Abstract
The GAL4-UAS system is a powerful tool for manipulating gene expression, but its application in C. elegans has not been described. Here we systematically optimize the system’s three main components to develop a temperature-optimized GAL4-UAS system (cGAL) that robustly controls gene expression in C. elegans across 15–25°C. We demonstrate its utility in transcriptional reporter analysis, site-of-action experiments and exogenous transgene expression, and provide a basic driver and effector toolkit.
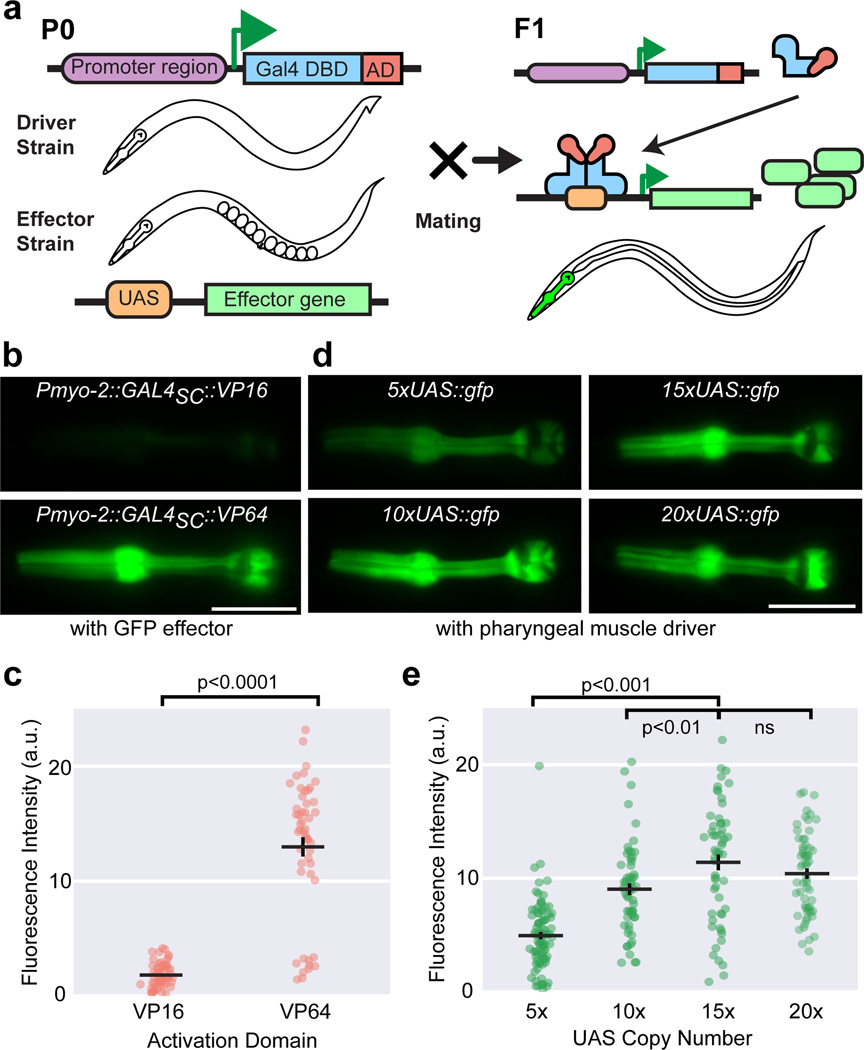
Bipartite gene expression systems offer fine spatial and temporal control of gene expression, and are crucial for dissecting gene function. The GAL4-UAS system, which has proven highly useful in Drosophila1, uses the Saccharomyces cerevisiae Gal4p protein, comprising a DNA-binding domain (DBD) and a transcriptional activation domain (AD). Gal4p homodimers bind a 17-nucleotide upstream activating sequence (UAS), resulting in transcription of a downstream gene2. Cell-specific promoters placed upstream of GAL4 constitute a ‘driver’ construct, while UAS sites placed upstream of the gene of interest constitute an ‘effector’ construct. Incorporation of these constructs into separate transgenics creates standardized driver and effector lines, which can be crossed together, generating progeny with desired expression patterns (Fig. 1a).
Figure 1. Optimization of activation domain and UAS copy number.
(a) Schematic of the GAL4-UAS bipartite gene expression system. DBD: DNA-binding domain; AD: activation domain. Representative images (b) and quantification (c) of GFP fluorescence in animals carrying transgene arrays of VP16 or VP64 activation domains fused to the DBD of S. cerevisiae Gal4p, driving a 15xUAS:gfp effector (syIs300) using the Pmyo-2 pharyngeal muscle promoter. Two independent lines for each driver, n = 58, 50. Bars are mean ± SEM, ****p < 0.0001, two-sample t-test with Welch’s correction. Representative images (d) and quantification (e) of GFP fluorescence in animals carrying transgene arrays of varying UAS copy number on GFP effector constructs, driven by the same integrated Pmyo-2::GAL4SC::VP64 driver (syIs301). 2–3 independent lines for each genotype, n = 89, 58, 56, 60, left to right. Bars are mean ± SEM. Scale bar is 20 µm. ** p < 0.01 and ***p<0.001. ns, not significant. One-way ANOVA with Tukey’s post-test. a.u., artificial units.
The bipartite nature of GAL4-UAS offers three major advantages over direct promoter-gene fusions. Large numbers of strains can be easily generated by crossing drivers and effector lines. Incorporating novel components into pre-existing driver and effector libraries requires minimal effort. Verified driver and effector strains become community reagents that foster experimental consistency and reproducibility. However, the GAL4-UAS system has not been successfully adapted for C. elegans. We optimized three components of the GAL4-UAS system to make it functional for C. elegans across 15–25°C, and demonstrated its robustness for tissue-specific gene expression, site-specific genetic rescue, and channelrhodopsin experiments.
Previous unpublished attempts using S. cerevisiae Gal4p DBD (residues 1–147, henceforth termed GAL4SC) with human herpes virus activation domain (VP16)3 as the driver components had poor performance in C. elegans. Since stronger activation domains improve performance of the GAL4 driver4, we tested VP645, which has four tandem copies of VP16. We fused GAL4SC to VP16 and VP64 and placed them both under the pharyngeal muscle promoter (Pmyo-2), designating them Pmyo-2::GAL4SC::VP16 and Pmyo-2::GAL4SC::VP64, respectively. Comparing their performance in a 15xUAS::gfp effector transgene strain (syIs300), the Pmyo-2::GAL4SC::VP64 driver yielded seven-fold more GFP fluorescence in pharyngeal muscles at room temperature (Fig. 1b–c). The fluorescence observed requires both driver and effector transgenes (Supplementary Fig. 1). We therefore adopted VP64 as our activation domain.
We optimized UAS copy number by comparing constructs with 5×, 10×, 15×, and 20× copies of UAS upstream of gfp at equal concentrations in a strain with a Pmyo-2::GAL4SC::VP64 driver (syIs301). We found successively increasing GFP fluorescence until 15xUAS copies (~2.3 fold versus 5xUAS, p<0.0001; ~1.3 fold vs. 10xUAS, p<0.01, One-way ANOVA, Tukey’s post-test), beyond which it appeared to saturate (~1.1 fold versus 20xUAS, p=0.5, One-way ANOVA, Tukey’s post-test; Fig. 1d–e). Thus, 15xUAS was adopted for effector constructs.
To determine the robustness of our GAL4-UAS system under standard C. elegans growth temperatures (15–25°C), we assayed our Pmyo-2::GAL4SC::VP64 driver and 15xUAS::gfp effector combination, and found its transcriptional efficacy depended heavily on temperature, performing well only at 25°C (Fig. 2b–c; ~67% drop at 20°C, ~80% drop at 15°C, p<0.0001, Two-way ANOVA with Tukey’s correction), consistent with findings in Drosophila6.
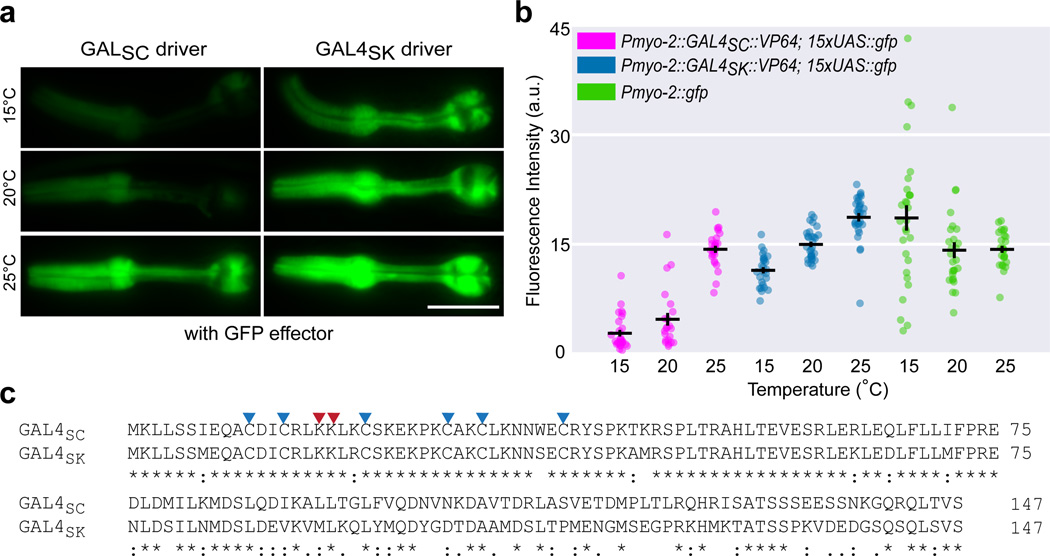
Figure 2. Designing a temperature-robust GAL4 driver via evolutionary analysis.
(a) Alignment of the DNA binding domains (residues 1–147) of Gal4p sequences from S. cerevisiae (GAL4SC) and S. kudriavzevii (GAL4SK). Two critical UAS-interacting lysine residues are marked with red arrowheads. Six cysteines forming the conserved Zn2Cys6 binuclear cluster are marked with blue arrowheads. Asterisk (*), colon (:), and period (.) indicate identical residues, strongly conserved residues and weakly conserved residues, respectively. Representative images (b) and quantification (c) of GFP fluorescence in animals carrying an integrated 15xUAS::gfp effector (syIs300), injected with identical concentrations of Pmyo-2::GAL4SC ::VP64 (syEx1434) or Pmyo-2::GAL4SK ::VP64 (syEx1436) at 15°C, 20°C, and 25°C. A direct fusion Pmyo-2::gfp was assayed as a benchmark. Scale bar is 20 µm. n = 28, 22, 25, 26, 30, 30, 29, 27, 26, left to right. Bars are mean ± SEM. All pairwise comparisons at each temperature are significant except Pmyo-2::GAL4SK::VP64 and Pmyo-2::gfp at 20°C, and Pmyo-2::GAL4SC::VP64 and Pmyo-2::gfp at 25°C. Two-way ANOVA, Tukey’s multiple comparison test. a.u., artificial units.
The temperature dependence of the canonical GAL4-UAS system and the optimal growth temperature of S. cerevisiae (30–34°C)6,7 suggested that activity of Gal4p from a specific yeast species may be adapted to its optimal growth temperature. Thus, a Gal4p from more cryophilic Saccharomyces yeast species might prove useful in designing a more robust driver at lower temperatures. We chose the Gal4p DBD from Saccharomyces kudriavzevii8 (Portuguese reference strain ZP591, residues 1–147, henceforth GAL4SK) because S. kudriavzevii has an optimal growth temperature (23–24°C)7 close to that of C. elegans and the key residues necessary for UAS binding9 are conserved between GAL4SC and GAL4SK (Fig. 2a).
We generated a new Pmyo-2 driver by replacing GAL4SC with GAL4SK, and compared its performance across 15–25°C using the same integrated 15xUAS::gfp effector strain (syIs300). The Pmyo-2::GAL4SK::VP64 driver lines exhibited more fluorescence than the Pmyo-2::GAL4SC::VP64 driver lines at room temperature; blinded researchers could accurately sort these two drivers using an epifluorescence dissecting microscope. We chose the brightest line from each for quantitation. At all temperatures, the Pmyo-2::GAL4SK::VP64 driver performed more robustly than the Pmyo-2::GAL4SC::VP64 driver, in some instances comparable to strains carrying a direct Pmyo-2::gfp fusion as a benchmark (Fig. 2c; Supplementary Fig. 2). Our GAL4SK driver had slightly higher fluorescence at 25°C (Fig. 2b–c), and exhibited only a ~20% drop at 20°C (versus ~67% for GAL4SC), and only a ~40% drop at 15°C (versus ~80% for GAL4SC). We adopted GAL4SK as our DBD of choice, in conjunction with VP64 and 15xUAS, to comprise our fully optimized GAL4-UAS system. We designate it cGAL to denote its original implementation in C. elegans and its engineered performance at cooler temperatures.
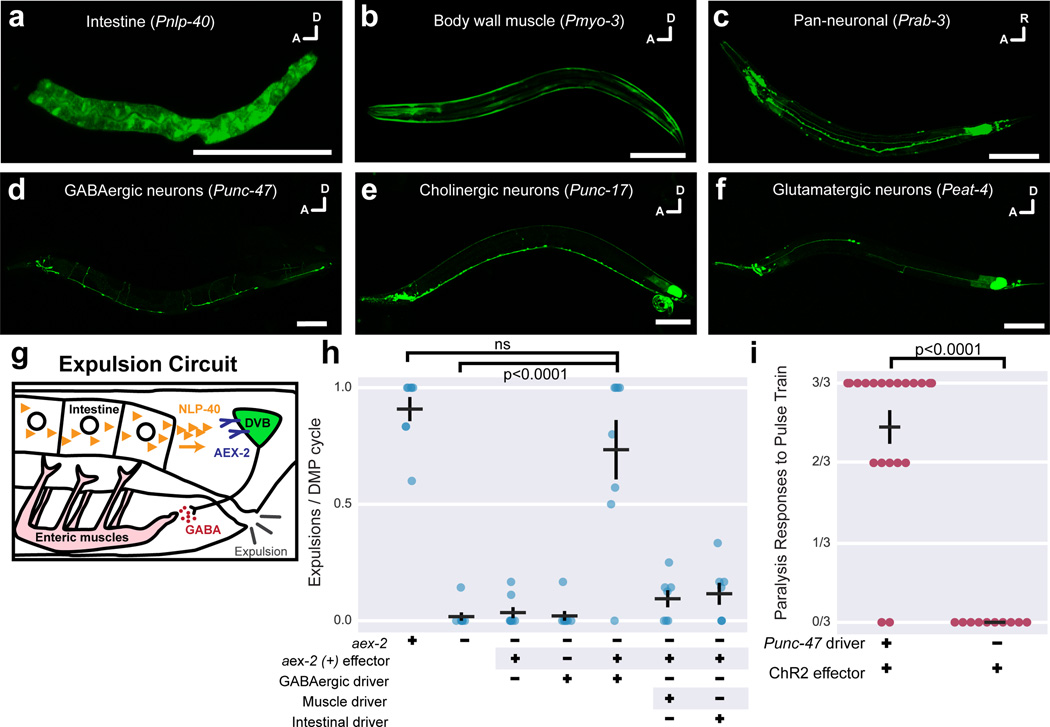
We tested whether cGAL would perform in other major tissues by generating new driver constructs containing various tissue-specific promoters (Pnlp-40, intestine; Pmyo-3, body wall muscles). When injected into a strain (syIs300) with an integrated 15xUAS::gfp effector, these new drivers produced robust and specific GFP fluorescence in expected tissues (Fig. 3a–b).
Figure 3. Functional studies with the cGAL system.
GFP effector expression in a variety of major tissues (a–f), each driven by the indicated cGAL driver. In (c–f), both driver and effector constructs (15xUAS::gfp::let-858 3’UTR) are built in the pPD117.01 vector. (g) The signaling pathway in the circuit controlling the final expulsion step in the defecation motor program. AEX-2 is necessary in two GABAergic neurons DVB and AVL (AVL not shown) to mediate expulsion. (h) Quantification of expulsion events per defecation cycle in animals with indicated genotypes. aex-2 rescue experiments with GABAergic driver, Punc-47; muscle driver, Pmyo-3; intestinal driver, Pnlp-40. Bars are mean ± SEM. n = 8, 8, 8, 8, 8, 7, 7 from left to right. **** p<0.0001. ns, not significant. One-way ANOVA with Tukey’s post-test. (i) Quantification of paralysis in animals having channelrhodopsin (ChR2) specifically expressed in GABAergic neurons (Punc-47 driver) and exposed to blue light in the presence of all-trans retinal. Each dot represents mean response to three blue light exposures from an individual animal. Bars are mean ± SEM. n = 20 and 10, from left to right. **** p < 0.0001, Mann-Whitney test.
However, our initial pan-neuronal (Prab-3) and GABAergic (Punc-47) drivers displayed poor, mosaic expression in the nervous system along with intense ectopic GFP fluorescence in the posterior gut (data not shown). We speculated that our vector backbone (a derivative of pPD49.26 containing the unc-54 3’UTR) might be responsible. We therefore switched to the vector pPD117.01 containing the let-858 3’UTR and a 5’ decoy sequence (comprising a decoy intron and ORF) upstream of the MCS (Methods; Supplementary Note 1–2); these modifications reduce ectopic expression and improve expression in a broad range of tissues (A. Fire, personal communication). We generated new Prab-3 and Punc-47 driver constructs in the pPD117.01 backbone and injected them into a strain (syIs390) with a new 15xUAS::gfp::let-858 3’UTR effector (also made using pPD117.01; Supplementary Note 3–4). These strains not only showed decreased ectopic fluorescence in the posterior gut, but also dramatically increased expression in the entire nervous system or GABAergic neurons, respectively (Fig. 3c–d; data not shown). Cholinergic (Punc-17) and glutamatergic (Peat-4) neuronal drivers gave similarly strong and specific results in the syIs390 effector strain (Fig. 3e–f). Thus, cGAL is robust across a variety of tissues and we recommend using the pPD117.01 vector for driver and effector construction.
We then applied cGAL to functional studies by examining the rhythmic defecation motor program (DMP), which contains three sequential muscle contractions10. The final enteric muscle contraction occurs when the intestine releases neuropeptide NLP-40, which binds its receptor AEX-2 on GABAergic neurons, triggering an expulsion event11,12 (Fig. 3g). We applied cGAL to confirm the site-of-action of aex-2 in GABAergic neurons for expulsion. We generated an effector line (syEx1444) with 15xUAS::aex-2(+) cDNA in the aex-2(sa3) background. This strain was then combined with a Pnlp-40 driver, Punc-47 driver, or Pmyo-3 driver to create aex-2(sa3) mutants expressing aex-2(+) cDNA specifically in the intestine, GABAergic neurons, or body wall muscles, respectively.
In aex-2(sa3) mutants, expulsion was nearly eliminated. Only the presence of both GABAergic driver and 15xUAS::aex-2(+) cDNA effector rescued the expulsion defect of aex-2(sa3) mutants; neither alone could (Fig. 3h). Expression of aex-2(+) cDNA in either body wall muscles or intestine did not rescue (Fig. 3h). These results recapitulate a study using conventional promoter-cDNA fusion transgenes12. Therefore, cGAL can be used for tissue-specific rescue experiments.
Next, we tested whether cGAL could be applied to gain-of-function experiments. Channelrhodopsin is a light-sensitive cation channel that depolarizes cells in the presence of all-trans retinal and blue light13. Activation of GABAergic neurons in C. elegans causes worms to adopt a flaccid, paralyzed state. We injected the GABAergic driver construct into a strain (syIs341) carrying an integrated 15xUAS::hChR2(H134R)::eyfp::let-858 3’UTR effector to express channelrhodopsin specifically in GABAergic neurons. In the presence of all-trans retinal, animals were subjected to a three pulse train of 475 nm blue light. Each excitation caused immediate, limp paralysis only in animals possessing both GABAergic driver and effector, but not in animals with either component alone (Fig. 3i, Supplementary Video 1–2 and data not shown). cGAL thus confers the ability to control exogenous transgene expression to dissect neural circuits in C. elegans.
Lastly, we built a basic cGAL driver and effector toolkit (Supplementary Table 1–2). For drivers, we constructed strains and constructs for major tissues, major neurotransmitter cell types, and some individual sensory neurons. For effectors, we have integrated strains for cell labeling (GFP, mKate2, GFP-H2B and mCherry-H2B), cell ablation (ICE), calcium indicator (GCaMP6s), neuronal activation (ChR2), neuronal inhibition (HisCl1) and synaptic inhibition (TeTx). All effectors are integrated and at least one line for each integrated effector was confirmed functional (Supplementary Fig. 3 and Supplementary Video 1–5).
One limitation of cGAL is the absence of Gal80p, a negative regulatory component used to repress Gal4p as a ‘NOT’ gate14. Gal80p recognizes the C-terminus of Gal4p15, absent in our drivers. Addition of the minimal C-terminal functional domain that interacts with S. kudriavzevii Gal80p might address this issue.
A similar bipartite expression system, Q16, has yet to be widely adopted in C. elegans, perhaps due to lack of sufficient strains. As with Drosophila1,17, the cGAL system could be combined with Q or other binary expression systems16,18–20 for control of multiple effectors.
cGAL can already greatly facilitate genetic site-of-action and genome-wide overexpression experiments, but we envision that a growing collection of drivers and effectors will allow analysis of genetic, developmental, and neural networks with increased rate and rigor.
METHODS
Maintenance of C. elegans Strains
Strains were maintained on NGM plates with E. coli OP50 as the food source at room temperature as originally described21, unless noted otherwise. Bristol strain N2 is the wild-type reference strain. The strains used in this study are detailed in the Supplementary Note 5–6 and all the integrated drivers and effectors are listed in Supplementary Table 1.
Molecular Biology
Plasmids were constructed by standard molecular cloning techniques with either restriction enzyme cleavage and DNA ligation or Gibson assembly using enzymes from New England Biolabs (Beverly, MA). The coding region of GAL4SK, residues 1–147 of Gal4p from Saccharomyces kudriavzevii (the Portuguese reference strain ZP591, a gift from C. T. Hittinger), was PCR amplified from genomic DNA using 5’ primer ggaGCTAGCatgaagctgttgtcttcaatgg and 3’ primer cggGAATTCcggcgatacactcaactgactttggc. The synthetic ScaI-17mer sequence (CGGAGTACTGTCCTCCG)22 was used for the UAS site and was placed upstream of the pes-10 basal promoter in all effector constructs. All constructs were built in either the pSM1 vector (a gift from C. Bargmann), a derivative of pPD49.26, which contains the unc-54 3’UTR, or the vector pPD117.01 (a gift from A. Fire), which contains the let-858 3’UTR and a 5’ decoy minigene upstream of the MCS for promoter insertion (Supplementary Note 1–4 for vector maps and sequences). We generated effector constructs for cell labeling (GFP, mKate2, GFP-H2B and mCherry-H2B), cell ablation (ICE)23, calcium reporter (GCaMP6s)24, neuronal activation (ChR2)25, neuronal inhibition (HisCl1)26 and synaptic transmission blocking (TeTx)27. Other details about plasmids and oligos used in the study are documented in Supplementary Table 2.
Transformation
Transgenic animals were generated using standard microinjection techniques28. Unless noted otherwise, 100 ng/µL total DNA injection samples were prepared, with either the pBluescript II KS+ plasmid or 1 kb DNA ladder, from New England Biolabs (Beverly, MA), as carrier. Extrachromosomal arrays were integrated into the genome via X-ray irradiation. Most of the integrants were outcrossed at least three times with the wild type strain N2. Full details about transgenic C. elegans strains in this study are listed in the Supplementary Note 5 and Supplementary Table 1.
Fluorescence Imaging
Approximately 25 animals per line were imaged and quantified for the optimization process of the cGAL system, using the myo-2 promoter (Pmyo-2). Briefly, L4 or young adults animals grown at corresponding temperatures (15°C, 20°C, 25°C or room temperature) were selected and imaged with Leica DMI600 inverted microscope equipped with a 40× oil objective and an Andor iXon Ultra 897 EMCCD camera, using Metamorph software (Molecular Devices). Images were captured with the same exposure time (20 ms) and the average fluorescence in the pharynx for each animal was analyzed. The representative fluorescent images in Fig. 3a–f showing the application of cGAL in different tissues were collected with a Zeiss LSM710 confocal microscope with a 20× objective.
Defecation Motor Program Assay
L4 animals raised at room temperature were picked one day before the assay. During the assay, which was performed at 20°C, each individual worm was picked to a new NGM plate seeded with OP50 and an 18×18 mm coverslip was placed over the animal for better optics. After a two-minute acclimation period, each animal was videotaped using a Zeiss Stemi SV11 coupled to a Unibrain Fire-i 501b camera for five minutes and the number of pBoc and expulsion events were scored. Each pBoc indicated the initiation of each defecation cycle. The ratio of expulsions over pBocs was used to quantify the expulsion phenotype for each animal (n = 7–8 for each genotype).
Optogenetics
One day before the assay, L4 animals raised at room temperature from each strain were picked individually onto NGM plates, seeded with 100 µL OP50 containing 500 µM all-trans retinal (Sigma). During the assay, which was performed at 20°C, animals were recorded using a Zeiss Stemi SV11 coupled to a Unibrain Fire-i 501b camera. Channelrhodopsin was activated using blue light generated from a Lumen Dynamics X-Cite series 120 lamp and a standard GFP filter set. Blue 475 nm light intensity was measured to be 0.2 mW/mm2. After an initial 10-second acclimation period, three light pulses, each 2 seconds in duration, were delivered to each worm at intervals of 20 seconds. The researcher doing the assay was blinded to the genotype of the animals.
Histamine Chloride Inhibition
Histamine plates were made by adding histamine dihydrochloride (Sigma) to the standard NGM recipe for a final concentration of 10 mM. Animals were picked onto plates with a small amount of food. After 30 minutes, worms were recorded using a Zeiss Stemi SV11 coupled to a Unibrain Fire-i 501b camera.
Statistics
For all descriptions of number of animals ‘n’ used, numbers correspond to each column from left to right. All bars are mean ± SEM.
Figure 1c: Two-sample Student’s t-test with Welch’s correction, as the Pmyo-2::GAL4SC::VP16 strain displayed much smaller variance. n = 58, 50.
Figure 1e: One-way ANOVA, with Tukey’s post-test for multiple comparisons. n = 89, 58, 56, 60.
Figure 2c: Two-way ANOVA, comparing genotype and temperature, with Tukey’s post-test for multiple comparisons. n = 28, 22, 25, 26, 30, 30, 29, 27, 26.
Figure 3h: One-way ANOVA, with Tukey’s post-test for multiple comparisons. n = 8, 8, 8, 8, 8, 7, 7.
Figure 3i: Mann-Whitney test. n = 20, 10.
Supplementary Material
Acknowledgments
We are grateful to H. Korswagen and A. Fire for sharing unpublished results and to C. T. Hittinger, D. Sieburth, E. M. Jorgensen, C. Bargmann and A. Fire for reagents. N.P. thanks C. Bargmann for her support. We thank M. Bao, Y. M. Kim, D. Leighton, J. DeModena and G. Medina for technical assistance, D. Angeles-Albores for advice on figure preparation and thoughtful discussion, and WormBase for technical support. We also thank M. Kato, H. Schwartz and other members of the Sternberg lab for editorial comments on the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Some imaging was performed at the Caltech Biological Imaging Facility, with the support of the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation. H.W. is supported by the Della Martin Fellowship. J.L. was supported by NIH grant T32GM007616. This work is supported by the Howard Hughes Medical Institute, with which P.W.S. is an investigator.
Footnotes
ACCESSION CODES
The coding sequence of Gal4p from Saccharomyces kudriavzevii is in GenBank (GU299177.1). The plasmids for cGAL driver (pHW393) and effector (pHW394) are available from Addgene (www.addgene.org, Plasmid #85583 and #85584, respectively).
DATA AVAILABILITY
The source data used for the plots in the figures are included in the paper and its supplementary information files. Other data are also available from the corresponding author upon request.
AUTHOR CONTRIBUTIONS
H.W., J.L., and P.W.S. conceived the project. H.W. and J.L. performed the experiments, analyzed the data and wrote the paper. S.G. helped with molecular cloning and strain handling. E.M.S. devised the idea of trying Gal4p from yeast species with lower growth temperatures. C.M.C. and N.P. contributed reagents.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 2.Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 4.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beerli RR, Segal DJ, Dreier B, Barbas CF. Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand AH, Manoukian AS, Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 7.Salvadó Z, et al. Temperature adaptation Markedly Determines evolution within the genus Saccharomyces. Appl. Environ. Microbiol. 2011;77:2292–2302. doi: 10.1128/AEM.01861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hittinger CT, et al. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–58. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 10.Thomas JH. Genetic analysis of defecation in Caenorhabditis elegans. Genetics. 1990;124:855–872. doi: 10.1093/genetics/124.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, et al. Neuropeptide secreted from a pacemaker activates neurons to control a rhythmic behavior. Curr. Biol. 2013;23:746–754. doi: 10.1016/j.cub.2013.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahoney TR, et al. Intestinal signaling to GABAergic neurons regulates a rhythmic behavior in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2008;105:16350–16355. doi: 10.1073/pnas.0803617105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 14.Lee T, Luo L. Mosaic Analysis with a Repressible Cell Marker for Studies of Gene Function in Neuronal Morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 15.Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 16.Wei X, Potter CJ, Luo L, Shen K. Controlling gene expression with the Q repressible binary expression system in Caenorhabditis elegans. Nat. Methods. 2012;9:391–395. doi: 10.1038/nmeth.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Valle Rodríguez A, Didiano D, Desplan C. Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods. 2012;9:47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoier EF, Mohler WA, Kim SK, Hajnal A. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 2000;14:874–886. [PMC free article] [PubMed] [Google Scholar]
- 19.Voutev R, Hubbard EJA. A ‘FLP-Out’ system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis MW, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 21.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster N, Jin JR, Green S, Hollis M, Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y, Brockie PJ, Mellem JE, Madsen DM, Maricq AV. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron. 1999;24:347–361. doi: 10.1016/s0896-6273(00)80849-1. [DOI] [PubMed] [Google Scholar]
- 24.Chen T-W, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 26.Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2770–2775. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 28.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.