Abstract
Aims
Impaired sarcoplasmic reticulum (SR) Ca2+ cycling and depressed contractility, a hallmark of human and experimental heart failure, has been partially attributed to increased protein phosphatase 1 (PP-1) activity, associated with down-regulation of its endogenous inhibitor-1. The levels and activity of inhibitor-1 are reduced in failing hearts, contributing to dephosphorylation and inactivation of key calcium cycling proteins. Therefore, we investigated the mechanisms that mediate decreases in inhibitor-1 by post-transcriptional modification.
Methods and Results
Bioinformatics revealed that 17 human microRNAs may serve as modulators of inhibitor-1. However, real-time PCR analysis identified only one of these microRNAs, miR-765, as being increased in human failing hearts concomitant with decreased inhibitor-1 levels. Expression of miR-765 in HEK293 cells or mouse ventricular myocytes confirmed suppression of inhibitor-1 levels through binding of this miR-765 to the 3′-untranslated region of inhibitor-1 mRNA. To determine the functional significance of miR-765 in Ca2+ cycling, pri-miR-765 as well as a non-translated nucleotide sequence (miR-Ctrl) were expressed in adult mouse ventricular myocytes. The inhibitor-1 expression levels were decreased, accompanied by enhanced PP-1 activity in the miR-765 cardiomyocytes, and these reflected depressed contractile mechanics and Ca2+ transients, compared with the miR-Ctrl group. The depressive effects were associated with decreases in the phosphorylation of phospholamban and SR Ca2+ load. These miR-765 negative inotropic effects were abrogated in inhibitor-1-deficient cardiomyocytes, suggesting its apparent specificity for inhibitor-1.
Conclusions
miR-765 levels are increased in human failing hearts. Such increases may contribute to depressed cardiac function through reduced inhibitor-1 expression and enhanced PP-1 activity, associated with reduced SR Ca2+ load.
Keywords: Heart failure, MicroRNA-765, Protein phosphatase 1, Inhibitor-1, Calcium cycling, Cardiomyocyte contractility
Introduction
Decreased cardiac contractile performance is universally characterized by aberrant calcium homeostasis, which contributes to the pathophysiological development of human and experimental heart failure. The SERCA2a/phospholamban (PLN) regulatome including HAX-1, SUMO, S100, protein phosphatase 1 (PP-1), and the histidine-rich calcium-binding protein has appeared as a crucial signal axis in modulating cardiac contractility.1 The PP-1, associated with this Ca2+ regulatome, is controlled by inhibitor-1 (inhibitor-1), an ~27 kDs endogenous protein.2 Inhibitor-1 is inactive in its dephosphorylated state, but cAMP-dependent phosphorylation of this protein during beta-adrenergic stimulation results in activation of inhibitor-1 and potent inhibition of PP-1.3 Current evidence indicates that the inhibitor-1–PP-1 complex is a crucial negative regulator of sarcoplasmic reticulum (SR) Ca2+ cycling. Ca2+ is transported into the SR by SERCA2a, whose activity is regulated by PLN to induce relaxation, and then Ca2+ is released from the SR through the ryanodine receptor complex (RyR2), resulting in muscle contraction. PLN is the reversible regulator of SERCA2a. In the dephosphorylated state, PLN inhibits the Ca2+ affinity of SERCA2a, and phosphorylation of PLN relieves this inhibition. PP-1 is the main phosphatase responsible for dephosphorylating PLN.1,3 Importantly, perturbations in PP-1 activity have been implicated in the pathogenesis and development of heart failure.3
In human and experimental heart failure, PP-1 activity is increased, and this is partially attributed to inhibitor-1 dysregulation. Indeed, reduced expression and phosphorylation of inhibitor-1 at Thr35 by protein kinase A (PKA) have been detected in failing hearts, consistent with enhanced PP-1 activity.4 Studies in mouse models have shown that inducible expression of active inhibitor-1 in the adult heart results in enhanced basal cardiac function and protection against ischaemia–reperfusion injury,5 suggesting that inhibitor-1 has beneficial effects in maintaining heart function under both basal and stress conditions. Furthermore, recent studies have shown that long-term expression of active inhibitor-1 through the ageing process or in a rat model of heart failure can maintain hyperdynamic cardiac function.6 The beneficial effects of long-term increases of active inhibitor-1 were also confirmed in a large model of heart failure showing increases in contractile parameters and reduced infarct size post-myocardial infarction.7 Thus, inhibitor-1 protein or activity levels are important determinants of heart function and survival, but the molecular mechanisms regulating inhibitor-1 expression levels are not currently known.
MicroRNAs have been well defined as post-transcriptional regulators that can silence and regress gene translation via binding to complementary sequences on target mRNA transcripts.8 Numerous studies have demonstrated that the cardiac-deregulated microRNA expression and distribution are associated with the occurrence of compromised heart function. Using genetically modified animal models, microRNAs have been identified as important players in various cardiac pathological events including cardiac hypertrophy, arrhythmia, and ischaemia–reperfusion. Although a few microRNAs have been reported to disturb calcium homeostasis via targeting sodium–calcium exchanger,9 junctophilin-2,10 or SERCA2a,11 the role of microRNAs in regulating and tuning up contractile function remains overall elusive.
In this study, we provide evidence that enhanced miR-765 expression in human failing hearts is associated with decreases in inhibitor-1 expression levels. Indeed, miR-765 bound to the 3′-untranslated region (3′-UTR) of inhibitor-1 mRNA and reduced its expression, resulting in increased PP-1 activity and depressed contractility as well as Ca2+ transients. Interestingly, these miR-765-induced effects were abrogated in the absence of inhibitor-1 expression. Taken together, our findings implicate miR-765 as a regulator of inhibitor-1 protein levels and contractile performance and suggest that down-regulation of miR-765 may serve as a potential therapeutic treatment for cardiac dysfunction.
Methods
For details regarding methods, refer to the Supplementary materials online, Methods.
Human myocardial tissue
The current investigation conforms to the principles outlined in the Declaration of Helsiniki. Briefly, failing heart samples were acquired from seven patients (4 females, 2 males, and 1 with gender that cannot be tracked), whose ages ranged from 48 to 69 years. Cardiac dysfunction was caused by ischaemic heart disease (IHD), idiopathic dilated cardiomyopathy (IDC), and congestive heart failure (CHF). The average EF of the patients was 20±3%, which can be defined as heart failure with reduced ejection fraction (HFrEF). All heart samples were obtained from explanted hearts at the time of cardiac transplantation. As controls, seven non-failing hearts (5 females and 2 males in the age range of 52–61 years) were obtained from donors who had normal cardiac function and died from neurological diseases or road traffic accidents, as previously described.12
Stem–loop polymerase chain reaction and real-time polymerase chain reaction
Total mRNA was isolated form human heart tissue, using an miRNeasy Mini Kit (Qiagen) according to the manufacturer’s instruction. The expression of mature microRNAs in human heart tissue was quantified by using stem–loop PCR, using human microRNA-specific primers (Supplementary material online, Table S2) and the universal primers from QIAGEN (Cat No. 218193). Human U6 gene expression was used as an internal control. The real-time PCR was run in a C1000 Touch Thermal Cycler (Bio-Rad). Relative expression of mRNA was calculated using the comparative threshold cycle (Ct) method, as previously described.13 The fold change of the microRNA expression level was indicated by a heat map which was generated by the software Gene Cluster and TreeView (version 1.60). The same protocol was also applied to rat or mouse PCR, using different primers as listed in the Supplementary materials online,
For the inhibitor-1 mRNA expression levels, we used glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control, and the corresponding primers are listed in the Supplementary materials online, Methods.
Luciferase reporter assay
A human, mouse, or rat inhibitor-1 3′-UTR segment of ~100 bp and its respective mutant non-complementary nucleotides were amplified by footprint two-step PCR,13 and then cloned into the pMiR-REPORT™ luciferase microRNA expression reporter vector (Ambion) at the SpeI and HindIII sites. Human embryonic kidney (HEK) 293AD cells were co-transfected in 6-well plates using DharmaFECT Duo Transfection Reagent (Thermo Scientific Inc.), according to the protocol of the manufacturer, with 0.4 μg of the 3′-UTR luciferase reporter vector and 0.16 μg of the control vector pMIR-β-gal (Ambion, Inc.). For each well, 100 nM miR-765 mimic or miR-Ctrl mimic was used. Cell lysates were prepared 48 h later. Luciferase activity was measured using a luminometer and expressed as relative light units using a luciferase assay kit (Promega). β-Galactosidase activity was measured with a commercially available kit (Promega).
Mouse ventricular cardiomyocyte isolation and culture
The animals were handled in accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85–23, revised 1996) and the National Research Council Guide for the Care and Use of Laboratory Animals: 8th Edition, published by The National Academies Press, 2011, Washington, DC. All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Cincinnati (Protocol No. 04-04-19-02). Wild-type (FVBN/n) or inhibitor-1 knockout (I-1 KO) mouse14 cardiomyocytes were isolated according to the protocols developed by the Alliance for Cellular Signaling and modified by us.15
Contractile parameter and calcium kinetic measurements
Cardiomyocytes that adhered to the coverslips were bathed in temperature- (37 °C) equilibrated Krebs–Henseleit–Buffer (KHB) containing 1mM Ca2+ for 20min. The contractile function and calcium transient were recorded and analysed as previously described.16
Western blots
Proteins were analysed by western blots as previously described.16
Statistics
Data were expressed as the mean±SEM. Comparisons were evaluated by Student’s t-test for two groups. Multiple comparisons among three or more groups were performed using one-way analysis of variance (ANOVA), and Bonferroni exact test was conducted for post-hoc analyses (SPSS 13.0. IBM Co., Armonk, NY, USA). A P-value of <0.05 was considered statistically significant.
Results
Profiling candidate microRNAs targeting inhibitor-1 in human ventricular heart
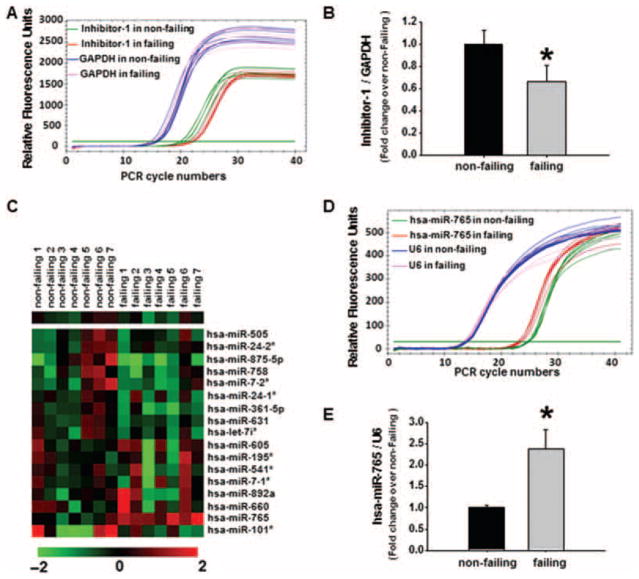
Previous studies have shown that the transcript and protein levels of inhibitor-1 are significantly reduced in human failing hearts.4 We also confirmed the down-regulation of inhibitor-1 at the transcriptional level, using real-time PCR analysis. The inhibitor-1 mRNA levels in human failing hearts decreased to 66% of those present in non-failing samples (Figure 1A and B), consistent with previous observations.17 To investigate whether this down-regulation could be mediated by microRNAs, we used the online database MicroCosm Target, which predicts microRNA targets. We identified 17 human-specific microRNAs as potential regulators of inhibitor-1 mRNA (Supplementary material online, Table S1). Synthetic mimics or inhibitors of these 17 predicted microRNAs were then transduced into HEK293 cells to confirm their potential regulatory effects on inhibitor-1 mRNA expression. Among these microRNA mimics, the hsa-miR-765 mimic exhibited the maximal inhibitory effect on inhibitor-1 mRNA expression (~70% reduction) (Supplementary material online, Figure S1A). Furthermore, silencing of the endogenous microRNAs, that regulated inhibitor-1 expression, resulted in enhanced inhibitor-1 mRNA levels, and the greatest increase was observed with an inhibitor specific for hsa-miR-765 (~60% increase) (Supplementary material online, Figure S1B). We then studied the expression levels of these candidate microRNAs by real-time PCR in human failing and non-failing hearts (Figure 1C). Among these candidates, a pronounced enhancement of hsa-miR-765 expression was detected in failing heart samples (Figure 1C). Indeed, hsa-miR-765-conjugated SYBR® green fluorescence signal appeared in the earlier PCR cycles in failing heart sample (Figure 1D), and there was a 2.3-fold increase in hsa-miR-765 expression in the failing heart group compared with non-failing samples (Figure 1E). These results provide evidence that miR-765 is expressed in human hearts and its expression levels increase in failing hearts.
Figure 1.
Expression levels of inhibitor-1 mRNA and its potential regulatory microRNAs in human hearts. Total mRNA was isolated and purified from human failing hearts (HFrEF) and non-failing hearts. (A) Representative real-time PCR curves illustrating inhibitor-1 and glyceraldehyde phosphate dehydrogenase (GAPDH) mRNA expression in non-failing and failing heart samples. (B) Quantitative analysis of inhibitor-1 expression levels relative to the non-failing heart group after normalization to GAPDH levels. (C) Heat map illustrating expression of candidate microRNAs regulating inhibitor-1 in non-failing and failing hearts. (D) Representative real-time PCR curves illustrating hsa-miR-765 and U6 mRNA expression in non-failing and failing heart samples. (E) Quantitative analysis of hsa-miR-765 expression levels relative to the non-failing group after normalization to U6 levels. (n=7 hearts per group; *P <0.05 vs. the non-failing group).
Regulation of inhibitor-1 expression and protein phosphatase-1 activity by a microRNA-765 mimic
Computational prediction (MicroCosm Targets, Version 5.0) indicated that miR-765 may target to a seed sequence (5′-UUCCUUCU-3′) in the 3′-UTR of human inhibitor-1, according to the Watson–Crick base-paring rule. Therefore, we investigated whether miR-765 can affect inhibitor-1 expression levels by transfection of a human-derived cell line, HEK293, with synthetic miR-Ctrl mimic and miR-765 mimic. As shown in the Supplementary material online, Figure S2A, after 48 h transfection of HEK293 cells, there is no significant alteration in the expression of the PP-1 level among all microRNA-treated groups. However, the expression levels of inhibitor-1 decreased by 30% in miR-765 mimic-transfected cells compared with the miR-Ctrl mimic group (Supplementary material online, Figure S2A and B). Correspondingly, PP-1 activity was enhanced in the presence of the miR-765 mimic (Supplementary material online, Figure S2C). These results suggest that miR-765 may act as an endogenous negative regulator of inhibitor-1 gene transcription, reflecting regulation of PP-1 activity.
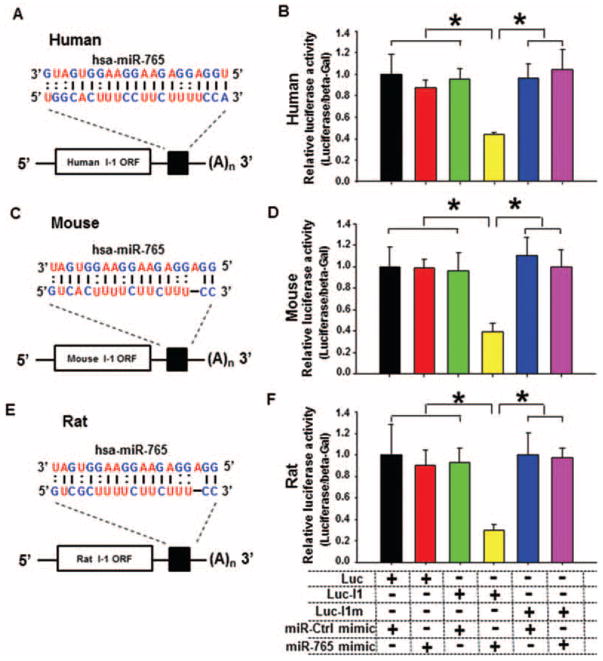
MicroRNA-765 targets the 3′-untranslated sequence of inhibitor-1 mRNA
It is well accepted that microRNAs degrade or translationally inhibit their target proteins through the nucleotide sequences at the 3′-UTR. Therefore, the 3′-UTR segment of human inhibitor-1 mRNA, containing the seed sequence for miR-765 (Figure 2A), was cloned and incorporated into the luciferase reporter vector (Luc-I1). To determine the specificity of this 3′-UTR for miR-765, the region containing the DNA-binding sequence for miR-765 was mutated to eliminate its covalent binding and then incorporated into the luciferase reporter vector construct (Luc-I1m). These vectors (Luc-I1 or Luc-I1m) were then transfected into HEK293 cells and the luciferase reporter activities were assessed upon treatment with miR-Ctrl or miR-765 mimic. Interestingly, when HEK293 cells were transfected with Luc-I1 vector and treated with miR-765 mimic, the luciferase activity was decreased by 56% (Figure 2B). These suppressive effects were abrogated when the binding sites for miR-765 were mutated (Luc-I1m) (Figure 2B). Control experiments with the luciferase vector, which did not contain the 3′-UTR of inhibitor-1, indicated no differences in luciferase activity in the presence of either miR-Ctrl or miR-765 mimic (Figure 2B). Collectively, these data suggest that miR-765 can down-regulate expression of human inhibitor-1 by targeting the 3′-UTR of this gene transcript.
Figure 2.
Luciferase assays of the microRNA-765 (miR-765)-binding site on the human, mouse, and rat inhibitor-1 3′-untranslated region (UTR). (A–C) Predicted miR-765-binding sequences on the 3′-UTRs of human (A), mouse (B), and rat (C) inhibitor-1 mRNAs. (D–F) Luciferase activity analysis indicated that human (D), mouse (E), and rat (F) inhibitor-1 are authentic target of miR-765 (n=5–6 preparations per group; *P <0.05)/
To explore whether other species contain putative hsa-miR-765-binding sites within their 3′-UTR of inhibitor-1, we performed bioinformatics analysis. This revealed that mouse and rat inhibitor-1 mRNA contained putative sites (Figure 2C and E), with the highest degree of homology to human (Figure 2A). Indeed, luciferase activity assays with the mouse or rat 3′-UTR sequences, incorporated into Luc-I1 vectors, indicated decreased activity in HEK293 cells transfected with either mouse Luc-I1 (62%) (Figure 2D) or rat Luc-I1 (70%) (Figure 2F). These data suggested that mouse and rat inhibitor-1 mRNA expression may also be regulated by hsa-miR-765. The different degrees of luciferase inhibition in HEK293 cells transfected with mouse Luc-I1 or rat Luc-I1 vectors (Figure 2D and F) may be due to distinct base distribution18 around the common 3′-UTR seed sequence (5′-CUUCU-3′) of these species. This 3′-UTR dissimilarity may contribute to differences in luciferase activity between mouse and rat.
Exogenous expression of mature human microRNA-765 in mouse cardiomyocytes
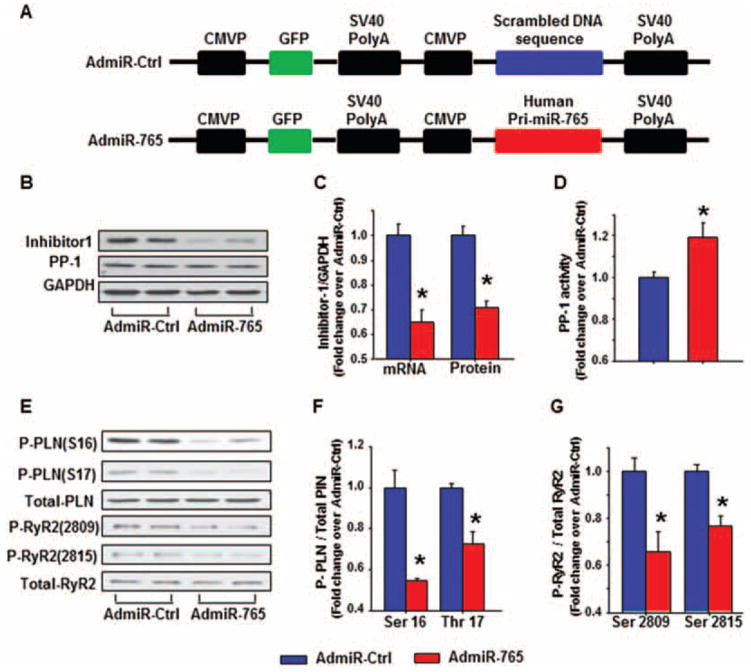
Given the fact that miR-765 can down-regulate expression of inhibitor-1 and ablation of inhibitor-1 can suppress cardiac function in vivo,19 we sought to determine whether miR-765 overexpression can also produce similar negative inotropic effects in cardiomyocytes. However, since this microRNA is expressed exclusively in human hearts, and bioinformatics along with luciferase reporter assays indicated that the expression levels of mouse or rat inhibitor-1 might be also regulated by miR-765 (see above), we used mouse and rat cardiomyocytes to explore the effect and the underlying mechanism of miR-765 on cardiac function. For these studies, we generated an adenovirus containing the human pri-miRNA-765- (AdmiR-765) encoding gene and an adenovirus containing the non-translated nucleotide sequence as control (AdmiR-Ctrl) (Figure 3A). Both viruses also contained the green fluorescent protein (GFP) gene. Nearly 100% of ventricular cardiomyocytes appeared infected after 48 h, as indicated by green fluorescence (Supplementary material online, Figure S3A). To determine further whether human pri-miRNA-765 can be converted into its mature form in mouse cardiomyocytes by rodent-associated post-transcriptional regulatory mechanisms, RNA samples were harvested and reverse transcription was performed to generate mature miR-765. As shown by electrophoresis in the Supplementary material online, Figure 3B, there was no mature miR-765 generated in the AdmiR-Ctrl-infected group, while mature miR-765 was detected in the AdmiR-765 infected group. These results suggest that mature human miR-765 can be generated in mouse cardiomyocytes.
Figure 3.
Expression of human microRNA-765 (miR-765) in mouse cardiomyocytes. (A) Schematic diagram of the recombinant adenoviral vector. The primary human miR-765 gene as well as a scrambled DNA sequence were amplified by PCR and incorporated into CMVP (cytomegalovirus promoter) downstream in the Adeasy-1/shuttle backbone vector. (B) Representative immunoblots illustrating inhibitor-1, protein phosphatase-1 (PP-1), and glyceraldehyde phosphate dehydrogenase (GAPDH) in mouse cardiomyocytes. (C) Quantitative results of total inhibitor-1 mRNA and protein expression levels in mouse cardiomyocytes. (D) PP-1 activity was assessed in mouse cardiomyocytes. (E) Representative immunoblots illustrate the phosphorylation of phospholamban (PLN) and ryanodine receptor 2 (RyR2) in mouse cardiomyocytes. (F) Quantitative analysis of the phosphorylation levels of PLN at Ser16 and Thr17, normalized to their respective total PLN protein levels. (G) Quantitative analysis of the phosphorylation levels of RyR2 at Ser2809 and Ser2815, normalized to their respective total RyR2 protein levels (n=5–6 hearts/group per group; *P <0.05 vs. Admir-Ctrl).
In addition, we examined the expression levels of inhibitor-1 in AdmiR-765-infected adult mouse cardiomyocytes. In the presence of AdmiR-765, the inhibitor-1 mRNA expression level was reduced by 35% (Figure 3C), while protein levels were decreased by 28% (Figure 3B and C) in mouse cardiomyocytes, compared with the control group. Accordingly, PP-1 activity was increased by 19% in AdmiR-765 mouse cardiomyocytes (Figure 3D). Similar alterations of inhibitor-1 expression levels and PP-1 activity were observed in isolated rat cardiomyocytes post-AdmiR-765 infection (Supplementary material online, Figure S4A–E). These findings suggest that the post-transcriptional suppressive effect of hsa-miR-765 on inhibitor-1, associated with increases in PP-1 activity, can also be observed in mouse and rat cardiomyocytes, similar to human hearts.
Calcium cycling protein phosphorylation
Earlier studies indicated that PP-1 is the major phosphatase regulating PLN dephosphorylation. To assess whether expression of miR-765 and the associated decreases in inhibitor-1 expression and increased PP-1 activity resulted in any alterations of PLN phosphorylation, quantitative immunoblotting was performed, using the infected mouse cardiomyocytes. As shown in Figure 3E and F, AdmiR-765 decreased the phosphorylation levels of PLN at residues Ser16 (PKA site) and Thr17 (Ca2+/calmodulin-dependent kinase site), compared with AdmiR-Ctrl in mouse cardiomyocytes. Besides PLN phosphorylation, altered phosphorylation of RyR2 and troponin I also serve as important molecular events that govern cardiac contractility through regulating SR Ca2+ release and modulating myofilament Ca2+ sensitivity, respectively.5 Western blot showed that AdmiR-765 also decreased the RyR2 phosphorylation at both Ser2809 and Ser2815 (Figure 3E and G). Similar effects on reduced phosphorylation of PLN and RyR2 were also observed in rat cardiomyocytes (Supplementary material online, Figure S4F–H). Importantly, miR-765 had no effect on PKA phosphorylation of troponin I (Supplementary material online, Figure S5).
Contractility of cardiomyocytes in the presence of microRNA-765
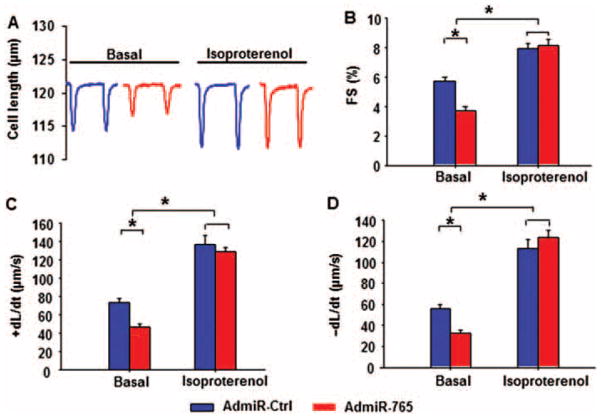
To evaluate the functional role of miR-765 in cardiac contractility, adult mouse cardiomyocytes were infected with AdmiR-Ctrl and AdmiR-765 in parallel. Under basal conditions, fractional shortening (FS%) (Figure 4A and B), the rate of contraction (+dL/dt) (Figure 4C), and the rate of relaxation (−dL/dt) (Figure 4D) in mouse cardiomyocytes were reduced by 35, 36, and 42%, respectively, by AdmiR-765 compared with AdmiR-Ctrl. Similar findings were obtained in adult rat cardiomyocytes (Supplementary material online, Figure S6A–D). Mouse cardiomyocytes were then exposed to beta-adrenergic receptor stimulation by isoproterenol, which induced positive inotropic effects in both groups of cardiomyocytes. All contractile parameters in the AdmiR-Ctrl group, including FS%, +dL/dt, and −dL/dt, were significantly enhanced to 139, 186, and 202% by isoproterenol stimulation (Figure 4A–D). Isoproterenol also increased FS%, +dL/dt, and −dL/dt in the AdmiR-765 treated group, compared with basal levels. Notably, the maximally stimulated parameters in the AdmiR-765 cardiomyocytes were increased to the same extent as those in the AdmiR-Ctrl cells (Figure 4A–D). These findings indicate that miR-765 depresses basal contractile parameters in isolated cardiomyocytes but this negative inotropic effect can be abrogated by isoproterenol stimulation.
Figure 4.
Contractile parameters of isolated adult mouse cardiomyocytes in the presence of microRNA-765 (miR-765). Isolated mouse left ventricular myocytes were suspended in 1.0 mmol/L Ca2+ Tyrode solution and field stimulated at 0.5Hz. (A) Representative cell-shortening traces of cardiomyocytes treated with AdmiR-Ctrl or AdmiR-765. Fractional shortening (FS%) (B), rates of shortening (+dL/dt), (C) and rates of relaxation (−dL/dt) (D) under resting conditions (Basal) and in the presence of 100 nM isoproterenol. (Basal, AdmiR-Ctrl=87 cells from six hearts; AdmiR-765=103 cells from 6 hearts; isoproterenol, AdmiR-Ctrl=45 cells from five hearts; AdmiR-765=50 cells from five hearts; a minimum of nine cells were used/heart) (*P <0.05)
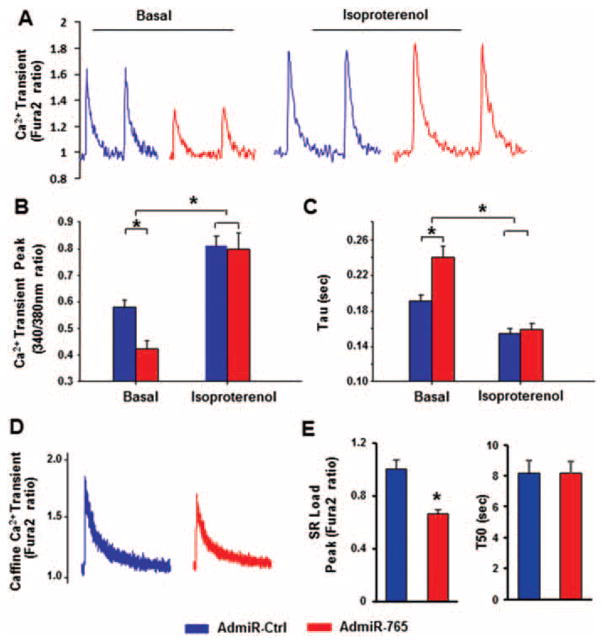
Regulation of intracellular Ca2+ transients by microRNA-765
The increase of cytosolic calcium or calcium transient is an important intracellular event induced by depolarization of the action potential on the cardiomyocyte membrane, and this subsequently activates calcium-sensitive contractile proteins, leading to cell shortening. In agreement with the above contractility results, expression of miR-765 resulted in a decrease (~27%) in the Ca2+ transient amplitude, compared with the AdmiR-Ctrl group under basal non-stimulated conditions (Figure 5A and B). Similar findings were observed in rat cardiomyocytes (Supplementary material online, Figure S6E and F). In response to isoproterenol stimulation, the calcium amplitude was increased in both AdmiR-Ctrl and AdmiR-765 groups, compared with their basal conditions, and the peak value was enhanced to the same extent between the AdmiR-765 group and the AdmiR-Ctrl group (Figure 5A and B). Furthermore, the time constant (Tau) of the calcium transient decay was prolonged in AdmiR-765, compared with the control group under basal conditions. In the presence of isoproterenol, this Ca2+ decay time was significantly shortened in both groups (Figure 5C). These data suggest that miR-765 depresses the amplitude of the intracellular calcium transient and prolongs its return to the diastolic level, under both basal and adrenergic stimulation conditions. Furthermore, the caffeine-induced calcium transient peak was decreased compared with AdmiR-Ctrl cardiomyocytes (Figures 5D and E), indicating a lower SR Ca2+ content. The caffeine-induced 50% time constant of calcium transient decay, which mainly reflects the calcium extrusion by sodium/calcium exchanger (NCX), was not different between AdmiR-Ctrl and AdmiR-765 cardiomyocytes (Figure 5E).
Figure 5.
Effects of microRNA-765 (miR-765) on cardiomyocytes calcium transients under basal and isoproterenol-stimulated conditions. Intracellular calcium transients were assessed by Fura2, a ratio metric fluorescent dye, which binds to free intracellular calcium. (A) Representative tracings of Ca2+ transients under basal conditions and in the presence of isoproterenol; Ca2+ transient peak (B); and time constant of the calcium transient decay (Tau) (C). (D) Representative curves for caffeine-induced calcium release fromthe sarcoplasmic reticulum (SR) under basal condition. (E) Peak caffeine-induced calcium release from the SR (SR load) is indicated by the Fura2 ratio of 340/380 nm, and the prolongation of time to 50% baseline is indicated by T50. (Basal, AdmiR-Ctrl=56 cells from six hearts; AdmiR-765=50 cells from six hearts; isoproterenol, AdmiR-Ctrl=50 cells from five hearts; AdmiR-765=48 cells from five hearts; a minimum of seven cells were used per heart) (*P <0.05).
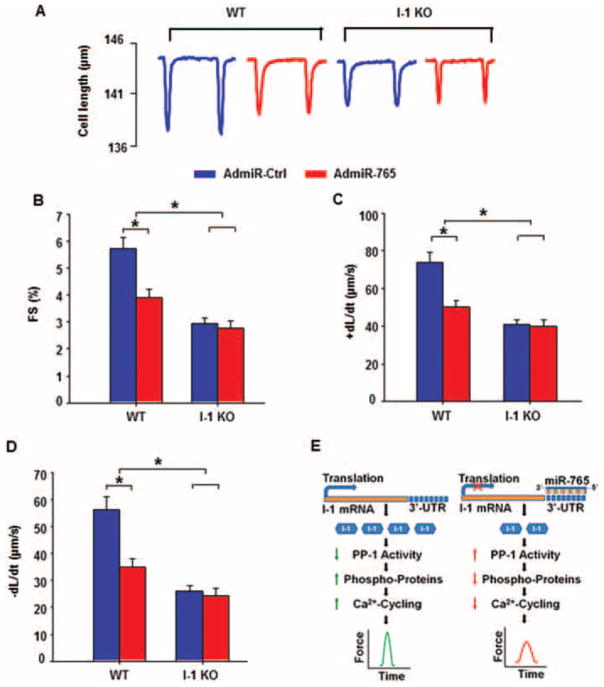
Inhibitor-1 ablation abrogates the contractile effects of microRNA-765 in cardiomyocytes
It has been suggested that a microRNA-induced cellular or biological phenotype may reflect the combined effects by its multiple targets.20 Thus, we utilized the online microRNA predicting software ‘MicroCosm Target’ to identify the potential targets of miR-765. As shown in Supplementary material online, Figure S7, there were 888 targets (647 unique targets) in human, 739 targets (305 are unique) in mouse, and 653 targets (284 are unique) in rat, according to the algorithms of this program. Further analysis on the basis of the online programs ‘Gene Ontology’ and ‘KEGG Pathway’ showed that among these miR-765 targets, there are 88 gene transcripts that are homologous among human, rat, and mouse (Supplementary material online, Table S3). Notably, these potential targets include protein phosphatase 5 (PP5) and FK506-binding protein, which have been reported to regulate cardiac function.21,22 Thus, to determine the specificity of inhibitor-1 in mediating the effects of miR-765 in cardiomyocyte contractility, we adenovirally infected wild-type (WT) and I-1 KO cardiomyocytes with miR-765. Under control conditions, or in the presence of AdmiR-Ctrl, the contractile function was significantly lower in I-1 KO cardiomyocytes, compared with the WT group, as evidenced by decreased FS%, +dL/dt, and −dL/dt (Figures 6A–D), in agreement with previous findings in this model.19 Expression of AdmiR-765 suppressed the contractile parameters in WT cardiomyocytes, while it had no effects in I-1 KO cardiomyocytes (Figure 6A). Importantly, there were no differences in contractile parameters between AdmiR-Ctrl and AdmiR-765 groups in KO cells (Figure 6B–D), indicating that the miR-765-elicited negative inotropic response was abrogated in the absence of inhibitor-1. As a positive control, to show that the depressed contractility in inhibitor-1-deficient cardiomyocytes can be further decreased, we used clenbuterol, a negative inotropic compound classified as a beta2-adrenergic receptor agonist.23 In the presence of clenbuterol, contractility was decreased not only in the WT but also the IO-1 KO cells. The maximally inhibited contractile parameters were similar between WT and KO cardiomyocytes, indicating that inhibitor-1 ablation did not elicit maximal depression in cardiac function (Supplementary material online, Figure S8).
Figure 6.
Effects of microRNA-765 (miR-765) overexpression on contractile function in inhibitor-1-deficient mouse cardiomyocytes. (A) Representative tracings for contraction in wild-type (WT) and inhibitor-1 knockout (I-1 KO) mouse cardiomyocytes post-AdmiR-Ctrl or AdmiR-765 infection. (B–D) Quantitative analysis of fractional shortening (FS%) (B), rates of shortening (+dL/dt) (C), and rates of relaxation (−dL/dt) (D). (E) Proposed mechanism of miR-765 regulation of cardiomyocyte contractility (WT infected with AdmiR-Ctrl=44 cells from four hearts; WT infected with AdmiR-765=53 cells from four hearts; I-1 KO infected with AdmiR-Ctrl=40 cells from four hearts; I-1 KO infected with AdmiR-Ctrl=47 cells from four hearts; a minimum of eight cells was used per heart) (*P <0.05).
Discussion
The current study reveals that the levels of hsa-miR-765 are increased in failing human hearts, characterized by reduced EF, and demonstrates for the first time that this microRNA can post-transcriptionally regulate inhibitor-1 expression, resulting in increased PP-1 activity, dephosphorylation of key SR Ca2+-handling proteins, and depressed cardiomyocyte contractile function (Figure 6E). Previous studies have shown that microRNAs play crucial roles in the pathogenesis and development of heart disease24 through post-transcriptional regulation of cardiomyocyte viability, inflammation, development of hypertrophy, and fibrosis.25 A small number of studies have also shown that microRNAs can target calcium-handling proteins in cardiomyocytes and, thus, alter contractility. Specifically, miR-24 expression was found to increase in failing ventricular cells, and overexpression of miR-24 in cardiomyocytes decreased junctophilin-2 expression and suppressed excitation–contraction coupling.10 In addition, miR-25 expression, whose specific target is SERCA2a, was higher in human failing hearts, and inhibition of this microRNA improved the impaired cardiac contractile function.11
The increases in PP-1 activity in failing human hearts have raised the possibility that inhibitor-1 expression might be dysregulated during the pathological development of remodelling.26 Actually, inhibitor-1 mRNA and protein levels are reduced by 60% in failing human hearts,27 and these may contribute to increased PP-1 activity and deteriorated contractile function.4 Accordingly, constitutive activation of inhibitor-1 (inhibitor-1c) can improve Ca2+ handling, contractility, and, most importantly, reverse remodelling in small animal models with heart failure.5,19,28 These promising studies have recently been extended into a pre-clinical heart failure model, indicating that long-term expression of active inhibitor-1 can preserve contractility and reduce scar size.7
To determine regulatory mechanisms in inhibitor-1 expression, computational algorithms were used, and 17 human-specific miR-NAs were identified that may serve as post-transcriptional regulators of this gene. However, only hsa-miR-765 was up-regulated in ventricular samples from patients with heart failure. Furthermore, the specificity of hsa-miR-765 for inhibitor-1 was confirmed by luciferase reporter assays of the 3′-UTR of inhibitor-1 mRNA and enzymatic analysis of PP-1 activity, using HEK293 cells. MicroRNA-765 is a 21 nucleotide non-coding RNA molecule, whose encoding gene is located in the long arm of chromosome 1 at position 23.1 (1q23.1) in Homo sapiens. The putative hsa-miR-765-binding site within the 3′-UTR of inhibitor-1 was similar between human, rat, and mouse, which allowed us to carry out further experiments in rodent cardiomyocytes and to elucidate the functional role of miR-765 in cardiac contractility and Ca2+ cycling. Indeed, expression of miR-765 in either mouse or rat cardiomyocytes resulted in down-regulation of inhibitor-1 at the post-transcription level and enhanced PP-1 activity. Accordingly, contractile parameters and the calcium transient were depressed in response to miR-765 overexpression. The molecular mechanisms underlying this suppressive effect by miR-765 involved dephosphorylation of PLN at Ser16 and Thr17 as well as of RyR2 at Ser2809 and Ser2815. The miR-765-induced negative inotropic effect was abrogated when the increased PP-1 activity was counteracted by maximal increases in PKA activation during beta-adrenergic receptor stimulation by isoproterenol. Previous studies have also indicated that alterations in inhibitor-1 expression or activity are associated with altered phosphorylation of PLN5 as well as RyR2.29,30 Dephosphorylation of PLN is expected to increase inhibition of SERCA2a activity, SR Ca2+ load, and contractility. However, dephosphorylation of RyR2 may decrease SR calcium leak, which may constitute an important compensatory mechanism in the face of reduced SR Ca2+ transport to maintain Ca2+ load and contractile function.
As noted recently, a single microRNA can target multiple mRNAs,20 and the induced cellular or biological phenotype may be the combined result of effects from all its targets.20 Notably, we used a loss-of-function approach to elucidate the role of inhibitor-1 in miR-765-mediated inotropic effects. We expressed miR-765 in the I-1 KO model to determine the specificity of the miR-765 effects. Contractile function was compromised in I-1 KO ventricular myocytes, in agreement with previous findings in KO mice.19 Importantly, although miR-765 could reduce function in WT cardiomyocytes, it had no further effect on the reduced contractility of the inhibitor-1-deficient cardiomyocytes. These findings suggest that inhibitor-1 serves as a specific molecular target in mediating the negative inotropic responses of miR-765 in cardiomyocytes. However, the in vivo effects of miR-765 in regulation of the inhibitor-1–PP-1 signalling axis in the heart remain to be determined. It is interesting to propose that inhibition of endogenous miR-765 may improve the depressed function of failing hearts but, since this microRNA is exclusively expressed in H. sapiens, such studies are not currently feasible. To this end, generation of a mouse model with cardiac-specific and inducible expression of hsa-miR-765 may provide a system to address the role of miR-765 in vivo and assess the benefits of its inhibition in restoring the depressed function in heart disease.
In summary, our data indicate that overexpression of miR-765 results in reduced inhibitor-1 expression, which subsequently suppresses cardiomyocyte contractile function and calcium cycling through regulation of the PP-1 signalling axis. In light of recent findings on the beneficial effects elicited by increased inhibitor-1 activity (inhibitor-1c) in pre-clinical studies,7 it is interesting to propose that down-regulation of miR-765 may serve as an additional therapeutic target to inhibit the increased PP-1 activity and improve calcium handling in failing hearts.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institutes of Health [grants HL26057 and HL64018 to E.G.K].
Footnotes
Conflict of interest: none declared.
Additional Supporting Information may be found in the online version of this article:
Figure S1 Inhibitor-1 mRNA expression in HEK293 cells after exposure to synthetic mimics or inhibitors of microRNAs predicted to regulate inhibitor-1 mRNA levels.
Figure S2 Expression of inhibitor-1 in HEK293 cells treated with the miR-765 mimic.
Figure S3 Expression of human miR-765 in mouse cardiomyocytes.
Figure S4 Expression of human miR-765 in rat cardiomyocytes.
Figure S5 Cardiac troponin I phosphorylation in isolated cardiomyocytes in response to miR-765 overexpression.
Figure S6 Contractility and calcium transients of isolated adult rat cardiomyocytes in the presence of miR-765.
Figure S7 Bioinformatics software (MicroCosm Target) predicts potential genes targeted by miR-765.
Figure S8 Effects of clenbuterol on contractile parameters in inhibitor-1-deficient mouse cardiomyocytes.
Table S1 Human-specific microRNAs as potential regulators of I-1 mRNA.
Table S2 Human microRNA-specific primers for stem loop PCR.
Table S3 miR-765-targeted gene transcripts that are homologous among human, rat and mouse.
References
- 1.Kranias EG, Hajjar RJ. Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res. 2012;110:1646–1660. doi: 10.1161/CIRCRESAHA.111.259754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Nicolaou P, Hajjar RJ, Kranias EG. Role of protein phosphatase-1 inhibitor-1 in cardiac physiology and pathophysiology. J Mol Cell Cardiol. 2009;47:365–371. doi: 10.1016/j.yjmcc.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Armouche A, Pamminger T, Ditz D, Zolk O, Eschenhagen T. Decreased protein and phosphorylation level of the protein phosphatase inhibitor-1 in failing human hearts. Cardiovasc Res. 2004;61:87–93. doi: 10.1016/j.cardiores.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Nicolaou P, Rodriguez P, Ren X, Zhou X, Qian J, Sadayappan S, Mitton B, Pathak A, Robbins J, Hajjar RJ, Jones K, Kranias EG. Inducible expression of active protein phosphatase-1 inhibitor-1 enhances basal cardiac function and protects against ischemia/reperfusion injury. Circ Res. 2009;104:1012–1020. doi: 10.1161/CIRCRESAHA.108.189811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pritchard TJ, Kawase Y, Haghighi K, Anjak A, Cai W, Jiang M, Nicolaou P, Pylar G, Karakikes I, Rapti K, Rubinstein J, Hajjar RJ, Kranias EG. Active inhibitor-1 maintains protein hyper-phosphorylation in aging hearts and halts remodeling in failing hearts. PLoS One. 2013;8:e8071. doi: 10.1371/journal.pone.0080717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fish KM, Ladage D, Kawase Y, Karakikes I, Jeong D, Ly H, Ishikawa K, Hadri L, Tilemann L, Muller-Ehmsen J, Samulski RJ, Kranias EG, Hajjar RJ. AAV9 inhibitor-1c delivered via direct coronary infusion in a porcine model of heart failure improves contractility and mitigates adverse remodeling. Circ Heart Fail. 2013;6:310–317. doi: 10.1161/CIRCHEARTFAILURE.112.971325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aurora AB, Mahmoud AI, Luo X, Johnson BA, van Rooij E, Matsuzaki S, Humphries KM, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca2+ overload and cell death. J Clin Invest. 2012;122:1222–1232. doi: 10.1172/JCI59327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu M, Wu HD, Li RC, Zhang HB, Wang M, Tao J, Feng XH, Guo YB, Li SF, Lai ST, Zhou P, Li LL, Yang HQ, Luo GZ, Bai Y, Xi JJ, Gao W, Han QD, Zhang YY, Wang XJ, Meng X, Wang SQ. Mir-24 regulates junctophilin-2 expression in cardiomyocytes. Circ Res. 2012;111:837–841. doi: 10.1161/CIRCRESAHA.112.277418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlquist C, Jeong D, Rojas-Muñoz A, Kho C, Lee A, Mitsuyama S, van Mil A, Park WJ, Sluijter JP, Doevendans PA, Hajjar RJ, Mercola M. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508:531–535. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaanine AH, Jeong D, Liang L, Chemaly ER, Fish K, Gordon RE, Hajjar RJ. JNK modulates FOXO3a for the expression of the mitochondrial death and mitophagy marker BNIP3 in pathological hypertrophy and in heart failure. Cell Death Dis. 2012;3:265. doi: 10.1038/cddis.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Zhang X, Ren XP, Chen J, Liu H, Yang J, Medvedovic M, Hu Z, Fan GC. MicroRNA-494 targeting both proapoptotic and antiapoptotic proteins protects against ischemia/reperfusion-induced cardiac injury. Circulation. 2010;122:1308–1318. doi: 10.1161/CIRCULATIONAHA.110.964684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allen PB, Hvalby O, Jensen V, Errington ML, Ramsay M, Chaudhry FA, Bliss TV, Storm-Mathisen J, Morris RG, Andersen P, Greengard P. Protein phosphatase-1 regulation in the induction of long-term potentiation: heterogeneous molecular mechanisms. J Neurosci. 2000;20:3537–3543. doi: 10.1523/JNEUROSCI.20-10-03537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrano GR, Fraser I, Han H, Ni Y, O’Connell T, Yan Z, Stull JT. Navigating the signalling network in mouse cardiac myocytes. Nature. 2002;420:712–714. doi: 10.1038/nature01306. [DOI] [PubMed] [Google Scholar]
- 16.Qian J, Vafiadaki E, Florea SM, Singh VP, Song W, Lam CK, Wang Y, Yuan Q, Pritchard TJ, Cai W, Haghighi K, Rodriguez P, Wang HS, Sanoudou D, Fan GC, Kranias EG. Small heat shock protein 20 interacts with protein phosphatase-1 and enhances sarcoplasmic reticulum calcium cycling. Circ Res. 2011;108:1429–1438. doi: 10.1161/CIRCRESAHA.110.237644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Armouche A, Rau T, Zolk O, Ditz D, Pamminger T, Zimmermann WH, Jäckel E, Harding SE, Boknik P, Neumann J, Eschenhagen T. Evidence for protein phosphatase inhibitor-1 playing an amplifier role in beta-adrenergic signaling in cardiac myocytes. FASEB J. 2003;17:437–439. doi: 10.1096/fj.02-0057fje. [DOI] [PubMed] [Google Scholar]
- 18.Witkos TM1, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carr AN, Schmidt AG, Suzuki Y, del Monte F, Sato Y, Lanner C, Breeden K, Jing SL, Allen PB, Greengard P, Yatani A, Hoit BD, Grupp IL, Hajjar RJ, DePaoli-Roach AA, Kranias EG. Type 1 phosphatase, a negative regulator of cardiac function. Mol Cell Biol. 2002;22:4124–4135. doi: 10.1128/MCB.22.12.4124-4135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorn GW., 2nd Decoding the cardiac message: the 2011 Thomas W. Smith Memorial Lecture. Circ Res. 2012;110:755–763. doi: 10.1161/CIRCRESAHA.111.256768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gergs U, Boknik P, Buchwalow IB, Fabritz L, Gründker N, Kucerova D, Matus M, Werner F, Schmitz W, Neumann J. Modulation of cardiac contractility by serine/threonine protein phosphatase type 5. Int J Cardiol. 2012;154:116–121. doi: 10.1016/j.ijcard.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Song YH, Qian LS, Ma XT, Wu KF. Telomerase: obviously activated in the accelerated phase of chronic myeloid leukemia. Haematologica. 2000;85:1222–1224. [PubMed] [Google Scholar]
- 23.Siedlecka U1, Arora M, Kolettis T, Soppa GK, Lee J, Stagg MA, Harding SE, Yacoub MH, Terracciano CM. Effects of clenbuterol on contractility and Ca2+ homeostasis of isolated rat ventricular myocytes. Am J Physiol Heart Circ Physiol. 2008;295:H1917–H1926. doi: 10.1152/ajpheart.00258.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adam O, Löhfelm B, Thum T, Gupta SK, Puhl SL, Schäfers HJ, Böhm M, Laufs U. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- 25.Abdellatif M. Differential expression of microRNAs in different disease states. Circ Res. 2012;110:638–650. doi: 10.1161/CIRCRESAHA.111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann J, Eschenhagen T, Jones LR, Linck B, Schmitz W, Scholz H, Zimmermann N. Increased expression of cardiac phosphatases in patients with end-stage heart failure. J Mol Cell Cardiol. 1997;29:265–272. doi: 10.1006/jmcc.1996.0271. [DOI] [PubMed] [Google Scholar]
- 27.Wittköpper K, Dobrev D, Eschenhagen T, El-Armouche A. Phosphatase-1 inhibitor-1 in physiological and pathological beta-adrenoceptor signalling. Cardiovasc Res. 2011;91:392–401. doi: 10.1093/cvr/cvr058. [DOI] [PubMed] [Google Scholar]
- 28.Pathak A, del Monte F, Zhao W, Schultz JE, Lorenz JN, Bodi I, Weiser D, Hahn H, Carr AN, Syed F, Mavila N, Jha L, Qian J, Marreez Y, Chen G, McGraw DW, Heist EK, Guerrero JL, DePaoli-Roach AA, Hajjar RJ, Kranias EG. Enhancement of cardiac function and suppression of heart failure progression by inhibition of protein phosphatase 1. Circ Res. 2005;96:756–766. doi: 10.1161/01.RES.0000161256.85833.fa. [DOI] [PubMed] [Google Scholar]
- 29.Wittköpper K, Fabritz L, Neef S, Ort KR, Grefe C, Unsöld B, Kirchhof P, Maier LS, Hasenfuss G, Dobrev D, Eschenhagen T, El-Armouche A. Constitutively active phosphatase inhibitor-1 improves cardiac contractility in young mice but is deleterious after catecholaminergic stress and with aging. J Clin Invest. 2010;120:617–626. doi: 10.1172/JCI40545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Armouche A, Wittköpper K, Degenhardt F, Weinberger F, Didié M, Melnychenko I, Grimm M, Peeck M, Zimmermann WH, Unsöld B, Hasenfuss G, Dobrev D, Eschenhagen T. Phosphatase inhibitor-1-deficient mice are protected from catecholamine-induced arrhythmias and myocardial hypertrophy. Cardiovasc Res. 2008;80:396–406. doi: 10.1093/cvr/cvn208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.