Abstract
Vibrio parahaemolyticus is a Gram-negative pathogen that causes food-borne gastroenteritis. A major virulence determinant of the organism is a type III secretion system (T3SS2) encoded on a pathogenicity island, Vp-PAI. Vp-PAI gene expression is regulated by two transcriptional regulators, VtrA and VtrB, whose N-terminal regions share homology with an OmpR-family DNA-binding domain. VtrA activates the gene expression of VtrB, which in turn activates Vp-PAI gene expression; however, the mechanism of this transcriptional activation by VtrA is not well understood. In this study, we determined that VtrA is a membrane protein with a transmembrane (TM) domain, which was required for its transcriptional regulatory activity. Although the N-terminal region of VtrA alone is insufficient for its transcriptional regulatory activity, forced oligomerization using the leucine-zipper dimerization domain of yeast GCN4 conferred transcriptional regulatory activity and a greater affinity for the promoter region of vtrB. A ToxR-based assay demonstrated that VtrA oligomerizes in vivo. We also showed that bile, a host-derived activator of VtrA, induces the oligomerization of VtrA, which requires the C-terminal domain. The promoter region of vtrB contained repetitive T-rich DNA elements, which are important for vtrB transcriptional activation and are conserved among T3SS2-possessing Vibrio species. These findings propose that VtrA is active as oligomers, which may facilitate its N-terminus binding the target DNA, thus enhancing its transcriptional regulatory activity.
Introduction
Vibrio parahaemolyticus is a Gram-negative halophilic bacterium that inhabits marine and estuarine environments [1]. The bacterium is a major food-borne pathogen that causes acute, seafood-associated gastroenteritis in humans worldwide [2–4]. Only a small subset of V. parahaemolyticus is associated with human infection. This association is closely related to the presence of a pathogenicity island, Vp-PAI, on the smaller of the two V. parahaemolyticus chromosomes [5–7]. The Vp-PAI region encodes one of the two type III secretion systems (T3SSs) of the organism, T3SS2 [5]. T3SSs are protein export systems that enable bacteria to secrete and translocate proteins, known as effectors, into the cytoplasm of host cells [8]. In addition to the epidemiological association, the intestinal pathology of V. parahaemolyticus is dependent on T3SS2 in several animal models [9–11]. Therefore, V. parahaemolyticus T3SS2 is thought to play an essential role in the enterotoxicity of the organism. The T3SS2 gene cluster is classified into two distinct phylogroups, T3SS2α and T3SS2β, which are distributed not only in V. parahaemolyticus but also in non-toxigenic V. cholerae and V. mimicus [12–14]. The T3SS of non-toxigenic V. cholerae is suggested to also be involved in the pathogenicity of the organism [12, 15].
The expression of Vp-PAI genes, including T3SS2 genes, is dependent on two transcriptional regulators, VtrA and VtrB, which are encoded in the Vp-PAI region [11, 16]. The N-terminal regions of VtrA and VtrB share homology with winged-helix-turn-helix (wHTH) DNA-binding domains of OmpR-family proteins including Escherichia coli OmpR and V. cholerae ToxR [16]. The N-terminal region of VtrA is capable of binding to the upstream region of vtrB, where VtrA positively regulates vtrB transcription. Therefore, it is thought that VtrA and VtrB constitute a regulatory cascade in which VtrA acts as an upstream regulator. A transcriptome analysis of V. parahaemolyticus has shown that the VtrA-VtrB regulatory cascade controls more than 60 genes, most of which are exclusively located in the Vp-PAI region. Vp-PAI gene expression is vigorously induced by bile in a vtrA-dependent manner, which contributes to the intestinal pathogenesis of V. parahaemolyticus [17]. Bile elevates the production of VtrB, while the protein level of VtrA is not affected. Therefore, it is likely that bile activates transcriptional regulatory activity of VtrA. A recent study reported that VtrC, a co-transcribed protein with VtrA, is required for the activation of T3SS2 by bile salts [18]. VtrC has been also shown to form a complex with the C-terminal domain of VtrA, which binds to bile salts. Thus, it has been proposed that the VtrA/VtrC complex senses bile salts to activate the DNA-binding domain of VtrA, but its mechanism is yet to be determined.
VtrA consists of 253 amino acids with a conserved DNA-binding region at the N-terminal side, described above. Despite a critical role of VtrA in the regulation of Vp-PAI genes and the pathogenesis of V. parahaemolyticus, its mechanism of transcriptional activation has been not well understood. Although VtrA was predicted to possess a putative transmembrane (TM) domain [16], it has not yet been determined whether VtrA is localized in the membrane. In this study, we experimentally determined that VtrA is a membrane protein with a TM domain. We show that VtrA forms oligomers via its TM and C-terminal domains, facilitating its N-terminal DNA-binding activity, and thereby enhancing its transcriptional regulatory activity. Furthermore, we found a T-rich DNA element in the vtrB promoter region that is conserved in other T3SS2-possessing Vibrio species and is essential for vtrB transcriptional activation by VtrA.
Materials and methods
Bacterial strains and plasmids
V. parahaemolyticus strain RIMD2210633 (a T3SS2α-positive clinical isolate) [5] was the wild-type strain in this study. V. parahaemolyticus vtrA deletion strain derived from the wild-type strain carries a stop codon at residue 30 and a deletion from nucleotides 88 to 471 in the vtrA gene [16]. To generate a Δvp0820 deletion mutant, a four-primer PCR technique with pYAK1, an R6Kori suicide vector containing sacB, was used as described previously [19]. V. parahaemolyticus strain TH3996 [13], and V. cholerae strains RIMD2214343 and RIMD2214428 [14] were obtained from the Laboratory for Culture Collection, Research Institute for Microbial Diseases, Osaka University. E. coli DH5α and SM10λpir strains were used for general manipulation of plasmids and their mobilization into V. parahaemolyticus, respectively. E. coli MC4100 was used for β-galactosidase assays. The strains and plasmids used in this study are listed in S1 and S2 Tables, respectively.
Bacteria fractionation
V. parahaemolyticus harboring pBAD-vtrA, pBAD-vtrA-FLAG or their derivatives were grown in Luria-Bertani (LB) media containing 0.5% NaCl in the presence of 0.01% (w/v) L-arabinose to an OD600 of 2.0. Then, the bacterial cells were harvested by centrifugation at 3,000 × g for 15 min. The pellets were re-suspended with 200 μl of the periplasting buffer (200 mM Tris-HCl at pH 7.5, 20% sucrose, 1 mM EDTA, and 20 mg/ml lysozyme) and incubated for 5 min at room temperature. The cells were osmotically shocked by the addition of 200 μl of ice-cold distilled water and incubated for 5 min on ice. After centrifugation at 12,000 × g for 2 min, the spheroplasts were treated with 500 U/ml benzonase for 5 min at room temperature, and sonicated to induce lysis. The sample was centrifuged twice at 12,000 × g for 2 min to remove unlysed cells. The supernatants were centrifuged at 100,000 × g for 1 h, and the resulting supernatants were collected as the cytoplasmic fraction. The membrane fractions were dissolved in SDS loading buffer. Each fraction was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis using anti-HA monoclonal (Medical & Biological Laboratories, Nagoya, Japan), anti-FLAG monoclonal (Sigma-Aldrich, St. Louis, MO, USA), anti-DnaK and anti-OmpA antibodies.
β-galactosidase assays
V. parahaemolyticus strains harboring pHRP309-derivative reporter plasmids were grown in LB broth with 0.5% NaCl at 37°C for 6 h. For the bile stimulation, V. parahaemolyticus cells were grown at 37°C for 3 h, and then incubated with 0.04% bile for another 3 h. E. coli strains harboring pHRP309-derivative reporter plasmids were grown in LB broth with 1% NaCl at 37°C for 3 h. In V. parahaemolyticus and E. coli strains carrying pBAD-vtrA or its derivatives, the expression of both HA-VtrA and its truncated forms was induced by addition of L-arabinose to a final concentration of 0.001% (w/v). The β-galactosidase activity of cell lysates was measured using Miller's method with the substrate o-nitrophenyl-β-D-galactopyranoside [20].
A ToxR-based transcriptional reporter assay
A ToxR-based transcriptional reporter assay was performed on V. parahaemolyticus ΔvtrA Δvp0820 carrying pHRP309-PompU and either pBAD-toxR, pBAD-toxRN or pBAD-toxRN-vtrATM-C. The resulting strains were grown in LB media with 0.5% NaCl in the presence of 0.001% (w/v) L-arabinose at 37°C for 6 h. For the bile stimulation, V. parahaemolyticus cells were grown at 37°C for 3 h, and then incubated with 0.04% bile for another 3 h. The β-galactosidase activity of cell lysates was measured as described above.
Protein expression, purification and cross-linking
The 6×Histidine-tagged N-terminal region of VtrA (His-VtrAN: aa 1–133) or its leucine-zipper dimerizing domain (ZIP)-fused proteins were expressed and purified from E. coli BL21(DE3) harboring pET28a expression plasmids containing the genes for VtrAN or its ZIP-fusions as described previously [21]. For cross-linking, 1 μg of each purified protein was incubated with 7.5 mg/ml dimethyl adipimidate (DMA) in phosphate-buffered saline containing 50 mM triethanolamine for 1 h at room temperature. Then, the protein was mixed with an equal volume of 2× SDS-loading buffer and boiled for 5 min. These samples were subjected to SDS-PAGE, followed by staining with Coomassie brilliant blue (CBB).
Electrophoretic mobility shift assays
The DNA fragments corresponding to the 284-bp upstream region of vtrB were amplified by PCR using 5’-biotinylated primers. The PCR products were separated by agarose gel electrophoresis and purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA). This product was used for the biotinylated DNA probe. Increased concentrations of His-VtrAN or ZIP-fused VtrAN proteins were incubated with 4 nM biotinylated DNA probe in the reaction buffer (10 mM Tris-HCl at pH 7.5, 100 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, 0.2% Nonidet P-40, 10% glycerol and 100 ng/ml bovine serum albumin) for 30 min at room temperature. Reaction mixtures were then separated by 6% non-denaturing polyacrylamide gel electrophoresis in TGE buffer (25 mM Tris, 190 mM glycine and 1 mM EDTA) containing 100 mM KCl at 4°C. The DNA probe was electroblotted to a positively charged nylon membrane, UV cross-linked, probed with horseradish peroxidase (HRP)-conjugated streptavidin and developed using the ECL Western blotting kit (GE Healthcare, Buckinghamshire, UK) according to the manufacturer's instructions.
Cytotoxicity assays
V. parahaemolyticus strains were grown in LB medium supplemented with 0.5% NaCl in the presence or absence of 0.04% bile for 3 h and then used for infections. HeLa cells were seeded at 1 × 104 cells per well in 96-well plates and grown for 48 h. The cells were then infected with V. parahaemolyticus at a multiplicity of infection of 10 in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) without serum for 4.5 h. The cytotoxicity assay was performed using the CytoTox96 Non-Radioactive Cytotoxicity Assay Kit (Promega, Madison, WI) as previously described [19, 21].
Mapping of transcriptional start site by 5′ random amplification of cDNA ends (5′-RACE)
Total RNA from V. parahaemolyticus was isolated using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The 5’ end of the vtrB transcript was determined using the SMARTer RACE 5’/3’ Kit (Takara Bio, Shiga, Japan) according to the manufacturer's instructions. Nucleotide sequencing was performed with the BigDye v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and the ABI PRISM 3100 genetic analyzer (Applied Biosystems). The putative −35 and −10 promoter elements were predicted using the software GENETYX ver. 11 (GENETYXS, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed using the two-tailed Student’s t-test or a one-way ANOVA followed by Dunnett’s multiple comparison test. A p-value < 0.05 was considered statistically significant.
Results
VtrA is a transmembrane protein localized in the membrane via its TM domain
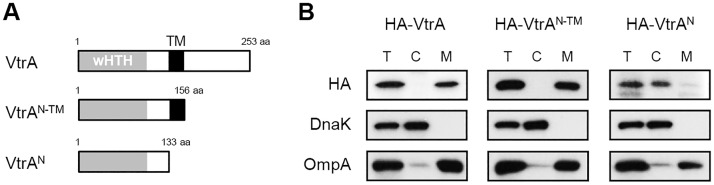
We first predicted the subcellular localization of VtrA in silico using a topological analysis program TMHMM v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). VtrA was predicted as a bitopic membrane protein with its N-terminus (aa 1–133) in the cytosol, a single TM region located between amino acid residues (aa) 134 and 156, and its C-terminus (aa 157–253) in the periplasm (Fig 1A). To verify whether VtrA was located in the membrane, a derivative of the arabinose-inducible plasmid, pBAD18-Cm, encoding an N-terminal, hemagglutinin (HA)-tagged VtrA (pBAD-vtrA) was introduced into a V. parahaemolyticus vtrA deletion strain (ΔvtrA). The resulting strain ΔvtrA/pBAD-vtrA was fractionated into cytosolic and membrane fractions. HA-tagged VtrA was detected in the membrane fractions, indicating that VtrA is localized in the membrane of V. parahaemolyticus (Fig 1B). Because VtrA possesses a putative TM region, we next investigated whether this region is involved in the membrane targeting of VtrA. We constructed two pBAD-vtrA derivatives encoding truncated forms of the HA-tagged protein. The first, pBAD-vtrAN, encodes only the N-terminal cytosolic domain of VtrA (aa 1–133; VtrAN). The second, pBAD-vtrAN-TM, encodes the N-terminal cytosolic domain and putative TM region but lacks the C-terminal putative periplasmic region of VtrA (aa 1–156; VtrAN-TM). Subcellular fractionation showed that VtrAN-TM was still localized in the membrane fractions, whereas VtrAN was enriched in the cytosolic fractions (Fig 1B). Similar results were observed in V. parahaemolyticus cells expressing C-terminal 3×FLAG-tagged VtrA and its truncated forms (S1 Fig). Taken together, these results indicate that VtrA is a membrane protein whose localization to the membrane depends on the TM domain.
Fig 1. VtrA is a membrane-bound protein with a TM domain.
(A) Schematic representations of full-length and truncated VtrA. wHTH, winged-helix-turn-helix domain; TM, putative transmembrane domain. (B) Subcellular localization of VtrA in V. parahaemolyticus. V. parahaemolyticus expressing HA-VtrA and its truncated derivatives were fractionated into cytosol (C) and membrane (M) fractions. Each of fractions and total cell lysates (T) were subjected to immunoblot analysis for HA to detect VtrA and its truncated derivatives. DnaK and OmpA were detected as controls for the cytosol and membrane, respectively.
N-terminal DNA-binding domain of VtrA alone is insufficient for transcriptional regulatory activity
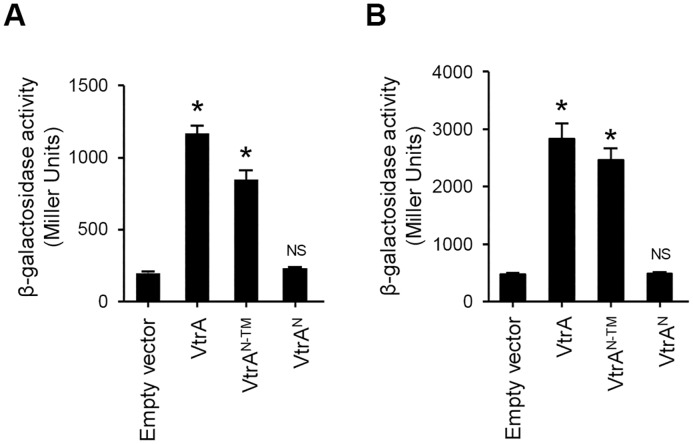
In a previous report, it has been shown that the N-terminal cytosolic region of VtrA can bind upstream regions of vtrB, activating its transcription [16]. Using a lacZ reporter gene assay, we next investigated whether the N-terminal region of VtrA was sufficient to activate vtrB transcription. A pHRP309-derived lacZ transcriptional fusion reporter, pHRP309-PvtrB [16], containing 284-bp upstream promoter region of vtrB (PvtrB), was introduced into V. parahaemolyticus strains ΔvtrA/pBAD-vtrA, ΔvtrA/pBAD-vtrAN and ΔvtrA/pBAD-vtrAN-TM, and vtrB transcription was evaluated by measuring the β-galactosidase activity. PvtrB-lacZ transcription was induced in ΔvtrA/pBAD-vtrAN-TM as well as in ΔvtrA/pBAD-vtrA, but not in ΔvtrA/pBAD-vtrAN (Fig 2A). Additionally, to avoid the potential effects of other factors in V. parahaemolyticus, we evaluated the VtrA-mediated transcriptional activation by heterologous expression in E. coli. VtrA and its truncated forms were expressed in E. coli MC4100 cells harboring pHRP309-PvtrB and were evaluated for their ability to activate vtrB transcription. PvtrB-lacZ expression was significantly induced in an E. coli strain expressing full-length VtrA, compared with a strain bearing an empty vector (Fig 2B). The heterologous expression of VtrAN-TM in E. coli likewise induced the lacZ expression, whereas E. coli expressing VtrAN showed no increase in the lacZ expression. Together, these data demonstrated that VtrAN alone is insufficient for the transcriptional activation of vtrB, suggesting that the other domains of VtrA are also involved.
Fig 2. Transcription of vtrB is activated by VtrA and VtrAN-TM but not VtrAN.
β-galactosidase activity from the PvtrB-lacZ transcriptional reporter of V. parahaemolyticus ΔvtrA (A) and E. coli MC4100 (B) upon expression of VtrA and its truncated derivatives. The values represent the mean ± standard deviations (SD) for a minimum of three independent experiments. *, p < 0.01; NS, not significant, compared with empty vector, determined by one-way ANOVA followed by Dunnett’s multiple comparison test.
Oligomerization of the N-terminal cytosolic DNA-binding domain is important for the transcriptional activation by VtrA
Many bacterial transcription factors function as oligomers in their activated states [22]. These include various membrane-bound transcriptional regulators whose cytosolic DNA-binding domains oligomerize through interactions between their TM and/or periplasmic domains [23–27]. Given that the transcriptional regulatory activity of VtrA is abolished by the deletion of its TM and C-terminal domains, we hypothesized that VtrA activation might also be mediated by oligomerization through these domains. To explore this hypothesis, we constructed an additional pBAD-vtrA derivative encoding a VtrA variant in which the TM domain was replaced with a TM domain composed of polyleucine (pBAD-vtrA-PL; S2A Fig). The TM domain composed of polyleucine is known to have a less tendency to oligomerize in E. coli [27, 28]. VtrA-PL was expressed in E. coli MC4100 cells harboring pHRP309-PvtrB and evaluated for its ability to activate vtrB transcription. The expression of VtrA-PL did not significantly induce PvtrB-lacZ expression, in contrast to the expression of VtrA (S2B Fig). Thus, this result suggested that the TM domain plays major roles in transcriptional activation of VtrA, through its oligomerization.
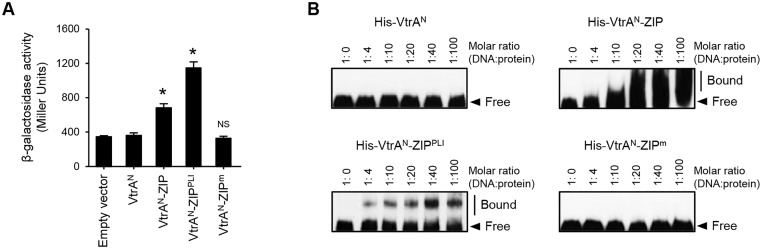
If the transcriptional regulatory activity of VtrA is activated via oligomerization, the forced oligomerization of VtrAN should exhibit transcriptional activation of vtrB. To examine this hypothesis, we used the ZIP domain of Saccharomyces cerevisiae transcriptional activator GCN4 [29] to force oligomerization of VtrAN. VtrAN was fused to a wild-type ZIP, a ZIP tetrameric mutant domain (ZIPPLI), and a ZIP monomeric mutant (ZIPm), yielding dimeric, tetrameric or monomeric VtrAN. We confirmed the oligomeric properties of VtrAN-ZIP fusion proteins. The 6×Histidine (His)-tagged VtrAN, VtrAN-ZIP, VtrAN-ZIPPLI and VtrAN-ZIPm proteins were purified, chemically crosslinked, and visualized by CBB staining of SDS-PAGE gels (S3 Fig). Without crosslinking, all ZIP-fused VtrAN migrated at ~26 kDa, consistent with the molecular weight of the monomer. Cross-linked VtrAN-ZIP and VtrAN-ZIPPLI migrated at ~50 kDa and ~110 kDa, respectively. By contrast, cross-linked VtrAN-ZIPm migrated at ~26 kDa, indicating its monomeric structure. Thus, these results confirmed that these fusion proteins form the desired oligomeric states. We further found that the expression of the VtrAN-ZIP fusion protein (VtrAN-ZIP) in E. coli MC4100 cells harboring pHRP309-PvtrB induced significant lacZ expression, in contrast to the expression of VtrAN alone (Fig 3A). Interestingly, the expression of VtrAN fused to tetrameric ZIPPLI (VtrAN-ZIPPLI) showed increased expression of lacZ, compared to that of dimeric VtrAN-ZIP. By contrast, VtrAN fused to monomeric ZIPm (VtrAN-ZIPm) failed to induce the lacZ expression. Taken together, the forced oligomerization of VtrAN conferred the expected transcriptional activation of vtrB, suggesting that the oligomerization of the cytosolic DNA-binding domain is important for the transcriptional regulatory activity of VtrA.
Fig 3. Oligomerization of the N-terminal DNA-binding region of VtrA is important for the transcriptional regulatory activity of VtrA.
(A) β-galactosidase activity from PvtrB-lacZ transcriptional reporter of E. coli MC4100 upon expression of VtrAN and its ZIP-fusions. The values represent the mean ± SD for a minimum of three independent experiments. *, p < 0.01; NS, not significant, compared with VtrAN by one-way ANOVA followed by Dunnett’s multiple comparison test. (B) Electrophoretic mobility shift assays using VtrAN and its ZIP fusions. Increased concentrations (0, 16, 40, 80, 160 and 400 nM) of each protein were incubated with 4 nM biotinylated DNA probe corresponding to 284-bp upstream region of vtrB, and then subjected to native PAGE. The biotinylated probe was transferred to a nylon membrane, UV cross-linked, and detected using HRP-conjugated streptavidin.
Forced oligomerization of VtrA enhances its DNA-binding activity
To investigate the mechanism underlying the oligomerization-mediated activation of VtrA, we examined the DNA-binding activity of VtrAN-ZIP fusion proteins using an electrophoretic mobility shift assay (EMSA). DNA fragments corresponding to the 284-bp upstream region of vtrB were incubated with His-VtrAN and its ZIP-fusions at various DNA/protein molar ratios (from 1: 0 to 1: 100), and then subjected to native gel electrophoresis. Neither His-VtrAN nor His-VtrAN-ZIPm (monomeric) induced a shift of the DNA probe at a molar ratio of 1:100 (Fig 3B). Note that the binding of His-VtrAN to the probe was observed at higher concentrations of His-VtrAN (S4 Fig). By contrast, His-VtrAN-ZIP induced the mobility shift even at a molar ratio of 1:10. Furthermore, His-VtrAN-ZIPPLI bound to the probe at lower concentration than His-VtrAN-ZIP. Therefore, the forced oligomerization of VtrAN conferred a greater affinity for the promoter region of vtrB. These results suggest that the oligomerization of VtrA facilitates its DNA-binding activity, thus enhancing the transcriptional regulatory activity of VtrA.
VtrA forms oligomers in vivo
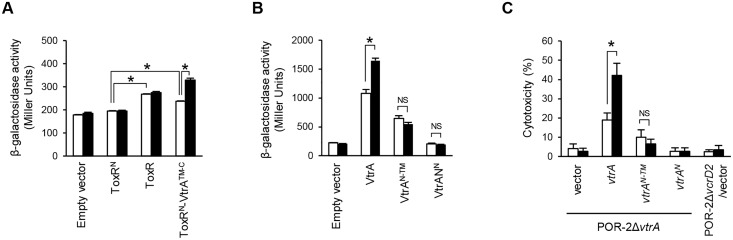
To examine whether VtrA forms oligomers in V. parahaemolyticus, we employed a ToxR-based transcriptional reporter assay [30]. V. cholerae ToxR is a membrane-spanning transcriptional regulator that activates the transcription of its target genes including ctx and ompU, in a manner dependent on the dimerization of its cytoplasmic DNA-binding domain [31, 32]. We generated a chimeric construct in which the TM and C-terminal putative periplasmic domains of VtrA (aa 134–253; VtrATM-C) were fused to the DNA-binding domain of V. cholerae ToxR (ToxRN), and evaluated whether the ToxRN-VtrATM-C fusion could activate the ompU promoter (PompU) in V. parahaemolyticus. V. parahaemolyticus has a ToxR homologue, VP0820, which bears homology with V. cholerae ToxR [33]. To exclude the potential effects of VP0820 on our assay, a deletion strain in the ΔvtrA background was constructed (ΔvtrA Δvp0820) and used for the PompU-lacZ reporter assay. Using a V. parahaemolyticus ΔvtrA Δvp0820 strain carrying a PompU-lacZ reporter plasmid, we confirmed that the expression of full-length ToxR increased PompU-lacZ transcription, in contrast to that of ToxRN (Fig 4A). The expression of the ToxRN-VtrATM-C fusion showed increased PompU-lacZ transcription, thus indicating the oligomerization of VtrATM-C in vivo.
Fig 4. Oligomerization of VtrA in V. parahaemolyticus during its active states.
(A) β-galactosidase activity from the PompU-lacZ transcriptional reporter of V. parahaemolyticus ΔvtrA Δvp0820 upon expression of ToxR, N-terminal domain of ToxR (ToxRN) and ToxRN-fused VtrATM-C, in the absence (white bar) or presence of 0.04% bile (black bar). *, p < 0.01; NS, by Student’s t-test. (B) β-galactosidase activity from the PvtrB-lacZ transcriptional reporter of V. parahaemolyticus ΔvtrA upon expression of VtrA and its truncated derivatives, in the absence (white bar) or presence of 0.04% bile (black bar). *, p < 0.01; NS, not significant, by Student’s t-test. (C) HeLa cells were infected with the indicated stains of V. parahaemolyticus unstimulated (white bar) or stimulated with bile (black bar) for 4.5 h. The cytotoxicity was evaluated using the lactate dehydrogenase release assay. *, p < 0.01; NS, not significant, by Student’s t-test. The values represent the mean ± SD for a minimum of three independent experiments (A—C).
Bile induces oligomerization of VtrA, which is dependent on the C-terminal domain
Bile is a host-derived factor that induces Vp-PAI gene expression in a vtrA-dependent manner [17]. The inducing activity of bile results in the elevation of vtrB transcription without affecting expression levels of VtrA, suggesting that the transcriptional regulatory activity of VtrA is activated by bile. It was reported that the C-terminal region of VtrA forms a complex with VtrC, which senses bile acids to activate the DNA-binding domain of VtrA through an undefined mechanism [18]. We observed that the bile treatment elevated PvtrB-lacZ transcription in V. parahaemolyticus ΔvtrA/pBAD-vtrA, but not in V. parahaemolyticus ΔvtrA/pBAD-vtrAN-TM (Fig 4B), supporting that the C-terminal domain of VtrA is required for the bile-mediated activation of VtrA. Because we observed that transcriptional regulatory activity of VtrAN is activated via oligomerization (Fig 3), we hypothesized that bile induces oligomerization of VtrA, resulting in the induction of VtrA transcriptional regulatory activity. To test this hypothesis, we examined the effect of bile on the transcriptional activation by ToxRN-VtrATM-C in V. parahaemolyticus ΔvtrA Δvp0820 carrying a PompU-lacZ reporter plasmid. As expected, the treatment of bile enhanced the PompU-lacZ transcription by a ToxRN-VtrATM-C fusion (Fig 4A). These results suggest that bile activates VtrA by enhancing oligomerization of VtrA in V. parahaemolyticus.
One virulence characteristic of V. parahaemolyticus is its T3SS2-dependent cytotoxicity in eukaryotic cultured cells. To address whether the oligomeric property of VtrA is correlated with the T3SS2-dependent cytotoxicity during infection, we evaluated the cytotoxicity of V. parahaemolyticus expressing truncated forms of VtrA. In HeLa cells, a V. parahaemolyticus ΔvtrA deletion strain derived from a T3SS1-defective strain POR-2 (POR-2ΔvtrA) did not exhibit cytotoxicity, which is similar to previous results observed in Caco-2 cells (Fig 4C) [19]. Complementation with plasmid-borne vtrA in a ΔvtrA strain (POR-2ΔvtrA/pBAD-vtrA) reconstituted the cytotoxicity, whereas complementation with vtrAN (POR-2ΔvtrA/pBAD-vtrAN) did not. Bile is known to stimulate the T3SS2-dependent cytotoxicity [17]. POR-2ΔvtrA/pBAD-vtrA exhibited elevated cytotoxicity when stimulated with bile, which reflects the bile-induced T3SS2-related gene expression. In contrast, complementation with vtrAN-TM in the ΔvtrA strain (POR-2ΔvtrA/ pBAD-vtrAN-TM) did not exhibit significant cytotoxicity regardless of the stimulation with bile. Collectively, these observations indicate that the C-terminal domain of VtrA plays an essential role in T3SS2 gene expression in response to bile.
T-rich DNA elements in the vtrB promoter region are important for vtrB transcriptional activation by VtrA and conserved among T3SS2-possessing Vibrio species
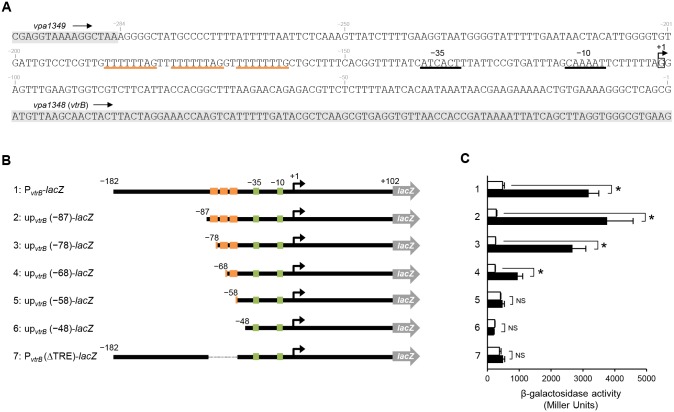
To analyze how VtrA recognizes the promoter region of vtrB, we mapped the transcriptional start site of vtrB using 5′-RACE and identified that the +1 nucleotide of the vtrB mRNA is 102 bp upstream of the ATG start codon (Fig 5A). The presence of putative −35 and −10 promoter elements was predicted in the upstream region. In addition, we found an 8-bp T-rich DNA element (5’-TTTTTTWG-3’) near the −35 promoter element, which was located 57 bp upstream of the transcription start site and repeated three times. To investigate whether the repetitive elements are involved in transcriptional activation of vtrB by VtrA, we prepared a series of truncated versions of the upstream region of vtrB (Fig 5B). These fragments were inserted into the pHRP309 plasmid, yielding upvtrB-lacZ transcriptional fusion reporter plasmids. Using these lacZ fusion reporters, we evaluated the transcriptional activation by VtrA in E. coli (Fig 5C). VtrA induced lacZ transcription from upvtrB-lacZ fusion reporters containing more than one T-rich element. The level of transcription is dependent on the number of T-rich elements, and the maximum lacZ transcription was observed in E. coli bearing the upvtrB (−87)-lacZ fusion, which contains all of three T-rich elements. By contrast, no significant lacZ transcription was observed from the upvtrB (−58)-lacZ and upvtrB (−48)-lacZ reporters, both of which contain no T-rich element. Similarly, in E. coli bearing the PvtrB (ΔTRE)-lacZ fusion, in which T-rich elements were deleted, VtrA did not significantly induce lacZ transcription. Thus, the T-rich DNA element is essential for the transcriptional activation of vtrB by VtrA.
Fig 5. T-rich elements in the vtrB promoter region are required for transcriptional activation by VtrA.
(A) The nucleotide sequence of the vtrB promoter region. The transcriptional start site of the vtrB, determined by 5’-RACE, is indicated as +1. Putative −35 and −10 elements are underlined. Repetitive T-rich elements are underlined in orange. Gray shading indicates the regions of coding sequence of vpa1349 and vpa1348. (B) Schematics representing lacZ fusion reporters of the upstream promoter region of vtrB (PvtrB) and its truncated forms (upvtrB). Putative −35 and −10 promoter elements are shown in green. T-rich elements are indicated in orange. (C) β-galactosidase activity from PvtrB-lacZ and upvtrB-lacZ transcriptional reporters of V. parahaemolyticus ΔvtrA carrying an empty vector (white bar) or a VtrA expression plasmid (black bar). The values represent the mean ± SD for a minimum of three independent experiments. *, p < 0.01; NS, not significant, by Student’s t-test.
Vibrio T3SS2 gene clusters are classified into two phylogroups, T3SS2α and T3SS2β, and are distributed among Vibrio species, including V. parahaemolyticus and non-toxigenic V. cholerae [13, 14]. Comparison of the amino acid sequences of VtrA from V. parahaemolyticus RIMD2210633 (T3SS2α-type), V. parahaemolyticus TH3996 (T3SS2β-type), V. cholerae RIMD2214343 (T3SS2α-type) and V. cholerae RIMD2214428 (T3SS2β-type) showed that the VtrA of V. parahaemolyticus RIMD2210633 is more closely related to the VtrA of V. cholerae RIMD2214343 than to the VtrAs of V. parahaemolyticus TH3996 and V. cholerae RIMD2214428. We aligned the nucleotide sequences of the upstream region of vtrB from these four strains and found that all four strains contain the repetitive T-rich elements (S5A Fig). To examine whether VtrA from V. parahaemolyticus RIMD2210633 recognizes the promoter region of vtrB from other T3SS2-possessing vibrios, we evaluated the transcriptional regulatory activity of VtrA toward vtrB of V. parahaemolyticus TH3996, V. cholerae RIMD2214343 and V. cholerae RIMD2214428. The upstream promoter regions of vtrB (PvtrB) from these strains were inserted into pHRP309 plasmid, and the resulting PvtrB-lacZ reporter plasmids were introduced into E. coli MC4100 harboring pBAD-vtrA. As shown in S5B Fig, VtrA from V. parahaemolyticus RIMD2210633 induced lacZ transcription from all PvtrB-lacZ fusion reporters. Thus, these results suggested that VtrA recognizes the promoter structure of vtrB which is conserved among T3SS2-possessing Vibrio species.
Discussion
In the present study, we experimentally determined that VtrA is a membrane protein with a central TM domain. VtrA also contains an N-terminal wHTH domain and C-terminal putative periplasmic domain, thus assuming a bitopic membrane protein. VtrA is a transcriptional regulator that plays a central role in the expression of the Vp-PAI genes of V. parahaemolyticus. The N-terminal wHTH domain of VtrA has homology with the conserved DNA binding domains of OmpR-family proteins. The OmpR-family contains many members that function as transcriptional regulators, including E. coli OmpR, V. cholerae ToxR and Salmonella Typhimurium HilA. E. coli OmpR is a response regulator of the EnvZ/OmpR two-component system with its DNA-binding domain at the C-terminal side [34]. On the other hand, V. cholerae ToxR and Salmonella Typhimurium HilA contain their DNA-binding domains on their N-terminal sides, which is similar to VtrA. V. cholerae ToxR is a single-span membrane protein with a TM domain and periplasmic domain at its C-terminus, whereas Salmonella Typhimurium HilA lacks a membrane-spanning region [35]. Given the defined localization of VtrA in the membrane and the presence of the TM domain, VtrA is a membrane-bound regulator belonging to a group of ToxR-like protein.
ToxR-like proteins such as V. cholerae ToxR, V. cholerae TcpP and E. coli CadC are generally active as dimers [23, 25, 27]. We showed that VtrA also forms oligomers through its TM and C-terminal domains in vivo using a ToxR-based assay. Our β-galactosidase assays using VtrA truncated variants and VtrA-PL showed that TM domain plays a predominant role in transcriptional activation, implying the oligomerization via the TM domain interactions. We also sought to define the detailed oligomeric state of VtrA in V. parahaemolyticus cells by the chemical cross-linking using DMA. However, despite considerable experimental efforts, we could not successfully detect any obvious band corresponding to VtrA oligomer in V. parahaemolyticus at this stage. It is thus uncertain about the detailed oligomeric state of VtrA in vivo, and this issue requires further studies for resolution.
V. cholerae ToxR interacts with the downstream, co-transcribed protein ToxS, which is required for the transcriptional regulatory activity of ToxR [36]. ToxS is hypothesized to stabilize ToxR and enhance the dimerization of its periplasmic domain. Recently, Li et al. reported that VtrC, encoded downstream of vtrA, is required for the transcriptional regulatory activity of VtrA and suggested that VtrC might stabilize VtrA in V. parahaemolyticus [18]. This feature of VtrA and VtrC is similar to that of V. cholerae ToxR and ToxS. It was reported that the expression of ToxR activates ctx transcription in E. coli even in the absence of ToxS, although ToxS is required for lower levels of ToxR to activate transcription, which is associated with an increase in the stability of ToxR [36, 37]. Similarly, we observed that the PBAD-based expression of VtrA could activate the PvtrB-lacZ transcription in E. coli, even without VtrC. Given that the expression of VtrA in E. coli was relatively overexpression base on the PBAD promoter, it is conceivable that VtrA activates transcription even without VtrC in E. coli, as is the case with ToxR. This further supports the functional parallel between VtrA /VtrC and ToxR /ToxS.
Bile is a host-derived factor that strongly induces the expression of the Vp-PAI genes of V. parahaemolyticus, and its stimulation is thought to be responsible for the virulence of V. parahaemolyticus during intestinal infection. It was recently reported that the periplasmic domain of VtrC and the C-terminal domain of VtrA form a complex where VtrC recruits structural elements from VtrA to complete a β-barrel with a hydrophobic inner chamber that binds bile salts [18]. In this study, we observed that the C-terminal putative periplasmic domain of VtrA was necessary for the bile-induced vtrB transcriptional activation, supporting that the C-terminal domain of VtrA participates in sensing bile. Given that oligomerization of VtrA induced its transcriptional regulatory activity, our observation that bile induces VtrA oligomerization could explain the mechanism by which the transcriptional activity of VtrA is activated without affecting its protein levels. In V. cholerae, the periplasmic domain of ToxR is required for leuO and ompU upregulation in response to bile, implicating the periplasmic domain of ToxR in sensing environmental stimuli [38]. Bile also stimulates V. cholerae TcpP transcriptional regulatory activity by mediating intermolecular disulfide bond formation between its periplasmic domains [39]. E. coli CadC is also known to form an intramolecular disulfide bond within its periplasmic domain, which is important in the function of CadC [40]. However, unlike these ToxR-like proteins, VtrA lacks cysteine residues in its C-terminal domain. Otherwise, bile might affect the conformation of the C-terminal domain of VtrA, bringing the adjacent TM and cytoplasmic DNA-binding domains into close proximity, thus facilitating the oligomerization and DNA-binding activity of VtrA.
The promoter region of vtrB contained repetitive 8-bp T-rich DNA elements near the putative −35 promoter element, which are essential for vtrB transcriptional activation by VtrA. Interestingly, the VtrA-mediated transcriptional activation of vtrB depended on the number of repetitive elements in the vtrB promoter region. OmpR-family proteins generally recognize direct repeats in the promoter regions of their target genes through dimerization, allowing these DNA-binding proteins to bind DNA repeats efficiently [41, 42]. Although the precise VtrA-binding site within the vtrB promoter region is not yet known, it might be possible that the oligomerization of VtrA promotes the binding to these repetitive DNA elements within the vtrB promoter region. It has been proposed that T3SS2-possessing Vibrio species may have horizontally acquired the T3SS2α or T3SS2β gene clusters during their evolution [13, 14]. The repetitive DNA elements in the vtrB promoter region were also found in T3SS2β-positive V. parahaemolyticus, T3SS2α-positive Vibrio cholerae and T3SS2β-positive Vibrio cholerae. Our β-galactosidase assays revealed that VtrA shows vtrB transcriptional activation in T3SS2β-positive V. parahaemolyticus and T3SS2-positive Vibrio cholerae strains, suggesting that the mechanism of target recognition by VtrA is also evolutionarily conserved among T3SS2-possessing Vibrio species.
In conclusion, we have presented experimental evidence showing that VtrA is a membrane-bound regulator and is active as oligomers. This provides new insights into the mechanism by which VtrA transcriptional regulatory activity is activated, including in response to bile. Further studies will be needed to explore the molecular mechanisms of VtrA oligomerization in detail.
Supporting information
Subcellular localization of C-terminal 3×FLAG-tagged VtrA (VtrA-FLAG) and its truncated forms in V. parahaemolyticus. V. parahaemolyticus expressing VtrA-FLAG and its truncated derivatives were fractionated into cytosolic (C) and membrane (M) fractions. Total cell lysates (T) and each fraction were subjected to immunoblot analysis for FLAG to detect VtrA and its truncated forms. DnaK and OmpA were detected as controls for the cytosol and membrane, respectively.
(TIF)
(A) Schematic representations of VtrA and VtrA containing a polyleucine TM domain (VtrA-PL). (B) β-galactosidase activity from the PvtrB-lacZ transcriptional reporter of and E. coli MC4100 upon expression of VtrA or VtrA-PL. The values represent the mean ±SD for a minimum of three independent experiments.
(TIF)
Each of the His-tagged VtrAN, VtrAN-ZIP, VtrAN-ZIPPLI and VtrAN-ZIPm proteins was treated (+) with dimethyl adipimidate (DMA) or was untreated (−), separated by SDS-PAGE and visualized by Coomassie brilliant blue staining. The migration positions of the molecular weight markers are indicated on the left side of the panel.
(TIF)
Electrophoretic mobility shift assays using VtrAN at concentrations higher than those shown in Fig 3B. Indicated concentrations of His VtrAN were incubated with 4 nM biotinylated DNA probe corresponding to a 284-bp upstream region of vtrB. The DNA probe was detected using HRP-conjugated streptavidin.
(TIF)
(A) ClustalW [1] multiple sequence alignments of the upstream sequences of vtrB from V. parahaemolyticus strain RIMD2210633 (Vp_2210633; T3SS2α-positive), V. parahaemolyticus strain TH3996 (Vp_TH3996; T3SS2β-positive), V. cholerae strain RIMD2214243 (Vc_2214234; T3SS2α-positive) and V. cholerae strain RIMD2214428 (Vc_2214428; T3SS2β-positive). Repetitive T-rich elements are indicated by gray shading. (B) Transcriptional activity of VtrA at the upstream promoter regions of vtrB from Vp_TH3996, Vc_2214243 and Vc_2214428 was evaluated by measuring β-galactosidase activity using lacZ transcriptional reporter in E. coli MC4100 carrying an empty vector (white bar) or a VtrA expression plasmid (black bar). Data represent the mean ± SD of a minimum of three independent experiments. *, p < 0.01, by Student’s t-test.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank members of the Iida lab for their support and helpful discussions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by Grants-in-Aid for Scientific Research (Grant numbers 26460526 and 17K08828 to S.M.) from the Japan Society for the Promotion of Science, URL: http://www.jsps.go.jp/english/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Thompson F. L., Iida T., and Swings J.. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004;68: 403–431. doi: 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blake PA, Weaver RE, Hollis DG. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34: 341–367. doi: 10.1146/annurev.mi.34.100180.002013 [DOI] [PubMed] [Google Scholar]
- 3.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis. 2000;181: 16661–1666. [DOI] [PubMed] [Google Scholar]
- 4.Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev. 2007;20: 39–48. doi: 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from V. cholerae. Lancet 2003;361: 743–749. doi: 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- 6.Sugiyama T, Iida T, Izutsu K, Park KS, Honda T. Precise region and the character of the pathogenicity island in clinical Vibrio parahaemolyticus strains. J Bacteriol. 2008;190: 1835–1837. doi: 10.1128/JB.01293-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izutsu K, Kurokawa K, Tashiro K, Kuhara S, Hayashi T, Honda T, et al. Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect Immun. 2008;76: 1016–1023. doi: 10.1128/IAI.01535-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68: 415–438. doi: 10.1146/annurev-micro-092412-155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiyoshi H, Kodama T, Iida T, Honda T. Contribution of Vibrio parahaemolyticus virulence factors to cytotoxicity, enterotoxicity, and lethality in mice. Infect Immun. 2010;78: 1772–1780. doi: 10.1128/IAI.01051-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie JM, Rui H, Zhou X, Iida T, Kodoma T, Ito S, et al. Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 2012;8: e1002593 doi: 10.1371/journal.ppat.1002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodama T, Hiyoshi H, Okada R, Matsuda S, Gotoh K, Iida T. Regulation of Vibrio parahaemolyticus T3SS2 gene expression and function of T3SS2 effectors that modulate actin cytoskeleton. Cell Microbiol. 2015;17: 183–190. doi: 10.1111/cmi.12408 [DOI] [PubMed] [Google Scholar]
- 12.Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, et al. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci USA. 2005;102: 3465–3470. doi: 10.1073/pnas.0409918102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada N, Iida T, Park KS, Goto N, Yasunaga T, Hiyoshi H, et al. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun. 2009;77: 904–913. doi: 10.1128/IAI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada N, Matsuda S, Matsuyama J, Park KS, de los Reyes C, Kogure K, et al. Presence of genes for type III secretion system 2 in Vibrio mimicus strains. BMC Microbiol. 2010;10: 302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin OS, Tam VC, Suzuki M, Ritchie JM, Bronson RT, Waldor MK, et al. Type III secretion is essential for the rapidly fatal diarrheal disease caused by non-O1, non-O139 Vibrio cholerae. MBio 2011;2: e00106–11. doi: 10.1128/mBio.00106-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kodama T, Gotoh K, Hiyoshi H, Morita M, Izutsu K, Akeda Y, et al. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One 2010;5: e8678 doi: 10.1371/journal.pone.0008678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park KS, Dryselius R, et al. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One 2010;5: e13365 doi: 10.1371/journal.pone.0013365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P, Rivera-Cancel G, Kinch LN, Salomon D, Tomchick DR, Grishin NV, et al. Bile salt receptor complex activates a pathogenic type III secretion system. Elife 2016;5: e15718 doi: 10.7554/eLife.15718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kodama T, Rokuda M, Park KS, Cantarelli VV, Matsuda S, Iida T, et al. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol. 2007;9: 2598–2609. doi: 10.1111/j.1462-5822.2007.00980.x [DOI] [PubMed] [Google Scholar]
- 20.Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; 1972. pp. 352–355. [Google Scholar]
- 21.Matsuda S, Okada N, Kodama T, Honda T, Iida T. A cytotoxic type III secretion effector of Vibrio parahaemolyticus targets vacuolar H+-ATPase subunit c and ruptures host cell lysosomes. PLoS Pathog 2012;8: e1002803 doi: 10.1371/journal.ppat.1002803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Hijum SA, Medema MH, Kuipers OP. Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiol Mol Biol Rev. 2009;73: 481–509. doi: 10.1128/MMBR.00037-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ottemann KM, Mekalanos JJ. The ToxR protein of Vibrio cholerae forms homodimers and heterodimers. J Bacteriol. 1996; 178: 156.–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauntlett JC, Gebhard S, Keis S, Manson JM, Pos KM, Cook GM. Molecular analysis of BcrR, a membrane-bound bacitracin sensor and DNA-binding protein from Enterococcus faecalis. J Biol Chem. 2008;283: 8591–8600. doi: 10.1074/jbc.M709503200 [DOI] [PubMed] [Google Scholar]
- 25.Goss TJ, Seaborn CP, Gray MD, Krukonis ES. Identification of the TcpP-binding site in the toxT promoter of Vibrio cholerae and the role of ToxR in TcpP-mediated activation. Infect Immun. 2010;78: 4122–4133. doi: 10.1128/IAI.00566-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalia AB, Lazinski DW, Camilli A. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio 2014;5: e01028–13. doi: 10.1128/mBio.01028-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindner E, White SH. Topology, dimerization, and stability of the single-span membrane protein CadC. J Mol Biol. 2014;426: 2942–2957. doi: 10.1016/j.jmb.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 1993;262: 1401.–. [DOI] [PubMed] [Google Scholar]
- 30.Fink A, Sal-Man N, Gerber D, Shai Y. Transmembrane domains interactions within the membrane milieu: principles, advances and challenges. Biochem Biophys Acta. 2012;1818: 974–983. doi: 10.1016/j.bbamem.2011.11.029 [DOI] [PubMed] [Google Scholar]
- 31.Miller VL, Taylor RK, Makalanos JJ. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 1987;48: 271.–. [DOI] [PubMed] [Google Scholar]
- 32.Goss TJ, Morgan SJ, French EL, Krukonis ES. ToxR recognizes a direct repeat element in the toxT, ompU, ompT, and ctxA promoters of Vibrio cholerae to regulate transcription. Infect Immun. 2013;81: 884–895. doi: 10.1128/IAI.00889-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Z, Kumagai K, Baba K, Mekalanos JJ, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175: 3844–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenney LJ. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol. 2002;5: 135–141. [DOI] [PubMed] [Google Scholar]
- 35.Bajaj V, Hwang C, Lee CA. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18: 715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x [DOI] [PubMed] [Google Scholar]
- 36.DiRita VJ, Mekalanos JJ. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 1991;64: 29.–. [DOI] [PubMed] [Google Scholar]
- 37.Miller VL, DiRita VJ, Mekalanos JJ. Identification of toxS, a regulatory gene whose product enhances ToxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 1989;171:1288.–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, Bina JE. Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. J Bacteriol. 2015;197: 3499–3510. doi: 10.1128/JB.00419-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, et al. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci USA. 2013;110: 2348–2353. doi: 10.1073/pnas.1218039110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetsch L, Koller C, Dönhöfer A, Jung K. Detection and function of an intramolecular disulfide bond in the pH-responsive CadC of Escherichia coli. BMC Microbiol. 2011;11: 74 doi: 10.1186/1471-2180-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez-Hackert E, Stock AM. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol. 1997;269: 301.–. doi: 10.1006/jmbi.1997.1065 [DOI] [PubMed] [Google Scholar]
- 42.Blanco AG, Sola M, Gomis-Rüth FX, Coll M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 2002;10: 701.–. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Subcellular localization of C-terminal 3×FLAG-tagged VtrA (VtrA-FLAG) and its truncated forms in V. parahaemolyticus. V. parahaemolyticus expressing VtrA-FLAG and its truncated derivatives were fractionated into cytosolic (C) and membrane (M) fractions. Total cell lysates (T) and each fraction were subjected to immunoblot analysis for FLAG to detect VtrA and its truncated forms. DnaK and OmpA were detected as controls for the cytosol and membrane, respectively.
(TIF)
(A) Schematic representations of VtrA and VtrA containing a polyleucine TM domain (VtrA-PL). (B) β-galactosidase activity from the PvtrB-lacZ transcriptional reporter of and E. coli MC4100 upon expression of VtrA or VtrA-PL. The values represent the mean ±SD for a minimum of three independent experiments.
(TIF)
Each of the His-tagged VtrAN, VtrAN-ZIP, VtrAN-ZIPPLI and VtrAN-ZIPm proteins was treated (+) with dimethyl adipimidate (DMA) or was untreated (−), separated by SDS-PAGE and visualized by Coomassie brilliant blue staining. The migration positions of the molecular weight markers are indicated on the left side of the panel.
(TIF)
Electrophoretic mobility shift assays using VtrAN at concentrations higher than those shown in Fig 3B. Indicated concentrations of His VtrAN were incubated with 4 nM biotinylated DNA probe corresponding to a 284-bp upstream region of vtrB. The DNA probe was detected using HRP-conjugated streptavidin.
(TIF)
(A) ClustalW [1] multiple sequence alignments of the upstream sequences of vtrB from V. parahaemolyticus strain RIMD2210633 (Vp_2210633; T3SS2α-positive), V. parahaemolyticus strain TH3996 (Vp_TH3996; T3SS2β-positive), V. cholerae strain RIMD2214243 (Vc_2214234; T3SS2α-positive) and V. cholerae strain RIMD2214428 (Vc_2214428; T3SS2β-positive). Repetitive T-rich elements are indicated by gray shading. (B) Transcriptional activity of VtrA at the upstream promoter regions of vtrB from Vp_TH3996, Vc_2214243 and Vc_2214428 was evaluated by measuring β-galactosidase activity using lacZ transcriptional reporter in E. coli MC4100 carrying an empty vector (white bar) or a VtrA expression plasmid (black bar). Data represent the mean ± SD of a minimum of three independent experiments. *, p < 0.01, by Student’s t-test.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.