Significance
We describe the spatial and temporal profiles of soybean and Arabidopsis seed methylomes during development. CHH methylation increases globally from fertilization through dormancy in all seed parts, decreases following germination, and targets primarily transposons. By contrast, CG- and CHG-context methylation remains constant throughout seed development. Mutant seeds lacking non-CG methylation develop normally, but have a set of up-regulated transposon RNAs suggesting that the CHH methylation increase may be a failsafe mechanism to reinforce transposon silencing. Major classes of seed genes have similar methylation profiles, whether they are active or not. Our results suggest that soybean and Arabidopsis seed methylomes are similar, and that DNA methylation does not play a significant role in regulating many genes important for seed development.
Keywords: seed development, DNA methylation, soybean, Arabidopsis, transposon
Abstract
We profiled soybean and Arabidopsis methylomes from the globular stage through dormancy and germination to understand the role of methylation in seed formation. CHH methylation increases significantly during development throughout the entire seed, targets primarily transposable elements (TEs), is maintained during endoreduplication, and drops precipitously within the germinating seedling. By contrast, no significant global changes in CG- and CHG-context methylation occur during the same developmental period. An Arabidopsis ddcc mutant lacking CHH and CHG methylation does not affect seed development, germination, or major patterns of gene expression, implying that CHH and CHG methylation does not play a significant role in seed development or in regulating seed gene activity. By contrast, over 100 TEs are transcriptionally de-repressed in ddcc seeds, suggesting that the increase in CHH-context methylation may be a failsafe mechanism to reinforce transposon silencing. Many genes encoding important classes of seed proteins, such as storage proteins, oil biosynthesis enzymes, and transcription factors, reside in genomic regions devoid of methylation at any stage of seed development. Many other genes in these classes have similar methylation patterns, whether the genes are active or repressed. Our results suggest that methylation does not play a significant role in regulating large numbers of genes important for programming seed development in both soybean and Arabidopsis. We conclude that understanding the mechanisms controlling seed development will require determining how cis-regulatory elements and their cognate transcription factors are organized in genetic regulatory networks.
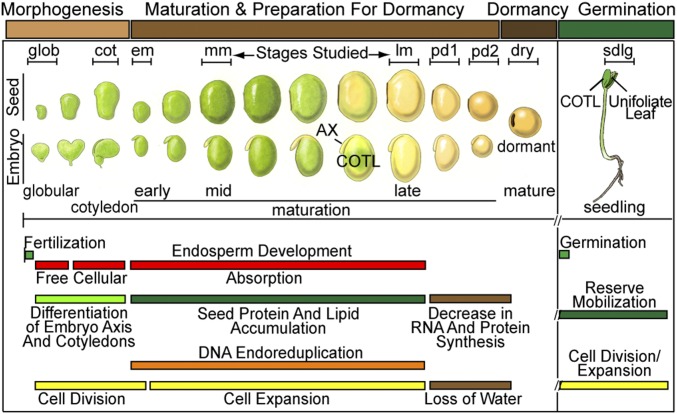
Seeds are derived from a double-fertilization process that leads to the differentiation of the seed coat (SC), endosperm, and embryo (EMB), the major regions of the seed that have distinct genetic origins and functions (1–3). The maternally derived SC differentiates from the ovule integuments that surround the embryo sac, transfers nutrients from the maternal plant to the developing EMB, and protects the seed during development and dormancy. The EMB and endosperm, by contrast, are descendants of the fertilized egg and central cell, respectively. The endosperm nourishes the EMB early in development, and in dicots, such as soybean and Arabidopsis, is absorbed and only present as a vestigial cell layer, the aleurone, in the mature seed. The EMB forms the two major embryonic organs: (i) an axis (AX), containing shoot and root meristems, which will give rise to the mature plant after seed germination; and (ii) the cotyledon (COTL), a terminally differentiated organ, which specializes in storage reserve production and senesces following germination (Fig. 1). Seeds shift into a maturation program following AX and COTL differentiation, that includes: (i) cessation of cell division, (ii) accumulation of storage reserves, and (iii) preparation for desiccation and dormancy (1, 2, 4) (Fig. 1). During this period, COTL cells enlarge and undergo a unique endoreduplication process that may facilitate the synthesis of highly prevalent seed-storage proteins that are utilized as an energy source during germination and early seedling (sdlg) growth (5, 6) (Fig. 1). At the end of maturation, programed water loss (i.e., desiccation) occurs, metabolic and developmental processes are suspended, a dormancy period begins that can last for several millennia (7), and the quiescent seed awaits an optimum environment for germination and sdlg growth (1) (Fig. 1).
Fig. 1.
Schematic representation of soybean seed stages and major developmental events. Adapted from refs. 38 and 48. Seed and EMB images are not drawn to scale. Brackets indicate stages investigated. Abbreviations are defined in Table 1.
DNA methylation plays a critical role in endosperm development (3, 8). DEMETER (DME) encodes a 5-methylcytosine glycosylase, and is expressed specifically in the central cell of the EMB sac (3, 9). DME removes methylated bases from maternal genes within the central cell that are then expressed preferentially in the endosperm following fertilization, in comparison with their sperm-derived-methylated paternal counterparts. In addition, because DME is not expressed in the egg, the EMB is hypermethylated relative to the endosperm (10). Mutations in DME that disrupt these parent-of-origin (i.e., imprinting) methylation events result in abnormal endosperm development and seed abortion (3). By contrast, imprinting caused by differential methylation of paternal and maternal alleles does not appear to occur within the EMB, although there is conflicting evidence for the preferential activity of maternal and paternal genomes during early embryogenesis (3, 11–13). The extent to which DNA methylation events play a role in seed formation at all stages of development, and within different seed regions and tissue layers, remains largely unexplored.
We used soybean and Arabidopsis seeds to address the following questions: (i) Are there global DNA methylation changes during seed development from fertilization through dormancy and germination? (ii) Do seed regions and tissues have different methylation patterns? (iii) Is DNA methylation maintained during COTL cell endoreduplication? (iv) Are DNA methylation events in seed development conserved in different plants?
We applied whole-genome bisulfite (BS) sequencing (BS-Seq) and laser capture microdissection (LCM) to profile the DNA methylation landscape during seed development. We observed that DNA methylation profiles are similar in soybean and Arabidopsis seeds, which diverged ∼90 Mya (14). Global CHH methylation increases throughout the entire seed from differentiation to dormancy, targets all classes of transposable elements (TEs), and decreases in postgermination COTLs and sdlg. In addition, DNA methylation patterns in all sequence contexts are maintained during endoreduplication. Mutant Arabidopsis seeds lacking CHG and CHH methylation (15) develop and germinate normally, and have gene expression profiles that are mostly congruent with wild-type seeds. By contrast, 106 transposons are de-repressed transcriptionally in mutant seeds, suggesting that the increase in CHH methylation during seed development may be a failsafe mechanism to reinforce TE silencing. Finally, no significant DNA methylation changes occur around many genes known to be important for seed formation—including storage protein genes, fatty acid biosynthesis genes, and several major transcription factor (TF) genes—and many of these genes are in genomic regions devoid of DNA methylation at any stage of development. We conclude that the next major challenge to understanding seed development is to determine how cis-regulatory elements encoded in the genome and their cognate TFs that activate and repress gene activity are organized into regulatory networks that are required to “make a seed.”
Results
Single-Base Resolution Soybean Seed Methylomes Throughout Development.
We profiled the soybean DNA methylation landscape at single-base resolution from nine seed stages using BS-Seq to obtain a comprehensive methylation profile from postfertilization through dormancy and germination (Fig. 1 and Table 1). To compare the methylomes of different seed regions, subregions, and tissues at distinct developmental stages, we hand-dissected the AX COTL and SC from early-maturation (em) and midmaturation (mm) seeds, and used LCM to isolate: (i) AX, COTL, and SC from cotyledon (cot) seeds; (ii) SC tissue layers [parenchyma (PY) and palisade (PA)] from em seeds; and (iii) AX regions [plumule (PL), PY, vascular (VS), and root tip (RT)] from em seeds. Finally, we used LCM to obtain two em tissues showing differential endoreduplication (Fig. 1) within the same embryonic organ [COTL abaxial (ABPY) and adaxial (ADPY) PY tissues]. Collectively, our BS-Seq datasets provide a comprehensive spatial and temporal profile of the DNA methylation landscape across the entire soybean genome throughout all of seed development (Dataset S1).
Table 1.
Development stage, region, and tissue abbreviations
| Abbreviation | Description |
| Soybean seed stage and postgermination | |
| glob | Globular |
| cot | Cotyledon |
| em | Early maturation |
| mm | Midmaturation |
| lm | Late maturation |
| pd1 | Early predormancy |
| pd2 | Late predormancy |
| dry | Dry seed |
| sdlg | Whole seedling |
| Arabidopsis seed stage | |
| glob | Globular |
| lcot | Linear cotyledon |
| mg | Mature green |
| pmg | Postmature green |
| dry | Dry seed |
| Seed regions, subregions, and tissues | |
| ABPY | Abaxial parenchyma |
| ADPY | Adaxial parenchyma |
| AL | Aleurone |
| AX | Axis |
| COTL | Cotyledon |
| EMB | Embryo |
| HG | Hourglass |
| PA | Palisade |
| PL | Plumule |
| PY | Parenchyma |
| RT | Root tip |
| SC | Seed coat |
| sdlg-COTL | Seedling cotyledon |
| VS | Vascular |
In total, we generated ∼8 billion Illumina BS-Seq reads from all seed stages, regions, organs, and tissues, obtaining in each case 11–27× coverage of the ∼1-Gb soybean genome (Dataset S1). We assayed 273–287 million cytosines, representing 94–98% of all cytosines, at an average sequence depth of 5–13× per cytosine (SI Materials and Methods and Dataset S2). We checked the conversion efficiency of the BS treatment by examining the conversion of C-to-T in both the unmethylated chloroplast genome and a λ genome that was added to our samples as an internal control (SI Materials and Methods). We observed an average BS conversion efficiency of unmethylated C-to-T greater than 99.5% for both the chloroplast and λ genomes, indicating high conversion efficiency for our BS treatment (Dataset S1). The BS-Seq data from biological replicates of whole seeds and seed parts were in excellent agreement with each other (correlation coefficients > 0.99) (Fig. S1A). Additionally, we observed that 9%, 12%, and 79% of the seed methylomes were present in CG, CHG, and CHH contexts (where H = A, C, T), which was similar to the proportion of CG, CHG, and CHH sites in the soybean genome (Fig. S1B). We calculated the bulk methylation levels to determine the extent to which the soybean seed genome was methylated (10) (SI Materials and Methods), and observed: (i) an average methylation level of 12% for all detected cytosines, and (ii) average methylation levels of 57%, 36%, and 2% in the CG, CHG, and CHH contexts across all samples, respectively (Dataset S2), values similar to those obtained in other soybean methylome studies (16–18). Taken together, these results indicate that our datasets represent unbiased, deep representation, and highly reproducible profiles of soybean seed methylomes.
CHH Methylation Levels Increase During Soybean Seed Development.
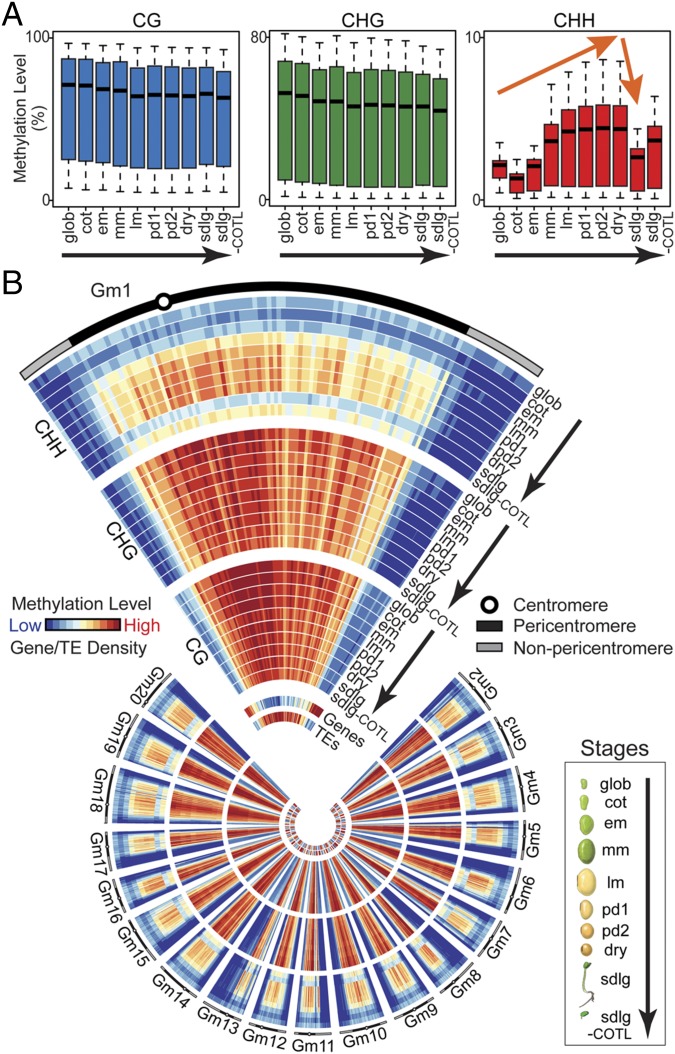
To determine whether global DNA methylation changes occurred during soybean seed development, we calculated the bulk methylation levels (SI Materials and Methods) for CG, CHG, and CHH contexts in 500-kb windows across the genome from whole seeds at the globular (glob), cot, em, mm, late maturation (lm), early predormancy (pd1), late predormancy (pd2), and dry stages, representing the differentiation, maturation, and dormancy phases (Fig. 1). The box plots show that there were no significant global DNA methylation changes in either CG or CHG contexts between: (i) adjacent seed stages or (ii) postfertilization glob and dry seeds (t test, P < 0.001 and >1.5-fold increase), suggesting that CG and CHG sites were either methylated at fertilization when seed development begins (i.e., before the glob stage) or before (Fig. 2A and Dataset S3). By contrast, global CHH methylation levels increased more than threefold from postfertilization (glob, cot) through maturation (em, mm, lm), and then plateaued from lm through desiccation (pd1 and pd2) and dormancy (dry) (Fig. 2A and Dataset S3). Most of the increase occurred after the cessation of cell division between the em and lm stages when the seed had ∼3 × 106 cells and was enlarging due to cell expansion (19). This is consistent with the mechanism of CHH methylation via the RNA-directed DNA methylation (RdDM) pathway that can occur “de novo” and independently of DNA replication (20).
Fig. 2.
Genome-wide methylation changes during soybean seed development and germination. DNA methylation levels in 500-kb windows across the genome are represented as box plots (A) and chromosome heat maps (B). The highest methylation levels (%) for heat maps tracks are 96, 80, and 9 for CG, CHG, and CHH contexts, respectively. Gene and TE tracks represent gene and TE densities in 500-kb windows across the genome. Gm, Glycine max. See Table 1 for abbreviations.
It was possible that the elevation in CHH bulk methylation level during seed development was due to: (i) accumulated methylation of the same cytosine sites in different cells, (ii) methylation of new cytosine sites, or (iii) both. To distinguish between these possibilities, we determined the absolute number of cytosine sites in the CHH context that were methylated at each developmental stage (SI Materials and Methods), and compared these values with the bulk CHH methylation levels (Fig. S2 A and B). The number of methylated CHH sites increased significantly (Fisher’s exact test, P < 0.001 and >1.5-fold increase) from the cot to em stage, and then leveled off in subsequent stages (Fig. S2 A and B). By comparison, the bulk CHH level increased during the same developmental period, but continued to increase through the lm stage (Fig. 2 and Fig. S2B). These results indicate that the increase in CHH methylation during seed development is due to the addition of new methylated CHH sites across the genome, and the accumulated methylation of the same CHH sites in different seed cells as shown by the heuristic model (Fig. S2B).
CHH Methylation Levels Drop During Soybean Seed Germination.
We determined whether the seed CHH methylation level was maintained after germination, by profiling sdlg and sdlg-COTL methylomes, representing the (i) transitional state from a dormant seed to a rapidly growing sdlg and (ii) COTL in different functional states (i.e., dormant seeds and metabolically active seed leaves), respectively (Fig. 1). The sdlg we assayed contained the developing root, elongating hypocotyl, postgermination COTL, and emerging leaves (Fig. 1 and SI Materials and Methods). In comparison with the dry seed, both the bulk CHH methylation levels and the number of methylated CHH sites dropped significantly in the sdlg during germination (t test and Fisher’s exact test, P < 0.001 and >1.5-fold change) and in germinating sdlg-COTL, although to a lesser extent in the latter (t test and Fisher’s exact test, P < 0.001 and <1.5-fold change) (Fig. 2A and Dataset S3). By contrast, CG and CHG methylation levels in the postgermination sdlg and sdlg-COTL were maintained and did not change significantly (Fig. 2A and Dataset S3). These data suggest that CG and CHG sites are methylated in seed AX tissues that give rise to the sdlg (e.g., PL, PY, RT) and are maintained following germination. By contrast, the hypomethylation of CHH sites in the sdlg compared with the dormant seed might indicate that either: (i) methylated CHH sites within the AX become diluted as they divide, increase in number, and differentiate within the germinating sdlg; (ii) CHH sites within the AX were hypomethylated within the dry seed before germination (i.e., not methylated during seed development); or (iii) both. The different results obtained between CG- and CHG-context methylation, and CHH-context methylation in the sdlg following seed germination might also reflect the distinct mechanisms by which CG, CHG, and CHH sites are methylated: the former (CG and CHG) utilizing hemimethylated cytosines or replication-dependent histone variants as guides during replication, whereas the latter (CHH) occur de novo without the use of a methylated cytosine on one DNA strand (21, 22).
CHH Methylation Levels Change Within All Soybean TE Classes During Seed Development and Germination.
The developmental changes in CHH-context methylation levels occurred predominantly in the pericentromeric regions of each soybean chromosome, where the majority of TEs were located (Fig. 2B and Fig. S3A). We divided the TEs by chromosome location, class, and length and determined that the genome-wide changes in seed and sdlg CHH-context methylation occurred within TEs, irrespective of location, size, or type in the genome (Fig. S3 B–D). For example, the temporal increases and decreases in CHH-context methylation occurred in DNA transposons (e.g., Mutator), retrotransposons (e.g., Gypsy and Copia), and all transposon size classes (Fig. S3 B–D). In addition, these changes also occurred in TEs that were located in the TE-rich pericentromeric region and the gene-rich chromosomal arms (Fig. S3 B and C). These results suggest that the mechanisms responsible for the developmental changes in CHH methylation levels are coordinated within TEs across the genome, and occur in parallel with developmental events that take place during seed development and germination (Figs. 1 and 2).
CHH Methylation Changes Occur Throughout the Entire Soybean Seed.
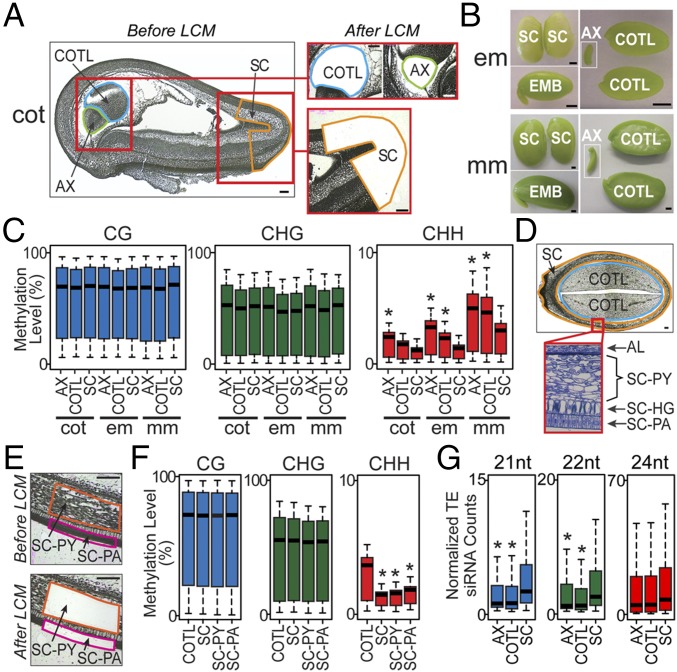
We isolated seed AX, COTL, and SC regions at different developmental stages using both LCM (cot stage) and manual dissection (em and mm stages) to determine whether CHH-context methylation changes occurred throughout the entire seed (Fig. 3 A and B). It was necessary to use LCM for cot-stage seeds because the AX, COTL, and SC regions were too small to be dissected by hand (Fig. 3A).
Fig. 3.
Comparison of methylomes between soybean seed parts and SC layers. (A) Paraffin sections of cot-stage SC, embryonic AX, and embryonic COTL before and after capture by LCM. (B) Whole-mount pictures of SC, AX, and COTL from em- and mm-stage EMB and seeds. (C) Box plots of DNA methylation levels in 500-kb windows across the genome in different seed parts. Asterisks indicate significant comparisons between SC and other seed parts at the same stage (t test, P < 0.001 and fold change > 1.5). (D) Paraffin cross-section of an em-stage seed (Upper), and a plastic section of SC layers (Lower), which is the red boxed region shown in the whole-seed section. (E) Paraffin cross-sections of em-stage SC-PA and SC-PY layers before and after capture by LCM. (F) Box plots of LCM-captured em-stage COTL, SC, SC-PA, and SC-PY DNA methylation levels in 500-kb windows across the genome. Asterisks indicate significant comparisons between SC, SC layers, and the COTL (t test, P < 0.001 and fold change > 1.5). (G) Box plots of TE siRNA levels (reads per million mapped reads) in em-stage SC, AX, and COTL. Asterisks indicate statistically significant comparisons between SC and other seed parts (t test, P < 0.001 and fold change > 1.5). [Scale bars, 100 µm (A, D, and E) and 1 mm (B).] See Table 1 for abbreviations.
The CG- and CHG-context bulk methylation levels were similar in all seed parts and did not change during seed development (Fig. 3C), analogous to the results obtained with whole seeds (Fig. 2). By contrast, the CHH-context methylation levels within the AX, COTL, and SC increased significantly (t test, P < 0.001 and >1.5 fold change) (Dataset S3) from the cot to mm stages (Fig. 3C), paralleling changes that were observed with the seed as a whole (Fig. 2A). In addition, the CHH methylation levels differed significantly (t test, P < 0.001, >1.5-fold change) between the AX, COTL, and SC regions at all developmental stages (Fig. 3C and Dataset S3). In general, the SC had the lowest level of CHH methylation, whereas the AX had the highest (Fig. 3C). The temporal and spatial differences in AX, COTL, and SC CHH-context methylation levels were also reflected within major TE classes scattered across the genome (Fig. S3E). Together, these results indicate that the biological events responsible for the temporal increase in CHH-context methylation during seed development are coordinated spatially within all major seed regions, and that the maternally derived SC layer is hypomethylated in comparison with the embryonic AX and COTL regions.
Individual Soybean SC Tissue Layers Are Hypomethylated.
The SC consists of different tissue layers—hourglass, PA, and PY—of which the PY is the most prominent and constitutes most of the SC (Fig. 3 D and E). We used LCM to capture em-stage SC-PA and SC-PY tissue layers to determine whether the CHH-context hypomethylation occurred throughout the entire SC or was unique to a given layer (e.g., major PY layer). As a control, we used LCM to capture the entire SC and COTL from em-stage seeds (Fig. 3D). No bulk methylation differences were observed between the SC-PA and SC-PY layers in any cytosine context (CG, CHG, and CHH) (Fig. 3F). By contrast, the SC-PA and SC-PY CHH-context methylation levels were both significantly lower relative to the COTL (t test, P < 0.001 and >1.5-fold change) (Dataset S3), and similar to results obtained with the SC as a whole (Fig. 3 C and F). Taken together, these results indicate that the methylation levels of the entire SC reflect those within individual tissue layers, including CHH-context hypomethylation relative to the AX and COTL regions of the seed.
The Soybean SC Layer Contains Elevated Levels of siRNAs.
We isolated small RNAs from em-stage AX, COTL, and SC, and used RNA-Seq to determine whether the CHH-context hypomethylation of the SC relative to other regions of the seed resulted in an elevated level of siRNAs derived from TEs (SI Materials and Methods). Twenty-four–nucleotide siRNA levels were similar in all seed regions (Fig. 3G). By contrast, both 21-nt and 22-nt siRNAs were elevated significantly (t test, P < 0.001 and >1.5-fold change) in the SC compared with the AX and COTL regions. These results suggest that: (i) CHH-context hypomethylation of TEs within the SC (Fig. S3E) might result in reduced TE silencing and TE movement, which would have little effect on subsequent development as the SC does not contribute to the postgerminating sdlg; (ii) elevated 21-nt and 22-nt siRNAs could mitigate this possibility by posttranscriptional silencing of SC TEs (23); and (iii) 21-nt and 22-nt SC siRNAs might move to other parts of the seed, such as the AX and COTL, which have higher CHH-context methylation levels (Fig. 3 C and F), and reinforce TE silencing (10) analogous to what occurs between vegetative and sperm cells within the pollen grain (23, 24).
CHH-Methylation Levels Within Soybean AX Subregions and Tissues Differ.
We used LCM to capture tissues of the em AX that give rise to sdlg following germination. These included: the (i) PL, (ii) PY and VS, and (iii) RT that participate in forming the seed leaves, hypocotyl, and root of the germinating sdlg, respectively (Fig. S4A). The CG- and CHG-context bulk methylation levels for the AX-PL, AX-PY, AX-RT, and AX-VS tissues were similar and not significantly different from those observed within the germinating sdlg (Fig. 2A and Fig. S4B). By contrast, the bulk CHH-context methylation level of the RT was significantly higher than that of the AX-PL, AX-PY, or AX-VS tissues, indicating that the AX as a whole represents the average of different CHH-context methylation levels in specific AX tissues. This situation differs from that observed with the SC (Fig. 3F), and suggests that the decrease in CHH-context methylation observed in the postgermination sdlg might be a consequence of (i) hypomethylated CHH sites within seed AX tissues remaining unmethylated following germination (e.g., AX-PL, AX-PA, and AX-VS), and (ii) preexisting hypermethylated CHH sites being diluted as sdlg cells divide (e.g., AX-RT), both of which were predicted by our seed development and germination results (Fig. 2A). By contrast, the similar levels of CG- and CHG-context methylation in the sdlg compared with the developing seed probably result from preexisting methylated sites in seed AX tissue layers remaining methylated in the postgermination sdlg.
DNA Methylation and Endoreduplication Are Coupled During Soybean Seed Development.
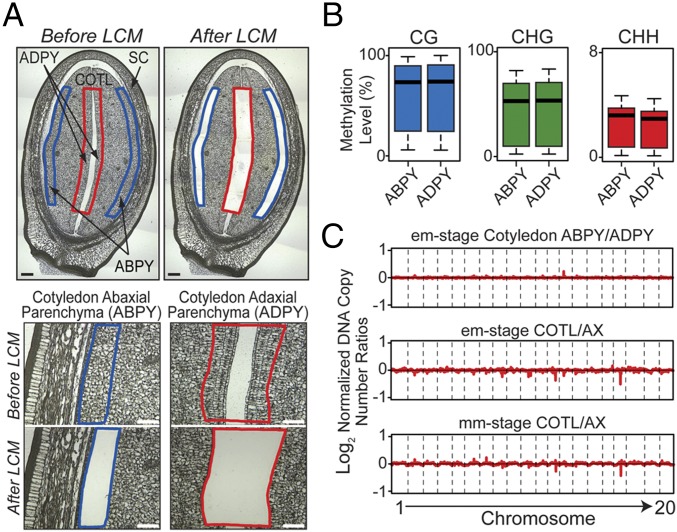
Because the increase in CHH-context DNA methylation coincided with the onset of COTL endopolyploidization (Figs. 1 and 2), we asked whether the DNA methylation landscape was maintained following endoreduplication in soybean seeds. We used LCM to capture em-stage COTL ABPY and ADPY tissues (Fig. 4A), taking advantage of the observation that COTL ABPY and ADPY tissues differ in endoreduplication timing: ABPY undergoes endoreduplication at the em stage, while ADPY does not (25). No significant differences were observed in the bulk methylation levels between endoreduplicating ABPY and nonendoreduplicating ADPY tissues in all three cytosine contexts (Fig. 4B and Dataset S3). In addition, >96% of cytosine sites across the genome retained their methylation status in endoreduplicating ABPY and nonendoreduplicating ADPY tissues. Confirming these observations, both the (i) bulk CG-, CHG-, and CHH-context methylation levels and (ii) cytosine site methylation status did not differ between mm-stage endoreduplicated COTL and nonendoreduplicated AX regions (5) (Fig. 3C and Dataset S3). Taken together, these data indicate that the methylation landscape is maintained following endoreduplication in COTL cells.
Fig. 4.
DNA methylation levels in endoreduplicated and nonendoreduplicated soybean COTL regions. (A) Paraffin cross-sections of em-stage COTL endoreduplicated ABPY and nonendoreduplicated ADPY before and after capture by LCM. (Scale bars, 100 µm.) (B) Box plots of DNA methylation levels in 500-kb windows across the genome in em-stage COTL ABPY and ADPY regions. (C) Log2 ratios of normalized DNA reads in 500-kb windows across all 20 chromosomes from: (i) em-stage ABPY and ADPY COTL regions, and (ii) em and mm seed parts (COTL and AX). See Table 1 for abbreviations.
We compared the DNA sequence coverage along the entire soybean genome for: (i) em-stage COTL ABPY and ADPY tissues, (ii) em-stage COTL and AX regions, and (iii) mm-stage COTL and AX regions, and did not observe any major differences in genome coverage indicating that there was uniform DNA replication along the genome in endoreduplicating cells (Fig. 4C). That is, all DNA sequences in the genome were replicated to the same extent with no selective amplification. These results indicate that DNA methylation is maintained in all sequence contexts during endoreduplication and is highly coordinated with DNA synthesis in the absence of cell division.
Many Genes Important for Soybean Seed Development Are Present in Genomic Regions That Are Not Methylated.
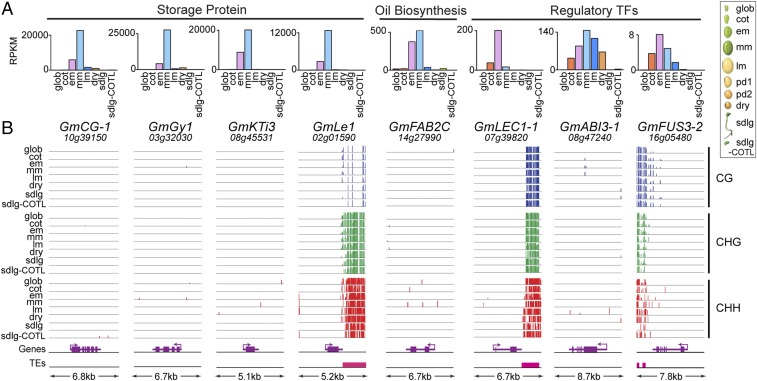
We scanned the seed methylomes in 5-kb sliding windows from postfertilization through dormancy and germination to characterize the methylation landscape surrounding ∼75 genes known to be important for seed development, and determined whether the regulation of these genes was associated with localized methylation changes in any cytosine context (Dataset S4). These included genes encoding: (i) storage proteins (e.g., glycinin and β-conglycinin), (ii) oil biosynthesis proteins (e.g., stearoyl-acyl–carrier-protein desaturase), (iii) transcription factors [e.g., LEAFY COTYLEDON1 (LEC1) and FUSCA3 (FUS3)], and (iv) germination-enhanced proteins (e.g., chlorophyll A/B binding protein and isocitrate lyase). Many laboratories, including our own, demonstrated that these genes are highly regulated and under transcriptional control (1, 26, 27), as suggested by the RNA-Seq data presented here (Fig. 5A, Fig. S5 A, C, and D, and Dataset S4).
Fig. 5.
Methylation levels and mRNA accumulation patterns of major soybean seed-specific gene classes during seed development and germination. (A) mRNA accumulation patterns. RPKM represents reads per kilobase per million sequences, and were taken from the Goldberg-Harada soybean (i) whole seed RNA-Seq dataset, GEO accession no. GSE29163 (37), and (ii) COTL-specific RNA-Seq dataset GSE29134 (sdlg-COTL). (B) Methylation levels of CG-, CHG-, and CHH-context sites are shown in genome browser view (vertical lines). Gene structures, transcription directions (arrows), and TEs are shown below each genome browser view. Adjacent genes are not shown. The size of each genomic region, including 2 kb of 5′ and 3′ flanking regions, is shown at the bottom. GmABI3-1, abscisic acid insensitive3-1; GmCG-1,β-conglycinin-1; GmFAB2C, stearoyl-ACP desaturase 2C; GmFUS3-2, FUSCA 3-2; GmGy1, glycinin 1; GmKTi3, Kunitz trypsin inhibitor 3; GmLEC1-1, Leafy Cotyledon 1-1; GmLe1, lectin 1. See Table 1 for developmental stage abbreviations. Gm, Glycine max.
Surprisingly, almost half of the seed and germination genes we investigated were localized within genomic regions designated as demethylated valleys (DMVs) (28), that averaged <5% bulk methylation level in any cytosine sequence context across all stages (Fig. 5, Fig. S5, and Dataset S4). Some of these regions extended for >50 kb but, on average, were 16 kb (Dataset S4). The methylation status of these regions did not change from fertilization through dormancy and germination, whereas the seed and germination genes in these regions were highly regulated (Fig. 5, Fig. S5, and Dataset S4). The remainder of genes we investigated fell into three categories: (i) genes with methylated TEs in their 5′ and 3′ flanking regions, (ii) genes with gene body methylation, and (iii) genes with gene body methylation and TEs in their 5′ and 3′ flanking regions (Fig. 5, Fig. S5, Dataset S4). In each case, however, no significant CG-, CHG-, or CHH-context methylation changes were observed within or flanking the seed and germination genes, although these genes were highly regulated during development (Fig. 5 and Fig. S5). In addition, previous cis-element analysis of the GmLe1 and GmKTi1 genes that have methylated flanking TEs (Fig. 5B and Fig. S5E) showed that these TEs were not required for regulation during seed development (29–31). Taken together, these data suggest that regulation of many important seed and germination genes is primarily due to transcriptional events that are independent of DNA methylation changes, in agreement with our observations three decades ago using more primitive technology (26).
The Methylation Landscape of Soybean Seeds Is Conserved in Arabidopsis.
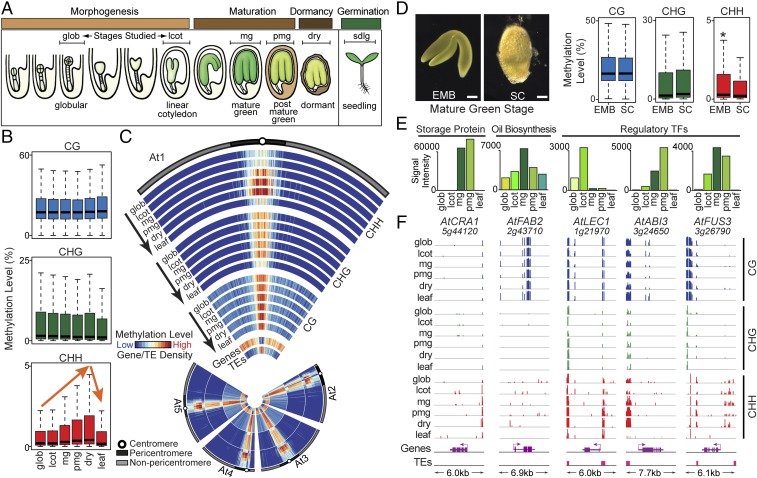
We carried out a series of BS-Seq experiments with Arabidopsis seeds at stages comparable with those studied in soybean to determine whether our methylation observations were specific for soybean seeds or found generally in higher plants (Datasets S1–S3). Stages investigated included those undergoing: (i) morphogenesis and differentiation [glob and linear cot (lcot) stages], (ii) maturation [mature green (mg) and postmature green (pmg) stages], (iii) dormancy (dry seeds), and (iv) postgermination (leaves from 3-wk-old plants) (Fig. 6A). We obtained 45–100× coverage of the 120-Mb Arabidopsis genome for each seed stage and part investigated (Dataset S1), and observed that: (i) the base composition of our reads reflected that of the Arabidopsis genome (Fig. S1C), and (ii) on average, the bulk methylation levels of cytosines in the CG, CHG, and CHH contexts were 24.6%, 7.7%, and 1.6%, respectively, (Dataset S2),values similar to those obtained in other Arabidopsis methylome studies (32).
Fig. 6.
Genome-wide methylation changes during Arabidopsis seed development. (A) Arabidopsis seed stages and major developmental events. Adapted from ref. 38). Seed and EMB images are not drawn to scale. Brackets indicate stages investigated. Box plots (B) and chromosome heat maps (C) of DNA methylation levels in 100-kb windows across the genome. The highest methylation levels were 96%, 80%, and 10% for CG, CHG, and CHH contexts, respectively. Gene and TE tracks represent densities of genes and TEs in 100-kb windows along the genome. (D) Box plots of DNA methylation levels in 100-kb windows across the genome in mg-stage EMB and SC. (Scale bars, 0.1 mm.) Asterisk indicates a significant comparison between EMB and SC (t test, P < 0.001 and fold change > 1.5). mRNA accumulation patterns (E) and genome browser views of DNA methylation levels (F) for major seed-specific gene classes. Transcript signal intensities were obtained from microarray analysis (38). Methylation levels of CG-, CHG-, and CHH-context sites are shown in genome browser view (vertical lines). Gene structures, transcription directions (arrows) and TEs are shown below each genome browser view. Adjacent genes are not shown. The size of each genomic region, including 2 kb of 5′ and 3′ flanking regions, is shown at the bottom. AtCRA1, Cruciferin 1. Names of other genes are defined in the legend to Fig. 5. At, Arabidopsis thaliana. See Table 1 for abbreviations of seed stages.
Surprisingly, the methylation events observed during Arabidopsis seed development were indistinguishable from those observed in soybean seeds. These included: (i) no significant changes in CG- and CHG-context bulk methylation levels during seed development and germination (Fig. 6 B and C and Dataset S3); (ii) a significant increase and decrease in both CHH-context bulk methylation levels and methylated sites (t test and Fisher’s exact test, P value < 0.001 and >1.5-fold change) within pericentromeric region TEs during maturation and germination, respectively (Fig. 6 B and C, Fig. S2 C and D, and Dataset S3); and (iii) CHH-context hypomethylation of the mg-stage SC relative to the EMB (t test, P < 0.001 and >1.5-fold change) (Fig. 6D and Dataset S3). Finally, genes encoding major classes of proteins important for seed development (e.g., storage proteins, oil biosynthesis, TFs) were: (i) localized within DMV regions with <5% bulk methylation levels across all of seed development (e.g., AtCRA1), (ii) methylated within their gene bodies (e.g., AtFAB2) or flanking regions (AtLEC1, AtABI3, AtFUS3), and (iii) either regulated in the absence of any detectable methylation events or changes that were not correlated with their expression programs (Fig. 6 E and F), similar to what was observed in soybean (Fig. 5 and Fig. S5). Together, these data suggest that the methylation landscape of soybean and Arabidopsis seeds is highly conserved despite a divergence of ∼90 My (14), and that the programmed changes in CHH-context methylation during development and between different seed regions may be a common feature of dicot seeds.
Seed Development Occurs Normally in a Mutant Arabidopsis Line Lacking CHG and CHH Methylation.
To explore the possible role, if any, that the increase in CHH-context methylation plays in seed development, we took advantage of the Arabidopsis ddcc mutant (drm1drm2cmt2cmt3) (33) that is deficient in all methyltranserases [DOMAINS REARRANGED METHYLTRANSFERASE1 (DRM1), DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2), CHROMOMETHYLASE2 (CMT2), and CHROMOMETHYLASE3 (CMT3)] required for non-CG–context methylation (15). We reasoned that the ddcc mutant would provide an excellent test of the functional relevance of seed CHH-context methylation because CHG-context methylation levels do not change in soybean and Arabidopsis seeds from fertilization through dormancy and germination.
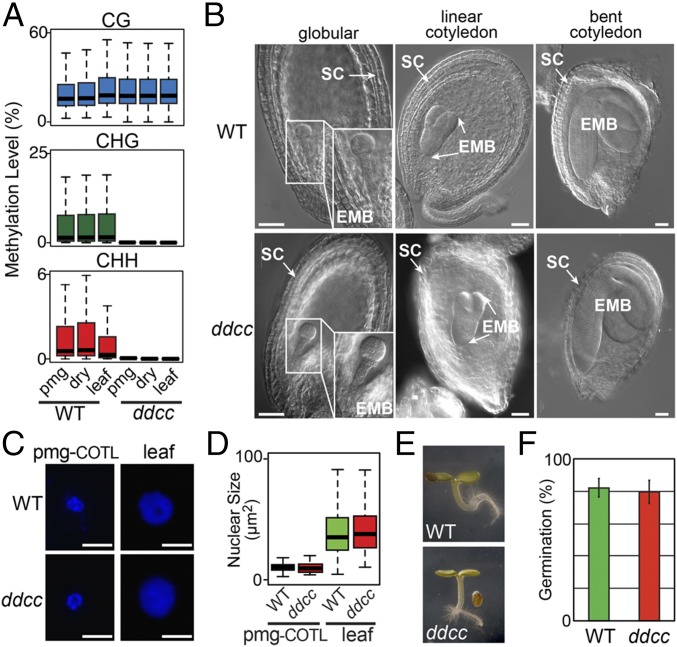
We did not detect any CHG- or CHH-context methylation in ddcc pmg seeds, dry seeds, or postgermination leaves in comparison with wild-type (Fig. 7A), as expected from knocking out genes required for non-CG–context methylation (15). ddcc seeds developed normally with no detectable morphological differences from wild-type at our level of resolution (Fig. 7B). In addition, ddcc COTL nuclei underwent normal shrinkage in pmg seeds, which is a marker for the desiccation events that occur at the end of seed development (Fig. 7 C and D), and then regained their size following germination (34). Finally, ddcc and wild-type seeds had the same levels of germination (Fig. 7 E and F). Taken together, these data suggest that non-CG methylation does not play a significant role in Arabidopsis seed development, and that the absence of programmed CHH-methylation changes does not affect seed morphogenesis, maturation, dormancy, or germination.
Fig. 7.
Comparison between Arabidopsis wild-type and ddcc seed development and germination. (A) Box plots of methylation levels in 100-kb windows across the wild-type and ddcc genomes. (B) Nomarski photographs of wild-type and ddcc seeds at different developmental stages. (Scale bars, 50 µm.) (C) DAPI-stained nuclei of pmg-COTL and leaves. (Scale bars, 5 µm.) Comparison of nuclear sizes (110 nuclei) (D), 4-d sdlg morphologies (E), and germination percentages (F) for wild-type and ddcc seeds. Five replicates with 50 seeds each were used in the germination assays.
Gene Expression Is Not Affected Significantly in Arabidopsis ddcc Seeds.
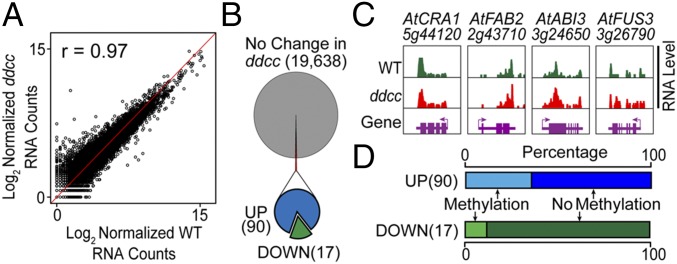
We compared the transcriptomes of Arabidopsis ddcc and wild-type pmg seeds to determine whether the loss of non-CG methylation, and CHH-context changes in particular, affected seed gene expression. Quantitative and qualitative mRNA levels in ddcc and wild-type seeds were not significantly different from each other (Fig. 8 A and B), including mRNAs encoding storage proteins (e.g., AtCRA1), fatty acids (e.g., AtFAB2), and major regulators of seed development (e.g., AtABI3, AtFUS3) (Fig. 8C). We obtained a 0.97 correlation coefficient between pmg ddcc and wild-type seed transcriptomes, which was the same as that obtained between either ddcc or wild-type biological replicates (Fig. S6A). Although the vast majority of the 19,638 mRNAs detected in ddcc pmg seeds were present in wild-type seeds at the same levels, we did detect a small number of ddcc mRNAs that were either up-regulated (90 genes) or down-regulated (17) [false-discovery rate (FDR) < 0.001 and >fivefold change] (Fig. 8B and Dataset S5). Gene ontology analysis showed that down-regulated genes were enriched for response to stress, while the up-regulated genes were associated with abiotic stress response and cellular component organization/anatomical structure formation (Dataset S6). We examined the 5′ flanking regions of the 107 differentially expressed ddcc genes, and found that ∼70% had no CHG- or CHH-context methylated sites in corresponding wild-type genes (Fig. 8D and Dataset S5). This suggests that mostly indirect effects were responsible for change in gene activity observed in ddcc pmg seeds. Together, these results suggest that non-CG methylation, including elevated CHH-context methylation levels, does not play a major role in regulating seed gene activity.
Fig. 8.
Comparison between Arabidopsis wild-type and ddcc pmg seed transcriptomes. (A) Correlation between wild-type and ddcc seed mRNA accumulation levels. (B) Differentially expressed genes in ddcc seeds. DOWN, down-regulated; UP, up-regulated. (C) Genome browser view of major seed-specific mRNA accumulation patterns in ddcc and wild-type seeds. Arrows show the transcription directions. Gene names are defined in the legends to Figs. 5 and 6. (D) Methylation status of 5′ flanking 1-kb region of differentially expressed genes.
Many TE mRNAs Are Up-Regulated in Arabidopsis ddcc Seeds.
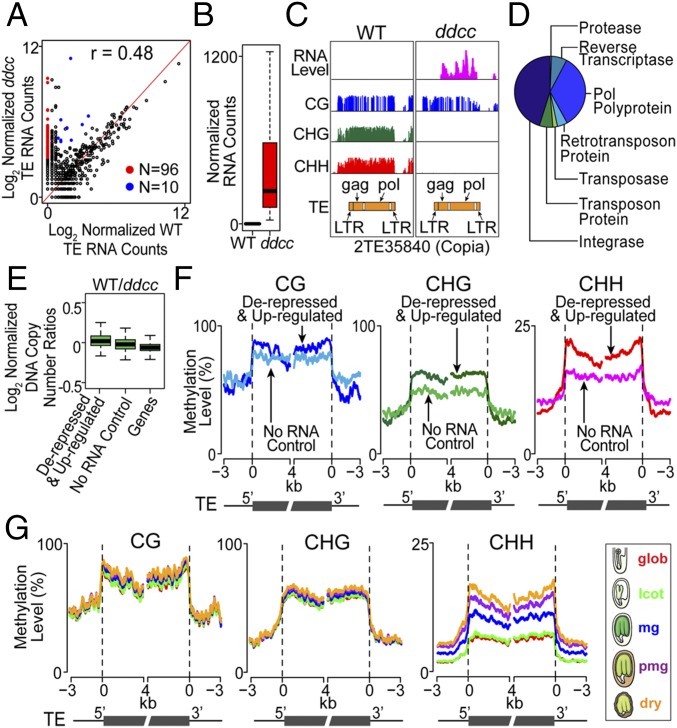
We investigated whether any Arabidopsis TEs were de-repressed at the RNA level in pmg ddcc seeds as a consequence of non-CG methylation loss (Fig. 7A). Qualitative and quantitative differences were observed between pmg ddcc and wild-type TE RNAs (0.48 correlation coefficient) (Fig. 9A), by contrast with the biological replicate controls (Fig. S6B). We found that 96 TEs were de-repressed transcriptionally from a silenced state, while 10 TEs were up-regulated >sixfold (FDR < 0.01) (Fig. 9 A–C and Dataset S7). TE RNAs were transcribed from different retrotransposon and DNA TE families, although TE transcripts were most numerous from retrotransposon classes (Fig. S7 A and B). TE RNAs encoded proteins responsible for copy number increase and transposition, including transposase, integrase, and reverse transcriptase (Fig. 9D). We compared the copy number of de-repressed and up-regulated TEs in ddcc and wild-type genomes (35), and did not detect any significant differences, implying that these TEs did not undergo transposition events within the seed generation that we investigated (Fig. 9E).
Fig. 9.
TE transcriptional activity in Arabidopsis wild-type and ddcc pmg seeds. (A) Correlation between wild-type and ddcc seed TE RNA accumulations levels. Red and blue dots represent de-repressed and up-regulated TEs, respectively. (B) Box plots comparing de-repressed and up-regulated TE RNA accumulation levels in wild-type and ddcc seeds. (C) Genome browser view of the methylation pattern and RNA accumulation profile of a de-repressed Copia TE in pmg wild-type and ddcc seeds. LTR, long terminal repeat. (D) Major protein classes involved in TE transposition encoded by 106 de-repressed and up-regulated ddcc seed TE RNAs (SI Materials and Methods). (E) TE copy number changes between ddcc and wild-type seeds. Box plots show log2 ratios of normalized read depths from ddcc versus wild-type TEs. The gene control includes all genes in the Arabidopsis genome. (F) Methylation levels across 106 de-repressed and up-regulated TEs in wild-type seeds. The no RNA control used in E and F represent 106 randomly selected TEs which have (i) no detectable RNA wild-type reads and (ii) similar TE family distribution and lengths compared with the 106 de-repressed and up-regulated TEs. (G) Methylation levels across 106 de-repressed and up-regulated TEs in Ws-0 wild-type seeds during Arabidopsis seed development.
We investigated the methylation landscape of the 106 ddcc de-repressed and up-regulated TEs in the wild-type pmg seed genome to determine how loss of non-CG methylation resulted in their transcriptional activation in ddcc seeds. We randomly selected 106 silenced TEs (i.e., no detectable RNA-Seq reads) with a similar distribution of TE classes and lengths as controls. The up-regulated and de-repressed TEs had significantly lower CG densities (t test, P < 0.001) compared with control TEs, by contrast with CHG and CHH densities, which did not differ from the control TE set (Fig. S7C and Dataset S8). CG-, CHG-, and CHH-context bulk methylation levels were significantly higher in the de-repressed and up-regulated TEs compared with the repressed controls, suggesting that there were insufficient methylated CG sites to prevent TE transcription in ddcc seeds (Fig. S7D and Dataset S8). Examination of the CG- and CHG-context methylation levels across the de-repressed and up-regulated TE bodies showed that they were similar to the TE controls (Fig. 9F). By contrast, the distribution of CHH-context methylation across the 106 de-repressed and up-regulated TEs differed significantly from the control set, and showed that there was a prominent increase in CHH methylation levels at the TE ends where the promoter sequences reside (Fig. 9F). The CHH-context methylation levels at the promoter sites and along the entire TE bodies increased significantly from fertilization through dormancy during seed development, by contrast to CG- and CHG-context methylation, which did not change (Fig. 9G). Together, these data imply that de-repression of TEs in ddcc pmg seeds might be caused primarily by the loss of highly methylated CHH-context sites within TE promoter regions, and suggest that the programmed increase in CHH-context methylation during soybean and Arabidopsis seed development might be a failsafe mechanism to ensure TE silencing.
Discussion
We determined that there is a programmed increase in CHH-context methylation during seed development from the glob stage through dormancy in both soybean and Arabidopsis seeds. The majority of this upswing occurs during the maturation phase in all major seed parts—SC, COTL, and AX—and is maintained during endoreduplication of the COTL genome. Although the SC layer is hypomethylated in CHH sites relative to other seed parts, the increase in CHH-context methylation occurs simultaneously throughout the seed. By contrast, CG- and CHG-context methylation does not change significantly during the same period of seed development, or in any specific seed part. Following germination, CHH-context methylation drops precipitously in germinating COTL and the emerging sdlg. Thus, the change that occurs in seed CHH-context methylation appears to be conserved in dicots, and is regulated with respect to space and time during the development of the seed. Very recently, an independent analysis of our Arabidopsis whole-seed data results in a similar conclusion (36).
The increase in seed CHH-context methylation occurs across the entire soybean and Arabidopsis genomes, targets primarily TEs, and is neutral with respect to TE class. Both DNA transposons and retrotransposons are targeted, as well as TEs that are either clustered in heterochromatic pericentromere regions or dispersed among genes in euchromatic chromosome arms. This implies that there is a coordinated targeting of CHH-context sites in TEs during seed development, most likely being directed by RdDM and non-RdDM pathway methylases DRM1, DRM2, CMT2, and CMT3, respectively (15, 20). Inspection of the soybean (GSE29163) (37) and Arabidopsis (GSE680) (38) seed transcriptome databases indicates that mRNAs encoding these methylases are present when the CHH-context methylation events occur both temporally and spatially during seed development, supporting this premise (Dataset S9). Following germination the decrease in CHH-context methylation is most likely caused by the methylation status of different dry seed AX regions that give rise to the sdlg. For example, methylated CHH sites within the AX-RT become diluted as the root cells divide and increase in number within the germinating sdlg. By contrast, hypomethylated sites within the AX-PY and AX-PL retain their status as these regions give rise to the sdlg hypocotyl and leaf, respectively.
What role does the CHH-context methylation increase play in seed development? We investigated this issue by using an Arabidopsis mutant that is defective in both RdDM and non-RdDM pathways, and has no detectable non-CG methylation. ddcc seeds develop normally, have no detectable morphological defects, and germinate with frequencies indistinguishable from wild-type. In addition, the absence of non-CG context methylation does not appear to affect seed gene activity significantly because the mRNA profiles of ddcc and wild-type seeds are congruous with each other at both the qualitative and quantitative levels, including genes critical for seed development.
The simplest hypothesis for the role of increasing CHH-context methylation in seed development may be that it is a failsafe mechanism for reinforcing TE silencing in seeds. We favor this hypothesis because the programmed increase in CHH-context methylation during seed development targets TEs across the genome, and over 106 TEs rich in CHH sites at their ends (i.e., promoter regions) are de-repressed or up-regulated (>sixfold) at the RNA level in ddcc seeds. Although we found no evidence for these TEs moving, or increasing in copy number, in the generation we investigated, the up-regulated TE RNAs encode the requisite proteins (e.g., transposase, reverse transcriptase) that might unleash these TEs in subsequent generations (35). If this were the case, the consequences could be devastating for the seed and lead to lethality either before or after germination, or detrimental effects could accumulate more gradually over multiple generations. This hypothesis is consistent with the prevailing role for non-CG context methylation in higher plants (20).
One of the most intriguing aspects of our results is the observation that a large number of highly regulated soybean and Arabidopsis genes involved in major seed regulatory and metabolic events are localized in regions that are depleted of methylation (<5%) in any cytosine context during development, regardless of whether these genes are active or repressed. These include major regulatory genes, as well as genes encoding storage proteins and oil biosynthesis enzymes that are critical for maturation and germination. These seed genomic regions are similar to the DMVs observed in mammalian genomes during the differentiation of stem cells (28), and suggest strongly that genes present in these regions (e.g., storage protein genes) are not regulated directly by methylation changes, a conclusion that we made over three decades ago (26). Other highly regulated seed genes lie within regions containing heavily methylated TEs, or have methylation within their gene bodies, or both (Figs. 5B and 6F, Fig. S5, and Dataset S4). However, similar to genes in the seed DMVs, the methylation patterns of genes in these regions do not vary, whether the genes are active or not, indicating that methylation changes do not play a major role in regulating genes in these genomic regions as well (Figs. 5B and 6F, Fig. S5, and Dataset S4). This conclusion is enhanced by the fact that the loss CHG- and CHH-context methylation in pmg ddcc seeds does not significantly affect seed gene activity or development. We conclude that the major challenge for understanding the mechanisms that control seed development is to uncover cis-regulatory elements and corresponding TFs that control the spatial and temporal expression of essential seed genes, and determine how they are integrated into the genetic regulatory networks (39) that program seed development from generation to generation.
Materials and Methods
Specific details are contained within SI Materials and Methods.
Plant Material and LCM.
The growth conditions and staging of soybean seeds [Glycine max (L.) cv. Williams 82] and Arabidopsis seeds [wild-type (Ws-0); ddcc (Col-0)] are detailed in SI Materials and Methods, following the procedures of Goldberg et al. (40) (soybean) and Le et al. (38) (Arabidopsis). AX, SC, and COTL were dissected manually from soybean seeds at the em and mm stages. LCM (41) was used to capture soybean: (i) cot-stage seed parts (AX, SC and COTL), (ii) em-stage seed parts (SC and COTL), (iii) em-stage COTL parenchyma tissues (ABPY and ADPY), (iv) em-stage SC layers (PA and PY), and (v) em-stage AX subregions and tissues (PL, PY, RT, and VS). EMB and SC were hand dissected from Arabidopsis seeds at the mg stage. Sample preparation procedures for the LCM experiments are detailed in SI Materials and Methods.
BS-Seq Library Construction, Methylome Sequencing, Data Processing, and Sequence Analysis.
Genomic DNA was isolated from soybean and Arabidopsis whole seeds and hand-dissected seed parts using the DNEASY Plant Mini kit (Qiagen). DNA from seed tissue captured by LCM was isolated using the QIAMP FFPE DNA isolation kit (Qiagen). DNA was prepared for BS-Seq library preparation and methylome sequencing following the methods of Hsieh et al. (10) and Lister et al. (42), with modifications (SI Materials and Methods). DNA sequences were aligned to the soybean genome (version Wm82.a1; https://www.soybase.org) (43) or Arabidopsis genome (version TAIR10; https://www.arabidopsis.org/index.jsp) (44) using BS Seeker software (45), allowing up to two base mismatches. The sequencing depth for each cytosine in the reference genome was defined as the total number of detected cytosines (methylated C) or thymines (unmethylated C) across all reads. The specific procedures that we used to determine whether an individual cytosine site was methylated, as well as the bulk methylation level for a given genomic feature (e.g., region, gene, TE) are described in detail in SI Materials and Methods.
RNA-Seq Library Construction, Sequencing, Data Processing, and Analysis.
RNA was isolated from soybean whole seeds, seed parts, and sdlg using the Concert Plant RNA Reagent (Invitrogen) and treated with RNase-free DNase I (Ambion). Poly-A+ RNA was selected using oligo-dT25 magnetic beads (Dynabeads). Poly-A+ RNA was prepared for RNA-Seq library construction using the Illumina mRNA-Seq Sample Prep Kit (Illumina). For Arabidopsis pmg seeds, total RNA was used to generate double-stranded cDNA using the Ovation RNA-Seq System V2 (Nugen), and RNA-seq libraries were constructed using the Illumina TruSeq DNA Sample Prep Kit (Illumina). Bowtie (46) was used to map sequenced reads to: (i) the soybean genome (version Wm82.a1) and cDNA models (version Wm82.a1.v1.1) (https://www.soybase.org) (43) or (ii) the Arabidopsis genome (version TAIR10) and cDNA models (https://www.arabidopsis.org/index.jsp) (44). The EdgeR package (v3.18.1) (47) was used to identify differentially expressed RNAs.
Arabidopsis ddcc Seed Analysis.
Arabidopsis ddcc seeds were obtained from Steve Jacobsen, University of California, Los Angeles (15). Detailed information for characterization of (i) seed morphology, (ii) nuclear size, (iii) seed germination, and (iv) differentially expressed RNAs is presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Steve Jacobsen and his laboratory for Arabidopsis ddcc mutant seeds, excellent suggestions for how to analyze methylated transposable elements, and help with characterizing seed nuclei. This work was supported by a grant from the National Science Foundation Plant Genome Program (to R.B.G., M.P., and J.J.H.), and a National Institutes of Health Training Grant in Genomic Analysis and Interpretation T32HG002536 (to B.H.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [soybean methylome (accession nos. GSE34637, GSE37893, GSE37895, GSE41061, and GSE57762); Arabidopsis methylome (accession nos. GSE57755, GSE68131, and GSE68132); soybean transcriptome (accession nos. GSE29134, GSE29163, and GSE37895); and Arabidopsis transcriptome (accession no. GSE76447)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716758114/-/DCSupplemental.
References
- 1.Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: Zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- 2.Becker MG, Hsu S-W, Harada JJ, Belmonte MF. Genomic dissection of the seed. Front Plant Sci. 2014;5:464. doi: 10.3389/fpls.2014.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gehring M, Satyaki PR. Endosperm and imprinting, inextricably linked. Plant Physiol. 2017;173:143–154. doi: 10.1104/pp.16.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devic M, Roscoe T. Seed maturation: Simplification of control networks in plants. Plant Sci. 2016;252:335–346. doi: 10.1016/j.plantsci.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Dhillon SS, Miksche JP. DNA, RNA, protein and heterochromatin changes during embryo development and germination of soybean (Glycine max L.) Histochem J. 1983;15:21–37. doi: 10.1007/BF01006069. [DOI] [PubMed] [Google Scholar]
- 6.Larkins BA, et al. Investigating the hows and whys of DNA endoreduplication. J Exp Bot. 2001;52:183–192. [PubMed] [Google Scholar]
- 7.Sallon S, et al. Germination, genetics, and growth of an ancient date seed. Science. 2008;320:1464. doi: 10.1126/science.1153600. [DOI] [PubMed] [Google Scholar]
- 8.Bauer MJ, Fischer RL. Genome demethylation and imprinting in the endosperm. Curr Opin Plant Biol. 2011;14:162–167. doi: 10.1016/j.pbi.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park K, et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci USA. 2016;113:15138–15143. doi: 10.1073/pnas.1619047114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing MQ, et al. Global analysis reveals the crucial roles of DNA methylation during rice seed development. Plant Physiol. 2015;168:1417–1432. doi: 10.1104/pp.15.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raissig MT, Bemer M, Baroux C, Grossniklaus U. Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet. 2013;9:e1003862. doi: 10.1371/journal.pgen.1003862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nodine MD, Bartel DP. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature. 2012;482:94–97. doi: 10.1038/nature10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant D, Cregan P, Shoemaker RC. Genome organization in dicots: Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc Natl Acad Sci USA. 2000;97:4168–4173. doi: 10.1073/pnas.070430597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz RJ, et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013;23:1663–1674. doi: 10.1101/gr.152538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song QX, et al. Genome-wide analysis of DNA methylation in soybean. Mol Plant. 2013;6:1961–1974. doi: 10.1093/mp/sst123. [DOI] [PubMed] [Google Scholar]
- 18.Kim KD, et al. A comparative epigenomic analysis of polyploidy-derived genes in soybean and common bean. Plant Physiol. 2015;168:1433–1447. doi: 10.1104/pp.15.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg RB, Hoschek G, Tam SH, Ditta GS, Breidenbach RW. Abundance, diversity, and regulation of mRNA sequence sets in soybean embryogenesis. Dev Biol. 1981;83:201–217. doi: 10.1016/0012-1606(81)90467-x. [DOI] [PubMed] [Google Scholar]
- 20.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19:320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawashima T, Berger F. Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet. 2014;15:613–624. doi: 10.1038/nrg3685. [DOI] [PubMed] [Google Scholar]
- 23.Martínez G, Panda K, Köhler C, Slotkin RK. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants. 2016;2:16030. doi: 10.1038/nplants.2016.30. [DOI] [PubMed] [Google Scholar]
- 24.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Nielsen NC. 2004. Endoreduplication during soybean seed development. PhD dissertation (Purdue University, West Lafayette, IN)
- 26.Walling L, Drews GN, Goldberg RB. Transcriptional and post-transcriptional regulation of soybean seed protein mRNA levels. Proc Natl Acad Sci USA. 1986;83:2123–2127. doi: 10.1073/pnas.83.7.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comai L, Dietrich RA, Maslyar DJ, Baden CS, Harada JJ. Coordinate expression of transcriptionally regulated isocitrate lyase and malate synthase genes in Brassica napus L. Plant Cell. 1989;1:293–300. doi: 10.1105/tpc.1.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindstrom JT, Vodkin LO, Harding RW, Goeken RM. Expression of soybean lectin gene deletions in tobacco. Dev Genet. 1990;11:160–167. doi: 10.1002/dvg.1020110206. [DOI] [PubMed] [Google Scholar]
- 30.de Paiva G. 1994. Transcriptional regulation of seed protein genes. PhD dissertation (University of California, Los Angeles)
- 31.Yadegari R. 1996. Regional specification and cellular differentiaiton during early plant embryogenesis. PhD dissertation (University of California, Los Angeles)
- 32.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Zanten M, et al. Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc Natl Acad Sci USA. 2011;108:20219–20224. doi: 10.1073/pnas.1117726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marí-Ordóñez A, et al. Reconstructing de novo silencing of an active plant retrotransposon. Nat Genet. 2013;45:1029–1039. doi: 10.1038/ng.2703. [DOI] [PubMed] [Google Scholar]
- 36.Kawakatsu T, Nery JR, Castanon R, Ecker JR. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 2017;18:171. doi: 10.1186/s13059-017-1251-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danzer J, et al. Down-regulating the expression of 53 soybean transcription factor genes uncovers a role for SPEECHLESS in initiating stomatal cell lineages during embryo development. Plant Physiol. 2015;168:1025–1035. doi: 10.1104/pp.15.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le BH, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–985. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg RB, Hoschek G, Ditta GS, Breidenbach RW. Developmental regulation of cloned superabundant embryo mRNAs in soybean. Dev Biol. 1981;83:218–231. doi: 10.1016/0012-1606(81)90468-1. [DOI] [PubMed] [Google Scholar]
- 41.Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 44.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 45.Chen P-Y, Cokus SJ, Pellegrini M. BS seeker: Precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203–208. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25–R34. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le BH, et al. Using genomics to study legume seed development. Plant Physiol. 2007;144:562–574. doi: 10.1104/pp.107.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saski C, et al. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 50.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 52.Sanger F, et al. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 53.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du J, et al. SoyTEdb: A comprehensive database of transposable elements in the soybean genome. BMC Genomics. 2010;11:113–119. doi: 10.1186/1471-2164-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 58.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katari MS, et al. VirtualPlant: A software platform to support systems biology research. Plant Physiol. 2010;152:500–515. doi: 10.1104/pp.109.147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee T-F, et al. RNA polymerase V-dependent small RNAs in Arabidopsis originate from small, intergenic loci including most SINE repeats. Epigenetics. 2012;7:781–795. doi: 10.4161/epi.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moissiard G, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yadegari R, et al. Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell. 1994;6:1713–1729. doi: 10.1105/tpc.6.12.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.