Significance
The mechanisms that regulate organismal growth and coordinate it with the availability of nutrients were unknown until a few decades ago. We now know that one pathway—the mechanistic target of rapamycin (mTOR) pathway—is the major nutrient-sensitive regulator of growth in animals and plays a central role in physiology, metabolism, the aging process, and common diseases. This work describes the development of the mTOR field, from its origins in studies into the mechanism of action of the drug rapamycin to our increasingly sophisticated understanding of how nutrients are sensed.
Keywords: mTOR, rapamycin, nutrients, Rag GTPase, growth
Abstract
In my PNAS Inaugural Article, I describe the development of the mTOR field, starting with efforts to understand the mechanism of action of the drug rapamycin, which ∼25 y ago led to the discovery of the mTOR protein kinase. I focus on insights that we have contributed and on work that has been particularly influential to me, as well as provide some personal reflections and stories. We now appreciate that, as part of two distinct complexes, mTORC1 and mTORC2, mTOR is the major regulator of growth (mass accumulation) in animals and is the key link between the availability of nutrients in the environment and the control of most anabolic and catabolic processes. Nutrients signal to mTORC1 through the lysosome-associated Rag GTPases and their many regulators and associated cytosolic and lysosomal nutrient sensors. mTOR signaling is deregulated in common diseases, like cancer and epilepsy, and mTORC1 is a well-validated modulator of aging in multiple model organisms. There is significant excitement around using mTORC1 inhibitors to treat cancer and neurological disease and, potentially, to improve healthspan and lifespan.
It is hard to believe that it has been almost 25 y since I developed the silver-stained gel showing I had purified mTOR. At the time, I was an MD-PhD student in Solomon H. Snyder’s laboratory at the Johns Hopkins Medical School, and I had no idea that mTOR would become a quasi-obsession for me and that I would still be working on it to this day. Nor would I have believed that the mTOR pathway would receive the recognition it has and eventually even be derided for “doing everything.” In fact, when I started my laboratory at the Whitehead Institute and Massachusetts Institute of Technology (MIT), I was advised to stop working on mTOR—advice that I seriously considered, but ultimately ignored because I did not know how to do anything else.
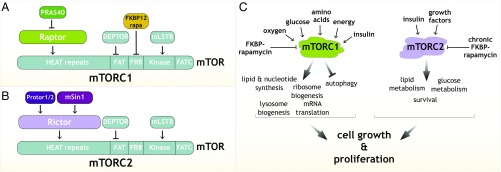
We now appreciate that the mTOR protein kinase is the key to answering one of the central questions in biology: How do organisms regulate their growth (mass accumulation) and coordinate it with the availability of nutrients? mTOR, as the catalytic subunit of two distinct protein complexes, mTORC1 and mTORC2, is the major regulator of growth in animals and controls most anabolic and catabolic processes in response to nutrients and nutrient-induced signals, like insulin (Fig. 1). As such, it plays critical roles in physiology, metabolism, and aging, and is deregulated in common diseases, including cancer and epilepsy (reviewed in ref. 1). Pharmacological and genetic data show that inhibition of mTORC1 increases lifespan in multiple model organisms, making it the best-validated aging regulator, and raising the tantalizing possibility that suppressing it might promote healthspan and longevity in humans. mTOR inhibitors are already used as immunosuppressive and anticancer agents in the clinic and are in development for many more indications, including the rejuvenation of tissues such as the immune system (2).
Fig. 1.
(A and B) Established components of mTORC1 (A) and mTORC2 (B). (C) Schematic showing the signals sensed by mTORC1 and mTORC2 and the processes they regulate to control growth.
I decided to use my PNAS Inaugural Article to write about the development of the mTOR field and to provide some personal recollections that highlight work that has been particularly influential to me. I suppose one writes such pieces when one has been around for a while. This appears to be the case, even though I am still surprised when someone refers to me as senior or I am asked by young scientists to talk about my career.
In the fall of 1992, I went to see Sol Snyder about a thesis project. I remember the meeting well, as I would meet with Sol one-on-one very few times during my time in his laboratory. Sol sat in a comfy office chair in the balled-up way that those of us in his laboratory found impossible to mimic, and he was quiet, knowing the power of silence (we assumed it was a trick he learned during his psychiatry training). I was nervous and blurted out that I wanted to talk about potential projects. After a bit, he said, “Well, David, we work on the brain.” That seemed like a great start, as I wanted to do neuroscience, but then more silence followed, and, as I was to learn, that meant the conversation was over. I left unsettled because the brain was obviously a big topic, meaning I was project-less. That conversation though was likely the most important scientific interaction of my career, as Sol was giving me the freedom to do whatever I wanted, which allowed me to develop my own research direction at a relatively young age. I never did end up working on the brain, but I do take some comfort in having originally purified mTOR from brains.
At that time, others in the laboratory were studying the effects of the immunosuppressant FK506 on neurons and using a structurally related molecule, rapamycin, as a negative control. We were fortunate to have rapamycin because, back then, it was not commercially available, and Sol had obtained it from Suren N. Sehgal at Wyeth-Ayerst (Fig. 2A). Sehgal—widely considered the father of rapamycin and its unrelenting champion until his death in 2003—had purified the compound in 1975 from bacteria found in soil collected on Easter Island (3). Sehgal had very kindly sent us a large amount, but just as importantly, he had also sent a book titled “Rapamycin Bibliography” with a little note wishing us luck. That book became my inspiration. It consisted mostly of abstracts describing the remarkable antifungal, immunosuppressive, and anticancer effects of rapamycin. It was clear that rapamycin inhibited the proliferation of a wide variety of cells ranging from lymphocytes and cancer cells to various species of yeast and preferentially delayed the G1 phase of the cell cycle (3–5). I had just finished the first 2 y of medical school and had learned how the immunosuppressant cyclosporin A was revolutionizing organ transplantation. At that point, I still thought I was going to be a practicing physician, so the medical applications of rapamycin were exciting to me and inspired me to determine how rapamycin works.
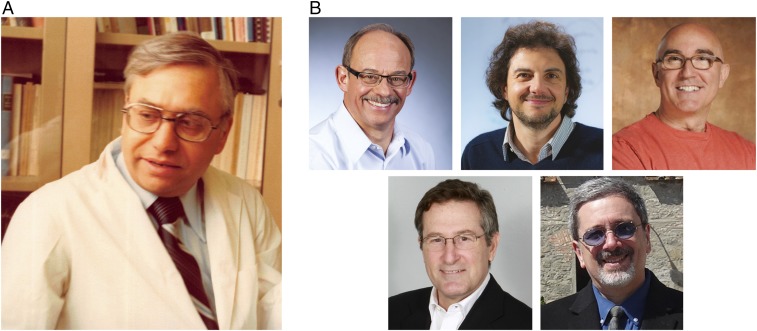
Fig. 2.
(A) Photograph of Suren Sehgal, the father of rapamycin. (B) Photographs of the discoverers of mTOR and TOR1/2. (B, Upper) Robert T. Abraham (Left), David M. Sabatini (Center), and Stuart L. Schreiber (Right). (B, Lower) Michael N. Hall (Left) and George P. Livi (Right). Ajai Sehgal kindly provided the photograph of Suren Sehgal.
Early insights into the mechanism of action of rapamycin came from the finding that FK506 and rapamycin inhibit different aspects of immune cell function and antagonize each other when given at the same time, suggesting that, despite their structural similarity, they have distinct targets (6–8). Stuart L. Schreiber made a major advance when he discovered that FK506 and rapamycin bind to the same site on FKBP12 (8), the founding member of the FKBP family of immunophillins (proteins that bind immunosuppressants). He proposed that both molecules act through a gain-of-function mechanism in which FKBP12–FK506 and FKBP12–rapamycin bind to distinct protein targets. Soon thereafter, he found that FKBP12–FK506 inhibits the phosphatase Calcineurin, which dephosphorylates and hence activates the T-cell transcription factor NFATc, and thus explained the cell biological and signaling effects of FK506 (9).
We had a saying in the Snyder laboratory that you could not purify what you did not know existed, which meant I needed an assay to detect the target of FKBP–rapamycin during its purification. Sol had pioneered the use of radiolabeled ligands to characterize neurotransmitter receptors, and a senior student, David S. Bredt, had recently purified nitric oxide synthase using an assay containing labeled arginine to track its activity (10, 11). Radioactivity-based assays being on my mind, I made versions of FKBP12 and FKBP25 (which because of its high affinity for rapamycin was also considered a potential mediator of its effects) that I could label with radioactive phosphate. I mixed the labeled proteins with tissue extracts, added a chemical cross-linker, and analyzed the samples by SDS/PAGE and autoradiography, the hope being that the small FKBP proteins would migrate at higher molecular weights when they became cross-linked to a protein target(s) in the presence of rapamycin. At first, I found nothing, but after I diluted and fractionated brain extracts, I was able to cross-link, in a rapamycin-dependent fashion, FKBP12, but not FKBP25, to two proteins that clearly cofractionated with each other: a large one I named RAFT1 (rapamycin and FKBP12 target 1) and a smaller one I called RAFT2. I never obtained sufficient amounts of RAFT2 to identify it, but I did manage to purify enough RAFT1 for our collaborator Paul Tempst to sequence—a nontrivial task in those days—several peptides derived from it, which enabled me to eventually clone its ∼9-kb cDNA (12). Around the same time, Stuart Schreiber identified the same protein, and named it FRAP (FKBP–rapamycin associated protein) (13), as we both wanted to emphasize that it bound rapamycin only in the presence of FKBP12. A few months later, Robert T. Abraham also reported the same protein, and called it mTOR (14), the name that, after some haggling and back and forth with the HUGO Gene Nomenclature Committee, most of us now use.
Abraham coined the mTOR name because sequence analysis showed that RAFT1/FRAP/MTOR shares homology, particularly in its kinase domain, to the proteins encoded by the TOR1/DRR1 and TOR2/DRR2 genes of budding yeast discovered about a year earlier by Michael N. Hall and George P. Livi, working independently. While Schreiber, Abraham, and I had taken a biochemical approach to identify the physical target of rapamycin, Hall and Livi had used genetics to identify genes that impact the rapamycin sensitivity of yeast. Consistent with Schreiber’s gain-of-function model, they found in genetic screens that loss of the yeast homolog of FKBP12 made cells resistant to rapamycin (15, 16). In the same paper, Hall also reported two additional rapamycin-resistant mutants that he called TOR1 and TOR2 (target of rapamycin 1 and 2) (16), and he went on to isolate and sequence the TOR2 gene (17), the first TOR gene identified in any system, followed soon thereafter by his characterization of TOR1 (18). Livi also discovered the same genes, but called them DRR1 and DRR2 (dominant rapamycin resistance 1 and 2) (19). That biochemical and genetic studies in distinct systems converged on clearly homologous gene products gave great confidence that mTOR/TOR was the pharmacologically relevant target of rapamycin and laid the foundation for much of the work that followed. Fig. 2B contains photographs of those who discovered mTOR and TOR1/2.
It is unfortunate that Livi is rarely recognized for his early contributions to the TOR field, perhaps because his names for TOR1 and TOR2 did not become popular. I recently had the pleasure of speaking with him—the first time we have interacted—and enjoyed hearing about his early efforts at SmithKline Beecham to understand the mechanism of action of rapamycin. Hall continues to be a pioneer of the field, and I am happy to consider him a friend and gracious colleague. In 2001, we co-organized in the south of France the first meeting focused on mTOR/TOR and repeated it every few years for >10 y. These meetings led to many collaborations and memorable adventures, including one where Hall and I became lost in a forest and a search party was dispatched, but not before I had an unfortunate encounter with an electric fence.
During my early work on mTOR, I was clueless about scientific competition and politics, and I am not sure I would have pursued the purification of mTOR had I known Schreiber was doing so as well. Anyone even a bit sophisticated would have known that his laboratory was seeking the rapamycin target, but it did not even cross my mind, and in retrospect, I was fortunate that our respective papers on mTOR were published at the same time. In fact, I did not even realize anyone else had also discovered mTOR until a journalist who was writing a story about our in-press paper faxed us a copy of Schreiber’s embargoed paper. I immediately sent Schreiber our paper, and we eventually spoke by phone, and he invited me to visit his laboratory at Harvard, memorably saying that if he was in town he was in the laboratory. When I asked where to meet, he said that if I walked around the Harvard Sciences area, I would find a Porsche and that I should knock on the nearby door. That July 4th, I was in Cambridge visiting my brother Bernardo, who is a neuroscientist, and we found the Porsche and the door and spent several fascinating hours with Schreiber hearing about his work. We left in awe and I remember thinking it was crazy to compete against Schreiber. Over the years, we have kept in touch, and I have served on the thesis committees of several of his students, and we now see each other frequently, as our laboratories are across the walkway that separates the Whitehead and Broad Institutes.
Over the years, I have also gotten to know very well Abraham, who went on to study how mTOR signals to downstream effectors and played a key role in translating the basic science of mTOR to the clinic. Other than once trying to exhaust me to death by cajoling me into my first and thankfully last cross-country skiing experience, he is among the kindest scientists I know, and has given me generous advice and support literally from the time I was in graduate school until now.
In parallel with efforts to identify the target of rapamycin, many laboratories were trying to understand its function by studying how rapamycin inhibits cell proliferation. Very early studies into the mechanism of rapamycin toxicity in the pathogenic yeast Candida albicans showed that rapamycin suppresses various metabolic processes, including protein synthesis (20). Subsequent work in human cells by John Blenis, George Thomas, Erwin W. Gelfand, and others showed that rapamycin inhibits the phosphorylation of the ribosomal protein S6 and the initiation of mRNA translation, establishing mTOR as a central regulator of anabolic metabolism and mass accumulation at the cellular level (21–25). These studies, particularly the one from Gelfand in 1995 (25), showed that rapamycin inhibits proliferation as a secondary consequence of reducing protein synthesis and growth, thereby demonstrating that mTOR controls cell size independently of the cell cycle, in contrast to what many had previously thought. Soon after, Hall reported that rapamycin also inhibits mRNA translation and growth in Saccharomyces cerevisiae (26), although for reasons that are still not clear, in yeast it does not cause the reduction in cell size that is a hallmark of its effects on mammalian cells. Importantly, using a temperature-sensitive allele, Hall found that TOR1 inactivation mimicked the effects of rapamycin on yeast growth, which directly implicated the TOR1 gene product as a key growth regulator. In animals, evidence connecting the target of rapamycin pathway to growth came first from genetic studies in the fruit fly Drosophila melanogaster. These showed that flies with reduced dTOR are smaller, specifically because of reduced cell size rather than cell number (27). Subsequent work in mice by my laboratory, particularly a collaborative study with Andrew S. Peterson describing the first loss-of-function allele of mTOR (28), and other laboratories established mTOR as a central regulator of cell, organ, and organismal size in mammals (reviewed in ref. 1).
From my work with RAFT2, I knew that mTOR was in a protein complex (12), and so after I arrived at the Whitehead Institute, I focused on identifying mTOR-interacting proteins. At this time, it was already clear that mTOR is a protein kinase because Schreiber had shown it can autophosphorylate (29) and Abraham had identified its first substrate, the translational regulator 4E-BP1 (30). I was certain that the binding partners of mTOR would be key to understanding its regulation and how it signals downstream. Two postdocs, Do-Hyung Kim and Dos D. Sarbassov, took on this project, but initially we were unsuccessful because of an unlucky choice of detergent. Dos eventually figured out that Triton X-100, a detergent routinely used to lyse mammalian cells and that is usually harmless to protein–protein interactions, unexpectedly disrupts the binding of mTOR to its partners. He identified a detergent (CHAPS) that does not do this, and soon after Do-Hyung discovered, as did Joseph Avruch, the first mTOR-interacting protein, which Do-Hyung named Raptor (regulatory associated protein of mTOR) (31, 32). Raptor is the defining component of what we now call mTOR complex 1 (mTORC1) (Fig. 1A).
We went on to identify the other components of mTORC1: DEPTOR, PRAS40, and GβL/mLST8, which turned out to be the RAFT2 protein I had chased unsuccessfully as a graduate student (33–35) (Fig. 1A). Importantly, in his initial report on Raptor (32), Do-Hyung showed that it regulates cell size and S6 Kinase, which we had discovered a number of years before is an mTOR substrate (36), and that FKBP12–rapamycin directly inhibits the kinase activity of mTORC1. These results established mTORC1 as the growth regulator targeted by rapamycin, and subsequent work by many laboratories connected it to most major anabolic and catabolic processes, like protein synthesis and autophagy, and the regulation of lifespan (reviewed in ref. 1). Over the years, diverse aspects of mTORC1 biology have been of interest to us. Postdoctoral fellow Tony Kang solved the first structure of mTORC1 in collaboration with Thomas Walz (37), and Peggy Hsu, an MD-PhD student, identified many new substrates for it, and in the process revealed cellular processes previously not connected to mTORC1 (38). David A. Guertin, a yeast geneticist who had bravely joined my biochemistry laboratory to do mouse genetics, generated mice lacking raptor, which he, and subsequently Shomit Sengupta, an MIT student from Texas, used to establish that mTORC1 is a growth regulator in animals (39, 40). David generously deposited the mice he generated carrying a floxed allele of raptor in a public repository, allowing dozens of laboratories to study mTORC1 function in vivo independently of us.
Soon after our discovery of Raptor, Hall reported the identification of budding yeast TORC1 and its Raptor homolog, which he called Kog1 (41). Perhaps reflecting a different time in science, I had contacted him to discuss our ongoing work on mTOR-interacting proteins and had sent Hall the unpublished Raptor cDNA so his laboratory could work with the human complex. Interestingly, in his initial report on TORC1, Hall also discovered that the yeast protein encoded by TOR2, but not TOR1, exists in a distinct complex he named TORC2 (41). Unlike fungi, other organisms have only one mTOR gene, so it was unknown whether a TORC2-like complex existed in animals, until Dos Sarbassov, still working in my laboratory, identified Rictor (Raptor-independent companion of mTOR) (42), which turned out to be the defining component of mTORC2 (Fig. 1B). Dos went on to identify its first substrate, the kinase Akt/PKB, a key effector of insulin signaling that immediately linked mTORC2 to glucose homeostasis and diabetes, adipogenesis, and cancer (43), and David Guertin revealed that mTORC2 is necessary for the development of tumors with activated PI3K signaling (44). When we identified mTORC2 as the long-sought kinase for Akt/PKB, I received a kind congratulatory note from Dario R. Alessi and Philip Cohen, which was very meaningful to me because they pioneered the study of how insulin activates Akt/PKB and identified the serine-473 phosphorylation site on Akt/PKB that mTORC2 turns out to phosphorylate (45).
Interestingly, while in yeast, rapamycin inhibits TORC1 but not TORC2, we found that in mammals, chronic rapamycin treatment—as happens in patients—blocks the assembly of mTORC2, so that over time, it inhibits both mTORC1 and mTORC2 signaling in many tissues (43). Based on genetic and pharmacological experiments in mice, Dudley W. Lamming, a postdoc with a long-standing interest in aging, proposed that the unexpected inhibition of mTORC2 by chronic rapamycin treatment explains some of the adverse effects of the drug (46). His findings have prompted an ongoing search for truly specific mTORC1 inhibitors that could be given chronically, as drugs for slowing the aging process would have to be.
It soon became clear that there are even more peculiarities to rapamycin. It not only suppresses mTORC2 when given chronically, but it also turns out to differentially inhibit the phosphorylation of established mTORC1 substrates, with some, like S6 Kinase, being dramatically affected, and others, like ULK1, barely so. This insight came largely from the use of the first specific ATP-competitive inhibitors of the mTOR kinase domain: Torin1, which Carson C. Thoreen, an MIT graduate student, developed in collaboration with Nathanael S. Gray (47), and PP242, which Kevan M. Shokat generated (48). In contrast to rapamycin, which we now recognize as an allosteric partial inhibitor of the mTORC1 kinase, Torin1 and PP242 profoundly inhibit the phosphorylation of all mTORC1 substrates and are now widely used to study mTORC1 signaling (47–49). In yeast, rapamycin strongly inhibits all TORC1-dependent processes that have been examined, suggesting that it may not have the partial inhibitory effects it does in mammalian systems.
Although perhaps seeming to be just a small technical advance, Dos’ identification of a detergent that preserves the stability of the mTOR complexes was an inflection point in our study of mTOR. His cell lysis buffer became the standard one in the field of mTOR biochemistry and enabled us to identify mTORC1 and mTORC2 and eventually most of the other core components of the mTOR pathway, including the Rag GTPases that transmit nutrient signals to mTORC1.
Ultimately, growth control is the process of linking the availability of nutrients in the environment to biomass production, so for me, the most fascinating aspect of mTORC1 has always been that it is regulated by nutrients. In an often-overlooked paper from 1995 (my laboratory is as guilty as any other), Alfred J. Meijer discovered that in cultured hepatocytes, amino acids activate S6 Kinase in a rapamycin-sensitive fashion (50), the first demonstration I am aware of in any system that what would eventually become known as mTORC1/TORC1 senses nutrients. Joseph Avruch generalized these results to other cell types and also identified leucine and arginine as key activators of mTORC1 signaling (51). In yeast, Hall found that TOR1 inactivation and rapamycin treatment mimic several of the effects of starvation, and Kim T. Arndt showed that a TOR1-controlled phosphatase responds to quality of the carbon source in the culture media (26, 52).
These studies were tantalizing, but how mTORC1 or TORC1 senses nutrients was a complete black box until the thesis work of Yasemin Sancak, an MIT student from Turkey. Ultimately, work she initiated would not only lead to an understanding of how nutrients are sensed, but also of how the mTORC1 pathway is organized so that it can respond to many other inputs besides nutrients, including growth factors and various forms of stress. She identified the heterodimeric Rag GTPases as mTORC1-interacting proteins that bind to it when cells are stimulated with nutrients, particularly amino acids (53). Using nucleotide-loading mutants of the Rag GTPases, she determined, as did Kun-Liang Guan, that they are necessary and sufficient for mTORC1 to sense nutrients (53, 54). When Alejo Efeyan, a postdoctoral fellow from Spain, joined the laboratory, he generated knockin mice with a constitutively active allele of RagA that prevents mTORC1 from becoming inhibited by nutrient starvation. These animals develop normally, but once born and separated from the maternal supply of nutrients, they do not survive periods of fasting because they cannot switch from an anabolic to a catabolic state (55). These results were the first evidence that nutrient sensing by mTORC1 is necessary for maintaining organismal homeostasis when food is scarce, likely the environmental condition under which humans evolved and most animals spend most of their lives.
We had no doubt that the Rag GTPases were important, but exactly how they regulate mTORC1 was frustratingly mysterious, because in vitro they do not stimulate its kinase activity. We eventually answered this question, but we would have done so much sooner if 10 y earlier I had followed the advice of my father, a prominent cell biologist (David D. Sabatini). Soon after I discovered mTOR, he suggested that I determine its subcellular localization. I was quite dismissive of this suggestion, but I did develop an antibody to mTOR and showed via immunofluorescence that in human cells it stained cytoplasmic puncta that looked like small vesicles. I never bothered to understand what the puncta were, and I eventually lost the antibody and moved on. In the following years, mTOR went through a “Where’s Waldo?” period, with papers claiming it was at many different locations, including the nucleus and mitochondria. Tim R. Peterson, an MD-PhD student, decided to nail down its localization and screened every available mTOR antibody to identify the only one whose staining pattern by immunofluorescence disappeared when he suppressed mTOR expression. This antibody revealed mTOR to be at subcellular structures very similar to those I had failed to define earlier. More excitingly, he and others in the laboratory found that within minutes of starving cells for amino acids or glucose, mTOR left the punctate structures and became diffuse throughout the cytoplasm (53, 55). We eventually concluded that the puncta are lysosomes and that nutrients signal through the Rag GTPases to promote the movement of mTORC1 to the lysosomal surface (53, 56).
But how does this translocation impact the activity of mTORC1? To answer this key question, it was necessary to consider the Rheb GTPase, a Ras-related GTPase that is essential for mTORC1 activation in all model organisms except, oddly, budding yeast (57). Work in mammalian cells and flies showed that the TSC protein complex inhibits Rheb by serving as its GTPase activating protein (GAP) and that multiple stimuli, including insulin and energy stress, signal to mTORC1 through TSC and Rheb (58–70). In humans, loss-of-function mutations in TSC components cause the overgrowth syndrome tuberous sclerosis (reviewed in ref. 71) and, in flies, increases in cell size that depend on dTOR activity (72).
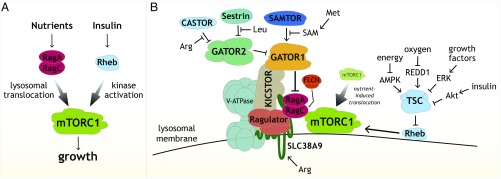
Everything began to fall into place when we found that GTP-loaded Rheb directly stimulates the kinase activity of mTORC1 and that Rheb also localizes, at least in part, to lysosomes (34, 53). We proposed that the Rag and Rheb GTPases are two arms of a coincidence detector mechanism that ensures that mTORC1 becomes activated only when nutrient and growth factor conditions are both appropriate (53). In this model, the function of the Rag GTPases is to localize mTORC1 to lysosomes when nutrients are present so that Rheb can stimulate its kinase activity if insulin (and energy) is also available (Fig. 3A). In one of my favorite experiments, we obtained strong support for such a pathway architecture by forcing mTORC1 onto the lysosomal surface and finding that, while mTORC1 became insensitive to nutrient starvation, it retained its capacity to sense the presence and absence of insulin (56). Satisfyingly, our model has readily incorporated new data from other laboratories. For example, Brendan D. Manning found that upon insulin withdrawal, the TSC complex translocates to the lysosomal surface to inhibit Rheb, providing for the first time a mechanistic understanding of how insulin regulates TSC and Rheb, and thus mTORC1 (73).
Fig. 3.
(A) Coincidence detector model for how mTORC1 integrates signals from nutrients and growth factors to regulate growth. The Rag GTPases promote the localization of mTORC1 to the lysosomal surface in response to nutrients, and, at the lysosome, the Rheb GTPase activates its kinase activity in response to insulin and energy levels. (B) Schematic showing components of the nutrient-sensing pathway upstream of mTORC1, including the many multiprotein complexes that regulate the Rag GTPases as well as the amino acid sensors Sestrin2, CASTOR1, and SLC38A9, and the SAM sensor SAMTOR.
The connection of mTORC1 to lysosomes has had a profound impact on how we think about these organelles. Through the efforts of Yasemin Sancak, Tim Peterson, and Roberto Zoncu, who has been the only card-carrying cell biologist in my laboratory, we showed that lysosomes are a scaffolding platform on which mTORC1 becomes activated and also an active participant in the amino acid sensing process, as mTORC1 can sense amino acids in the lysosomal lumen (53, 56, 74). These discoveries gave rise to the field of lysosomal signaling, which has been greatly boosted by the work of Andrea Ballabio on the control of lysosomal biogenesis by the TFEB transcription factor (75), which he found in a collaborative study with us is controlled by mTORC1 at the lysosomal surface (76). In his own laboratory, Roberto recently discovered that mTORC1 senses cholesterol through a pathway that also controls its translocation to lysosomes (77), extending the range of nutrients these organelles sense beyond the amino acids and glucose we have focused on. I remember that when I first presented a seminar on mTORC1 and lysosomes, I was told that it did not make any sense because the “lysosome is just a garbage can,” something that few would say today.
Once we figured out the function of the Rag GTPases, we turned to understanding how nutrients, particularly amino acids, regulate them. This has turned out to be a fascinating story and much more complicated than I ever imagined, particularly as it became clear that the Rag GTPases respond not only to lysosomal amino acids but also to cytosolic ones. Over a period of 12 years, a remarkable group of graduate students—Yasemin Sancak, Liron Bar-Peled, Shuyu Wang, Zhi Y. Tsun, Lynne Chantranupong, Rachel L. Wolfson, Robert A. Saxton, and Greg A. Wyant—discovered and characterized many multiprotein complexes that, in response to nutrients, regulate the nucleotide state of the Rag GTPases, including Ragulator, GATOR1, GATOR2, FLCN-FNIP, and KICSTOR (56, 78–88). So far, we have identified 26 proteins, most of them previously molecularly uncharacterized, that comprise the nutrient-sensing arm of the pathway, telling us that cells devote a significant amount of protein space to regulating mTORC1 via nutrients (Fig. 3B). We now appreciate that in cancer and neurological diseases like epilepsy, many of these proteins are mutated to cause mTORC1 hyperactivation, opening the door to their rational treatment (reviewed in ref. 1).
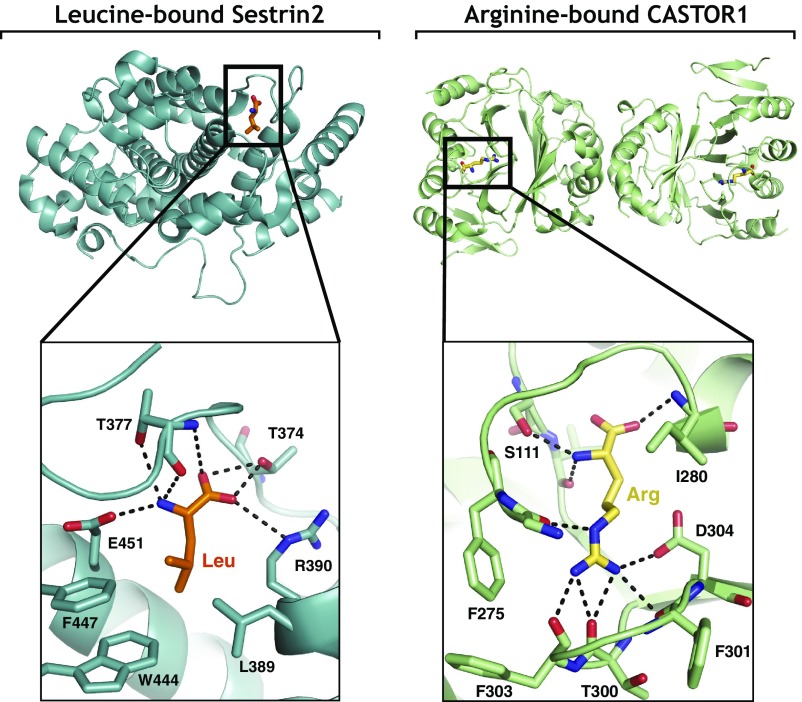
Most excitingly, we have finally identified what has been the holy grail of the pathway for us—the proteins that bind nutrients and sense their presence (Fig. 3B). We now know that SLC38A9 is a sensor of lysosomal arginine, and Sestrin2 and CASTOR1 of cytosolic leucine and arginine, respectively (83, 84, 87, 88). It was moving for me to see the amino acid binding pockets in the Sestrin2 and CASTOR1 structures that Robert Saxton solved in collaboration with Thomas U. Schwartz (82, 85) (Fig. 4). After so many years of chasing these sensors, we could finally see in atomic detail exactly how nature had connected mTORC1 to nutrients. Interestingly, while the Rag GTPase and GATOR components of the nutrient-sensing pathway are relatively well conserved (89), most other components are not, including the amino acid sensors, suggesting that different organisms evolved to detect distinct nutrients that are perhaps limiting in their environmental niche or of special importance to them. A test of this idea will have to await the discovery of nutrient sensors upstream of TORC1 in other organisms besides animals. We have also become quite interested in the evolution of nutrient sensors, and one theme that is emerging is that the sensors we have so far identified appear to have their origins in prokaryotic enzymes.
Fig. 4.
Views of the amino acid binding pocket of Sestrin2 bound to leucine [Protein Data Bank (PDB) ID code 5DJ4] (Left) and of the amino acid binding pocket of CASTOR1 bound to arginine (PDB ID code 5I2C) (Right).
Our interest in nutrient sensing and the increasing appreciation that the mTORC1 pathway directly regulates many metabolic pathways—an insight that is almost entirely the result of Brendan Manning’s work in mammalian cells (reviewed in ref. 90)—has led us to study small-molecule metabolism. A significant part of the laboratory now studies metabolic pathways important for cell growth and proliferation, and having students and postdocs with these interests has led to the cross-fertilization of ideas and techniques between laboratory members working on mTOR and metabolism and to a richer and more stimulating atmosphere within the laboratory. My friends in the field laugh at me when every few years I claim I am done with mTOR, saying it is too competitive or that there is nothing left to discover. I have yet to follow through on this, and about half my laboratory continues to work on the mTOR pathway. Whenever I feel like calling it quits, the laboratory conveniently makes a great discovery that piques our interest in a new facet of the pathway. In the last 10 years, these discoveries have been mostly around nutrient sensing and the connection of mTORC1 to lysosomes. More recently, they have concerned SAMTOR (Fig. 3B), a S-adenosylmethionine sensor for the pathway (91), which links mTORC1 to methionine levels and potentially to the beneficial effects of methionine restriction on health and lifespan (92). We have yet to figure out how mTORC1 detects glucose, why the pathway evolved to sense both lysosomal and cytosolic amino acids, or how, as a megadalton complex, mTORC1 quickly moves to lysosomes to dock on the Rag GTPases. So, while mTOR may not regulate everything, there are enough mysteries in how it senses everything to keep us occupied for the foreseeable future.
Acknowledgments
I thank all past and current members of my laboratory for contributing to the remarkable understanding we now have of how mTOR regulates growth in response to nutrients. I thank R. A. Saxton, D. D. Sabatini, B. L. Sabatini, L. F. Pernas, P. P. Budde, R. T. Abraham, and B. D. Manning for insightful comments on earlier versions of this article; R. A. Saxton for preparing Fig. 4; Whitehead Institute for Biomedical Research, National Institutes of Health, Howard Hughes Medical Institute, Koch Institute for Integrative Cancer Research, and Broad Institute of Harvard and MIT for providing long-standing funding support of our mTOR-related work; and A. Sehgal, R. T. Abraham, M. N. Hall, G. P. Livi, and S. L. Schreiber for providing the photographs. I am most grateful to Solomon H. Snyder for having faith that I would figure things out when I was a student in his laboratory and thus giving me the freedom to pursue my own research direction. He inspired his laboratory members to think big and explore new topics, and his influence is with me every day in the way I run my own laboratory.
Footnotes
The author declares no conflict of interest.
References
- 1.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mannick JB, et al. mTOR inhibition improves immune function in the elderly. Sci Transl Med. 2014;6:268ra179. doi: 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- 3.Vézina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 4.Eng CP, Sehgal SN, Vézina C. Activity of rapamycin (AY-22,989) against transplanted tumors. J Antibiot (Tokyo) 1984;37:1231–1237. doi: 10.7164/antibiotics.37.1231. [DOI] [PubMed] [Google Scholar]
- 5.Martel RR, Klicius J, Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol. 1977;55:48–51. doi: 10.1139/y77-007. [DOI] [PubMed] [Google Scholar]
- 6.Dumont FJ, Staruch MJ, Koprak SL, Melino MR, Sigal NH. Distinct mechanisms of suppression of murine T cell activation by the related macrolides FK-506 and rapamycin. J Immunol. 1990;144:251–258. [PubMed] [Google Scholar]
- 7.Dumont FJ, et al. The immunosuppressive macrolides FK-506 and rapamycin act as reciprocal antagonists in murine T cells. J Immunol. 1990;144:1418–1424. [PubMed] [Google Scholar]
- 8.Bierer BE, et al. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc Natl Acad Sci USA. 1990;87:9231–9235. doi: 10.1073/pnas.87.23.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, et al. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 10.Bredt DS, Snyder SH. Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc Natl Acad Sci USA. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pert CB, Snyder SH. Opiate receptor: Demonstration in nervous tissue. Science. 1973;179:1011–1014. doi: 10.1126/science.179.4077.1011. [DOI] [PubMed] [Google Scholar]
- 12.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 13.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 14.Sabers CJ, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 15.Koltin Y, et al. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol. 1991;11:1718–1723. doi: 10.1128/mcb.11.3.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 17.Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 18.Helliwell SB, et al. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cafferkey R, et al. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh K, Sun S, Vézina C. Rapamycin (AY-22,989), a new antifungal antibiotic. IV. Mechanism of action. J Antibiot (Tokyo) 1979;32:630–645. doi: 10.7164/antibiotics.32.630. [DOI] [PubMed] [Google Scholar]
- 21.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 22.Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 23.Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo CJ, et al. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- 25.Terada N, Takase K, Papst P, Nairn AC, Gelfand EW. Rapamycin inhibits ribosomal protein synthesis and induces G1 prolongation in mitogen-activated T lymphocytes. J Immunol. 1995;155:3418–3426. [PubMed] [Google Scholar]
- 26.Barbet NC, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hentges KE, et al. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc Natl Acad Sci USA. 2001;98:13796–13801. doi: 10.1073/pnas.241184198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown EJ, et al. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 30.Brunn GJ, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 31.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 33.Kim DH, et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 34.Sancak Y, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Peterson TR, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Sengupta S, Peterson TR, Laplante M, Oh S, Sabatini DM. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature. 2010;468:1100–1104. doi: 10.1038/nature09584. [DOI] [PubMed] [Google Scholar]
- 41.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 42.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 43.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 44.Guertin DA, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alessi DR, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 46.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang SA, et al. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 51.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 52.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 53.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Efeyan A, et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 58.Garami A, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 59.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 60.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 61.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 63.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 64.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 65.Shaw RJ, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 66.Stocker H, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 67.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 69.Lee DF, et al. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 70.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci USA. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwiatkowski DJ, Manning BD. Molecular basis of giant cells in tuberous sclerosis complex. N Engl J Med. 2014;371:778–780. doi: 10.1056/NEJMcibr1406613. [DOI] [PubMed] [Google Scholar]
- 72.Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 2002;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menon S, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Settembre C, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castellano BM, et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chantranupong L, et al. The sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014;9:1–8. doi: 10.1016/j.celrep.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bar-Peled L, et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolfson RL, et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543:438–442. doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature. 2016;536:229–233. doi: 10.1038/nature19079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang S, et al. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–194. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wolfson RL, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saxton RA, et al. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016;351:53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsun ZY, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chantranupong L, et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell. 2016;165:153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wyant GA, et al. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosome and use protein as a nutrient. Cell. doi: 10.1016/j.cell.2017.09.046. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wolfson RL, Sabatini DM. The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 2017;26:301–309. doi: 10.1016/j.cmet.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu X, et al. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017 doi: 10.1126/science.aao3265. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Orentreich N, Matias JR, DeFelice A, Zimmerman JA. Low methionine ingestion by rats extends life span. J Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]