Abstract
Identifying and characterizing highly accessible chromatin regions assists in determining the location of genomic regulatory elements and understanding transcriptional regulation. In this chapter we describe an approach to map accessible chromatin features in plants using the Assay for Transposase Accessible Chromatin, combined with high throughput sequencing (ATAC-seq), which was originally developed for cultured animal cells. This technique utilizes a hyperactive Tn5 transposase to cause DNA cleavage and simultaneous insertion of sequencing adapters into open chromatin regions of the input nuclei. The application of ATAC-seq to plant tissue has been challenging due to the difficulty of isolating nuclei sufficiently free of interfering organellar DNA. Here we present two different approaches to purify plant nuclei for ATAC-seq: the INTACT method (Isolation of Nuclei TAgged in Specific Cell Types) to isolate nuclei from individual cell types of the plant, and tissue lysis followed by sucrose sedimentation to isolate sufficiently pure total nuclei. We provide detailed instructions for transposase treatment of nuclei isolated using either approach, as well as subsequent preparation of ATAC-seq libraries. Sequencing-ready ATAC-seq libraries can be prepared from plant tissue in as little as one day. The procedures described here are optimized for Arabidopsis thaliana but can also be applied to other plant species.
Keywords: ATAC-seq, INTACT system, chromatin, nucleus, transposition, nucleosome, transcription factor, enhancer
1 Introduction
Plants are sessile organisms that must precisely regulate their transcription in response to their environment, as well as for proper development, growth, and homeostasis. Transcription is associated with regions of relatively open chromatin, in which cis-regulatory elements such as enhancers and promoters can recruit transcription factors and RNA polymerase II to transcribe DNA [1]. Binding of transcription factors to DNA generally results in the depletion of nucleosomes, rendering these regions hypersensitive to nucleases. Characterizing such regulatory regions throughout the genome has therefore relied on methods that combine enzymatic digestion of nuclear DNA and high-throughput sequencing, such as microccocal nuclease sequencing (MNase-seq, see Chapter 10) and DNase I Hypersensitivity sequencing (DNase-seq) [2, 3]. Alternatively, regulatory regions can be inferred by Chromatin Immunoprecipitation sequencing (ChIP-seq, see Chapter 5) where antibodies are used to pull down transcription factors or histone marks associated with active transcription [4].
An improved method for identifying accessible regions of chromatin and transcription factor binding is the Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq) [5,6]. This method uses a hyperactive Tn5 transposase to integrate preloaded sequencing adapters into regions of open chromatin (Fig. 1A). ATAC-seq is a fast protocol with simple library amplification steps and requires very small amounts of starting material, making it a vast improvement over alternative methods. However, a drawback of this protocol is that the hyperactive Tn5 transposase also targets sources of extranuclear genetic material, including the genomes of mitochondria and chloroplasts. This decreases the proportion of reads that map to the nuclear genome, reducing the amount of information that can be used to identify regulatory regions of open chromatin. Such extranuclear reads must be discarded at the start of the data analysis process, diminishing the efficiency of the assay both in terms of cost and in effective use of materials. To gain the maximum efficiency of this powerful procedure, input material free from extranuclear genetic material, such as purified nuclei, is the ideal input for ATAC-seq
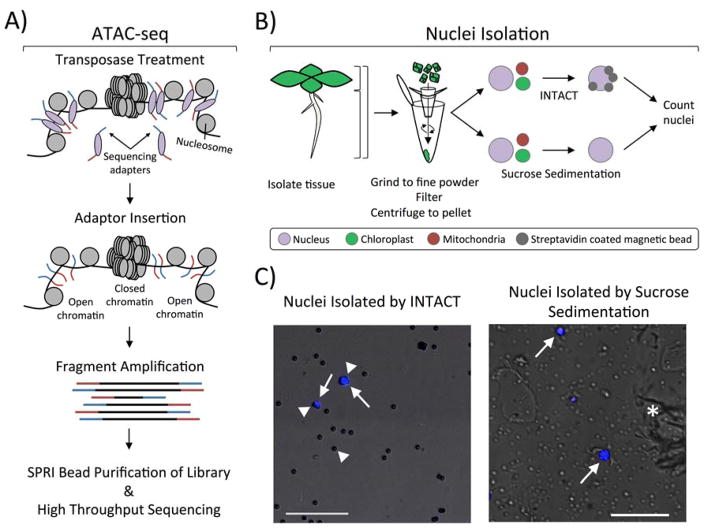
Figure 1. ATAC-seq profiling using nuclei isolated by INTACT or sucrose sedimentation.
A) Overview of the ATC-seq procedure. Nuclei are incubated with sequencing adapter-loaded Tn5 transposase, which diffuses into the nucleus to interact with chromatin. Sequencing adapters are inserted into open chromatin regions, and the fragmented DNA is amplified wherever the sequencing adapters were inserted. This generates a library of DNA fragments in which each end represents an insertion site. The amplified libraries are purified and sequenced with next generation sequencing. B) Two different methods for purifying nuclei from Arabidopsis thaliana can be used: 1) INTACT for isolating nuclei from specific cell types, and 2) sucrose sedimentation to isolate total nuclei from input tissue. The two methods have the same initial steps: tissue is collected from a specific part of the plant (root, leaf, or the entire plant), ground to a fine powder, resuspended, filtered, and centrifuged to pellet nuclei and cellular debris. Nuclei isolation using tissue that expresses INTACT transgenes uses streptavidin coated magnetic beads to affinity purify biotinylated nuclei out of the resuspended pellet. This allows for the isolation of nuclei from specific cell-types that express the nuclear tagging fusion (NTF) and the biotin ligase BirA, resulting in very low contamination by organellar genomes. Alternatively, total nuclei can be isolated from tissue by resuspending the nuclei/debris pellet in a buffer with Triton X-100 to lyse organelles and centrifuging through a dense sucrose layer. Nuclei isolated from both procedures are stained with DAPI and quantified using a hemocytometer. C) Fluorescent microscope images of nuclei (white arrows) stained with the DNA-binding dye DAPI (blue) isolated either through INTACT or sucrose sedimentation. INTACT isolated nuclei are identified by their DAPI-fluorescence and binding to multiple beads (white arrowhead). Beads are easily visualized by increasing transmission of white light while viewing the nuclei in the DAPI channel. Sucrose sedimentation isolated nuclei (white arrows) are DAPI-stained objects around 4–6 μm in diameter, although they can vary in size and shape depending on starting tissue. Much more cellular debris (white asterisk) is observed in sucrose sedimentation-isolated nuclei as compared to INTACT-purified nuclei, but this should not impact the procedure described here. Each picture contains a 50 μm scale bar shown at the bottom left.
In this chapter, we describe the use of two different methods to isolate either total nuclei from tissues or nuclei from specific cell types of Arabidopsis thaliana (Fig. 1B). To isolate total nuclei from plant tissue we use extraction buffers with a non-ionic detergent to lyse organelles, followed by sucrose sedimentation to further purify the nuclei [7]. This method of nuclei isolation can be done in any lab on most plant tissues. However, these partially purified nuclei still contain some organellar DNA in addition to nuclear DNA, which reduces the efficiency of Tn5 transposition to nuclear DNA and results in fewer sequencing reads that map to nuclear DNA. In addition, we describe the Isolation of Nuclei TAgged in specific Cell Types (INTACT) method to isolate nuclei from tissue or from specific cell types [8]. This system uses two transgenes for nuclear targeting for affinity purification: 1) the Nuclear Tagging Fusion (NTF) construct, which encodes a fusion of WPP nuclear envelope-targeting domain, a Green Fluorescent Protein (GFP), and the Biotin Ligase Recognition Peptide (BLRP); and 2) an E. coli biotin ligase (BirA), which biotinylates the BLRP tag. The BirA is expressed from a constitutive promoter while the NTF is expressed either from a constitutive or cell type-specific promoter. The specificity of the NTF promoter determines which cell types will have biotinylated nuclei and can then be isolated by affinity purification with streptavidin-coated magnetic beads [9]. A key advantage of the INTACT approach is not only that the isolated nuclei have less organellar DNA contamination, but also that this method can be used to selectively isolate nuclei from specific cell types. While INTACT is a powerful technique, it does require that stable transgenic lines containing BirA and NTF cassettes for the cell type of interest are available, which are time-consuming to generate and can be limiting for many species. Even so, the protocol described here, particularly ATAC-seq using sucrose sedimentation-purified nuclei, can readily be adapted for chromatin profiling in any plant species.
2 Materials
2.1 Equipment
Porcelain 50 mL mortar and pestle, or equivalent.
Liquid nitrogen.
Metal lab spoon.
DynaMag 2 magnetic rack for 1.5 mL tubes (e.g. Life Technologies, catalog no. 12321D).
DynaMag 15 magnetic rack for 15 mL tubes (e.g. Life Technologies, catalog no. 12301D).
MagWell 96 well magnetic separator plate (e.g. EdgeBio, catalog no. 57624)
Nylon cell strainers with 70 μm pores.
Long-stem analytical funnel.
Pipet-Aid.
Sterile 10 mL plastic serological pipettes.
Eppendorf tubes, 1.5 mL.
PCR tubes, 0.2 mL.
Falcon tubes, 15 and 50 mL.
Nutator platform rotator.
Hemocytometer (e.g. Hausser Bright Line hemocytometer, Fisher Scientific)
Microcentrifuge and refrigerated centrifuge with rotor for 15 mL tubes.
Cold room, 4 °C.
Molecular biology grade water.
Sterile disposable filter unit, 500 mL.
Sterile 0.2 μm syringe filter.
Sterile 10 mL plastic syringe.
Thermal cycler
Real-Time PCR machine
A 64-bit computer with at least 1 TB hard disk and 16 Gb of memory for ATAC-seq data analysis.
Fluorescent microscope.
2.2 Stock Solutions and Reagents
Complete, EDTA-free Protease Inhibitors (e.g. Roche).
Stock solution of 2 M spermidine. Prepare by dissolving 2.904 g spermidine powder in 10 mL water. Aliquot 1mL of solution per 1.5 mL Eppendorf tube and store at −20 °C.
Stock solution of 200 mM spermine. Prepare by dissolving 0.4047 g spermine powder in 10 mL of water. Aliquot 1 mL of solution per 1.5 mL Eppendorf tube and store at −20 °C.
Stock solution of incomplete Nuclei Purification Buffer (NPBi): 20 mM MOPS, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, adjusted to pH 7 with 2M KOH. Filter sterilize the solution and degas under vacuum for 10 minutes. Store at 4 °C for up to 3 months.
Stock solution of 10% Triton X-100.
Stock solution of 10X DAPI. Prepare by dissolving 10 mg DAPI powder in 5 mL water, for a final concentration of 2 μg/μL. Filter sterilize the solution and store at 4 °C in the dark for several months. To stain nuclei with DAPI, dilute the 10X DAPI solution to 1X using water (final concentration of 0.2 μg/μL), and use within 2–3 hours.
2.3 Purification of Tagged Nuclei using INTACT
Plant material: tissue from transgenic plants expressing both NTF and BirA in the cell type of interest. INTACT transgenic lines targeting the root epidermal hair and non-hair cell types, as well as INTACT plasmid vectors are available from the Arabidopsis Biological Resource Center at Ohio State University.
M-280 Streptavidin Dynabeads (e.g. Life Technologies).
Nuclei Purification Buffer (NPB): 20 mM, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, 1X Roche Complete protease inhibitors, adjusted to pH 7 with 2M KOH. Prepare by adding spermidine, spermine, and Roche Complete protease inhibitors to NPBi just before starting the INTACT nuclei purification procedure. Keep solution on ice, and use within 1 hour of preparation.
Nuclei Purification Buffer containing 0.1% Triton X-100 (NPBt): 20 mM MOPS pH 7, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, 0.1% (v/v) Triton X-100. Prepare by adding spermidine, spermine, and Triton X-100 to NPBi just before starting the INTACT nuclei purification procedure. Keep solution on ice, and use within 1 day of preparation.
2.4 Purification of Total Nuclei using Sucrose Sedimentation
Plant material: fresh or frozen plant tissue.
Stock solution of 1M Tris-HCl pH 8
Stock solution of 1M MgCl2
Stock solution of 2M sucrose.
Nuclei Purification Buffer (NPB): 20 mM MOPS pH7, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.2 mM spermine, 1X Roche Complete protease inhibitors. Prepare by adding spermidine, spermine, and Roche Complete protease inhibitors to NPBi just before starting the nuclei purification procedure. Keep solution on ice, and use within 1 hour of preparation.
Nuclei Extraction Buffer 2 (NEB 2): 0.25 M Sucrose, 10 mM Tris-HCl pH 8, 10 mM MgCl2, 1% Triton X-100, 1X Roche Complete Protease Inhibitors. Prepare solution just before use, keep on ice, and use within 1 hour of preparation.
Nuclei Extraction Buffer 3 (NEB 3): 1.7 M Sucrose, 10 mM Tris-HCl pH 8, 2 mM MgCl2, 0.15% Triton X-100, 1X Roche Complete Protease Inhibitors. Prepare solution just before use, keep on ice, and use within 1 hour of preparation.
2.5 Tagmentation of Chromatin by Tn5 transposase
Nextera Library Kit (Illumina, FC-121-1030).
MinElute PCR Purification kit (Qiagen).
2.6 Sequencing Library Preparation
ATAC Primer 1 (AATGATACGGCGACCACCGAGATCTACACTCGTCGGCAGCGTCAGATGTG)
ATAC barcoded Primer 2 (CAAGCAGAAGACGGCATACGAGATNNNNNNNNGTCTCGTGGGCTCGGAGATGT); N’s indicate the 8-base index sequence. Each library to be pooled for sequencing should be amplified with a different barcoded primer 2. See Supplementary Table 1 of [5] for all primer sequences.
NEBNext High-Fidelity 2X PCR Master Mix (NEB).
Solution of 20X EvaGreen dye (Biotium).
Solution of 50X ROX dye (Invitrogen).
MinElute PCR Purification kit (Qiagen).
Agencourt Ampure XP PCR Purification beads (Beckman Coulter).
100% ethanol.
Horizontal electrophoresis gel box and power source.
302 nm ultraviolet transilluminator.
NEBNext Library Quantification kit for Illumina (NEB)
3 Methods
Users should either begin at section 3.1 for affinity purification of nuclei using INTACT, or at section 3.2 for isolation of total nuclei. In either case, the purified nuclei are used for tagmentation by Tn5 transposase in step 3.3. All procedures are carried out at room temperature (25 °C) unless otherwise specified.
3.1 Purification of Tagged Nuclei Using INTACT
Grind tissue (3 g of roots or 0.5 g of leaves) to a fine powder in liquid nitrogen using a mortar and pestle. Using a nitrogen-cooled metal lab spoon, quickly transfer the frozen tissue powder to another mortar containing 10 mL of ice-cold Nuclei Purification Buffer (NPB). Thoroughly resuspend the powder in NPB by grinding it with a new, cold pestle (see Note 1).
Use a 10 mL serological pipette to draw up the tissue suspension and filter it through a 70 μm nylon cell strainer, placed in the center of a long stemmed funnel. Collect the flow-through in a chilled 15 mL tube on ice.
Spin down the nuclei at 1,200 x g for 10 minutes at 4 °C. Use a 10 mL serological pipet and then a 1 mL pipette tip to carefully remove as much of the supernatant as possible without disturbing the pellet.
Gently resuspend the pellet in 1 mL of ice-cold NPB. Transfer the crude nuclei suspension to a 1.5 mL tube. Keep on ice.
Wash the appropriate amount of Streptavidin M280 Dynabead suspension (25 μL for nuclei from 3 g of roots or 10 μL for 0.5 g of leaves) with 1 mL of ice-cold NPB in a 1.5 mL tube. Collect the beads on the DynaMag2 magnetic rack. Discard the supernatant and resuspend the beads with ice-cold NPB to their original volume (e.g. 25 μL). Keep on ice.
Add the washed and resuspended beads to the 1 mL of resuspended nuclei from Step 4. Rotate on a nutator in a 4 °C cold room for 30 minutes. Work in the 4 °C cold room for Steps 7–14.
Transfer the 1 mL bead-nuclei mixture to a 15 mL tube and slowly add to it 13 mL of ice-cold NPBt. Mix gently and place on a nutator for 30 seconds.
Place the 15 mL tube in the DynaMag 15 magnetic rack for 2 minutes to capture the nuclei-beads along the walls of the tube.
Slowly remove the NPBt supernatant with a serological pipette, making sure not to disturb the beads on the side walls of the tube. Gently resuspend the beads with 14 mL of ice-cold NPBt, mix gently, and place on a nutator for 30 seconds.
Place the 15 mL tube in the DynaMag 15 magnetic rack for 2 minutes to capture the nuclei and beads.
Repeat Steps 9 and 10 one more time, for a total of three washes.
Slowly remove the NPBt supernatant with a serological pipette. Resuspend the beads in 1 mL of ice-cold NPBt. Remove 25 μL of this nuclei-bead suspension to a 0.6 ml tube on ice for counting captured nuclei with a hemocytometer.
Transfer the remaining nuclei-bead suspension to an ice-cold 1.5 mL tube. Place the 1.5 mL tube in the DynaMag 2 magnetic rack to capture the beads along the walls of the tube.
Carefully remove the NPBt supernatant and resuspend the nuclei-beads in 20 μL of ice-cold NPB. Keep on ice until the nuclei are counted and ready for tagmentation. (see Note 2).
To view and quantify nuclei under a light microscope, add 1 μL of diluted DAPI solution (0.2 μg/μL) to each 25 μL aliquot of nuclei from Step 12. Mix well, and place on ice for 5 minutes in the dark.
Use a hemocytometer to count the DAPI-stained, bead-bound nuclei and determine the total yield. Purified nuclei should appear as shown in Figure 1C (see Note 3).
Use the calculated total yield to determine the volume of resuspended nuclei from Step 14 needed to obtain 50,000 nuclei for the ATAC-seq reaction. Transfer this volume of resuspended nuclei to a new 0.2 mL tube, and keep on ice. Immediately proceed to Section 3.3.
3.2 Purification of Total Nuclei Using Sucrose Sedimentation
Grind 0.1 to 1 g of plant tissue to a fine powder in liquid nitrogen using a mortar and pestle (see Note 4).
Using a nitrogen-cooled metal lab spoon, quickly transfer the frozen tissue powder to another mortar containing 10 mL ice-cold NPB. Thoroughly resuspend the powder in NPB by grinding it with a new, cold pestle.
Use a 10 mL serological pipette to draw up the tissue suspension and filter it through a 70 μm nylon cell strainer, placed in the center of a long stemmed funnel. Collect the flow-through into a 15 mL tube on ice.
Centrifuge the tube at 1,200 x g for 10 minutes at 4 °C.
Gently remove the supernatant and gently but thoroughly resuspend the pellet in 1 mL of ice-cold NEB2 buffer. Transfer this suspension to a new 1.5 mL microcentrifuge tube.
Spin the resuspended nuclei at 12,000 x g for 10 minutes at 4 °C.
Carefully remove the supernatant and resuspend the pellet thoroughly in 300 μL of NEB3 buffer.
Add 300 μL of ice-cold NEB3 to a new 1.5 mL microcentrifuge tube. Carefully layer the resuspended pellet from Step 7 on top of the fresh NEB3. Centrifuge at 16,000 x g for 10 minutes at 4 °C (see Note 5).
Carefully remove the supernatant and resuspend the nuclei pellet in 1 mL of cold NPB. Keep these nuclei on ice.
Remove 25 μL of this nuclei suspension and move to a fresh 0.6 ml tube on ice. To this add 1 μL of diluted DAPI solution (0.2 μg/μL). Mix well and place on ice for 5 minutes in the dark.
Use a hemocytometer to quantify the DAPI-stained nuclei and determine the total yield. Purified nuclei should appear as shown in Fig. 1C (see Note 6).
Use the calculated total yield to determine the volume of resuspended nuclei from Step 9 needed to obtain 50,000 nuclei for the ATAC-seq reaction. Transfer this volume of the resuspended nuclei to a new 0.2 mL tube, and keep on ice. Immediately proceed to Section 3.3.
3.3 Tagmentation with Tn5 Transposase
Prepare the transposition reaction master mix in a 0.2 mL PCR tube on ice according to Table 1 and mix well. The volumes given in Table 1 are for a single reaction with 50,000 nuclei.
If the nuclei were isolated using the Sucrose Sedimentation procedure, pellet 50,000 nuclei from Subheading 3.2 Step 9 by spinning the appropriate volume of nuclei at 1,500 x g for 7 minutes at 4 °C. Remove the supernatant, and resuspend the nuclei in 50 μL of ice-cold transposition reaction mix prepared in step 1. Move the reaction to a 0.2 mL PCR tube on ice. If the nuclei were isolated using the INTACT procedure, move 50,000 bead-bound nuclei from Subheading 3.1 Step 14 into a 0.2 mL tube and capture the beads on the tube wall in a MagWell 96 well magnetic plate on ice. Remove the supernatant, and resuspend the bead-bound nuclei in 50 μL of ice-cold transposition reaction mix. Keep on ice.
Place the transposition reaction in a thermal cycler block pre-warmed to 37 °C and incubate for 30 minutes with occasional gentle mixing to keep the nuclei in suspension.
Purify the transposed DNA using the Qiagen MinElute PCR purification kit according the manufacturer’s instructions. Elute DNA in 11 μL of elution buffer EB, provided in the kit. DNA can now be stored at −20 °C until future use, or used immediately for PCR amplification.
Table 1.
Transposition reaction mix
| Component | Volume (μL) |
|---|---|
| 2X TD Buffer | 25 |
| Water | 22.5 |
| TDE1 Transposase | 2.5 |
| Total | 50 |
3.4 PCR Amplification of the DNA Library
Prepare the PCR amplification mix in a 0.2 mL tube on ice according to Table 2. Mix well, and perform PCR cycling as described in Table 3 (see Note 7).
Once the thermal cycler reaches 4 °C, remove the samples and place them on ice.
To determine the number of additional PCR cycles needed to adequately amplify the DNA library, prepare the qPCR Library Amplification Mix described in Table 4 in a 0.2 mL PCR tube. Keep the mixture on ice.
Perform thermal cycling in the qPCR machine according to Table 5.
To determine the optimal number of cycles needed to amplify the remaining 45 μL of each library from Step 2, view the linear fluorescence versus cycle number plot on the qPCR machine once the reaction is finished. The cycle number at which the fluorescence for a given reaction is at 1/3 of its maximum is the number of additional cycles (N) that each library requires for adequate amplification (see Note 8).
Run the remaining 45 μL of each PCR reaction from Step 2 according to Table 6.
Purify the libraries by mixing Ampure XP beads with the reaction products at a 1.5:1 ratio of beads:PCR sample by volume (see Note 9). Incubate at room temperature for 5 minutes.
Place the 0.2 mL tube on the MagWell 96 well magnetic plate for 1 minute to capture the Ampure beads, and discard the supernatant.
With the tube still in the magnetic plate, wash the beads twice for 30 seconds each with 200 μL of 80% ethanol without disturbing the bead pellet. After the last wash, allow the beads to dry for 5 minutes to remove all traces of ethanol (see Note 10).
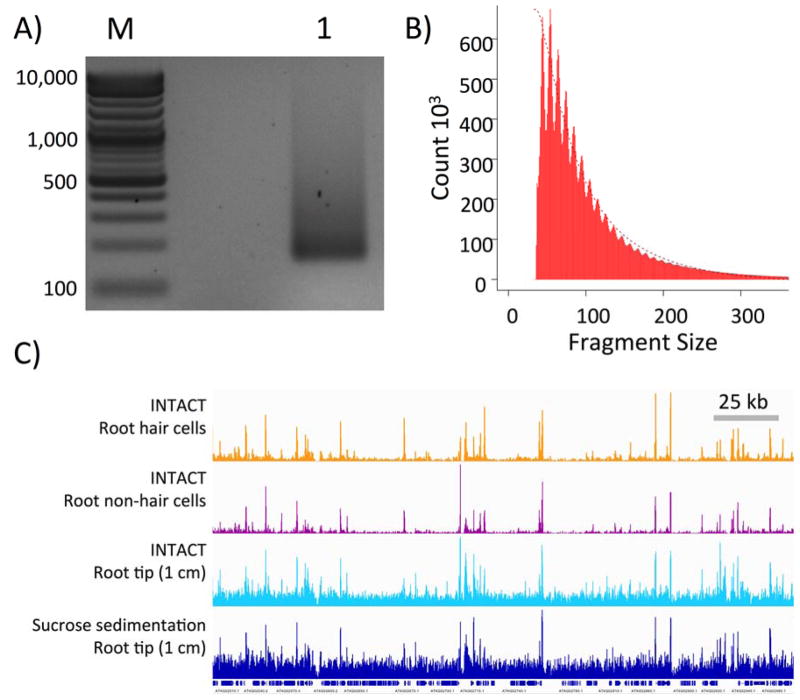
Remove the tube from the magnet and resuspend the bead pellet in 20 μL 10 mM Tris pH 8. Incubate at room temperature for 2 minutes, capture the beads on the magnet, and transfer the supernatant into a fresh 0.2 mL PCR tube on ice. A small aliquot of the library, 1–2 μL, can be run on a 2% agarose gel to visualize the abundance and size distribution of amplified libraries (Fig. 2A) (see Note 11). The purified libraries can now be stored at −20 °C.
Quantify the molar concentrations of the libraries using the NEBNext Library Quantification kit for Illumina, according to manufacturer’s directions. Alternatively, other qPCR-based library quantification kits can be used to determine the concentration of the amplified libraries.
Once quantified, the libraries are ready for pooling and high-throughput sequencing on the Illumina platform (see Note 12).
The quality of the sequencing reads, alignment to the genome, fragment size distribution (Fig. 2B), and downstream analyses can be performed as described in Note 13. A genome browser shot of the typical Arabidopsis ATAC-seq data from libraries made using the procedures described here can be seen in Fig. 2C.
Table 2.
Transposed DNA Amplification mix
| Component | Volume (μL) |
|---|---|
| Transposed DNA (from Subheading 3.3 step 4) | 10 |
| Water | 10 |
| 25 μM ATAC Primer 1 | 2.5 |
| 25 μM ATAC barcoded Primer 2* | 2.5 |
| 2X NEBNext High Fidelity PCR Mix | 25 |
| Total | 50 |
A different barcoded Primer 2 should be used for each library that is to be pooled into a single sequencing run.
Table 3.
Thermal Cycling Conditions for Transposed DNA Amplification
| Cycle number | Temperature (°C) | Time |
|---|---|---|
| 1 | 72 | 5 min |
| 98 | 30 sec | |
| 5 cycles | 98 | 10 sec |
| 63 | 30 sec | |
| 72 | 1 min | |
| 4 | Hold |
Table 4.
qPCR Library Amplification Mix
| Component | Volume (μL) |
|---|---|
| Amplified library (from Subheading 3.4 step 2) | 5 |
| Water | 0.45 |
| 25 μM ATAC Primer 1 | 0.5 |
| 25 μM ATAC barcoded Primer 2 | 0.5 |
| 20X Evagreen dye | 0.75 |
| 50X ROX dye* | 0.30 |
| 2X NEBNext High Fidelity PCR Mix | 7.5 |
| Total | 15 |
ROX concentration may vary depending on qPCR instrument. The amount described here is optimized for the ABI Step-One-Plus instrument.
Table 5.
qPCR Cycling Conditions to Determine Additional Library Amplification Cycles
| Cycle number | Temperature (°C) | Time |
|---|---|---|
| 1 | 98 | 30 sec |
| 20 cycles | 98 | 10 sec |
| 63 | 30 sec | |
| 72 | 1 min |
Table 6.
Final Library Amplification
| Cycle number | Temperature (°C) | Time |
|---|---|---|
| 1 | 98 | 30 sec |
| N cycles | 98 | 10 sec |
| 63 | 30 sec | |
| 72 | 1 min | |
| 4 | Hold |
Figure 2. ATAC-seq library preparation and high-throughput sequencing.
A) An amplified ATAC-seq library purified with Ampure XP beads (lane “1”) was resolved in a 2% agarose gel stained with ethidium bromide. Lane “M” is the molecular weight marker lane. Amplified library fragments generally range in size from 180 bp to several kb in size. The size distribution of the resolved gel may vary somewhat, but the final product should be free of adapter dimers (distinct band around 125 bp) and primer dimers (distinct band around 80 bp). See Note 11. B) Insert sizes of ATAC-seq paired-end reads from 50,000 nuclei isolated by INTACT from non-hair cells calculated using the InsertSizeMetrics option from Picard Tools (Note 13). The distribution shows periodicity of helical pitch of DNA for fragments smaller than 200 bp. Fragments containing one or more nucleosomes, related to insert periodicity increasing in 150 bp, were not observed using the transposase:nuclei and bead:DNA ratios described in this protocol. C) Integrated Genome Viewer snapshot of four different libraries sequenced on the Illumina platform. The tracks shown are of ATAC sequencing reads from INTACT isolated nuclei from root hair cells (orange), root non-hair cells (purple), root tip (cyan), and sucrose sedimentation isolated nuclei from 1 cm root tip (navy). Gene tracks are shown below the ATAC-seq tracks and a 25 kb scale bar is show
Acknowledgments
This work was supported by the National Science Foundation Grant no. 1238243. We thank Paja Sijacic and Shannon Torres for helping to optimize the protocol for nuclei isolation and for suggestions on the manuscript.
Footnotes
This protocol is optimized for 3 g of root or 0.5 g of leaf tissue from Arabidopsis thaliana. Ground leaf tissue contains more debris, relative to roots, and therefore requires a lower amount of starting material to obtain highly purified nuclei. INTACT may also be performed on fresh tissue by chopping the tissue in NPB as opposed to grinding to a fine powder using liquid nitrogen. However, this approach does require the use of fresh tissue. The number of samples that can be run through INTACT purification simultaneously is mainly limited by the capacity of the DynaMag 15 magnetic rack used for nuclei capture. Up to four separate samples can be processed in parallel using one DynaMag 15 magnetic rack.
Using an INTACT line with nuclei labeled in the root epidermal non-hair cell type, approximately 200,000 purified nuclei can be obtained from 3 g of roots. Larger amounts of tissue can be used for purifying nuclei from less abundant cell types, and this generally only requires adjustments to the amount of streptavidin beads used and the volume of solution used for bead capture. See [9] for more details on variations in the INTACT procedure.
After isolating the bead bound nuclei, keep the sample on ice while quantifying the nuclei from the aliquot in Subheading 3.1 Step 12. Do not freeze the isolated nuclei before doing tagmentation and library preparation. Freezing and thawing of isolated nuclei can disrupt protein-DNA interactions.
After DAPI staining, nuclei purified by INTACT can be easily identified and counted using a hemocytometer. The ideal setup for visualizing nuclei is under a mix of dim white light and DAPI channel fluorescence. The dim white light allows for visualization of the hemocytometer grid and the beads, and the DAPI fluorescence allows for the visualization of nuclei. A sample image of isolated bead-bound nuclei is shown in Fig. 1C. A nucleus is identified as a circle that fluoresces in the DAPI channel and has several beads clustered around it. Minimal cellular debris or contaminating unbound nuclei should be observed in the final product. These contaminants may be further reduced by using fewer beads and by increasing the volumes of NPB and NPBt used during purification as described in Note 1.
We have successfully used as few as 20,000 to as many as 200,000 INTACT-purified nuclei in this procedure without altering any other parameters of the protocol presented here.
This protocol is optimized for less than 1 g of root or 0.5 g of leaf tissue. Ground leaf tissue contains more debris relative to roots, and therefore requires a lower amount of starting material to obtain purified nuclei. As with the INTACT protocol, sucrose sedimentation of nuclei may also be performed on fresh tissue by chopping the tissue in NPB as opposed to grinding to a fine powder using liquid nitrogen. However, this approach does require the use of fresh tissue. We recommend starting with the minimum amount of tissue needed to obtain the required number of nuclei (e.g. 50,000 per ATAC-seq reaction).
Proper separation of nuclei from other cellular debris requires the nuclei to pass through the sucrose cushion during centrifugation. The NEB3 resuspended nuclei should therefore be placed gently on top of NEB3 layer present in the tube. After centrifugation, the contaminating organelles and debris may be visible at the top of the tube. If leaf tissue was used, the top layer will become greener after centrifugation and the pellet will become noticeably less green than it was prior to centrifugation.
After DAPI staining, nuclei purified by sucrose sedimentation can be identified and quantified using a hemocytometer. A mixture of DAPI-channel fluorescence and white light illumination allows the stained nuclei and the hemocytometer grid to be seen simultaneously. A sample image of isolated nuclei is shown in Fig. 1C. A nucleus is identified as a punctate circle with strong DAPI fluorescence. The nucleus is typically ~5 μm in size and can be easily identified at 200X and 400X magnifications. Cellular debris may be observed in the final preparation, but this generally does not affect the outcome of the ATAC-seq procedure. To reduce cellular debris contamination, starting tissue can be chopped with a razor blade (see Note 4) and/or additional NEB3 wash steps may also be done by repeating Subheading 3.2 steps 7–9 for a second sucrose cushion centrifugation.
Ensure that all work surfaces, pipettes, and reagents needed for amplification and library preparation are free of DNA contamination. For library amplification, unique barcoded adapters are used for each sample if multiple libraries are to be sequenced in an individual flow cell lane. The sequences of all primers can be found in the supplementary material of [5].
The number of PCR cycles needed to amplify ATAC libraries is determined by the PCR reaction in Subheading 3.4 step 5. We recommend using the minimum number of cycles necessary to obtain a sufficient molar amount of library for Illumina sequencing. This must be determined empirically and will also depend on the number of libraries to be pooled for sequencing.
The ratio of Ampure XP PCR Purification beads to PCR volume determines the size of purified DNA fragments isolated. The 1.5 Ampure bead to PCR reaction ratio results in the isolation of DNA fragments shown in Fig. 2A. Using ratios that have higher proportions of beads may result in purification of sequencing adapters and PCR primers, which can negatively affect sequencing.
A drying time of 5 minutes is generally sufficient to remove all traces of ethanol from the beads, but this time may vary based on humidity and room temperature. Georgia is very humid in the summer. Ensure that all ethanol has evaporated before moving on to the next step. Do not allow beads to dry to the extent that the pellet begins to crack.
Libraries can generally be visualized by agarose gel electrophoresis followed by ethidium bromide staining. Sensitivity can be greatly increased by staining the gel with Sybr green stain or using an Agilent Bioanalyzer or equivalent instrument, if available.
The libraries that we have prepared using this method generally present as a DNA smear starting at ~180 bp and ranging to greater than 1 kb, with peak intensity between ~180 – 500 bp (See Figure 2A). The original publication on ATAC-seq [5] reported a nucleosome-like periodicity in the library size distribution, but we have not observed this phenomenon as assayed by either electrophoresis or estimation of fragment size distribution based on distance between paired-end sequencing reads, as shown in Fig. 2B. This lack of observed nucleosome fractions may be due to size selection of library fragments by Ampure XP beads and the low transposase to nuclei ratio described in this protocol.
Paired-end sequencing is recommended in order to maximize the number of transposase integration events that can be observed in a given sample and to allow measurement of the length of the sequenced fragments (Fig. 2B).
To identify open chromatin regions in Arabidopsis, users should aim to obtain at least 10–20 million reads per library that map to the nuclear genome. For transcription factor footprinting the number of nuclear genome-mapping reads should be increased to at least 100 million per library.
When using sucrose sedimentation for nuclei purification, users should expect ~50% of reads to map to the nuclear genome, while the use of INTACT purification will increase this number to > 90%.
Sequencing reads are checked for overall quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) or equivalent. The reads are aligned to the TAIR10 Arabidopsis thaliana genome (https://www.arabidopsis.org/download/index-auto.jsp?dir=%2Fdownload_files%2FGenes%2FTAIR10_genome_release) using Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). The resulting SAM file is converted to a binary BAM file, which is sorted and indexed using Samtools (http://samtools.sourceforge.net/). The quality of the resulting BAM file, including fragment size distribution, is analyzed using Picard Tools (https://broadinstitute.github.io/picard/). Alignment data is visualized using the Integrated Genome Viewer (http://software.broadinstitute.org/software/igv/). For ease of visualization, BAM files were converted to BigWig files using DeepTools BamPECoverage tool (http://deeptools.readthedocs.io/en/latest/index.html). Downstream analyses of ATAC-seq data include calling peaks with HOMER (http://homer.salk.edu/homer/index.html), editing BED files with bedtools (http://bedtools.readthedocs.io/en/latest/) and identifying transcription factor footprints using pyDNase (http://pythonhosted.org/pyDNase/).
References
- 1.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ken Z. Micrococcal Nuclease Analysis of Chromatin Structure. Current Protocols in Molecular Biology. 2005;21(1):1–17. doi: 10.1002/0471142727.mb2101s69. [DOI] [PubMed] [Google Scholar]
- 4.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29 1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gendrel A, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nature Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- 8.Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Deal RB. Epigenome Profiling of Specific Plant Cell Types Using a StreamLined INTACT Protocol and ChIP-seq. Mehods Mol Biol. 2015;1284:3–25. doi: 10.1007/978-1-4939-2444-8_1. [DOI] [PubMed] [Google Scholar]