Significance
The endoplasmic reticulum (ER) forms membrane contact sites (MCSs) with other organelles such as mitochondria, endosomes, and peroxisomes in eukaryotic cells. The MCS plays a pivotal role in exchanging cellular materials such as ions and lipids. More importantly, nonvesicular lipid trafficking occurring at the ER-mitochondria MCS is essential for the biogenesis of the mitochondrial membrane. In yeast, the ER-mitochondria encounter structure (ERMES) complex comprising the ER proteins Mmm1 and cytosolic Mdm12 and the mitochondria proteins Mdm34 and Mdm10 provides a tethering force between the ER and the mitochondria and mediates lipid trafficking. Here, we present two crystal structures of Mmm1 and the Mdm12–Mmm1 complex. Based on these structures, we propose the model by which the Mdm12–Mmm1 complex contributes to phospholipid trafficking at the ER-mitochondria MCS.
Keywords: Mmm1, Mdm12–Mmm1 complex, ERMES, phospholipid trafficking, membrane contact site
Abstract
The endoplasmic reticulum (ER)-mitochondria encounter structure (ERMES) comprises mitochondrial distribution and morphology 12 (Mdm12), maintenance of mitochondrial morphology 1 (Mmm1), Mdm34, and Mdm10 and mediates physical membrane contact sites and nonvesicular lipid trafficking between the ER and mitochondria in yeast. Herein, we report two crystal structures of the synaptotagmin-like mitochondrial lipid-binding protein (SMP) domain of Mmm1 and the Mdm12–Mmm1 complex at 2.8 Å and 3.8 Å resolution, respectively. Mmm1 adopts a dimeric SMP structure augmented with two extra structural elements at the N and C termini that are involved in tight self-association and phospholipid coordination. Mmm1 binds two phospholipids inside the hydrophobic cavity, and the phosphate ion of the distal phospholipid is specifically recognized through extensive H-bonds. A positively charged concave surface on the SMP domain not only mediates ER membrane docking but also results in preferential binding to glycerophospholipids such as phosphatidylcholine (PC), phosphatidic acid (PA), phosphatidylglycerol (PG), and phosphatidylserine (PS), some of which are substrates for lipid-modifying enzymes in mitochondria. The Mdm12–Mmm1 structure reveals two Mdm12s binding to the SMP domains of the Mmm1 dimer in a pairwise head-to-tail manner. Direct association of Mmm1 and Mdm12 generates a 210-Å-long continuous hydrophobic tunnel that facilitates phospholipid transport. The Mdm12–Mmm1 complex binds all glycerophospholipids except for phosphatidylethanolamine (PE) in vitro.
Membrane contact sites (MCSs) play an essential role in subcellular communication by exchanging cellular materials and information (1, 2). Among the various endoplasmic reticulum (ER)-mediated MCSs reported to date (3), the ER-mitochondria contact site has been the most extensively studied, and an involvement in ion homeostasis, mitochondrial dynamics such as membrane fission and fusion, and cooperative lipid synthesis has been reported (4–9). Most importantly, lipid trafficking occurring at the ER-mitochondria MCS is essential for the biogenesis of the mitochondrial membrane, since mitochondria are not connected with the vesicular transport machinery, and essential lipids required for the composition of mitochondrial membrane must therefore be supplied directly from the ER (10–12).
Formation of the MCS is the result of direct interaction between protein components located at two distinct subcompartments to be adjoined. In yeast, ER-mitochondria contact sites are primarily mediated by the ER-mitochondria encounter structure (ERMES) complex that comprises four proteins: the cytosolic component mitochondrial distribution and morphology 12 (Mdm12); the ER membrane protein maintenance of mitochondrial morphology 1 (Mmm1); and two mitochondria outer membrane proteins, Mdm34 and Mdm10 (13). Additionally, mitochondria anchoring Gem1, a Ca2+-binding small GTPase, directly associates with the ERMES complex and regulates its size and number (14–16). ERMES components are also regulated by Rsp5 E3 ubiquitin ligase, and ubiquitination is required for efficient mitophagy (17).
Accumulated evidence suggests that Mdm12, Mmm1, and Mdm34 share a synaptotagmin-like mitochondrial lipid-binding protein (SMP) domain (7, 18–20), suggesting that the ERMES complex not only tethers two connecting membranes but also acts as a transfer vehicle to exchange phospholipids between the ER and mitochondria (21). Indeed, ERMES mutants have an altered phosphatidylserine (PS)-to-phosphatidylethanolamine (PE) conversion rate (13, 22), suggesting that the ERMES complex might be critically involved in phospholipid trafficking at ER-mitochondria contact sites, although its direct involvement in converting PS to PE still remains contentious (23). Recent studies have highlighted alternative lipid trafficking pathways involving vacuoles, which reciprocally supply mitochondria with phospholipids (24–26). Furthermore, the ER membrane protein complex (EMC) comprising conserved Emc1–Emc6 proteins performs a comparable role in lipid transfer from the ER to mitochondria by mediating tethering between these organelles (26). In addition to lipid trafficking, other functions of the ERMES complex have been reported, including mitochondrial protein assembly (27) and import (28), maintenance of mitochondrial DNA (15, 29, 30), mitochondria inheritance (31), and mitophagy (17, 32–34).
Previously, we determined the crystal structure of Saccharomyces cerevisiae Mdm12 at 3.1 Å resolution and revealed that Mdm12 forms a dimeric SMP structure that binds phospholipids inside a hydrophobic channel, with a preference for glycerophospholipids harboring a positively charged head group (20). Another study determined a 17 Å resolution electron microscopy (EM) structure of the Mdm12–Mmm1 (SMP domain) complex, revealing an elongated tubular structure with an Mdm12-Mmm1-Mmm1-Mdm12 arrangement (19, 35). Despite these structure studies, the molecular-level mechanism by which the SMP domains of Mdm12, Mmm1, and Mdm34 are directly organized and facilitate phospholipid trafficking without consuming energy at the ER-mitochondria contact site remains unknown. Additionally, exactly how Mmm1, an ER component of the ERMES complex, recognizes specific phospholipids in the ER membrane remains elusive, as does the mechanism by which phospholipids selected by Mmm1 are transported into Mdm12, as a direct binding partner of the ERMES complex.
In the present study, we determined crystal structures of the Mmm1 SMP domain and the Mdm12–Mmm1 binary complex, and discuss the resultant molecular-level insight into how the Mmm1 SMP domain contributes to the organization of the ERMES components, as well as phospholipid trafficking.
Results
Structure Determination of Mmm1.
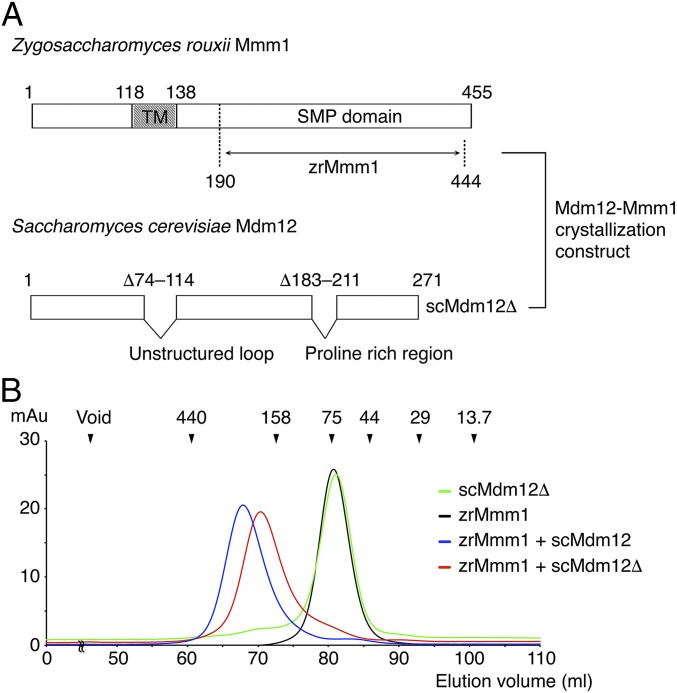
The Mmm1 protein is predicted to comprise a single transmembrane domain near its N terminus that anchors it to the ER membrane, an unstructured region consisting of around 50 residues, and an SMP domain at the C terminus (Fig. 1A and Fig. S1). The N-terminal region of Mmm1 is located in the ER lumen, while the SMP domain is localized in the cytosol and directly interacts with Mdm12, a cytosolic component of the ERMES complex. Despite significant effort to purify Mmm1 proteins, size-exclusion chromatography (SEC) experiments revealed that the SMP domain of S. cerevisiae Mmm1 (scMmm1) aggregated in solution unless in a complex with Mdm12 (20). Extensive screening for solubility and homogeneous dispersal in solution for Mmm1 orthologs, together with limited proteolysis analysis, revealed that the Mmm1 SMP domain of Zygosaccharomyces rouxii (zrMmm1, residues 190–444) was soluble even when not complexed with Mdm12 (Fig. 1B). The SMP domain of zrMmm1 shares 76% sequence identity with that of scMmm1. The zrMmm1 proteins eluted from the gel-filtration column at a volume corresponding to the molecular weight of a dimer, suggesting that the recombinantly expressed zrMmm1 SMP domain forms a homodimer in solution. Interestingly, the SEC experiment confirmed that zrMmm1 was able to interact with scMdm12 when coexpressed in Escherichia coli cells despite the organismal discrepancy (Fig. 1B). Diffraction-quality crystals of zrMmm1 were grown in the P3221 space group at 4 °C over a period of 1 wk, and the structure was solved using selenomethionine-substituted crystals by the single-wavelength anomalous dispersion method (Fig. S2). The final model of zrMmm1 was refined with data from native crystals to 2.8 Å resolution.
Fig. 1.
Domain structure and direct interaction of Mmm1 and Mdm12. (A) Diagrams showing the domain structure of Z. rouxii Mmm1 and S. cerevisiae Mdm12. Mmm1 has a transmembrane (TM) domain in the middle of the protein chain that is required for anchoring the ER membrane, and the SMP domain is at the C terminus. Full-length scMdm12 covers the overall SMP domain. The Mmm1 construct used in this study is indicated with an arrow (Z. rouxii Mmm1 residues 190–444, referred to as zrMmm1). To obtain diffraction-quality crystals of the Mdm12–Mmm1 complex, two unstructured regions were omitted in the scMdm12 construct (Δ74–114 and Δ183–211, referred to as scMdm12Δ). (B) SEC profiles of scMdm12Δ (green), zrMmm1 (black), and complexes of zrMmm1 and scMdm12 (blue) and zrMmm1 and scMdm12Δ (red). Experimental details are provided in Materials and Methods. Protein standards used in the experiment are indicated above the chromatogram. mAu, milliabsorbance unit.
Structure of the zrMmm1 SMP Domain.
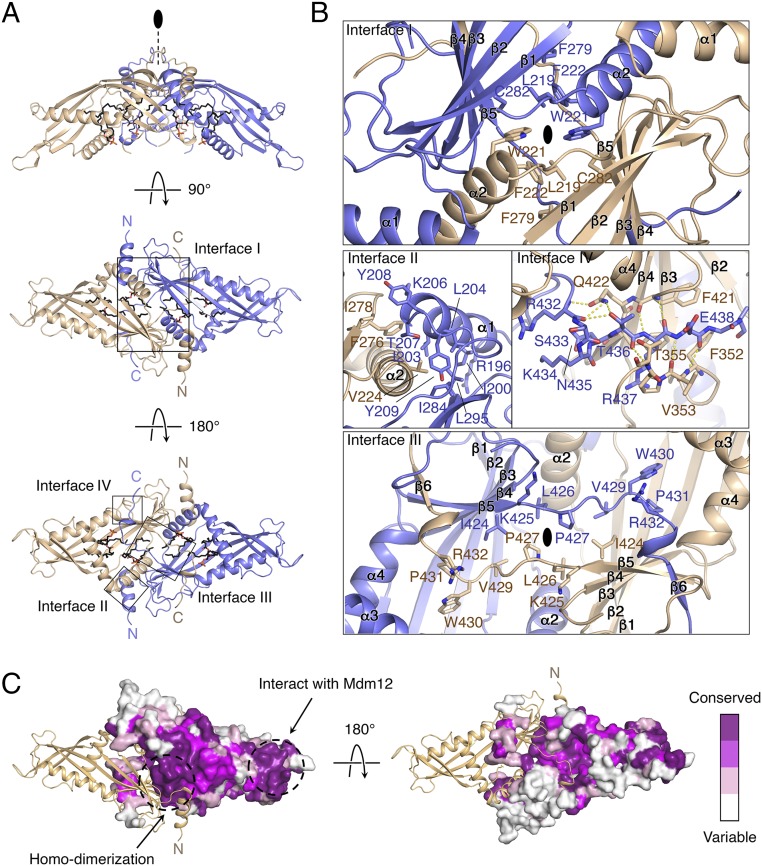
Crystals of zrMmm1 contained one zrMmm1 molecule in the asymmetric unit. However, zrMmm1 forms a tight dimer with a crystal symmetry-related molecule via a twofold rotation arrangement. The dimeric organization of zrMmm1 was confirmed by previous biochemical experiments, and is consistent with other SMP domain structures (20, 36–38). Overall, the dimeric zrMmm1 SMP structure resembles a compact diamond with dimensions of 50 × 60 × 120 Å, and each component consists of four helices and six extended and twisted antiparallel β-strands that assemble into a typical SMP structure with an extended hydrophobic channel (Fig. 2A and Figs. S1 and S2). In a previous study, we suggested that the N terminus (residues 198–214) of the Mmm1 SMP domain dimer might be involved in the twofold interface and might be structurally similar to that of E-SYT2 based on sequence similarity (20). Consistent with our prediction, the twofold interface of the zrMmm1 dimer is composed of two helices in a face-to-face arrangement reminiscent of that in the E-SYT2 structure (Fig. 2B, interface I and Fig. S3A). In particular, three hydrophobic residues (Leu219, Trp221, and Phe222) stabilize the twofold axis through van der Waals interactions.
Fig. 2.
Crystal structure of the zrMmm1 SMP domain. (A) Ribbon diagrams of zrMmm1 viewed in three orientations. The crystal structure of the SMP domain of zrMmm1 was determined by Se single-wavelength anomalous dispersion phasing and refined to 2.8 Å resolution. The protein adopts a dimeric SMP structure consisting of four helices and six strands in each monomer. Phospholipids bound to zrMmm1 are shown in black stick representation. Four dimeric interfaces for self-association are highlighted with black boxes. (B) Close-up view of the highlighted boxes (interfaces I–IV). Key residues that contribute to the self-association of zrMmm1 are shown in ball-and-stick representation. Oxygen and nitrogen atoms are colored red and blue, respectively. Yellow dotted lines indicate intermolecular H-bonds. (C) Molecular surface view of zrMmm1. The surface is colored according to sequence conservation from white (variable) to dark purple (conserved) as calculated by the Consurf server (consurf.tau.ac.il) (42) using 35 different yeast orthologs. To show the orientation of zrMmm1, one molecule of the zrMmm1 dimer is drawn in ribbon representation. Highly conserved regions indicated by dotted circles are essential for self-association or interaction with Mdm12.
Upon comparing the SMP domains of E-SYT2 and Mdm12, it was immediately apparent that two extra structural elements absent in the Mdm12 and E-SYT2 domains are present at the N and C termini of zrMmm1 (Fig. 2B and Fig. S3). These structural elements presumably make an important contribution to the tight association between subunits of the zrMmm1 dimer, since over 3,400 Å2 of solvent-accessible surface area is buried upon self-association. The N terminus of zrMmm1 adopts an α-helix (α1) and a well-ordered loop that contacts the head region of the other molecule of the dimer (interface II). In particular, the N-terminal helix comprising residues 196–207 wraps around the twofold axis helix of the opposing molecule in an antiparallel domain-swapped manner (Fig. 2B, interface II). The highly conserved C terminus of zrMmm1 exhibits a long, extended loop that crosses over the two molecules and essentially mediates the self-association of the zrMmm1 dimer, as well as phospholipid binding (Fig. 2 B and C, interface III). In more detail, the extended loop consisting of residues 425–432 forms an antiparallel β-strand–like strap structure that zips up the opposing twofold central helices, and eventually covers the concave surface at the center of the dimeric SMP domain (Fig. 2B, interface III). This loop also contains the absolutely conserved Trp430 and Arg432 residues that are essential for the recognition of phospholipids, as discussed below. Additionally, the C terminus of zrMmm1 adopts a short 310 helix (residues 433–435), followed by antiparallel β-strands, and is incorporated between β5 and an 11-residue loop (residues 347–357) from the opposing molecule of the dimer through the formation of an extensive hydrogen-bonding network (Fig. 2B, interface IV).
In summary, the extensive interfaces that are lacking in E-SYT2 and Mdm12 provide the driving force for the tight self-association observed in the zrMmm1 dimer. Consistently, SEC and native PAGE revealed that the dynamic distribution between monomer and dimer observed for Mdm12 and the SMP domain of E-SYT2 was not a feature of zrMmm1 (20).
The zrMmm1 Dimer Binds Glycerophospholipids.
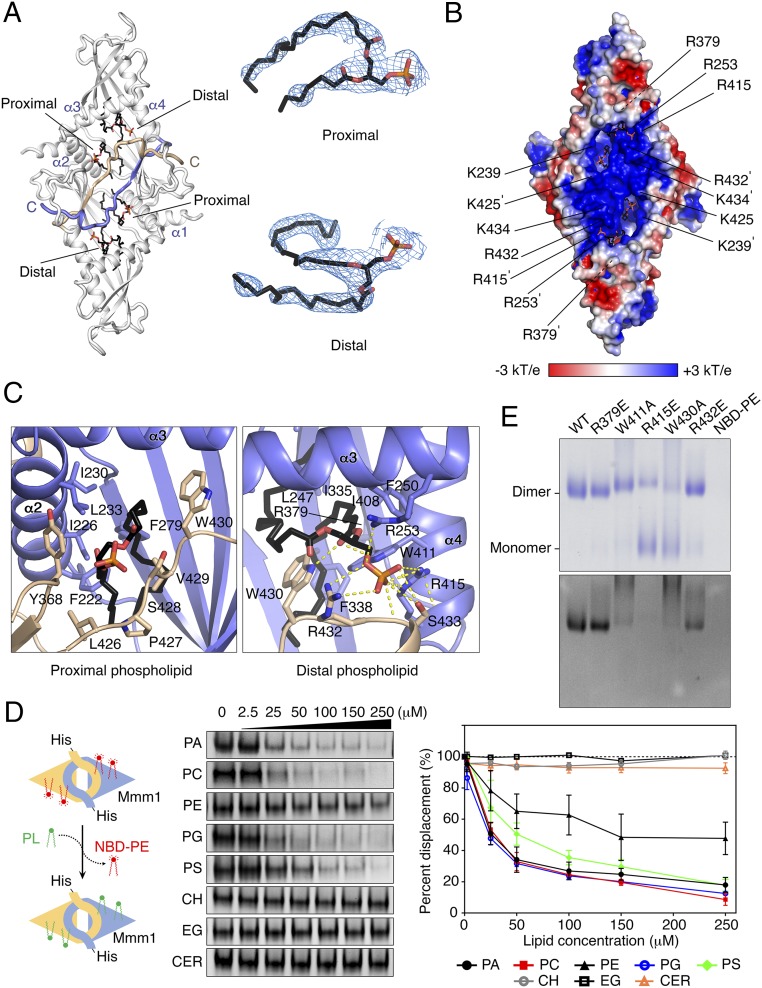
The crystal structure revealed that recombinant zrMmm1 expressed in bacteria contained glycerophospholipids bound in the hydrophobic channel formed from the SMP domain (Fig. 3A). Based on the observed electron density, we concluded that two glycerophospholipids were bound to each zrMmm1 molecule in two distinct regions: One phospholipid binds at the dimeric interface (proximal), and the other molecule is located in the middle (distal) part of the SMP channel. As mentioned above, the zrMmm1 dimer formed from symmetry-related molecules in the crystal, and the two phospholipids superimposed precisely over the two molecules of the zrMmm1 dimer, suggesting that the phospholipids are specifically recognized by zrMmm1 and were not the result of nonspecific binding. The head groups of two glycerophospholipids are located within a concave surface generated by helices α2–α4, and are solvent-exposed and disordered in the structure, suggesting that zrMmm1 does not possess clear selectivity for particular phospholipids, consistent with Mdm12 and E-SYT2 (20, 38) (Fig. 3 B and C and Fig. S3). However, unlike in other SMP domain proteins, the phosphate group and carboxyl oxygen of the distal phospholipid can be clearly seen in the structure, and are systematically coordinated by the conserved Arg253, Arg415, Trp411, Trp430, Arg432, and Ser433 through an extensive hydrogen-bonding network (Fig. 3C). Among these, three residues (Trp430, Arg432, and Ser433) are from the opposing molecule in the dimer, suggesting that lipid coordination in zrMmm1 requires homodimerization.
Fig. 3.
zrMmm1 binds to glycerophospholipids. (A, Left) Overall structure of zrMmm1 (gray) bound to two phospholipids (black) viewed from the concave surface of the SMP domain. One molecule of zrMmm1 binds two phospholipids in two distinct regions, referred to as proximal and distal phospholipids (details are provided in the main text). Highly conserved C-terminal loops in the zrMmm1 dimer that are important for specific and tight lipid conjugation are colored yellow and blue. (A, Right) Molecular structures of the two phospholipids bound to zrMmm1 are shown with Fo-Fc electron density difference maps calculated in the absence of phospholipids (2.8 Å resolution, contoured at 2.0 σ). (B) Electrostatic surface representation of zrMmm1 viewed in the same orientation as in A. The electrostatic potential was calculated with the APBS program (39), and colored from −3 (red) to +3 (blue) kT/e (k, Boltzmann’s constant; T, temperature; e, charge of an electron). (C) Ribbon diagram showing a close-up view of the coordination of bound phospholipids (black) by the SMP domains of the zrMmm1 dimer (blue and yellow). The dimeric organization of zrMmm1 is clearly essential for the specific interactions with the phosphate ion of the distal phospholipid. (D) In vitro phospholipid displacement experiment using fluorescently labeled NBD-PE (details are provided in Materials and Methods). (Left and Center) NBD-PE preloaded His-zrMmm1 was incubated with natural phospholipid ligands (PA, PC, PE, PG, and PS) and nonphospholipid ligands (CER, ceramide; CH, cholesterol; EG, ergosterol) at increasing concentrations, and the quantity of NBD-PE displaced by natural ligands was measured as the diminishment in NBD-PE fluorescence. (Right) Graph indicates quantification data. Experiments were performed in triplicate. Means ± SD are shown. (E) To probe interactions between wild-type (WT) or mutant (R379E, W411A, R415E, W430A, and R432E) zrMmm1 and phospholipids, proteins indicated in each lane were incubated with NBD-PE for 2 h on ice. Mixtures were separated by blue native PAGE, and binding was analyzed by Coomassie staining (Top) and fluorescence detection (Bottom).
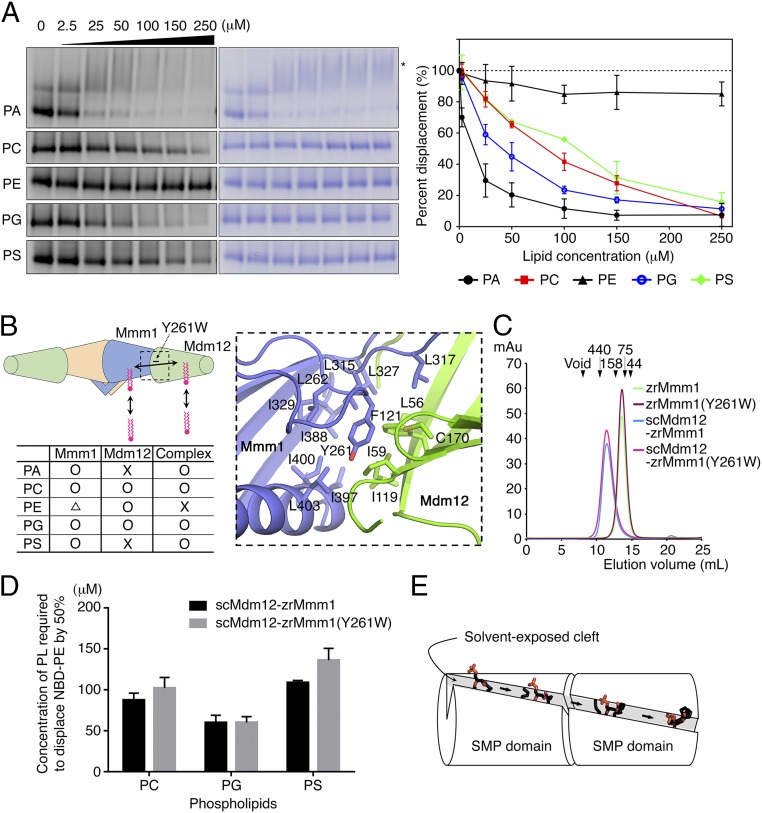
To examine if zrMmm1 shows preferential binding to certain phospholipids in solution, we performed lipid displacement experiments using 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-,3-benzoxadiazol-4-yl) (NBD)-PE, as reported in our previous study (20). First, we confirmed the binding between NBD-PE and purified zrMmm1 using native PAGE and fluorescence detection (Fig. 3D), and found that NBD-PE bound to zrMmm1 could be easily displaced by phosphatidylglycerol (PG), phosphatidic acid (PA), PS, or phosphatidylcholine (PC), but only relatively weakly by PE, even at high concentrations (Fig. 3D). However, the NBD-PE on Mmm1 could not be displaced by the nonphospholipid cholesterol, ergosterol, or ceramide, even at high concentrations (Fig. 3D). Based on these results, we conclude that zrMmm1 can bind efficiently to any glycerophospholipid. A previous structural study suggested that Mdm12 binds preferentially to PC or PE, both of which have a positively charged head group in common, via their negatively charged surfaces (20). Analysis of the electrostatic surface potential of zrMmm1 using the Adaptive Poisson–Boltzmann Solver (APBS) program (39) revealed a strong positively charged region in the vicinity of the bound phospholipid head group (Fig. 3B). Unlike Mdm12, the positively charged residues of zrMmm1 might be critically responsible for screening phospholipids themselves, not for the selection of certain head groups of phospholipids.
Next, we mutated key residues involved in lipid coordination and measured binding between zrMmm1 mutants and NBD-PE using blue native PAGE and fluorescence methods. As shown in Fig. 3E, R415E, W411A, and W430A variants completely lost the ability to bind NBD-PE, while the negative control R379E could still bind NBD-PE. Interestingly, two bands consistent with the monomer and dimer of zrMmm1 were observed with the R415E and W430A mutants, supporting our structural analysis and conclusion that self-association of zrMmm1 is required for lipid conjugation, and suggesting that lipid binding might enhance the stability of the dimeric form.
Structure Determination of the Mdm12–Mmm1 Complex.
Mmm1 specifically interacts with the Mdm12 component of the ERMES complex (19, 20). In our previous study, we proposed a putative model for the Mdm12–Mmm1 complex involving dimerization via the SMP domains in a tail-to-tail manner. In this model, the conserved long C-terminal helices of the SMP domains lie adjacent to each other in a twofold rotation arrangement, resulting in an extended arch-shaped structure (20). However, one of the concerns raised from this model was the lack of direct evidence for the tail-to-tail junction, and contacts between the self-associated Mdm12 molecules could be an artifact of crystallization (i.e., the result of crystal contacts rather than physiologically relevant molecular interfaces). Additionally, the potential interface between Mdm12 and Mmm1 in this model is exposed to solvent, suggesting that it is energetically unfavorable for hydrophobic glycerophospholipids to cross the solvent region in the Mdm12 and Mmm1 interface.
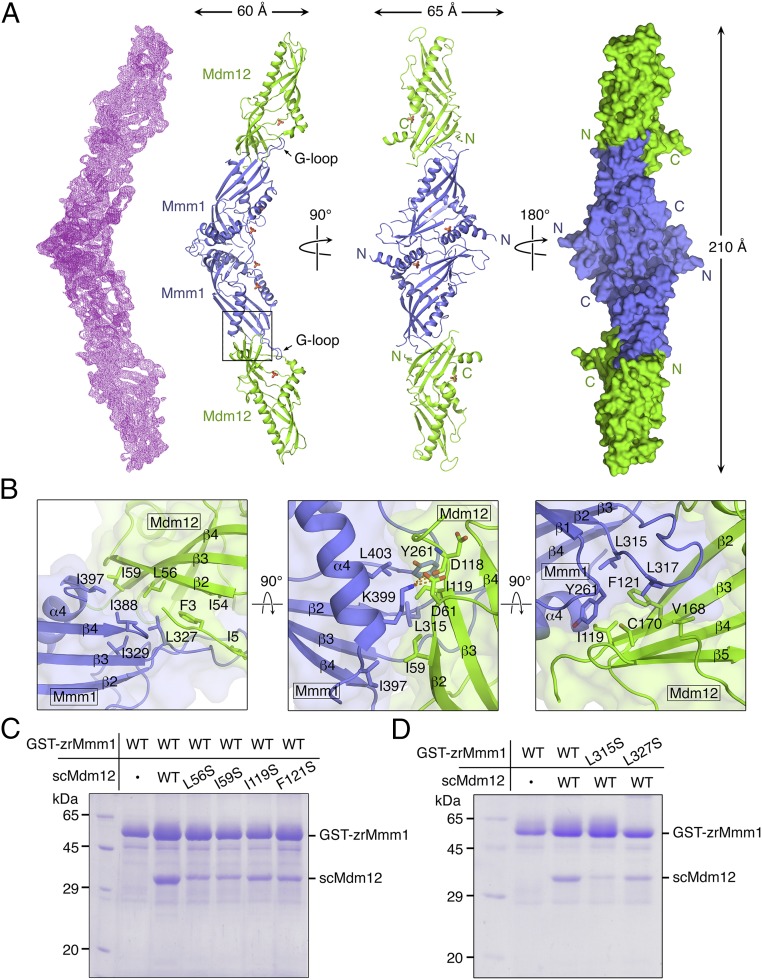
To further investigate how phospholipids could be transferred through the SMP domains of Mdm12 and Mmm1, we determined the crystal structure of the Mdm12–Mmm1 complex. Initially, we obtained crystals of the S. cerevisiae Mdm12–Mmm1 complex and hybrid complex of scMdm12–zrMmm1, but all were of low crystallographic quality. Through extensive screening, we eventually obtained diffraction-quality crystals of truncated scMdm12Δ, in which both the unstructured loop (residues 74–114) and proline-rich region (residues 184–211) were excluded, in complex with zrMmm1 (Fig. 1A). The ability of scMdm12Δ to interact with zrMmm1 was assessed by SEC experiments (Fig. 1B). However, crystals only diffracted to low resolution (∼5 Å). To overcome this, we attempted dehydration of crystals using a higher percentage of precipitant, and the diffraction quality was dramatically improved (details are provided in Materials and Methods). Dehydrated crystals of the scMdm12Δ–zrMmm1 complex diffracted to 3.8 Å synchrotron radiation, and the structure was determined by the molecular replacement method. Crystals contained one heterotetramer organized in an scMdm12Δ-zrMmm1-zrMmm1-scMdm12Δ arrangement in the asymmetric unit (Fig. 4A). The Mdm12 modification needed for crystallization did not affect the overall structure or binding to Mmm1 compared with wild-type Mdm12 (rmsd of 1.5 Å for all Cα atoms). The overall conformation of zrMmm1 and scMdm12Δ was not significantly changed upon formation of the complex (rmsd of 0.9 Å and rmsd of 1.5 Å, respectively). No apparent electron density corresponding to the hydrocarbon chain of glycerophospholipids was observed in the complex structure except for the phosphate group of phospholipids, but this might be due to the relatively low resolution of the complex structure or to treatments such as crystal dehydration.
Fig. 4.
Overall architecture of the scMdm12Δ–zrMmm1 complex. (A) Figures showing the overall architecture of the scMdm12Δ (green)–zrMmm1 (blue) complex. The structure of the scMdm12Δ–zrMmm1 complex was determined by the molecular replacement method and refined to 3.8 Å resolution. The 2Fo-Fc electron density map (Left, calculated with data to 3.8 Å resolution and contoured at 1.0 σ) and the surface representation of the crystallographic asymmetric unit of the scMdm12Δ–zrMmm1 complex (Right) are shown. Phosphate ions are shown as ball-and-stick models with red for oxygen and orange for phosphorus atoms. (B) Binding interface between zrMmm1 and scMdm12Δ in three orientations. Residues involved in the interaction are shown in ball-and-stick representation. (C) Role of scMdm12 residues in the interaction with zrMmm1 assessed through GST pull-down experiments using scMdm12 mutants (L56S, I59S, I119S, and F121S). (D) SDS/PAGE showing the results of a reciprocal test of the effect of mutations in zrMmm1 (L315S and L327S) on the interaction with scMdm12. WT, wild type.
Architecture and Organization of the scMdm12Δ–zrMmm1 Complex.
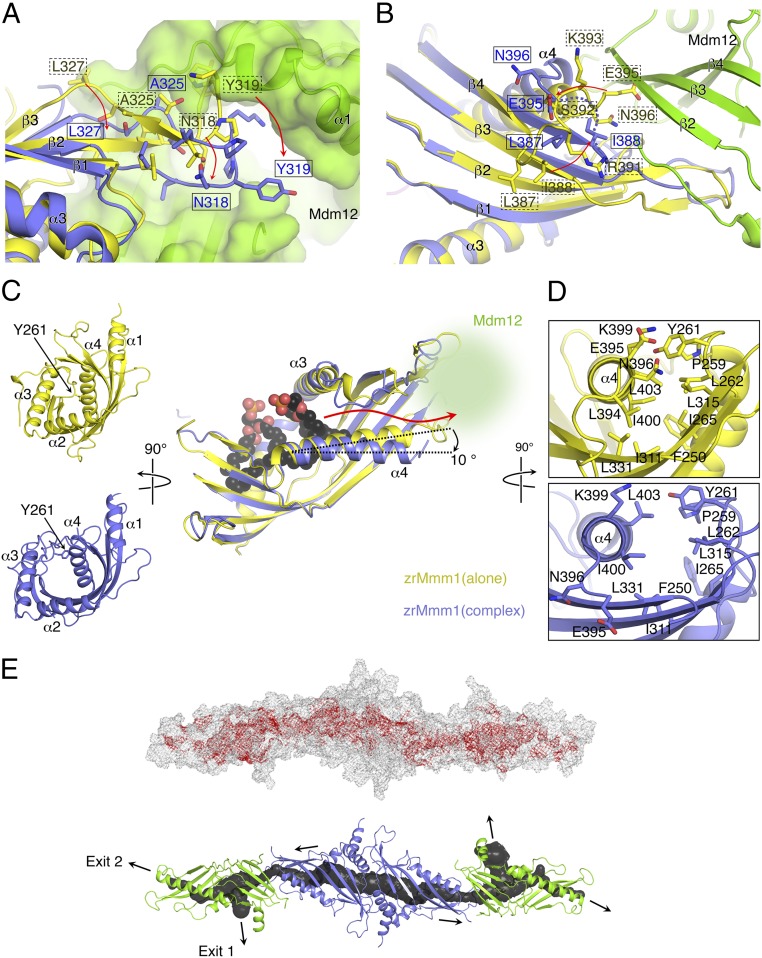
The overall structure of the scMdm12Δ–zrMmm1 complex closely resembles the EM structure described in a previous study (19) (Fig. S4A). The scMdm12Δ–zrMmm1 complex adopts an elongated curved and tubular structure with dimensions of 60 × 65 × 210 Å. The zrMmm1 dimer is located at the center, with scMdm12Δ monomers bound at each end (Fig. 4A and Fig. S4A). Consistent with the previously reported model (19), scMdm12Δ and zrMmm1 are organized in a head-to-tail manner, with the N terminus of scMdm12Δ (referred to as the head) that is proximal to the dimeric interface in the scMdm12 dimer associating with the distal end (referred to as the tail) of the homodimeric interface of the zrMmm1 SMP domain. The interaction between scMdm12Δ and zrMmm1 appears to be strong, and buries 1,012 Å2 of surface-accessible surface area. The truncated residues of the unstructured loop and proline-rich region of Mdm12 are not involved in the interaction. In the crystal structure of Mdm12 alone, the N terminus (residues 1–7) adopts a β-strand that is involved in self-association by forming a domain-swapped structure with the opposing molecule of the dimer (20). However, no such conformation of Mdm12 was observed in the complex structure. Rather, the N terminus of scMdm12Δ forms an extended loop structure and lies adjacent to the β2 strand of scMdm12Δ itself.
The highly conserved β2 and β3 strands, the extended hairpin loop [referred to as the guide loop (G-loop)] generated between β2 and β3, and the α4 helix of zrMmm1 contribute to interactions with the β2 and β3 strands of scMdm12Δ (Fig. 4B). In particular, the hydrophobic amino acids Leu315, Leu317, Leu327, Ile388, and Ile397 in zrMmm1 form extensive and coordinated nonpolar contacts with the side chains of Phe3, Ile5, Leu56, Ile59, Ile119, Phe121, and Cys170 of scMdm12Δ (Fig. 4B). In addition, Lys399 of zrMmm1 forms a salt bridge and H-bonds with the side chain of Asp61 and the main chain of Asp118 of scMdm12Δ. To confirm whether these residues are involved in the interaction, we generated a series of zrMmm1 mutants and scMdm12 proteins (with GST fused at the N terminus of zrMmm1) and examined their binding ability using GST pull-down experiments. Single-residue mutants of scMdm12 (L56S, I59S, I119S, and F121S) lost appreciable affinity for zrMmm1 (Fig. 4C). Likewise, single-site mutants of zrMmm1 (L315S or L327S) interacted with scMdm12 in a less stable manner (Fig. 4D). Furthermore, to confirm the effect of the L315S mutation in solution, we titrated purified native and L315S mutant tag-free zrMmm1 proteins with purified scMdm12 over a wide protein concentration range and analyzed their interactions using native PAGE. As shown in Fig. S4B, wild-type zrMmm1 interacted with scMdm12 and formed a heterotetramer in a concentration-dependent manner, while the L315S mutant did not interact with scMdm12 at even higher concentrations, suggesting that the observed hydrophobic contacts are critical for the Mdm12–Mmm1 interaction.
The scMdm12Δ–zrMmm1 Complex Has an Extended Hydrophobic Tunnel Mediating Lipid Trafficking.
Structural comparison between zrMmm1 and scMdm12 alone, and as part of the scMdm12Δ–zrMmm1 complex, revealed that the structure of zrMmm1 was changed slightly upon complex formation. Interestingly, the structural changes appear to be functionally relevant regarding phospholipid trafficking between the two distinct SMP domains. First, the G-loop of zrMmm1 undergoes a conformational change to form a more extended form that can plug into the scMdm12Δ head region and completely covers the solvent-exposed concave surface of scMdm12Δ (Fig. 5A). Second, the β4 strand of zrMmm1 is extended by two residues (Leu387 and Ile388) in the complex, and these residues are part of a flexible loop and are solvent-exposed in the structure of zrMmm1 alone. By interacting with scMdm12Δ, Ile388 is projected inward toward the center of the SMP domain and contributes to the formation of a hydrophobic boundary at the junction of the two SMP domains (Fig. 5B). Third, the conserved loop formed between β4 and α4, which are well ordered in the structure of zrMmm1 alone, becomes disordered upon forming a complex with scMdm12Δ. In particular, three hydrophilic residues (Arg391, Ser392, and Lys393) are not visible in the scMdm12Δ–zrMmm1 complex (Fig. 5B). Finally, the α4 helix of zrMmm1 and the loop formed between α3 and β1 are pushed outward, generating a wider space inside the cavity that might be important for phospholipid trafficking (Fig. 5 C and D). Taken together, the formation of the scMdm12Δ–zrMmm1 complex generates a continuous hydrophobic tunnel ∼210 Å long through the elongated SMP domains of scMdm12Δ and zrMmm1, which could conceivably translocate phospholipids harboring nonpolar hydrocarbon chains between two components without consuming energy (Fig. 5E). These results strongly indicate that the Mdm12–Mmm1 complex acts as a lipid-transferring vehicle in addition to tethering molecules to physically connect two distinct subcompartments.
Fig. 5.
Direct association of zrMmm1 and scMdm12Δ generates a hydrophobic tunnel for phospholipid trafficking. (A) Ribbon diagram showing superposition of zrMmm1 (yellow) and the scMdm12Δ (green)–zrMmm1 (blue) complex. To analyze structural changes in zrMmm1 upon association with scMdm12Δ, the structure of zrMmm1 was aligned with the zrMmm1 structure in the scMdm12Δ–zrMmm1 complex. The scMdm12Δ is shown in surface representation. The G-loop of zrMmm1 undergoes conformational changes following interaction with scMdm12Δ, forming an extended structure that covers the solvent-exposed region of scMdm12Δ. Residues of zrMmm1 undergoing this structural reorganization are shown, and their directions are indicated with red arrows. (B) Structural changes in zrMmm1 occurring upon association with scMdm12Δ further highlighted (more information is provided in the main text) in a diagram colored the same as in A. The dotted line indicates zrMmm1 residues that become disordered upon forming the complex. (C, Right) Direct association of zrMmm1 and scMdm12Δ moves the α4 helix of zrMmm1 by ∼10° outward, vacating enough space to accommodate and transfer phospholipids. Phospholipids bound to zrMmm1 are shown in surface-filling representation. The red arrow indicates the putative pathway of phospholipids from zrMmm1 to scMdm12Δ. (C, Left) Ribbon diagrams compare the overall structure of zrMmm1 in the apo (yellow) and complexed (blue) forms viewed from the left side of the figure (C, Right). Loops, including Tyr261, in the complexed form are shifted outward, generating an open space in the process. The scMdm12Δ is omitted for clarity. The overall color scheme is the same as in A. (D) Structures of zrMmm1 in the apo (yellow) and complexed (blue) forms viewed from the right side of the picture (C, Right). (E) Overall structure of the scMdm12Δ–zrMmm1 complex shown in meshed line (Top) and ribbon (Bottom) representations. (Top) Red mesh representing hydrophobic amino acids inside the tunnel was superimposed on the figure. (Bottom) Channel (cavity) through the scMdm12Δ–zrMmm1 complex was analyzed by Mole 2.0 (43), and is shown in black tubule representation. Black arrows indicate the putative pathway for phospholipid trafficking.
The scMdm12–zrMmm1 Complex Binds All Glycerophospholipids Except for PE in Vitro.
To identify differences in binding priority to phospholipids between the scMdm12–zrMmm1 complex and zrMmm1 or scMdm12 alone, we performed a lipid displacement experiment using the scMdm12–zrMmm1 complex. Interestingly, NBD-PE bound to the scMdm12–zrMmm1 complex could be displaced only by PA, PG, PC, or PS (Fig. 6A). In the case of PA, high concentrations resulted in band shifts above those of the NBD-PE preloaded scMdm12–zrMmm1 complex alone on native PAGE. No such changes have been observed using NBD-PE–preloaded scMdm12 alone (20). However, high concentrations of PA also resulted in similar band shifts of NBD-PE–preloaded zrMmm1 alone, indicating that PA binding to zrMmm1 might affect the overall conformation of zrMmm1 or the scMdm12–zrMmm1 complex.
Fig. 6.
The scMdm12–zrMmm1 complex does not bind PE in vitro, and acts as a lipid transfer module. (A) In vitro phospholipid displacement experiments using the scMdm12–zrMmm1 complex. NBD-PE–preloaded scMdm12–zrMmm1 complexes were mixed with increasing concentrations of phospholipids (PA, PC, PE, PG, and PS). Decreasing fluorescence was used to measure NBD-PE displacement by each phospholipid. (Left) Fluorescence and Coomassie staining of clear-native PAGE gels are shown side by side. (Right) Graph shows quantification data. Experiments were performed in triplicate, independently. Means ± SD are given. (B, Left) Schematic diagram shows possible routes for phospholipid access to Mdm12 or Mmm1 in the Mdm12–Mmm1 complex. The table below shows a summary of the results of the phospholipid displacement experiment using Mmm1, Mdm12–Mmm1 complex (from this study), and Mdm12 (20). The symbols X, △, and ○ indicate that the fluorescence reduction rate is within the range of 0–35%, 35–70%, and 70–100% at 250 μM, respectively, of each phospholipid. (B, Right) Ribbon diagram highlights the role of the zrMmm1 Y261 residue at the interface between scMdm12Δ and zrMmm1. (C) SEC analysis shows that the Y261W mutant of zrMmm1 can still associate with scMdm12. Molecular weight standards are indicated above the chromatogram. mAu, milliabsorbance unit. (D) In vitro phospholipid displacement experiment with the scMdm12–zrMmm1 complex (wild-type and Y261W mutant). The graph indicates the concentration of a phospholipid required to reduce the NBD-PE fluorescence signal by 50%. The bar graph shows means ± SD (n = 3). (E) Schematic representation highlights the role of the SMP domain in phospholipid transport. The SMP domains in the two distinct subunits directly associate with each other, generating a successive hydrophobic tunnel through which phospholipid transfer can occur from one subunit to the other.
One of the most striking differences between zrMmm1 and the scMdm12–zrMmm1 complex was the absence of scMdm12-zrMmm1 binding to PE (Fig. 6A). Even though both scMdm12 alone and zrMmm1 alone bound to PE with noticeable efficiency (20) (Figs. 3D and 6B), the scMdm12–zrMmm1 complex did not bind PE at all, suggesting that the association between scMdm12 and zrMmm1 affects the binding preferences of zrMmm1 and scMdm12 to phospholipids. Although the tests were performed using purified proteins in vitro, these results could have important biological implications. The PE component of the mitochondrial membrane might not be directly transferred from the ER but might be synthesized within the mitochondrial matrix via the conversion of PS to PE. Furthermore, the PE generated outside mitochondria via the Kennedy pathway might not be efficiently transferred to mitochondria for unknown reasons (40). Consistent with this, the scMdm12–zrMmm1 complex did not engage in PE binding in vitro.
PS transfer to mitochondria is required for the synthesis of PE in mitochondria. Because scMdm12 alone could not bind PS (20) (Fig. 6B), we inferred that the PS that displaced NBD-PE from scMdm12 in the scMdm12–zrMmm1 complex might have been directly transferred from zrMmm1. To verify this, we generated an Y261W mutant of zrMmm1. The Y261 residue is located at the interface between zrMmm1 and scMdm12 and is involved in generating a hydrophobic channel. However, the residue does not directly contribute to the interaction between scMdm12 and zrMmm1 (Figs. 5C and 6B). We hypothesized that the conversion of Tyr to Trp would sterically hinder the transfer of phospholipids between zrMmm1 and scMdm12. As expected, the mutation did not affect the association between scMdm12 and zrMmm1(Fig. 6C), and PS binding by the zrMmm1 (Y261W) mutant was similar to that of wild-type zrMmm1 (Fig. S5). However, in contrast to the wild type, the NBD-PE bound to the zrMmm1(Y261W)–scMdm12 complex was slowly displaced by PS (Fig. 6D), suggesting that the bulky side chain of Trp sterically impeded PS transfer from zrMmm1 to scMdm12 (Fig. 6E). We also tested whether the mutation affected the displacement of NBD-PE from the zrMmm1(Y261W)–scMdm12 complex by PC and PG, and observed that PC, but not PG, resulted in slightly slow displacement (Fig. 6D). Since scMdm12 alone could efficiently bind to PC and PG unlike PS (20) (Fig. 6B), the effect of the mutation might not be significant in vitro. In summary, from these observations, we confirmed that the direct association of SMP domains in the scMdm12–zrMmm1 complex generates a hydrophobic tunnel for lipid trafficking.
Discussion
SMP domains in ERMES and tubular lipid-binding superfamily complexes are believed to have a common role in binding and transferring lipids (41). However, molecular recognition of specific phospholipids by SMP domains is not conserved among SMP-containing proteins. For example, scMdm12 has a binding preference for phospholipids harboring positively charged head groups, while the SMP domain of zrMmm1 broadly binds to most phospholipids, although zrMmm1 preferentially binds to PS, PA, PG, and PC. In addition, our previous work revealed that scMdm12 binds one molecule of phospholipid (20), while the zrMmm1 SMP domain binds two phospholipids in distinct regions (Fig. S3B). Interestingly, the phosphate group of the distal phospholipid is specifically coordinated by conserved residues in zrMmm1 (Fig. 3C). Specifically, two pairs of Arg-Trp residues (Arg415/Trp411 and Arg432/Trp430 from the opposing molecule of the zrMmm1 dimer), which are absolutely conserved among other Mmm1 orthologs, form an extensive H-bonding network with the phosphate ion and carboxyl oxygen of the phospholipid (Fig. 3C). From this observation, we proposed that the Arg and Trp residues act as a filter for screening phospholipids among the pool of cellular lipids. This represents a unique feature of Mmm1 because most SMP domains bind hydrocarbon chains of phospholipids through nonpolar contacts with hydrophobic residues inside the cavity of the SMP domain.
Regarding phospholipid trafficking at the ER-mitochondria contact site, it is well established that PC is synthesized from PS via PE through the action of two enzymes that are distinctly located in the ER and mitochondria. The conversion of PS to PE is catalyzed by enzymes resident in mitochondria, whereas PA, an important intermediate in the formation of PG and cardiolipin in mitochondria, is synthesized in the ER (11). PS, PA, and PG must therefore be transferred from the ER, their site of synthesis, to mitochondria. Furthermore, PC synthesized in the ER must be eventually translocated to mitochondria for maintenance of membrane integrity. Because Mmm1 is the only ER resident protein among ERMES components, and since Mmm1 might be involved in phospholipid selection from the ER, the specific and favored recognition of phospholipids by Mmm1 might help to facilitate efficient lipid trafficking. In this study, we structurally and biochemically demonstrated that zrMmm1 alone and the scMdm12–zrMmm1 complex preferentially bind to phospholipids. This apparent selective extraction of phospholipids, facilitated by the surface charge and phospholipid filter of Mmm1, might be critical to the initiation of cooperative phospholipid synthesis at ER-mitochondria contact sites.
The proximal surfaces of membrane proteins are often positively charged, and we therefore suggest that the positively charged concave inner surface in the SMP domain of zrMmm1 might interact closely with the ER membrane. The concave structure of zrMmm1 might complement membrane curvature in terms of shape and size. In addition, the adjacent circumference of a positively charged patch composed of hydrophobic residues, including Y245, W238, P354, P357, and Y406, with the side chains of these residues exposed to the surface of zrMmm1, indicates that these residues might play a role in tight docking to the ER membrane (Fig. S6 A and B). Interestingly, we observed that unlike the head groups of phospholipids bound to Mdm12, which are distal from the concave surface of Mdm12, the head groups of phospholipids bound to zrMmm1 project into the concave surface of zrMmm1 (Fig. S6C). Moreover, the concave surface in the scMdm12–zrMmm1 complex precisely conforms to that generated by zrMmm1, strongly supporting the possibility that the concave inner surface of zrMmm1 binds to a convex membrane region.
Mmm1 interacts with Mdm34 through Mdm12 via relatively weak or transient interactions (19, 20). Additionally, we previously suggested that the N terminus of Mdm34 might be involved in the interaction with Mdm12 (20). Based on these findings, we propose two putative models for the organization of the ERMES complex. First, the N terminus of Mdm34 might interact with the N terminus of Mdm12 via β-strand swapping, as shown in the Mdm12 dimer (20). Second, the head of the Mdm34 SMP domain might interact with the tail of the Mdm12 SMP domain, as shown in the Mdm12–Mmm1 interaction (Fig. 4A). At present, it remains difficult to test these models because the interaction is likely to be transient. Interestingly, the scMdm12Δ–zrMmm1 structure demonstrates that it is possible to generate a continuous hydrophobic tunnel through both the head and tail of Mdm12 (Fig. 5E), suggesting that the head and tail of Mdm12 might interact directly with the head of Mdm34. Future work is required to address exactly how the SMP domain of Mdm34 is organized in the Mmm1–Mdm12–Mdm34 ternary complex.
In conclusion, the Mdm12–Mmm1 complex establishes a molecular basis for protein-mediated MCSs between the ER and mitochondria, and for phospholipid trafficking through the ERMES complex.
Materials and Methods
Plasmid Construction.
The DNA fragment encoding the SMP domain of Mmm1 (Z. rouxii, residues 190–444) was generated by PCR amplification from genomic DNA and cloned into the pET28b-SMT3 expression vector with BamHI and SalI restriction enzymes. To construct scMdm12Δ, residues 74–114 and residues 183–211 from full-length Mdm12 were substituted to GGSGG (E73-GGSGG-S115) and GG (D182-GG-S212), respectively, and cloned into the pCDF-duet vector with NdeI and XhoI. All mutants were generated by PCR-based mutagenesis, and mutations were confirmed by DNA sequencing.
Protein Expression and Purification.
All proteins in this study were expressed by transforming the expression plasmids into E. coli BL21 (DE3) bacterial cells. Cells were grown to an OD600 nm of ∼0.7 at 37 °C with vigorous shaking and induced overnight at 18 °C with 0.3 mM isopropyl-β-d-thiogalactoside. Cells were collected by centrifugation at 3,200 × g for 15 min; resuspended in buffer A containing 25 mM sodium phosphate (pH 7.8), 400 mM sodium chloride, and 10 mM imidazole; and flash-frozen in liquid nitrogen for later use. The zrMmm1 proteins were purified by Ni2+-immobilized metal affinity chromatography (Ni2+-IMAC). His6-SMT3 tags were removed by adding Ulp1 protease at a ratio of 1:1,000 (wt/wt), and proteins were dialyzed overnight against buffer B comprising 25 mM Tris⋅HCl (pH 7.5), 150 mM sodium chloride, and 5 mM β-mercaptoethanol at 4 °C. Digested proteins were passed through an Ni2+-chelating column a second time to remove SMT3 tags and undigested protein, followed by SEC with a Superdex 200 (16/60) column (GE Healthcare) preequilibrated with buffer C comprising 25 mM Tris⋅HCl (pH 7.5), 150 mM sodium chloride, and 5 mM DTT.
For the scMdm12Δ–zrMmm1 complex, pET28b-SMT3-zrMmm1 and pCDF-duet-scMdm12Δ plasmids were simultaneously transformed into E. coli BL21 (DE3) cells. The scMdm12Δ–zrMmm1 complex proteins were purified using Ni2+-IMAC. After Ulp1 digestion, proteins were further purified by HiTrap Q HP (GE Healthcare) and Superdex 200 columns in buffer C. Purified zrMmm1 and scMdm12Δ–zrMmm1 complex proteins were concentrated to 12.5 mg/mL and 5 mg/mL, respectively, using Amicon ultra-15 centrifugal filters (Merck Millipore), and were flash-frozen at −80 °C for later use.
For selenomethionine-substituted proteins, the zrMmm1 plasmid was transformed and expressed in the E. coli B834 (DE3) methionine auxotrophic strain. Cells were grown in M9 minimal media supplemented with l-selenomethionine, and proteins were purified as described above. Additional methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank staff of the 5C beamline at the Pohang Accelerator Laboratory for assistance with synchrotron facilities. This research was supported by the Cell Logistics Research Center (Grant 2016R1A5A1007318), the Basic Research Program (Grant NRF-2015R1D1A1A01058016), and the Global Ph.D. Fellowship Program Grant NRF-2014H1A2A1020322 (to H.J.) from the National Research Foundation of Korea. This work was also supported by the Institute for Basic Science (Grant IBS-R022-D1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank www.pdb.org (PDB ID codes 5YK6 and 5YK7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715592114/-/DCSupplemental.
References
- 1.Elbaz Y, Schuldiner M. Staying in touch: The molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Helle SC, et al. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Phillips MJ, Voeltz GK. Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol. 2016;17:69–82. doi: 10.1038/nrm.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: Molecular hubs for the regulation of mitochondrial biology. J Cell Sci. 2010;123:1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornmann B. The molecular hug between the ER and the mitochondria. Curr Opin Cell Biol. 2013;25:443–448. doi: 10.1016/j.ceb.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Lang A, John Peter AT, Kornmann B. ER-mitochondria contact sites in yeast: Beyond the myths of ERMES. Curr Opin Cell Biol. 2015;35:7–12. doi: 10.1016/j.ceb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Csordás G, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatsuta T, Scharwey M, Langer T. Mitochondrial lipid trafficking. Trends Cell Biol. 2014;24:44–52. doi: 10.1016/j.tcb.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Flis VV, Daum G. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb Perspect Biol. 2013;5:a013235. doi: 10.1101/cshperspect.a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 13.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci USA. 2011;108:14151–14156. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murley A, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stroud DA, et al. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011;413:743–750. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Belgareh-Touzé N, Cavellini L, Cohen MM. Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy. Autophagy. 2017;13:114–132. doi: 10.1080/15548627.2016.1252889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26:1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AhYoung AP, et al. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci USA. 2015;112:E3179–E3188. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong H, Park J, Lee C. Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex. EMBO Rep. 2016;17:1857–1871. doi: 10.15252/embr.201642706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012;125:49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima R, Endo T, Tamura Y. A phospholipid transfer function of ER-mitochondria encounter structure revealed in vitro. Sci Rep. 2016;6:30777. doi: 10.1038/srep30777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TT, et al. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13:880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elbaz-Alon Y, et al. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30:95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Hönscher C, et al. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev Cell. 2014;30:86–94. doi: 10.1016/j.devcel.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Lahiri S, et al. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 2014;12:e1001969. doi: 10.1371/journal.pbio.1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellenrieder L, et al. Separating mitochondrial protein assembly and endoplasmic reticulum tethering by selective coupling of Mdm10. Nat Commun. 2016;7:13021. doi: 10.1038/ncomms13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meisinger C, et al. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J. 2007;26:2229–2239. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J Cell Biol. 2003;163:503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao K, Klionsky DJ. Participation of mitochondrial fission during mitophagy. Cell Cycle. 2013;12:3131–3132. doi: 10.4161/cc.26352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böckler S, Westermann B. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell. 2014;28:450–458. doi: 10.1016/j.devcel.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 35.AhYoung AP, Lu B, Cascio D, Egea PF. Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies. Biochem Biophys Res Commun. 2017;488:129–135. doi: 10.1016/j.bbrc.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beamer LJ, Carroll SF, Eisenberg D. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- 37.Qiu X, et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat Struct Mol Biol. 2007;14:106–113. doi: 10.1038/nsmb1197. [DOI] [PubMed] [Google Scholar]
- 38.Schauder CM, et al. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510:552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bürgermeister M, Birner-Grünberger R, Nebauer R, Daum G. Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2004;1686:161–168. doi: 10.1016/j.bbalip.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Alva V, Lupas AN. The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim Biophys Acta. 2016;1861:913–923. doi: 10.1016/j.bbalip.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 42.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sehnal D, et al. MOLE 2.0: Advanced approach for analysis of biomacromolecular channels. J Cheminform. 2013;5:39. doi: 10.1186/1758-2946-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, editor. Methods in Enzymology. Vol 276. Academic; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.