2α directly activates the Steap4 promoter, the 2.7-kb promoter sequence of mouse Steap4 was analyzed. Luciferase reporter assays showed that TNF-α activated the transcriptional activity of NF-κB but not Steap4 promoter luciferase activity (Fig. S3D), whereas HIF-2α but not HIF-1α increased the transcriptional activity of the Steap4 promoter (Fig. 3H). HIF-2α is critical in the regulation of iron responsive genes during iron deficiency (17); however, STEAP4 was not an iron-responsive gene (Fig. S3E). Furthermore, superrepressor IκBα (SR-IκBα) repressed TNF-α–activated NF-κB activity, but not HIF-2α–activated STEAP4 promoter activity (Fig. S3 F and G), suggesting that NF-κB signaling was not involved in the HIF-2α–mediated activation of STEAP4. Bioinformatics analysis found two putative hypoxia response elements (HREs) in the 2.7-kb proximal promoter of the Steap4 gene (Fig. 3I). A 5′ deletion demonstrated that the HREs were essential for the HIF-2α–mediated activation of Steap4 (Fig. 3I). An in vivo ChIP assay showed that HIF-2α binds to the proximal promoter of Steap4 gene as well as the known HIF-2α target gene DMT1 (Fig. 3 J and K). Together, these data demonstrate that Steap4 is a direct target gene of HIF-2α.
Fig. 3.
Steap4 is a direct target gene of HIF-2α. (A) qPCR analysis in HIOs under normoxic or hypoxic conditions. (B) Immunofluorescent costaining for STEAP4 and pimonidazole in the colon tissue from DSS-treated or control mice. (Magnification: Upper Left, 20×; Upper Middle, 20×; Upper Right, 40×; Lower Left, 40×; Lower Middle, 40×; Lower Right, 40×.) (C) qPCR analysis of Steap4 expression in intestinal tissues from VhlF/F (n = 5), VhlΔIE (n = 5), VhlF/F/Hif-2αF/F (n = 5), and VhlΔIE/Hif-2αΔIE (n = 5) mice. (D) Immunofluorescent staining for STEAP4 in the colon tissue from VhlF/F or VhlΔIE mice (n = 3). (Magnification: Upper Left, 20×; Upper Right, 20×; Lower Left, 40×; Lower Right, 40×.) (E) qPCR analysis in the colon tissues from wild-type (n = 3), Hif-1αLSL/LSL (n = 4), and Hif-2αLSL/LSL (n = 4) mice. (F) qPCR analysis and (G) immunofluorescent staining in the colon tissues from Hif-2αF/F (n = 3–5) or Hif-2αΔIE mice (n = 3–6) treated with or without 3% DSS for 3 d. (Magnification: Left, 20×; Right, 20×.) (H) Luciferase promoter activity assay. HCT116 cells were transiently transfected with the Steap4 promoter luciferase construct, and cotransfected with empty vector (EV), HIF-1α, or HIF-2α expression plasmids. (I) Schematic diagram of mouse Steap4 gene promoter depicting two potential putative HREs. Luciferase-reporter constructs under the control of the proximal 5′-flanking region of the mouse Steap4 gene (−2.7, −1.0, or −0.65 kb). (J and K) In vivo ChIP assays were performed on tissue extracts from VhlF/F and VhlKO mice using primers amplifying the proximal 5′-flanking region of the mouse Dmt1 or Steap4 gene. *P < 0.05, **P < 0.01, and ***P < 0.001. NS, not significant. One-way ANOVA followed by Dunnett’s multiple comparisons test or Student’s t test.
Characterization of Mice with Intestinal Epithelial-Specific Overexpression of STEAP4.
To investigate the role of Steap4 in vivo, transgenic mouse lines with intestinal epithelial cell specific overexpression of GFP-tagged human STEAP4 were generated using the Villin promoter. Western blot analysis and ex vivo fluorescent imaging of the tissues from these transgenic mice confirmed the expression of GFP in the intestines (Fig. S4 A and B). Through qPCR analysis, two founder lines that express STEAP4 at moderate and high levels were expanded (Fig. S4C). Western blot analysis of the intestines demonstrated that STEAP4 was expressed primarily in the mitochondria (Fig. S4 D and E). Immunofluorescent staining confirmed that the expression of STEAP4 protein was in the intestinal epithelial cells (Fig. S4E). Moreover, the transgenic mice had increased oxidoreductase activity in the intestine, demonstrating that the fusion protein was functional (Fig. S4F). These results demonstrate that we have successfully generated a mouse model with functional STEAP4 overexpression in the intestine.
STEAP4 Overexpression Aggravates Acute Experimental Colitis in Mice.
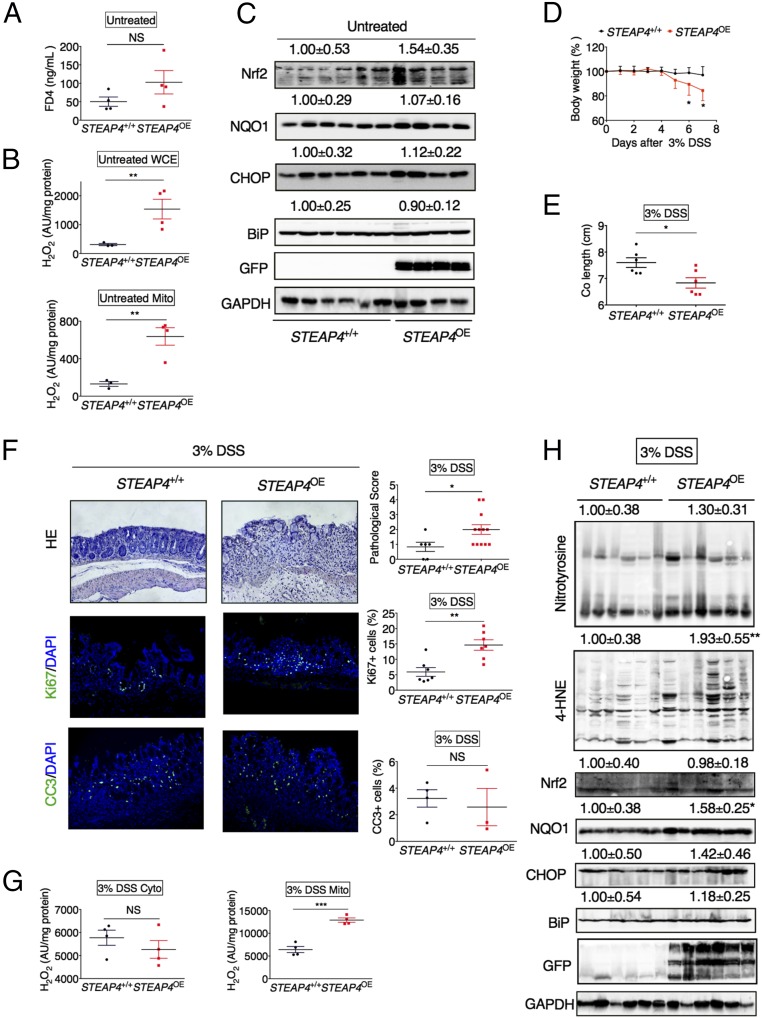
To study the physiological functions of STEAP4, the STEAP4 transgenic mice with moderate expression of human STEAP4 levels were used. The body weight and colon length of the STEAP4 transgenic mice were similar to their wild-type littermates (Fig. S5A) and were histologically indistinguishable from wild-type 9-mo-old littermates (Fig. S5 B and C). However, gene expression of the cytokine TNF-α and the chemokine Cxcl1 were slightly increased in the colons of STEAP4 2-mo-old transgenic mice (Fig. S5D). The increase in TNF-α did not lead to significant changes in epithelial barrier (Fig. 4A), but whole-cell, and specifically mitochondrial ROS levels, were highly induced in the STEAP4 transgenic mice compared with littermate control mice (Fig. 4B). However, the oxidative stress and endoplasmic reticulum stress-response proteins were not basally increased in the STEAP4 transgenic mice compared with littermate control mice (Fig. 4C).
Fig. 4.
Overexpression of STEAP4 in the intestine enhances susceptibility to experimental colitis. (A) Serum FD4, (B) H2O2 levels in colonic whole-cell extract (WCE) and mitochondrial (Mito) fraction, and (C) Western blot analysis of colonic tissues in 2-mo-old STEAP transgenic (STEAP4OE) (n = 4) and littermate control (STEAP4+/+) mice (n = 4–6). (D) Body weight, (E) colon lengths, (F) H&E and immunofluorescent staining showing complete loss of epithelium in STEAP4OE but not STEAP4+/+ mice, and pathological score of colon tissues following 7 d of 3% DSS treatment in STEAP4OE (n = 3–12) and STEAP4+/+ mice (n = 3–6). (Magnification: 20×.) (G) H2O2 levels in cytosolic (cyto) and mitochondrial (Mito) fraction and (H) Western blot analysis of intestinal tissues from STEAP4OE (n = 4–6) and STEAP4+/+ (n = 4–6) mice following 7 d of 3% DSS treatment. Numbers above the blots indicate the mean value and SD normalized with GAPDH. *P < 0.05, **P < 0.01, and ***P < 0.001. NS, not significant. Student’s t test.
To study the role of STEAP4 in an inflammatory environment, two models of acute colitis were assessed: DSS-induced chemical colitis and S. Typhimurium -induced infectious colitis. DSS treatment led to significantly more reduction in body weight and colon length in STEAP4 transgenic mice compared with wild-type littermates (Fig. 4 D and E). Histological analysis by H&E staining and pathological score further confirmed that overexpression of STEAP4 led to more severe colitis (Fig. 4F). Immunofluorescence staining revealed that cell proliferation, but not apoptosis, was increased in colon tissues from transgenic mice compared with control mice (Fig. 4F). ROS levels from mitochondrial but not cytosolic extracts were highly elevated compared with wild-type control mice (Fig. 4G). Western blot analysis showed that the levels of 4-hydroxynonenal (4-HNE), a major end product of lipid peroxidation, and NQO1, an oxidative stress-response protein, were increased in the colon tissues from DSS-treated STEAP4 overexpressing mice compared with their wild-type littermates (Fig. 4H). Similarly, the STEAP4 transgenic mice were highly susceptible to S. Typhimurium-induced colitis as assessed by body weight loss, colon length, and histological analysis (Fig. S6). These results demonstrate that STEAP4 promotes the intestinal inflammatory response through increased oxidative stress.
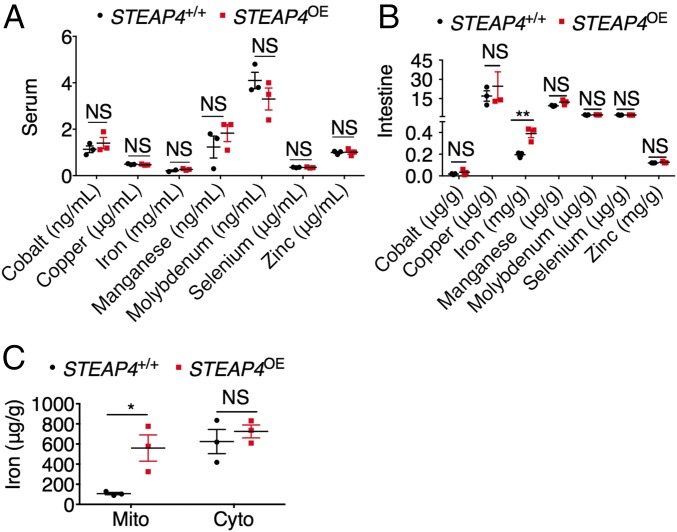
Overexpression of STEAP4 in the Intestine Increases Mitochondrial Iron Content.
STEAP4 is a metalloreductase (12), but serum mineral analysis did not demonstrate a significant increase in systemic levels of any divalent metals that were assessed (Fig. 5A). Interestingly, only local intestinal iron was increased in STEAP4 transgenic mice (Fig. 5B). Subcellular iron assay demonstrated a specific increase in the mitochondrial fraction (Fig. 5C), consistent with STEAP4 mitochondrial expression. This finding is in contrast to the increased iron levels in both mitochondrial fraction and cytosolic fraction of intestine tissues from VhlΔIE mice, as these mice have increase in STEAP4 as well as the plasma membrane iron importer DMT-1 (16) (Fig. S7). These data confirm that STEAP4 is a mitochondrial ferrireductase important in intestinal iron homeostasis.
Fig. 5.
Overexpression of STEAP4 in the intestine leads to mitochondrial iron accumulation. Mineral elements analysis of (A) serum and (B) intestinal tissues, and (C) iron content in mitochondrial (Mito) or cytosolic (Cyto) fraction of intestinal tissues from STEAP4 transgenic (STEAP4OE) (n = 3) and littermate control (STEAP4+/+) (n = 3) mice. *P < 0.05 and **P < 0.01. NS, not significant; two-way ANOVA test.
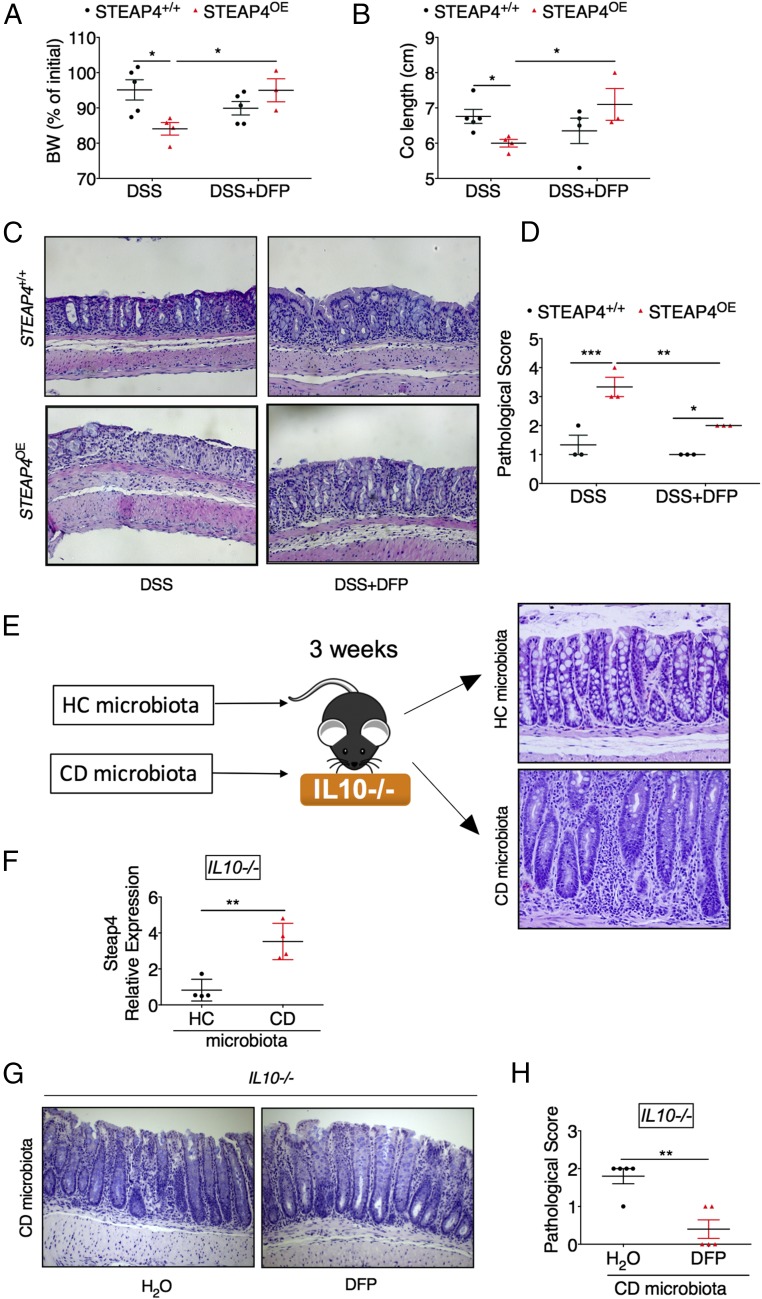
Mitochondrial Iron Chelation Reduces Experimental Colitis.
In STEAP4 transgenic mice we demonstrate a clear increase in mitochondrial iron; however, assessing mitochondrial iron levels following colitis is difficult due to confounding effects of a large number of inflammatory infiltrates in the colon and blood contamination from the injured site. To understand if mitochondrial iron dysregulation is a pathophysiologically relevant mechanism, deferiprone (DFP) was assessed. DFP is a clinically approved reagent that can chelate mitochondrial iron (18–21). DFP effectively protected the body weight loss, preserved colon length, and reduced histological changes and pathological score in DSS-treated STEAP4 transgenic mice (Fig. 6 A–D). Although in this acute model of colitis DFP did not protect STEAP4+/+ mice significantly, it is noteworthy to point out that the disease severity in STEAP4+/+ mice was mild and less than STEAP4-overexpressing mice. However, wild-type mice with more severe colitis following DSS were significantly protected by DFP treatment (Fig. S8). Moreover, to assess the role of mitochondrial iron dysregulation in a more clinically relevant model of colitis, we developed a novel microbiota humanized IL10−/− model of chronic colitis that accurately reproduces the host–microbiota interaction observed in IBD patients (22) (Fig. 6E). STEAP4 was highly increased in the colon tissues from germ-free IL10−/− mice colonized with microbiota of a CD patient (severe colitis) compared with those colonized with microbiota of a healthy control (HC, histologically normal) (Fig. 6 E and F). Importantly, DFP reduced the severity of colitis in the IL10−/− mice colonized with CD microbiota (Fig. 6 G and H). These data suggest that mitochondrial iron plays a critical role in the gut dysbiosis-driven intestinal pathology in patients with IBD.
Fig. 6.
Mitochondrial iron restriction reduces the severity of colitis. (A) Body weight change, (B) colon lengths, and (C) H&E staining showing complete loss of epithelium after DSS but not DSS+DFP in STEAP4 transgenic (STEAP4OE) mice, and (D) pathological score of colon tissues for STEAP4OE and littermate control (STEAP4+/+) mice following 7 d of 3% DSS with or without 1 mg/mL DFP treatment. (E) Schematic diagram for establishing a novel microbiota humanized IL10−/− model of chronic colitis. Germ-free IL10−/− mice were inoculated with microbiota from HC or CD patients and 3 wk later H&E staining indicates extensive colitis in CD microbiota treated mice. (F) Gene expression in colons from IL10−/− mice inoculated with microbiota from HC or CD patients. (G) Pathological score and (H) H&E staining of colon tissues from IL10−/− mice at 3 wk after inoculation with microbiota from a CD patient and treated with or without 1 mg/mL DFP treatment for 2 wk. (Magnification: C, 10×; E and G, 20×.) *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t test or two-way ANOVA test.
STEAP4 Is Critical for Colon Tumorigenesis Following Inflammation.
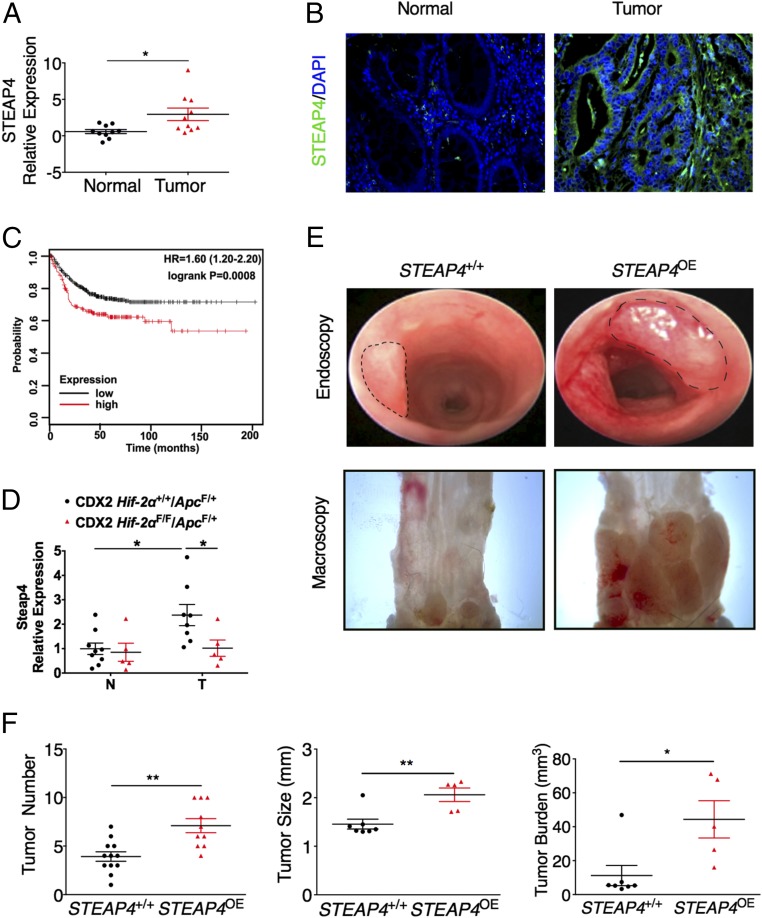
Inflammation is a high-risk factor for CRC (23), and mitochondrial defects are observed early in dysplastic regions of UC patients (24). In the colon tissue from STEAP4 transgenic mice, the cell proliferation rate is significantly increased after DSS treatment. Moreover, except for STEAP1, the expression of all of the other STEAP family proteins examined were increased in the tumor tissues compared with normal adjacent tissues (Fig. 7A and Fig. S9A). Immunofluorescence staining showed that STEAP4 is significantly increased in the tumor epithelial cells compared with normal epithelial cells (Fig. 7B). Kaplan–Meier survival curves were generated and stratified using datasets submitted to the Gene Expression Omnibus (GEO) (25). Increased expression of STEAP4, but not the other STEAP family proteins predicted worse patient survival (Fig. 7C and Fig. S9B). Together, these data suggest that STEAP4 may play a role in colon tumor development and cancer progression.
Fig. 7.
STEAP4 is critical for colon tumorigenesis. (A) qPCR analysis and (B) representative immunofluorescent staining images of STEAP4 in colon tissues from 10 paired colorectal tumors and their adjacent normal colon tissues. (C) Kaplan–Meier survival curves of STEAP4 gene in colorectal tumors patients. (D) Steap4 expression in CRCs and adjacent normal tissues from CDX2 Hif-2α F/F/ApcF/+ mice and their littermate controls is shown. Representative (E) endoscopy and gross images of colon tumors, and (F) tumor number, tumor size, tumor burden induced by AOM-DSS protocol in the STEAP4 transgenic (STEAP4OE) and littermate control (STEAP4+/+) mice. (Magnification: B, 40×; E, Upper, 5×, Lower, 8×.) *P < 0.05 and **P < 0.01. Student’s t test.
To further understand if HIF-2α is involved in the regulation of STEAP4 in CRC, a colon-specific disruption of HIF-2α in the colon-specific sporadic CRC model was assessed (26). Interestingly, disruption of HIF-2α significantly reduced tumoral STEAP4 expression (Fig. 7D). To determine if the increased STEAP4 expression contributes to the increase in colon tumorigenesis, transgenic mice with STEAP4 overexpression were treated with one dose of azoxymethane (AOM) and three cycles of 1.5% DSS to establish colitis-associated colon tumors. The tumor number, tumor size, and tumor burden were significantly increased in transgenic mice compared with their littermate controls (Fig. 7 E and F). Immunofluorescence staining revealed that cell proliferation determined by Ki67 staining, DNA damage determined by γ-H2AX staining, and granulocytic cell infiltration determined by myeloperoxidase (MPO) staining—but not apoptosis determined by cleaved caspase 3 staining—were increased in colorectal tumor tissues from transgenic mice compared with control mice (Fig. S10A). Furthermore, 3,3′-diaminobenzidine (DAB)-enhanced Perls’ iron staining revealed an increased deposition of iron in the tumor tissues from transgenic mice compared with control mice (Fig. S10B). Similar to colitis, NQO1was also increased in the colon tumor tissues from STEAP4-overexpression mice (Fig. S10C). However, prosurvival ERK, AKT, and STAT3 signaling pathways were not changed (Fig. S10C). These data indicate that STEAP4-mediated iron metabolism and oxidative stress are critical in the colorectal tumor development.
Mitochondrial Iron Homeostasis Is Critical for Colon Tumorigenesis Following Inflammation.
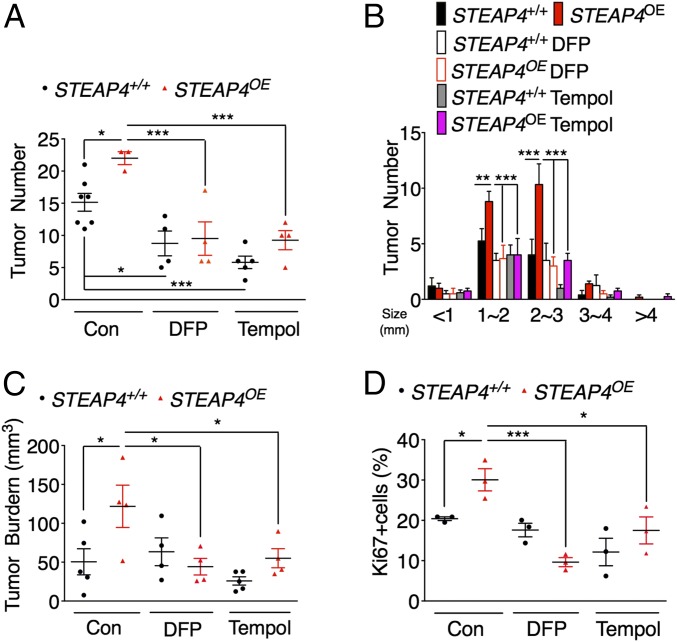
Similar to inflamed areas, mitochondrial iron is difficult to measure in tumors; therefore, to further confirm the role of mitochondrial iron and oxidative stress in colon tumorigenesis, we treated STEAP4 transgenic and littermate wild-type mice with AOM-DSS in the presence or absence of DFP or antioxidant Tempol. Strikingly, both DFP and Tempol significantly decreased tumor number, tumor burden, and cell proliferation in wild-type and STEAP4 transgenic mice (Fig. 8 and Fig. S11). Together, these results suggest that overexpression of STEAP4 via mitochondrial iron promotes the susceptibility of mice to CRC by increasing mitochondrial iron and oxidative stress in the intestine.
Fig. 8.
Mitochondrial iron restriction reduces colon tumorigenesis in STEAP4 transgenic mice. (A) Colon tumor numbers and (B) tumor number grouped by size, (C) tumor burden and (D) quantification of Ki67 staining of colon tumors from STEAP4 transgenic (STEAP4OE) and littermate control (STEAP4+/+) mice treated with AOM-DSS protocol in the presence or absence of 1 mg/mL DFP, 0.064% Tempol in the drinking water. *P < 0.05, **P < 0.01, and ***P < 0.001; two-way ANOVA test.
Discussion
Altered metabolic programming is now considered a major feature in both inflammation and tumor development. It was long thought that mitochondria are less functional in inflamed regions and in tumors, as most of the ATP is generated through glycolysis. However, the intermediate metabolites produced by mitochondria are essential for anabolic pathways to generate lipids, amino acids, and nucleic acids. Many of the anabolic mitochondrial enzymes require iron. However, it is not clear what impact iron has on metabolic reprogramming. Here we identified that the ferrireductase STEAP4 is highly increased in the colon tissues from IBD and CRC patients. Intestinal epithelial cell-specific overexpression of STEAP4 in mice elevated the levels of intestinal mitochondrial iron, oxidative stress, and enhanced the susceptibility of these mice to experimental colitis and CRC. Mitochondrial iron chelation protects mice from colitis and CRC, suggesting a critical role of mitochondrial iron homeostasis in inflammation and cancer. Moreover, our data in a novel microbiota-induced colitis model is consistent with recent data linking microbiota–mitochondrial cross-talk, and provides a molecular mechanism by which microbiota can alter mitochondrial function during inflammation (27). Direct detection of mitochondrial iron in vivo is currently challenging. Conventional colorimetrical ferrozine-based assays are not specific enough to detect iron in isolated mitochondria from injured colonocytes. A more sensitive inductively coupled plasma mass spectrometry (ICP-MS) method cannot distinguish heme and nonheme iron from the tissue, which leads to confounding issues from bleeding that often occur in inflamed and tumor colon tissues. However, using DFP, a mitochondrial iron chelator, our data suggest that mitochondrial iron plays a critical role in the mechanism of intestinal inflammation and cancer.
STEAP4 was first identified as a novel gene induced by TNF-α during adipose differentiation (28). STEAP4 has 60% similarity to three other proteins, designated STEAP1, STEAP2, and STEAP3 (11). Overexpression of all STEAP proteins, except STEAP1, which lacks an N-terminal NAD(P)H binding-reductase motif, resulted in increased ferrireductase and cupric reductase activity in human embryonic kidney cells (11). Among these proteins, STEAP4 has the highest activity, suggesting its important role in iron and copper homeostasis. However, STEAP4 deficiency exhibited an aggravated inflammatory response in both macrophages (14) and adipose tissues (13) without other obvious abnormalities. It is proposed that in adipose tissue STEAP4 integrates inflammatory cytokine signaling with lipid and glucose metabolism (13). STEAP4 deficiency contributes to metabolic disorders, including mild hyperglycemia, dyslipidemia, atherosclerosis, and fatty liver disease. Here we found that STEAP4 overexpression in the intestine increases mitochondrial iron and aggravates experimental colitis, indicating that STEAP4 functions as a rheostat for inflammatory response in a tissue-specific manner. Mice with intestinal epithelial-specific overexpression of STEAP4 did not have spontaneous colitis, which indicates that mitochondrial iron accumulation alone is not enough to drive colitis. These data are in line with the complexity of IBD as several major defects are needed for an injurious inflammatory progression. Our data show that mitochondrial iron handling is altered in IBD, and in mice with these defects leads to epithelial cell injury and degeneration. Interestingly, recent work also demonstrates a more far-reaching role of mitochondrial ROS in altering intestinal immunity, and our work provides an ideal mechanistic target for these effects (29).
All four STEAP proteins are increased in prostate cancer and play important role in prostate cancer development and progression (30–34). STEAP4 increases ROS in prostate cancer cells through its iron reductase activity (15). This supports our hypothesis that STEAP4 provides ferrous iron to mitochondria for maintaining normal respiratory function and producing free radicals (35). Iron-catalyzed free radicals increase protein nitrosation (36), lipid peroxidation (37), and oxidative DNA damage during colitis and colon tumorigenesis (38). Here we show that overexpression of STEAP4 enhances the levels of NQO1, 4-HNE, and γ-H2AX in experimental colitis and CAC. Furthermore, the antioxidant Tempol and mitochondrial iron chelator DFP have similar inhibitory effects on the pathogenesis of colon tumors in mice with STEAP4 overexpression, suggesting that STEAP4 may be the cause of increased oxidative stress through providing excess mitochondrial iron during the pathogenesis of colitis and CRC.
In summary, we have identified a mitochondrial iron regulatory protein that is induced in both colitis and CRC, which links hypoxia-induced iron metabolic changes to inflammation and tumorigenesis. The hypoxia/HIF-2α/STEAP4/mitochondrial iron/mitochondrial ROS axis promotes colitis and colon cancer development. The localization of STEAP4 in the mitochondria and differential expression in normal and cancer tissues make STEAP4 a potential candidate as a therapeutic target in CRC. Moreover, our work clearly provides evidence for clinically approved iron chelators in iron overload syndromes to be used in IBD and CRC.
Materials and Methods
Generation of GFP Human STEAP4-Expressing Transgenic Mice.
We generated transgenic mice expressing human STEAP4 under control of the Villin promoter. The human STEAP4 in pcDNA4/HisMax-TOPO vector (a kind gift from Fahri Saatcioglu, University of Oslo, Norway) was subcloned into the pEGFP-C1 to generate the pEGFP-STEAP4 vector that encodes STEAP4 with EGFP sequences on the N-terminal end of the protein. The EGFP-STEAP4 fragment was subcloned into the MluI and KpnI site of the pUC12.4-kb-villin plasmid to generate the pUC12.4k-Villin-EGFP-STEAP4 plasmid (39). After digestion with Pme I, the 16-kb transgene was used to create STEAP4 transgenic mice at the University of Michigan. The mice were back-crossed eight generations onto C57BL/6J background. For all studies, littermate control mice were used.
Animals and Treatments.
VhlF/F, VhlΔIE, VhlF/F/Hif-2αF/F, VhlΔIE/Hif-2αΔIE, Hif-2αF/F, Hif-2αΔIE, Hif-1αLSL/LSL, Hif-2αLSL/LSL CDX2 Hif-2α+/+/ApcF/+, and CDX2 Hif-2αF/F/ApcF/+ mice were described previously (26, 40). For chemical-induced colitis, 6- to 8-wk-old mice were administered 3% (wt/vol) DSS (36–50 kDa; MP Biomedicals) in the drinking water. For pathogen-induced colitis, 6- to 8-wk-old mice were orally given 7.5 mg streptomycin per mouse and 24 h later were infected with 1 × 107 colony-forming units of S. Typhimurium SL1344. To establish the humanized gnotobiotic mouse colitis model, germ-free IL10−/− mice were inoculated with the gut microbiotas from a CD patient or HC subject and housed for 3 wk, as described previously (22). Humanized gnotobiotic mice were treated with or without 1 mg/mL DFP in the drinking water in last 2 wk. The AOM-DSS study was done as previously described (26). At the end of the experiment, high-resolution mouse endoscopy was performed to identify colon adenomas as previously described (26). Next, 1 mg/mL DFP and 0.064% Tempol were given in the drinking water. All mice were maintained in standard cages in a light and temperature-controlled room and were given standard chow and water ad libitum. All animal studies were carried out in accordance with Institute of Laboratory Animal Resources guidelines and approved by the University Committee on the Use and Care of Animals at the University of Michigan.
Gut Permeability Assay.
Colon permeability was assessed with FITC-labeled dextran (FD4; Sigma-Aldrich) as previously described (41).
Histology, Immunofluorescence Staining, and Enhanced Perls’ Iron Staining.
Paraffin-embedded tissue sections (5 μm) were deparaffinized in xylene and rehydrated in ethanol gradient. All histological scoring was done by a blinded gastrointestinal pathologist, as previously described (40). Immunofluorescence analysis was performed with antibodies for STEAP4 (11944-1-AP; Proteintech), E-cadherin (33-4000; Invitrogen), GFP (SC-9996; Santa Cruz Biotechnology), Ki67 (VP-RM04, Clone SP6; Vector Labs), cleaved caspase 3 (CC3, 9664; Cell Signaling Technology), γ-H2AX (2577; Cell Signaling Technology), and MPO (RB-373-A0; Thermo Scientific), as previously described (40). Pimonidazole staining for hypoxia was described previously (42). Enhanced Perls’ iron staining was carried out in paraffin-embedded sections stained with Perls’ Prussian blue and enhanced with DAB and H2O2 (43).
Transmission Electron Microscopy.
For ultrastructural analysis, mouse intestines were excised and fixed in a solution of 2% paraformaldehyde and 2% glutaraldehyde in PBS for 2 h at room temperature. After washing in PBS, the tissue samples were postfixed in osmium tetroxide for 45 min at room temperature. Dehydration of the samples was accomplished by transferring the samples through a series of graded ethanol and then 100% propylene oxide. The tissue was then infiltrated by transferring the samples into increasing concentrations of Epon to propylene oxide solutions; 1:3, 1:1, and 3:1, then 100% Epon and finally embedded. Sections were made with a Leica EM UC7 μLtramicrotome (Leica), stained for 15 min with 7% (saturated) aqueous uranyl acetate, washed, stained with lead citrate, and examined with a JEOL JEM 1400 plus transmission electron microscope (JEOL).
Quantitative Real-Time RT-PCR.
RNA isolation and qPCR was performed as previously described (26). Primers are listed in Table S1.
Mitochondria Isolation.
Mitochondria isolation was adapted from a previous report (44). Tissues were homogenized with 300 μL SH buffer (pH 7.2) containing 250 mM sucrose and 10 mM Hepes. After incubation on ice for 5 min, the homogenate was centrifuged at 1,000 × g for 10 min at 4 °C. The supernatant was kept as the cytosolic fraction and the pellet was resuspended in 500 μL SH buffer, and was centrifuged at 700 × g for 10 min at 4 °C. The resulting supernatant was centrifuged at 9,000 × g for 10 min at 4 °C to pellet crude mitochondria.
Determination of Serum and Tissue Minerals, Mitochondrial Iron, ATP, and H2O2 Levels.
Serum and tissue minerals were quantitated by ICP-MS at The Diagnostic Center for Population and Animal Health, Michigan State University. Mitochondrial and cytosolic nonheme iron in intestinal tissue was quantitated as previously described (45). Mitochondrial ATP was determined with ATP assay kit (ThermoFisher). Tissue H2O2 was determined using ROS-Glo H2O2 Assay kit (Promega).
Unbiased Colonic Mitochondrial Proteomics Analysis.
Mice were treated with 3% DSS for 3 d, and then colonic mitochondria were extracted. Mitochondrial proteins were digested into peptides with trypsin, labeled with a tandem mass tag labeling kit (ThermoFisher) according to the manufacturer’s instructions, and followed with mass spectrometry analysis and protein identification.
Western Blot Analysis.
Western analysis was performed as previously described (20). Antibodies against STEAP4 (NB100-68162) were from Novus; Ltf (sc-53498), Lcn2 (sc-515876), GFP (SC-9996), NQO1 (sc-32793), CHOP (sc-7351), BiP (sc-376768), and GAPDH (SC-25778) were from Santa Cruz Biotechnology; VDAC1 (4866), Nrf2 (12721), p-ERK (4370P), t-ERK(9102), p-AKT(4060), t-AKT(9272), p-STAT3 (9145S), and t-STAT3(9139P) were from Cell Signaling Technology; and nitrotyrosine (05-233) and 4-HNE (AB5605) were from Millipore. Ponceau staining was used for normalization of total proteins.
HIOs Culture, Colitis, and Colorectal Tumor Tissues.
HIO culture was described previously (40). HIOs were treated with 21% (normoxia) or 1% oxygen (hypoxia) for 24 h and RNAs were extracted. Human UC, CD, and adjacent normal tissues were obtained from individuals undergoing biopsy, and these samples all had active disease. Human colorectal tumor and adjacent normal tissues were obtained from individuals undergoing colorectal tumor removal surgery. The Institutional Review Board of the University of Michigan approved the use of these materials.
Ex Vivo Tissue Fluorescent Imaging.
Fluorescence indicating GFP for excised colons was visualized by IVIS spectrum imaging system (Caliper Life Sciences) following the instructions of the manufacturer.
Luciferase Assay.
CRC-derived HCT116 cells were obtained from ATCC and maintained at 37 °C in 5% CO2 and 21% O2. Cells were cultured in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic. Cells were seeded into a 24-well plate at a cell density of 5 × 104 cells per well. Steap4 promoter luciferase reporter constructs were generated using primers listed in Table S1. The luciferase reporters were cotransfected with HIF-1α, HIF-2α, or empty vector into cells with polyethylenimine (PEI; Polysciences Inc.). Cells were lysed in reporter lysis buffer (Promega), and firefly luciferase activity was measured and normalized to β-galactosidase (β-gal) activity 48 h after transfection.
ChIP Assay.
ChIP assays were performed as previously described (46). qPCR was assessed using primers listed in Table S1.
Nitroblue Tetrazolium Assay.
Duodenal tissues were removed, opened lengthwise, and rinsed with 150 mM NaCl. Slices (full width of duodenum by 1–2 mm) were taken ∼1 cm from the pylorus and incubated for 5 min at 37 °C in 200 µL of 1 mM Nitroblue tetrazolium in incubation buffer [125 mM NaCl, 3.5 mM KCl, and 16 mM Hepes/NaOH (pH 7.4)]. After incubation, tissues were rinsed twice with 150 mM NaCl and photographed with a dissecting microscope.
Meta-Analysis of CRC Samples.
CRC gene-expression datasets GSE12945, GSE14333, GSE17538, GSE31595, GSE33114, GSE37892, GSE39582, and GSE41258 with survival were identified in the GEO using the search keywords “colorectal,” “cancer,” and “microarray” (www.ncbi.nlm.nih.gov/geo/). Only publications providing raw data, clinical survival information, and containing at least 30 patients were included. The gene chips were MAS 5.0 normalized in the R statistical environment (www.R-project.org) using the Bioconductor package Affy (www.bioconductor.org). The most reliable probe sets for each gene were selected using Jetset and survival analysis using Cox proportional hazards regression was performed, as described previously (25).
Statistical Analysis.
Data are expressed as mean ± SD. P values were calculated by independent t test, one-way and two-way ANOVA. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Bradley Nelson for excellent technical support in the electron microscopy experiments. X.X. was supported by NIH Grant 1K01DK114390-01 and a Research Scholar Award from the American Gastroenterological Association. H.N.-K. and N.K. were supported by the Crohn’s and Colitis Foundation of America. B.G. was supported by Grant NVKP_16-1-2016-0037 of the National Research, Development, and Innovation Office of Hungary. Y.M.S. was supported by NIH Grants CA148828 and DK095201.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.S.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712946114/-/DCSupplemental.
References
- 1.Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756–1770. doi: 10.1016/S0140-6736(16)32126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn’s disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 3.Santhanam S, et al. Mitochondrial electron transport chain complex dysfunction in the colonic mucosa in ulcerative colitis. Inflamm Bowel Dis. 2012;18:2158–2168. doi: 10.1002/ibd.22926. [DOI] [PubMed] [Google Scholar]
- 4.Roediger WE. The colonic epithelium in ulcerative colitis: An energy-deficiency disease? Lancet. 1980;2:712–715. doi: 10.1016/s0140-6736(80)91934-0. [DOI] [PubMed] [Google Scholar]
- 5.Hess J, et al. Ischaemic colitis due to mitochondrial cytopathy. Lancet. 1995;346:189–190. [PubMed] [Google Scholar]
- 6.Ho GT, et al. MDR1 deficiency impairs mitochondrial homeostasis and promotes intestinal inflammation. Mucosal Immunol. 2017 doi: 10.1038/mi.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgan SP, Taylor CT. Hypoxia: An alarm signal during intestinal inflammation. Nat Rev Gastroenterol Hepatol. 2010;7:281–287. doi: 10.1038/nrgastro.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunelle JK, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci USA. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shay JE, Celeste Simon M. Hypoxia-inducible factors: Crosstalk between inflammation and metabolism. Semin Cell Dev Biol. 2012;23:389–394. doi: 10.1016/j.semcdb.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ten Freyhaus H, et al. Stamp2 controls macrophage inflammation through nicotinamide adenine dinucleotide phosphate homeostasis and protects against atherosclerosis. Cell Metab. 2012;16:81–89. doi: 10.1016/j.cmet.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellen KE, et al. Coordinated regulation of nutrient and inflammatory responses by STAMP2 is essential for metabolic homeostasis. Cell. 2007;129:537–548. doi: 10.1016/j.cell.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HY, et al. Hepatic STAMP2 alleviates high fat diet-induced hepatic steatosis and insulin resistance. J Hepatol. 2015;63:477–485. doi: 10.1016/j.jhep.2015.01.025. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, et al. STAMP2 increases oxidative stress and is critical for prostate cancer. EMBO Mol Med. 2015;7:315–331. doi: 10.15252/emmm.201404181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue X, et al. Hypoxia-inducible factor-2α activation promotes colorectal cancer progression by dysregulating iron homeostasis. Cancer Res. 2012;72:2285–2293. doi: 10.1158/0008-5472.CAN-11-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor M, et al. Hypoxia-inducible factor-2α mediates the adaptive increase of intestinal ferroportin during iron deficiency in mice. Gastroenterology. 2011;140:2044–2055. doi: 10.1053/j.gastro.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elincx-Benizri S, et al. Clinical experience with deferiprone treatment for Friedreich ataxia. J Child Neurol. 2016;31:1036–1040. doi: 10.1177/0883073816636087. [DOI] [PubMed] [Google Scholar]
- 19.Sohn YS, Breuer W, Munnich A, Cabantchik ZI. Redistribution of accumulated cell iron: A modality of chelation with therapeutic implications. Blood. 2008;111:1690–1699. doi: 10.1182/blood-2007-07-102335. [DOI] [PubMed] [Google Scholar]
- 20.Filosa A, et al. Long-term treatment with deferiprone enhances left ventricular ejection function when compared to deferoxamine in patients with thalassemia major. Blood Cells Mol Dis. 2013;51:85–88. doi: 10.1016/j.bcmd.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Cloonan SM, et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagao-Kitamoto H, et al. Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell Mol Gastroenterol Hepatol. 2016;2:468–481. doi: 10.1016/j.jcmgh.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukas M. Inflammatory bowel disease as a risk factor for colorectal cancer. Dig Dis. 2010;28:619–624. doi: 10.1159/000320276. [DOI] [PubMed] [Google Scholar]
- 24.Ussakli CH, et al. Mitochondria and tumor progression in ulcerative colitis. J Natl Cancer Inst. 2013;105:1239–1248. doi: 10.1093/jnci/djt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sztupinszki Z, Győrffy B. Colon cancer subtypes: Concordance, effect on survival and selection of the most representative preclinical models. Sci Rep. 2016;6:37169. doi: 10.1038/srep37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue X, et al. Iron uptake via DMT1 integrates cell cycle with JAK-STAT3 signaling to promote colorectal tumorigenesis. Cell Metab. 2016;24:447–461. doi: 10.1016/j.cmet.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mottawea W, et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat Commun. 2016;7:13419. doi: 10.1038/ncomms13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moldes M, et al. Tumor necrosis factor-alpha-induced adipose-related protein (TIARP), a cell-surface protein that is highly induced by tumor necrosis factor-alpha and adipose conversion. J Biol Chem. 2001;276:33938–33946. doi: 10.1074/jbc.M105726200. [DOI] [PubMed] [Google Scholar]
- 29.Formentini L, et al. Mitochondrial ROS production protects the intestine from inflammation through functional M2 macrophage polarization. Cell Rep. 2017;19:1202–1213. doi: 10.1016/j.celrep.2017.04.036. [DOI] [PubMed] [Google Scholar]
- 30.Hubert RS, et al. STEAP: A prostate-specific cell-surface antigen highly expressed in human prostate tumors. Proc Natl Acad Sci USA. 1999;96:14523–14528. doi: 10.1073/pnas.96.25.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porkka KP, Helenius MA, Visakorpi T. Cloning and characterization of a novel six-transmembrane protein STEAP2, expressed in normal and malignant prostate. Lab Invest. 2002;82:1573–1582. doi: 10.1097/01.lab.0000038554.26102.c6. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, et al. STAMP1 is both a proliferative and an antiapoptotic factor in prostate cancer. Cancer Res. 2010;70:5818–5828. doi: 10.1158/0008-5472.CAN-09-4697. [DOI] [PubMed] [Google Scholar]
- 33.Machlenkin A, et al. Human CTL epitopes prostatic acid phosphatase-3 and six-transmembrane epithelial antigen of prostate-3 as candidates for prostate cancer immunotherapy. Cancer Res. 2005;65:6435–6442. doi: 10.1158/0008-5472.CAN-05-0133. [DOI] [PubMed] [Google Scholar]
- 34.Korkmaz CG, et al. Molecular cloning and characterization of STAMP2, an androgen-regulated six transmembrane protein that is overexpressed in prostate cancer. Oncogene. 2005;24:4934–4945. doi: 10.1038/sj.onc.1208677. [DOI] [PubMed] [Google Scholar]
- 35.Okada S. Iron-induced tissue damage and cancer: The role of reactive oxygen species-free radicals. Pathol Int. 1996;46:311–332. doi: 10.1111/j.1440-1827.1996.tb03617.x. [DOI] [PubMed] [Google Scholar]
- 36.Seril DN, et al. Dietary iron supplementation enhances DSS-induced colitis and associated colorectal carcinoma development in mice. Dig Dis Sci. 2002;47:1266–1278. doi: 10.1023/a:1015362228659. [DOI] [PubMed] [Google Scholar]
- 37.Carrier J, Aghdassi E, Cullen J, Allard JP. Iron supplementation increases disease activity and vitamin E ameliorates the effect in rats with dextran sulfate sodium-induced colitis. J Nutr. 2002;132:3146–3150. doi: 10.1093/jn/131.10.3146. [DOI] [PubMed] [Google Scholar]
- 38.Phillips LL. Effect of free radicals on chromosomes of barley. Science. 1956;124:889–890. doi: 10.1126/science.124.3227.889. [DOI] [PubMed] [Google Scholar]
- 39.Madison BB, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 40.Xue X, et al. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie L, et al. Hypoxia-inducible factor/MAZ-dependent induction of caveolin-1 regulates colon permeability through suppression of occludin, leading to hypoxia-induced inflammation. Mol Cell Biol. 2014;34:3013–3023. doi: 10.1128/MCB.00324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ragnum HB, et al. The tumour hypoxia marker pimonidazole reflects a transcriptional programme associated with aggressive prostate cancer. Br J Cancer. 2015;112:382–390. doi: 10.1038/bjc.2014.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meguro R, Asano Y, Iwatsuki H, Shoumura K. Perfusion-Perls and -Turnbull methods supplemented by DAB intensification for nonheme iron histochemistry: Demonstration of the superior sensitivity of the methods in the liver, spleen, and stomach of the rat. Histochem Cell Biol. 2003;120:73–82. doi: 10.1007/s00418-003-0539-y. [DOI] [PubMed] [Google Scholar]
- 44.Frezza C, Cipolat S, Scorrano L. Organelle isolation: Functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 45.Anderson ER, et al. Intestinal HIF2α promotes tissue-iron accumulation in disorders of iron overload with anemia. Proc Natl Acad Sci USA. 2013;110:E4922–E4930. doi: 10.1073/pnas.1314197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue X, Shah YM. Hypoxia-inducible factor-2α is essential in activating the COX2/mPGES-1/PGE2 signaling axis in colon cancer. Carcinogenesis. 2013;34:163–169. doi: 10.1093/carcin/bgs313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.