Significance
G-protein–coupled receptors (GPCRs) are the largest group of membrane receptors in eukaryotes, are involved in a plethora of cellular processes, and are implicated in many human diseases. Several individual GPCRs were shown to interact with a small gene group called receptor activity-modifying proteins (RAMPs). In this study, we present genomic- and expression-based observations suggesting that GPCRs interact with RAMPs in a more widespread manner than previously appreciated. Mapping of GPCR−RAMP global interactions can potentially point to specific signaling assays needed for deorphanization of orphan receptors. In addition, such interactions could affect testing of drugs targeting a RAMP-interacting GPCR.
Keywords: coevolution, G-protein–coupled receptor, phylogenetic analysis, receptor activity-modifying protein, signal transduction
Abstract
Receptor activity-modifying proteins (RAMPs) are widely expressed in human tissues and, in some cases, have been shown to affect surface expression or ligand specificity of G-protein–coupled receptors (GPCRs). However, whether RAMP−GPCR interactions are widespread, and the nature of their functional consequences, remains largely unknown. In humans, there are three RAMPs and over 800 expressed GPCRs, making direct experimental approaches challenging. We analyzed relevant genomic data from all currently available sequenced organisms. We discovered that RAMPs and GPCRs tend to have orthologs in the same species and have correlated phylogenetic trees to the same extent, or higher than other interacting protein pairs that play key roles in cellular signaling. In addition, the resulting RAMP−GPCR interaction map suggests that RAMP1 and RAMP3 interact with the same set of GPCRs, which implies functional redundancy. We next analyzed human transcriptomes and found expression correlation for GPCRs and RAMPs. Our results suggest global coevolution of GPCRs and RAMPS and support the hypothesis that GPCRs interact globally with RAMPs in cellular signaling pathways.
G-protein–coupled receptors (GPCRs) are essential for transmembrane signal transduction, which governs a plethora of basic molecular processes. GPCRs are also highly druggable therapeutic targets (1). Receptor activity-modifying proteins (RAMPs) change ligand specificity, trafficking, and posttranslational modification of several GPCRs (2–4). For example, RAMPs facilitates the transport of calcitonin receptor-like receptor (CALCRL) to the plasma membrane (3), and the ligand specificity of the calcitonin receptor is altered in the presence of RAMPs (5, 6). Previous bioinformatics studies addressed RAMP structural conformations and the identification of functional residues (7–9). Most reports of GPCR−RAMP interactions have focused on family B (secretin family) GPCRs, which make up only about 21 of more than 800 GPCRs in the human genome. However, whether GPCR−RAMP interactions are a common and global feature in the human GPCR gene family is an open question and one with direct therapeutic implications.
One approach to elucidate protein−protein interactomes is to carry out a phylogenetic analysis of protein sequences (10). This approach has previously shown excellent correspondence with protein interaction maps derived from a yeast two-hybrid system (11) and affinity purification (12). In a global phylogenetic analysis, the evolution history of pairs (or more) of proteins is compared according to the working hypothesis that interacting proteins must coevolve in nature. The analysis is based on the supported assumption (10) that proteins with shared function that provides a fitness advantage to an organism would be passed along together to its offspring. These shared-function proteins would also have similar phylogenetic pattern changes, since mutations in one protein would tend to be coupled with mutations in the other. It is important to note that protein coevolution analysis could detect direct as well as indirect interactions and that there is no trivial, inherent way in the analysis to distinguish between the two. Therefore, highly coevolved protein pairs could be members in the same pathway, or they could be components of the same complex.
Previously, several cases of coevolution of receptor genes and their endogenous protein−ligand genes were reported (13), but large-scale coevolution of different signal transduction components was not examined. Phylogenetic analysis to conclude coevolution could be implemented by two complementary approaches. The first searches for ortholog genes across species with the assumption that interacting proteins would tend to coexist in genomes (10). The second compares evolution history of each pair member while assuming similar phylogenetic trees (i.e., similar mutational rate) for interacting proteins (14) (Fig. 1A). Coexpression networks can also be used to infer biological knowledge. Interacting proteins have higher correlation of expression across tissues than random gene pairs in human, mouse, yeast, and Escherichia coli (15). Thus, the coevolution and coexpression analyses presented here provide complementary information about RAMP−GPCR interactions.
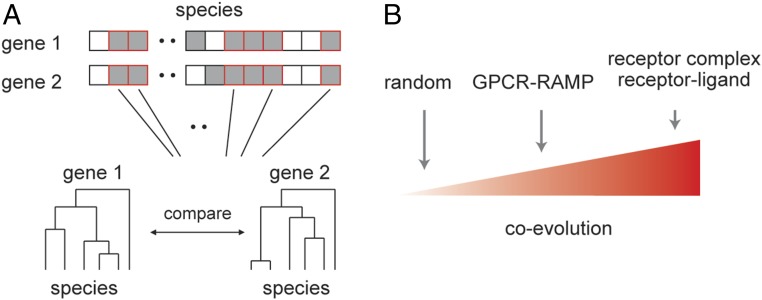
Fig. 1.
Estimates of coevolution. (A) Estimates of coevolution can rely on the number of species (gray squares) two proteins share. On top of that, across species that have both proteins (red-framed squares), one can search for a finer coevolution where the phylogenetic trees of gene pairs are compared either by the phylogeny comparison algorithm or by pair-wise correlation. (B) Our hypothesis was that GPCR−RAMP would show an intermediate degree of coevolution compared with random gene pairs and directly interacting gene pairs.
We hypothesized that, if human RAMPs interact globally with GPCRs, we would be able to identify a signal of coevolution for GPCR−RAMP pairs, above random pairs of genes. If, on the other hand, RAMP regulation over GPCR activity is restricted to a small number of genes in humans, then averaged coevolution would be similar to that expected by chance. Furthermore, if RAMPs have relatively subtle effects on GPCR activity (16), then we predict that the coevolution strength would be somewhere between the random (low) and that of previously known interacting gene pairs (high) that participate in signal transduction. Such genes are, for example, members of the same receptor complex and genes that have a receptor–endogenous ligand relationship (Fig. 1B). As was predicted, we identified a global GPCR−RAMP coevolution signal that is intermediate in strength between the signal expected by chance and the signal measured across receptor complex members or across receptor−ligand pairs. We further strengthened this conclusion by showing higher coexpression between GPCRs and RAMPs across human tissues. Our genomic- and transcriptomic-based results support the hypothesis that RAMPs globally interact with GPCRs.
Results
GPCRs and RAMPs Have Orthologs in the Same Species.
To compile a comprehensive database of orthologous genes for all human GPCRs and RAMPs across all currently sequenced genomes, we downloaded all Orthologous Matrix (OMA) groups and gene sequences from the OMA database (17) (omabrowser.org/oma/home/, downloaded in March 2017). OMA groups, as defined by the OMA database, are sets of genes from various organisms, which are all bona fide orthologous to each other. This high confidence grouping strategy is required for reconstructing phylogenetic species trees. Olfactory GPCRs were excluded from our analysis (but presented in Supporting Information as noted below), as they compose a group so large that it would dominate the outcome of a global phylogenetic analysis of GPCRs. Out of 1,970 organisms, 44 had an OMA group member gene to at least one human GPCR and one human RAMP (Table S1 for a list of organisms). These organisms all belonged to the Eukaryota domain, Metazoa kingdom, and Chordata phylum, and the frequencies in which different classes appeared in our analysis were 79%, 9%, and 7% for Mammalia, Actinopteri, and Aves, respectively (for frequencies according to order and family, see Fig. S1). Lists with organisms in OMA groups orthologous for the three RAMPs are provided in Table S2.
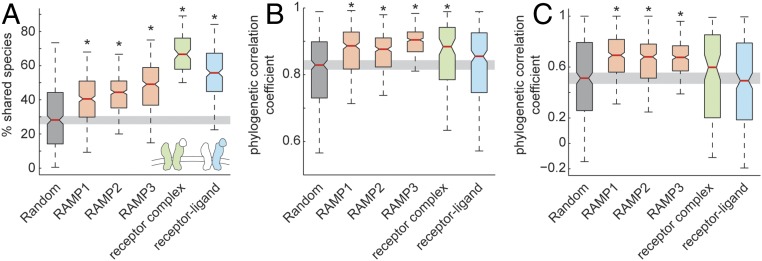
We calculated the percent of shared species between OMA groups of gene pairs as the fraction of the shared species from the joint list of species for both genes in the species pair (see equation in Materials and Methods). These percentages were calculated for 1,000 randomly chosen protein-coding gene pairs, GPCRs (330; all protein coding GPCR genes, excluding olfactory GPCRs, Dataset S1) and either RAMP1, RAMP2, or RAMP3, receptor and endogenous peptide or protein−ligand genes (370 pairs; see Table S3 for full list), and genes composing subunits in the same receptor (127 pairs; see Table S4 for full list). The lists with receptor−ligand pairs and receptor subunits were compiled from data downloaded from the Database of Interacting Proteins, University of California, Los Angeles (dip.doe-mbi.ucla.edu/dip/Main.cgi). The pairs of all three RAMPs with GPCRs showed significantly 1.6- to 1.8-fold higher percentages of shared species than expected by chance. Receptor complex and receptor−ligand pairs showed significantly 2.4- and 2.1-fold higher percentages of shared species than expected by chance, respectively (Fig. 2A). Percent of shared species between olfactory GPCRs and each of the three RAMPs was not different from that expected by chance (Fig. S2A). Calculated percent shared species, as well as all other measurements reported in the study, are shown in Dataset S1.
Fig. 2.
Shared species and similar phylogenetic trees between GPCRs and RAMPs. (A) Boxplot showing median (red line), 95% confidence interval (notch edge), 25th and 75th percentiles (box edges), and 2 SDs (whiskers) for the fraction of shared species from the total number of species in percentages, for protein pairs (Materials and Methods). This boxplot is shown for 500 randomly chosen gene pairs, RAMP1, RAMP2, or RAMP3 against GPCRs, protein pairs that are members of the same receptor complex, and receptor−ligand protein pairs. (B) Boxplot (as in A) for phylogenetic correlation coefficient across all organisms, normalized by 18S rRNA sequence distance, for the same groups shown in A. (C) Boxplot (as in A) for phylogenetic correlation coefficient across mammalians alone, not normalized by 18S rRNA sequence distance. Asterisks denote that the median differs from that of randomly chosen protein pairs, with *P < 0.05, with Bonferroni correction.
GPCRs and RAMPs Have Correlated Phylogenetic Trees.
We next estimated gene pair coevolution by comparing the phylogenetic trees of each gene in the pair across organisms that have orthologs for both examined genes. The average number of examined organisms across which we have built and compared the phylogenetic trees was 16.5 with SD 7.2. The phylogenetic trees were built only for GPCR−RAMP pairs that had more than five shared species (96% of pairs). First, we aligned the multiple orthologous amino acid sequences in a progressive and local manner using the BLOSUM50 scoring matrix (18) and then we calculated a matrix of pairwise sequence distances using Jukes−Cantor algorithm (maximum likelihood estimate of the number of substitutions between two sequences). At this point, two methods for comparing phylogenetic trees were used. The first tree comparison method compares the manner in which each tree partitions its nodes (species) and is an implementation of an established phylogeny comparison algorithm (19) (Materials and Methods). This method showed a statistically significant similarity of GPCR−RAMP trees compared with random (Fig. S2B). The second method of tree comparison was a calculation of Pearson correlation coefficient and P value between the distance matrices. This comparison was made in two complementary ways: first, across all organisms in which orthologous genes were identified while normalizing by the “tree of life” with the 18S rRNA gene (Fig. 2B) and, second, across only mammalians without normalizing to the tree of life (Fig. 2C). See Materials and Methods and Fig. S3 for the effect of normalizing by the tree of life. Across all organisms with orthologous genes, the pairs of all three RAMPs with GPCRs, and those of receptor complexes, showed significantly higher correlation coefficients than that expected by chance. Pairs of receptor−ligand were not different from those expected by chance. Across mammalians alone, and without normalization by 18S rRNA distance, the pairs of the three RAMPs with GPCRs, but not those of receptor complexes and receptor−ligand pairs, showed significantly higher correlation coefficient than that expected by chance. Coevolution estimates (“percent shared species” and “phylogenetic tree correlation”) showed a similar pattern across GPCR−RAMP pairs (Fig. S4). As shown in Fig. 3, we further annotated GPCR class according to “G protein-coupled receptors,” set 139, from HUGO Gene Nomenclature Committee (HGNC) (https://www.genenames.org/).
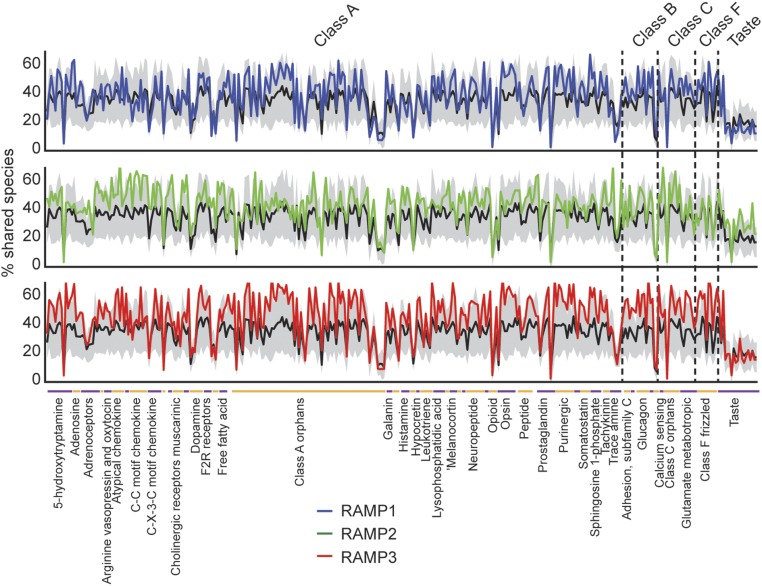
Fig. 3.
Percentage of shared species across GPCR superfamily and RAMP1, RAMP2, and RAMP3. Mean (black line) surrounded by SEM (gray area) percentages of shared species for each GPCR with 1,000 randomly chosen genes. Also shown are the percentages for each of the RAMPs against all GPCRs (blue, green, and red). GPCR families are shown separately, and subfamilies of four members or more are shown as labels on the x axis. Genes are organized along the x axis as in Dataset S1.
RAMP1 and RAMP3 Coevolved with Similar GPCRs but Differently from RAMP2.
Having observed global coevolution between GPCR and RAMP genes, we next asked whether different RAMPs coevolved with different sets of GPCRs. RAMP1 and RAMP3 show structural resemblance at the extracellular region (Fig. 4 A and B), which is required for transport of RAMP1–CALCRL to the plasma membrane (3). For this reason, we hypothesized that RAMP1 and RAMP3 would act in a redundant manner and that each RAMP would interact with the same GPCRs. To test this hypothesis, we first normalized by the mean percent shared species of each GPCR with 1,000 randomly chosen genes, because each GPCR would share species with other genes to a different extent, according to its evolutionary age and conservational pressure. As predicted, we found that all three RAMPs showed higher percentages of shared species across the GPCR superfamily compared with random pairs (Fig. 3 and Dataset S1).
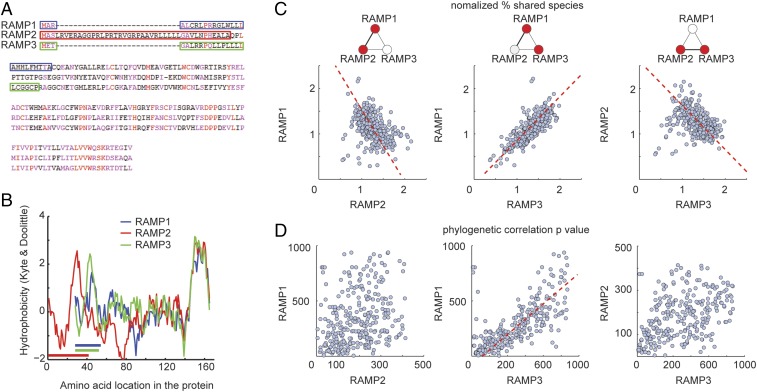
Fig. 4.
Similar estimated GPCR interactions between RAMP1 and RAMP3. (A) Amino acid multiple alignment using progressive method for the three RAMPs shows that RAMP2 N terminus is distinguished from those of RAMP1 and RAMP3. Highly conserved positions appear in red, and conserved positions appear in magenta. Colored rectangles indicate the position of the signal peptide according to UniProt. (B) Hydrophobicity plots (Kyte−Doolittle method), aligned by the C terminus, for the three RAMPs show similar hydrophobicity profiles for RAMP1 and RAMP3, distinguished from that of RAMP2. Colored horizontal lines indicate the position of the signal peptide according to UniProt. (C) Scatter plots for normalized shared species between RAMP1 and all GPCRs versus RAMP2 and all GPCRs and similarly for RAMP1 versus RAMP3 or RAMP2 versus RAMP3. Each dot corresponds to one GPCR−RAMP pair. Pearson’s r = –0.73, P < 0.001 for RAMP1 versus RAMP2; r = 0.88, P < 0.001 for RAMP1 versus RAMP3; and r = −0.78, P < 0.001 for RAMP2 versus RAMP3. (D) Scatter plots for phylogenetic correlation P value between RAMP1 and all GPCRs versus RAMP2 and all GPCRs and similarly for RAMP1 versus RAMP3 or RAMP2 versus RAMP3. Pearson’s r = 0.02, P > 0.05 for RAMP1 versus RAMP2; r = 0.83, P < 0.01 for RAMP1 versus RAMP3; and r = 0.11, P > 0.05 for RAMP2 versus RAMP3. Red dashed line corresponds to linear regression fit for cases of significant Pearson correlation.
We next compared the pattern of normalized shared species between each of the RAMPs and all GPCRs. RAMP1 and RAMP2 showed a significant reverse pattern such that GPCRs that have high percentages of shared species with RAMP1 have low percentages of shared species with RAMP2, and vice versa. A similar effect was observed for RAMP2 and RAMP3. RAMP1 and RAMP3, on the other hand, showed a significantly similar pattern, so that GPCRs that have high percentages of shared species with RAMP1 have high percentages of shared species with RAMP3 as well (Fig. 4C). Comparison of phylogenetic trees between RAMPs and GPCRs for the three different RAMPs showed a corresponding effect, where a significantly correlated resemblance in phylogenetic trees was identified only for RAMP1 and RAMP3 (Fig. 4D). This correlation means that GPCRs that had mutational rates similar to RAMP1 also had mutational rates similar to RAMP3. This effect corroborates our hypothesis and may imply redundancy between RAMP1 and RAMP3 at the functional level.
Coexpression of RAMPs and GPCRs Across Tissues.
We next searched for evidence supporting global interactions at the RNA expression level in vivo across human tissues. We hypothesized that, generally, transcripts of interacting proteins would tend to be coexpressed across different tissues. RAMP−GPCR interaction has not generally been considered to be a requirement for a functioning GPCR, and, based on previous data, a GPCR usually functions properly as a transmission unit by itself, or, in some cases, as a dimer or oligomer. However, the presence of one of the RAMPs may alter ligand specificity as well as the temporal and spatial pattern of ligand-induced signaling. Therefore, as in the phylogenetic analysis, we expected a less stringent coexpression of GPCRs and RAMPs compared with receptor−ligand and other control receptor complex pairs, albeit higher than expected by chance.
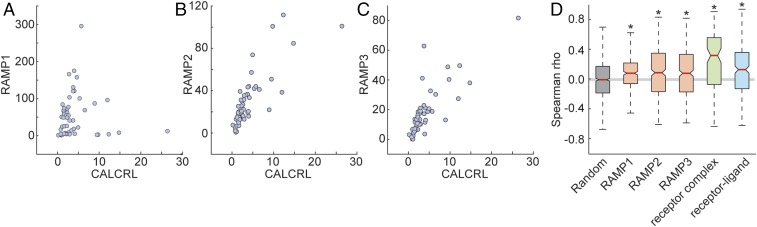
To examine this issue, we downloaded and analyzed all tissue expression data from the Genotype-Tissue Expression (GTEx) project database. The RNA-seq dataset downloaded from https://gtexportal.org/home (GTEx_Analysis_v6p_RNA-seq_RNA-SeQCv1.1.8_gene_rpkm.gct.gz) comprised 8,555 samples from 53 human tissues provided by 544 donors. To account for different statistical properties of gene expression among tissues, the data were quantile-normalized (20), and expression correlation was calculated for each of the examined groups (random pairs, RAMP1−GPCR, RAMP2−GPCR, RAMP3−GPCR, receptor−ligand, receptor complex) across the 53 tissues. Fig. 5 A–C, shows the tissue expression correlation between each of the three RAMPs and the example gene CALCRL. As mentioned in Introduction, interactions between CALCRL and each of the RAMPs has been well documented. As was predicted, random pairs showed averaged correlation coefficient of zero, and GPCR−RAMP pairs, receptor−ligand, and receptor complex gene pairs showed higher correlation coefficient than expected by chance (Fig. 5D and Fig. S5 for different methods of data normalization).
Fig. 5.
GPCR−RAMP coexpression across human tissues. (A–C) Scatter plot of normalized tissue expression data (detailed in Materials and Methods) for (A) RAMP1, (B) RAMP2, and (C) RAMP3 versus CALCRL. Spearman correlation coefficient is 0.21, 0.84, and 0.82 for A–C, respectively. (D) Boxplot (as in Fig. 2) of coexpression, measured as Spearman correlation coefficient, across 8,555 samples (including 53 human tissues) between protein pairs, using data from the GTEx database (www.gtexportal.org/home/). Asterisks denote that the median differs from that of randomly chosen protein pairs, with 95% confidence.
Discussion
Previous studies have shown that 11 GPCRs, primarily those in class B, interact with one or more RAMPs (5) (Table S5). Our aim here was to address the question of whether or not other additional GPCRs, including some of the ∼320 nonolfactory class A GPCRs, are also regulated by RAMPs. We took a global approach to evaluate the possibility of coevolution of GPCRs with RAMPs in the human genome. Coevolution of two genes implies that the proteins they encode interact in some way, either directly or indirectly. First, we created phylogenies based on all currently available sequenced organisms and used various methods to estimate coevolution of GPCRs and RAMPs. Our findings show that GPCRs and RAMPs substantially coevolved, suggesting that there is a functional interaction between the two families of membrane proteins.
All three methods that we employed for estimation of gene coevolution—percent shared species, tree comparison (19), and pair-wise tree correlation—showed a similar effect of identifiable coevolution among each of the RAMPs and all nonolfactory GPCRs. The percent shared species analysis provides the most general estimate of coevolution, because it shows the tendency of a gene pair to exist in the same species. The receptor−ligand and receptor complex pair controls showed higher values in the percent shared species measurements than GPCR−RAMP pairs. Across species that displayed both genes, the tree comparison and pair-wise tree correlations were carried out. Interestingly, the mutational rate (i.e., phylogenetic tree shape) was more correlated, or in-synch, for GPCR−RAMP pairs in all Eukaryota, as well as specifically in mammals. The correlation of mutational rate might imply some phenotypic implications for the role of GPCR−RAMP interactions, particularly in mammals.
Our coevolution data also suggest that RAMP1 and RAMP3 coevolved with a similar set of GPCRs. However, RAMP2 appears to have coevolved with a distinct set of GPCRs that is different from the GPCRs that coevolved with RAMP1 and RAMP3. RAMP1 and RAMP3 show structural similarity at the extracellular region (3). Moreover, a previous study (8) shows that mammalian RAMP1 and RAMP3 proteins evolve less than RAMP2, and have fewer residues with functional divergence in comparison with RAMP2. These findings agree with our observation that RAMP1 and RAMP3 coevolve with similar GPCRs. Importantly, we show here that RAMP1 and RAMP3 share a higher similarity of amino acid sequence than either of them share with RAMP2. Nevertheless, all RAMPs share a similar principal structure when compared on the level of extracellular domains linked to a single transmembrane segment (7).
A recent study analyzed the evolutionary dynamics of GPCR and G-protein sequences by measuring the overlap in protein repertoires in different organisms (21). This study is similar to ours in that it utilizes genomic data to dissect protein interaction. Particularly, the cases of GPCR−G protein interaction, as well as GPCR−RAMP potential interaction, include a large group (i.e., GPCRs) and a small group (i.e., G proteins or RAMPs). An important distinction between the two cases is that the small group is composed of three genes in the RAMP case and 16 genes in the G-protein case. A group of three genes is too small for calculation of overlap in protein repertoires. In contrast, the percent shared species measurement and the phylogenetic tree comparison method used in our study are computed per gene pair and so are suitable for the GPCR−RAMP analysis.
In available human tissue databases, we found a corresponding correlation of message levels for GPCRs and RAMPs. We analyzed 8,555 human transcriptomes and identified higher gene coexpression across human tissues for GPCRs and RAMPs compared with that predicted by chance. Several GPCRs were previously shown to interact with RAMPs; these include the calcitonin and CALCRL, the secretin receptor (22), vasoactive intestinal peptide/pituitary adenylate cyclase-activating peptide receptor, parathyroid hormone receptors, and glucagon receptor (2, 23). All of these showed a higher than chance coevolution with at least one of the three RAMPs, as shown in Dataset S1. In addition, in agreement with our coexpression analysis with RNA-seq data, a coexpression analysis using data from microarray analysis shows that RAMP2 and CALCRL have high expression correlation in human tissues (9).
In vivo, protein−protein interactions can be probed with either sequence evolution analyses or direct biochemical assays. In large protein networks or families, such as GPCRs, it is impractical to study all possible biochemical interactions directly. Both genomic and experimental approaches are subject to limitations, including false detection (“false positives”) and misdetection (“false negatives”). For example, in a biochemical protein affinity assay, a pair of proteins might show a positive binding interaction even though no specific binding actually occurs in the cell. In a coexpression gene network, as an example of a gene expression-based analysis, a gene pair might show an interaction due to shared connections with other genes. Between the two approaches, sequence evolution analysis has a higher capability to detect global effects in an unbiased manner, since it estimates interactions in identical conditions for each gene pair. Computational approaches also have the advantage that they are independent of the quality and validation of antibody affinity or other technical parameters that are needed to carry out direct experimentation.
In our computational analysis, we have estimated global GPCR−RAMP interaction parameters based on both coevolution and RNA expression. On the other hand, as mentioned above, coevolution analysis cannot distinguish between cases of direct protein−protein interactions and cases of interrelated function due to membership in the same pathway. Further studies are required to map the specific interaction type of each interacting GPCR−RAMP pair. Our phylogenetic and coexpression measures across gene pairs (in Dataset S1) could serve as a list of candidates for further studies.
CALCRL is evolutionarily more ancient than the RAMP family of proteins. While CALCRL and RAMP can function as a heterocomplex in more modern species, CALCRL in more evolutionarily ancient animals may function without RAMPs. This situation could cause the percentage of shared species to be modest even in cases of interacting protein pairs. Coevolution and coexpression of protein pairs may be a better indicator of interaction in this particular scenario. Indeed, when examining coevolution coefficient, coevolution P value, and coexpression coefficient, the resulting rank of CALCRL with each RAMP is within the upper third, and, with RAMP2 and RAMP3, it is within the upper tenth.
There are about 80 orphan receptors in the GPCR gene family (according to HGNC, https://www.genenames.org/) to which no known endogenous ligand has yet been functionally linked. Our observation that some of the RAMPs may interact with some those orphan receptors points to the possibility that RAMP−receptor interactions should be considered in deorphanization strategies. GPCR deorphanization trials could potentially be enhanced with signaling assays that include both a given RAMP and a putative ligand. It is possible that, under these conditions (i.e. existence of a RAMP and a ligand), novel receptor−ligand interactions would be found.
In summary, our results support the hypothesis that a global GPCR−RAMP interaction map exists. Given that GPCR signaling is a fundamental and essential process across organisms, tissues, and cell types, the need for a detailed GPCR−RAMP interaction map cannot be overestimated. In principle, a full map would detail which GPCR interacts with which RAMP and would provide information about the corresponding molecular impact, including changes in ligand specificity, protein translocation, and trafficking or other factors.
Materials and Methods
Percent of Shared Species.
To calculate percent shared species among gene pairs, we first constructed lists of species for each gene, in which one can find orthologs. Percent of shared species for a pair of genes a and b [PSS(a,b)] was calculated by the intersection of species lists for gene a (Sa) and gene b (Sb), divided by their union, multiplied by 100,
Intersection and union are operations from set theory.
Phylogeny Comparison.
In this study, we compared phylogenetic trees using two methods. First, we compared trees by calculating the correlation between two genes based on pair-wise distances among species. The second method we used is an established phylogeny comparison algorithm (19). This algorithm is based on pruning tree branches and comparing the resulted species partitions. The full algorithm can be found in ref. 19. Briefly, given a pair of phylogenetic trees, the algorithm arbitrarily chooses one tree, goes through each of the edges (i.e., branches), and prunes the tree in this spot. This pruning produces two subtrees, which determine a specific partition of the leaf nodes (this is the first partition for the first tree). Similarly, all of the possible partitions produced by edge pruning of the second tree are generated and compared with the first partition of the first tree. A score representing the similarity of each partition pairs is calculated. The same procedure is performed for all of the other edges of the first tree. Next, the optimal match between branch pruning-derived partitions of the two trees is attained with the Munkres algorithm (also known as the Hungarian algorithm). Lastly, a global similarity score based on the optimal branches match is calculated.
Normalizing to the Tree of Life.
When one compares phylogenetic patterns of two genes across species of varying classifications, the major determinant of the comparison would be the global phylogenetic distance between the species (Fig. S3 A–C). To reduce the global phylogenetic distance effect and directly compare the phylogenetic patterns of specific gene pairs, one has to normalize the trees by the general tree of life (10). In our study, we compared phylogenetic patterns between genes using matrices of amino acid sequence distance and normalized by the 18S rRNA matrix. Normalizing distance matrices by the 18S rRNA matrix (Fig. S3 E and F) introduces higher correlation among the compared genes than do the nonnormalized matrices. However, the normalization procedure indeed reduced the global phylogenetic distance by mixing the interspecies and intraspecies clusters (Fig. S3G). Hence, normalization to the tree of life corrects for the interspecies basal distances and enables the specific protein pair to govern the correlation strength.
Coexpression Analysis.
RNA-seq data from 53 tissues provided by 544 donors, with a total of 8,555 samples, were downloaded from https://gtexportal.org/home (GTEx_Analysis_v6p_RNA-seq_RNA-SeQCv1.1.8_gene_rpkm.gct.gz). Downloaded data were provided as reads per kilobase per million mapped reads (RPKM). Only samples passing quality control were included in the dataset. Read counts and RPKM values were produced with RNA-SeqC; importantly, reads were mapped to a single gene (see https://gtexportal.org/home documentation for more information). For transcripts per kilobase million (TPM) normalization, the RPKM of each gene was divided by the total RPKM for the sample and multiplied by 106. For rank normalization, each gene in each sample was ranked in descending order (highest RPKM having the highest numeric rank). For quantile normalization, gene expression across an individual sample was fit to the averaged distribution observed across samples. Before implementing the Spearman correlation analysis, the median normalized expression (RPKM, TPM, rank, or quantile) per tissue was calculated to account for differences in the number of samples per tissue type. Spearman correlation coefficient was calculated for each RAMP/nonolfactory GPCR pair (Dataset S1) as well as ligand−receptor and receptor subunit pairs (Tables S3 and S4).
Code Availability.
Code is available, upon request, from sbarbash@rockefeller.edu.
Supplementary Material
Acknowledgments
This work was supported by a Rothschild postdoctoral fellowship (to S.B.), Bengt Winblad’s Foundation for Young Scientists, the Margaretha af Ugglas Foundation, the Gun och Bertil Stohnes Foundation (T.P.), the Nicholson Short-Term Exchange fellowship (to E.L.), the Robertson Therapeutic Development Fund, the Crowley Family Fund, and the Danica Foundation (T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.C.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713074114/-/DCSupplemental.
References
- 1.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 2.McLatchie LM, et al. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 3.Parameswaran N, Spielman WS. RAMPs: The past, present and future. Trends Biochem Sci. 2006;31:631–638. doi: 10.1016/j.tibs.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Weston C, et al. Modulation of glucagon receptor pharmacology by receptor activity-modifying protein-2 (RAMP2) J Biol Chem. 2015;290:23009–23022. doi: 10.1074/jbc.M114.624601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay DL, Pioszak AA. Receptor activity-modifying proteins (RAMPs): New insights and roles. Annu Rev Pharmacol Toxicol. 2016;56:469–487. doi: 10.1146/annurev-pharmtox-010715-103120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hay DL, Poyner DR, Sexton PM. GPCR modulation by RAMPs. Pharmacol Ther. 2006;109:173–197. doi: 10.1016/j.pharmthera.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 7.Benítez-Páez A. Sequence analysis of the receptor activity-modifying proteins family, new putative peptides and structural conformation inference. In Silico Biol. 2006;6:467–483. [PubMed] [Google Scholar]
- 8.Benítez-Páez A, Cárdenas-Brito S. Dissection of functional residues in receptor activity-modifying proteins through phylogenetic and statistical analyses. Evol Bioinform Online. 2008;4:153–169. doi: 10.4137/ebo.s705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foord SM, Topp SD, Abramo M, Holbrook JD. New methods for researching accessory proteins. J Mol Neurosci. 2005;26:265–276. doi: 10.1385/JMN:26:2-3:265. [DOI] [PubMed] [Google Scholar]
- 10.Pazos F, Ranea JA, Juan D, Sternberg MJ. Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J Mol Biol. 2005;352:1002–1015. doi: 10.1016/j.jmb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 12.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 13.Moyle WR, et al. Co-evolution of ligand-receptor pairs. Nature. 1994;368:251–255. doi: 10.1038/368251a0. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhardwaj N, Lu H. Correlation between gene expression profiles and protein-protein interactions within and across genomes. Bioinformatics. 2005;21:2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- 16.Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol. 2009;10:819–830. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altenhoff AM, et al. The OMA orthology database in 2015: Function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–D249. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nye TM, Liò P, Gilks WR. A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics. 2006;22:117–119. doi: 10.1093/bioinformatics/bti720. [DOI] [PubMed] [Google Scholar]
- 20.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 21.Flock T, et al. Selectivity determinants of GPCR-G-protein binding. Nature. 2017;545:317–322. doi: 10.1038/nature22070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harikumar KG, Simms J, Christopoulos G, Sexton PM, Miller LJ. Molecular basis of association of receptor activity-modifying protein 3 with the family B G protein-coupled secretin receptor. Biochemistry. 2009;48:11773–11785. doi: 10.1021/bi901326k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christopoulos A, et al. Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem. 2003;278:3293–3297. doi: 10.1074/jbc.C200629200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.