Abstract
Inactivation of the adenomatous polyposis coli (APC) tumor suppressor is frequently found in colorectal cancer. Loss of APC function results in deregulation of the Wnt/β-catenin signaling pathway causing overexpression of the c-MYC oncogene. In lymphoma, both p19ARF and ribosomal proteins RPL11 and RPL5 respond to c-MYC activation to induce p53. Their role in c-MYC-driven colorectal carcinogenesis is unclear, as p19ARF deletion does not accelerate APC loss-triggered intestinal tumorigenesis. To determine the contribution of the RP-MDM2-p53 pathway to APC loss-induced tumorigenesis, we crossed mice bearing MDM2C305F mutation, which disrupts RPL11- and RPL5-MDM2 binding, with Apcmin/+ mice, which are prone to intestinal tumor formation. Interestingly, loss of RP-MDM2 binding significantly accelerated colorectal tumor formation while having no discernable effect on small intestinal tumor formation. Mechanistically, APC loss leads to overexpression of c-MYC, RPL11 and RPL5 in mouse colonic tumor cells irrespective of MDM2C305F mutation. However, notable p53 stabilization and activation were observed only in Apcmin/+;Mdm2+/+ but not Apcmin/+;Mdm2C305F/C305F colon tumors. These data establish that the RP-MDM2-p53 pathway, in contrast to the p19ARF-MDM2-p53 pathway, is a critical mediator of colorectal tumorigenesis following APC loss.
Keywords: MDM2, p53, APC, RPL11, c-MYC
Introduction
Colorectal cancer is the third most common cancer and the third leading cause of cancer-associated death worldwide20, 22. Adenomatous polyposis coli (APC) loss or inactivation is sufficient to induce colorectal tumorigenesis, and inactivation of APC through mutation occurs in an estimated 80% of adenomatous polyposis2, 14, 25, 26, 37. APC loss is also the cause of familial adenomatous polyposis (FAP)14. This striking importance of APC in colorectal cancer has led APC to be named the “gatekeeper” of colonic carcinogenesis26.
It has been established that a major function of APC is to degrade cytosolic β-catenin, thus preventing the formation of the nuclear β-catenin/T-cell factor-4 (TCF4) transcriptional complex and switching off Wnt signaling pathway transduction33. The proto-oncogene c-MYC has been identified as a target of the Wnt signaling pathway16, 40, 48. In addition, Sansom et al. discovered that deletion of c-MYC diminishes the tumorigenic capacity of APC deficiency in the small intestine, establishing c-MYC as a critical mediator of intestinal neoplasia following APC loss41. As a transcription factor, the pro-tumorigenic functions of c-MYC have been attributed to its ability to regulate a wide spectrum of gene expression programs6, 7, 39. c-MYC target genes are believed to promote cell proliferation, tumorigenesis, and cell transformation5, 21, 24, 43, 45, 50. Notably, elevated expression of c-MYC can also induce apoptosis18. The biological meaning of this function is not yet fully understood, but is thought to result from a protective mechanism to counteract the effects of oncogenic activation and avoid propagation of transformed cells.
Mutations in p53, known as a “the guardian of the genome” and an overarching tumor suppressor, are the second most common genomic alteration in colorectal cancers2. Loss of p53 activation has been shown to be important in the progression of c-MYC-driven cancers. One recent study showed that p53-inactivating mutations are a leading cause of the relapse of MYC-driven medulloblastoma, and restoration of p53 activity reduces tumor growth and prolongs survival17. On its own, c-MYC overexpression in B cells induces lymphomagenesis1, and in c-MYC-driven lymphomas, p53 deletion significantly accelerates tumor growth44. The role of p53 in APC loss-driven cancers is less clear, but some reports have shown that p53 loss can enhance the incidence and invasiveness of tumors in Apcmin/+ mice15. Nevertheless, the intricate interactions between c-MYC and p53 have yet to be clarified in the context of APC loss-driven tumor environments.
The regulation of p53 is complex, but it is well established that murine double minute 2 (MDM2) is the primary negative regulator of p53. MDM2 regulates p53 both by binding and inhibiting the transactivation domain of p53 and also serving as an E3 ubiquitin ligase for p53, causing its proteosomal degradation19, 38. Upon various cellular stresses, transducer proteins bind and inhibit MDM2, thereby stabilizing p53.
c-MYC expression causes p53 stabilization and activation through two primary pathways. c-MYC induces p19ARF, which binds and inhibits MDM2 3, 51, 52. Additionally, c-MYC serves as a regulator of ribosomal biogenesis. When active, c-MYC induces the transcription of ribosomal proteins (RPs), which also bind and inhibit MDM2 23, 29, 30. This results in dual p19ARF-MDM2-p53 and RP-MDM2-p53 signaling pathways responsive to c-MYC overexpression. Deletion of p19ARF or disruption of the RP-MDM2-p53 pathway by MDM2C305F mutation causes significant acceleration of c-MYC-driven lymphomas8, 30. In the Eμ-Myc mouse model system these pathways are non-redundant, as concomitant disruption of both further accelerates c-MYC-driven lymphoma progression31.
Although the deletion of c-MYC diminishes APC loss-driven intestinal tumorigenicity41, the elimination of p19ARF does not increase intestinal tumor formation in ApcMin/+ mice13, indicating that p19ARF signaling is not essential in this situation. Whether or not the RP-MDM2-p53 response pathway is important in preventing APC loss-induced tumor formation has not been explored. In this work, we sought to determine whether the RP-MDM2-p53 pathway could respond to APC loss-induced c-MYC upregulation to induce p53. To test the importance of this pathway, we crossed ApcMin/+ mice with mice bearing an MDM2C305F mutation, which disrupts the binding of RPL11 and RPL5 to MDM2, and analyzed intestinal tumorigenesis.
Results
MDM2C305F mutation has no discernable effect on APC loss-induced small intestinal tumors
Since the loss of APC induces c-MYC expression through the Wnt signaling pathway16, and the RP-MDM2-p53 pathway has been shown to be important in preventing c-MYC-induced lymphomagenesis30, we sought to determine whether this pathway could also contribute to APC loss-induced tumor prevention. To this end, we crossed ApcMin/+ mice, which are prone to developing intestinal tumors, with Mdm2C305F/C305F (hereafter known as Mdm2m/m) mice, thus disrupting the RP-MDM2-p53 pathway.
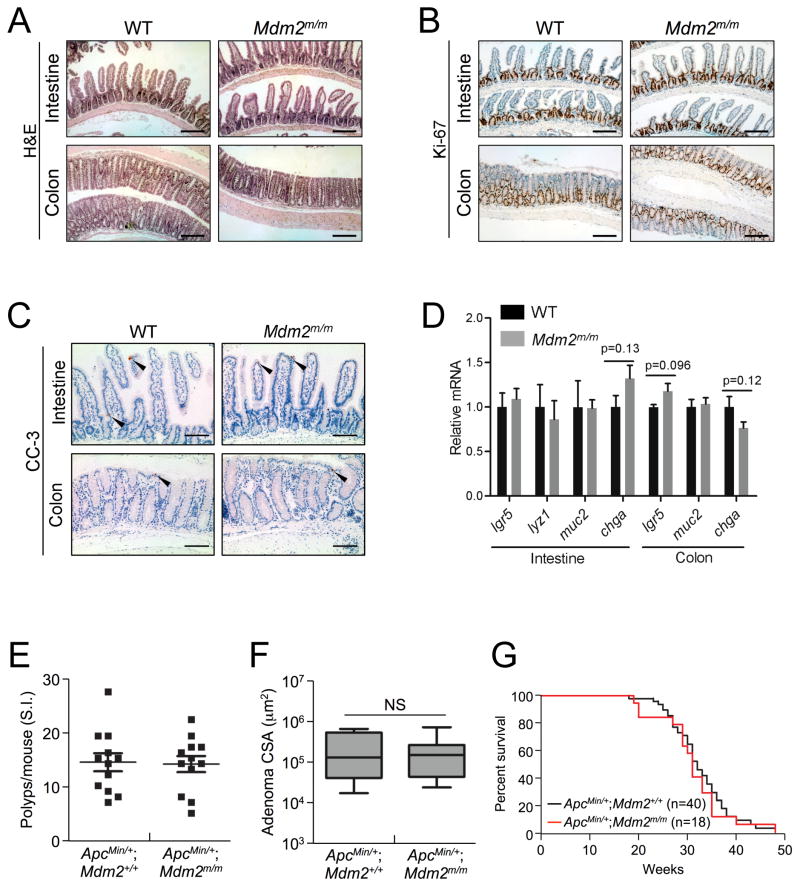
Because Mdm2m/m mice have been shown to have a normal lifespan30, we wanted to first examine normal intestinal tissues from wild type (WT) and Mdm2m/m mice to ascertain whether the MDM2C305F mutation affects normal intestinal homeostasis. We harvested small intestine and colon tissue from eight month-old mice and performed both hematoxylin and eosin (H&E) staining and immunohistochemical (IHC) staining to detect Ki-67 and cleaved caspase-3 (CC-3). Mdm2m/m mice have a normal intestinal epithelium compared to WT mice, with normal appearance of villi and crypts (Figure 1A). In addition, the Ki-67 and CC-3 staining of the intestinal epithelium did not differ between WT and Mdm2m/m mice (Figures 1B–C), indicating a similar intestinal proliferative and apoptotic rate between the two genotypes. Furthermore, we examined the expression of several intestinal cell markers, including an intestinal stem cell marker (lgr5), Paneth cell marker (lyz1), Goblet cell marker (muc2) and enteroendocrine cell marker (chga) by quantitative real time PCR (qRT-PCR)42. No significant differences in the expression of any of these markers were detected between WT and Mdm2m/m mice in either the small intestine or the colon (Figure 1D).
Figure 1. MDM2C305Fmutation has no discernable effect on APC loss-induced small intestinal tumors.
A. Hematoxylin and eosin (H&E) staining of small intestine and colon isolated from eight month-old WT and Mdm2m/m mice. Scale bar = 200μm.
B. Ki-67 immunohistochemical (IHC) staining of proliferating cells in small intestine and colon isolated from eight month-old mice. Proliferating cells are located at the bottom of crypts in both the small intestine and colon. Scale bar = 200μm.
C. IHC staining of cleaved caspase-3 (CC-3) to probe for apoptotic cells (indicated by arrows) in small intestine and colon tissue isolated from eight month-old mice. Scale bar = 100μm.
D. mRNA expression of lgr5, lyz1, muc2 and chga in small intestine or colon from eight month-old WT or Mdm2m/m mice was analyzed by qRT-PCR. n=3 for each genotype. Error bars, ±SEM.
E. Small intestines were isolated from 15 week-old mice, and polyp numbers per mouse were counted under a dissection microscope (n=12 for each genotype).
F. The average cross sectional areas (CSA) of H&E stained small intestinal adenomas were determined quantitatively using ImageJ.
G. Kaplan-Meier survival curves for ApcMin/+; Mdm2+/+ (n=40) and ApcMin/+; Mdm2m/m (n=18) mice are shown. The median survival times did not differ significantly between the two genotypes.
We next compared APC loss-induced tumor initiation in these mice by analyzing small intestinal polyp development in 15 week-old ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m littermates. Upon gross examination of the small intestine, we did not observe a difference in the incidence of small intestinal polyps in the presence of the MDM2C305F mutation (Figure 1E). We performed histological analysis of the small intestinal tumor tissue and found that the average size of the tumors also did not differ between ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m mice (Figure 1F). In agreement with this observation, between the two genotypes there was no significant difference in overall lifespan (Figure 1G). Hence, the MDM2C305F mutation does not affect APC loss-induced small intestinal tumorigenesis, which is a major contributor to mortality in ApcMin/+ mice.
MDM2C305F mutation accelerates APC loss-induced colorectal cancer
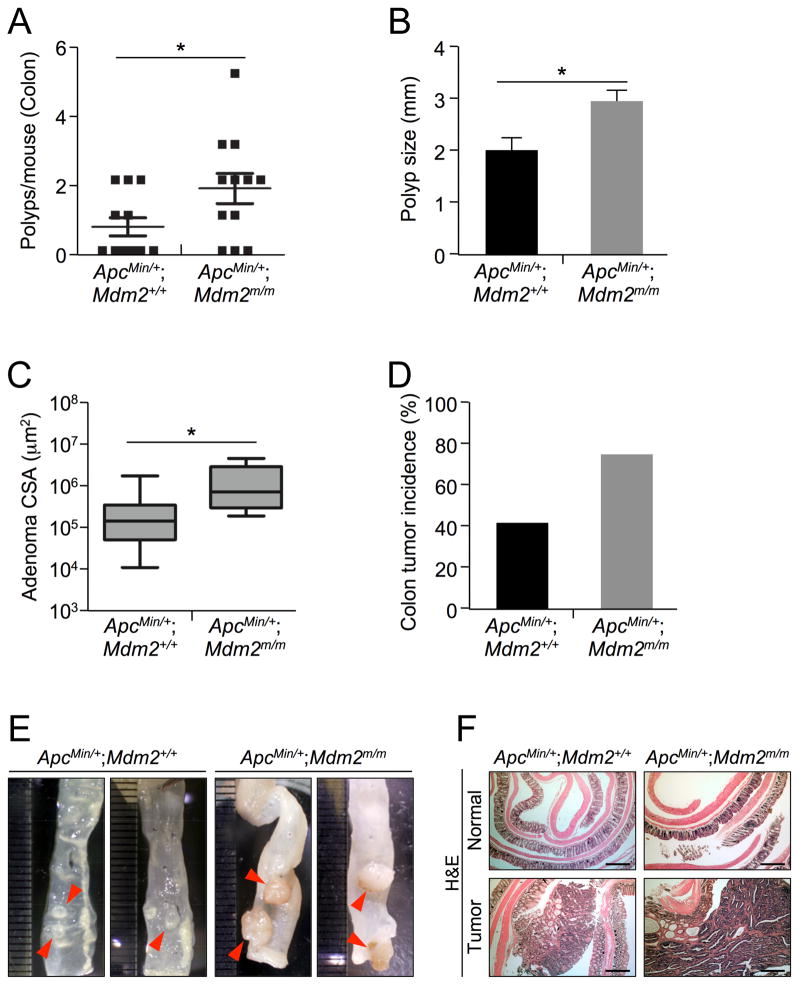
Although the overall survival and small intestinal tumor formation of ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m mice did not differ significantly, we noticed an increased frequency of rectal prolapse and bleeding in the ApcMin/+;Mdm2m/m mice. More than 30% of ApcMin/+;Mdm2m/m mice presented with rectal prolapse and bleeding compared to less than 10% of ApcMin/+;Mdm2+/+ mice. Rectal prolapse and bleeding are indicative of increased colonic tumor formation, so we examined colonic polyps in ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m mice. We observed significantly more polyps in the colon of ApcMin/+;Mdm2m/m mice than in their ApcMin/+;Mdm2+/+ counterparts (on average 0.66 polyps per mouse in ApcMin/+;Mdm2+/+ vs. 1.75 per mouse in ApcMin/+;Mdm2m/m) (Figure 2A), further suggesting increased colon tumor initiation. After histopathological examination, we determined that the average size of these polyps was significantly larger in ApcMin/+;Mdm2m/m mice (2.00 mm in ApcMin/+;Mdm2+/+ vs. 2.95 mm in ApcMin/+;Mdm2m/m) (Figure 2B). Furthermore, the average size of the adenomas found in the tumor bearing colons was also larger in ApcMin/+;Mdm2m/m mice (Figure 2C). Although both ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m mice developed small intestinal tumors with almost 100% penetrance, on average only 40% of ApcMin/+;Mdm2+/+ mice developed colon adenocarcinomas, compared to almost 75% of ApcMin/+;Mdm2m/m mice (Figure 2D). The sizes of colon polyps in ApcMin/+;Mdm2m/m mice were variable, but there were many more large polyps (>4 mm) than in ApcMin/+;Mdm2+/+ mice (Figures 2E). Although the architecture of the normal colonic epithelium in ApcMin/+;Mdm2m/m mice was similar to that of ApcMin/+;Mdm2+/+ mice, the architecture of the colon adenocarcinomas in the ApcMin/+;Mdm2m/m mice showed higher grade complex glandular structures than the colon adenocarcinomas from ApcMin/+;Mdm2+/+ mice of same age (Figure 2F). Taken together, these evidences suggest that the disruption of the RP-MDM2-p53 pathway by MDM2C305F mutation accelerates APC loss-driven colonic tumor initiation and growth.
Figure 2. MDM2C305F mutation increases prevalence of APC loss-induced colon cancer.
A. Colon tissue was isolated from 15 week-old mice, and polyp numbers per mouse were counted under a dissection microscope. Error bars, ±SEM; *P<0.05.
B. The average size of colonic polyps in each genotype (n=12 mice). Apcmin/+; Mdm2m/m mice had significantly more large (>4mm) polyps. Error bars, ±SEM; *P<0.05.
C. The average cross sectional areas (CSA) of H&E stained colon adenomas were determined quantitatively using ImageJ. Error bars, ±SEM; *P<0.05.
D. The incidence of colon tumors in each genotype (n=12), measured by percentage of mice with observed colon tumors.
E. Pictures show the colonic polyps from Apcmin/+; Mdm2+/+ or Apcmin/+; Mdm2m/m mice under a dissection microscope. Adenomas from Apcmin/+; Mdm2m/m mice appeared significantly larger.
F. Representative H&E stained sections from Apcmin/+; Mdm2+/+ or Apcmin/+; Mdm2m/m mouse colon (Normal) and colon adenoma (Tumor) tissue. Scale bar = 500μm.
MDM2C305F mutation promotes proliferation and inhibits apoptosis in APC loss-induced colon cancers
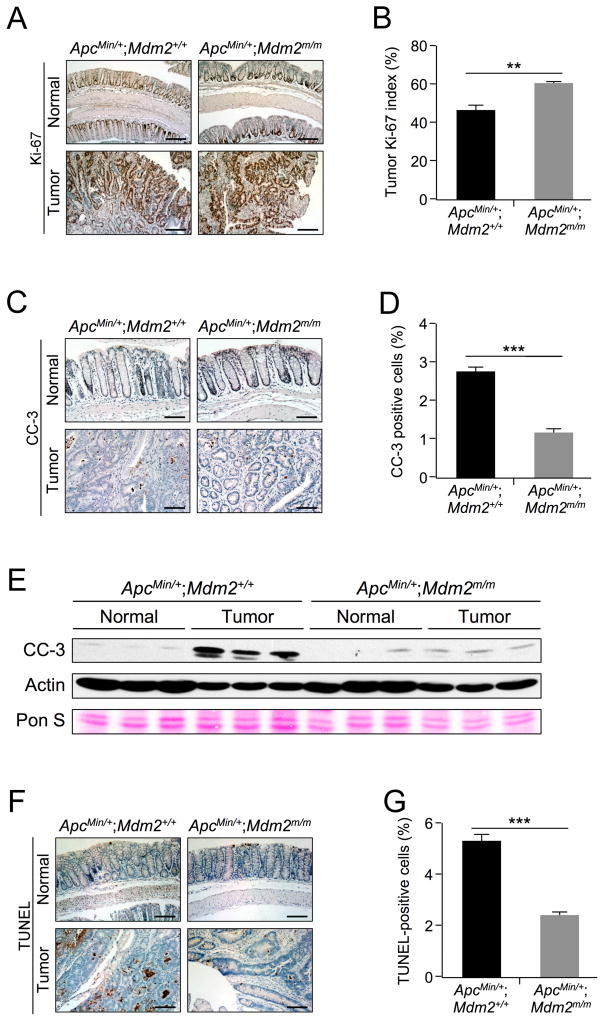
We sought to further characterize the colon tumors. To this end, we first performed Ki-67 staining in normal and tumor samples (Figure 3A–B). We observed significantly more cells with positive Ki-67 staining in ApcMin/+;Mdm2m/m tumors compared to their WT MDM2 counterparts (46% in ApcMin/+;Mdm2+/+ vs. 60% in ApcMin/+;Mdm2m/m), indicating increased proliferation upon MDM2C305F mutation. We also performed IHC staining to detect CC-3 in colon and tumor tissues (Figure 3C–D). We observed a significantly lower percentage of cells with CC-3 staining in ApcMin/+;Mdm2m/m tumors as compared to ApcMin/+;Mdm2+/+ tumors (2.7% of cells in ApcMin/+;Mdm2+/+ vs. 1.1% in ApcMin/+;Mdm2m/m), indicating a decreased percentage of cells undergoing apoptosis. Next, we detected average overall CC-3 levels in these tissues by western blot (Figure 3E), and observed a similarly muted CC-3 signal in ApcMin/+;Mdm2m/m tumors. Because of the difference in average actin levels between normal colon and tumor tissues, we included Ponceau S staining (Pon S) as an additional loading control. To confirm the CC-3 results, we performed terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays (Figure 3F–G) and observed a significantly decreased TUNEL signal in ApcMin/+;Mdm2m/m tumors (5.3% of cells in ApcMin/+;Mdm2+/+ vs. 2.4% in ApcMin/+;Mdm2m/m). This result is consistent with decreased levels of apoptosis in these cells. Together, these results indicate that the MDM2C305F mutation promotes proliferation and inhibits apoptosis in colon cancers induced by APC loss.
Figure 3. MDM2C305F mutation promotes proliferation and inhibits apoptosis in APC loss-induced colon cancers.
A. Ki-67 immunohistochemical staining of proliferating cells in colon (Normal) and colonic adenoma (Tumor) tissue isolated from 15 week-old mice. In normal colon, the proliferating cells are located at the bottom of crypts, whereas tumor cells are highly proliferative. Scale bar = 200μm.
B. Percentage of Ki-67-positive cells in colonic adenomas was calculated quantitatively from five images using ImageJ. Error bars, ±SEM; **P<0.01.
C. Apoptosis was measured by IHC staining of cleaved caspase-3 (CC-3) in colon (Normal) and colonic adenoma (Tumor) tissue. Scale bar = 100μm.
D. Percentage of CC-3 positive cells in colonic adenomas was calculated quantitatively from five images using ImageJ. Error bars, ±SEM; ***P<0.001.
E. Western blotting was performed with protein lysates isolated from colonic polyps (Tumor) or adjacent normal colon tissues (Normal). n=3 for each group. Pon S = Ponceau S staining.
F. Apoptosis was measured by TUNEL staining in colon (Normal) and colonic adenoma (Tumor) tissue. In normal colon, apoptotic cells are located at the tips of the villi. Scale bar = 125μm.
G. Percentage of TUNEL positive cells from colonic adenomas was calculated quantitatively from five images using ImageJ. Error bars, ±SEM; ***P<0.001.
APC loss-induced colon cancers express high levels of c-MYC and RPL11
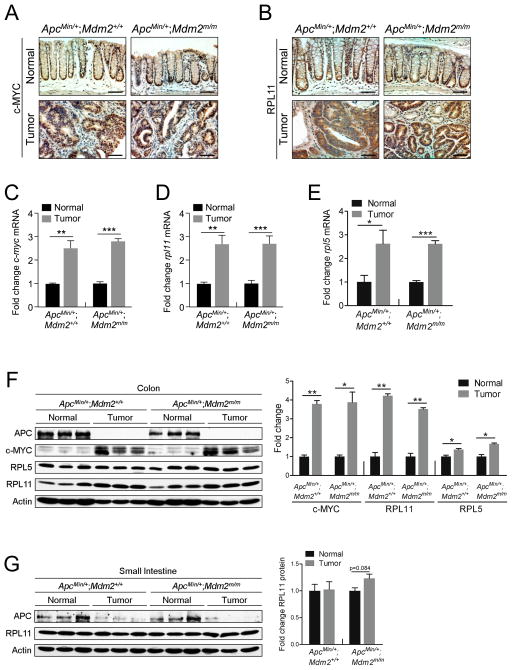
Because the MDM2C305F mutation disrupts the binding of RPL11 and RPL5 to MDM227, 30, and RPL11 and RPL5 are transcriptional targets of c-MYC4, 32, we analyzed c-MYC, RPL11 and RPL5 levels to determine if the MDM2C305F mutation affects APC loss-induced c-MYC signaling. First, we performed IHC staining for c-MYC in normal and tumor tissues (Figure 4A). As expected, there was an increase in c-MYC detected in tumor tissue compared to normal tissue and there was no difference in c-MYC levels between the two genotypes. This pattern was also observed in IHC staining for RPL11, a downstream target of c-MYC (Figure 4B). In order to evaluate the expression of c-MYC, RPL11 and RPL5 more precisely, we performed qRT-PCR and western blotting using samples from normal colon or colonic adenomas. Consistent with IHC staining, we observed an approximately 2.5-fold increase in c-myc, rpl11 and rpl5 mRNAs in tumor samples of either genotype (Figure 4C–E).
Figure 4. APC loss-induced colon cancers express high levels of c-MYC and RPL11.
A. IHC staining was performed in colon (Normal) and colonic adenomas (Tumor) to detect the expression of c-MYC. Scale bar = 75μm.
B. IHC staining was performed in colon (Normal) and colonic adenomas (Tumor) to detect the expression of RPL11. Scale bar = 75μm.
C. qPCR analysis of mRNA expression of c-myc in colon (Normal) or colonic adenomas (Tumor) from Apcmin/+; Mdm2+/+ and Apcmin/+; Mdm2m/m mice. n=3 for each genotype. Error bars, ±SEM; **P<0.01, ***P<0.001.
D. qPCR analysis of mRNA expression of rpl11 in colon (Normal) or colonic adenomas (Tumor) from Apcmin/+; Mdm2+/+ and Apcmin/+; Mdm2m/m mice. n=3 for each genotype. Error bars, ±SEM; **P<0.01, ***P<0.001.
E. qPCR analysis of mRNA expression of rpl5 in colon (Normal) or colonic adenomas (Tumor) from Apcmin/+; Mdm2+/+ and Apcmin/+; Mdm2m/m mice. n=3 for each genotype. Error bars, ±SEM; *P<0.05, ***P<0.001.
F. Western blotting analysis of protein lysates isolated from colon (Normal) or adjacent colonic adenomas (Tumor) with three mice per genotype. APC expression was undetectable in colonic adenomas, implying the loss of both wild type alleles. The bar graph illustrates the quantification of the western blot. Error bars, ±SEM; *P<0.05, **P<0.01.
G. Western blotting analysis of protein lysates isolated from small intestine (Normal) or adjacent adenomas (Tumor) with three mice per genotype. c-MYC expression was undetectable in small intestinal tissue. The bar graph illustrates the quantification of the western blot. Error bars, ±SEM.
By western blot, we were able to confirm the loss of APC in tumor tissue of both ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m mice, as well as observe a comparable increase in c-MYC, RPL11 and RPL5 abundance (Figure 4F). These results indicate that the MDM2C305F mutation does not ultimately affect the expression of upstream c-MYC, RPL11 and RPL5 signaling in colon cancers and that APC loss still induces relatively high levels of these proteins independent of MDM2 mutational status.
To investigate the apparent discrepancy in tumor formation after MDM2C305F mutation between small intestine (Figure 1) and colon (Figure 2), we also analyzed c-MYC and RPL11 levels in normal small intestine and small intestinal adenoma tissues (Figure 4G). Although we were unable to detect c-MYC in these tissues, we found that RPL11 levels were unchanged after small intestinal tumor formation, in contrast to what we observed in colon tumors. This provides a potential explanation for the disparity of tumor formation rates in colon and small intestine of ApcMin/+;Mdm2m/m mice compared to ApcMin/+;Mdm2+/+ mice, as there is likely to be little RP-MDM2-p53 signaling action in small intestinal tissues. Although c-MYC has been shown to be an oncogene critical in mediating colorectal tumorigenesis following APC deletion, in our hands the role of RPL11 in mediating c-MYC-induced tumorigenesis appears to be colon tissue specific.
MDM2C305F mutation attenuates p53 activation in colon tumors
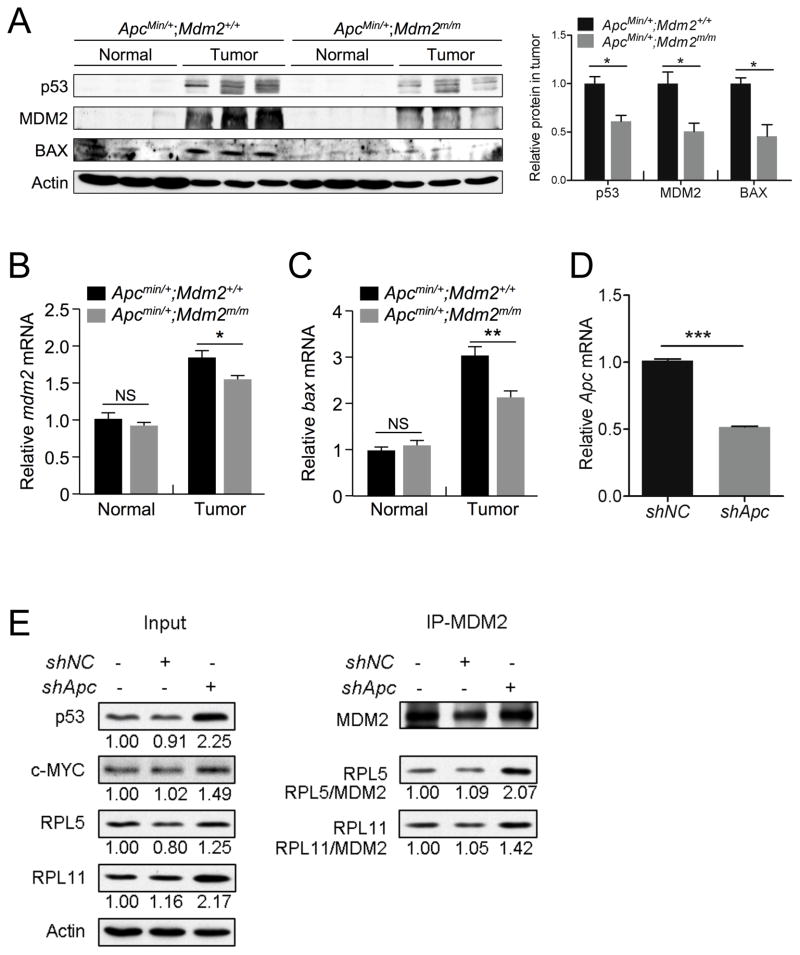
Because the MDM2C305F mutation disrupts the binding of RPL11 and RPL5 to MDM2, thus abrogating ribosomal stress-mediated p53 induction, we wanted to analyze p53 levels and activity in ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m mouse colon tumors to determine whether p53 induction correlated with the observed increase in proliferation and decrease in apoptosis in ApcMin/+;Mdm2m/m tumors. We found that in ApcMin/+;Mdm2m/m mouse colon tumors, p53 protein levels were notably lower than in tumors from ApcMin/+;Mdm2+/+ mice (Figure 5A). The protein levels of BAX, a p53 target important for induction of apoptosis47, and of MDM2, also a p53 target, followed the same trend. We also compared p53 transcriptional activation in these tissues. In normal tissues, we observed no significant difference between the relative levels of mdm2 and bax mRNA; however in tumor tissues, there was significantly less transcription of mdm2 and bax (Figure 5B–C) in ApcMin/+;Mdm2m/m mice compared to ApcMin/+;Mdm2+/+ mice, indicating attenuated p53 induction upon MDM2C305F mutation.
Figure 5. MDM2C305F mutation attenuates p53 activation in APC loss-induced colon cancers.
A. Western blotting analysis of protein lysates isolated from colon (Normal) or adjacent colonic adenomas (Tumor) with three mice per genotype. p53 and MDM2 were undetectable in normal colon. The bar graph illustrates the relative protein levels in tumor samples from Apcmin/+;Mdm2+/+ and Apcmin/+;Mdm2m/m mice. Error bars, ±SEM; *P<0.05.
B. qPCR analysis of relative mRNA expression of mdm2 in colon (Normal) or colonic adenomas (Tumor) from Apcmin/+;Mdm2+/+ and Apcmin/+;Mdm2m/m mice. n=3 for each genotype. Error bars, ±SEM; NS=not significant, *P<0.05.
C. qPCR analysis of relative mRNA expression of bax in colon (Normal) or colonic adenomas (Tumor) from Apcmin/+;Mdm2+/+ and Apcmin/+;Mdm2m/m mice. n=3 for each genotype. Error bars, ±SEM; NS=not significant, **P<0.01.
D. qPCR analysis of relative mRNA expression of Apc in HCT116 cells infected with lentivirus containing either a negative control scrambled shRNA (shNC) or Apc shRNA (shApc). Error bars, ±SEM; ***P<0.001.
E. HCT116 cells were infected with lentivirus containing shRNA targeting either Apc (shApc) or a scrambled control (shNC). Following selection with puromycin, immunoprecipitation (IP) of MDM2 was performed, followed by western blotting with the indicated antibodies. Quantification of protein expression was performed using ImageJ, and the relative protein expression is indicated under the blots. The amount of RPL11 or RPL5 immunoprecipitated (relative to MDM2 IP protein levels) is also indicated under each IP blot.
To more precisely determine whether APC loss will increase signaling to the RP-MDM2-p53 pathway, we knocked down Apc expression in the colonic tumor cell line HCT116, which has wild type p53. We infected HCT116 cells with a lentivirus containing shRNA targeting Apc, and after puromycin selection we confirmed a decrease in Apc expression by qRT-PCR (Figure 5D). As expected, knockdown of Apc induced the expression of c-MYC, RPL11 and RPL5 (Figure 5E). Additionally, p53 was stabilized. We next performed immunoprecipitation (IP) of MDM2 and probed for RPL11 and RPL5 binding. After Apc knockdown, there was a clear increase in both RPL11-MDM2 and RPL5-MDM2 binding, suggesting that APC loss triggers RP-MDM2 interaction to activate p53 in colon tumors (Figure 5E). Taken together, these results collectively suggest that the RP-MDM2-p53 pathway is important for the prevention of APC loss-induced colonic tumors.
Discussion
The roles of APC deletion and p53 inactivation in intestinal tumorigenesis are not completely understood. Although p53 is widely known as an overarching tumor suppressor protein, APC has been established as the most important intestinal tumor suppressor. Large scale sequencing of intestinal tumors has indicated that APC and p53 are the most frequently mutated genes at 81% and 60% of these tumors, respectively2.
In addition, several mouse models have established that upon Wnt signaling pathway activation, the major consequence of APC loss, p53 plays critical tumor suppressive functions9, 46. It is possible that activation of the Wnt signaling target c-MYC drives growth and proliferation, stimulating p53 to perform its tumor suppressive functions but at the same time pressuring its inactivation, allowing for tumor maintenance. Here, we confirm that p53 is indeed induced by APC deletion (Figure 5). While inactivating mutations of APC are considered to be a first step for colonic carcinogenesis, p53 is thought to act as a final barrier to carcinoma formation26. Our results are consistent with this notion.
We also demonstrate that inactivation of the c-MYC responsive RP-MDM2-p53 pathway through MDM2C305F mutation allows for increased APC loss-driven colon tumorigenesis. ApcMin/+;Mdm2m/m mice display increased tumor size and incidence compared to their ApcMin/+;Mdm2+/+ counterparts (Figure 2), which correlates with decreased p53 protein abundance and activation (Figure 5).
The importance of p19ARF- and RP-dependent p53 activation is tissue specific
This work and others also clarify that there are distinct and tissue-specific roles for the RP-MDM2-p53 and p19ARF-MDM2-p53 pathways in tumorigenesis. Although each of these pathways have been shown to be independently critical for the prevention of c-MYC-driven lymphoma31, the role of these pathways in APC loss-driven, c-MYC-dependent tumorigenesis is more ambiguous. It has been previously established that p19ARF loss does not accelerate or promote intestinal tumors upon APC loss13, but our work indicates that loss of RP-MDM2 interaction can sensitize mice to APC loss-driven colonic tumorigenesis (Figure 2). In this study we observed no difference in survival between ApcMin/+;Mdm2+/+ and ApcMin/+;Mdm2m/m mice; however, in both genotypes there were many more tumors in the small intestine than colon, which is consistent with previous reports34, 35. Observed differences in colon tumor formation depending on MDM2 mutational status may not have an effect on survival because of the relatively higher tumor burden in the small intestine compared to the colon. In future studies, it would be informative to cross MDM2C305F mice with mice expressing a colon tissue specific Apc deletion, such as the CDX2P-CreERT2 Apcflox/flox model10, to determine whether differential RP-MDM2-p53 pathway activation could contribute to a difference in tumor burden and overall lifespan.
Because RPL11 is a primary ribosomal protein responder from c-MYC to p5311, 12, disruption of RPL11-MDM2 binding by MDM2C305F mutation is likely to have little effect on p53 activation if RPL11 protein abundance does not change. It appears from our data that c-MYC signaling to RPL11 is much more prominent in colon tissue than in small intestine (Figure 4F–G), but the reason for this is presently unclear. Although this study demonstrates that there are tissue-specific roles for each of the RP-MDM2-p53 and p19ARF-MDM2-p53 signaling pathways, it also raises questions about why p19ARF signaling is less important in APC loss-driven c-MYC activation.
One possible reason for the difference in activation of these two pathways during colonic tumorigenesis could be due to a difference in threshold of c-MYC response. As a global transcriptional amplifier28, c-MYC can trigger different programs depending on certain levels of expression. Both in vitro and in vivo studies have demonstrated that p19ARF induction requires a high level of c-MYC expression3, 36. On the other hand, c-MYC serves as a direct regulator of ribosomal biogenesis via the transcriptional control of RNA and protein components of ribosomes, the gene products required for the processing of ribosomal RNA, the nuclear export of ribosomal subunits, and the initiation of mRNA translation49. It is possible that RP-MDM2-p53 activation requires a lower level of c-MYC activation than that of the p19ARF-MDM2-p53 pathway. In mouse model systems where c-MYC is directly overexpressed (e.g. Eμ-Myc), rather than upregulated by a change in upstream signaling (e.g. Apcmin/+), p19ARF is critical in prevention of tumorigenesis8. The presence of the RP-MDM2-p53 pathway could explain why deletion of p19ARF does not sensitize mice to APC deletion-induced intestinal tumorigenesis.
In this study, we demonstrated that RPL11 and RPL5 are upregulated by the APC-MYC axis, correlating with p53 induction and apoptosis. We postulate that the RP-MDM2-p53 pathway is a fail-safe mechanism for c-MYC-dependent tumorigenesis because it can be executed directly after c-MYC activation. It is clear from the ApcMin/+ mouse model and others that the interplay of these pathways is tissue specific, and more work will need to be done to establish the biological and contextual significance for these tumor suppressive pathways.
Materials and Methods
Ethics Statement
This investigation has been conducted in accordance with ethical standards, the Declaration of Helsinki, national and international guidelines, and has been approved by the authors’ institutional review board.
Mouse experiments
All mice were bred and maintained on a 12 hour light and dark cycle. Mdm2C305F mutant mice were generated as previously described30. Apcmin mice on C57BL/6J background were purchased from The Jackson Laboratory (Stock Number J002020). All mice were handled in strict accordance with protocol (10-045) approved by the Institutional Animal Care and Use Committee at The University of North Carolina at Chapel Hill.
Histological analysis
Intestines were dissected and flushed gently with cold phosphate buffered saline (PBS) and rolled into a compact circle. They were then fixed in 10% formalin overnight, dehydrated in 50% ethanol and stored in 70% ethanol until they were transferred to the Histology Research Core Facility at the University of North Carolina at Chapel Hill (UNC) for paraffin embedding. Sections (4 μm) were cut and then processed for hematoxylin and eosin (H&E) staining or for immunohistochemical (IHC) staining.
For IHC staining, sections were deparaffinized in SafeClear II (Fisher Scientific) and rehydrated in gradient alcohols (100%, 95%, 85%, 70%, H2O). Antigen retrieval was performed by boiling slides in 10mM sodium citrate buffer, pH 6.0. Endogenous peroxidase activity was quenched with 3% H2O2 in methanol for 15 minutes. To develop the staining, we used VECTASTAIN Elite ABC kit (PK6100, Vector Laboratories) following the manufacturer’s instructions. Slides were counterstained with Harris’ hematoxylin (Sigma) and then dehydrated and mounted in Permount (Fisher Scientific). The following primary antibodies were used: rabbit monoclonal anti Ki-67 (NeoMarkers, #RM-9106), rabbit polyclonal anti cleaved-Caspase3 (Cell Signaling, #9661), rabbit polyclonal anti c-MYC (Santa Cruz, N262), and rabbit polyclonal anti RPL11 (homemade). All primary antibodies were incubated overnight.
For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays, sections were deparaffinized following the IHC protocol, and then stained with the Apoptosis Detection Kit (S7100, Millipore) following the manufacturer’s instructions.
Quantitative real time PCR
Total RNA was prepared from mouse tissues using Trizol® Reagent (Invitrogen, #15596-026). RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Scientific, NanoDrop™ 2000c) and quality was assessed by agarose gel electrophoresis. cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen, 18080-051). qRT-PCR was performed with SYBR Green probes using the Applied Biosystems 7900HT Fast Real-Time PCR system. Thermal cycling conditions were 50°C for 2 minutes, 95°C for 5 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Target gene transcript levels were normalized to β-actin transcript levels obtained in each sample via the subtraction of the Ct value of β-actin from the Ct value for each target gene. Results were expressed as the fold-change in transcript levels. Primers used for qRT-PCR were as follows:
β-Actin, 5′-GGCTGTATTCCCCTCCATCG-3′ and 5′-CCAGTTGGTAACAATGCCATGT-3′;
c-Myc, 5′-TGAGCCCCTAGTGCTGCAT-3′ and 5′-AGCCCGACTCCGACCTCTT-3′;
Rpl11, 5′-CAATATCTGCGTCGGGGAGA-3′ and 5′-TTCCGCAACTCATACTCCCG-3′;
Rpl5, 5′-AGCATTGACGGTCAGCCTGGTG-3′ and 5′-CTGACCCATGATGTGCTTCCGATG-3′;
Bax, 5′-GGACAGCAATATGGAGCTGCAGAGG-3′ and 5′-GGAGGAAGTCCAGTGTCCAGCC-3′;
Mdm2, 5′-TGTGTGAGCTGAGGGAGATG-3′ and 5′-CACTTACGCCATCGTCAAGA-3′.
Lgr5, 5′-CAAGCCATGACCTTGGCCCTG-3′ and 5′-TTTCCCAGGGAGTGGATTCTATT-3′
Lyz1, 5′-GGAATGGATGGCTACCGTGG-3′ and 5′-CATGCCACCCATGCTCGAAT-3′
Muc2, 5′-CGGTTCCAGAACCATACCTG-3′ and 5′-GGTCAGCAGCCTCTCACATT-3′
Chga, 5′-CACAGCAGCTTTGAGGATGA-3′ and 5′-ATGGGGGACTCTTGGTTAGG-3′
Protein analysis
For western blotting, proteins were extracted from tissues as previously described30. Briefly, mouse tissue was homogenized and lysed in 0.5% NP-40 lysis buffer. Proteins were detected by using either Pico or Dura enhanced chemiluminescence (ECL) systems (Thermo Scientific, SuperSignal™ West Dura Substrate). The following primary antibodies were used: rabbit polyclonal anti c-MYC (N262; Santa Cruz), mouse monoclonal anti p53 (NCL-505; Novocastra), mouse monoclonal anti MDM2 (2A10, homemade), rabbit polyclonal anti cleaved-caspase 3 (#9661; Cell Signaling), rabbit polyclonal anti APC (NBP2-15422; Novus Biologicals), rabbit polyclonal anti Bax (554104, BD Biosciences), rabbit polyclonal anti RPL11 and RPL5 (made in house as previously described30).
For protein analysis of HCT116 cells (purchased and authenticated from the UNC Tissue Culture Facility), we homogenized and lysed cells in 0.1% NP40 lysis buffer. For each sample, 2.5 mg of protein extract was incubated with antibodies against MDM2 (4B11) overnight at 4 °C before incubation with protein A beads for 60 minutes. Immunoprecipitates were washed three times in ice-cold lysis buffer, resuspended in SDS loading buffer, and subjected to Western blot analysis.
Lentiviral vector and infection
For shRNA transduction, a pLKO.1-puro vector (Addgene) containing shRNA targeting human Apc (GAAAGTGGAGGTGGGATATTA) and a scrambled control (CAACAAGATGAAGAGCACCAA) were used. We made the viral particles in HEK-293T cells using pspAX2 and pMD2.G plasmids. Twenty-four hours after infection, positively infected HCT116 cells were selected with puromycin. Stable shRNA knockdown cell lines were used for further analysis.
Statistical analysis
Results are represented as mean ± standard error of the mean. Differences in tumor size measurements and tumor numbers were evaluated for significance using the unpaired t test. Quantitative PCR data and immunohistochemistry quantification differences were also evaluated for significance using the unpaired t test. Variances were not significantly different within treatment groups. A p value <0.05 was considered significant for all analyses. Significant differences between experimental groups were: *P < 0.05, **P < 0.01, or ***P < 0.001. Each experiment was performed at least two times. At least three mice per group were used for every experiment. No blinding or randomization were used. Calculations were performed using the GraphPad Prism 5 software.
Acknowledgments
This research was supported by grants from the National Institutes of Health (CA127770, CA 100302 and CA167637) and Natural Science Foundation of China (NSFC) to Y.Z., and the National Institute of General Medical Sciences (5T32 GM007092) to N.R.T.
Footnotes
Conflict of Interest
The authors declare that they have no competing financial interests.
References
- 1.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen D, Kon N, Zhong J, Zhang P, Yu L, Gu W. Differential effects on ARF stability by normal versus oncogenic levels of c-Myc expression. Molecular cell. 2013;51:46–56. doi: 10.1016/j.molcel.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Seminars in cancer biology. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in medicine. 2013:3. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eilers M, Eisenman RN. Myc’s broad reach. Genes & development. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes & development. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elyada E, Pribluda A, Goldstein RE, Morgenstern Y, Brachya G, Cojocaru G, et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470:409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Sentani K, Wiese A, Sands E, Green M, Bommer GT, et al. Sox9 Induction, Ectopic Paneth Cells, and Mitotic Spindle Axis Defects in Mouse Colon Adenomatous Epithelium Arising From Conditional Biallelic Apc Inactivation. The American Journal of Pathology. 2013;183:493–503. doi: 10.1016/j.ajpath.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nature cell biology. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fumagalli S, Ivanenkov VV, Teng T, Thomas G. Suprainduction of p53 by disruption of 40S and 60S ribosome biogenesis leads to the activation of a novel G2/M checkpoint. Genes & development. 2012;26:1028–1040. doi: 10.1101/gad.189951.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson SL, Boquoi A, Chen T, Sharpless NE, Brensinger C, Enders GH. p16(Ink4a) inhibits histologic progression and angiogenic signaling in min colon tumors. Cancer biology & therapy. 2005;4:1389–1394. doi: 10.4161/cbt.4.12.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 15.Halberg RB, Katzung DS, Hoff PD, Moser AR, Cole CE, Lubet RA, et al. Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3461–3466. doi: 10.1073/pnas.050585597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 17.Hill RM, Kuijper S, Lindsey JC, Petrie K, Schwalbe EC, Barker K, et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer cell. 2015;27:72–84. doi: 10.1016/j.ccell.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 19.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS letters. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 20.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA: a cancer journal for clinicians. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 21.Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PloS one. 2011;6:e26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 23.Kim TH, Leslie P, Zhang Y. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget. 2014;5:860–871. doi: 10.18632/oncotarget.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YH, Girard L, Giacomini CP, Wang P, Hernandez-Boussard T, Tibshirani R, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2006;25:130–138. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- 25.Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 26.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 27.Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Molecular and cellular biology. 2007;27:1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Littlewood TD, Kreuzaler P, Evan GI. All things to all people. Cell. 2012;151:11–13. doi: 10.1016/j.cell.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 30.Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng X, Carlson NR, Dong J, Zhang Y. Oncogenic c-Myc-induced lymphomagenesis is inhibited non-redundantly by the p19Arf-Mdm2-p53 and RP-Mdm2-p53 pathways. Oncogene. 2015;34:5709–5717. doi: 10.1038/onc.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 34.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 35.Moser AR, Dove WF, Roth KA, Gordon JI. The Min (multiple intestinal neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. The Journal of cell biology. 1992;116:1517–1526. doi: 10.1083/jcb.116.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, et al. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 38.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor. 1993:53. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 39.Sabo A, Amati B. Genome recognition by MYC. Cold Spring Harbor perspectives in medicine. 2014:4. doi: 10.1101/cshperspect.a014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes & development. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- 42.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 43.Schlosser I, Holzel M, Hoffmann R, Burtscher H, Kohlhuber F, Schuhmacher M, et al. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–524. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- 44.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes & development. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic acids research. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y, et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer cell. 2013;23:93–106. doi: 10.1016/j.ccr.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Toshiyuki M, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 48.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 49.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature reviews Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 50.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome biology. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 52.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & development. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]