Significance
The formation of skeletal muscle fibers during embryogenesis and adult injury-induced muscle repair occurs through the fusion of myoblasts. We recently discovered myomixer, a muscle-specific micropeptide required for myoblast fusion in mice. Myomixer and myomaker, another muscle-specific membrane protein, are sufficient to induce fusion of nonmuscle cells. Here, we extend these findings and demonstrate the requirement of myomixer for zebrafish myoblast fusion. We also demonstrate a striking functional conservation of myomixer in zebrafish, elephant shark, and turtle among other species. Comparison of conserved regions of myomixer in these species led to the identification of peptide domains essential for myomixer function. Our findings provide further understanding of the mechanistic basis of myomixer function and muscle cell fusion during development and regeneration.
Keywords: myomaker, fusogenic, zebrafish, myogenesis, micropeptide
Abstract
Skeletal muscle formation requires fusion of mononucleated myoblasts to form multinucleated myofibers. The muscle-specific membrane proteins myomaker and myomixer cooperate to drive mammalian myoblast fusion. Whereas myomaker is highly conserved across diverse vertebrate species, myomixer is a micropeptide that shows relatively weak cross-species conservation. To explore the functional conservation of myomixer, we investigated the expression and function of the zebrafish myomixer ortholog. Here we show that myomixer expression during zebrafish embryogenesis coincides with myoblast fusion, and genetic deletion of myomixer using CRISPR/Cas9 mutagenesis abolishes myoblast fusion in vivo. We also identify myomixer orthologs in other species of fish and reptiles, which can cooperate with myomaker and substitute for the fusogenic activity of mammalian myomixer. Sequence comparison of these diverse myomixer orthologs reveals key amino acid residues and a minimal fusogenic peptide motif that is necessary for promoting cell–cell fusion with myomaker. Our findings highlight the evolutionary conservation of the myomaker–myomixer partnership and provide insights into the molecular basis of myoblast fusion.
Skeletal muscle is a multinucleated tissue that forms from fusion of mononucleated myoblasts during embryogenesis. Myoblast fusion depends on a complex series of events, including cell–cell recognition and adhesion, cytoskeletal reorganization, and, ultimately, membrane merger (1, 2). A variety of proteins have been shown to participate in myoblast fusion, but the molecular basis of muscle specificity and the complete identity of all components of the process have not been fully established.
Recently, we discovered a muscle-specific transmembrane protein, called myomaker, which is expressed at the onset of muscle formation and is essential for myoblast fusion in mice (3, 4) and zebrafish (5–7). Remarkably, myomaker can also promote the fusion of fibroblasts to myoblasts, but cannot promote fibroblast–fibroblast fusion, suggesting the existence of additional muscle-specific components of the cell fusion machinery (3, 8). Insight into the identity of a myomaker partner was provided by the discovery of a muscle-specific micropeptide, which we named myomixer (9), also known as myomerger (10) and minion (11). Like myomaker, myomixer expression coincides with the timing of myoblast fusion in cultured myoblasts and mouse embryos, and loss-of-function of myomixer in mice or cultured myoblasts completely prevents fusion (9–11). Moreover, coexpression of myomixer and myomaker in fibroblasts allows their autonomous fusion (9–11). As obligate partners, myomaker and myomixer may engage the more general cellular machinery required for membrane merger, including components of the actin cytoskeleton (1–3, 11).
In contrast to myomaker, which is highly conserved from mammals to zebrafish, myomixer is relatively divergent at the amino acid sequence level, sharing only 36% amino acid identity between these species. To investigate the function of zebrafish myomixer and begin to define the regions of the protein that control membrane merger, we performed genetic and structure–function studies of zebrafish myomixer. Here, by deletion of myomixer with CRISPR/Cas9, we show that myomixer is required for zebrafish myoblast fusion. We also identify additional distantly related myomixer orthologs from genomes of turtle and elephant shark, which like zebrafish myomixer can substitute for mouse myomixer to promote cell fusion together with myomaker in cultured cells. By comparison of amino acid sequences across these diverse species, we identify a unique protein motif required for the fusogenic activity of myomixer. Together, our findings provide insights into the molecular basis of myoblast fusion and reveal an evolutionarily conserved role for the myomaker–myomixer duo in muscle formation.
Results
Myomixer Is Transiently and Specifically Expressed in the Developing Zebrafish Myotome.
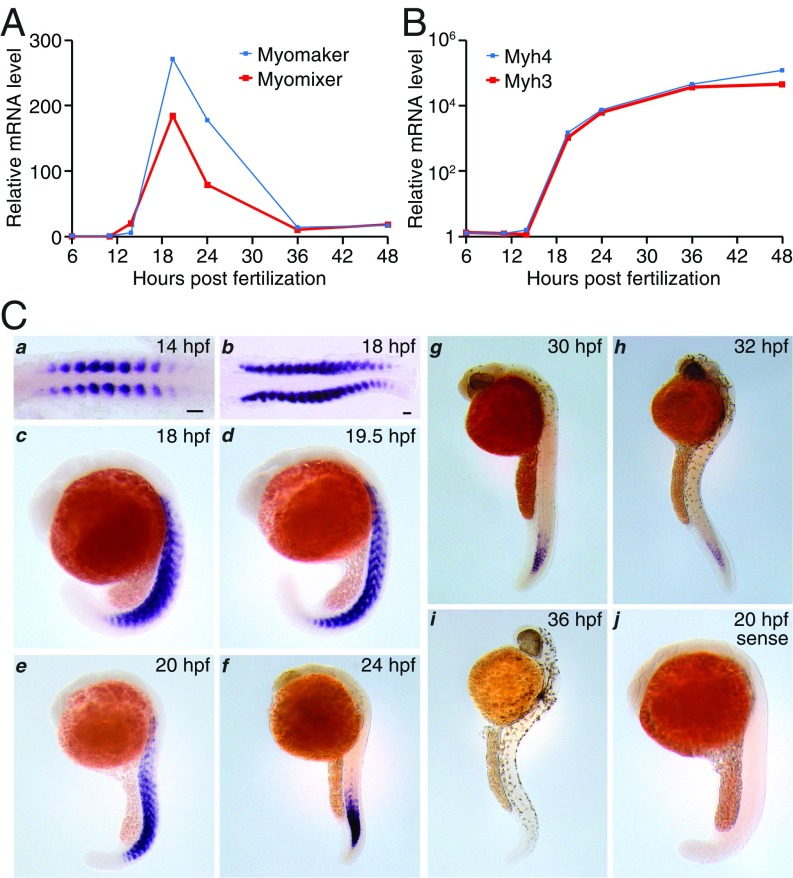
Previously, we identified a putative zebrafish ortholog of mammalian myomixer proteins (9). To investigate the expression of this putative myomixer ortholog, we designed several pairs of quantitative PCR (qPCR) primers to amplify the predicted myomixer ORF using cDNA from zebrafish embryos at 19.5 h post fertilization (hpf) and observed PCR products of predicted sizes (SI Appendix, Fig. S1). No PCR products were detected in control reactions without the reverse transcriptase, indicating the absence of genomic DNA contamination in the cDNA samples (SI Appendix, Fig. S1). Our qPCR analyses revealed that myomixer expression was initiated between 12 and 14 hpf, reached its peak level at 19.5 hpf, gradually decreased thereafter, and disappeared at 36 hpf (Fig. 1A). This temporal expression profile coincided with myogenesis and resembled that of myomaker (Fig. 1A), suggesting a potential functional correlation between these two regulators of myoblast fusion during zebrafish muscle development. As expected, the expression of two muscle structural genes, Myh3 and Myh4, continued to increase after 19.5 hpf during zebrafish development (Fig. 1B).
Fig. 1.
Characterization of myomixer expression during zebrafish embryogenesis. (A and B) Gene expression in timed zebrafish embryos determined by quantitative PCR. (C) In situ hybridization at different developmental stages of zebrafish embryos with antisense (a–i) and sense (j) probes. hpf, hours post fertilization. (Scale bars, 50 µm.)
Consistent with the qPCR results, in situ hybridizations of zebrafish embryos showed intense expression of myomixer exclusively in the developing somites, concomitant with muscle differentiation. Specifically, myomixer mRNA was detected in all developing myotomes in 14-hpf (10 somites), 18-hpf (18–19 somites), and 19.5-hpf (21 somites) embryos (Fig. 1 C, a–d). As somitogenesis proceeded along the anteroposterior axis, myomixer expression gradually disappeared from the differentiated anterior somites and shifted to the more caudal somites (Fig. 1 C, e–h), until it was no longer detected at 36 hpf (Fig. 1 C, i). Myomixer expression was absent in craniofacial muscles and the paraxial mesoderm (Fig. 1 C, c–i). The specificity of the antisense probe was confirmed by a sense probe that did not detect any signal (Fig. 1 C, j). Although our qPCR analyses revealed expression of both myomixer and myomaker at 14 hpf (Fig. 1A), a previous study reported the absence of myomaker expression before 15 hpf by in situ hybridization (5). To clarify the timing of myomaker expression, we performed in situ hybridization of myomaker in zebrafish embryos. The expression of myomaker (SI Appendix, Fig. S2A), like that of myomixer (Fig. 1 C, a), was detected in the developing somites of 14-hpf embryos, consistent with the qPCR data (Fig. 1A). In later stages of myogenesis, myomaker and myomixer also exhibited similar spatial and temporal expression patterns (compare Fig. 1 C, c–i, and SI Appendix, Fig. S2 B–G′). The synchronized expression of myomixer and myomaker in the developing myotomes further supports a functional correlation between these two proteins in zebrafish muscle development.
Interestingly, the spatiotemporal expression pattern of myomixer resembles that of MyoD (12), a muscle-specific transcription factor that binds E-box sequences (CANNTG) in the promoter regions of its target genes (13). Accordingly, we identified three conserved E-boxes in the promoters of the zebrafish and fugu myomixer genes (SI Appendix, Fig. S3), suggesting that myomixer may be transcriptionally regulated by MyoD during myogenesis. In this regard, it is worth noting that mouse myomixer was initially identified as a transcriptional target of MyoD (14).
Myomixer Is Required for Myoblast Fusion During Zebrafish Myogenesis.
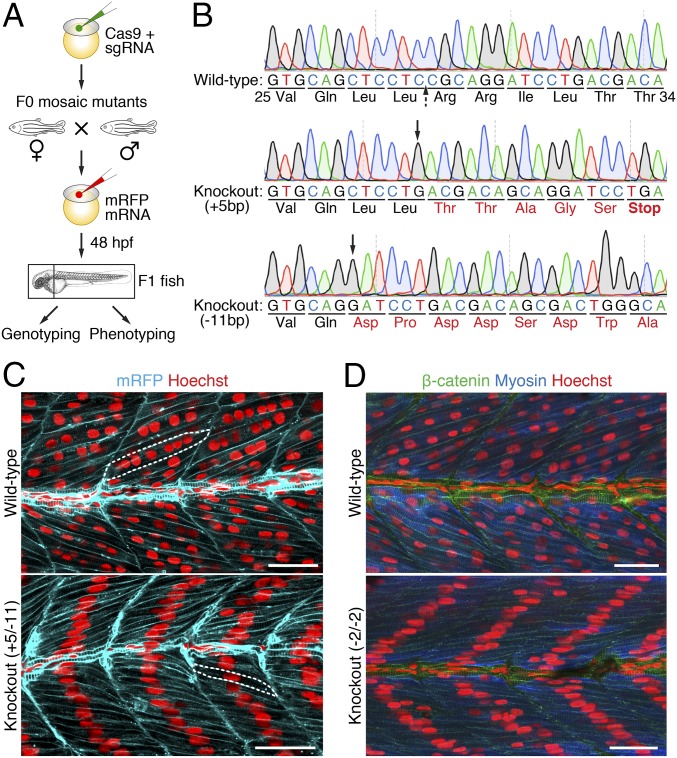
To assess the potential requirement of myomixer in muscle formation in vivo, we inactivated the gene during zebrafish embryogenesis using CRISPR/Cas9 mutagenesis. Embryos were injected at the one-cell stage with Cas9 protein and one of two myomixer single-guide RNAs (sgRNAs) (Fig. 2A). Ten injected embryos were used for genotyping at 24 hpf, whereas others were allowed to develop to adulthood (F0). Sequencing of the PCR products amplified from embryonic genomic DNA revealed high efficiency of genome editing and disruption of the myomixer gene by both sgRNAs, resulting in truncated proteins (SI Appendix, Fig. S4). Due to the mosaicism of the adult F0 fish, we intercrossed six pairs of injected F0 fish to generate F1 knockout (KO) fish for phenotypic analysis (Fig. 2A). Genotyping of F1 embryos at 48 hpf using DNA extracted from the head tissues revealed indels that disrupted the ORFs of both alleles of myomixer (Fig. 2B). The occurrence of such transheterozygous myomixer KO embryos was between 25 and 90% among the six mating pairs, demonstrating a high efficiency of CRISPR/Cas9 mutagenesis of germ cells. The trunk regions of such transheterozygous myomixer KO embryos were then immunostained to assess the muscle phenotype.
Fig. 2.
Ablation of myomixer in zebrafish abolishes myoblast fusion. (A) Schematic diagram of the experimental design. (B) DNA sequences of the myomixer coding region in wild-type and F1 knockout (KO) zebrafish. A region of wild-type (Val25–Thr34) and frame-shifted myomixer ORFs is shown. The Cas9 cleavage site is indicated by a dashed arrow, the starting positions of indels are indicated by black arrows. Amino acid mutations are in red. (C) Confocal images of 48-hpf wild-type and KO (+5/−11) embryos expressing a membrane-localized mRFP and stained with Hoechst to visualize the nuclei. Note the multinucleated muscle fibers (one of which is outlined) in the wild-type and the mononucleated muscle fibers (one of which is outlined) in the KO embryos. (D) Confocal images of 48-hpf wild-type and KO (−2/−2) embryos stained with anti–β-catenin, anti-fast muscle myosin, and Hoechst. (Scale bars, 25 µm.)
The zebrafish myotome is composed of two distinct populations of muscle fibers, slow and fast (15, 16). At the end of the segmentation period (24 hpf), slow muscle fibers form a superficial monolayer on the surface of the myotome and remain mononucleated, whereas fast muscle fibers in deeper layers have undergone myoblast fusion and thus are multinucleated. The transheterozygous myomixer KO embryos showed normal differentiation of the mononucleated slow muscle fibers, stained with a slow muscle myosin antibody and a slow muscle nuclear marker at 48 hpf (SI Appendix, Fig. S5). To visualize the fast muscle fibers, we labeled 48-hpf embryos with a fast muscle myosin antibody, a nuclear dye Hoechst, and a cell membrane marker, such as β-catenin or a membrane-localized red fluorescent protein (mRFP). Strikingly, each fast muscle fiber in myomixer KO embryos contained a single nucleus aligned at the center of the fiber, compared with the multiple nuclei in each myofiber of wild-type embryos, demonstrating a myoblast fusion defect (Fig. 2 C and D). Myomixer KO myofibers showed normal expression of myosin, as revealed by the fast myosin heavy chain antibody (Fig. 2D), indicating a specific defect in myoblast fusion independent of muscle differentiation. The myoblast fusion defect in myomixer KO fast fibers resembled that observed in myomaker KO or knockdown embryos (5–7), consistent with the coordinated function of myomixer and myomaker in fusion of fast muscle cells.
Zebrafish Myomixer Is a Membrane Micropeptide That Induces Cell Fusion.
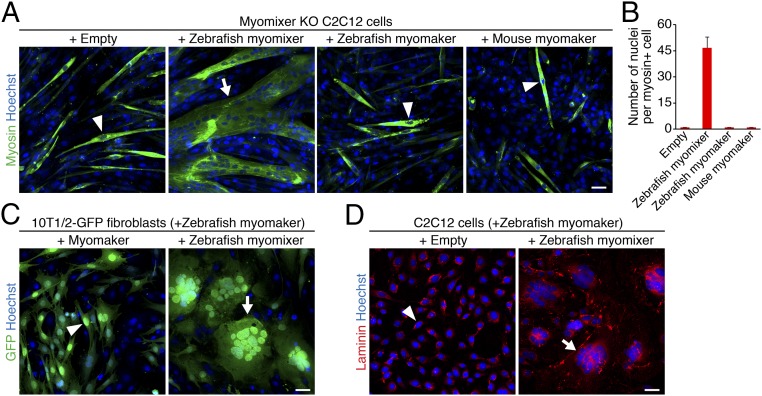
Zebrafish myomixer is a 75-amino acid micropeptide, sharing only 36% identity (27 amino acids) with mouse myomixer. To examine whether zebrafish myomixer can functionally replace its mouse ortholog, we first generated myomixer knockout (KO) C2C12 myoblasts using CRISPR/Cas9 mutagenesis and then infected the cells with retrovirus encoding zebrafish myomixer. Two days after retroviral infection, C2C12 cells were switched to differentiation medium (DM) for 1 wk to allow myotube formation. Strikingly, expression of zebrafish myomixer completely rescued the fusion defect, evidenced by the presence of large multinucleated myosin-positive syncytia and the absence of mononucleated myosin-positive cells (Fig. 3 A and B), whereas myomixer KO cells infected with an empty retrovirus failed to fuse (9) (Fig. 3 A and B). Of note, overexpression of either mouse or zebrafish myomaker failed to rescue the fusion of myomixer KO C2C12 cells, indicating that myomaker depends on myomixer for myoblast fusion (Fig. 3 A and B).
Fig. 3.
Zebrafish myomixer induces cell fusion. (A) Myomixer KO C2C12 cells infected with retroviruses expressing zebrafish myomixer, myomaker, or mouse myomaker were differentiated for 1 wk and stained with anti-myosin and Hoechst. Note that myomixer, but not myomaker, rescued the fusion defect in the myomixer KO cells. (B) Quantification of the fusion index in A, shown as the average number of nuclei in a myosin+ cell. For each genotype, the nuclei in myosin+ cells of seven 10× fields were counted. Error bars stand for SE of mean. (C) The 10T1/2-GFP fibroblasts were infected with retroviruses expressing zebrafish myomaker with or without zebrafish myomixer for 2 d to induce cell fusion. Note that cell fusion was induced when both myomaker and myomixer were expressed. (D) Proliferating C2C12 cells were infected with retroviruses expressing zebrafish myomaker with or without zebrafish myomixer for 2 d to induce cell fusion. Cells were stained with anti-laminin and Hoechst. Arrowheads indicate mononucleated cells; arrows indicate multinucleated cells. (Scale bars, 50 µm.)
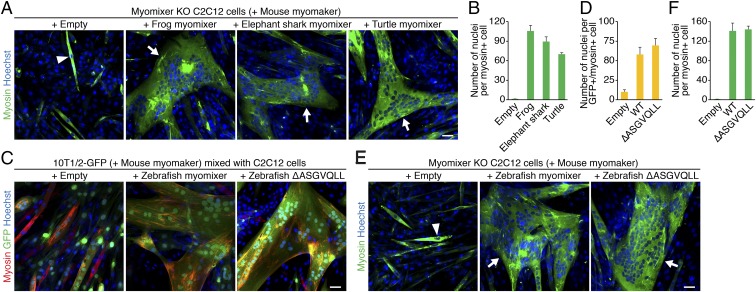
The functional dependence between myomaker and myomixer was further revealed by the observation that coexpression of zebrafish myomaker and myomixer was sufficient to generate syncytia of 10T1/2 fibroblasts (Fig. 3C), as well as of C2C12 myoblasts under culture conditions that did not promote differentiation (Fig. 3D). Fibroblasts induced to fuse by zebrafish myomaker together with myomixer exhibited a morphology resembling bird nests filled with eggs (Fig. 3C), similar to the syncytia formed by coexpression of mouse myomixer and myomaker (9). Consistent with the fusogenic function of myomixer, Western blot analysis revealed the association of zebrafish myomixer with the membrane fraction of human kidney 293 cells transfected with a plasmid encoding zebrafish myomixer (SI Appendix, Fig. S6). N-Cadherin was used as a positive control for membrane proteins, whereas α-Tubulin and Gapdh served as positive controls for cytosolic proteins (SI Appendix, Fig. S6). We conclude that despite the relatively weak homology between zebrafish and mouse myomixer, both proteins possess the ability to induce fusion of nonfusogenic cells together with myomaker.
A Conserved AxLyCxL Motif Is Essential for Myomixer Function.
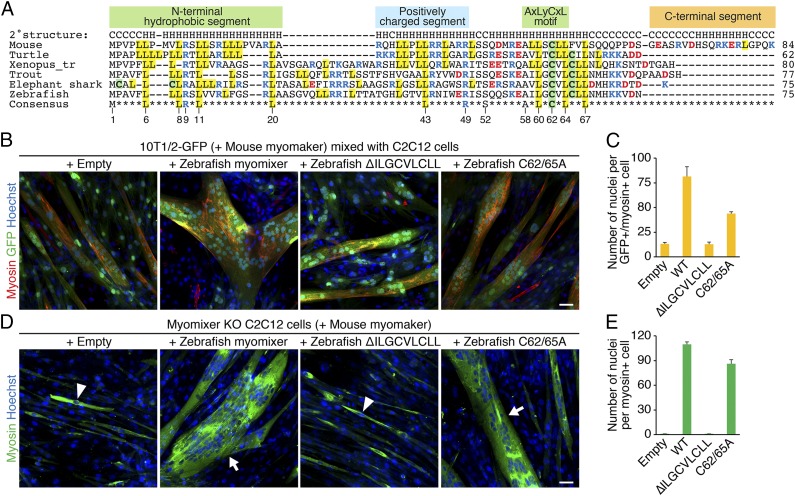
Tblastn searches using the zebrafish myomixer against whole-genome shotgun sequences in NCBI identified homologs in other vertebrate species such as reptile (turtle), amphibian (frog), and fish, but not in invertebrate species (Fig. 4A). None of these genes have been predicted in their genomes, presumably because the myomixer peptides are short, and ORFs shorter than 100 amino acids are typically not annotated. Cross-species alignment of myomixer sequences revealed conservation of only a few residues distributed in three distinct motifs (Fig. 4A): an N-terminal hydrophobic domain, which is predicted to be the membrane anchoring region; a C-terminal hydrophobic AxLyCxL motif, in which x denotes leucine, valine, or isoleucine, and y denotes serine, threonine, or glycine; and several charged residues in the middle region, which have been shown to bind myomaker (9).
Fig. 4.
Functional analyses of zebrafish myomixer mutant proteins. (A) Cross-species alignment of myomixer proteins. Basic residues are shown in blue, and acidic residues are shown in red. Cysteines are highlighted in green, and leucines are highlighted in yellow. Identical residues in all orthologs are shown at the bottom. The numbers below the consensus sequence refer to the amino acid positions in zebrafish myomixer. (B) The 10T1/2-GFP fibroblasts were infected by retroviruses expressing mouse myomaker with or without wild-type or mutant zebrafish myomixer and mixed with C2C12 cells for heterologous fusion following 1 wk of differentiation. Cells were stained with anti-myosin and Hoechst. (C) Quantification of the fusion index in B. (D) Myomixer KO C2C12 cells expressing mouse myomaker with or without wild-type or mutant zebrafish myomixer were induced to fuse in DM for 1 wk. Cells were stained with anti-myosin and Hoechst. (E) Quantification of the fusion index in D. Arrowheads indicate mononucleated cells; arrows indicate multinucleated cells. (Scale bars, 50 µm.)
To test the functional significance of the AxLyCxL motif, we created retroviral constructs, in which the LLSCLL or ILGCVLCLL amino acids were deleted from mouse or zebrafish myomixer, referred to as ∆LLSCLL and ∆ILGCVLCLL, respectively. The 10T1/2 fibroblasts stably expressing GFP were infected with retroviruses expressing myomaker and different versions of myomixer and mixed with equal numbers of C2C12 cells 2 d postinfection. Cells were switched to DM to induce heterologous fusion and chimeric myotube formation. While wild-type mouse and zebrafish myomixer proteins robustly enhanced myomaker-mediated fusion between 10T1/2 fibroblasts and C2C12 cells (9) (Fig. 4 B and C), myomixer mutants ∆LLSCLL and ∆ILGCVLCLL failed to do so (Fig. 4 B and C and SI Appendix, Fig. S7 A and B). Consistently, the zebrafish ∆ILGCVLCLL mutant also failed to rescue the fusion of myomixer KO C2C12 cells (Fig. 4 D and E). By Western blot analysis, we confirmed the stable expression and membrane localization of the mouse ∆LLSCLL mutant (SI Appendix, Fig. S7B). We note, however, that upon prolonged exposure, we often detected wild-type and mutant myomixer proteins in the cytoplasmic fractions, which we interpret to indicate relatively weak association with membranes. Interestingly, although Cys62 in the AxLyCxL motif is highly conserved across the vertebrate species (Fig. 4A), mutating both Cys62 and 65 to Ala (C62/65A) in zebrafish myomixer only partially affected its fusogenic activity. Specifically, expressing myomixer-C62/65A rescued the fusion defect in myomixer KO C2C12 cells (Fig. 4 D and E) and induced heterologous fusion between 10T1/2 and C2C12 cells (Fig. 4 B and C), albeit at a compromised level compared with the wild-type myomixer protein. These results are consistent with that observed with a mouse myomixer C52A mutation (9). Taken together, we conclude that the AxLyCxL hydrophobic motif found in all myomixer orthologs is essential for myomixer function, although the Cys residue within the AxLyCxL motif is not critical.
Turtle Myomixer Is the Shortest Myomixer Homolog.
We also identified a myomixer homolog from elephant shark (Callorhinchus milii), the slowest evolving species of all known vertebrates (17). Notably, expression of either elephant shark or frog (Xenopus tropicalis) myomixer rescued the fusion defect of myomixer KO C2C12 cells (Fig. 5 A and B). One unique feature of the fish and frog myomixer orthologs is a 15-amino acid bridging region close to the N terminus (Fig. 4A). Sequences in this region among these species are highly variable, suggesting a nonessential role of this region. Supporting this notion, disruption of this region in zebrafish myomixer by removing the ASGVQLL fragment did not affect myomixer function in either the 10T1/2-C2C12 heterologous fusion assay (Fig. 5 C and D) or myomixer KO C2C12 rescue experiments (Fig. 5 E and F). Finally, expression of turtle (Chrysemys picta bellii) myomixer, a 62-amino acid micropeptide which lacks the fish/frog-specific bridging region and the unique mammalian C terminus region, also rescued the fusion defect of myomixer KO C2C12 cells (Fig. 5 A and B). In summary, among these identified homologs, the turtle myomixer is the shortest micropeptide that can induce cell fusion and reveal the minimal domains required for myomixer function.
Fig. 5.
Functional analyses of myomixer homologs in different vertebrate species. (A) Myomixer KO C2C12 cells expressing mouse myomaker with or without myomixer from different vertebrate species were induced to fuse in DM for 1 wk. Cells were stained with anti-myosin and Hoechst. (B) Quantification of the fusion index in A. (C) The 10T1/2-GFP fibroblasts were infected by retroviruses expressing mouse myomaker with or without wild-type and mutant zebrafish myomixer and mixed with C2C12 cells for heterologous fusion following 1 wk of differentiation. (D) Quantification of the fusion index in C. (E) Myomixer KO C2C12 cells expressing mouse myomaker with or without wild-type or mutant zebrafish myomixer were induced to fuse in DM for 1 wk. Cells were stained with anti-myosin and Hoechst. (F) Quantification of the fusion index in E. Arrowheads indicate mononucleated cells; arrows indicate multinucleated cells. (Scale bars, 50 µm.)
Discussion
The results of this study demonstrate the essentiality of myomixer for vertebrate myoblast fusion and highlight the evolutionary conservation of the fusion mechanism and its dependence on the partnership of myomixer and myomaker. Together, these two muscle-specific membrane proteins are capable of driving fusion of plasma membranes, even in nonfusogenic fibroblasts. According to our current understanding of the fusion process, we propose that myomaker serves as a transmembrane anchor for recruitment of myomixer and other components of the fusion machinery. Whereas myomaker is required in both cells whose membranes are destined to fuse, it appears that myomixer is required in only one of the two fusogenic cells (10, 11).
Myomixer is a small micropeptide with remarkably little cross-species homology. While the myomixer orthologs from divergent vertebrate species share only modest amino acid homology, they all share several general domains, including an N-terminal hydrophobic domain, which is likely to function as a membrane anchoring region; a C-terminal hydrophobic AxLyCxL motif; and several charged residues in between that are required for binding myomaker (9). Our mutational analyses indicate that the AxLyCxL motif is required for membrane fusion, but the mechanism whereby this domain influences membrane merger remains to be determined.
Despite the essential role for myomixer and myomaker in vertebrate myoblast fusion, we have not identified homologs of myomixer or myomaker in any invertebrates where myoblast fusion also takes place. Such species-specificity of fusogenic proteins contrasts with the conserved function of actin-propelled membrane protrusions in promoting cell–cell fusion in species ranging from insects to mammals (1, 2, 18–23). Both fusogenic proteins and actin-propelled membrane protrusions are indispensable for cell membrane fusion, demonstrated by reconstitution studies in cultured cells (24), suggesting that cell–cell fusion is controlled by both evolutionarily conserved and species-specific mechanisms. We speculate that in addition to using the common cellular machinery to bring cell membranes into close proximity, cells destined to fuse have adopted different fusogenic proteins during evolution, resulting in species and tissue specificity. Indeed, most known fusogenic proteins for cell–cell fusion appear to be species- and tissue-specific at the primary sequence level. For example, the placenta trophoblast fusogen syncytin has been identified in placental mammals (25–27); the Caenorhabditis elegans epidermal cell fusogens EFF-1 and AFF-1 have orthologs mainly in nematodes (28, 29); and the ancient gamete fusogen HAP2 is present in several unicellular and multicellular plant and animal species (30–34). Structural analyses to date revealed striking similarities between cellular fusogens and viral proteins (25, 26, 35, 36), suggesting that the former may be acquired via horizontal gene exchange with viruses. Whether the fusogenic micropeptide myomixer was acquired by an ancient vertebrate ancestor via gene exchange with a virus awaits future investigation. In this regard, the strong functional conservation of myomixer in vertebrates revealed by this study provides a unique tool for understanding its mechanism of action and evolutionary origin.
Materials and Methods
Zebrafish Husbandry.
Zebrafish (Danio rerio) were maintained on a 14-h light, 10-h dark cycle at 28 °C. Embryos were obtained by natural spawning. Fertilized eggs were raised in embryo medium at 28.5 °C and staged according to the standard protocol (37). To prevent pigmentation, embryos were kept in E3 medium with 0.00001% (wt/vol) methylene blue after fertilization for 5 d. Animal work described in this manuscript has been approved and conducted under the oversight of the UT Southwestern Institutional Animal Care and Use Committee.
CRISPR-Cas9 Mutagenesis and Microinjection.
Two independent myomixer sgRNAs were designed using ZiFiT Targeter version 4.2 (zifit.partners.org/ZiFiT/ChoiceMenu.aspx): sgRNA-1 TCTGGTTGTCCGACTCTTCGG and sgRNA-2 GGCGTGCAGCTCCTCCGCAGG. sgRNAs were synthesized from PCR templates using the MEGAscript T7 Transcription Kit (AM1333; Invitrogen) and purified with the MEGAclear Kit (AM1908; Invitrogen). Two hundred embryos were injected at the single-cell stage with a mixture of 250 ng/µL sgRNA and 500 ng/µL Cas9 protein (CP01; PNABio). Ten embryos were randomly collected to evaluate the targeting efficiency by PCR and sequence analysis. To visualize the cell boundary, F1 single-cell embryos were injected with 100-pg capped membrane-localized red fluorescent protein (RFP) mRNA, synthesized from a plasmid shared by the Ciruna laboratory (38).
Whole-Mount in Situ Hybridization.
Whole-mount in situ hybridization using digoxygenin-labeled riboprobes was performed using standard protocols (39). Briefly, chorion was removed from zebrafish embryos at different developmental stages by Pronase (P6911; Sigma) or manual operation. Embryos were fixed in 4% paraformaldehyde overnight at 4 °C and stored in methanol at −20 °C until use. To generate the riboprobes for myomixer, the following primers were used to amplify the sequence containing myomixer 5′UTR, ORF, and 3′UTR, which was then cloned into the pGEM-T Easy Vector (A1360; Promega): 5′-CAGAAACAGAGCGGTGACTTCATCCC-3′ (forward) and 5′-GCGAACCGATCTGTCCTCAAGTCTG-3′ (reverse). To generate the riboprobes for myomaker, the following primers were used to amplify the full-length myomaker ORF and clone into the pGEM-T Easy Vector: 5′-TGAGTCGCAATGGGAGCGTTTATCG-3′ (forward) and 5′-GCATCATACACAGCAGCAGAGGGT-3′ (reverse). Digoxigenin-labeled RNA probes were synthesized from linearized plasmids using the T7 RNA polymerases (P207B; Progema). Embryos were incubated first with hybridization buffer containing a specific RNA probe and then with anti-digoxigenin alkaline phosphatase Fab fragments (1093274; Roche). Color development was performed with the NBT (N6639; Sigma)/BCIP (B8503; Sigma) labeling mix. Stained embryos were mounted in 100% glycerol, and images were obtained using a camera mounted on the LEICA M205 FA microscope.
Whole-Mount Immunohistofluorescent Staining.
Zebrafish whole-mount immunostaining was performed according to standard methods (40). Primary antibodies used were: anti-fast muscle myosin (F310, 1:20 dilution; Developmental Studies Hybridoma Bank), anti-slow muscle myosin (F59, 1:50 dilution; Developmental Studies Hybridoma Bank), anti-Prox1 (ab209849, 1:500 dilution; Abcam), and anti–β-catenin (ab6302, 1:200 dilution; Abcam). Secondary antibodies used were: anti-mouse IgG Alexa-647 and anti-rabbit IgG Alexa-568 (1:5,000 dilution; Molecular Probes). Nuclei were visualized by Hoechst staining (1:200 dilution; Molecular Probes). Embryos were mounted in ProLong Gold Antifade Mountant (P36930; Molecular Probes). Confocal images were collected using a Nikon A1R confocal microscope. Images were acquired with the NIS–Elements Acquisition software and processed using ImageJ and Adobe Photoshop CS6.
Cell Culture, Retroviral Expression, and Cell Fusion Experiments.
The 10T1/2 fibroblasts and C2C12 cells were maintained in 10% FBS with 1% penicillin/streptomycin in DMEM.
Myomixer coding sequences were synthesized by Integrated DNA Technologies and inserted into the pMXs-Puro Retroviral Vector (#RTV-012; Cell Biolabs) (41) with the In-Fusion HD Cloning Plus Kit (#638910; Clontech). The inserted sequences were verified by sequencing. Plasmids were amplified in Stable3 cells in overnight cultures, followed by plasmid preparation using the NucleoBond Xtra Maxi Columns (#740414.10). A retrovirus plasmid carrying an N-terminally tagged Signal FLAG (SF) myomaker was used to overexpress myomaker (3), except for in the homologous fusion experiments of 10T1/2 or C2C12 cells, where the untagged myomaker was used. A retrovirus expressing GFP was used to infect and stably label the 10T1/2 fibroblasts.
Platinum-E cells (#RV-101; Cell Biolabs) were plated on a 10-cm cell culture dish at a density of 5 × 106 cells per dish 12 h before transfection. Fifteen micrograms of total retroviral plasmid DNA were transfected using FuGENE 6 (#E2692; Promega). Two days after transfection, the viral medium was filtered through a 0.45-μm cellulose syringe filter and mixed with polybrene (Sigma) at a final concentration of 6 μg/mL. Before infection, 10T1/2 fibroblasts or C2C12 myoblasts were washed twice with PBS. For fusion experiments, virus-infected 10T1/2 cells were mixed with C2C12 cells (5 × 104 C2C12 cells and 5 × 104 fibroblasts per well) and plated onto a six-well plate. One day after plating, the cells were switched to myoblast differentiation medium (2% horse serum in DMEM with 1% penicillin/streptomycin) for 1 wk with a medium change at day 3 of differentiation.
Supplementary Material
Acknowledgments
We thank Dr. Xiaoying Bai for sharing the zebrafish facility. We are grateful to Jose Cabrera for graphics and Jennifer Brown for editorial assistance. We thank Alexis Houghton, Andres Ramirez-Martinez, and Akansha Shah for technical assistance and other members of the E.N.O. and E.H.C. laboratories for technical advice and comments. This work was supported by NIH Grants AR-067294, HL-130253, and DK-099653 (to E.N.O. and R.B.-D.) and AR-053173 and GM-098816 (to E.H.C.); Grant 1-0025 from the Robert A. Welch Foundation (to E.N.O.); National Established Investigator Award 13EIA14560083 from the American Heart Association (to E.H.C.); and a Faculty Scholar Award from the Howard Hughes Medical Institute (to E.H.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715229114/-/DCSupplemental.
References
- 1.Kim JH, Jin P, Duan R, Chen EH. Mechanisms of myoblast fusion during muscle development. Curr Opin Genet Dev. 2015;32:162–170. doi: 10.1016/j.gde.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abmayr SM, Pavlath GK. Myoblast fusion: Lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Millay DP, et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature. 2013;499:301–305. doi: 10.1038/nature12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millay DP, Sutherland LB, Bassel-Duby R, Olson EN. Myomaker is essential for muscle regeneration. Genes Dev. 2014;28:1641–1646. doi: 10.1101/gad.247205.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landemaine A, Rescan PY, Gabillard JC. Myomaker mediates fusion of fast myocytes in zebrafish embryos. Biochem Biophys Res Commun. 2014;451:480–484. doi: 10.1016/j.bbrc.2014.07.093. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Roy S. Myomaker is required for the fusion of fast-twitch myocytes in the zebrafish embryo. Dev Biol. 2017;423:24–33. doi: 10.1016/j.ydbio.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 7.Di Gioia SA, et al. Moebius Syndrome Research Consortium A defect in myoblast fusion underlies Carey-Fineman-Ziter syndrome. Nat Commun. 2017;8:16077. doi: 10.1038/ncomms16077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millay DP, et al. Structure-function analysis of myomaker domains required for myoblast fusion. Proc Natl Acad Sci USA. 2016;113:2116–2121. doi: 10.1073/pnas.1600101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi P, et al. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017;356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn ME, et al. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat Commun. 2017;8:15665. doi: 10.1038/ncomms15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, et al. The microprotein Minion controls cell fusion and muscle formation. Nat Commun. 2017;8:15664. doi: 10.1038/ncomms15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberg ES, et al. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- 13.Kophengnavong T, Michnowicz JE, Blackwell TK. Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations. Mol Cell Biol. 2000;20:261–272. doi: 10.1128/mcb.20.1.261-272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong AP, et al. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev Cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stickney HL, Barresi MJF, Devoto SH. Somite development in zebrafish. Dev Dyn. 2000;219:287–303. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1065>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 16.Jackson HE, Ingham PW. Control of muscle fibre-type diversity during embryonic development: The zebrafish paradigm. Mech Dev. 2013;130:447–457. doi: 10.1016/j.mod.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesh B, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sens KL, et al. An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J Cell Biol. 2010;191:1013–1027. doi: 10.1083/jcb.201006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amalaka S, et al. Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development. 2011;138:1551–1562. doi: 10.1242/dev.057653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin P, et al. Competition between blown fuse and WASP for WIP binding regulates the dynamics of WASP-dependent actin polymerization in vivo. Dev Cell. 2011;20:623–638. doi: 10.1016/j.devcel.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan R, et al. Group I PAKs function downstream of Rac to promote podosome invasion during myoblast fusion in vivo. J Cell Biol. 2012;199:169–185. doi: 10.1083/jcb.201204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin NY, et al. Dynamin and endocytosis are required for the fusion of osteoclasts and myoblasts. J Cell Biol. 2014;207:73–89. doi: 10.1083/jcb.201401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH, et al. Mechanical tension drives cell membrane fusion. Dev Cell. 2015;32:561–573. doi: 10.1016/j.devcel.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shilagardi K, et al. Actin-propelled invasive membrane protrusions promote fusogenic protein engagement during cell-cell fusion. Science. 2013;340:359–363. doi: 10.1126/science.1234781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 26.Blond JL, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohler WA, et al. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 29.Sapir A, et al. AFF-1, a FOS-1-regulated fusogen, mediates fusion of the anchor cell in C. elegans. Dev Cell. 2007;12:683–698. doi: 10.1016/j.devcel.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson MA, et al. Arabidopsis hapless mutations define essential gametophytic functions. Genetics. 2004;168:971–982. doi: 10.1534/genetics.104.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat Cell Biol. 2006;8:64–71. doi: 10.1038/ncb1345. [DOI] [PubMed] [Google Scholar]
- 32.von Besser K, Frank AC, Johnson MA, Preuss D. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development. 2006;133:4761–4769. doi: 10.1242/dev.02683. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Genes Dev. 2008;22:1051–1068. doi: 10.1101/gad.1656508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele RE, Dana CE. Evolutionary history of the HAP2/GCS1 gene and sexual reproduction in metazoans. PLoS One. 2009;4:e7680. doi: 10.1371/journal.pone.0007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Vargas J, et al. Structural basis of eukaryotic cell-cell fusion. Cell. 2014;157:407–419. doi: 10.1016/j.cell.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 36.Fédry J, et al. The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell. 2017;168:904–915.e10. doi: 10.1016/j.cell.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 38.Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature. 2006;439:220–224. doi: 10.1038/nature04375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 40.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 4th Ed Univ of Oregon Press; Eugene, OR: 2000. [Google Scholar]
- 41.Kitamura T, et al. Retrovirus-mediated gene transfer and expression cloning: Powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.