Significance
Sunlight is a powerful but fluctuating energy source used by photosynthetic organisms as a substrate for biochemistry. To optimize photosynthetic efficiency, the distribution of energy input between the two photosystems is regulated through state transitions. This mechanism is common to all organisms of the green lineage and depends on the concerted response of the cytochrome b6f complex and the Stt7 kinase to changing redox conditions along the electron transport chain. The identification of a cytochrome b6f stromal motif involved in state transitions gives insight into the mechanism of Stt7 kinase activation.
Keywords: cytochrome b6f complex, structure–function, random mutagenesis, Stt7 kinase, state transitions
Abstract
The cytochrome (cyt) b6f complex and Stt7 kinase regulate the antenna sizes of photosystems I and II through state transitions, which are mediated by a reversible phosphorylation of light harvesting complexes II, depending on the redox state of the plastoquinone pool. When the pool is reduced, the cyt b6f activates the Stt7 kinase through a mechanism that is still poorly understood. After random mutagenesis of the chloroplast petD gene, coding for subunit IV of the cyt b6f complex, and complementation of a ΔpetD host strain by chloroplast transformation, we screened for impaired state transitions in vivo by chlorophyll fluorescence imaging. We show that residues Asn122, Tyr124, and Arg125 in the stromal loop linking helices F and G of cyt b6f subunit IV are crucial for state transitions. In vitro reconstitution experiments with purified cyt b6f and recombinant Stt7 kinase domain show that cyt b6f enhances Stt7 autophosphorylation and that the Arg125 residue is directly involved in this process. The peripheral stromal structure of the cyt b6f complex had, until now, no reported function. Evidence is now provided of a direct interaction with Stt7 on the stromal side of the membrane.
Compared with other biological controls of light harvesting, such as the pH-dependent component of nonphotochemical quenching (qE), state transitions are energy-conserving: the excess photon energy is not dissipated as heat (1, 2), but redirected from one photosystem (PS) to the other (3). State transitions therefore greatly contribute to the optimization of the maximal quantum yield of photosynthesis (4–6), probably through the poising of cyclic (PSI-dependent) vs. linear (PSI+PSII-dependent) electron flows in limiting light conditions (7, 8). In this process, the relative absorption cross-sections of PSI and PSII are dependent on the redox state of intersystem electron carriers, plastoquinones (PQ) (9), and cyt b6f complex (10), and on the reversible phosphorylation of light harvesting complexes II (LHCII) (11, 12). Depending on their phosphorylation state, these mobile protein–pigment complexes can migrate between the stacked (grana) and nonstacked (stroma lamellae) regions of the thylakoid membrane, where PSII and PSI localize, respectively (13). LHCII migration and excitonic connectivity to either PSI or PSII therefore balance excitation between photosystems. In Chlamydomonas reinhardtii, when the PQ pool is reduced, the serine-threonine kinase Stt7 (14) is activated and phosphorylates LHCII, causing their migration toward nonappressed thylakoid membranes and attachment to PSI (state II). On the contrary, when the PQ pool is oxidized, LHCII are dephosphorylated by a phosphatase [protein phosphatase 1 (PPH1)/thylakoid-associated phosphatase 38 (TAP 38) in Arabidopsis thaliana (15, 16)] and dock to PSII (state I).
Our knowledge of the mechanism linking Stt7 kinase activation to the redox state of the PQ pool is presently incomplete, apart from certain key elements: cyt b6f with a functional Qo site and PQH2 binding are required (10, 17, 18), the PetO subunit of cyt b6f is phosphorylated by Stt7 during PQ pool reduction (19), and the lumenal domain of Stt7 interacts directly with the Rieske-ISP subunit of cyt b6f and contains two conserved cysteine residues (20). However, the Stt7 kinase domain (20) and Stt7-dependent LHCII phosphorylation sites (21) are located on the stromal side of the membrane. Despite the recent characterization of the interaction between these two proteins (20, 22–27), the mechanism linking PQH2 binding at the lumenal Qo site to the activation of the stromal kinase domain of Stt7 remains unknown.
In this work, we used the technique of error-prone PCR for the mutagenesis of the chloroplast-encoded petD gene coding for subunit IV of cyt b6f. We probed the regions of subunit IV putatively interacting with Stt7 (22, 24) by screening in vivo for impaired state transitions and found mutants blocked in state I. Furthermore, we show that cyt b6f and Stt7 interact directly through their stromal domains and that Arg125suIV is involved in Stt7 activation.
Results
Random Mutagenesis, Chloroplast Transformation, and Sequencing of petD Variants.
The petD sequence coding for the cyt b6f subunit IV region going from the PEWY motif (28) to the C-terminal, and comprising helices F and G, was targeted for random mutagenesis (RM), whereas helix E, buried in the cyt b6f core, remained untouched (see Fig. 5 for a structural view). This petD fragment was randomly mutagenized by error-prone PCR and its variants reconstructed into plasmids (SI Appendix, Text S1). After amplification in Escherichia coli, the plasmid library was used to transform the chloroplast genome of a ΔpetD host strain. RM mutants described in this study were generated through two independent mutagenesis experiments (SI Appendix, Table S1). The diversity introduced in the petD gene was assessed by sequencing the target sequence in both E. coli and C. reinhardtii. Sequencing chromatograms did not show any SNPs or ambiguity in the petD sequences retrieved from C. reinhardtii; that is, when a mutated nucleotide appeared in the chromatogram, the signals corresponding to the other nucleotides stayed within the baseline noise (typically <1% for the sequences reported in this work; see SI Appendix, Fig. S1). Therefore, no petD sequence heterogeneity was observed, suggesting that in a given transformant, only one variant of petD was retained and replicated. In both rounds, no obvious bias was found in the positions of mutations along the sequence; mutations were diverse and covered the entire region of petD targeted for mutagenesis.
Fig. 5.
Simplified drawing of the cyt b6f monomer from C. reinhardtii and possible interactions with Stt7. The second cyt b6f monomer would be located on the right-hand side of the figure. Atomic coordinates of cofactors, stromal loops and amino acid side chains shown here are from PDB 1Q90 (33). Helices A, B, and C of cyt b6 and the transmembrane helix of the Rieske-ISP were removed for the sake of simplicity. The subunit IV (magenta) region targeted for random mutagenesis extends from the PEWY motif of the Qo site to the C-terminal through helices F and G exposed at the periphery of the complex. The Stt7 kinase interacts with the Rieske-ISP (20), F and G helices of subunit IV (22, 27) and the stromal loop of subunit IV (this work). (Inset) Stromal regions of subunit IV and cyt b6 (here in their ribbon representations), showing the structural link between Arg125suIV and the Qi site through the C-terminal of cyt b6 and the Arg207cyt b6 residue that interacts with heme ci.
The conditions chosen for error-prone PCR (SI Appendix, Text S1) introduced a higher mutational frequency in trial B than in trial A. This was confirmed by counting the mutations in the petD fragment recovered in E. coli and C. reinhardtii clones (see “Distribution” columns in SI Appendix, Table S1). Expectedly, the relative amount of nonmutated sequences was also smaller in trial B than in trial A for both E. coli and C. reinhardtii (see “% Non-mutated” columns in SI Appendix, Table S1). Interestingly, we observed that in each of the two different RM experiments, C. reinhardtii cells retained mutated copies of petD less often than E. coli cells, and when they did, they contained fewer mutations. This was a result of the difference in selection pressure exerted on the two organisms. Whereas E. coli transformants were selected on ampicillin-containing medium after integration of an intact AmpR cassette, C. reinhardtii transformants were selected on photoautotrophic growth after integration of a petD variant. Not all versions of randomly mutagenized petD genes are expected to restore a proper synthesis and folding of subunit IV and assembly of functional cyt b6f complexes. The more mutations are introduced in an essential photosynthetic gene, the more likely they are to be deleterious and lost through phototrophic selection.
Screening for C. reinhardtii Transformants Impaired for State Transitions.
For our screen, chlorophyll fluorescence images were recorded over the course of 10 min of anaerobic adaptation in the dark (SI Appendix, Fig. S2A), using an imaging system described in ref. 29. Anoxic reduction of the PQ pool was detected as an increase of F0′, and fluorescence quenching associated with the formation of state II (qT) was measured as a decrease of Fm′ in the WT. In the stt7-1 strain, blocked in state I, no fluorescence quenching was observed after 10 min. All the >2,000 C. reinhardtii clones obtained after transformation were screened for state transitions, and most of them showed a decrease in Fm′ similar to the WT. Sixteen clones, however, showed very stable Fm′ values after anaerobic adaptation, as in stt7-1, suggesting they were affected in state transitions. SI Appendix, Fig. S2A shows a typical state transition kinetics curve for a petD random mutant affected for state transitions compared with the WT and stt7-1 reference strains.
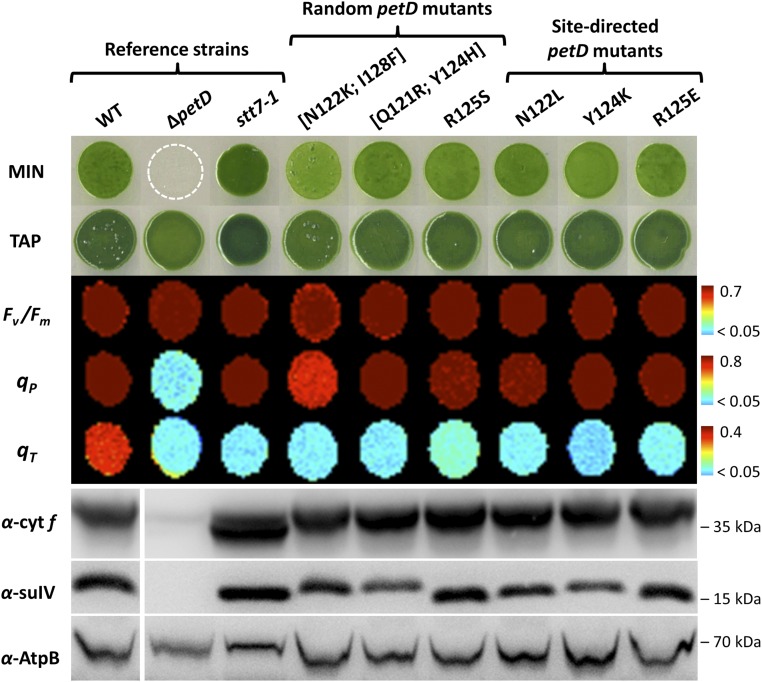
Fig. 1 illustrates some of the growth phenotypes and functional data obtained for three random petD mutants in comparison with the three reference strains [the site-directed (SD) petD mutants are discussed in SD Mutagenesis of the Stromal fg Loop of Subunit IV]. All strains show similar heterotrophic growth (TAP conditions) and maximal quantum efficiency of PSII photochemistry (Fv/Fm ≥ 0.7). The host strain ΔpetD does not grow in photoautotrophic conditions (minimum medium, MIN) and shows a very low photochemical quenching (qP < 0.05) when grown in heterotrophic conditions (TAP). In all mutants selected on minimal medium, photoautotrophic growth and qP were similar to WT (qP ≥ 0.8), suggesting a normal linear electron flow through PSII and downstream electron carriers in the photosynthetic chain, including the cyt b6f complex. However, the qT component of nonphotochemical quenching measured on anaerobic adaptation in the dark, high in the WT (qT ≥ 0.4), was greatly reduced in the two control strains lacking either cyt b6f or Stt7, as well as in the three random petD mutants shown here (qT ≤ 0.05).
Fig. 1.
Random and SD petD mutants are impaired for state transitions despite normal photosynthetic growth, electron transfer, and cyt b6f subunit accumulation. Growth on minimal (MIN, 100 μmolphotons m−2⋅s−1, 2% CO2) and acetate (TAP, 20 μmolphotons m−2⋅s−1) media, maximum quantum efficiency of PSII [Fv/Fm = (Fm − F0)/Fm], photochemical quenching of chlorophyll fluorescence [qP = (Fm − F)/Fm − F0)], qT component of nonphotochemical quenching measured upon anaerobic adaptation in the dark [qT = (Fm − Fm′)/Fm′, see SI Appendix, Fig. S2] and cyt b6f subunits accumulation of the three reference strains (WT, ΔpetD, stt7-1) and six petD mutants affected for state transitions. An antibody against AtpB was used as a loading control. SI Appendix, Table S2 mutants 19 ([Asn122Lys; Ile128Phe]), 13 ([Gln121Arg; Tyr124His]), and 28 (Arg125Ser) were obtained by random mutagenesis. Mutants 22 (Asn122Leu), 27 (Tyr124Lys) and 30 (Arg125Glu) were obtained by SD mutagenesis. All false color images and fluorescence parameters were calculated from chlorophyll fluorescence measurements of cells grown on MIN, except for the ΔpetD strain grown on TAP.
Among the 16 selected mutants that showed normal photochemistry but were blocked in state I, nine of them (italicized in SI Appendix, Table S2) were affected in the region coding for the loop linking helices F and G of cyt b6f subunit IV (hereafter designated as fg loop), exposed at the stromal side of the membrane. These RM mutants also often contained additional mutations on either side of this stromal loop that are reported in columns 2 and 4 of SI Appendix, Table S2. To tease out certain phenotypic ambiguities that emerged from the comparison of the RM mutants, we generated single-point mutations along the loop (from Asn118 to Arg125) by SD mutagenesis.
SD Mutagenesis of the Stromal fg Loop of Subunit IV.
Similar to RM, SD mutants were obtained by chloroplast transformation of the ΔpetD host strain with SD mutagenized petD, followed by photoautotrophic selection of clones. To avoid structural destabilization of the loop, Pro123, Pro127, and residues pointing toward the core of the protein were not touched, whereas residues putatively involved in protein–protein interactions were targeted. For each residue, for example, Arg125, two amino acids with similar steric properties were substituted, bearing either a hydrophobic side chain (e.g., mutant 29 in SI Appendix, Table S2, Arg125Leu) or a side chain of “opposite” chemical property (e.g., mutant 30, Arg125Glu). Similar to the RM mutants, SD mutants grew under photoautotrophic conditions, and some of them, especially those containing mutations at residues Asn122, Tyr124, and Arg125, showed chlorophyll fluorescence parameters identical to the stt7-1 strain, strongly affected in qT, but not in qP (Fig. 1 and SI Appendix, Figs. S2B and S4). Obtaining SD point mutants was important to discern the effect of individual mutations on the phenotype of certain RM mutants that contained more than one mutation. For example, for positions 121 and 124, although a marked decrease of qT was observed in mutant 13 ([Gln121Arg; Tyr124His]), it was not nearly as drastic in mutant 24 ([Tyr124His; Phe149Leu]), and not observed at all in mutant 14 ([Val104Ala; Gln121Arg; Phe149Ser]; SI Appendix, Table S2). The Gln121Arg SD mutant 17 showed that changing Gln121 did not affect qT, whereas SD mutant 27 (Tyr124Lys) revealed a critical role for Tyr124: substitution to Lys induced a loss of qT, whereas substitution to the aromatic residue Phe (mutant 26, Tyr124Phe) had no effect on qT, as observed in the RM mutant 25 ([Tyr124Phe; Ile128Thr; Thr137Ala; Leu159Ser]). Further characterization focused on SD mutants 22, 27, and 30 (Asn122Leu, Tyr124Lys, and Arg125Glu, respectively) that exhibited the most robust “locked-in-state-I” phenotype of all the SD subunit IV mutants.
Characterization of Modified cyt b6f Complexes.
In ΔpetD, totally devoid of subunit IV, the rate of synthesis of cyt f is strongly decreased, resulting in very little accumulation of cyt f (30), also observed in Fig. 1. In complemented strains, the 34.3-kDa cyt f subunit is tagged with 6-His at its C-terminal and shows a slower electrophoretic mobility on denaturing gels than in the stt7-1 strain, as previously reported for the f297C strain (31). Small differences are observed in the migration profiles of the 17.4-kDa subunit IV and can be attributed to slight changes in peptide mobility induced by single-point mutations. In the petD RM and SD mutants, the accumulation of subunit IV and cyt f was similar to WT levels (see SI Appendix, Fig. S5 for 50% and 10% loading controls).
The electron transfer rate through the cyt b6f complex was assessed in control and mutant strains (SI Appendix, Figs. S6 and S7). The transmembrane electrogenic phase of electron transfer between hemes bL and bH, occurring after quinol oxidation at the Qo site, was measured as an electrochromic shift of carotenoids, giving an absorbance increase at 520 nm. Electron transfer rates in mutants Asn122Leu, Tyr124Lys, and Arg125Glu were very similar to the WT.
Changes in Photosystems Antenna Size Associated with State Transitions.
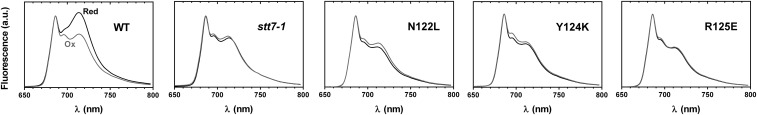
At 77 K, the light energy absorbed by chlorophyll in the mobile LHCII proteins is not reemitted at 682 nm, as it would be if these protein complexes were disconnected from the photosystems (32), but is transferred either to PSII and reemitted at 686 and 696 nm, or to PSI and reemitted at 713 nm. In the WT, the relative amplitudes of the fluorescence peaks change, with PSII contribution being larger in oxidizing conditions (state I) and PSI fluorescing mostly in reducing conditions (state II). It therefore reflects a change in excitonic connectivity of mobile LHCII to either PSI or PSII that depends on the Stt7 kinase. Similar to that observed in the stt7-1 strain, the Asn122Leu, Tyr124Lys, and Arg125Glu mutants were blocked in state I despite reducing conditions, with LHCII constitutively connected to PSII (Fig. 2).
Fig. 2.
Subunit IV fg loop mutants do not change photosystem antenna sizes between oxidizing and reducing conditions. Low-temperature (77 K) fluorescence emission spectra of the subunit IV mutants compared with the WT and stt7-1 strains. Cells were placed in oxidizing (Ox, gray curve: 10 µM DCMU, moderate light, 30 min) or reducing (Red, black curve: dark anoxia, 30 min) conditions. Spectra were normalized at 685 nm on PSII contribution.
Phosphorylation of Thylakoid Polypeptides.
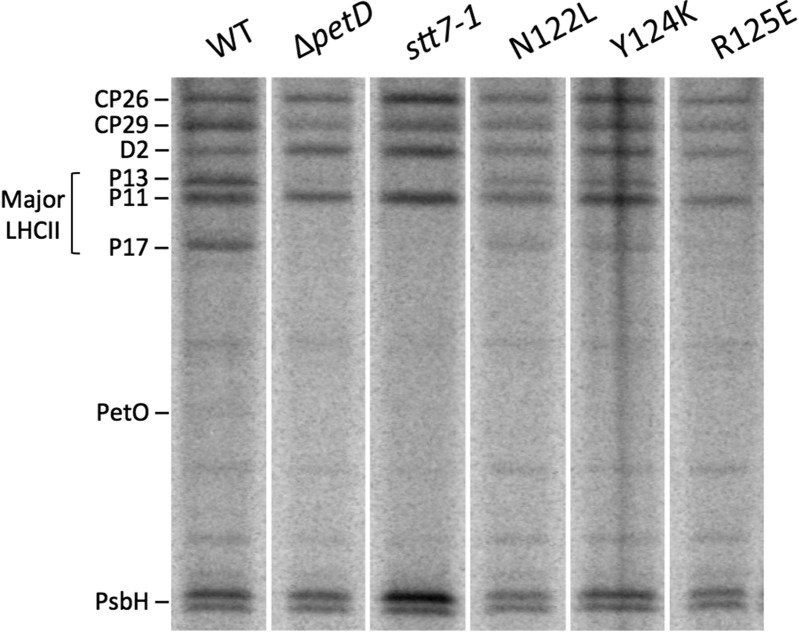
Incorporation of radiolabeled orthophosphate [33PO43−] into proteins during PQ pool reduction was measured in vivo. Intact cells of SD mutants and reference strains were preincubated for 30 min under dark anoxic conditions. During nonphotochemical reduction of the PQ pool, thylakoid polypeptides incorporate radiolabeled phosphate that is detected by autoradiography after membrane preparation and fractionation by PAGE on an 8 M urea 12–18% gradient gel (Fig. 3 and see SI Appendix, Fig. S8 for loading controls). Contrary to the polypeptides that are constitutively phosphorylated in the various strains (CP26, CP29, D2, P11, PsbH), P13 and P17 were phosphorylated in the WT, but not in ΔpetD or in stt7-1, as reported earlier (6, 10). Among the SD mutants, which accumulate WT levels of the Stt7 kinase (SI Appendix, Fig. S9), Asn122Leu and Tyr124Lys showed low phosphorylation of P13 and P17, and Arg125Glu showed none. Similar phosphorylation profiles were observed in another comparative state I/state II 33P-labeling experiment (SI Appendix, Fig. S10).
Fig. 3.
Subunit IV fg loop mutants show low levels of Stt7-dependent LHCII phosphorylation. Autoradiogram of in vivo 33P-labeled thylakoid polypeptides, extracted from cells placed in reducing conditions (5 µM FCCP, dark, 30 min) and separated by SDS/PAGE on an 8 M urea 12–18% gradient gel.
Cyt b6f–Stt7 Interaction Studies.
A GAL4-based yeast two-hybrid assay was carried out to probe the interaction between subunit IV and Stt7 (SI Appendix, Text S2 and Fig. S11). The WT and double-mutant ([Tyr124Lys; Arg125Glu]) subunit IV fg loops (residues 113–128) were expressed as fusions to the Gal4 DNA-binding domain and used as bait proteins. The stromal kinase domain of Stt7 was cut into four fragments that were expressed as fusions to the Gal4 activation domain and used as prey proteins: A, residues 123–238; B, 244–379; C, 397–483; and D, 484–600. Whereas no interaction was detected between the WT fg loop of subunit IV and Stt7 fragments C (397–483) and D (484–600), an interaction was detected between the WT fg loop and Stt7 fragments AB (123–379) and B alone (244–379). As expected, this interaction was lost in the double mutant ([Tyr124Lys; Arg125Glu]) fg loop. We further addressed the question of the biochemical interaction between cyt b6f and Stt7 in a context in which the folding of subunit IV and assembly into a functional cyt b6f complex is preserved.
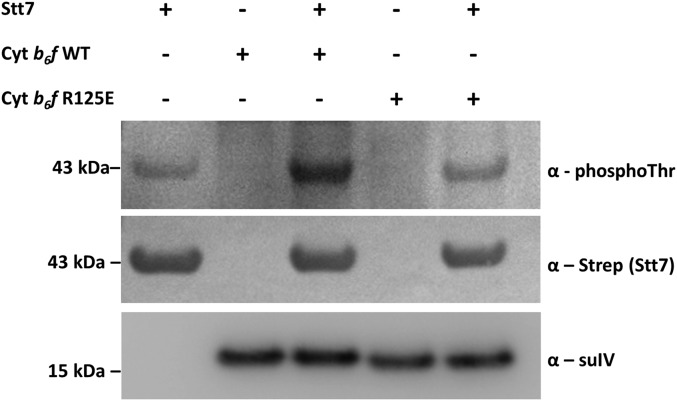
Intact and functional cyt b6f complexes were purified from C. reinhardtii thylakoid membranes following ref. 33. The recombinant kinase domain of Stt7 (Stt7-KD, residues 139–495) was produced in E. coli and purified as described in the SI Appendix, Text S3 and Fig. S12. Stt7-KD was active in vitro, as seen by its ATP hydrolysis activity (SI Appendix, Table S3). Stt7-KD was incubated for 30 min in the presence of ATP and MgCl2 with either the WT or Arg125Glu purified cyt b6f complexes. Whereas Stt7 showed a low level of autophosphorylation when incubated alone with ATP and MgCl2 (Fig. 4, lane 1), addition of WT cyt b6f greatly increased its phosphorylation (lane 3). In stark contrast, incubation of Stt7-KD with cyt b6f containing the Arg125Glu substitution (lane 5) showed only the basal signal of Stt7 autophosphorylation, similar to lane 1. This in vitro reconstitution shows that the cyt b6f complex and the stromal kinase domain of Stt7 interact directly and that Arg125 of subunit IV is involved in Stt7 autophosphorylation.
Fig. 4.
Purified cyt b6f complex increases autophosphorylation of the Stt7 kinase domain depending on the presence of Arg125suIV. Immunoblots of Stt7-KD autophosphorylation reactions in the presence of WT or Arg125Glu cyt b6f complex. An antibody against phospho-threonine (Cell Signaling) was used to detect Stt7-KD autophosphorylation. Antibodies against Strep-tag (recognizing the C-terminal tag of Stt7-KD) and subunit IV were used as loading and transfer controls.
Discussion
Design of an Optimized Protocol for the Random Mutagenesis of Chloroplast Genes and Screening for Modified Photosynthetic Functions.
We have targeted a ∼300-bp fragment of petD, corresponding to ∼100 amino acids in subunit IV. The number of single amino acid substitutions in a 100-amino acid protein is 19 × 100 = 1,900. In total, among our ∼2,000 transformants, we have obtained ∼1,200 random mutants bearing, on average, ≥2 mutations (rightmost column of SI Appendix, Table S1). When looking for a loss-of-function phenotype, single amino acid substitutions are sufficient, with no need of obtaining all possible combinations of ≥2 substitutions. Therefore, the number of genetic variants generated in this work was adequate for screening for a loss of function.
Previous studies reported the random mutagenesis of chloroplast DNA. Mutagenic agents such as 5-fluorodeoxyuridine can be used to mutagenize the entire chloroplast genome (34), and degenerate oligonucleotides can introduce a given number of mutations in the sequence spanned by the oligonucleotides (35, 36). Although both these methods contain obvious caveats, the technique described here allows for specific targeting of a chloroplast DNA sequence without any size limitation other than the maximal length of amplification by PCR. In our case, considering the rather low efficiency of chloroplast transformation, and because selection on phototrophy discards nonfunctional gene variants, we were able to increase the mutation rate of the error-prone PCR to enrich the transformation plates in putative mutant clones. We did so until we reached quite a high mutational frequency for a protein structure–function study (3.5 mutations/300 bp for RMB; see SI Appendix, Table S1). High mutation rates are used more often for the directed evolution of enzymes, where many substitutions can be introduced (37). As a broader perspective, this approach can be used for the study of many other chloroplast-encoded proteins and opens opportunities for the directed evolution of enzymes in transplastomic cell lines.
Identification of a cyt b6f Stromal Loop Involved in State Transitions.
After screening for a specific mutant phenotype, it was possible to gain functional insight from the comparison of sequences retrieved from the RM mutants, and we further confirmed the attribution of functional changes to point mutations by using SD mutagenesis. Given that the catalytic site of the Stt7 kinase is found in the stroma (20), we narrowed our target of investigation to the subunit IV fg loop starting from Asn118 and extending to Arg125. Protein sequence alignment showed a high degree of conservation between subunit IV from various organisms, but a strong divergence with cyt b from bacteria and mitochondria (SI Appendix, Fig. S13). We identified NKFQNPxRR as a conserved motif (where x corresponds to aromatic residues tyrosine or phenylalanine). This site, rich in lysine (K), arginine (R), and proline (P), to a lesser extent, strongly resembles a kinase binding site such as that of Stt7 (21), except that it contains no phosphorylatable residue (threonine or serine for Stt7).
Mutations at residues Asn122, Tyr124, and Arg125 produced mutants that are blocked in state I, despite a reduced PQ pool and normal cyt b6f assembly and electron transfer activity. Upon a switch from oxidizing to reducing conditions, these mutants showed no quenching of PSII fluorescence emission associated with state transitions, no modification of the relative antenna sizes of PSII and PSI, and very low levels of phosphorylation of LHCII antenna proteins. This function was not expected for the peripheral stromal structure of the cyt b6f complex.
Insight into the Role of cyt b6f Complex in Stt7 Kinase Activation.
The present work shows that the stromal region of cyt b6f is involved in the activation of Stt7. Yeast two-hybrid experiments indicated that the stromal fg loop of subunit IV interacts with the kinase domain of Stt7 in the region between Leu244Stt7 and Pro379Stt7. Substitutions of fg loop residues Tyr124suIV and Arg125suIV abolished this interaction, showing that they are involved in the binding of the kinase to the stromal side of the complex. Furthermore, in vitro reconstitution experiments demonstrated that the autophosphorylation activity of the kinase domain was enhanced in the presence of purified cyt b6f complex, through an interaction dependent on Arg125suIV. The Stt7 region between Leu244Stt7 and Pro379Stt7 identified by two-hybrid assays contains two important kinase regulatory motifs, HRD and DLG, as well as the activation loop and the α5-hairpin (38). Several acidic Asp and Glu residues are present in this region and could form interactions with Arg125suIV in vivo. The absence of the lumenal domain [with the two regulatory Cys residues (20)] and the transmembrane domain of Stt7 did not prevent cyt b6f-mediated autophosphorylation of the kinase. In our attempt to define the minimal functional length of Stt7, and going a step further than the truncation at residue 549 (27), we narrowed the kinase domain down to even before Ser533Stt7, a phosphorylation site identified by phosphoproteomics but not involved in state transitions (20, 21). In addition to the PThr and PSer detected in previous phosphoproteomics studies (21, 39), our results suggest there are other phosphorylatable residues in the catalytic domain of Stt7. These residues could be important for the activation of the kinase in vivo.
On the basis of previous studies showing the crucial role of the lumenal Qo site in state transitions (17, 28) and topology analysis showing that the kinase domain (20) and the LHCII phosphorylation sites (21) are found in the stroma, a signal transduction mechanism across the thylakoid membrane was postulated. The Chl a molecule, bound between helices F and G of subunit IV and protruding into the Qo site, was proposed to play a role in this signal transduction (22, 24, 25, 40). Redox changes of the lumenal cysteine residues of Stt7, caused by Qo site catalysis, were also suggested to induce Stt7 activation through transient dimerization (26) or conformational changes of its transmembrane domain (27). The present study identifies the stromal fg loop of subunit IV within the pattern of interactions between the cyt b6f and Stt7. We propose that the primary activation of Stt7 depends on its interaction with the stromal side of cyt b6f complex, whereas its release for LHCII phosphorylation could be controlled by plastoquinol occupancy and turnover at the Qo site, as suggested previously (41).
We found here that Arg125 from the subunit IV fg loop plays a key role in the activation of the state transition kinase Stt7. The electron density map of the cyt b6f complex from C. reinhardtii shows that the guanidinium group of Arg125 and the C-terminal carboxyl group of cyt b6 are at a distance suitable to form a salt-bridge or an H-bond (see Fig. 5 and ref. 33). It suggests that Arg125suIV has two possible interacting partners: either the terminal carboxyl of the cyt b6 or a negatively charged residue of Stt7 (Glu or Asp) in the region between Leu244Stt7 and Pro379Stt7. As shown in Fig. 5, Inset, the cyt b6 C-terminal end is a short strand away from the conserved Arg207 that interacts with one of the propionates of heme ci and shapes part of the Qi pocket. In our previous work on heme ci, we reported a discrepancy between its redox potentials estimated from in vivo and in vitro measurements and some peculiar electronic properties of this cofactor, depending on the pH and/or the binding of NQNO (42, 43). We attributed these unusual features to conformational heterogeneity at the stromal side of the cyt b6f complex. It remains to be explored whether the dual interaction of Arg125suIV with either Stt7 or cyt b6 could account for such structural heterogeneity.
Materials and Methods
Cell Growth Conditions.
For photoautotrophic growth, C. reinhardtii cells were grown on Tris-phosphate (MIN, pH 7.2) medium in the light (100 μmolphotons m−2⋅s−1) in air supplemented with 2% CO2. For heterotrophic growth, cells were grown on Tris acetate-phosphate (TAP, pH 7.2) medium in very low to moderate light (10–40 μmolphotons m−2⋅s−1).
Plasmids and Strains.
Plasmid pWQH6 (24), carrying the 3′-end sequence of petA with a 6-His tag, the intergenic petA-petD region, and the entire petD sequence, was used for mutagenesis and chloroplast transformation experiments. The mt+ acetate-requiring ΔpetD deletion strain (30) was used as the recipient strain for chloroplast transformation. The WT reference strain was obtained by chloroplast transformation of the ΔpetD strain with plasmid pWQH6 and selection on the restoration of photoautotrophic growth. The petD random and SD mutants were obtained the same way, apart from the use of pWQH6 plasmids carrying mutations on the petD sequence. The WT reference strain and petD mutant strains therefore all express a petA gene coding for a His-tagged cyt f. The stt7-1 strain (14), containing a deletion of the gene coding for the Stt7 kinase, was used as a control.
Mutagenesis of the petD Gene.
Plasmid pWQH6 and various primers were used for random and SD mutageneses of petD (SI Appendix, Table S4). For RM, error-prone PCR was performed using kits GeneMorph II EZClone (Agilent Technologies, “RMA” in SI Appendix, Table S1), and Diversify PCR (Clontech, “RMB”; see SI Appendix, Table S1 and see SI Appendix, Text S1 for details on the protocols).
Chloroplast Transformation and Mutant Selection.
Plasmid DNA was precipitated on nanometer-scale gold particles using the Seashell Technology S550d gold DNA protocol. ΔpetD cells on MIN medium were transformed by gold particle bombardment at 7 bars under vacuum. After transformation, cells were left to recover in the dark for up to 24 h and transformants were incubated in air + 2% CO2 under low light (40–50 μmolphotons m−2⋅s−1) for phototrophic selection.
Screening for State Transition Mutants.
Transformation plates were left aerated in the dark for up to 15 min to induce PQ pool oxidation. The plates were then transferred to anaerobic conditions (N2 flush) and the maximal fluorescence yield of PSII was recorded, using a fluorescence camera similar to the one described in ref. 29, during a 10-min period in the dark with 12 saturating light pulses.
77 K Chlorophyll Fluorescence Emission Spectra.
Chlorophyll was excited with a LED at ∼455 nm, and fluorescence was selected with a red-colored Kodak Wratten filter and collected with an Ocean Optics USB2000 CCD spectrometer. C. reinhardtii cells were placed in an aluminum-made sample holder and snap frozen at 77 K in liquid nitrogen. Sample, LED, and spectrometer were coupled via a Y-shaped fiberoptics.
In Vivo Phosphorylation of Antenna Proteins.
Cells grown to around 2 × 106 cells⋅mL−1 were harvested and resuspended in a phosphate-depleted minimal medium containing 2 μCi mL−1 33Pi. Samples were then treated as described in ref. 44, and thylakoid membranes were isolated as in ref. 45. Phosphorylation patterns of thylakoid membrane polypeptides were then analyzed on a 12–18% polyacrylamide gel containing 8 M urea, which was then dried and autoradiographed.
In Vitro Assays with Purified cyt b6f Complexes and Recombinant Stt7-KD.
Cyt b6f complexes were purified as in ref. 33, and recombinant Stt7-KD (residues 139–495) was purified following a protocol adapted from ref. 27, as described in the SI Appendix, Text S3 and Fig. S12. The ADP-Glo Kinase Assay (Promega) was used to measure the in vitro activity of Stt7-KD (see SI Appendix, legend of Table S3). To test the interaction between the two proteins, 4.5 µM purified Stt7-KD and 1.5 µM purified WT or Arg125Glu cyt b6f complex were incubated together with 1 mM ATP and 20 mM MgCl2 for 30 min. Samples were denatured in a buffer containing LDS and DTT at 70 °C for 15 min and separated on a 10% Bis-Tris gel by SDS/PAGE. After transfer on a nitrocellulose membrane, antibodies against phospho-threonine (Cell Signaling), Strep-tag (recognizing the C-terminal tag of Stt7-KD), and subunit IV were used to detect the proteins.
Supplementary Material
Acknowledgments
We are thankful for Michel Goldschmidt-Clermont’s kind gift of the Stt7 antibody. We thank Daniel Picot for his structural expertise and discussion of the model. Pascal Arnoux and Bernard Genty are acknowledged for stimulating inputs. L.D. acknowledges the IRTELIS PhD Grant from CEA, the F.Z. laboratory was supported by the “Initiative d’Excellence” program (Grant “DYNAMO,” ANR-11-LABEX-0011-01), X.J. acknowledges a grant from the Agence Nationale de la Recherche (ChloroPaths: ANR-14-CE05-0041-01), and J.A. was supported by CNRS and INSIS (Institut des Sciences de l’Ingénierie et des Systèmes) Energie CNRS Grant PhotoModes 72749.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.-D.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713343114/-/DCSupplemental.
References
- 1.Nagy G, et al. Chloroplast remodeling during state transitions in Chlamydomonas reinhardtii as revealed by noninvasive techniques in vivo. Proc Natl Acad Sci USA. 2014;111:5042–5047. doi: 10.1073/pnas.1322494111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ünlü C, Drop B, Croce R, van Amerongen H. State transitions in Chlamydomonas reinhardtii strongly modulate the functional size of photosystem II but not of photosystem I. Proc Natl Acad Sci USA. 2014;111:3460–3465. doi: 10.1073/pnas.1319164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nawrocki WJ, Santabarbara S, Mosebach L, Wollman FA, Rappaport F. State transitions redistribute rather than dissipate energy between the two photosystems in Chlamydomonas. Nat Plants. 2016;2:16031. doi: 10.1038/nplants.2016.31. [DOI] [PubMed] [Google Scholar]
- 4.Canaani O, Barber J, Malkin S. Evidence that phosphorylation and dephosphorylation regulate the distribution of excitation energy between the two photosystems of photosynthesis in vivo: Photoacoustic and fluorimetric study of an intact leaf. Proc Natl Acad Sci USA. 1984;81:1614–1618. doi: 10.1073/pnas.81.6.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canaani O, Malkin S. Distribution of light excitation in an intact leaf between the two photosystems of photosynthesis. Changes in absorption cross-sections following state 1-state 2 transitions. Biochim Biophys Acta. 1984;766:513–524. [Google Scholar]
- 6.Fleischmann MM, et al. Isolation and characterization of photoautotrophic mutants of Chlamydomonas reinhardtii deficient in state transition. J Biol Chem. 1999;274:30987–30994. doi: 10.1074/jbc.274.43.30987. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi H, Clowez S, Wollman FA, Vallon O, Rappaport F. Cyclic electron flow is redox-controlled but independent of state transition. Nat Commun. 2013;4:1954. doi: 10.1038/ncomms2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumas L, Chazaux M, Peltier G, Johnson X, Alric J. Cytochrome b6f function and localization, phosphorylation state of thylakoid membrane proteins and consequences on cyclic electron flow. Photosynth Res. 2016;129:307–320. doi: 10.1007/s11120-016-0298-y. [DOI] [PubMed] [Google Scholar]
- 9.Allen JF, Bennett J, Steinback KE, Arntzen CJ. Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature. 1981;291:25–29. [Google Scholar]
- 10.Wollman FA, Lemaire C. Studies on kinase-controlled state transitions in photosystem II and b6f mutants from Chlamydomonas reinhardtii which lack quinone-binding proteins. Biochim Biophys Acta. 1988;933:85–94. [Google Scholar]
- 11.Bennett J. Phosphorylation of chloroplast membrane polypeptides. Nature. 1977;269:344–346. [Google Scholar]
- 12.Horton P, Allen JF, Black MT, Bennett J. Regulation of phosphorylation of chloroplast membrane polypeptides by the redox state of plastoquinone. FEBS Lett. 1981;125:193–196. [Google Scholar]
- 13.Andersson B, Anderson JM. Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim Biophys Acta. 1980;593:427–440. doi: 10.1016/0005-2728(80)90078-x. [DOI] [PubMed] [Google Scholar]
- 14.Depège N, Bellafiore S, Rochaix JD. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- 15.Shapiguzov A, et al. The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:4782–4787. doi: 10.1073/pnas.0913810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 2010;8:e1000288. doi: 10.1371/journal.pbio.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vener AV, Van Kan PJM, Gal A, Andersson B, Ohad I. Activation/deactivation cycle of redox-controlled thylakoid protein phosphorylation. Role of plastoquinol bound to the reduced cytochrome bf complex. J Biol Chem. 1995;270:25225–25232. doi: 10.1074/jbc.270.42.25225. [DOI] [PubMed] [Google Scholar]
- 18.Zito F, et al. The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 1999;18:2961–2969. doi: 10.1093/emboj/18.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamel P, Olive J, Pierre Y, Wollman FA, de Vitry C. A new subunit of cytochrome b6f complex undergoes reversible phosphorylation upon state transition. J Biol Chem. 2000;275:17072–17079. doi: 10.1074/jbc.M001468200. [DOI] [PubMed] [Google Scholar]
- 20.Lemeille S, et al. Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 2009;7:e45. doi: 10.1371/journal.pbio.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemeille S, Turkina MV, Vener AV, Rochaix JD. Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol Cell Proteomics. 2010;9:1281–1295. doi: 10.1074/mcp.M000020-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zito F, Vinh J, Popot JL, Finazzi G. Chimeric fusions of subunit IV and PetL in the b6f complex of Chlamydomonas reinhardtii: Structural implications and consequences on state transitions. J Biol Chem. 2002;277:12446–12455. doi: 10.1074/jbc.M110914200. [DOI] [PubMed] [Google Scholar]
- 23.de Vitry C, Ouyang Y, Finazzi G, Wollman FA, Kallas T. The chloroplast Rieske iron-sulfur protein. At the crossroad of electron transport and signal transduction. J Biol Chem. 2004;279:44621–44627. doi: 10.1074/jbc.M406955200. [DOI] [PubMed] [Google Scholar]
- 24.de Lacroix de Lavalette A, Finazzi G, Zito F. b6f-associated chlorophyll: Structural and dynamic contribution to the different cytochrome functions. Biochemistry. 2008;47:5259–5265. doi: 10.1021/bi800179b. [DOI] [PubMed] [Google Scholar]
- 25.Hasan SS, Cramer WA. Internal lipid architecture of the hetero-oligomeric cytochrome b6f complex. Structure. 2014;22:1008–1015. doi: 10.1016/j.str.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapiguzov A, et al. Activation of the Stt7/STN7 kinase through dynamic interactions with the cytochrome b6f complex. Plant Physiol. 2016;171:82–92. doi: 10.1104/pp.15.01893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh SK, et al. Trans-membrane signaling in photosynthetic state transitions: Redox- and structure-dependent interaction in vitro between Stt7 kinase and the cytochrome b6f complex. J Biol Chem. 2016;291:21740–21750. doi: 10.1074/jbc.M116.732545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zito F, Finazzi G, Joliot P, Wollman FA. Glu78, from the conserved PEWY sequence of subunit IV, has a key function in cytochrome b6f turnover. Biochemistry. 1998;37:10395–10403. doi: 10.1021/bi980238o. [DOI] [PubMed] [Google Scholar]
- 29.Johnson X, et al. A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth Res. 2009;102:85–93. doi: 10.1007/s11120-009-9487-2. [DOI] [PubMed] [Google Scholar]
- 30.Kuras R, Wollman FA. The assembly of cytochrome b6/f complexes: An approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 1994;13:1019–1027. doi: 10.1002/j.1460-2075.1994.tb06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choquet Y, Zito F, Wostrikoff K, Wollman FA. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell. 2003;15:1443–1454. doi: 10.1105/tpc.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garnier J, Maroc J, Guyon D. Low-temperature fluorescence emission spectra and chlorophyll-protein complexes in mutants of Chlamydomonas reinhardtii: Evidence for a new chlorophyll-a-protein complex related to photosystem I. Biochim Biophys Acta. 1986;851:395–406. [Google Scholar]
- 33.Stroebel D, Choquet Y, Popot J-L, Picot D. An atypical haem in the cytochrome b(6)f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 34.Wurtz EA, Boynton JE, Gillham NW. Perturbation of chloroplast DNA amounts and chloroplast gene transmission in Chlamydomonas reinhardtii by 5-fluorodeoxyuridine. Proc Natl Acad Sci USA. 1977;74:4552–4556. doi: 10.1073/pnas.74.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer N, Hippler M, Sétif P, Jacquot JP, Rochaix JD. The PsaC subunit of photosystem I provides an essential lysine residue for fast electron transfer to ferredoxin. EMBO J. 1998;17:849–858. doi: 10.1093/emboj/17.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naver H, Boudreau E, Rochaix JD. Functional studies of Ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell. 2001;13:2731–2745. doi: 10.1105/tpc.010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Currin A, Swainston N, Day PJ, Kell DB. Synthetic biology for the directed evolution of protein biocatalysts: Navigating sequence space intelligently. Chem Soc Rev. 2015;44:1172–1239. doi: 10.1039/c4cs00351a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo J, et al. Structure of the catalytic domain of a state transition kinase homolog from Micromonas algae. Protein Cell. 2013;4:607–619. doi: 10.1007/s13238-013-3034-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergner SV, et al. STATE TRANSITION7-dependent phosphorylation is modulated by changing environmental conditions, and its absence triggers remodeling of photosynthetic protein complexes. Plant Physiol. 2015;168:615–634. doi: 10.1104/pp.15.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasan SS, Stofleth JT, Yamashita E, Cramer WA. Lipid-induced conformational changes within the cytochrome b6f complex of oxygenic photosynthesis. Biochemistry. 2013;52:2649–2654. doi: 10.1021/bi301638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finazzi G, Zito F, Barbagallo RP, Wollman FA. Contrasted effects of inhibitors of cytochrome b6f complex on state transitions in Chlamydomonas reinhardtii: The role of Qo site occupancy in LHCII kinase activation. J Biol Chem. 2001;276:9770–9774. doi: 10.1074/jbc.M010092200. [DOI] [PubMed] [Google Scholar]
- 42.Alric J, Pierre Y, Picot D, Lavergne J, Rappaport F. Spectral and redox characterization of the heme ci of the cytochrome b6f complex. Proc Natl Acad Sci USA. 2005;102:15860–15865. doi: 10.1073/pnas.0508102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zito F, Alric J. Cytochrome Complexes: Evolution, Structures, Energy Transduction, and Signaling. Springer; Dordrecht, The Netherlands: 2016. Heme ci or cn of the cytochrome b6f complex, a short retrospective; pp. 295–306. [Google Scholar]
- 44.Wollman FA, Delepelaire P. Correlation between changes in light energy distribution and changes in thylakoid membrane polypeptide phosphorylation in Chlamydomonas reinhardtii. J Cell Biol. 1984;98:1–7. doi: 10.1083/jcb.98.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chua NH, Bennoun P. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: Wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA. 1975;72:2175–2179. doi: 10.1073/pnas.72.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.