Abstract
Here, we describe a protocol to generate expandable and multipotent induced cardiac progenitor cells (iCPCs) from mouse adult fibroblasts using forced expression of Mesp1, Tbx5, Gata4, Nkx2.5, Baf60c (MTGNB) along with activation of Wnt and JAK/STAT signalling. This method does not use iPS cell factors and thus differs from cell activation and signaling-directed reprogramming to cardiac progenitors. The protocol describes in detail the method to isolate and infect primary fibroblasts, induce reprogramming and observe emergence of iCPC colonies, expand and characterize reprogrammed iCPCs by immunostaining, flow cytometry and gene expression, differentiate iCPCs in vitro into cardiac lineage cells, including cardiomyocytes, smooth muscle cells, endothelium, and test the embryonic potency of iCPCs via injection into the cardiac crescent of mouse embryos. A scientist experienced in cell-molecular biology and embryology can reproduce this protocol in 6–8 weeks. iCPCs may be useful for studying cardiac biology, drug discovery and regenerative medicine.
INTRODUCTION
Transdifferentiation technology using lineage-specific defined factors has generated a variety of terminally differentiated cell types, including neurons1 and hepatocytes2, without the necessity of going through an intermediate pluripotent cell state. More recently, master regulators of cell fate, as well as culture conditions adapted for expansion in vitro of native tissue-specific stem cells have been exploited to reprogram fibroblasts into proliferative progenitor cells of neural3, hepatic4, oligodendrocyte5 and hematopoietic lineages6. Direct reprogramming into cardiomyocytes has also been accomplished7–12. However, due to the lack of consensus on master regulators of the cardiac progenitor cell state and culture conditions required to stabilize cardiac progenitor cells (CPCs) in vitro, reprogramming into expandable CPCs proved challenging.
Development of the Protocol
We recently reported that adult mouse fibroblasts can be directly reprogrammed into expandable induced cardiac progenitor cells (iCPCs)13. iCPCs are cardiac mesoderm-restricted progenitors that can be extensively passaged, and show multipotency toward cardiovascular lineages (cardiomyocytes, smooth muscle cells and endothelium) in vitro as well as after transplantation into the embryonic cardiac crescent or into the adult post-myocardial infarction heart. iCPCs hold potential advantages over pluripotent stem cell (PSC)-derived cells as they do not require pluripotent precursor cells. This may be beneficial if iCPCs are used for cell therapy due to there being a reduced tumorigenic risk. Also, iCPC reprogramming is more efficient compared to reprogramming to the induced pluripotent stem cell (iPSC) state followed by differentiation to CPCs14. iCPCs hold promise as they are expandable and have a greater potency for differentiation and repair compared to directly reprogrammed induced cardiomyocytes (iCM), which are not expandable, or to adult heart-derived CPCs that undergo age-related senescence. iCPCs can generate large quantities of desired cardiovascular cell lineages required for drug discovery, and they may serve as a model system for unraveling cardiovascular disease. Overall, iCPC reprogramming technology potentially has broad applications for understanding the molecular mechanism(s) involved in reprogramming, for studying cardiac development and physiology, for modeling cardiovascular diseases and for advancing drug discovery and cardiac regenerative medicine.
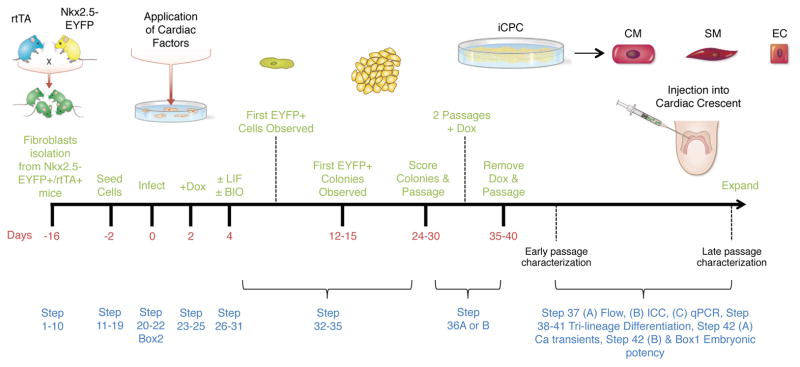
We hypothesized that fibroblasts could be reprogrammed into proliferative and multipotent iCPCs using knowledge of embryonic cardiovascular development and defined factor-mediated reprogramming. Towards this end, we generated a doxycycline-inducible lentivirus library of 22 factors to screen for factors that could reprogram fibroblasts into iCPCs. We used a unique Nkx2.5-EYFP reporter system in which EYFP is specifically expressed at the cardiac progenitor cell stage (E7.5 – E9.5) and is turned off during later stages of cardiac development, including the adult heart15. We devised a two-stage screening strategy. In Stage 1, we isolated adult fibroblasts from Nkx2.5-EYFP/rtTA double transgenic mice (which do not express Nkx2.5-EYFP), and screened for defined factors and signaling molecules that activated the Nkx-reporter and produced proliferative EYFP+ colonies. In Stage 2, we assessed whether the resulting EYFP+ colonies could be stably expanded without forced expression of cardiac factors. Using this rigorous screening approach, we discovered that five cardiac factors (Mesp1, Tbx5, Gata4, Nkx2.5, Baf60c), along with activation of Wnt and JAK-STAT signaling, resulted in complete reprogramming of adult mouse fibroblasts into iCPCs. Figure 1 details the stages involved in reprogramming mouse fibroblasts into iCPCs, and their characterization.
Figure 1. Experimental design.
Illustration depicting various steps and stages in reprogramming adult mouse fibroblasts into iCPCs, characterization of iCPCs and potency testing in vitro as well as in mouse embryos.
iCPCs are cardiac mesoderm-restricted progenitors that express CPC transcription factors (TFs), including Nkx2.5, Gata4, Irx4 (Figure 2), and cell surface markers, including Cxcr4, Flk1 and cKit. iCPCs can differentiate in vitro into alpha-actinin-, alpha-MHC-, cardiac actin-, MLC-2a-, and MLC-2v-expressing cardiomyocytes, as well as SM-MHC-positive smooth muscle cells and CD31-expressing endothelial cells (Figure 3).
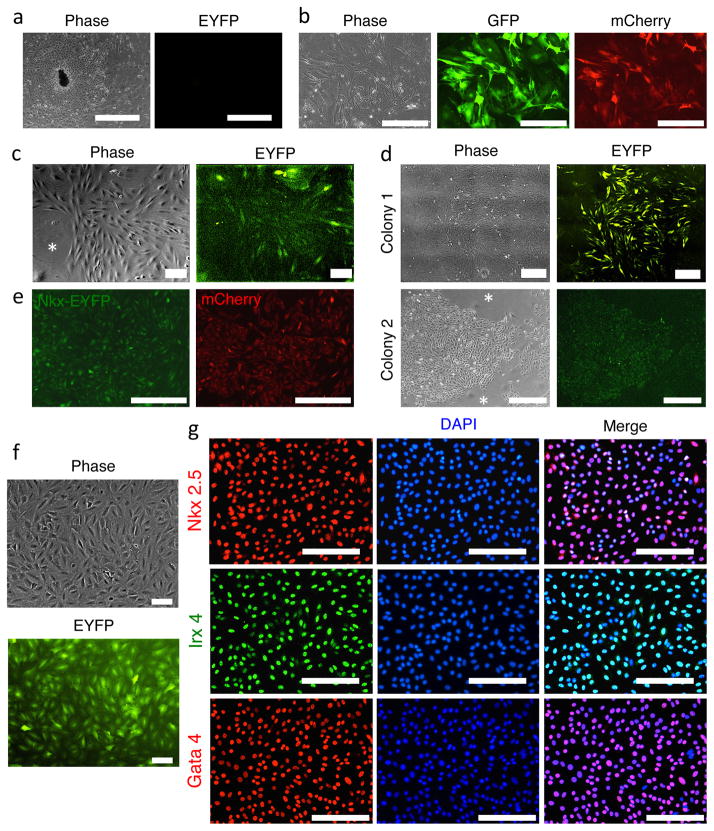
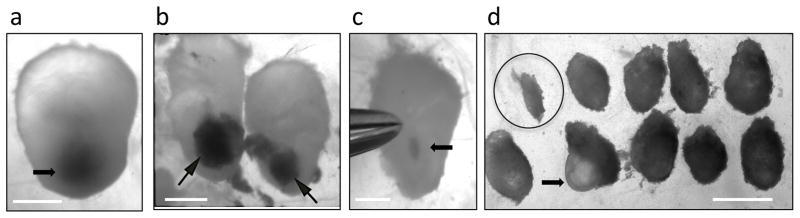
Figure 2. Transduction of adult fibroblasts with cardiac factors, identification of reprogramming cells based on morphological changes and characterization of iCPCs.
(a) Cardiac fibroblasts migrating out of adult heart tissue pieces during explant culture. The cardiac fibroblasts as well as heart tissue do not express Nkx-EYFP. (b) Cardiac fibroblasts infected with doxycyclin inducible GFP lentivirus were imaged 24 hr after doxycycline induction. >90% of infected cells showed GFP and mCherry fluorescence only upon doxycycline induction, indicating high infection efficiency and a working inducible system. (c) Morphologically distinct, proliferative, EYFP+ cells appear 10–14 days after infecting adult cardiac fibroblasts with cardiac defined factors and culturing in iCPC Induction Medium. Note the striking morphological difference between EYFP+ iCPCs and EYFP- fibroblasts (indicated by *) (d) 3–4 weeks after initiation of protocol cells undergoing iCPC reprogramming develop into 2 dimensional, proliferative EYFP+ colonies that reach 3–4mm in diameter. Note the striking morphological difference between EYFP+ iCPCs and EYFP- fibroblasts (indicated by *) (e) iCPC colonies show co-expression of Nkx-EYFP and mCherry indicating that cells expressing cardiac factors (indicated by IRES mCherry expression) are undergoing reprogramming (turning on Nkx-EYFP reporter). (f) iCPCs maintain Nkx-EYFP expression and can be expanded for upto 30 passages. (g) Immunocytochemistry showing stably reprogrammed iCPCs having nuclear localization of CPC transcription factors including Nkx2.5, Gata4 and Ix4. Scale bar=1000um in a, 400um in b and e, 100um in c and f, 500um in d, 200 um in g.
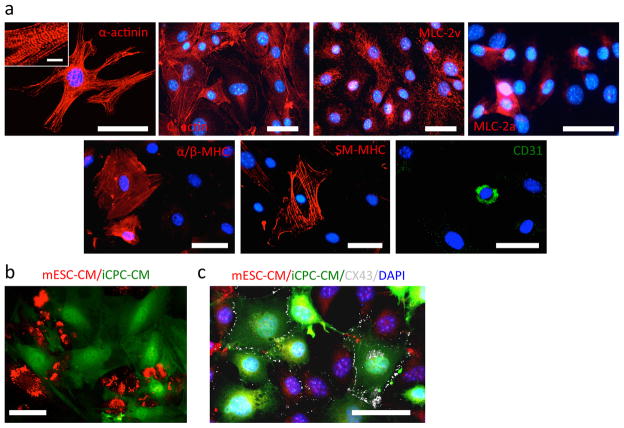
Figure 3. Differentiation of iCPCs into cardiac lineage cells.
(a) Immunocytochemistry showing iCPCs differentiate into cardiac lineage cells including cardiomyocytes (alpha-actinin, cardiac actin, MLC-2v, MLC-2a, alpha-MHC), smooth muscle cells (SM-MHC) and endothelial cells (CD 31). (b) Live imaging showing GFP+ iCPCs co-cultured with td-tomato+ CMs derived from mouse embryonic stem cells. Absence of detectable GFP+/td-tomato+ cells indicated cell fusion was unlikely during co-culture (c) Cx43 immunolabeling showing iCPC-CMs (green) and mESC-CMs (red) develop gap junctions upon co-culture. iCPC-CMs and mESC-CMs show synchronous calcium transients upon co-culture due to presence of gap junctions. Scale bar=50um in a and b, 5um in inset.
Embryonic Potency Test
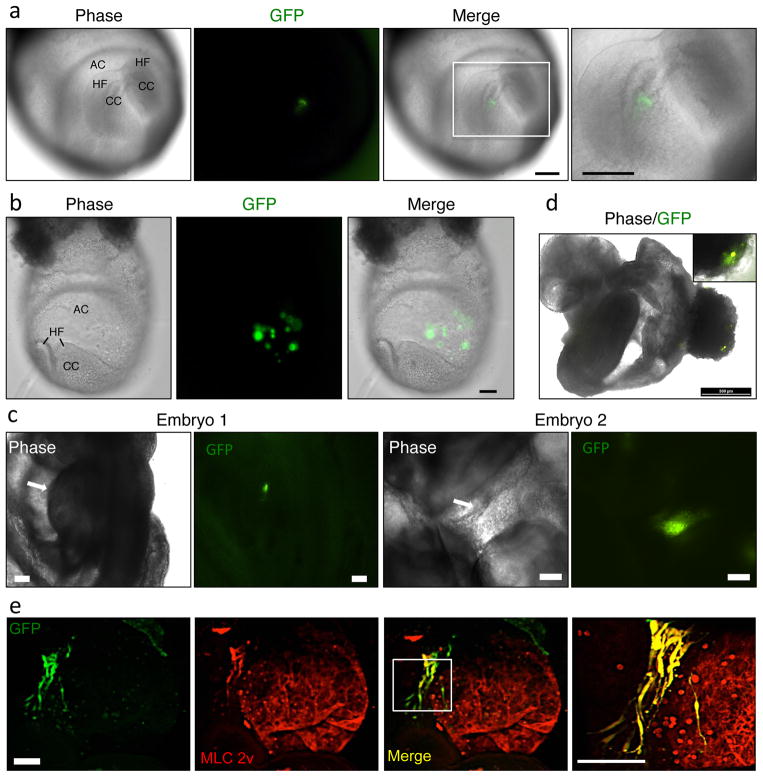
CPCs have been identified through the expression of various biomarkers in the developing embryo, in differentiating pluripotent stem cell cultures and in adult heart. They have been characterized extensively in vitro with respect to their gene and protein expression16–18. CPCs can be differentiated in vitro to yield cardiomyocytes, smooth muscle cells and endothelium, indicating that they are multipotent. However, differentiation is operationally defined19, and stem cell potency can only be fully evaluated in the native microenvironment of the organism. Unfortunately, the embryonic potency of most described CPC populations has not undergone such rigorous scrutiny. If iCPCs are bona fide progenitor cells of the cardiac lineage, we reasoned that they should respond to cardiomorphogenic signals present in the embryonic cardiac crescent and differentiate into cardiomyocytes, especially as our Nkx2.5-EYFP reporter identifies embryonic CPCs. Hence, we injected iCPCs into the cardiac crescent of mouse embryos, where native CPCs are present, and assessed their differentiation potential. iCPCs indeed integrated into the developing heart tube and differentiated into cardiomyocytes13.
Comparison to Other Methods
Direct reprogramming of fibroblasts to induced cardiomyocytes (iCMs) was accomplished in 2010 by Ieda et al8. They reported in vitro transdifferentiation of mouse neonatal cardiac fibroblasts into iCMs using three transcription factors (TFs) Gata4, Tbx5, and Mef2c (GMT)8. Subsequently, a number of groups reported iCM reprogramming in both mouse and human cells using various combinations of TFs as well as microRNAs7, 9–12. A detailed protocol to reprogram mouse cardiac neonatal fibroblasts into iCMs using GMT has previously been published20. Recently, direct reprogramming of human fibroblasts into iCMs has been achieved using small molecules only21. iCM reprogramming in vivo has also been demonstrated in the mouse heart via viral delivery of cardiac factors following myocardial infarction22, 23. However, the above reports also provided evidence that iCM reprogramming does not pass through a cardiac progenitor intermediate. iCMs like CMs are terminally differentiated non-proliferative cells. Combined with low in vitro reprogramming efficiency, this may not yield the required quantity of cells for regenerative medicine applications or drug discovery. On the contrary, reprogramming to a cardiac progenitor cell state has several advantages. Cardiac progenitors are multipotent and can differentiate not only into cardiomyocytes, but also smooth muscle and endothelial cells, which are the necessary compliment of cells required to fully reconstitute the damaged heart. Moreover, cardiac progenitors are proliferative and hence scalable to yield large quantitates of desired cells (progenitors or differentiated progeny. An initial study aiming to reprogram human fibroblasts to cardiac progenitors was unable to isolate and stabilize a proliferative, multipotent progenitor cell24.
We first reported lineage reprogramming of fibroblasts into expandable and multipotent cardiac progenitor cells13. Independently, Zhang et al. confirmed this finding using an alternate method25. Although both reprogramming methods generate cardiac progenitors, the approaches are fundamentally different. Specifically, Zhang et al. used mouse embryonic fibroblasts (MEFs) derived from a transgenic mouse line that was engineered to express Yamanaka-iPSC factors upon addition of doxycycline, called “secondary MEFs”. Further, they used transient expression of iPSC factors in secondary MEFs to induce a partially reprogrammed epigenetically unstable cell state, which could be directed towards a CPC state using signaling molecules that included Bmp4, Activin A, and Wnt activation (CHIR99021), while at the same time inhibiting signaling by FGF, VEGF, PDGF, (SU5402) and JAK/STAT (JI1). This approach is referred to as “cell activation and signaling-directed (CASD) reprogramming” and has been used by the same group to reprogram fibroblasts into various cell types, including cardiomyocytes, hepatocytes and neural stem cells20, 26. Thus, the CASD reprogramming system is not specific for the CPC state, but rather, it has the potential to generate cell types derived from all three germ layers depending upon the signaling pathways used. Recent reports have suggested that transdifferentiation to both cardiomyocytes and neural stem cells using CASD reprogramming transitions through an intermediate pluripotent state, which raises tumorigenic concerns27, 28. By contrast, we used adult fibroblasts from a cardiac progenitor specific Nkx2.5-EYFP reporter mouse, screened for cardiac-specific genes that induce iCPC reprogramming and then used Wnt and JAK/STAT activation to stabilize the reprogrammed iCPCs in vitro. Finally, Zhang et al. used FACS (Flk+/Pdgfr+) to identify reprogrammed cells, whereas we used morphological changes and Nkx2.5-EYFP expression to identify iCPCs.
Applications of the Protocol and Limitations
The current protocol uses lentiviruses that encode cardiac factors for reprogramming; these integrate into the genome which can produce undesired integration effects. However, the protocol can be adapted to test integration-free methods such as episomal vectors, Sendai viruses or modified mRNAs. The protocol can also be optimized for screening small molecules that improve reprogramming efficiencies or to replace defined factors. Using the in vitro differentiation protocol described here, the majority of iCPCs differentiate into cardiomyocytes. However, the differentiation protocol can be adapted to suit differentiation into smooth muscle cells and endothelial cells. This protocol uses mouse adult fibroblasts as the starting source for reprogramming. In our experience mouse fibroblast cultures can contain karyotypically abnormal cells, which may result in chromosomal abnormalities in resulting iCPCs. Hence, we recommend screening to identify karyotypically normal iCPC lines. Other investigators have reported that lineage reprogramming protocols optimized for mouse cells can require significant adaptation to enable reprogramming of human cells11, 12. The majority of direct reprogramming protocols have been discovered using mouse embryonic or neo-natal cells, which, even though they are epigenetically more amenable to reprogramming, are less clinically relevant, as comparable human cells are not available. Our protocol has been developed for reprogramming adult mouse fibroblasts, for which clinically relevant human cells are available. Hence, this protocol will facilitate reprogramming of human fibroblasts into iCPCs, which will be an important step in moving this technology to human cardiac regenerative medicine. The screening strategy and characterization criteria described in this protocol can be used to derive and benchmark novel CPC populations from embryos, pluripotent stem cells, as well as from the adult heart. The unique embryo injection model described here can be utilized to test the embryonic potency of other identified CPC populations, including CPCs derived from pluripotent stem cells29 and adult heart derived CPCs. In addition to testing potency of cardiac cells, this protocol can be used to test the potency of cells for other mesodermal cell types, as well as for definitive ectoderm, definitive endoderm, and extra-embryonic cell types by injecting them into respective test sites. A limitation of the whole embryo culture technique presented here is that it has been designed to culture embryos ex-vivo for 24–48 hr until they reach E9.5. We have not tested culturing embryos beyond E9.5 using this set-up.
Experimental Design
Starting cell source
This protocol was developed using adult mouse fibroblasts as a starting cell source for reprogramming. It is critical to have healthy and proliferative fibroblasts cultures for reprogramming. Hence, we recommend using fibroblasts that are between 0–3 passages. Also, the cardiac factor viruses used in this protocol are doxycycline inducible, and require co-expression of the reverse tetracycline trans-activator (rtTA) protein in order to be functional. It is important to confirm that the fibroblasts contain the rtTA transgene before initiating reprogramming experiments. Although rtTA can be introduced into wild type cells using a lentivirus constitutively overexpressing rtTA, we recommend using fibroblasts from mice homozygous or heterozygous for the rtTA transgene to achieve highest reprogramming efficiency.
Lentivirus optimization
Lentivirus production is a complex, multistage process that needs to be optimized before initiating reprogramming experiments. For efficient lentivirus production it is very important to start with high quality 293TN cells and DNA vectors. We recommend using low passage 293TN cells and confirming that DNA vectors are of high purity. Lentivirus, envelope and packaging vectors are prone to recombination. Hence, it is critical to check their integrity before starting lentivirus production (see Box 2 for more details). We recommend using the pSAM2 GFP lentivirus vector as a starting point to optimize virus production (Box 2 can be used as guide). Infect mouse fibroblasts with pSAM2 GFP lentivirus to confirm the efficiency of infection (Step 11–26) by checking for GFP expressing cells. Optimize parameters including 293TN seeding density, lipofectamine ratio, volume of virus supernatant used for infection, duration of virus infection, polybrene concentration etc to achieve at least 80% infection efficiency. If expected infection efficiency is not achieved, consider concentrating lentiviruses using commercially available kits.
BOX 2. Production of lentivirus particles ● TIMING 5 days.
Day 1 - Setting up HEK 293 TN cells for virus production ● TIMING 1–2 hr
-
1
Plate HEK 293 TN cells at a density of 4.5 x 106 cells per 100 mm dish in HEK medium.
△ CRITICAL STEP Do not use cells over passage 20. Check the alignment of the incubator shelf with a spirit level to make sure it is perfectly horizontal; this is important for ensuring even distribution of cells and efficient transfection.
?TROUBLESHOOTING
Day 2 – DNA transfection using Lipofectamine 2000 ● TIMING 1–2 hr
-
2
Check cell confluence before setting up transfections. Cells should be 85–90% confluent.
-
3
Add 1 ml of Opti-MEM medium each into two separate sterile transfection tubes. Add 7ug of pSAM2 lentivirus vectors (individual gene plasmids or pooled plasmids for 5-factor combination – Mesp1, Tbx5, Gata4, Nkx2.5, Baf60c), 10ug of lentiviral packaging vector psPAX2 (Addgene #12260), 5ug of lentiviral envelope vector pMD2.G (Addgene #12259) in one tube and 44 ul of Lipofectamine 2000 into second tube. Incubate at RT for 5–10 min.
△ CRITICAL STEP Lentiviral vectors are prone to recombination events due to the presence of long terminal repeats (LTRs). We recommend using Stbl3 strain of E. coli for all lentiviral vectors to reduce recombination events. However, before transfection, all lentivirus-associated vectors must be analyzed by restriction digestion to confirm absence of recombination. pSAM2 vectors should be digested with AflII and NotI (determine expected bands for each gene). psPAX2 should be digested with BamHI and EcoRI (expected bands: 4374bp, 3712bp, 1273bp, 1007bp, 337bp). pMD2.G should be digested with NotI and EcoRI (expected bands:3358bp, 1671bp, 795bp). Qiagen Endofree maxi prep should be used for DNA purification.
△ CRITICAL STEP For every batch of lentivirus production, at least 1 plate should be reserved for pSAM2 GFP lentivirus production. This will be used as control for every batch of reprogramming experiments using cardiac factors.
?TROUBLESHOOTING
-
4
Combine the two tubes and mix. Incubate at RT for 30–40 min.
-
5
Transfer 2ml Opti-MEM medium containing DNA and lipofectamine into a sterile 15 ml conical tube, add 3 ml of HEK Medium and mix thoroughly. This will be referred to as “transfection medium”, below.
△ CRITICAL STEP Reducing the total volume of transfection medium to 5ml and reducing serum concentration from 10% to 6% improves transfection efficiency to up to 90–95%.
-
6
Aspirate HEK Medium from plates and gently add 5ml of transfection medium from step 5 onto cells. Return plates to incubator and incubate for 16–20 hr.
△ CRITICAL STEP Add transfection medium slowly from the side of the dish to avoid detaching HEK cells.
Day 3 – Remove transfection medium ● TIMING 30 min
-
7
Remove transfection medium and feed cells with 5.5 ml of fresh HEK Medium. Return plates to incubator and incubate for 48–54 hr.
△ CRITICAL STEP Add fresh HEK Medium slowly from the side of the dish to avoid detaching HEK cells.
! CAUTION Transfection medium should be discarded in 10% bleach solution after removal and treated with ultraviolent light for at least 1 hr before discarding.
Day 5 – Harvest lentivirus particles and ● TIMING 1 hr
-
8
Analyze the cells in the microscope before harvesting medium. Most cells should be attached and have a ‘swollen appearance’. This morphology indicates high transfection efficiency and optimal virus production. Some floating cells will be observed.
-
9
Gently harvest the medium with a sterile 5ml pipette and collect in sterile 15 ml conical tube. Centrifuge at 400g for 5 min to pellet floating HEK cells.
-
10
Carefully remove the supernatant without disturbing the HEK pellet and filter through 0.45 um filter using a syringe. Lentiviral supernatant can be aliquoted in 1 or 5 ml sterile 15 ml conical tubes and immediately stored at − 80° C or used to infect fibroblasts.
△ CRITICAL STEP Lentivirus supernatant should be used within 2 freeze thaw cycles.
■ PAUSE POINT Lentivirus supernatant can be stored at − 80° C for at least 1 yr.
! CAUTION All disposable equipment that comes into contact with lentiviral supernatant should be discarded in 10% bleach and treated with ultraviolent light for at least 1 hr before discarding.
MATERIALS
REAGENTS
Addgene plasmids - psPAX2 (12260), pMD2.G (12259), pLenti GFP Puro (17448)
Agarose, Low melting point, Analytical (Promega, V2111)
Alexa 647 Rat IgG2g, k isotype control (BD,552849)
Alexa 647 Rat IgG2g, k isotype control (BioLegend, 400626)
BIO (Cayman Chemical, 13123)
BMP4 (R&D, 5020-BP-010)
Bovine Serum Albumin (BSA) (Sigma, A9418)
Breeder chow (Teklad Global 19% Protein Extruded Rodent Diet, Envigo Teklad, Indianapolis, IN)
Cysteine-HCl (Sigma, C-1276)
CFX96 Touch Real-Time PCR Detection System (Bio-Rad)
DAPI (ThermoFisher, D1306)
DMEM (ThermoFisher, 11885-092) – used to make HEK and fibroblasts culture medium
DMEM +4500 g/L glucose + NaPyruvate (110mg/mL) w/o NaHCO3 (Gibco-BRL 12800-017) – used for whole embryo culture CRITICAL
DMSO (Sigma, D2650-100ML)
Doxycycline (Sigma, D9891)
Double-distilled (tissue culture grade) sterile water (Sigma W-3500)
Endofree Maxi-prep kit (Qiagen, 12362)
Ethanol, 70% (vol/vol)
F2 generation of embryos from intercrosses of B6CBAF1/J (Jackson Laboratories)
Fetal Bovine Serum (FBS), heat-inactivated (ThermoFisher, 16140071)
Fetal Calf Serum (FCS), heat-inactivated (Gibco-BRL 16141-044) – used for embryo dissection medium and whole embryo culture medium
FGF2 (R&D, 3139-FB-025)
Formaldehyde, 16% (Polysciences, 18814-10) !CAUTION Hazardous chemical. Wear proper protect gear including lab coat, gloves etc. while handling.
Gelatin (Sigma, G9136)
Goat anti-mouse IgG (H+L), Alexa 488, 2mg/ml (ThermoFisher, A-11001)
Goat anti-mouse IgG (H+L), Alexa 568, 2mg/ml (ThermoFisher, A-11004)
Goat anti-mouse IgG (H+L), Alexa 647, 2mg/ml (ThermoFisher, A-21235)
Goat anti-mouse IgM (H+L), Alexa 568, 2mg/ml (ThermoFisher, A-21043)
Goat anti-mouse, IgG/IgM (H+L), Alexa 488, 2mg/ml (ThermoFisher, A-10680)
Goat anti-rabbit IgG (H+L), Alexa 568, 2mg/ml (ThermoFisher, A-11011)
Goat anti-rat IgG (H+L), Alexa 568, 2mg/ml (ThermoFisher, A-11077)
Goat serum (ThermoFisher, 16210-072)
HEK/293 TN cells (SBI, LV900A-1) !CAUTION The cell lines used in your research should be regularly checked to ensure they are authentic and are not infected with mycoplasma.
Hepes, free acid (Sigma H-3375)
iCPC defined factor plasmids, Addgene – Mesp1 (72687), Tbx5 (72688), Nkx2.5 (72689), Gata4 (72690), Baf60c (72691), Irx4 (72692), Mesp2 (72693), Gata6 (72694), SRF (72695), Isl1 (72696), Tbx20 (72697), GFP (78172).
Igloos (K3327 red; Bio-Serv, Flemington, NJ)
Isofluorane (MidWest Vet 558.09058.3) !CAUTION Anesthetic. Use under fume hood.
Isopropanol (ThermoFisher, A416-500) !CAUTION Highly flammable liquid and vapour.
IWP4 (Stemgent, 04-0036)
L-alanine (Sigma, A-7469)
L-asparagine (Sigma, A-0884)
L-aspartic acid (Sigma, A-8949)
L-glutamic acid (Sigma, G-1626)
L-Glutamine, 200mM (ThermoFisher, 25030-081)
L-proline (Sigma, P-5607)
LIF (Millipore, ESG1106)
Lipofectamine 2000 (ThermoFisher, 11668019)
Mouse anti-alpha actinin IgM (Sigma, A5044)
Mouse anti-alpha MHC IgG1,k, 1mg/ml (Thermo Scientific, ma1-26180)
Mouse anti-Cardiac actin IgG1, 1mg/ml (Sigma, A9357)
Mouse anti-connexin 43 IgG1, 250ug/ml (BD, 610061)
Mouse anti-GFP IgG2a (ThermoFisher, A-11120)
Mouse anti-Irx4 IgG/IgM (Abgent)
Mouse anti-MLC2a IgG, 1mg/ml (Synaptic Systems, 311011)
Mouse anti-Nkx2.5 IgG1, 0.5 mg/ml (R&D, MAB2444)
Mouse anti-Smooth muscle actin IgG (Thermo Scientific, MS113-P1)
NaHCO3 (Sigma S-8875)
NaOH, 5N, for adjusting pH (Sigma, 656046)
Nkx2.5-EYFP mice (C57BL/6J strain, Garry Lab, University of Minnesota) and rtTA mice (C57BL/6J strain, Jackson Laboratories, 006965) !CAUTION Any experiments involving live mice must conform to relevant Institutional and National regulations. All animal experiments in this protocol were performed in accordance to University of Wisconsin-Madison’s animal use guidelines.
Non-essential Amino Acids, 100X (ThermoFisher, 11140-050)
One shot Stbl3 chemically competent E.coli (ThermoFisher, C737303)
Opti-MEM Medium (ThermoFisher, 31985-088)
PE Rat IgG2a isotype control (BD, 553930)
Penicillin-Streptomycin (ThermoFisher, 15140-122) – this is used in cell culture media
Penicillin-Streptomycin, 5000 Units/ml, (Invitrogen, 15070-063) – this is used in embryo dissection and whole embryo culture medium
Phosphate-buffered saline (PBS; Sigma, P-4417) dissolved in double-distilled water and filter-sterilized for mouse uterine dissections; stored at 4°C. Also made with non-sterile distilled water and used in Embryo Blocking Solution.
Phosphate-buffered saline (PBS) (ThermoFisher, 10010049) – sterile, use for tissue culture
Polybrene (Sigma, H9268)
Prolong Gold Antifade Solution (ThermoFisher, P10144)
Rabbit anti-Gata4 IgG, 200ug/ml (Santacruz, sc-9053)
Rabbit anti-GFP IgG, 2mg/ml (ThermoFisher, A11122)
Rabbit anti-MLC2v IgG, 200ug/ml (ProteinTech Group, Inc., 10906-1-AP)
Rabbit anti-SMMHC IgG (Biomedical Technologies, BT-562)
Rat anti-CD31 IgG2a, 1mg/ml (BD, 553369)
Rat anti-cKit PE-cy7 IgG2b, k, 200ug/ml (BD, 558163)
Rat anti-Cxcr4-Alexa 647 IgG2b, k, 0.5mg/ml (Biolegend, 146503)
Rat anti-Flk-PE IgG2a, 200ug/ml (BD, 555308)
Rat serum, heat-inactivated; commercially available, but if too hemolyzed, must prepare in house 30
Rhod-2 (ThermoFisher, R1245MP)
RNA Aqueous kit (ThermoFisher, AM1931)
RNA Zap (ThermoFisher, AM9780)
Ssofast EvaGreen SuperMix (Bio-Rad, 1725201)
Triton X-100 (Sigma, X100)
Trypsin-EDTA (ThermoFisher, 25200-072)
VEGF (R&D, 493-MV-005)
EQUIPMENT
−20°C freezer
−80°C freezer
0.22 um filter medium (ThermoFisher, 09-740-22K)
100 mm tissue culture dish (Fisher, 08-772-E)
12-well tissue culture plate (Fisher, 07-200-82)
12mm glass coverslips (12-545-82)
15ml sterile conical tubes (VMR, 101093-696 (CS))
19-gauge syringe needle (1) (1.5″ long)
1ml 28.5 or 29.5 g hypodermic needles
24-well low attachment tissue culture plate (Sigma, CLS3473-24EA)
24-well tissue culture plate (Fisher, 09-761-146)
50 ml polypropylene conical tubes (VMR, 101093-574 (CS))
6-well tissue culture plate (Fisher, 07-200-83)
60 mm tissue culture dish (Fisher, 08-772-B)
96-well tissue culture plate (Fisher, 07-200-87)
Arkansas stone (Fine Science Tools, 29008-01)
Aspirator tube assemblies (Sigma A-5177-5EA) – the microcapillary holder is a useful piece for making the microflame, as well; replace the Sigma mouthpiece, which is awkwardly round, with “Mouthpiece, flat”
Cell strainer (Fisher, 08-771-2)
Clamp Holder (max grip ¾”; Fisher 05 754) to hold nylon clamp with extension rod so as to secure hypodermic needle for microflame onto ring stand
Clamp hosecock (Fisher 05 847) for regulating gas flow to microflame
Clamp, nylon, with extension stem (Fisher 058868) to hold hypodermic needle for microflame
Cloning cylinder (Sigma, C1059-1EA)
Cryo box (Fisher, 15-350-107)
Cryogenic vials, Nalgene, 5 mL capacity, sterile (Fisher 03-337-7H)
Depression slides for whole mount embryo staining (VWR, 470019-022)
Dissection microscope, total magnification 7.5 – 35.0X, equipped with trans- and epi-illumination
Electrode puller, Sutter, Model P-87, with a trough filament (0.45 mm – FT345B)
Erlenmeyer flasks (2), 1000 mL, sterile
EVOS AL Auto microscope (ThermoFisher, AMAFD1000)
Filter units, 1000 mL – 0.22 μm cellulose acetate (Fisher 09 740 40A)
Filter units, 500 mL – 0.22 μm cellulose acetate (Fisher, 09 740 28C)
Filter, 0.22 μm filter (Fisher 09-754-13)
Filter, 0.45 μm (Fisher 09-754-21)
Flint striker
Forceps, 1 pair, “Jewellers” straight (Fisher, 09 953E) – these are used to dissect uteri from female
Forceps, 2 pairs, fine, straight, #5 Dumoxel (Fine Science Tools, 11252-30) – these are used to remove conceptuses from implantation sites
Forceps, 2 pairs, robust, straight, Inox (Fine Science Tools, 11251-20) – these are used to reflect Reichert’s membrane
Forceps, rounded (for checking copulation plugs) (Fisher 08890)
Gas regulator for CO2 tank
Glass scorer, diamond tip (Fisher 11-315)
Glass tubing, thin-walled (O.D. 1.00 mm, I.D. 0.85 mm; 150 mm long) (World Precision Instruments, X838)
Gloves (Quest, G-4048)
Graduated cylinder, 1000 mL, tissue culture-dedicated, sterile
Graduated cylinder, 500 mL, tissue culture-dedicated, sterile
Haemocytometer (Sigma, Z359629-1EA)
Heated stage or 37°C heated chamber for inverted tissue culture microscope
Hemostat (“Halstead Mosquito Forceps”), 5″, straight (Fisher 13-812-8)
Image J software (https://imagej.nih.gov/ij/)
Incubator, water-jacketed set to 37°C and 6.2% CO2, dedicated for whole embryo culture
Instrument holder, Leica, for glass capillaries (optional; Leica Microsystems, 11520145)
Inverted tissue culture microscope (Nikon)
Kimwipes (Fisher, 06-666A)
Liquid Nitrogen cryostorage tank
Microflame (These are not commercially available but made in the lab)
Microforge (deFonbrune; circa 1950), for breaking off injection pipettes at desired bore widths
Microscope filters for epifluorescence (and excitations): GFP, DAPI, Texas Red, CY5, Phase contrast.
Mouthpiece, flat (HPI Hospital Products (a division of MEDTECH International, Inc. P.O.Box 162992, Altamonte Springs, FL 32716-2992; Tel: 1-407-880-6904; Fax: 1-407-880-6908.1501)
Epifluorescence microscope (Nikon)
Organ culture dishes (Fisher, 08 772 12)
Parafilm
Pasteur pipettes, 9″ sterile, non-absorbent cotton-plugged for bubbling CO2 gas to culture medium before storage at −86°C.
Pasteur pipettes, 9″, sterile
Pasteur pipettes, wide-bore, to accommodate larger embryos, cut via glass scorer, diamond tip
pH indicator strips, 5.0–10 (VWR, EM-9588-3); 6.5–10 (VWR, EM-9583-3).
pH meter
Pipettor, automatic, dedicated to tissue culture
Plastic-backed disposable paper
Roller apparatus (Fisher, 14-277-4; Fisher, 14-277-3) inserted into the incubator, set at 8° off the horizontal, and rotating continuously at 0.5 rpm
Safety goggles (used when making wide bore pipettes)
Sandpaper, very fine (any local hardware store).
Scissors, 1 pair, small, straight, 4.5″ (Fisher, 08-940)
Sharps container for used syringes and test tubes
Spirit level
Squeeze bottle with distilled water.
Stir apparatus, magnetic
Stir flea, tissue culture-dedicated
Storage boxes for injection pipettes (Fisher 15 350 55), containing an insert of flexible foam scored with a razor blade
Syringe needles, 21-gauge × 1.5″ long
Syringes, 10, 20 mL, sterile
Test tubes, 15 mL, sterile, polypropylene conical (Fisher, 05 538 53D)
Test tubes, 50 mL, sterile, polypropylene conical (Fisher, 14 959 49A)
Test tubes, disposable glass (Fisher, 14 961 32) inserted into the rollers as “adaptors” for embryo culture tubes
Test tubes, whole embryo culture (Falcon 2003, 12 × 75 mm, sterile, individually-wrapped, Fisher, 14 959A)
Timer (multi-channel)
Tissue culture 4°C refrigerator
Tissue culture centrifuge (Eppendorf, 5427 R)
Tissue culture hood, laminar flow
Tissue culture incubator
Tissue culture pipettes: 5ml (Fisher, 13-678-11D), 10ml (Fisher, 13-678-11E), 25ml (Fisher, 13-678-11)
Transfection tubes (Fisher, 14-959-1A)
Tubing, clear, to deliver CO2 gas to medium
Tubing, latex, OD, 3/16″; ID, 1/8″;W, 1/32″ (Sigma Z25, 577-7)
Tubing, rubber, 1/4 × 1/16″ (12 ft roll, Fisher 14178C)
Tubing, rubber, 5/32 × 3/64″ (12 ft roll, Fisher 14178A)
Vacuum apparatus
Water bath, 37°C and 56°C
Weigh boats (VWR, 89106-766)
REAGENT SETUP
0.1% (wt/vol) Gelatin Solution
This solution contains 0.1% (wt/vol) gelatin in PBS. Measure 0.5 gm of gelatin powder and add to 500 ml PBS. Heat and stir on hot plate for 15 min or until the gelatin granules dissolve. Allow solution to return to room temperature/25° C (RT) and then filter-sterilize using 0.2 um filter. Store gelatin solution at RT for up to several weeks or at 4° C for long-term storage.
Gelatin-Coated Dishes
Add enough 0.1% gelatin solution to cover the entire surface of tissue culture dishes/wells and place in incubator for at least 30 min before use, or overnight when possible. Dishes can be kept in incubator for up to 1week before use. Remove gelatin solution before seeding cells.
BIO (10mM stock solution)
Directly add 280 ul of sterile DMSO to 1mg BIO to make a 10 mM stock solution. Protect from light and store at −20° C for long-term storage. Store stock at 4° C for up to several weeks. Use 0.25 ul/ml for a final concentration of 2.5 uM.
Doxycycline (2mg/ml stock solution)
Add 20 mg of doxycycline hyclate powder to 10 ml PBS to make 2 mg/ml stock solution and filter-sterilize through a 0.45 um syringe filter. Protect from light and store long-term at −20° C or at 4° C for up to several weeks. Use 4 ul/ml for a final concentration of 8ug/ml.
BMP4 (100 μg/ml stock solution)
Reconstitute 10 ug of recombinant BMP4 in 100 ul sterile 4mM HCl solution containing 0.1% (wt/vol) BSA. Store at −80° C for up to 6 months. Use 0.5 ul/ml for a final working concentration of 50ng/ml.
FGF2 (100 μg/ml stock solution)
Reconstitute 25 ug of recombinant FGF2 in 250 ul sterile PBS containing 0.1% (wt/vol) BSA. Store at −80° C for up to 6 months. Use 0.3 ul/ml for a final concentration of 30ng/ml.
VEGF (5 μg/ml stock solution)
Reconstitute 5 ug of recombinant VEGF in 1ml sterile PBS containing 0.1% (wt/vol) BSA. Store at −80° C for up to 6 months. Use 2 ul/ml for a final concentration of 10ng/ml.
IWP4 (1.2 mM stock solution)
Add 3.36 ml sterile DMSO to a vial containing 2mg IWP4. Protect from light and store at −20° C for long-term storage or at 4° C for up to several weeks. Use 4.2 ul/ml of for a final concentration of 5 uM.
0.1% (vol/vol) Triton X-100 solution
Add 50 ul of Triton X-100 to 50 ml PBS (non-sterile). Vortex vigorously to mix. Store solution at 4° C up to 6 months.
DAPI Solution (5mg/ml)
Add 5 mg of DAPI powder to 1 ml PBS (non-sterile). Store at −20° C for long term or at 4° C for up to several months. Use 1 ul/ml (1:1000) dilution in immunostaining protocol.
Polybrene Solution (10mg/ml stock solution)
Add 100 mg of polybrene powder to 10 ml PBS and filter-sterilize using 0.45 um syringe filter. Dispense stock into 1 ml aliquots in sterile tubes and store at −20° C indefinitely. Add 0.8ul/ml for a final concentration of 8ug/ml. Thaw just before use and discard any unused stock – do not re-freeze.
HEK Culture Medium
This medium contains 10% heat-inactivated FBS (vol/vol), 1% penicillin/streptomycin (vol/vol), 1% non-essential amino acids (vol/vol), 1% L-glutamine (vol/vol) in DMEM. To make 500 ml of HEK medium, add 50ml heat-inactivated FBS, 5ml 1% penicillin/streptomycin, 5ml 100X non-essential amino acids, and 5ml 1% L-glutamine to 435 ml DMEM and filter-sterilize through 0.2 um filter. Store HEK culture medium at 4° C for up to 2 months. CRITICAL Optimal HEK growth must be confirmed for each new FBS lot prior to use.
Fibroblast Medium
This medium contains 10% heat-inactivated FBS (vol/vol), 1% penicillin/streptomycin (vol/vol), 1% non-essential amino acids (vol/vol), 1% L-glutamine (vol/vol) in DMEM. To make 500 ml of Fibroblasts Medium, add 50ml FBS, 5ml 1% penicillin/streptomycin, 5ml 100X non-essential amino acids, and 5ml 1% L-glutamine to 435 ml DMEM and filter-sterilize through 0.2 um filter. Store Fibroblast Medium at 4° C for up to 2 months. FBS lots must be tested for efficient cardiac differentiation using mESCs prior to use.
iCPC Induction Medium
This medium contains a final concentration of 8ug/ml doxycycline, 2.5 uM BIO and 103 units LIF/ml, all in Fibroblast Medium. To make 1 ml iCPC induction medium, add 4ul doxycycline, 1 ul LIF (103 units, add directly from vial), and 0.25 ul BIO solution to 1ml Fibroblast Medium. Make fresh.
iCPC Maintenance Medium
This medium contains 2.5 uM BIO and 103 units LIF/ml in Fibroblast Medium. To make 1 ml iCPC Maintenance Medium, add 1 ul LIF (103 units, add directly from vial) and 0.25 ul BIO solution to 1ml Fibroblast Medium. Make fresh.
iCPC Differentiation Medium
This medium contains 50 ng/ml BMP4, 30 ng/ml FGF2, 10ng/ml VEGF and 5 uM IWP4, all in Fibroblast Medium. To make 1 ml iCPC Differentiation Medium, add 0.5 ul BMP4 solution, 0.3 ul FGF2 solution, 2 ul VEGF solution and 4 ul IWP4 solution to 1 ml Fibroblasts Medium. Make fresh.
Differentiation Maintenance Medium
This medium contains 1% heat-inactivated FBS (vol/vol), 1% penicillin/streptomycin (vol/vol), 1% non-essential amino acids (vol/vol), 1% L-glutamine (vol/vol) in DMEM. To make 500 ml of iCPC Differentiation Maintenance Medium, add 5ml heat-inactivated FBS, 5ml 1% penicillin/streptomycin, 5ml 100X non-essential amino acids, 5ml 1% L-glutamine to 435 ml DMEM and filter-sterilize through 0.2 um filter. Store medium at 4° C for up to 2 months.
2X Freezing Medium
This medium contains Fibroblast Medium, DMSO and heat-inactivated FBS in 3:1:1 ratio (vol:vol). To make 10 ml of 2X Freezing Medium, mix 6 ml of Fibroblasts Medium, 2 ml of DMSO, and 2 ml of heat-inactivated FBS in a 50 ml conical tube and filter-sterilize through 0.2 um filter. Store medium at 4° C for up to 1 month. Add 0.5 ml of 2X Freezing Medium to 0.5 ml of Fibroblasts medium containing cells. Mix gently and immediately place cells in Cryo Box and place cells at −80° C overnight. Transfer cells to liquid nitrogen on the next day.
4% Formaldehyde Solution
This solution contains 16% formaldehyde and PBS in 3:1 ratio (vol/vol). To make 4ml solution of 4% formaldehyde solution, add 1ml of 16% formaldehyde to 3 ml PBS. Make fresh. Unopened 16% formaldehyde solution can be stored at RT. Once opened,16% formaldehyde should be immediately aliquoted and then stored at −20° C for up to 6 months.
Blocking Solution
This solution contains 5% BSA (wt/vol), 2% serum (vol/vol) and 0.1 % Triton X-100 (vol/vol) (optional). To make Blocking Solution, add 2.5 gm BSA, 1 ml goat or donkey serum, and 50ul Triton X-100 to a 50 ml conical tube and add PBS (non- sterile) to 50 ml mark. Gently mix and store at 4° C for up to 6 months. Do not add Triton X-100 when immunostaining for cell surface markers.
Flow Cytometry Blocking Solution
This solution contains 5% BSA (wt/vol) and 2% goat serum (vol/vol). To make Flow Cytometry Blocking Solution add 2.5 gm BSA and 1 ml goat serum to a 50 ml conical tube and add PBS (non-sterile) to 50 ml mark. Mix by hand and store at 4° C for up to 6 months.
Tissue Culture Incubator Conditions
37° C, 5% CO2 and high humidity. Make sure the tray in the incubator is always filled with deionized water.
Timed Matings of mice for Embryo Isolation
We use a 12-hour light/dark cycle and “reversed lighting” scheme (lights off/on 13.00/1.00) to obtain crescent-stage embryos mid-afternoon. Just before the lights go off (13.00), pro- and estrous females of breeding age are selected by examining the external genitalia for four signs: pink (pro-estrus)-to-white (estrus), swollen, gaping, and crinkled 31, 32. An individual pro- or estrous female is then placed with a male stud of breeding age. Several hours later (~16.30), copulation plugs are checked during the dark cycle with the aid of a red light. While the copulation plug is generally obvious by visual inspection with the F1 inbred hybrid strain (B6CBA/J), nevertheless, a rounded forceps is always used to probe the vaginal opening for evidence of a hard “plug”. Once copulation has taken place, the plug of semen persists for up to 14–16 hours. Metestrus, when vaginal discharge is present 32, may be mistaken for a copulation plug. However, while this discharge appears coagulated, it is not solid, as indicated by the forceps. If a plug is detected at 16.30, the plugged female remains with the stud male overnight. Check for plugs no later than 14–16 hours after beginning of the dark cycle, or you risk missing a plug due to dissolution. The next morning when the lights are on, check for more plugs; the early plugged females are housed separately from the later plugged ones, and dissected several hours in advance of the later plugs. Alternatively, if the males are not needed for mating over the next week, you may leave the plugged females with the males until the day of the experiment. If your female did not hold her litter and you are sure that you detected a plug, investigate lighting, movement in and out of the rooms when the lights are off, and noise/construction– eliminate all of these factors. Set up timed matings on two successive nights eight days in advance of your injection for several reasons, including loss of pregnancy, too many resorptions per litter or incorrect stages. In this way, you will be able to use the precious whole embryo culture medium on the second day.
Forceps for Embryo Dissection
Purchased forceps must be crafted after purchase 30. Briefly, the tines should be of equal length and thickness in frontal and sagittal profiles. Tines should come to a tapered point when viewed in frontal profile and should meet at their tips when gently squeezed together and viewed in sagittal profile. Before dissecting conceptuses, clean your forceps with soap and water, followed by rinsing in tap and distilled water and absolute alcohol. Do not flame forceps.
100X Amino Acid Solution
This solution is used in embryo dissection medium and whole embryo culture medium. The following amounts are for 1000 ml. To a sterile 1000 ml graduated cylinder, add a stir flea, and approximately 900 ml tissue culture-grade sterile double-distilled water. While stirring on a mixing plate, add 4.80 g cysteine-HCL, 3.60 g L-alanine, 6.00mg L-asparagine, 5.30 g L-aspartic acid, 4.60 g L-proline, 5.90 g L-glutamic acid and stir. The powders will not go into solution until pH 9.0 is achieved. Adjust to pH 9.0 by dispensing ≈ 550 drops (100 drops = 2.25 ml) 5N NaOH administered from a 9″ sterile Pasteur pipette. Do not filter-sterilize at this point, dispense solution into 10 ml aliquots, and store indefinitely at −80°C.
Embryo Dissection Medium
This medium is buffered for use in atmospheric oxygen. To make 1000ml embryo dissection medium, add ingredients in the following order to ~800 ml tissue-culture grade double-distilled water in a 1000ml-graduated cylinder with stir flea on a mixing plate: 13.5 g DMEM, 2.4g Hepes, 0.8g NaCl, 10 ml 100× Amino Acid Solution, 2ml 5000 Units/ml penicillin/streptomycin and 75ml of heat-inactivated fetal calf serum. Adjust pH to 7.4 with ≈ 25 drops 5 N NaOH delivered with a 9″ Pasteur pipette and monitor with either a pH meter or color pHast indicator strips. Bring to volume. Filter-sterilize through 0.22 μm cellulose acetate filter. Aliquot dissection medium into sterile 50 ml polypropylene tubes (do not overfill, as medium will expand upon freezing) and store indefinitely at −80°C. Warm medium to RT before use. Store unused thawed medium at 4°C up to 1 month.
Incomplete Whole Embryo Culture Medium (without serum)
This medium is buffered for use in tissue-culture incubator. To make 1000ml whole embryo culture medium (without serum) add ingredients in the following order to ~800 ml tissue-culture grade double-distilled water in a 1000ml-graduated cylinder with stir flea on a mixing plate: 13.5 g DMEM, 3.7g NaHCO3, 10 ml 100× Amino Acid Solution, 2ml 5000 Units/ml penicillin/streptomycin. Bring volume to 1000 ml. Divide in half and transfer 2 × 500ml to two sterile 1000 mL Erlenmeyer flasks. Adjust the pH in each 500 ml aliquot to 6.5–7.3 by bubbling CO2 directly into the flasks with the aid of tubing attached at one end to the gas cylinder and the other to a cotton-plugged 9″ Pasteur pipette. Filter-sterilize through 0.22 μm cellulose acetate filter. Aliquot dissection medium into sterile 50 ml polypropylene tubes (do not overfill, as medium will expand upon freezing) and store indefinitely at −80°C; when frozen, the color of the medium will be yellow (Figure 4a); when thawed, the color will turn red – mix well by gentle inversion. Unused thawed medium can be stored at 4°C up to 1 month.
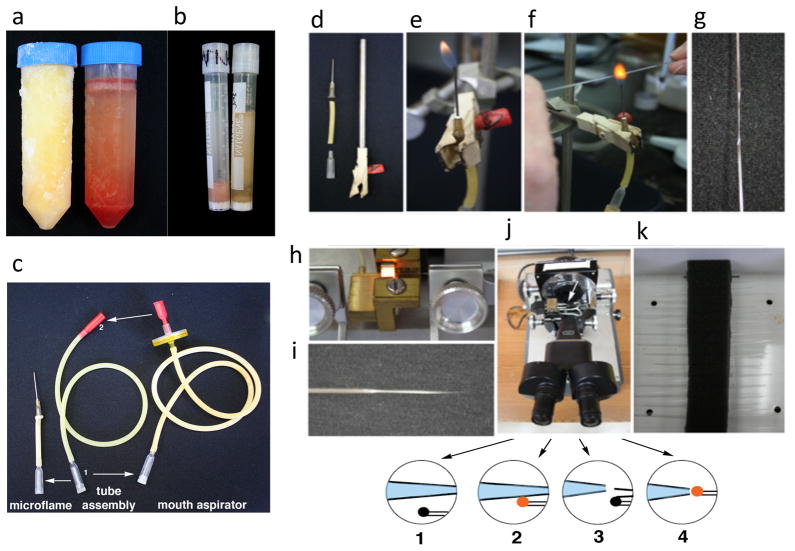
Figure 4. Generation of tools for embryo dissection and cell injection.
(a) Complete frozen Embryo Dissection Medium (right) and frozen Incomplete Whole Embryo Culture Medium (without serum) (left). Upon thawing, the Incomplete Whole Embryo Culture Medium will turn pink. (b) Comparison of non-hemolyzed (right) and hemolyzed (left) rat serum. (c) The mouth aspirator assembly (center) is bought from Sigma. Its major useful feature is the microcapillary holder (1). The holder can serve as an adaptor for the microflame (left), which consists of the adaptor minus the gasket, a piece of latex tubing and a cut-off 19-g hypodermic needle. A piece of wider-gauge latex tubing is then secured over the adaptor and connected to the gas outlet. The round mouthpiece (2) is replaced by a flat mouthpiece that is inserted into a disposable filter (0.22 or 0.45 μm) using parafilm to create a tight seal (not shown). The filter is then secured onto a longer piece of latex tubing followed by the microcapillary and its gasket, which securely holds an injection pipette. (d) The miciroflame (left) will be secured onto a clamp (right) and connected to the gas outlet (not shown). (e) The microflame is ignited. (f, g) A piece of glass microtubing is briefly softened at its center (f) and, when red hot, removed from the flame and pulled horizontally (g). (h, i) The pulled microcapillary is loaded onto an electrode puller, the trough filament of which is heated (h), and the glass is then pulled, creating two tapered glass capillaries (i, only one is shown). (j) The glass capillaries are cut to the right diameter (20 μm) on a microforge, in the following steps: (1) The glass bead (black) stuck to the platinum wire is made to approach the glass capillary at the desired diameter (measured with an eyepiece reticle). (2) The bead is placed onto the glass microcapillary and the heat is momentarily turned on. (3) Turning the filament off results in contraction of the glass bead, bringing with it the distal portion of the fused glass filament. (4) The end of the injection pipette is fire-polished by aligning the glass bead to its opening and briefly turning on the heat. (k) Once the capillaries are microforged, they can be stored in a Nalgene box into which a piece of flexible Styrofoam has been glued (Glue-Stick); the Styrofoam is scored with a razor blade, and the capillaries inserted up to three deep into the scored Styrofoam.
Complete Whole Embryo Culture Medium (with rat serum and heat-and gas- equilibration)
On the day of the cell injection, calculate the number of conceptuses that you expect to inject and culture. In your media calculations include an extra 1ml of medium to be distributed to two “holding” organ culture dishes to maintain the embryos during the injection procedure. For the first 24 hours, individual or pairs of embryos will be cultured in 1 ml culture medium. For the second 24 hours, individual embryos will be cultured in 1 ml culture medium. Label an appropriate number of whole embryo culture tubes for the first day and loosen caps. Prepare rat serum as described33 to avoid hemolysis. Commercially available rat serum may be used provided it is not hemolyzed (Figure 4b). Store rat serum in 1, 2 or 3 ml aliquots indefinitely at −80°C. Once thawed, rat serum cannot be re-frozen. Thaw rat serum in a small beaker of water (~10 minutes). Heat-inactivate thawed rat serum at 56°C for 30 minutes (exactly) to inactivate complement. Be sure that the water entirely covers the serum. Spin heat-inactivated rat serum for 5 minutes, 1625xg, room temperature, to bring unwanted solids to the bottom of the conical cryotube. With a sterile 9″ Pasteur pipette, transfer all but the bottommost serum into a sterile 50-mL polypropylene tube. It is important that you add the serum to the test tube before adding the incomplete culture medium, in case you spill some. Add an equal volume of Incomplete Whole Embryo Culture Medium (without rat serum) via a sterile 5mL disposable volumetric pipette attached to a dedicated automated pipettor. Distribute 1 mL of complete culture medium to each embryo culture tube, and 500 μl of extra culture medium to each of two organ culture dishes. With caps in the loose position, balance the culture tubes against each other in the incubator’s roller apparatus, placed at 8° off the horizontal, and roll at 0.5 rpm; the organ culture dishes remain stationary. Gas- and temperature-equilibrate the Complete Whole Embryo Culture Medium for at least 1 hour before placing embryos into the tubes. When equilibrated, the color of the medium will be fleshy-pink. Any unused medium can be stored for up to 24 h at 4°C and used the next day only. Medium can either be stored in the embryo culture tubes with caps closed securely (gas- and heat-equilibrate it the next day after loosening the caps), or stored in a large 50 ml sterile polypropylene tube.
Mouth Aspirator
This is used to inject cells into the cardiac crescent. The Mouth Aspirator is fixed to an injection pipette, which is used to inject cells of interest into the cardiac crescent. Sigma supplies mouth aspirator tube assemblies that come with a round mouth aspirator, about a foot of latex tubing, and a microcapillary holder. However, the only useful piece of this assembly is the capillary holder, which is used to make both the mouth aspirator and serve as an adaptor for the microflame. The rest of the aspirator assembly is not ideal, as the round mouthpieces are inferior to the flat ones in terms of suction control, and the latex tubing supplied with the mouth aspirator assemblies is too short for comfortable work. Hence, it is recommended to replace the round mouthpiece with the flat one, and replace the short tubing with approximately 2 feet of fresh tubing (OD, 3/16″; ID, 1/8″;W, 1/32″). At one end of the tubing, attach the microcapillary holder and at the other end of the tubing, attach a disposable 0.22 or 0.45 μm filter. Wrap the stem of the flat mouthpiece in paraffin film, and insert it snugly into the filter (Figure 4c).
Microflame
The microflame is used to stretch glass microcapillary tubing destined for injection. Cut off the beveled end of a 19-gauge hypodermic needle with a hemostat in the following way: secure the tip of the needle onto the hemostat and bend the tip until the metal breaks. Insert the base of the hypodermic needle into an adequate length of latex tubing (OD, 3/16″; ID, 1/8″;W, 1/32″). Remove the rubber gasket that holds a glass capillary and place the funnel portion of the assembly capillary holder into the latex tubing. Connect the wide end of the capillary holder into a piece of rubber tubing and connect the latter to the gas outlet. The needle is vertically supported in a clamp secured onto a ring stand. Place the hosecock clamp (optional) near the middle of the tubing, and tighten slightly to regulate the gas flow (Figure 4d–e).
Wide-bore Pasteur pipettes
Sterile 9″ long Pasteur pipettes are useful for the transfer of early gastrulating embryos but post-culture embryos are best transferred with wider bore pipettes. Score the wide part of a Pasteur pipette with a diamond tip glass scorer and break off with your fingers. The cut end of the wide bore pipettes should be flush, never jagged. Wide bore pipettes can be placed into a small metal canister and sterilized, though because embryo culture is short-term and the dissection and culture media contain antibiotics, autoclaving is not necessary. CAUTION! Be sure to wear safety goggles during this procedure.
Injection pipettes
Injection pipettes are used to inject cells into the cardiac region. They are made as described in box 1. See also Figure 4f–k.
Box 1. Manufacture of injection pipettes: 5 min per pipette.
They are made in three steps
Microflame Gently pre-stretch the thin-walled microcapillary tubing by placing the central portion over the microflame until it is red-hot. Remove the capillary from the flame and gently pull, but do not break into two pieces (Figure 4f–g). Do not fuse the glass, or you will not be able to inject cells.
Electrode puller Place the central pulled portion of the stretched microcapillary over the platinum trough filament of an electrode puller. Pull to produce a tapered pipette of varying diameter (~1 – 80 μm) (Figure 4h–I). The amount of pull, time and speed of pulling should be empirically determined with the aid of the instruction manual of your equipment.
Microforge You can either insert the pulled pipette into a Leitz instrument holder and insert onto the microforge or you can insert the pipette directly onto the microforge if it will accommodate a “naked” pipette. Align the glass platinum filament’s bead to just beneath the point where the capillary’s internal diameter is 20 μm, and make sure that both the bead and capillary are in the same focal plane. Place the bead onto the underside of the capillary, and heat the filament so that the glass bead is slightly red hot, thereby fusing the bead to the filament. Turn off the heating element of the microforge. The cooled glass bead will contract, resulting in a clean break of the microcapillary at 20 μm. To remove the excess glass from the bead, strike it off with a glass pipette and turn on the heat to melt the remainder into the glass bead. Then, if desired, flame-polish the end of the pipette by placing the capillary at an appropriate distance to the glass bead without any chance of fusing them (Figure 4j). Re-heat the glass bead until the end of the microcapillary softens. Turn off the filament. Injection tips should be smooth, never jagged. Microcapillaries can be stored in a purpose-built box containing an insert of flexible foam that has been glued to the bottom of the box and scored with a razor blade (Figure 4k).
Whole embryo culture incubator conditions Culture embryos in a humidified water-jacketed incubator set to 6.2% CO2 and 37°C. Alternatively, 5% CO2 may be used.
Embryo Blocking Solution
This is used for whole-mount immunostaining embryos. This solution contains 5% serum (vol/vol) and 0.1 % Triton X-100 (vol/vol). To make Embryo Blocking Solution, add 2.5 ml goat or donkey serum (or other animal, depending upon the species in which the antibodies were raised) and 50ul triton X-100 to a 50 ml polypropylene conical tube containing 47 ml of PBS (Sigma). Mix gently by inversion. Make fresh and use for 2–3 days with storage at 4° C.
PROCEDURE
Isolation of adult cardiac, lung and tail-tip fibroblasts from Nkx2.5-EYFP/rtTA transgenic mice TIMING 12–14 days
CRITICAL Confirm the genotype of mice by performing RT-PCR on genomic DNA to verify the presence of rtTA and Nkx-EYFP transgenes before tissue isolation. The following procedure can be used for male and female mice ranging from 1–3 months in age.
-
1|
Tissue Isolation. Anesthetize mice using isoflurane and place in tissue culture hood designated for primary tissue isolation. Disinfect the chest and tail area with 70% ethanol before proceeding to dissection.
! CAUTION Experiments using mice must be performed in accordance with the respective institutional and governmental regulations.
-
2|
Dissect heart, lung and/or tail from mice and place in cold sterile PBS in 10 cm tissue culture dish (separate for each organ) to wash off residual blood. For tail tissue, remove epidermis using scissors and forceps.
Δ CRITICAL STEP Thoroughly wash tissue to remove as much blood as possible. Use sterile equipment and aseptic technique to avoid contaminating fibroblast cultures. All isolation steps must be performed inside a tissue culture hood.
-
3|
Transfer washed tissue into a fresh sterile 10 cm tissue culture dish containing 3–4 ml of cold Fibroblast Medium (tilt dish) and mince tissue using scissors to obtain tissue pieces <1mm3 in size. Tissue should be minced immediately after harvest.
Δ CRITICAL STEP Mince quickly to obtain as small tissue chunks as possible. Smaller tissue chunks attach faster than large ones, and are optimal for explant culture. If fibroblasts are being derived from multiple sources, make sure to avoid tissue cross contamination. Do not re-use equipment and reagents for different tissue sources.
?TROUBLESHOOTING
-
4|
Cut the end of a 1ml pipette tip with sterile scissors to allow tissue pieces to be pipetted. Using this tip, transfer the minced tissue pieces into a sterile 15 ml conical tube. Wash the dish used for mincing 1–2 times with cold sterile PBS to collect all tissue pieces.
-
5|
Centrifuge the tube for 2 min at 200g. Aspirate off the supernatant and discard.
-
6|
Resuspend the pellet of tissue pieces in 5 ml of cold sterile PBS. Centrifuge for 2 min at 200g, aspirate the supernatant, and discard.
-
7|
Setting up explant culture. Resuspend heart/lung tissue pieces in 4 ml warm (37° C) Fibroblast Medium and tail tissue pieces in 2–3 ml warm Fibroblast Medium. Plate 5 ml of warm Fibroblast Medium per 10cm dish (gelatinized), to that add 1 ml of medium containing tissue pieces (total 6ml/dish). Thus, one adult heart/lung should be plated onto four 10cm dishes, and tail tissue pieces should be plated on two or three 10cm dishes depending on the size of the initial tail tissue dissected.
Δ CRITICAL STEP Make sure the tissue pieces are evenly distributed amongst the dishes and within individual dishes. Do not use more than 6 ml of Fibroblast Medium per dish; this is the optimal medium volume for tissue attachment and fibroblast migration.
?TROUBLESHOOTING
-
8|
Place dishes in incubator and culture for 7 days.
Δ CRITICAL STEP. Do not disturb dishes during this culture period as frequent movement hampers tissue attachment.
-
9|
At the end of 7 days, observe the explant culture plates in a tissue cul0ure microscope. Most viable tissue pieces should be attached and Nkx2.5-EYFP− fibroblasts should have migrated from tissue pieces (Figure 2a). Change medium at this stage and culture for additional 5–7days depending on rate of fibroblast migration.
?TROUBLESHOOTING
-
10|
Passage fibroblasts 1–2 times, proceed immediately to set up reprogramming experiments, or freeze for future use. We have not seen significant differences between freshly isolated, passaged or frozen fibroblasts with respect to reprogramming outcome or efficiency. However, the majority of our experiments were with frozen fibroblasts between passages 1–2. Cardiac and lung fibroblasts are more proliferative than tail-tip fibroblasts and show higher reprogramming efficiencies as compared to tail-tip fibroblasts.
■ PAUSE POINT Frozen fibroblasts can be stored for at least 4 yr in liquid nitrogen without losing viability.
Day (Minus) – 2 - Seeding mouse fibroblasts for lentivirus infection ● TIMING (2 days for fibroblasts in culture, 5–6 days for frozen fibroblasts)
CRITICAL The following procedure has been optimized for adult mouse cardiac, lung and tail-tip fibroblasts. However, we recommend starting with cardiac fibroblasts.
-
11|
Culture fibroblasts on gelatinized 10 cm dishes. If using previously frozen fibroblasts, culture 3–4 days in Fibroblast Medium prior to setting up for infection. Fibroblasts should not express Nkx-EYFP (Figure 2a).
-
12|
Aspirate medium from fibroblast dish and wash with 3–4 ml sterile PBS.
-
13|
Warm 0.25% trypsin-EDTA to 37°C, aspirate PBS, and add 1.5 ml of warm 0.25% trypsin-EDTA. Tilt plate from side to side to ensure that trypsin covers entire surface area of the plate. Place in tissue culture incubator for 5–10min.
Δ CRITICAL STEP Monitor plate under microscope every 3–4 min to detect cell detachment. Tap plates with the palm of your hands to facilitate detachment. Trypsinization should be stopped when >90% of cells have detached and can be observed floating. Prolonged trypsinization should be avoided as it reduces cell viability.
-
14|
Add 1.5ml of Fibroblast Medium to plate to quench trypsin. Mix and thoroughly wash the entire surface of the plate using a 5ml pipette to remove any undetached cells from plate.
-
15|
Place a 40 um cell strainer on top of a 50 ml sterile polypropylene conical tube and pass cells through filter to eliminate tissue pieces.
-
16|
Add 5 ml sterile PBS to dish, wash thoroughly and pass again through same cell strainer to collect cells in the 50 ml conical tube from step 5.
-
17|
Centrifuge cells at 200g for 5 min at RT. Aspirate supernatant and resuspend cell pellet in 2–3 ml Fibroblast Medium.
-
18|
Count cells using a haemocytometer and seed 50,000 cells/well in a gelatinized 12 well plate.
CRITICAL STEP Reprogramming using this protocol is optimal when starting with 50,000 cells/well (12-well plate).
-
19|
Place plate in incubator and culture for 2 days.
Day 0 – Lentiviral infection of mouse fibroblasts ● TIMING 2 days
-
20|
Add 0.8 ul/ml polybrene solution to lentivirus supernatant in sterile 15ml conical tube and mix thoroughly. Use fresh lentivirus supernatant or thaw frozen lentivirus in a 37° C water bath. Production of lentiviral supernatant is described in Box 2. As an alternative, commercially available lentiviral vector systems and even complete commercial lentiviral particle production are available, but we have not tested these systems.
Δ CRITICAL STEP Addition of polybrene enhances lentivirus transduction efficiency. Thawed lentivirus supernatant should be immediately used to infect fibroblasts. ! CAUTION All equipment, including pipettes and dishes, that comes in contact with lentiviral supernatant should be treated with 10% bleach followed by ultraviolent light for at least 1 hr before discarding.
-
21|
Aspirate medium from fibroblast wells and add 2.5 ml of lentiviral supernatant/well (supplemented with polybrene) to initiate virus infections.
Δ CRITICAL STEP Fibroblasts should be 90–95% confluent, at which lentivirus infection is optimal. Infecting less confluent cells may lead to cell death or reduced viability during subsequent reprogramming steps.
Δ CRITICAL STEP For each batch of infection using cardiac factors, one well should be infected with pSAM2 GFP lentivirus that was produced alongside cardiac factor viruses. This is an important control to account for batch-to-batch variability in lentivirus production.
-
22|
Return plates to incubator for 2 days.
Day 2 - Induction of reprogramming ● TIMING 15 min
-
23|
Remove lentivirus supernatant from cells using 5ml pipette and discard in 10% bleach solution. Wash cells with 1ml PBS/well using 5ml pipette and discard PBS in 10% bleach solution.
-
24|
Add 8ul/ml doxycycline solution to Fibroblast Medium and feed infected cells with 1ml medium per well.
-
25|
Return plates to incubator for 2 days.
Day 4 – Introduction of CPC culture conditions and splitting ● TIMING 1 hr
-
26|
Remove plate from incubator and observe well infected with control pSAM2 GFP lentivirus under the green channel of an epifluorescence microscope. >80% of the cells should appear GFP+. All pSAM2 vectors have IRES mcherry engineered downstream of GFP/cardiac factors and hence, the majority of GFP+ cells should have mcherry expression (Figure 2b). mcherry expression should also be detected in wells infected with cardiac factors. mcherry expression is brighter in lung and tail-tip fibroblasts relative to cardiac fibroblasts.
△ CRITICAL STEP For each batch of reprogramming experiments, it is important to confirm GFP expression in >80% of cells in the control well. This ensures that the cells have rtTA expression and lentivirus production was efficient. If the infection efficiency is <80%, we recommend starting with fresh cells/virus batch.
?TROUBLESHOOTING
-
27|
Aspirate medium from wells and wash with PBS.
-
28|
Aspirate PBS and add 0.5ml of warm 0.25% trypsin-EDTA to each well and return plate to incubator for 5–10 min.
△ CRITICAL STEP Monitor plate under microscope every 3–4 min to detect cell detachment. Tap plates with the palm of your hands to facilitate detachment. Trypsinization should be stopped when >90% of cells have detached and can be observed floating. Prolonged trypsinization should be avoided as it reduces cell viability.
-
29|
Quench trypsin by adding 0.5 ml of warm Fibroblast Medium to each well using 1ml pipette. Mix and thoroughly wash the entire surface of the well using a 1ml pipette to remove any undetached cells from well. Collect cells in sterile 15 ml conical tube. Wash each well with 1ml sterile PBS and collect in sterile 15 ml tube.
-
30|
Centrifuge cells at 200g for 4.5 min at RT. While cells are spinning, prepare iCPC Induction Medium.
-
31|
Aspirate supernatant and resuspend cells in 3ml of iCPC Induction Medium per starting well. Seed cells onto gelatinized 60 mm dishes and return to incubator.
△ CRITICAL STEP One infected well (12 well plate) should be split onto one 60 mm dish (1:5 to 1:6 split). This will result in a starting cell confluency of 30–40% and is the optimal cell density for iCPC reprogramming. Splitting cells sparsely at this stage will severely hamper reprogramming.
?TROUBLESHOOTING
Day 5 to Day 25/30 - Monitoring for appearance of iCPC colonies ● TIMING 3 – 4 weeks
-
32|
Change medium every 5–6 days by aspirating old medium and adding 3ml of fresh iCPC Induction Medium per 60 mm dish.
△ CRITICAL STEP. Frequent medium changes should be avoided as this may result in reduced reprogramming efficiency.
-
33|
Assess for the appearance of EYFP+ cells 3–5 days after doxycycline induction by placing cells in epifluorescence microscope (YFP filter). Typically, the first EYFP+ cells appear 3–5 days after doxycycline induction and have regular fibroblast morphology.
-
34|
Assess for the appearance of EYFP+, proliferative iCPC colonies two weeks after doxycycline induction. Fibroblasts undergoing reprogramming become proliferative, lose parental fibroblast morphology, exhibit a high nuclear-cytoplasmic ratio (iCPC morphology) and express Nkx-EYFP (Figure 2c). Cells undergoing iCPC reprogramming can be easily identified based on this dramatic morphological change. Allow iCPC colonies to expand for additional 10–16 days.
?TROUBLESHOOTING
-
35|
After 3–4 weeks of doxycycline induction, the 2-dimensional iCPC colonies reach 3–4 mm in size. They should be scored at this stage based on expression of EYFP and iCPC morphology (Figure 2d). iCPC colonies may express mcherry along with EYFP, indicating that cells undergoing reprogramming are overexpressing cardiac factors (Figure 2e). Typically, if you started from 50,000 cells, 5–15 iCPC colonies can be detected (0.01 – 0.03% reprogramming efficiency). The reprogramming efficiency will vary according to fibroblast source. Colonies can be split after scoring.
Day 25 to 60 - Expanding iCPC colonies and establishing stable iCPC cell lines
-
36|
Establish stable iCPC cell lines using iCPC colonies either by bulk culture (polyclonal) (A) or single cell clonal passaging (B).
(A) Establishing iCPC Bulk Cultures ● TIMING 2–3 weeks
Aspirate medium and wash cells with 3 ml sterile PBS.
-
Aspirate PBS and add 0.75ml of warm 0.25% trypsin-EDTA to each dish and return plate to incubator for 5–10 min.
△ CRITICAL STEP Monitor plate under in microscope every 3–4 min to detect cell detachment. Tap plates with the palm of your hands to facilitate detachment. Trypsinization should be stopped when >90% of cells have detached and can be observed floating. Prolonged trypsinization should be avoided as it reduces cell viability.
Quench trypsin by adding 0.75 ml of warm Fibroblast Medium to each well using 1ml pipette. Mix and thoroughly wash the entire surface of the well using a 1ml pipette to remove any undetached cells from well. Collect cells in 15 ml conical tube. Wash each well with 3ml PBS and collect in 15 ml tube.
Centrifuge cells at 200g for 4.5 min at RT.
-
Aspirate supernatant and resuspend cells in 6ml of iCPC Induction Medium per initial 60 mm dish. Seed cells on to gelatinized 100 mm dishes and return to incubator. Culture plates for 5–7 days (Passage 1).
△ CRITICAL STEP. One 60 mm dish containing iCPC colonies should be split onto one 100 mm dish (approximately 1:3 split).
Repeat Steps i-v (Passage 2). For Passage 2, cells can be spilt in 1:6 ratio on gelatinized 100 mm dish and cultured for 5–7 days. Remaining cells can be frozen using freezing medium (2–3 vials).
-
For passage 3, repeat steps i-iv. Aspirate supernatant and resuspend cells in 6 ml of iCPC Maintenance Medium using a 1:6 split ratio and plate cells on gelatinized 100 mm dish. For subsequent passages, iCPC can be split every 5–6 days using 1:6 to 1:8 split ratio as desired using iCPC Maintenance Medium. Cells maintain Nkx-EYFP expression and iCPC morphology for at least 30 passages (Figure 2f)
■ PAUSE POINT iCPC lines can be frozen down and stored in liquid nitrogen for several years.
?TROUBLESHOOTING
(B) Establishing iCPC single cell clones ● TIMING 4 – 5 weeks
-
Dip one side of a cloning cylinder (8mm × 8mm) in vacuum grease and place on top of an iCPC colony while watching through a microscope. Make sure the seal is tight.
△ CRITICAL STEP This procedure must be done in a picking hood or regular cell culture hood to avoid contamination.
Aspirate the culture medium inside the cloning cylinder and wash with 100 ul sterile PBS.
Aspirate PBS and add 40 ul of warm 0.25% trypsin-EDTA solution inside each cloning cylinder and place in incubator for 5–10 min till majority of cell have detached. During this time add 50 ul iCPC Induction Medium into desired number of wells of a gelatinized 96-well plate.
-
Add 100 ul iCPC Induction Medium into the cloning cylinder and triturate to release all cells from plate. Transfer cell suspension directly into 1 well of a 96 well plate containing 50 ul iCPC Induction Medium and place in incubator for 12 hrs.
△ CRITICAL STEP One iCPC colony is split in 1 well of a 96 well plate.
Aspirate medium and feed with fresh 200 ul iCPC Induction Medium. Culture for 1 week or until clones reach 95% confluency.
Aspirate the Induction Medium and wash with 100 ul sterile PBS.
Aspirate PBS and add 40 ul of warm 0.25% trypsin-EDTA solution into each well of a 96-well plate containing cells. Place in incubator for 5–10 min until majority of cell have detached. During this time, add 400 ul iCPC Induction Medium into equal number of wells of a gelatinized 24-well plate.
Add 100 ul iCPC Induction Medium into trypsin containing wells of a 96-well plate and triturate to release all cells from plate. Transfer cell suspension directly into 1 well of a 24 well plate containing 400 ul iCPC Induction Medium and place in incubator for 12 hrs.
-
Aspirate medium and feed with fresh 500 ul iCPC Induction Medium. Culture for 1 week or until clones reach 95% confluency.
△ CRITICAL STEP One 96 well is split into one 24 well (approximately 1:6 split)
Aspirate the Induction Medium and wash with 500 ul sterile PBS.
Aspirate PBS and add 100 ul of warm 0.25% trypsin-EDTA solution into each well of a 24-well plate containing cells. Place in incubator for 5–10 min until majority of cell have detached. During this time add 2 ml iCPC Induction Medium into desired number of wells of a gelatinized 6 well plate.
Add 500 ul iCPC Induction Medium into trypsin containing wells of a 24-well plate and triturate to release all cells from plate. Transfer cell suspension directly into 1 well of a 6-well plate containing 2 ml iCPC Induction Medium and place in incubator for 12 hrs.
-
Aspirate medium and feed with fresh 2ml iCPC Induction Medium. Culture for 1 week or until clones reach 95% confluency.
△ CRITICAL STEP One 24 well is split into one 6 well (approximately 1:4 split)
Aspirate the Induction Medium and wash with 500 ul PBS.
Aspirate PBS and add 100 ul of warm 0.25% trypsin-EDTA solution into each well of a 24-well plate containing cells. Place in incubator for 5–10 min until the majority of cell have detached. During this time, add 2 ml iCPC Induction Medium into equal number of wells of a gelatinized 6 well plate.
Add 500 ul iCPC Induction Medium into trypsin-containing wells of a 24-well plate and triturate to release all cells from plate. Transfer cell suspension directly into 1 well of a 6-well plate containing 2 ml iCPC Induction Medium and place in incubator for 12 hrs.
Aspirate medium and feed with fresh 2ml iCPC Induction Medium. Culture for 1 week or until clones reach 95% confluency.
Aspirate the culture medium and wash with 2ml sterile PBS.
Aspirate PBS and add 500 ul of warm 0.25% trypsin-EDTA solution into each well of a 6-well plate containing cells. Place in incubator for 5–10 min until majority of cell have detached.
Add 500 ul Fibroblast Medium into trypsin-containing wells of a 6-well plate and triturate to release all cells from plate. Transfer cells to a 15 ml sterile conical tube.
Centrifuge at 200g for 4.5 min at RT.
-
Aspirate supernatant and resuspend cells in 6 ml iCPC Maintenance Medium and plate cells on two 60 mm dishes. Culture for 1 week or until clones reach 95% confluency.
△ CRITICAL STEP One 6-well is split onto two 60 mm dishes (approximately 1:5 split). One plate can be frozen down and other can be used for further passaging and characterization. Only 50–60% of iCPC colonies form expandable iCPC cell lines.
■ PAUSE POINT iCPC clones can be frozen down and stored in liquid nitrogen for several years.
?TROUBLESHOOTING
Characterization of iCPCs in vitro
-
37|
Established iCPC lines cultured in iCPC Maintenance Medium can be characterized for expression of cell surface markers by flow cytometry (A), expression of CPC transcription factors by immunocytochemistry (B) and gene expression (C).
(A) Flow cytometry analysis for cell surface markers ● TIMING 1 day
Harvest iCPCs as described in Step 25 (i-iv) and re-suspend in 0.5 ml 4% formaldehyde solution. Incubate at RT for 12 min. 100,000 to 500,000 cells should be used for each flow experiment.
-
Centrifuge cells at 1000g for 8 min.
△ CRITICAL STEP Do not reduce centrifugation speed/time, as this may lead to loss of cells.
Aspirate supernatant and wash with 2ml sterile PBS. Repeat step (ii), above.
-
Resuspend cells in 100 ul of Flow Buffer and add 1ug conjugated cell surface antibody as described in Table 1. Incubate at 4° C for 45 min. We recommend using Cxcr4, Flk1 and cKit primary conjugated antibodies.
■ PAUSE POINT Fixed cells can be stored in PBS at 4° C for 2–3 weeks.
-
Wash twice following steps (ii–iii) above. Resuspend in 100 ul of Flow Buffer and perform flow cytometry analysis.
△ CRITICAL STEP Perform isotype controls for respective antibodies. mESCs (cKit) or mESCs differentiated for 5 days (Cxcr4, Flk1) can be used for positive control. Fibroblasts can be used as negative controls. Use controls to determine flow gates for the respective antibodies.
Table 1.
Antibodies used in the protocol
| Primary antibody | Species | Blocking buffer | Dilution | Secondary antibody (1:500) |
|---|---|---|---|---|
| Nkx2.5 | Mouse monoclonal IgG1 | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:100 | Goat anti-mouse IgG488/647 |
| Gata4 | Rabbit polyclonal IgG | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:200 | Goat anti-rabbit IgG 568 |
| Irx4 | Mouse IgG+IgM | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | Undiluted | Goat anti-mouse IgG+IgM 488 |
| Cxcr4-647 | RatIgG2g, κ | 5% (wt/vol) BSA, 2% (vol/vol) goat serum | 1:50 | NA |
| Flk1-PE | RatIgG2a | 5% (wt/vol) BSA, 2% (vol/vol) goat serum | 1:20 | NA |
| cKit-cy5 | RatIgG2g, κ | 5% (wt/vol) BSA, 2% (vol/vol) goat serum | 1:50 | NA |
| α-Actinin | Mouse monoclonal IgM | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:250 | Goat anti-mouse IgM568 |
| Cardiac actin | Mouse monoclonal IgG1 | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:400 | Goat anti-mouse IgG488/647 |
| α/β-MHC | Mouse monoclonal IgG1, κ | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:200 | Goat anti-mouse IgG488/647 |
| MLC-2v | Rabbit polyclonal IgG | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:200 | Goat anti-rabbit IgG633 |
| MLC-2a | Mouse monoclonal IgG | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:200 | Goat anti-mouse IgG488/647 |
| Cx43 | Mouse monoclonal IgG1 | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:100 | Goat anti-mouse IgG488/647 |
| SM-MHC | Rabbit polyclonal IgG | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:250 | Goat anti-rabbit IgG568 |
| SMA | Mouse monoclonal IgG | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:400 | Goat anti-mouse IgG488/647 |
| CD31 | Rat monoclonal IgG2a | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:400 | Goat anti-rat IgG 568 |
| GFP | Rabbit polyclonal IgG or mouse monoclonal IgG | 5% (wt/vol) BSA, 2% (vol/vol) goat serum, 0.1% (vol/vol) Triton X-100 | 1:200 | Goat anti-rabbit IgG488/568 or Goat anti-mouse 488/568/647 |
(B) Immunocytochemistry for CPC transcription factors ● TIMING 4 days
Day 1 - Plating cells on coverslips (1 hr). Place 12mm coverslips into a 24-well plate using sterile forceps and aseptic technique.
-
Plate 10,000 – 40,000 cells in 100–150ul of appropriate medium onto coverslip using a 200ul pipette. Place in tissue culture incubator for 45 min.
△ CRITICAL STEP 150ul medium is sufficient to cover the entire coverslip. Plating cells in 100–150ul medium containing 10% FBS facilitates rapid attachment exclusively to the coverslip and not to the 24-well plate. Use iCPC Maintenance Medium if plating undifferentiated iCPCs. Use Fibroblast Medium if plating cells differentiated from iCPCs.
?TROUBLESHOOTING
-
Add 350ul medium to each well containing cells and place in the incubator overnight
△ CRITICAL STEP Use iCPC Maintenance Medium if plating undifferentiated iCPCs. Use Fibroblast Medium if plating cells differentiated from iCPCs.
-
Day 2 - Immunostaining with primary antibody (2–3 hr). Aspirate medium and wash cells with 500 ul/well non-sterile PBS.
△ CRITICAL STEP Make sure the suction on the aspirator is set to minimum to prevent cell detachment and excessive drying for all successive steps.
-
Aspirate PBS and add 250ul/well of 4% formaldehyde in PBS per well. Incubate at RT for 12 min.
△ CRITICAL STEP Prepare fresh 4% formaldehyde solution for each staining experiment.
-
Aspirate formaldehyde and wash cells with 500ul PBS/well.
PAUSE POINT Fixed cells can be stored in PBS at 4° C for at least 1 month.
Aspirate PBS and permeabilize cells with 500ul 0.1% Triton X-100 solution/well for 6 mins.
Aspirate and add 250ul/well of appropriate blocking solution and incubate for 60–90 mins at RT.
-
Prepare enough primary antibody dilutions in appropriate Blocking Solution to make 250ul/well. Aspirate Blocking Solution and add primary antibody solution. Incubate at 4° C overnight. Antibody information can be found in Table 1. We recommend using Nkx2.5, Gata4 and Irx4 antibodies for characterization.
?TROUBLESHOOTING
Day 3 - Immunostaining with secondary antibody, DAPI and mounting (2–3 hr) Aspirate primary antibody solution and wash 3 times with 500ul PBS/well.
-
Prepare secondary antibody dilutions in appropriate blocking solution for 250ul/well. Add secondary antibody solution to wells and incubate for 90 min at RT in dark.
△ CRITICAL STEP Place in dark room to avoid quenching of secondary antibody. Antibody information can be found in Table 1.
Wash cells three times with 500ul PBS/well. During second wash, add DAPI solution to PBS (1:1000).
-
On a fresh glass slide, put a drop (10ul) of Prolong Gold Antifade Solution. Carefully remove coverslip from 24-well plate using forceps, drain excess PBS, and mount onto the glass slide (cells facing down). Place glass side in dark for 24 hr at RT. Then place at 4° C in dark.
△ CRITICAL STEP Cells need to be cured in the Antifade Solution for 24 hrs before imaging for best results.
-
Day 4 - Imaging. Image coverslips in epifluorescence or confocal microscope using appropriate lasers/filters. >95% of iCPCs express Nkx2.5, Gata4 and Irx4 (Figure 2g).
PAUSE POINT Coverslips that have been accurately mounted and cured using Antifade Solution can be stored at 4° C in dark for several months without losing signal intensity.
(C) Gene expression analysis ● TIMING 1–2 days
-
Isolate RNA from early/late passage iCPCs using RNA Aqueous Kit according to manufacturer’s instructions.
△ CRITICAL STEP The protocol is most efficient when working with at least 100,000 cells. If more cells are available, use 1–3 million cells. Work in an area designated for RNA preparation. Apply RNA Zap to bench and equipment before starting procedure to avoid degradation by RNAase.
■ PAUSE POINT Store RNA at −80° C for several months.
Measure RNA concentration/purity using a nanodrop and perform reverse transcription using iScript cDNA Synthesis Kit following manufacturer’s instructions.
-
Perform qPCR analysis using Ssofast EvaGreen Supermix and CFX96 Touch Real-Time PCR Detection System using manufacturer’s instructions. Potential cardiac genes that can be used are as follows: Nkx2.5, Tbx5, Gata4, Gata6, Irx4, Mesp1, Isl1, Hand1, Hand2, Tbx20, Baf60c, Mef2c; fibroblast genes are as follows: Thy1, Fsp1, Vim, Posn. Alternately, RNA can be used to perform genome wide gene expression via microarray or RNA Seq analysis.
△ CRITICAL STEP All qPCR experiments including primer design should be performed in accordance with the MIQE guidelines34.
Differentiation of iCPCs into cardiac lineage cells ● TIMING 4 – 8 weeks
-
38|
Dissociate iCPCs into single cells using 0.25% trypsin and seed at a density of 400,000 – 500,000 cells/ml in iCPC Differentiation Medium and add 0.5ml cell suspension to desired number of wells of a 24-well low attachment plate. Place in incubator and culture for 4–6 days. iCPCs aggregate into embryoid body-like spheres during this period.
-
39|
Transfer iCPC aggregates to gelatinized regular 24-well plates and culture for 24 hrs. Aggregates attach to plates during this culture period.
△ CRITICAL STEP Transfer aggregates from 1 low attachment well (24-well plate) to 1 gelatinized regular well (24-well plate).
-
40|
Carefully aspirate medium without detaching aggregates and add 0.5 ml of Differentiation Maintenance Medium to each well. Feed cells on a weekly basis. iCPC differentiated cells can be cultured for up to 6 months in Differentiation Maintenance Medium.
△ CRITICAL STEP Do not split cells after initiation of differentiation even if they become confluent.
-
41|
Immunostain iCPC-differentiated cells with cardiac lineage differentiation makers like cardiac actin, alpha-MHC, alpha-actinin, MLC-2v, SM-MHC, CD31 as detailed in step 27 option B. Table 1 lists the antibodies to use. Both intensity and pattern of staining improve with extended culture in Differentiation Maintenance Medium. We recommend differentiation for 3–4 weeks before immunostaining for best results. iCPCs differentiate into cardiomyocytes (CM), smooth muscle cells and endothelial cells (Figure 3a).
Downstream assays
-
42|
If you wish to co-cuture iCPC-CMs and mouse CMs and measure calcium transients, follow option A. To characterize iCPC potency in ex vivo mouse embryos, follow option B.
(A) Co-culture of iCPC-CMs with spontaneously contracting mouse CMs and measurement of calcium transients ● TIMING 4 weeks to set up culture and 2h to measure calcium transients
-
Setting up co-culture of iCPC-CMs and mouse CMs. Differentiate iCPCs as described in Steps 27–29 for 1–4 months. Infect differentiated iCPCs with pLentiGFP lentivirus (Lentivirus production described in Box 1 and infection procedure described in step 20–22).
△ CRITICAL STEP To distinguish iCPC derivatives from mouse CMs, iCPC derivatives must be infected with lentivirus constitutively expressing GFP.
Obtain contracting CMs either from mouse neonatal hearts (<3 days old) or purified from mESC differentiation. Mouse CMs may be infected with lentivirus constitutively expressing red fluorescent protein.
Seed 50,000 mouse CMs per well and 5,000 iCPC differentiated cells per well in 24- well plate in 0.5 ml Fibroblast Medium (iCPC:mouse CM ratio = 1:10). Mix cells and culture for 24 hrs.
-
Replace medium with 0.5 ml of Differentiation Maintenance Medium and culture for 3–4 weeks. Live imaging should reveal iCPCs (green) and mESC-CMs (Red) culture side-by-side as monolayer (Figure 3b). CMs1–5% of iCPC-derived CMs can be seen contracting after 3 weeks of co-culture and can be imaged using epifluorescence microscope. At this stage immunocytochemistry for Cx43 can be performed and should reveal abundant gap junctions between iCPC-CMs and mESC-CMs (Figure 3c).
?TROUBLESHOOTING
-
Measurement of calcium transients from iCPC-CMs. Replace medium with 1x Rhod-2 (see Manufacturer’s instructions) in Differentiation Maintenance Medium and place cells in incubator for 15–20 min. Use enough medium to evenly cover the cells (e.g., 0.5ml/24-well plate well)
△ CRITICAL STEP Perform calcium transient measurements after 3–4 weeks of co-culture. Use aseptic technique throughout this procedure.
In this De-Esterification Step, wash cells in PBS twice and feed cells with Differentiation Maintenance Medium. Place in incubator for 30 min to allow de-esterification of the dye.
-
Record calcium transients by placing 24-well dish with cells on an inverted epifluorescence microscope equipped with a CCD camera using Texas Red filter. Analyse videos of calcium transients using Image J software. Cells can be cultured as normal in Differentiation Maintenance Medium after analysis and can be reused for future experiments.
△ CRITICAL STEP. Use a heated stage or chamber (37° C) while recording transients, as activity progressively decreases when CMs are kept at room temperature. Cells should be analyzed immediately after the de-esterification Step, above.
(B) Characterization of iCPC potency in ex vivo mouse embryos TIMING 3–4 days
-
Infect iCPCs with pLentiGFP lentivirus (Lentivirus production, Box 2 and Infection Procedure, Steps 20–22) and passage as normal. The infection efficiency should be >95%. Enrich GFP+ iCPC by FACS if necessary. Adult cardiac fibroblasts infected with pLentiGFP lentivirus can be used as controls for this experiment.
△ CRITICAL STEP iCPC/cardiac fibroblasts must be infected with lentivirus constitutively expressing GFP to trace their progeny after injection into the cardiac crescent. We recommend using passage 10–15 iCPCs for this experiment.
-
Sacrifice pregnant dam and dissect uterine horns (2 min). Bring pregnant mice to dissection room on the 9th day of gestation (~8.0–8.5 dpc); dissect plugs identified several hours after pairing 8 days later at 14.00, when the range of stages may be headfold – 5-somite pairs 35.
△ CRITICAL STEP If possible, dissection room should be at ≤64°F or the embryos may lose viability.
-
On the space where you will sacrifice the mouse, place a paper towel, clean scissors, Jeweller’s forceps, and a carcass bag. Fill the base and the lid of a sterile 60 mm tissue culture dish with sterile PBS. At the dissection microscope, place the following items: clean and touched-up robust and fine forceps, the Arkansas stone and fine sandpaper for touching up forceps midway through a dissection, if necessary30, a small pile of sterile 35 mm dishes, and an appropriate amount of thawed Embryo Dissection Medium.
△ CRITICAL STEP The tissue culture dish should contain enough sterile PBS to cover the implantation sites, otherwise the refractive index of the tissue and medium will vary as the tissue is manipulated and make dissections difficult.
Euthanize pregnant females by cervical dislocation according to approved institutional protocol. A partial schematic guide to embryo dissections, with modifications that might be preferred by the investigator, is available 36, 37.
Sterilize the abdomen with ethanol, and make an incision orthogonal to the body axis at the level of the upper thigh of the mouse. With your hands, pull the skin on either side of the incision simultaneously toward the anus and head and open the peritoneal cavity with your scissors, cutting toward each thigh and the anus. With the tines of your scissors together, move the fat toward the anus and the visceral organs toward the head to expose the uterine horns.
-
Remove the uterine horns from the pregnant dam by gently grabbing each horn between decidual swellings; gently elevate them toward the ceiling, and make three cuts: one between each ovary and oviduct, and one at the cervix. The uterine organ should now be liberated. Snip away as much fat as possible from the uterine horns and place them into the lid of the 60 mm dish.
△ CRITICAL STEP The total dissection period from the time of removal of the uterine horns to reflexion of Reichert’s membrane begins now, and should take no longer than 30 minutes, and preferably 10–20 minutes.
-
Reflexion of uterine muscle and liberation of decidua (5 min). Transfer the uterus to the deep portion of the 60 mm tissue culture dish. Bring the dish with the uterine horns to the dissection microscope. Turn on both epi- and trans- illumination.
△ CRITICAL STEP Work at the lowest magnification possible (typically x7.5 total magnification), with both forceps and arms resting on glove boxes or other similar supports. The supports will transfer tension from your arms, thereby relaxing your wrists for dissection.
-
Locate the puffiest side of the first implantation site, closest to where the uterus was snipped from one of the ovaries, and with your robust forceps, tear through the two uterine muscle layers on one face and pick away at these until one decidual half is exposed (Supplementary Video 1).
△ CRITICAL STEP You must coordinate your forceps to work closely together. Do not pull or tear tissue with forceps far apart or you may rupture the cavities in your embryos. Use your robust forceps for tearing the uterine muscle and in later steps for manipulating the decidual swellings.
-
With one forceps, grip the uterus between the exposed implantation site and the next unexposed one. With the other forceps, squeeze the uterine tissue near the same site but closest to the exposed deciduum, tines together, and shell out the exposed deciduum. Repeat this with each implantation site (Supplementary Video 1).
△ CRITICAL STEP Do not place pressure on the implantation sites as you reflect the muscle as this will cause deflation of the conceptus inside.
-
With forceps, transfer all decidua to the deep part of a 35-mm dish containing Embryo Dissection Medium and count them as you do this (Supplementary Video 2).
△ CRITICAL STEP Grab each decidual swelling at its widest diameter; the conceptus is in the opposite pole.
-
Split decidual swellings to expose conceptuses (5 min). Increase the magnification slightly and gently stabilize the wider part of each deciduum between the tines of one forceps. With the other forceps, tines together, pierce the decidual dimple, located in the band of maternal blood, at a 60° angle to the bottom of the dish (Supplementary Video 3).
△ CRITICAL STEP Make sure that the tips of the tines of your impaling forceps travel through the entire deciduum and rest on the bottom of the dish.
Slightly separate the decidual halves with the same forceps. With the other forceps, clip the wider part of the deciduum that does not contain the embryo (Supplementary Video 3).
-
Rotate the deciduum 90 degrees, so that the wider part is at the top of your visual field. With palms facing down, hold your forceps and apply even but gentle pressure to each decidual wall; split the deciduum into two halves with a vertical downward motion. The embryo with its associated trophoblast will stay with one of the halves (Supplementary Video 3).
△ CRITICAL STEP If the decidual halves begins to tear, you must stop, and insert your forceps at the vertex of the split and snip the walls. Resume pulling as described above (Supplementary video 3)
Release conceptuses from the decidual halves and reflect Reichert’s membrane (10 min). With the tines of one forceps separated, impale the exposed decidual half and its embryo to the bottom of the dish and with the tines of the other forceps together, scrape the embryo out (Supplementary Video 4).
-
When all of the embryos have been scraped out, transfer them via a 9” sterile Pasteur pipette, maintaining them close to the pipette opening, into the lid of the 35 mm dish that contains dissection medium. Count as you transfer, making sure that the number of conceptuses transferred is the same as the number of dissected decidua, unless a deciduum was empty due to resorption (Figure 5).
△ CRITICAL STEP The ends of the Pasteur transfer pipettes should be smooth, not jagged, otherwise embryos may get stuck to the glass or damaged during transfer.
-
Using both pairs of fine forceps, pinch the trophoblast, associated parietal endoderm, and its Reichert’s membrane (all three tissues are collectively, and informally, called “Reichert’s membrane”) at the embryonic/extraembryonic junction. With one of the forceps, reflect Reichert’s membrane toward the embryo’s distal end, rounding that end, and allowing Reichert’s membrane to retract toward the extraembryonic region (Supplementary Video 5).
△ CRITICAL STEP Turn off the epi-illumination. This will allow Reichert’s membrane to be more readily visualized by trans-illumination only. Place your embryos side-by-side and eliminate any suspected small resorbing embryos (Figure 5). If you are not certain, reflect anyway.
Trim away enough of the reflected membrane so that the embryos can be staged38 (Supplementary Video 6).
-
Stage embryos (10 min). Place the reflected embryos into a 35 mm dish containing fresh dissection medium and stage them.35 If the embryos are at the appropriate stages (2–3 somite stage), you can use them immediately for cell injection, or place them in 500 μl of whole embryo culture medium in an organ culture dish and return to the incubator until cells are ready for injection.
■ PAUSE POINT The reflected embryos will be stable in this dish at room temperature (≤ 64°F) for about 90 minutes.
-
For any embryos that are not at the correct stage, two things can happen: (i) if they are too young, you may incubate them in organ culture dishes in whole embryo culture medium until they reach 2–3 somites a few hours later, or (ii) they may be fixed in 4% paraformaldehyde in preparation for other types of experiments, e.g., immunostaining 33.
△ CRITICAL STEP Twins and resorbing embryos should be discarded (Figure 5), if ≥30% of embryos are resorbed and/or resorbing, the entire litter should be discarded.
-
Injecting iCPCs into the cardiac crescent of living mouse embryos and ex vivo whole embryo culture (24 or 48 hr). Singularize cells from step i using 0.25% trypsin-EDTA, and suspend GFP+ iCPCs/cardiac fibroblasts at a density of 1000–1200 cells/ul in Fibroblast Medium and place on ice until embryo injections.
△ CRITICAL STEP Well-separated single cells are required for this procedure, and not cell clumps. Verify absence of cell clusters under microscope before proceeding and aspirate only well-separated cells.
-
Remove one embryo from the organ culture dish and place into center of the lid of a 60 mm tissue culture dish filled with Embryo Dissection Medium. Immediately place the organ culture dish and remaining conceptuses back to the incubator.
△ CRITICAL STEP Do not leave your conceptuses on the bench in whole embryo culture medium, as the medium is not buffered for atmosphere and the pH will quickly change.
Using a glass scorer, cut the injection pipette at its base so that it is half the original size and load it into the aspirator assembly.
-
Just prior to the injection, resuspend iCPCs or control fibroblasts and place 10 μl of the cells into the base of a 35 mm dish and cover it. Aspirate ~1 μl of cells into the injection pipette. This requires active aspiration rather than capillary action (Supplementary Video 7).
△ CRITICAL STEP In the dissection microscope, verify that iCPCs are not clumped. Verify that the medium and your cells have reached equilibration in the capillary after you cease to aspirate. If cells continue to enter, the opening of your injection pipette is too large and you must replace it.
-
While maintaining the aspirator’s mouthpiece in your mouth, its capillary in your injection hand, and the forceps in your other hand, orientate your embryo so that its cardiac crescent is facing upwards. Gently squeeze the embryo and aim your injection pipette into the cardiac crescent (Supplementary Video 8).
△ CRITICAL STEP It is useful to practice the injection on 1–2 embryos to gauge the batch of injection pipettes and your stability. The lid of the 60 mm dish is preferred over the base to perform injections, as the walls of the lid are much lower, and will not interfere with the injection.
-
Working at a magnification of ~x20, gently pierce the left or right half of the cardiac crescent with your injection pipette and blow cells into one half (Supplementary Video 8). The volume injected is about 0.05 μl (~50–100 cells), some of which will stream out of the injection hole; ~10–50 cells remain in the crescent, as verified by assessing GFP immunofluorescence post-injection (Figure 6a). You may repeat the injection on the other half of the crescent.
△ CRITICAL STEP During a perfect injection only the cardiac crescent inflates. If the crescent does not inflate, you might have penetrated the cardiac crescent too deeply and the cells may be injected in the amniotic cavity (Figure 6b; Supplementary Video 8), visible by inflation of the amniotic cavity and the pink colour of the medium (not seen in the black-and-white video); alternatively, the entire anterior half of the embryo might inflate, indicating that you have not penetrated the cardiac crescent but the space between the outer endoderm and the epiblast (not shown). In both cases, you may inject the other half of the crescent, as you are testing the potency of the cells in the cardiac region.
△ CRITICAL STEP Always culture 2–3 control unoperated conceptuses alongside injected embryos for every injection experiment.
Immediately after the injection, remove a culture tube from the incubator, and place the injected embryo into the tube with the aid of a sterile 9″ Pasteur pipette, and return the tube to the roller apparatus in the incubator as quickly as possible. Record the time at which each embryo goes into culture. Verify that your injection pipette is clear of clogs by gently blowing into the drop of medium that contains the cells and refill the capillary. Repeat the injection with the next embryo, and so on. Because rat serum is expensive, you may wish to culture pairs of injected embryos, in which case you must distinguish one from the other. This can be easily done without disturbing development by trimming the ectoplacental cone with scissor-action of two opposing hypodermic needles (Supplementary Video 9).
-
Culture injected embryos for 24 or 48 hrs. However, if culturing for 48 hours, you must transfer your embryos to fresh heat- and gas-equilibrated whole embryo culture medium at 24 hours using a wide-bore pipette. Because of increased growth, they can no longer be cultured in pairs, but as singlets. After the end of the whole embryo culture period, all embryos (injected and unoperated controls) should be scored for appropriate morphology39. Although they should be rare, as injection into the cardiac crescent does not affect development, abnormal embryos should be excluded from further analysis.
△ CRITICAL STEP Once all of the embryos are in the culture tubes, the incubator must not be opened at any time during the remainder of the culture period, as temperature and gas fluctuations compromise embryo growth and development.
-
Live imaging of embryos and whole-mount staining (30 min to 2 days). Using a wide-bore pipette, remove embryo from tube and place into a cell culture dish using minimal dissection medium required to keep the embryo hydrated. Perform live imaging using GFP and phase contrast filters to identify the location of the injected cells. The majority of the iCPCs should be seen within the contracting heart tube (Figure 6c). By contrast, the adult fibroblasts should not colonize the heart and may stick to the reflected trophoblast/parietal endoderm (Figure 6d). Put embryos back in the culture tube after imaging and proceed to next steps.
△ CRITICAL STEP Use a heated stage or chamber (37° C) while imaging live embryos, as viability and heart tube contractility both decrease when embryos are kept at room temperature. Embryos should be imaged immediately after removing from incubator.
-
Remove medium and wash embryos with 500ul PBS. Fix embryos in 4% Formaldehyde Solution for 1–2 hours at 4° C.
△ CRITICAL STEP. To remove medium use a 1ml pipette or wide-bore Pasteur pipettes. Do not use aspirator. Leave 20–30ul liquid in the tube to keep embryo wet at all times. Follow this for all subsequent wash steps.
-
Wash embryos 3 times with 500ul PBS. Dehydrate embryos in an increasing methanol gradient made in PBS (25%, 50%, 75%, 100%) (5 min per step at RT on a bench top rocker). Store at −20° C.
■ PAUSE POINT Embryos can be stored in 100% methanol at −20° C for up to 6 months.
Wash embryos twice with 100% methanol at RT. Rehydrate embryos reversing the Methanol gradient (75%, 50%, 25%, PBS) (5 min per step at RT on a bench top rocker). The dehydration-rehydration steps are required for permeabilizing the embryos.
Block embryos in Embryo Blocking Solution for 1hr at RT on benchtop rocker. Replace Blocking Solution after 1 hr and block for additional 1 hr (total blocking time is 2 hr)
Prepare the appropriate primary antibody dilutions in 500ul Embryo Blocking Solution/embryo and add to embryo tubes. We recommend staining with cardiac actin, MLC-2v, alpha/beta-MHC antibodies to identify CMs, and with GFP to label CPC-derivatives. Incubate at 4° C on rocker overnight.
Remove primary antibody solution and wash 5 times with Embryo Blocking Solution for 1 hr/wash at RT on rocker.
Prepare the appropriate secondary antibody dilutions in 500ul Embryo Blocking Solution/embryo and add to embryo tubes. Incubate at 4° C on rocker overnight.
Remove secondary antibody solution and wash 5 times with Embryo Blocking Solution for 1 hr/wash at RT on rocker.
-
Mount embryos on coverslips using melted agarose and place in depression slides. Use epifluorescence, confocal or multiphoton microscope for imaging whole mount embryos. Injected iCPCs should express markers of cardiac differentiation (Figure 6e).
△ CRITICAL STEP Imaging should be performed within 1 week after mounting as staining intensity will progressively decrease due to absence of anti-fade agents in agarose.
?TROUBLESHOOTING
Figure 5. Identification of resorbing embryos.
(a, b) Resorption in decidual swelling identified by the blood-filled site where the conceptus resides (arrows) in the intact deciduum (a) and decidual halves after splittiing (b). (c) Small conceptus (arrow) in the decidual half after splitting. (d) Small conceptus (circled) is notably smaller than its littermates, and is thus resorbing. Arrowhead indicates a conceptus whose Reichert’s membrane was reflected, in situ, within the decidual half. Scale bar 500um. All animal experiments were performed in accordance to University of Wisconsin-Madison’s animal use guidelines.
Figure 6. Injection of iCPCs into cardiac crescent for embryonic potency test.
(a) iCPCs labelled with GFP expressing lentivirus were injected into the cardiac crescent of mouse embryos. Images show a perfect injection where the cells are localized to the cardiac crescent. (b) Images show a faulty injection where the pipette pierced through the cardiac crescent and cells were injected into the amniotic cavity. (c) iCPCs injected into the cardiac crescent localize to the developing heart tube after 24 hr of whole embryo culture. Arrow indicates developing heart tube. (d) Adult cardiac fibroblasts injected into the cardiac crescent are excluded from the developing heart tube and localize to the ecto-placental cone (extra-embryonic tissue) after 24 hr of whole embryo culture. (e) iCPC-injected embryos were immunostained in whole-mount preparations for CM markers and GFP. Three-dimensional reconstruction images show iCPCs differentiated into CMs, as indicated by co-expression of CM marker MLC-2v and GFP. AC=amniotic cavity, HF=head fold, CC=cardiac crescent. Scale bar 100um in a,b,c,e. 500 um in d. All animal experiments were performed in accordance to University of Wisconsin-Madison’s animal use guidelines.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting table.
| Step | Problem | Possible reasons | Solution |
|---|---|---|---|
| Steps 1–3,7,8 | Low/no attachment of tissue pieces to the plates | Tissue pieces are too large | Mince tissue into pieces that are as small as possible (~1 mm in size). Smaller pieces tend to attach better |
| Too much medium used to set up explant culture | Tissue pieces float if more medium is used. Use no more than 6 ml of fibroblast medium per 100 mm dish | ||
| Checking for fibroblast outgrowth daily/frequently | Explant cultures should not be disturbed for 1 week after setup | ||
| Step 9 | No migrating fibroblasts | FBS lot is not suitable for explant culture | Test several FBS lots for explant culture and choose one that yields efficient fibroblast outgrowth |
| Steps 1–10 | Explant cultures become contaminated | Isolation performed in open air or improper aseptic technique used | All isolation steps must be performed in a sterile tissue culture hood. Follow aseptic technique when working in a culture hood |
| Step 26 and Box 1, step 3 | No GFP+ cells seen in control well or no mCherry+ cells seen in cardiac factor well after doxycycline induction | Absence of rtTA transgene in cells | Confirm genotype of mice or ensure that rtTA was introduced into cells before infecting with pSAM2 factors |
| LTR recombination of pSAM2 vectors | Check DNA via restriction digest to confirm the absence of LTR recombination | ||
| Step 26 and Box 1, steps 1–10 | Low infection efficiency/low GFP or mCherry expression | Inefficient virus production | Optimize virus production using pLenti GFP Puro plasmid and follow all steps exactly as detailed in Box 1 |
| Use of high-passage HEK cells | Use HEK cells below Passage 20 | ||
| Steps 32–35 | No proliferative iCPC colonies detected at 3–4 weeks | Inefficient virus production | Optimize virus production. Make sure that control GFP well has >80% infection efficiency for each batch of reprogramming |
| Initial cell density was too low | Starting cell density should be 50,000 fibroblasts per well of a 12-well plate. When transferred to a 60-mm dish, the cell confluency should not be lower than 25–30% on Day 5 | ||
| Frequent medium changes | Replace iCPC induction medium every 6–7 days until colonies form | ||
| Step 36A(vii) and 36B(xviii) | iCPC colonies do not expand upon passaging | Colonies were split too soon | Allow colony size to reach at least 3–4 mm in diameter or wait for 3–4 weeks after doxycycline induction |
| Split ratio too high | For early passages (1–3), we recommend a 1:3 split ratio. For Passage 4 onward, we recommend a 1:6 split ratio | ||
| Doxycycline was removed too early | Doxycycline induction should be maintained for at least 40 days or 2 passages. | ||
| Forgot to add LIF, BIO | LIF, BIO must be added at the appropriate concentration to expand iCPC colonies | ||
| Step 36B(xviii) | Not all iCPC colonies are expandable | Only 50–60% of the colonies give rise to expandable iCPC lines | We recommend picking 20–30 iCPC colonies while establishing clonal lines |
| Step 37A(iv) | No or low staining for CPC cell surface markers | Inadequate cell staining | Try staining for 1 h at 4 °C or at RT. Test antibodies on Day 5 for embryoid bodies differentiated from mESCs. Cell surface staining improves with passaging |
| Steps 37B(ii), 41 | No or low staining for CPC TFs/cardiac differentiation markers | Cells were seeded on coverslips at a very high density | Do not seed more than 40,000 cells per coverslip. Cell densities that are too high hinder staining. Follow instructions exactly as detailed in Step 37B |
| Antibodies not working | Check antibody staining on ES-differentiated cells. Order fresh antibodies | ||
| Step 41 | Lack of organized sarcomere staining in iCPC-differentiated CMs | Staining was performed <2 weeks after initiating differentiation | Sarcomere staining improves with extended culturing. We recommend culturing cells in differentiation maintenance medium for 3–4 weeks before staining for cardiac-lineage differentiation markers. Do not split cells after initiation of differentiation |
| Step 42A(iv) | No contracting iCPC-CMs observed during coculture with mESC-CMs | Improper iCPC-CM/mESC-CM ratio was used for coculture | Use at least a 1:10 ratio of iCPC-CM/mESC-CM. Coculture for 3 weeks before checking. Do not split cells during coculture |
| Step 42B(xxxvii) | No or low staining intensity detected for iCPC-injected embryos | Imaging embryos >1 week after mounting | Embryos should be imaged immediately after mounting, ideally within 4 d. As agarose does not contain anti-fade agents, the staining intensities decline with storage |
● TIMING
Step 1–10, isolation of adult mouse fibroblasts: 12–14 days
Steps 11–19, seed fibroblasts for lentivirus infection: 2 days
Steps 20–22, infect fibroblasts with cardiac factor lentiviruses: 2 days
Steps 23–25, induce expression of cardiac factor with doxycycline: 2 days
Steps 26–35, introduce CPC culture conditions and monitor for appearance of iCPC colonies: 3–4 weeks
Step 36A, establish bulk iCPC lines: 2–3 weeks
Step 36B, establish clonal iCPC lines: 4–5 weeks
Step 37A, analyze iCPC cell surface marker expression by flow cytometry: 1 day
Step 37B, analyze iCPC transcription factor expression by immunocytochemistry: 4 days
Step 37C, analyze iCPC gene expression by qPCR: 1–2 days
Steps 38–41, differentiate iCPCs into cardiac lineage cells: 4–8 weeks
Steps 42A, co-culture iCPCs with contracting CMs and measure calcium transients: 4 weeks
Steps 42B, characterize iCPC potency in ex vivo mouse embryos, 3–4 days
Box 1, manufacture injection pipettes: 5 min per pipette
Box 2, produce lentivirus particles: 5 days
ANTICIPATED RESULTS
After infecting fibroblasts with lentiviruses and inducing expression of cardiac factors by doxycycline treatment, the first EYFP+ proliferative colonies can be seen after 2 weeks (Figure 2b). These early iCPC colonies should continue to proliferate and by 3–4 weeks, they will reach 3–4mm in diameter (Figure 2d). At this time, they should be scored and passaged twice at 1:3 split ratio if establishing bulk iCPC lines or picked via cloning ring and passaged as individual clonal lines. iCPC reprogramming efficiency varies between 0.01% – 0.03% depending on starting fibroblasts. Doxycycline can be withdrawn after 2 passages for bulk cultures or 3 passages for clonal lines. iCPC lines should maintain Nkx-EYFP expression and continue to proliferate in iCPC Maintenance Medium with a doubling time of approximately 30 hr (Figure 2f). After passage 3, iCPCs can be split every 4–5 days at a split ratio of 1:6 to 1:8. At this stage, iCPCs can be analysed for expression of CPC TFs, including Nkx2.5, Gata4 and Irx4. >95% of iCPCs should express these TFs (Figure 2g). Heterogeneity with respect to staining intensity may be observed. Gene expression of iCPCs can be analysed using qPCR. We recommend using CPC markers, including Nkx2.5, Tbx5, Gata4, Gata6, Irx4, Mesp1, Isl1, Hand1, Hand2, Tbx20, Baf60c, Mef2c, and fibroblast-specific genes Thy1, Fsp1, Vim, Posn. The expression for CPC markers should be upregulated in iCPCs relative to fibroblasts. By contrast, fibroblast gene expression should be downregulated. Flow cytometry analysis of iCPCs should reveal homogeneous expression of Cxcr4. However, only a fraction of iCPCs will exhibit Flk1 and cKit labeling. In vitro characterization of iCPCs can be performed every 5 passages and should reveal increasing enrichment of cardiac markers with passaging. RNA Seq or microarray analysis can be performed to assess global gene expression of iCPCs and should reveal a shift from a fibroblast to a cardiac progenitor program. Late passage iCPCs (10–20 passages) should have higher enrichment for cardiac progenitor markers relative to early passage iCPCs (1–3 passages). Stably reprogrammed iCPCs (passage 5 onwards) can be differentiated into cardiac lineage cells by aggregation in iCPC Differentiation Medium followed by plating and culture in Differentiation Maintenance Medium. Immunostaining using cardiac lineage differentiation markers should reveal that iCPCs differentiate into three cardiac lineage cells: cardiomyocytes, smooth muscle cells and endothelial cells (Figure 3a). We recommend using alpha-actinin, cardiac actin, alpha/beta-MHC, MLC-2v, MLC-2a to identify cardiomyocytes, SM-MHC and SMA to identify smooth muscle cells and CD31 to identify endothelial cells differentiated from iCPCs. Sarcomere staining for iCPC-derived CMs gets progressively organized with extended culture in vitro. In the described differentiation protocol, the majority of the iCPCs differentiate into CMs (80–90%) whereas smooth muscle and endothelial cells represent 10–15% and 1–5% of iCPC-differentiated progeny, respectively. iCPCs should maintain cardiac lineage tri-potency with passaging. iCPC derived CMs should develop gap junctions and show synchronous calcium transients when co-cultured with mESC-CMs or neonatal CMs (Figure 3c). iCPCs can be injected into the cardiac crescent of mouse embryos to characterize their embryonic potency. Live imaging of iCPC-injected embryos following ex vivo whole embryo culture for 24 or 48 hours should reveal localization of iCPCs exclusively to the developing heart tube (Figure 6c). By contrast adult cardiac fibroblasts should be excluded from the heart tube (Figure 6d). Whole mount staining on iCPC-injected embryos with GFP and cardiac actin/MLC-2v/alpha-MHC antibodies should reveal that GFP labeled iCPCs stain for cardiac actin/MLC-2v/alpha-MHC, indicating their differentiation into CMs within the heart tube (Figure 6e). In contrast, the adult cardiac fibroblasts will not stain for CM markers, indicating their inability to differentiate into CMs. iCPCs can also be injected into the adult mouse heart following experimentally induced myocardial infarction, either via direct cell injection or using tissue engineered cardiac patches13, 14. iCPCs should engraft within scar tissue and differentiate into cardiomyocytes, smooth muscle cells, and endothelial cells in vivo. Taken together, researchers can use this protocol to reprogram mouse adult fibroblasts into expandable iCPCs that are multipotent and can differentiate into cardiac lineage cells both in vitro and in vivo.
Supplementary Material
Acknowledgments
The research was supported by National Institutes of Health grants U01HL099773, U01HL134764, R01 HL129798, and S10RR025644 (to T.J.K.); R01 HD042706 and R01 HD079481 (to K.M.D); T32 HD041921 (to A.M.D) and pre-doctoral fellowship R25GM083252 (to A.M.D.); and AHA pre-doctoral fellowship 12PRE9520035 (to P.A.L.)
Footnotes
Author Contributions
P.A.L conceived project, designed and performed experiments, analyzed data and wrote the final manuscript. K.M.D. and A.M.R. carried out animal husbandry, K.M.D. made all of the embryo reagents/tools, designed the potency experiments, performed all of the embryo dissections/manipulations/injections/potency testing, analyzed data, and wrote the embryonic potency testing section of the manuscript. A.M.R. recorded and edited the supplemental videos. T.J.K designed experiments, analyzed data, contributed to writing the manuscript, provided funding, and approved the final manuscript.
Competing Financial Interests
P.A.L, A.M.R, K.M.D declare no competing financial interests. T.J.K is founder and consultant for Cellular Dynamics International.
References
- 1.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 3.Han DW, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell stem cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Yu B, et al. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell stem cell. 2013;13:328–340. doi: 10.1016/j.stem.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Yang N, et al. Generation of oligodendroglial cells by direct lineage conversion. Nature biotechnology. 2013;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riddell J, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Protze S, et al. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. Journal of molecular and cellular cardiology. 2012;53:323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayawardena TM, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circulation research. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam YJ, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu JD, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem cell reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wada R, et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalit PA, et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell stem cell. 2016;18:354–367. doi: 10.1016/j.stem.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalit PA, Hei DJ, Raval AN, Kamp TJ. Induced pluripotent stem cells for post-myocardial infarction repair: remarkable opportunities and challenges. Circulation research. 2014;114:1328–1345. doi: 10.1161/CIRCRESAHA.114.300556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masino AM, et al. Transcriptional regulation of cardiac progenitor cell populations. Circulation research. 2004;95:389–397. doi: 10.1161/01.RES.0000138302.02691.be. [DOI] [PubMed] [Google Scholar]
- 16.Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- 17.Bondue A, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Oh H, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner RL. Extrinsic factors in cellular differentiation. The International journal of developmental biology. 1993;37:47–50. [PubMed] [Google Scholar]
- 20.Qian L, Berry EC, Fu JD, Ieda M, Srivastava D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc. 2013;8:1204–1215. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- 21.Cao N, et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352:1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
- 22.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islas JF, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Expandable Cardiovascular Progenitor Cells Reprogrammed from Fibroblasts. Cell stem cell. 2016;18:368–381. doi: 10.1016/j.stem.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu S, Wang H, Ding S. Reprogramming fibroblasts toward cardiomyocytes, neural stem cells and hepatocytes by cell activation and signaling-directed lineage conversion. Nature protocols. 2015;10:959–973. doi: 10.1038/nprot.2015.059. [DOI] [PubMed] [Google Scholar]
- 27.Maza I, et al. Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. Nat Biotechnol. 2015;33:769–774. doi: 10.1038/nbt.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bar-Nur O, et al. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat Biotechnol. 2015;33:761–768. doi: 10.1038/nbt.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson DO, et al. Irx4 Marks a Multipotent, Ventricular-Specific Progenitor Cell. Stem Cells. 2016 doi: 10.1002/stem.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Downs KM. In vitro methods for studying vascularization of the murine allantois and allantoic union with the chorion. Methods in molecular medicine. 2006;121:241–272. doi: 10.1385/1-59259-983-4:239. [DOI] [PubMed] [Google Scholar]
- 31.Champlin AK, Dorr DL, Gates AH. Determining the stage of the estrous cycle in the mouse by the appearance of the vagina. Biol Reprod. 1973;8:491–494. doi: 10.1093/biolreprod/8.4.491. [DOI] [PubMed] [Google Scholar]
- 32.Byers SL, Wiles MV, Dunn SL, Taft RA. Mouse estrous cycle identification tool and images. PLOS One. 2013;7:1–5. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Downs KM. Systematic localization of Oct-3/4 to the gastrulating mouse conceptus suggests manifold roles in mammalian development. Dev Dyn. 2008;237:464–475. doi: 10.1002/dvdy.21438. [DOI] [PubMed] [Google Scholar]
- 34.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical chemistry. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 35.Downs KM, Davies T. Staging of gastrulation in mouse embryos by morphological landmarks in the dissection microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- 36.Cockroft DL. In: Postimplantation Mammalian Embryos: A Practical Approach. Copp AJ, Cockroft DL, editors. IRL Press; Oxford: 1990. pp. 15–40. [Google Scholar]
- 37.Nagy A, Gertsenstein M, Vintersten K, Behringer RR. Manipulating the mouse embryo: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2003. [Google Scholar]
- 38.Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- 39.Downs KM, Gardner RL. An investigation into early placental ontogeny: allantoic attachment to the chorion is selective and developmentally regulated. Development. 1995;121:407–416. doi: 10.1242/dev.121.2.407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.