Abstract
Opportunistic pathogens inhabiting premise (i.e., building) plumbing (OPPPs, e.g., L. pneumophila, M. avium complex, P. aeruginosa, Acanthamoeba, and N. fowleri) are a significant and growing source of disease. Because OPPPs establish and grow as part of the native drinking water microbiota, they do not correspond to fecal indicators, presenting a significant challenge to common and effective monitoring strategies. Further, different OPPPs present distinct requirements for sampling, preservation, and analysis, creating a significant impediment to their parallel detection. The aim of this critical review is to synthesize the state of the science of monitoring OPPPs and to identify a path forward for their simultaneous detection and quantification in a manner commensurate with the need for reliable data to inform risk assessment and mitigation. Water and biofilm sampling procedures, as well as factors influencing sample representativeness and detection sensitivity, are critically evaluated with respect to the five representative bacterial and amoebal OPPPs noted above. Available culturing and molecular approaches are discussed in terms of their advantages, limitations, and applicability. Knowledge gaps and research needs are identified.
1. Introduction
Premise plumbing refers to the portion of potable water distribution systems beyond the property line and in buildings (NRC, 2006), which includes both hot water and cold water lines and devices such as water heaters, showers, faucets and filters (Figure 1). Premise plumbing is generally characterized as a wet, warm, periodically stagnant environment with low disinfectant residual and high surface area, which creates ideal conditions for microbial growth. Accordingly, opportunistic pathogens can become established in premise plumbing as part of the native microbiota, thereby presenting a major challenge for detection and monitoring since they violate the paradigm of traditional fecal indicator bacteria. As OPPPs are now the primary source of drinking water-related disease outbreaks, particularly in developed countries (CDC, 2013; Anaissie et al., 2002; Craun et al., 2010; Pruden et al., 2013), there is an urgent need for reliable detection and monitoring strategies. In particular, simultaneous monitoring of multiple OPPPs in a manner conducive to risk assessment and mitigation, as well as to support research seeking to advance a fundamental understanding of their behavior in premise plumbing and inform better control measures, would be ideal.
Figure 1.
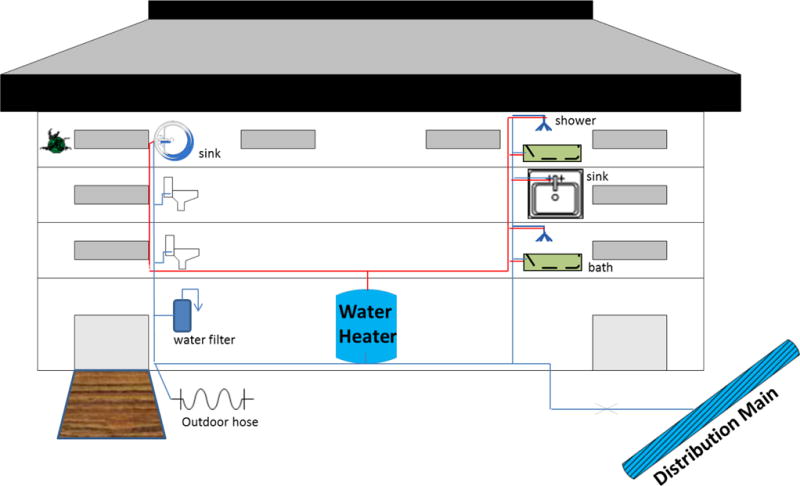
Schematic of premise plumbing and potential sampling sites for OPPPs
Here we focus on a representative cross section of OPPPs, including bacterial species such as Legionella pneumophila, nontuberculous mycobacteria (NTM), Pseudomonas aeruginosa and protozoans such as Acanthamoeba spp. and Naegleria fowleri. Among these, Legionella pneumophila, and other Legionella spp. that cause Legionnaires’ disease, have attracted the most attention as they cause a deadly form of pneumonia and account for the greatest proportion of OPPP-associated outbreak in the U.S., with an increasing incidence (CDC, 2011; Hubbs, 2014). NTM is comprised of several pathogenic strains of mycobacteria that cause chronic pulmonary disease in an estimated 30,000 people in the United States. (Winthrop et al., 2010), first noticed in immunocompromised populations but also affecting groups with other less-obvious risk factors (e.g., slender elderly women) (Falkinham, 2009). P. aeruginosa is especially problematic as an agent of nosocomial (hospital-acquired) infection, particularly neonates and burn patients, causing about 1400 deaths each year in the United States (Anaissie et al., 2002). Acanthamoeba can cause severe eye infections (i.e., keratitis) and granulomatous amebic encephalitis, the former being associated with poor contact lens hygiene (Kilvington et al., 2004). Naegleria fowleri is the only pathogenic species of the genus, known also as the “brain-eating amoeba,” causing the rare but highly fatal disease, primary amebic meningoencephalitis (PAMs) (Yoder et al., 2012). Of these, L. pneumophila, Mycobacterium avium and N. fowleri have been included on the drinking water contaminant candidate list 3 (CCL3) (EPA, 2009). A major impediment to advancing further policy action towards public health protection from OPPPs is the challenge that they pose to monitoring.
There is currently no consensus with respect to methodologies or protocols for monitoring OPPPs, particularly when seeking simultaneous detection. From a policy standpoint, a major obstacle is that existing microbiological monitoring practices focus on the main distribution system and do not extend across the property line, where conditions are particularly susceptible to the multiplication of OPPPs (NRC, 2006). This is problematic because insight is needed into the relationship between the municipal water characteristics and the potential for regrowth of OPPPs, as has recently been illustrated in the case of corrosive water being associated with a Legionnaire’s outbreak in Flint, MI (Schwake et al., 2016). From a practical standpoint, premise plumbing is extremely complex and can differ greatly between buildings, including diverse fixture types and materials, pipe surface to volume ratios, and water temperatures (NRC, 2006). Flow conditions within pluming can also vary dramatically from one point to another depending on the building layout (Inkinen et al., 2014). All these factors result in heterogeneity within and between premise plumbing environments, making it difficult to identify common sampling plans that are representative of the actual contamination by OPPPs and risk for disease transmission. Finally, OPPPs themselves are characterized by complex microbial ecology and physiology, which has led towards highly specific protocols that are not necessarily compatible for monitoring of multiple OPPPs.
This work presents a critical review of various sampling and methodological approaches employed for monitoring OPPPs. Advantages, limitations, and applicability of current monitoring methodologies are discussed in the context of their value and potential for informing prevention and mitigation of risk, outbreak response, and research. Key knowledge gaps and urgent research needs are identified that must be addressed to improve the science and practice of OPPPs monitoring.
2. Sampling Premise Plumbing and Sample Handling
Given the complexity of premise plumbing, key components of sampling considerations include site selection and sampling frequency, sample type (i.e., biofilm, water) and size, and flow patterns (e.g., first-flush, post flush). Moreover, sample handling before analytical testing, such as sample preservation, transport, and pretreatment (e.g., concentration) can also impact the recovery efficiency of targeted OPPPs.
2.1 Site Selection and Sampling Frequency
2.1.1 Site Selection
Currently, there are no generally accepted protocols for choosing sampling sites or frequency for OPPPs (Lucas et al., 2011). The selection of sampling sites for routine monitoring of L. pneumophila in healthcare facilities and in response to outbreaks has been described most extensively (Barbaree et al., 1987; Allegheny County Health Department, 2014; U.K. Department of Health, 2006; FRML, 2010; VHA, 2014). Sampling points are chosen based on vulnerability of certain sectors of the building to pathogen proliferation (e.g., dead-ends or areas with infrequent use) (U.K. Deaprtment of Health, 2006) as well as the susceptibility of the occupants to exposure risks (e.g., frequently occupied areas, intensive care, neonatal, and burn units) (ASHRAE, 2015; FRMHS, 2010). To gain a full systems perspective, recommended sampling sites for Legionella in healthcare facilities have included incoming water mains, water softeners, holding tanks, water heaters (at the inflows and outflows), faucets, and shower heads (Figure 1, CDC, 2015). Importantly, OPPPs commonly prefer warmer water and may be spread via aerosols (e.g., Legionella), rather than ingestion, making hot water systems particularly important monitoring targets. A minimal sample number has been required in some scenarios, e.g., sampling at least 10 outlets on the hot and cold water distribution system respectively for each building was endorsed by the U.S. Veterans’ Health Administration (VHA, 2014). Another guideline published by U.K. Department of Health suggests that samples should at least be taken from the cold water storage, the most distal outlet from the tank, the water heater flow (or the closest and furthest tap to the heater and associated recirculating systems), and drain valves, when available (U.K. Deaprtment of Health, 2006). Monitoring temperature and disinfectant levels of cold and/or hot water systems is a practical approach to identify points that are susceptible to OPPP colonization and candidates for testing. For example, the warmest point in a cold water system, or the coolest part of a hot water system are likely to pose the greatest risk to Legionella growth (U.K. Environmental Agency, 2005; U.K. Deaprtment of Health, 2006). Regardless, a detailed knowledge of water system layout together with a thorough understanding of conditions that are conducive to OPPP growth is essential to developing a sampling plan (ASHRAE, 2015).
Routine Legionella monitoring of non-healthcare related plumbing is generally not mandatory, except in France, where operators of all public buildings are required to complete regular monitoring for Legionella for any hot water distribution system (FRMHS, 2010). In general, cold water systems should not require routine monitoring for Legionella unless temperatures and stagnation times are elevated in the system (e.g., >25 °C, dead ends, storage tanks). However, it cannot be said that there is zero risk of Legionella contamination in cold water systems. In a cold water tap survey across the United States, nearly half the taps showed the presence of L. pneumophila sg1 in one sampling event, and 16% of taps were positive in more than one sampling event (Donohue et al., 2014). In Germany, a survey of German public buildings revealed 5.4 % of cold water samples to exceed the action value for Legionella (Volker et al., 2010). In hot water systems, it is recommended to sample water heaters set at low temperatures (e.g., <50° C), header tanks, water softeners, water heater drain-off points, and affected taps (pre-flush, post-flush) (U.K. Environmental Agency, 2005). For residential surveys of L. pneumophila, it has been recommended that at least 3 of 5 of the following sites be sampled: hot water tank, kitchen tap, bathroom tap, showerhead, and bath tub outlet (Stout et al., 1992).
Sampling plans are an essential aspect of responding to Legionella outbreaks, with the aim of identifying all potential sources of contamination and modeling exposures. Cold water systems should be sampled at the incoming supply, the outlet of each reservoir, and outlets closest/furthest to the reservoir. Hot water system should also be thoroughly investigated, including showers and taps used by infected people with Legionnaires’ disease or in proximal areas. Any other sources of exposure, including those susceptible of generating aerosols, also need to be considered: ice machines, evaporative cooling systems, spa pools, humidifier, decorative fountains, etc. (U.K. Environment Agency, 2005).
Published guidelines concerning sampling site selection strategies for OPPPs other than Legionella are scarce. A recently published health technical memorandum by U.K. Department of Health briefly suggested that water outlets supplying water for patients (direct and indirect contact) and staff hand-washing should be sampled for P. aeruginosa in augmented care units (U.K. Department of Health, 2013). With regard to NTM and amoebae, for which there is no surveillance guideline for their monitoring in premise plumbing, recent studies generally followed standard methods for microbiological tests or sampling guidelines. Taps and showers were the frequent targets (Falkinham, 2011; Feazel et al., 2009; Ichijo et al., 2014; Kilvington et al., 2004; Thomas et al., 2006), with cisterns, filters, garden hoses and ice machines also sampled in some cases (Covert et al., 1999; Falkinham, 2010, 2011; Thomas et al., 2014). In an epidemiological and environmental investigation of a PAM death, a garden hose, service line hose bib, and toilet tank were also included in household samples, all of which were positive for Naegleria fowleri (Cope et al., 2015).
2.1.2 Sampling Frequency
There is no universal direction with regard to when and how often to sample premise plumbing for microbiological analysis. Monthly, quarterly, semi-annual and annual samplings have been proposed for testing of Legionella and/or P. aeruginosa (U.K. Department of Health, 2013; Regierung, 2001; FRML, 2010; VHA, 2014). The U.S. VHA requires quarterly testing of their buildings’ hot and cold water distribution systems for L. pneumophila (VHA, 2014); while annual sampling of hot water systems is generally recommended by German health care centers and residential monitoring programs (Regierung, 2001). Sampling selected taps every 6 months is suggested for P. aeruginosa monitoring in augmented care units in the U.K. However, more frequent tests or further engineering survey might be needed in case of positive test results or clinical-evidenced suspicion of contamination (U.K. Department of Health, 2013). Sampling frequency can also differ based on water usages. For example, it has been suggested to perform 1 microbiological control per year per 100 beds, with a minimum of 4 controls per year for a healthcare facility in terms of monitoring P. aeruginosa and total coliforms in water for general use; while monthly monitoring of HPC, total coliforms, L. pneumophila, P. aeruginosa, and Staphylococcus aureus is required for water used for spa, bath and showers (FRMHS, 2005). Sampling frequency is also associated with temperature variation and disinfectant levels in the plumbing. Daily or monthly temperature control has been prescribed for healthcare buildings in many European countries, a fundamental strategy for controlling Legionella exposure risks (U.K. Department of Health, 2006; FRMHS, 2005).
The reality is that any given sampling event does not capture the true dynamics of OPPPs in the system. A study examining the variability of L. pneumophila in 84 taps over 5 consecutive days in an Italian hospital revealed significant variation of the bacterial load from day to day, although the pattern was similar across the wards monitored (Napoli et al., 2009). Thus, repeated samplings might be required in order to gain sufficient statistical power, especially when the data are for the purpose of research, risk assessment, or mitigation.
2.2 Sample Types: Bulk Water and Biofilm
Microbes and OPPPs generally reside within two niches in the premise plumbing environment: the biofilm and the bulk water (Falkinham et al., 2008; Flannery et al., 2006; Moore et al., 2006; Thomas et al., 2006; Wang et al., 2012). Biofilm is thought to serve as the ideal habitat for OPPPs, providing food and protection from disinfectants and other harmful agents. Amoebae have been detected at a higher frequency in biofilm than bulk water (Stockman et al., 2011), as the biofilm consists of a diverse and concentrated source of bacteria to graze upon (Huws et al., 2005; Paris et al., 2007). Interestingly, it is a common feature among bacterial OPPPs to survive phagocytosis and experience enhanced growth and virulence within the amoeba digestive vacuole (Thomas and Ashbolt, 2011). Such a “Trojan horse” phenomenon has been confirmed for Legionella, NTM and P. aeruginosa (Delafont et al., 2014; Greub and Raoult, 2004) and is thought to be obligate for Legionella replication under oligotrophic conditions (Declerck et al., 2009; Thomas and Ashbolt, 2011). However, the rich nutrient content of biofilm may allow nutritionally fastidious Legionella spp. to multiply extracellularly (Taylor et al., 2009).
Even if biofilm is ultimately proven to be the most important ecological niche for OPPPs, bulk water is more accessible than biofilm and remains the most targeted for OPPPs investigation. Arguably, bulk water is also the most relevant to actual dissemination and exposure (Buse et al., 2012; Schoen and Ashbolt, 2011). As such, guidelines for Legionella and P. aeruginosa control are often based on bulk water bacterial loads (U.K. Department of Health, 2006, 2013). In a study comparing 3,910 paired water/biofilm samples, concordant Legionella detection results were found among 81% of pairs, with only 2% of pairs only yielding positive detection in the biofilm (Ditommaso et al., 2010). Ecological niches of specific species might also affect their distributions in suspended versus attached phases. For example, across eight water distribution systems, higher detection frequency in biofilm relative to water samples was confirmed for M. intracellulare, but not M. avium (Falkinham et al., 2001). Further systematic comparisons between biofilm and water samples are needed for other OPPPs to justify sampling plans.
2.2.1 Sampling of Water
Three consecutive steps are generally employed for sampling plumbing water: turning on a tap or valve, adjusting the flow rate, and collecting a prescribed volume of water using a sterilized container. Sodium thiosulfate is often used to neutralize residual chlorine and other halogens when disinfectant is used in the source water (CDC, 2005). However, technical details during sampling and preservation beyond the basic steps are worthy of consideration, especially when simultaneous monitoring multiple OPPPs is the goal. Table 1 summarizes procedures for OPPPs sampling applied across several studies.
Table 1.
Sampling procedures for P. aeruginosa and L.pneumophila in premise pluming in different protocols and studies
| P. aeruginosa | L. pneumophila | ||||||
|---|---|---|---|---|---|---|---|
| Device | Doc Type | Ref | Device | Doc Type | Ref | ||
| Flow regime | First draw | Taps | Best practice guidance | (Health; and facilities, 2013) | Taps/Showers/Water heaters | Study Protocol |
(Martinelli et al., 2000) (Scotland, 2011) |
|
| |||||||
| Brief period of flow (eliminate cold water) | Taps | Study | (Borella et al., 2004) | ||||
|
| |||||||
| 1 min flow | Taps/Showers | Study | (Bargellini et al., 2011) | ||||
|
| |||||||
| 2 min flow | Taps (post-flush) | Best practice guidance | (Health; and facilities, 2013) | Faucets | Protocols | (Barbaree et al., 1987; Scotland, 2011) | |
|
| |||||||
| Temperature at equilibrium | Study | (Flannery et al., 2006; Moore et al., 2006) | |||||
|
| |||||||
| Volume of sample (mL) | 100 | Faucets | Studies | (Lavenir et al., 2008; Trautmann et al., 2001; van der Mee-Marquet et al., 2005) | Taps | Study | (Zhang et al., 2009) |
|
| |||||||
| 250 | Faucets | Studies | (Berthelot et al., 2006; Rogues et al., 2007) | ||||
|
| |||||||
| 500 | Faucets | Studies Protocol |
(Halabi et al., 2001) (Scotland, 2011) |
||||
|
| |||||||
| 1000 | General potable water system Residential taps/showers |
Protocols Study |
(Barbaree et al., 1987; Scotland, 2011) (Flannery et al., 2006; Mathys et al., 2008; Moore et al., 2006) |
||||
|
| |||||||
| 1000 – 2000 | Standards | ((APHA), 2012; (ISO), 2004) | |||||
|
| |||||||
| Showers/Taps | Study | (Borella et al., 2004) | |||||
|
| |||||||
| 10,000 | Incoming water | Protocol | (Barbaree et al., 1987) | ||||
Sample Volume
Sampling volumes applied for the detection of Legionella and P. aeruginosa, as two representative bacterial OPPPs, in drinking water systems have varied between 50 ml to 1L depending on the anticipated concentrations of target organisms (Ferroni et al., 1998; Halabi et al., 2001; Lavenir et al., 2008; van der Wielen and van der Kooij, 2013). Larger volumes (i.e., 5–10 L) were collected in case of low biomass level (e.g., distribution mains) in order to ensure sufficient recovery (U.K. Environment Agency, 2005; Barbaree et al., 1987). And even larger volumes, > 100 L, have been filtered for N. fowleri detection, yielding molecular and culture detections when smaller volumes (0.7 L) did not (Cope et al 2015). However, for some analytes and water qualities, larger volume sampling might be associated with concentration of potential inhibitory substances which might affect downstream analysis such as quantitative polymerase chain reaction (qPCR) amplification (Hata et al., 2011).
Pre-flushing or Post-flushing
Complex water usage patterns typify premise plumbing, resulting in temporal and spatial heterogeneity and periodically stagnant water. Higher numbers of microorganisms are likely to be detected following stagnation or infrequent use, as a result of disinfectant decay and microbial regrowth (Lautenschlager et al., 2010). Thus, one approach to maximize the likelihood of OPPPs detection is to sample taps after a period of stagnation (e.g., first draw of water taken in the morning). Taking the first sample (pre-flushing) after at least 2 hours of stagnation or during a period of minimum water usage has been recommended for P. aeruginosa detection in augmented care centers (U.K. Department of Health, 2013). Similarly, in a molecular survey of multiple OPPPs in building plumbing, imposing 8-h stagnation before sampling resulted in higher discrepancy between first-draw (pre-flushing) and post-flushing samples relative to sites without enforced stagnation (Wang et al., 2012). In case of a positive pre-flushing sample, it is suggested that a second sample be collected after running taps for 2 minutes in order to identify the source of contamination (U.K. Department of Health, 2013). Given these considerations, a major shortcoming of water microbiological quality monitoring as recommended by U.S. EPA regulations and other common guidelines for distribution system monitoring is the practice of thorough flushing and disinfection of taps prior to sampling, which is unlikely to detect water quality problems related to premise plumbing (NRC, 2006).
With regards to hot water systems, Western Pennsylvania Guidelines suggest sampling the first-draw (i.e., first 1 L) hot water from the outlet valve and prior to cold water system sampling (Allegheny County Health Department, 2014). However, some researchers recommend flushing the tap for 1–2 minutes prior to sampling in order to exclude cold water (Bargellini et al., 2011; Borella et al., 2005). Since it is not easy to determine when cold water will be completely flushed out, reaching temperature equilibrium has been used as a criteria to determine when to collect samples (Moore et al., 2006; von Baum et al., 2010; Wellinghausen et al., 2001). Additional steps might be needed if the goal is to solely sample the hot water. For instance, one study surveying Legionella occurrence in hot water systems in German single-family residences removed all devices from the taps, heated taps with a gas burner, and discarded the first 5 L water prior to water collection (Mathys et al., 2008). ASTM guideline and some researchers also suggested stratified sampling, which includes a first flush sample and a sample after temperature stabilization for Legionella in order to delineate the source of contamination (Bates et al., 2000; ASTM, 2008).
Aerator Removal
Faucet aerators represent potential risks for colonization of Legionella and other pathogens (CDC, 2003). Whether to remove aerators before sampling depends on the sampling objective. If the objective is to evaluate the risk and colonization of the outlet, the sample should be taken with the aerator and without disinfection. When the water flowing within the distribution system is the target, potential contamination from outlets should be minimized. CDC specifies that water samples for biological tests should be taken after removing aerators and disinfecting taps with sodium hypochlorite if the cleanliness of the tap is questionable (CDC, 2005). Other disinfection approaches for taps include ethanol or isopropanol disinfection (70% v:v), as well as flaming if the tap is made of metals (U.K. Environmental Agency, 2005; Mathys et al., 2008). Care should be taken when removing aerators as biofilm disruption and detachment might occur during dissembling.
Sample Preservation and Transportation
Handling and shipping samples usually takes from a few hours to several days, during which samples should be preserved accordingly in order to minimize cell cultivability impairment and/or potential regrowth. Recommended preservation procedures for Legionella enumeration varied considerably in different documents in terms of storing temperature and holding time. For example, The International Standards Organization (ISO) requires to refrigerate samples that cannot be processed within 24 h (ISO, 2004); while CDC and American Society for Testing and Materials ASTM recommend to refrigerate samples only if they cannot be processed within 72 h (CDC, 2005), although protection from extreme heat or cold (i.e., <3 °C and/or >30 °C) is required at all times (ASTM, 2008). The U.K. Environmental Agency recommends maintaining samples between 6 and 20°C, in order to prevent cells from entering a viable but not culturable (VBNC) state during shipping, and to commence concentration and incubation procedures within 48 h (U.K. Environmental Agency, 2005). Two studies have investigated the effect of holding time on Legionella cultivability and reported conflicting results. McCoy observed significant changes (>1 log10 unit) in 52% of water samples (n=42) with 6 ~120 h holding time at room temperature (McCoy et al., 2012). Another recent study analyzing 159 samples found little effect of holding time on Legionella counts, with root mean squared error increased by only about 3~8 % in overnight held samples at room temperature (Flanders et al., 2014).
On the other hand, water samples collected for P. aeruginosa culturing should be refrigerated (2~8 °C) within 2 h and processed within 24 h in order to prevent regrowth (U.K. Department of Health, 2013). On the contrary, little change in NTM numbers would be expected during shipping due to their slow grow and decay rates, as well as resistance to a wide range of environmental temperatures (Falkinham, 2009).
In terms of free-living amoeba, caution must be taken when handling and storing samples since common practices such as chilling and refrigeration will trigger the formation of cysts and VBNC state. Naegleria fowleri trophozoites are known to become VBNC when the temperatures is <10°C (Chang, 1978). Samples for N. fowleri cultivation should be stored and shipped at ambient temperature (i.e., non-chilled) and with adequate headspace since N. fowleri are thought to be highly aerobic (Kyle and Noblet, 1985). Thus, various shipping and handling requirements for each OPPP poses a significant challenge to monitoring multiple OPPPs simultaneously, likely requiring aliquoting of sub-samples in order to achieve optimal preservation conditions.
Concentration Methods
Sample concentration is typically required for detection of OPPPs, typically ranging from 50~1000 ml premise plumbing water. Filtration, centrifugation and immuno-magnetic separation (IMS) are three common concentration methods. The first two are non-specific approaches widely used for microbial analysis, whereas IMS is able to select target microbes from background microorganisms by using antibody-coated magnetic media (Allegra et al., 2011; Bedrina et al., 2013; Mull et al., 2013).
Filtration is suitable for water samples with sufficiently low turbidity to avoid clogging. Generally, water is filtered through a 0.22 or 0.45 μm membrane (Bartie et al., 2001; CDC, 2005), which later is transferred directly to media for culturing (U.K. Department of Health, 2013) or used for cell resuspension by sonicating or votexing the filter in a small volume of water (Bartie et al., 2001; CDC, 2005; Thomas et al., 2008). Membranes may also be shredded for DNA extraction. Membranes with larger pore-size (e.g., 1.2 μm) have sometimes been used for amoeba filtration, given the larger diameter of protozoan cells (Pernin et al., 1998), but more recently N. fowleri have been recovered from large volumes of water using ultrafiltration (Cope et al., 2015). Centrifugation can be applied for both clean water and non-filterable water with higher turbidity. Typically, water samples are centrifuged with a speed of 3000 g for 30 min or 6000g for 10 min (Bartie et al., 2001; Brindle et al., 1987), leaving pellets that can be resuspended in 2 to 20 ml of diluents after discarding the supernatants. Recovery efficiencies of filtration and centrifugation have been evaluated for Legionella and Mycobacterium across several studies, with filtration demonstrating similar or higher specific recovery rates relative to centrifugation (Boulanger and Edelstein, 1995; Brindle et al., 1987; Ta et al., 1995; Thomson et al., 2008). However, centrifugation is a better choice for recovery of Naegleria, from small-volume samples as trophozoites may lyse by during vacuum filtration (Pernin et al., 1998). Moreover, specific recovery rates can vary considerably when different kinds of membranes and centrifugation conditions are applied (Boulanger and Edelstein, 1995).
2.2.2 Sampling of Biofilm
In premise plumbing, biofilm samples are often collected from faucets, shower heads, drains, hoses and water filters (Charron et al., 2015; Falkinham, 2010; Liu et al., 2012; Proctor et al., 2016; Thomas et al., 2014; Wang et al., 2012). Anti-splash or spray nozzles are recommended to be removed from faucets and shower heads and disassembled prior to biofilm sampling in order to access the inner area and obtain representative biofilm (U.K. Environmental Agency, 2005; Feazel et al., 2009). However, this is difficult to apply consistently in all cases since often devices are not designed for ready disassembly. Biofilm can be removed from surface by scraping a known area of surface using a sterile knife or swab (Charron et al., 2015; Liu et al., 2012; Srinivasan et al., 2008). Thorough removal of biofilm may require multiple scrapings (Srinivasan et al., 2008) or further treatments such as sonicating removable parts (e.g., faucet gaskets) in a cold bath (Liu et al., 2012). Although some studies used epifluorescence microscopy to quantify the cell removal efficiencies (Percival et al., 1999), this technology is constrained for premise plumbing biofilm samples since most parts in premise plumbing are not removable.
Biofilm sampling sequence may affect how representative the samples are. When both biofilm and bulk water are sampling targets, biofilm samples should be taken first as water sampling and flushing will dislodge the microbes from biofilms. However, sampling biofilm first might release bacteria from the biofilm into the bulk phase and affect the representativeness of subsequent bulk water sample if the goal is to take the first-draw samples. On the other hand, when sampling biofilm in a sink, drain biofilm must be taken prior to contact with water from the tap.
2.3 Sample Preservation Strategies for Culture-based versus Molecular Methods
Different sample preservation strategies may be required when subject to culturing, molecular analysis, or both. Water samples concentrated by membrane filtration or centrifugation can be resuspended in media prior to culturing. Same (e.g., drinking water (Borella et al., 2005)) or similar non-nutrient medium (e.g., phosphate-buffered saline (Leoni et al., 2001)) are usually used for resuspension, in order to minimize the influence of shifting media conditions on regrowth potential and cultivability, with storage at 4 °C if plating is not performed immediately. When samples are intended for molecular analysis, membranes or resuspended media can be directly kept at -20 °C or lower until DNA/RNA extraction (Eichler et al., 2006). Similarly, culturing biofilm cells involves resuspension in appropriate media (Wullings et al., 2011); while cotton swab tips can be directly preserved in 70% ethanol (Feazel et al., 2009) at -20 °C or lower for molecular analysis.
3. Detection Methods
3.1 Culturing of OPPPs
Culturing is used to verify and recover viable cells, remaining the “gold standard” for identifying and typing infectious and life-threatening pathogens. Thus, it is not a surprise that the majority of current standard methods, regulations, and action limits for OPPPs are based on conventional culture methods. However, culturing of OPPPs is often criticized as labor-intensive and time-consuming, requiring specialized expertise to correctly identify target cells and failing to provide timely information in urgent situations. Culture-based techniques may also limited in their quantitative capacity.
3.1.1 Legionella spp
Legionella is a fastidious microorganism with highly-specific nutrient (e.g., iron, L-cysteine) and cultivation requirements. Labs around the world use different culturing protocols for Legionella recovery, with considerable variation noted in terms of pretreatment approaches, cultivation medium and incubation period. ISO (ISO11731-2:2004) recommends use of buffered charcoal yeast extract (BCYE) agar containing L-cysteine or selective GVPC agar (BCYE supplemented with glycine, vancomycin, polymyxin B, cycloheximide) for recovery of Legionella from filtered water samples. The results are compared to that of BCYE agar without L-cysteine for specificity confirmation (ISO, 2004). Meanwhile, the CDC advocates simultaneous use of four different kinds of media, including BCYE base media, two selective BCYE agars (i.e., PCV (BCYE supplemented with polymyxin B, cycloheximide, and vancomycin) and GVPC, and PCV without L-cysteine as a negative control (CDC, 2005). Other versions of Legionella cultivation medium include variation of antimicrobial supplements (Ta et al., 1995), dye supplements for colony staining (e.g., MWY medium) (Ta et al., 1995) or specific nutrients (e.g., ABCYE: BCYE with bovine serum albumin) to cater to the growth requirements of certain Legionella spp. such as L. micdadei (CDC, 2005).
Heat and acid treatments are routinely used to suppress growth of non-legionella species by taking advantage of Legionella’s tolerance of these extreme conditions. Heat treatment is typically performed between 50–59 °C for 3–30 min and pH treatment at ~2.2 for 3–15 min (De Luca et al., 1999; Reinthaler et al., 1993). Improved recovery frequency had been observed for both pretreatment methods compared to no treatment (Reinthaler et al., 1993). However, inconsistent results have been noted among different studies in terms of optimal combinations of pretreatment approaches and cultivation media (Bartie et al., 2003; De Luca et al., 1999; Leoni and Legnani, 2001). Factors such as sample matrices, Legionella concentration, and commensal flora could possibly affect sensitivity and specificity of applied culturing techniques. However, both heat treatment and acid treatments may not be 100% effective and can also lead to underestimation of Legionella counts as a result of cell cultivability damage (Leoni and Legnani, 2001).
Another approach for recovering Legionella in premise plumbing samples is to co-culture samples with amoebae (e.g., Acanthamoeba or Hartmanella). This approach may improve recovery frequency and detection limits for samples with low Legionella counts and/or VBNC cells by increasing their numbers and resuscitating cultivability (Conza et al., 2013; Garcia et al., 2007).
3.1.2 Mycobacterium spp
Mycobacterium spp. are a group of slow-growing, hydrophobic microorganisms ubiquitous in natural and engineered aquatic environments (Falkinham, 2009). Their cells tend to aggregate and adhere to surfaces, increasing difficulty for isolation and enumeration. Culturing techniques for Mycobacterium spp. typically include a pretreatment step to prevent bacterial and fungal overgrowth, use of nutrient-rich medium (e.g., M7H10 agar supplemented with 0.5% glycerol and 10% oleic-albumin enrichment) to support growth, and incubation at 35–37 °C for 10–21 days to allow enough time for recovery (Falkinham et al., 2008; Falkinham et al., 2001; Thomson et al., 2008). Although a variety of protocols have been used for mycobacteria recovery, a standard protocol has not yet been established.
A range of bacterial and fungal inhibitors, including cetylpyridinium chloride (CPC), NaOH, formaldehyde, and oxalic acid had been used for sample pretreatment (Falkinham et al., 2008; Falkinham et al., 2001; Thomson et al., 2008; Torvinen et al., 2004). Decontamination with CPC was considered the best approach for treated water (Neumann et al., 1997) and has been widely applied in many drinking water-related studies (Falkinham et al., 2001, 2008; Torvinen et al., 2004). However, these pretreatment methods are not always effective. Decontamination efficacy can vary dramatically upon selection of different inhibitors, which might be partially ascribed to characteristics of the water matrix and presence of various mycobacterial species. Moreover, decontamination steps can also result in loss of viable mycobacteria (Thomson et al., 2008).
Employment of different culture media also leads to various recovery efficiencies. Neumann et al. suggested that Lowenstein-Jensen medium and Ogawa egg yolk medium are superior to Ogawa whole-egg medium containing ofloxacin and ethambutol in terms of mycobacterial recovery in surface and treated water (Neumann et al., 1997). Thomson et al. obtained similar results with Middlebrook 7H10 and 7H11, and Lowenstein-Jensen slants after 3 week incubation of water samples. However, Lowenstein-Jensen slants initially appeared to be less sensitive when examined earlier (Thomson et al., 2008). Other factors such as temperature (Neumann et al., 1997) and incubation period (Thomson et al., 2008) can also affect recovery.
3.1.3 P. aeruginosa
P. aeruginosa is a Gram-negative, oxidase-positive bacterium that usually produces pyocyanin and fluorescein and hydrolyzes casein (U.K. Department of Health, 2013). Standard culturing methods for P. aeruginosa in drinking and/or recreational water include the membrane filtration and multiple-tube techniques, both of which take advantages of its pigment-producing characteristics (APHA, 2012). M-PA agar is used to recover P. aeruginosa from filter membranes, incubated at 41.5±0.5 °C for 72 h for presumptive tests. Colonies with 0.8~2.2 mm in diameter, flat in appearance with light outer rims and brownish to greenish-black centers are counted as positive numbers. Presumptive tests are followed by confirmation tests using milk agar for another round of cultivation at 35 °C for 24 h to assess atypical colonies, taking advantages of P. aeruginosa’s capability of hydrolyzing casein and producing a yellowish to green pigment. Multiple-tube techniques use asparagine broth for P. aeruginosa growth and green fluorescent pigment production in presumptive tests (35~37 °C for 24–48 h), followed by confirmation tests using acetamide broth or agar for 24–36 h cultivation. Acetamide can be deaminated by P. aeruginosa, resulting in phenol red indicator shifting from yellow orange to purple.
As described above, standard culture methods for P. aeruginosa are highly time-consuming. An alternative most probable number (MPN) method, commercialized by IDEXX laboratories (Maine, USA), provides semi-quantitative results within 24–48 h by using a bacterial enzyme detection reagent. This method is reported to perform equivalently to the membrane filtration method for examination of hospital water (Sartory et al., 2015) and was recently listed as an alternative method for P. aeruginosa detection in swimming pool water testing by the German Federal Environment Agency (IDEXX, 2015).
3.1.4 Acanthamoeba and Naegleria fowleri
Spreading concentrated water samples or placing a membrane filter on non-nutrient agar plates containing a lawn of Gram-negative bacteria (e.g., Escherichia coli or Enterobacter aerogenes) as a food source is a common approach for recovering amoebae from drinking water (Delafont et al., 2014; Tyndall et al., 1989). Plates are typically incubated at 28–44 °C for up to one month (Delafont et al., 2014; Marciano-Cabral et al., 2010; Thomas et al., 2006; Tyndall et al., 1989), being observed at regular intervals by microscopy to confirm the presence or absence of amoeba. Assays specifically targeting Naegleria spp. favor a higher incubation temperature (i.e., 42–44 °C), since Naegleria spp. are thermophiles (Ithoi et al., 2011; Marciano-Cabral et al., 2010; Tyndall et al., 1989. Mull et al., 2013). Identities of amoeba isolates are subsequently determined according to their morphology or using molecular methods (e.g., PCR) (Garcia et al., 2013). Acanthamoeba trophozoites and cysts have distinct features (e.g., double-walled cysts) that can be categorized into three different morphological groups based on cyst size and numbers of arms (Pussard and Pons, 1977). Naegleria-like trophozoites and cysts typically demonstrate eruptive-like formation of the pseudopodia and smooth cyst walls, respectively (Tyndall et al., 1989). Further confirmation of Naegleria species involves a test to induce trophozoites to flagellate by adding sterile distilled water for additional 1–2 h incubation (Ithoi et al., 2011; Visvesvara et al., 2007). However, full validation of either Acanthmoeba or Naegleria isolates to species level using culturing alone is not reliable, as morphology of trophozoites and cysts can be strongly affected by culturing conditions (Visvesvara et al., 2007). Therefore, other assays (molecular, enzymatic, or virulence tests) are needed for species identification.
3.2 Molecular methods
Molecular methods generally involve manipulation and analysis of DNA, RNA, or proteins. Compared to conventional cultivation methods, molecular methods have the advantage of low detection limit, high specificity and sensitivity, ability to detect viable but not cultivable (VBNC) cells, and short turnaround time (Girones et al., 2010). Such advantages are very attractive for pathogen monitoring, especially when results are urgent and for simultaneous detection and enumeration of multiple OPPPs.
A number of molecular techniques, including PCR, quantitative-PCR, sequencing, etc., have been developed for identifying and quantifying OPPPs (Table 2). These methods are nucleic acid-based, with DNA/RNA extraction as the first and the foremost step in analysis protocols. Obtaining sufficient high-quality DNA/RNA with minimal PCR inhibitors is prerequisite to downstream amplification, posing a particular challenge to drinking water given its inherently low biomass concentration. On the other hand, nucleic acid extraction efficiency can vary considerably across studies (Girones et al., 2010). Sample types and properties, cell characteristics, extraction procedures or DNA/RNA extraction methods can affect the quantity and quality of nucleic acid yields (Hwang et al., 2012). Some OPPPs may demand special treatment during the nucleic acid isolation process. It has been reported that the structures of Acanthamoeba cysts are resistant to DNA extraction reagents, requiring incubation with protease K prior to extraction to improve yields (Goldschmidt et al., 2008). Some molecular methods do not require extraction of nucleic acids. For example, fluorescence in situ hybridization (FISH) and flow cytometry methods utilize fluorescently-labeled DNA/RNA probes that can penetrate cells and bind to target nucleic acids without rupturing cell structures. The emitted fluorescence is captured by microscopy or flow cytometry. However, these techniques are often criticized for high detection limits, laborious procedures and low resolution with aggregated microbial cells (Buchbinder et al., 2002; Nocker et al., 2009; Wullings and van der Kooij, 2006). Such techniques are rarely encountered, typically in research contexts employing inoculation of high numbers of OPPPs to simulated drinking water systems (Declerck et al., 2009; Lehtola et al., 2007). Below we focus more specifically on nucleic acid-based molecular techniques that are more widely practiced for drinking water monitoring (Girones et al., 2010).
Table 2.
Examples of available analytical methods for OPPP detection in environmental samples
| Targeting genus | Species(if any) | Method | Sample characteristics | Detection or quantification limit | Notes | Ref. |
|---|---|---|---|---|---|---|
| Legionella | Culturing | Pool water and shower water | LOD: 5 cfu/l or pool water and 10 cfu/l for shower water | Agglutination tests were used following cultivation to distinguish L. pneumophila serogroup 1–6, L.bozemanii, L. dumoffii, L.gormanii, and L. micdedei | (Leoni et al., 2001) | |
| Culturing (ISO 11731) | Hot water from hotels | LOD:25 cfu/l | Agglutination tests were used to separate L. pneumophila isolates serogroup 1 and 2-14, as well as seven species of non- L. pneumophila legionellae | (Bargellini et al., 2011; Borella et al., 2005) | ||
| Legionella spp. & L. pneumophila | Culturing, PCR, &q-PCR | Drinking water from treatment plants | LOD (Legionella spp., direct culturing, without concentration): 1000 cfu/l LOD (L. pneumophila, q-PCR): 1000 gene copies/L |
Semiquantitative PCR was used for concentration assessment. PCR products were cloned and sequenced for genitive diversity exploration | (Wullings and van der Kooij, 2006) | |
| Legionella spp. & L. pneumophila | Culturing, PCR, &q-PCR | Hospital water | LOD (Legionella spp. culturing): 1 cfu/100 ml LOD (Legionella spp. q-PCR): 2.3 cfu/100 ml; LOQ (Legionella spp. q-PCR): 23 cfu/100 ml LOD and LOQ for L. pneumophila: <2.3 and 23 cfu/100 ml, respectively |
Correlated q-PCR and culturing results (P<0.001) with higher q-PCR enumeration number in relative to culturing; Correlated results between genus-specific and species-specific assays | (Wellinghausen et al., 2001) | |
| Legionella spp. & L. pneumophila | Culturing & q-PCR | Cooling towers Hot and cold water |
LOD for q-PCR and culturing: 750 GU/l for water samples from cooling tower and 190 GU/l for samples from hot and cold water systems. |
Greater discrepancy between q-PCR and culturing results for cooling tower samples compared to hot and cold water samples. | (Lee et al., 2011b) | |
| q-PCR | Spa water | LOD:40 GU/l LOQ: 1000 GU/l |
Results revealed weak correlation between culturing and q-PCR | (Guillemet et al., 2010) | ||
| Culturing, q-PCR, EMA-q-PCR | Hot water samples | LOD for EMA-q-PCR: 200 GU/ml for 1 ml of sample treated with EMA; 250GU/l for 1L of sample water | v-PCR counts were equal to or higher than those obtained by culture, and lower than or equal to conventional qPCR counts |
(Delgado-Viscogliosi et al., 2009) | ||
| Immunofluorescent labeling combined with solid-phase flow cytometry | LOD: 10-100 bacteria/l | Obtained numbers are higher than CFU counting | ||||
|
| ||||||
| Mycobacterium | Culturing | Drinking water samples | LOD: 10 cfu/l | After culturing, PCR amplification of the hsp-65 gene followed by enzyme restriction of the PCR product was used for identification |
(Falkinham et al., 2001) | |
| q-PCR | Cooling tower water | LOQ: 500 cells/l | (Adrados et al., 2011) | |||
| q-PCR and culturing | Drinking water and other environmental samples | LOD for q-PCR: 6 genome equivalents for M. chelonae LOQ for q-PCR:100 genome equivalents |
Higher concentration level but lower detection rates with q-PCR in relative to culturing method | (Radomski et al., 2013) | ||
| Mycobacterium avium subsp. paratuberculosis | q-PCR | Drinking water and biofilm | Assay LOD: 1.8 gene copy | (Beumer et al., 2010) | ||
|
| ||||||
| P. aeruginosa | Pseudalert®/Quanti-Tray® MPN Test, Culturing method | pool water samples, artificially contaminated samples | Comparable results between Pseudalert®/Quanti-Tray® MPN Test and ISO 16266 and MoDW Part 8 methods | (Sartory et al., 2015) | ||
| q-PCR | Clinical and environmental isolates | Duplex q-PCR assay with two targeted genes (ecfX and gyrB), requires simultaneous confirmation of P. aeruginosa by two genes | (Anuj et al., 2009) | |||
| q-PCR and culturing | Hospital faucets (water, aerator and drain swabs) | qPCR revealed 50% positivity for P. aeruginosa remaining in the water compared with 7% by culture | (Bedard et al., 2015) | |||
| q-PCR, PMA-q-PCR, standard cultivation-based technique | Drinking water and process water | LOD of q-PCR and PMA-q-PCR :102~103 cells/l | >80% samples yield accordant results by q-PCR, PMA-q-PCR, and cultivation based method. PMA-q-PCR reduced 4% false positive rates when compared to q-PCR. | (Gensberger et al., 2014) | ||
|
| ||||||
| Acanthamoeba | Culturing followed by morphological identification | Household water | Fungal overgrowth of the samples occurred more often with biofilm sample than water samples. | (Stockman et al., 2011) | ||
| Culturing followed by morphological identification and PCR/sequencing | Tap water | (Winck et al., 2011) | ||||
| q-PCR | Anthropogenic water and biofilms | Assay LOD (trophozoites): 3 cells for water samples, 10 cells for biofilm samples | Qvarnstrom assays outperforms Riviere assay for Acanthamoeba detection and quantification | (Chang et al., 2010) | ||
| PMA-q-PCR | Culture suspension, water samples from eyewash station, cooling tower, wastewater treatment plant | Detection range: 5~1.5×105 cells | (Chang et al., 2013) | |||
|
| ||||||
| Naegleria fowleri | q-PCR and MPN methods | Cooling water samples | LOQ for q-PCR: 320 cells/l | Samples with concentration <200 cells require culture method analysis; high q-PCR estimated numbers compared to MPN method | (Behets et al., 2007a) | |
| q-PCR and NNA-E. coli culturing method | N. fowleri cells spiked into water and biofilm samples | LOD in water samples: 5 cells in 250 ml water for a 66% detection rate and 10 cells for a 100% detection rate; DOL in biofilm samples: 1 cell for a 66% detection rate and 5cells for a 100% detection rate |
Culturing method is less sensitive compared to q-PCR method | (Puzon et al., 2009) | ||
| IMS-q-PCR | N. fowleri seeded lake water | LOD: 14 cells/l; | The methods has an average recover rate of 46% | (Mull et al., 2013) | ||
| ELISA | Environmental water samples | LOD: 2000 cells/l | Can detect N. fowleri at three morphological stages, with 97.4% sensitivity and 97% specificity | (Reveiller et al., 2003) | ||
3.2.1 PCR
PCR can selectively amplify signature genes (i.e., gene markers) from target microorganisms. Observation of PCR products following agarose gel electrophoresis can serve to verify the presence/absence of target organisms. Assay specificity depends on the specificity of primers and the stringency of the PCR (Nocker et al., 2009). PCR may also be combined with some downstream genotyping techniques for genetic diversity exploration or species identification (Huang and Hsu, 2011). For example, phylogenetic analysis of cloned and sequenced PCR-amplified Legionella 16S rRNA genes revealed a large diversity of uncultured Legionella spp., including L. bozemanii, L. worsleiensis, L. quateirensis, L. waltersii, and L. pneumophila in water from water treatment plants (Wullings and van der Kooij, 2006). Some studies used endonuclease digestion of PCR products for mycobacterial isolate identification (Cheunoy et al., 2005; Chimara et al., 2008). However, this technique requires establishment of restriction patterns for a collection of known mycobacterial species. If the typical restriction pattern is not present, other cultural, biochemical, and enzymatic characterization may also be necessary for further identification (Falkinham et al., 2001). Other variations of PCR-based methods, such as repetitive sequenced PCR (rep-PCR) fingerprinting, multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA), can also be used for detection of novel genotypes, or matching OPPP isolates between environmental and clinical samples (Falkinham, 2010, 2011; Kahlisch et al., 2010; Sobral et al., 2011).
3.2.2 Quantitative PCR (q-PCR)
q-PCR is advantageous relative to traditional PCR in that it provides quantitative information and a lower detection limit. It has been extensively applied for OPPP monitoring for both environmental and clinical samples (Adrados et al., 2011; Ahmed et al., 2014; Bedard et al., 2015; Behets et al., 2007a; Beumer et al., 2010; Bonetta et al., 2010; Madarova et al., 2010; Morio et al., 2008; Qin et al., 2003; Radomski et al., 2010). Given its consistency and quantitative capabilities, q-PCR has potential to be accepted as a standard method for pathogen monitoring. For instance, the Association Française de Normalisation (AFNOR) and the ISO have developed standards NF T90-471 and ISO/TS 12869:2012, respectively, for detection and quantification of Legionella spp. and L. pneumophila by q-PCR (AFNOR, 2015; ISO, 2012).
A variety of q-PCR assays for monitoring Legionella spp. and L. pneumophila in environmental samples have been established for research (Behets et al., 2007b; Nazarian et al., 2008; Wellinghausen et al., 2001) or commercial (Yaradou et al., 2007) purposes. Target genes for Legionella genus level include 16S rRNA (Wellinghausen et al., 2001; Wullings and van der Kooij, 2006) and 23S rRNA (Nazarian et al., 2008) genes, which contains regions conserved for all Legionella species; while the macrophage infectivity potentiator (mip) gene, associated with its virulence, is the most common bio-marker for differentiating L. pneumophila from other Legionella (Behets et al., 2007b; Wellinghausen et al., 2001).
q-PCR assays for the Mycobacterium genus often target the 16S rRNA gene (Adrados et al., 2011; Radomski et al., 2010) or other housekeeping genes (e.g., heat shock protein 65 gene hsp65 (Tobler et al., 2006)). In order to develop a reliable q-PCR assay to detect mycobacteria in water, Radomski (Radomski et al., 2010) tested 18 pairs of previously published primers in silico and in house, finally identifying 110F/I571R as the best candidate. Verification of the newly-developed q-PCR method demonstrated improved specificity, but lower sensitivity relative to two previously-published q-PCR methods. An increasing number of mycobacterial whole-genome data can facilitate identification of alternative genes for improved mycobacterial identification. A q-PCR method based on the atpE gene that codes ATP synthase subunit C was recently proposed for mycobacteria detection and quantification in environmental and clinical samples. This assay was demonstrated to provide adequate specificity and sensitivity, yielding results that correlated well with the Radomski assay (2010) (Radomski et al., 2013). q-PCR was also used to target species level mycobacteria in drinking water, including pathogenic species Mycobacterium avium complex (Chern et al., 2015; Whiley et al., 2015), Mycobacterium intracellulare (Chern et al., 2015), and M. avium subspecies paratuberculosis (Beumer et al., 2010; Chern et al., 2015; Rodriguez-Lazaro et al., 2005).
Commonly used q-PCR-targeted signature sequences for P. aeruginosa identification that have been applied to both clinical and environmental samples include regions of the 16S rRNA, 23S rRNA, gyrase subunit B (gyrB), exotoxin A (ETA), oprl and ecfX genes (Anuj et al., 2009; Lee et al., 2011a; Qin et al., 2003; Schwartz et al., 2006). However, these assays have been criticized for false positive and/or false-negative detection of P. aeruginosa, likely owing to wide sequences variation of P. aeruginosa strains and frequent genetic exchange with other microorganisms (Anuj et al., 2009; Choi et al., 2013; Qin et al., 2003; Schwartz et al., 2006). As a result, some researchers have proposed that a combination of two targets is more suitable for increased specificity and sensitivity of P. aeruginosa identification (Anuj et al., 2009; Qin et al., 2003). For instance, Anuj developed a duplex qPCR assay targeting the ecfX and gyrB genes. Although this assay can provide simultaneous confirmation of positive results, presence of only one gene might require supplement tests with a third gene, as a single positive result might represent either a cross-reaction with a non-P. aeruginosa species or otherwise a P. aeruginosa presenting a sequence variation within one of the target gene regions (Anuj et al., 2009). With advances in DNA sequencing technology and availability of increasing whole genome data, novel targets for P. aeruginosa may yet be identified. Choi et al. recently took advantage of comparative genomic tools and developed a new q-PCR assay targeting the O-antigen acetylase gene. The specificity of this assay was tested against 6 P. aeruginosa isolates, 18 different Pseudomonas species and 23 other reference pathogens (Choi et al., 2013). However, tests against environmental samples would be needed for further assay specificity validation considering variation of background microorganisms in different sample matrices.
There are two commonly applied q-PCR assays for Acanthamoeba, the Riviere (Riviere et al., 2006) and Qvarnstrom (Qvarnstrom et al., 2006) assays, both of which employ Taqman probes and target a fragment of the 18S rRNA gene at the genus level. These two assays are reported to have high specificity and a comparable detection limits when monitoring A. castellanii trophozoites in water and biofilm samples. However, the Qvarnstrom assay is reported to have higher positive detection rates and quantification numbers in terms of environmental sample monitoring (Chang et al., 2010).
Several q-PCR assays for N. fowleri have been developed in recent years to complement traditional culturing and MPN methods for detection in clinical and environmental samples. These assays generally target the 18S rDNA gene (Qvarnstrom et al., 2006), 5.8S gene and internal transcribed spacer (ITS) regions (Mull et al., 2013; Puzon et al., 2009) or the MP2Cl5 sequence (Behets et al., 2007a; Madarova et al., 2010). Puzon developed and applied a q-PCR assay for N. fowleri monitoring in water and biofilm samples, demonstrating higher sensitivity relative to culture methods in terms of spiked cell detection in biofilm and water (i.e., 5 cells in 250 ml water or biofilm) (Puzon et al., 2009). A recent study (Streby et al., 2015) systematically evaluated four previously published q-PCR assays in terms of their themodynamic stability, sensitivity, specificity, detection limits, humic acid inhibition effects, and performances with seeded environmental matrices by standardizing the reaction conditions. Results demonstrated that Qvarnstrom (Qvarnstrom et al., 2006) and Mull assays (Mull et al., 2013) have better performance in terms of detection and amplification limits, but lower specificity (93%); while the Puzon assay (Puzon et al., 2009) was reported to provide100% sensitivity and specificity, but a relatively higher detection limit. In other cases, q-PCR assays targeting Naegleria genus or a more broader free-living amoeba species followed by a melting curve analysis were used for discrimination and identification of N. fowleri and other closely related species (Behets et al., 2006; Robinson et al., 2006).
Head-to-head comparison of OPPP detection by q-PCR and culture of water and biofilm samples has been the subject of multiple studies (Table 2), with the general trend being higher detection frequency and cell numbers by q-PCR (e.g., Legionella (Wang et al., 2012; Wellinghausen et al., 2001; Wullings et al., 2011), mycobacteria (Hussein et al., 2009; Radomski et al., 2013), P. aeruginosa (Bedard et al., 2015), N. fowleri (Behets et al., 2007a)). This phenomenon is likely associated with higher sensitivity and lower detection limit of q-PCR. However, overestimation of OPPP numbers by q-PCR cannot be ruled out, as DNA from dead cells may also be amplified. This possibility may be particularly applicable to samples with high levels of disinfectant residual (Girones et al., 2010), though commonly disinfectants have decayed and microbes are in re-growth mode in premise plumbing. Systematic application of qPCR at multiple monitoring points to inform a mass balance is one approach to discerning information regarding the behavior of viable cells in premise plumbing. Another solution to decrease false positives resulting from DNA from dead cells is to target a longer DNA fragment instead of shorter pieces (McCarty and Atlas, 1993) or pretreatment with nucleic acid-binding dye (see section 3.2.3 for more details). Positive correlations have been observed between q-PCR results and culturing for Legionella under some circumstances (Guillemet et al., 2010; Wellinghausen et al., 2001), indicating the potential of using q-PCR as an alternative method for OPPP monitoring. However, in other cases, no correlations were found (e.g., Legionella (Wullings et al., 2011); mycobacteria (Hussein et al., 2009)). Discrepancies between q-PCR and culturing may also be associated with different sample types and characteristics (e.g., the number of VBNC cells and/or dead cells in samples). An international trial involving 6 participating countries demonstrated less discrepancy of log mean Legoinella number between q-PCR and culturing of plumbing water samples compared to cooling tower samples (Lee et al., 2011b).
Culture is still sometimes more effective for detecting low levels of target pathogens than q-PCR, since DNA extraction can result in substantial cell loss, while culturing under the right conditions can facilitate amplification of dilute cells. For example, one study isolated Mycobacterium spp. from 76% of tap water samples, while q-PCR only yielded about 21–36% detection rates, depending on the DNA extraction method (Radomski et al., 2013). Another study monitoring N. fowleri in water samples also recommended MPN for samples with low N. fowleri numbers (i.e., <200 cells/l) and suggested that DNA extraction and PCR volume limitations contributed to a high q-PCR quantification limit (Behets et al., 2007a). Moreover, the presence of PCR inhibitors may also affect q-PCR readings (Levi et al., 2003). For example, disinfectant residues present in drinking water sample can inhibit PCR amplification (Lee et al., 2011a).
3.2.3 Viable PCR/q-PCR
The major disadvantages of PCR/q-PCR lie in their inabilities to distinguish DNA from live or dead cells. This is problematic for samples with a large amount of dead cells, which is likely to occur in drinking water systems where disinfectants are present or high temperatures exist (e.g., water heaters). Therefore, development of a new molecular method capable of selectively amplifying nucleic acids from viable cells is would be extremely valuable for microbial analysis of premise plumbing.
PCR/q-PCR combined with nucleic acid-binding dye (e.g., ethidium monoazide bromide, propidium monoazide bromide) pretreatment is being explored for selectively monitoring viable OPPPs in drinking water. These intercalating dyes are intended to enter damaged cell membranes and covalently bind to DNA after photo activation, preventing downstream PCR amplification of DNA from membrane-compromised cells. Several studies have indicated promise of an EMA/PMA-based qPCR method for Legionella (Adela Yanez et al., 2011; Chen and Chang, 2010; Delgado-Viscogliosi et al., 2009; Inoue et al., 2015; Mansi et al., 2014; Slimani et al., 2012), mycobacteria (Lee et al., 2015; Nocker et al., 2007), P. aeruginosa (Gensberger et al., 2013, 2014), and Acanthamoeba (Chang et al., 2013) cells by testing against heat-treated, chlorine-treated cells and/or environmental water samples (e.g. drinking water, spa water, swimming pool water) as controls. In general, EMA/PMA-qPCR reduces the detection rates compared to conventional qPCR, but demonstrates equal or higher detection rates and cell counts relative to culturing due to inclusion of VBNC cells (Delgado-Viscogliosi et al., 2009; Inoue et al., 2015; Mansi et al., 2014). The effectiveness of EMA/PMA-qPCR is associated with a variety of factors, such as dye selection and dosage (Chen and Chang, 2010; Yanez et al., 2011), incubation time (Yanez et al., 2011), sample types (Inoue et al., 2015), target-cell loads and background flora (Gensberger et al., 2013; Slimani et al., 2012), as well as characteristics of target microorganisms (e.g., bacterial or eukaryotic cells (Fittipaldi et al., 2011)). Therefore, optimization processes and extra adaptation steps for each sample and/or microorganism type may be needed prior to application of EMA/PMA-qPCR. Further, intercalating dye treatment might not be effective for cells embedded in biofilm, since absolute dominance of non-target microorganisms may reduce the dye-quenching effect and complex biofilm composition and structures may also prevent dye penetration (Taylor et al., 2014). In addition, it has been reported that PMA-PCR/qPCR assays with longer amplicons (e.g., 400 bp) may lead to better suppression of dead-cell signal compared to short-amplicons (e.g., 100 bp), since PMA-induced damage is more likely to occur in longer amplicons (Contreras et al., 2011; Ditommaso et al., 2015).
3.2.4 High throughput DNA sequencing
High throughput DNA sequencing, also called next generation sequencing, is a catch-all term describing a variety of technologies that allow rapid and simultaneous sequencing of millions of nucleic acid fragments (Shendure and Ji, 2008). High throughput sequencing is a powerful tool for understanding the microbial ecology of premise plumbing, revealing diverse microbial community compositions in drinking water distribution systems across the globe (Chao et al., 2015; Delafont et al., 2014; Hong et al., 2010; Ji et al., 2015). A typical procedure for drinking water microbiome investigation includes application of universal primers targeting 16S rRNA genes for DNA amplification followed by high throughput sequencing, the result of which could demonstrate the presence or absence of genera containing potential OPPPs (Delafont et al., 2014; Gomez-Smith et al., 2015; Ji et al., 2015; Wang et al., 2013). Reported abundances of sequences belonging to these genera varied considerably across different samples. Typically, lower abundances have been observed for Legionella (e.g., 0–2.1% and 0–0.48% for Legionella spp. in drinking water (Wang et al., 2014) and biofilm samples (Wang et al., 2013), respectively) compared to Mycobacterium spp. (e.g., 25–78% in water main biofilm samples (Gomez-Smith et al., 2015); ~20% in pooled dataset of municipal drinking water samples from different U.S. cities (Holinger et al., 2014)). However, due to the short read length, high throughput sequencing based on 16S rRNA gene PCR products have limited taxonomic resolution, which inhibits the ability to differentiate species and therefore cannot verify detection of actual pathogens. Thus, more specific primers (e.g., mycobacterial functional genes) have recently been chosen prior to deep sequencing in order to increase the taxonomic resolution. PCR amplification of a 461-bp fragment of the mycobacterial heat shock protein (hsp65) gene followed by Illumina Miseq analysis were applied in water main samples contacting different pipe materials, demonstrating varying abundances of M. frederiksbergense, M. aurum, M. hemophilum, and M. lentiflavum (Gomez-Smith et al., 2015). High throughput sequencing can also be applied at the whole-genome level for high resolution characterization of OPPPs. For instance, Gomez-Valero et al. (Gomez-Valero et al., 2014) sequenced L. micdadei, L. hackeliae and L. fallonii (LLAP10) and compared them with existing Legionella genome data, revealing surprisingly dynamic genomes due to a large mobilome mainly comprising the type IV secretion system. Further, characterization of OPPP whole genomes by high-throughput sequencing might assist in identifying critical subset of genes (i.e., “pan-genome”) defining the pathogenic strains of each OPPP genus from non-pathogenic strains, improving molecular detection of virulence pathogens (Pruden et al., 2013).
3.3 Phenotypic assays
“Phenotypic assays” generally encompass techniques that take advantage of phenotypic properties, such as cell morphological, biochemical, serological, and physiological traits. Here we will mainly discuss immunoassays that have been developed and applied for OPPP detection in environmental samples.
Immunoassays developed for OPPPs typically rely on genus or species-specific monoclonal or polyclonal antibodies to recognize and quantify antigens. These assays can have various formats such as lateral flow assay (Helbig et al., 2006), enzyme-linked immunosorbent assay (ELISA) (Reveiller et al., 2003), immunochromatograpic test (Helbig et al., 2006), and can be combined with other detection methods (e.g., microscopy (Baba et al., 2012), cytometry (Fuechslin et al., 2010), sensors (Bekir et al., 2015; Enrico et al., 2013)). For instance, anti-L. pneumophila and anti-Legionella antibodies have been widely used in a number of immunoassays to detect L. pneumophila of various serogroups and/or distinguish L. pneumophila from other Legionella species (Delgado-Viscogliosi et al., 2005; Fuechslin et al., 2010). In general, Legionella antibodies are conjugated with reporter enzyme or fluorescent tags prior to specific antibody-antigen recognition processes, signals of which are captured after colormetric reactions (Helbig et al., 2006), or directly detected by microscopy (Baba et al., 2012), solid-phase cytometry (Parthuisot et al., 2011) or flow cytometry (Tyndall et al., 1985). In addition, immunomagnetic separation assays in which magnetic beads or particles were covered with antibodies have been used to magnetically recover or concentrate Legionella (Bedrina et al., 2013). Recovered cells can be subjected to downstream analyses such as q-PCR (Mull et al., 2013) or flow cytometry (Fuechslin et al., 2010; Keserue et al., 2012). Furthermore, another layer of staining with cell viability markers in addition to antibody labeling allows simultaneous detection of target microorganisms and live/dead discrimination. These techniques have recently been applied for live Legionella monitoring in drinking water samples (Delgado-Viscogliosi et al., 2005; Keserue et al., 2012). Other reported immunoanalytical methods include construction of miniaturized immonobiosensors (e.g., optic biosensor, impedimetric biosensor) with immobilized antibodies to provide rapid screening and on-site measurement of L. pneumophila and P. aeruginosa in contaminated water samples (Bekir et al., 2015; Enrico et al., 2013).
Advantages of immunoassays include rapid turnaround time (e.g., ~20 min to several hours), high specificity and sensitivity, and overall cost effectiveness (Lesnik, 2000). However, their application efficacy for OPPP monitoring in premise plumbing is still subject to the concentration level of target cells and availability of high quality antibodies. It has been reported that selected antibodies might not be able to cover the whole range of serogroups or species (Keserue et al., 2012) and validation of specificity and sensitivity against environmental background is often lacking (Toriyama et al., 2015; Visvesvara et al., 1987).
4. Conclusions and Research Needs for OPPP Detection Methods
OPPPs pose a particular challenge to monitoring given that they do not correspond to fecal indicators and they establish as part of the native microbiota of premise plumbing systems, which are highly complex and variable from site to site. Further, the ecology and physiology of OPPPs further challenges their detection, particularly when seeking to analyze multiple OPPPs in parallel. Unified approaches to OPPP monitoring and detection are urgently needed, especially in the context of informing proactive risk prevention, outbreak response, and research aimed at better understanding of their behavior and identifying improved means of monitoring and control. Identification and agreement upon effective monitoring practices for OPPPs also stands as an impediment to effective action towards development of policy aimed at preventing their proliferation beyond the property line and protecting public health.
Appropriate methodology for OPPP monitoring is dependent upon the intended goal. Given the high level of resolution needed to distinguish a true pathogen from related, non-pathogenic organisms, culture-based methods are likely to remain the gold standard, particularly for outbreak investigation. However, in the context of risk prevention and mitigation, molecular- and physiological-based methods hold the promise of supporting more extensive, timely, and economical evaluation of problematic sites that could be followed-up upon with culturing. Such an approach could help overcome challenges of applying a comprehensive and representative monitoring plan for complex premise-plumbing systems. For example, although regular application of q-PCR for monitoring L. pneumophila would not directly provide information about live/dead status, it would serve to characterize the baseline and identify “hot spots” that could be acted upon with culturing when there is suspicion of a problem. Essentially, such an approach could serve as a molecular “indicator,” much in the same way the fecal pathogen paradigm has held value as an indicator for fecal pathogens, though the indicators themselves are not pathogens. Approaches for improving the ability of molecular methods for distinguishing live/dead status are currently under development.
In the context of research, molecular-based methods, particularly next-generation DNA sequencing, is beginning to revolutionize understanding of OPPP microbial ecology in plumbing systems. As OPPP whole genomes are being sequenced, new insight is being gained into exactly what differentiates pathogenic from non-pathogenic forms. Thus, molecular-based methods can serve to identify improved markers for highly specific monitoring of virulent OPPP strains. Molecular methods will also improve understanding of their microbial ecology, including identification of key relationships and interactions that could be exploited for improved engineering control. In particular, the relationship between biofilm and bulk water and the role and importance of amoebal hosts in influencing replication and virulence of OPPPs would be extremely valuable.
Acknowledgments
The idea for this manuscript was originally conceived at an expert workshop sponsored by the Water Research Foundation as part of Project 4379 awarded to Virginia Tech. The key knowledge gaps were identified based on consensus at the workshop and subsequent literature review. The findings and conclusions in this report are those of the authors and do not necessarily represent those of the CDC or other sponsors.
References
- Adela Yanez M, Nocker A, Soria-Soria E, Murtula R, Martinez L, Catalan V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. Journal of Microbiological Methods. 2011;85(2):124–130. doi: 10.1016/j.mimet.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Adrados B, Julian E, Codony F, Torrents E, Luquin M, Morato J. Prevalence and concentration of non-tuberculous mycobacteria in cooling towers by means of quantitative PCR: A prospective study. Current Microbiology. 2011;62(1):313–319. doi: 10.1007/s00284-010-9706-2. [DOI] [PubMed] [Google Scholar]
- Ahmed W, Brandes H, Gyawali P, Sidhu JPS, Toze S. Opportunistic pathogens in roof-captured rainwater samples, determined using quantitative PCR. Water Research. 2014;53:361–369. doi: 10.1016/j.watres.2013.12.021. [DOI] [PubMed] [Google Scholar]
- Allegheny County Health Department. Updated guidelins for the control of Legionella in western Pennsylvania. Pittsburgh Regional Health Initiative; 2014. http://www.achd.net/infectd/pubs/pdf/2014_FINAL_Legionella_Guidelines_for_Western_PA.pdf Accesded in Dec, 2016. [Google Scholar]
- Allegra S, Girardot F, Grattard F, Berthelot P, Helbig JH, Pozzetto B, Riffard S. Evaluation of an immunomagnetic separation assay in combination with cultivation to improve Legionella pneumophila serogroup 1 recovery from environmental samples. Journal of Applied Microbiology. 2011;110(4):952–961. doi: 10.1111/j.1365-2672.2011.04955.x. [DOI] [PubMed] [Google Scholar]
- American Public Health Association (APHA) Standard methods for the examination of water and wastewater. 22nd. American Water Works Association and Water Environment Federation; Washington, DC: 2012. [Google Scholar]
- American Society for Testing and Materials (ASTM) ASTM D5952-08 Standard guide for inspection of water systems for Legionella and the investigation of possible outbreaks of Leginellosis (Legionnaires’ disease or pontiac fever) West Conshohocken, PA: 2008. [Google Scholar]
- American Society of Heating and Air-Conditioning Engineers (ASHRAE) Standard 188-2015 Legionellosis: Risk Management for Building Water Systems. ASHRAE; Atlanta, GA: 2015. [Google Scholar]
- Anaissie EJ, Penzak SR, Dignani MC. The hospital water supply as a source of nosocomial infections. Archives of Internal Medicine. 2002;162(13):1483–1492. doi: 10.1001/archinte.162.13.1483. [DOI] [PubMed] [Google Scholar]
- Anuj SN, Whiley DM, Kidd TJ, Bell SC, Wainwright CE, Nissen MD, Sloots TP. Identification of Pseudomonas aeruginosa by a duplex real-time polymerase chain reaction assay targeting the ecfX and the gyrB genes. Diagnostic Microbiology and Infectious Disease. 2009;63(2):127–131. doi: 10.1016/j.diagmicrobio.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Baba T, Inoue N, Yamaguchi N, Nasu M. Rapid enumeration of active Legionella pneumophila in freshwater environments by the microcolony method combined with direct fluorescent antibody staining. Microbes and Environments. 2012;27(3):324–326. doi: 10.1264/jsme2.ME11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaree JM, Gorman GW, Martin WT, Fields BS, Morrill WE. Protocol for sampling environmental sites for legionellae. Applied and Environmental Microbiology. 1987;53(7):1454–1458. doi: 10.1128/aem.53.7.1454-1458.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargellini A, Marchesi I, Righi E, Ferrari A, Cencetti S, Borella P, Rovesti S. Parameters predictive of Legionella contamination in hot water systems: association with trace elements and heterotrophic plate counts. Water Research. 2011;45(6):2315–2321. doi: 10.1016/j.watres.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Bartie C, Venter SN, Nel LH. Evaluation of detection methods for Legionella species using seeded water samples. Water SA. 2001;27(4):523–527. [Google Scholar]
- Bartie C, Venter SN, Nel LH. Identification methods for Legionella from environmental samples. Water Research. 2003;37(6):1362–1370. doi: 10.1016/S0043-1354(02)00220-8. [DOI] [PubMed] [Google Scholar]
- Bates MN, Maas E, Martin T, Harte D, Grubner M, Margolin T. Investigation of the prevalence of Legionella species in domestic hot water systems. New Zealand Medical Journal. 2000;113(1111):218–220. [PubMed] [Google Scholar]
- Bedard E, Laferriere C, Charron D, Lalancette C, Renaud C, Desmarais N, Deziel E, Prevost M. Post-outbreak investigation of Pseudomonas aeruginosa faucet contamination by quantitative polymerase chain reaction and environmental factors affecting positivity. Infection Control and Hospital Epidemiology. 2015;36(11):1337–1343. doi: 10.1017/ice.2015.168. [DOI] [PubMed] [Google Scholar]
- Bedrina B, Macian S, Solis I, Fernandez-Lafuente R, Baldrich E, Rodriguez G. Fast immunosensing technique to detect Legionella pneumophila in different natural and anthropogenic environments: comparative and collaborative trials. BMC Microbiology. 2013;13 doi: 10.1186/1471-2180-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behets J, Declerck P, Delaedt Y, Verelst L, Ollevier F. Quantitative detection and differentiation of free-living amoeba species using SYBR green-based real-time PCR melting curve analysis. Current Microbiology. 2006;53(6):506–509. doi: 10.1007/s00284-006-0241-0. [DOI] [PubMed] [Google Scholar]
- Behets J, Declerck P, Delaedt Y, Verelst L, Ollevier F. A duplex real-time PCR assay for the quantitative detection of Naegleria fowleri in water samples. Water Research. 2007a;41(1):118–126. doi: 10.1016/j.watres.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Behets J, Dederck P, Delaedt Y, Creemers B, Ollevier F. Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. Journal of Microbiological Methods. 2007b;68(1):137–144. doi: 10.1016/j.mimet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Bekir K, Bousimma F, Barhoumi H, Fedhila K, Maaref A, Bakhrouf A, Ben Ouada H, Namour P, Jaffrezic-Renault N, Ben Mansour H. An investigation of the well-water quality: immunosensor for pathogenic Pseudomonas aeruginosa detection based on antibody-modified poly(pyrrole-3 carboxylic acid) screen-printed carbon electrode. Environmental Science and Pollution Research. 2015;22(23):18669–18675. doi: 10.1007/s11356-015-5004-7. [DOI] [PubMed] [Google Scholar]
- Berthelot P, Chord F, Mallaval F, Grattard F, Brajon D, Pozzetto B. Magnetic valves as a source of faucet contamination with Pseudomonas aeruginosa? Intensive Care Medicine. 2006;32(8):1271–1271. doi: 10.1007/s00134-006-0206-6. [DOI] [PubMed] [Google Scholar]
- Beumer A, King D, Donohue M, Mistry J, Covert T, Pfaller S. Detection of Mycobacterium avium subsp. paratuberculosis in drinking water and biofilms by quantitative PCR. Applied and Environmental Microbiology. 2010;76(21):7367–7370. doi: 10.1128/AEM.00730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetta S, Bonetta S, Ferretti E, Balocco F, Carraro E. Evaluation of Legionella pneumophila contamination in Italian hotel water systems by quantitative real-time PCR and culture methods. Journal of Applied Microbiology. 2010;108(5):1576–1583. doi: 10.1111/j.1365-2672.2009.04553.x. [DOI] [PubMed] [Google Scholar]
- Borella P, Montagna MT, Romano-Spica V, Stampi S, Stancanelli G, Triassi M, Neglia R, Marchesi I, Fantuzzi G, Tato D, Napoli C, Quaranta G, Laurenti P, Leoni E, De Luca G, Ossi C, Moro M, Ribera D'Alcala G. Legionella infection risk from domestic hot water. Emerging Infectious Diseases. 2004;10(3):457–464. doi: 10.3201/eid1003.020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borella P, Montagna MT, Stampi S, Stancanelli G, Romano-Spica V, Triassi M, Marchesi I, Bargellini A, Tato D, Napoli C, Zanetti F, Leoni E, Moro M, Scaltriti S, D'Alcala GR, Santarpia R, Boccia S. Legionella contamination in hot water of Italian hotels. Applied and Environmental Microbiology. 2005;71(10):5805–5813. doi: 10.1128/AEM.71.10.5805-5813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger CA, Edelstein PH. Presicion and accuracy of recovery of Legionella pneumophila from seeded tap water by filtration and centrifugation. Applied and Environmental Microbiology. 1995;61(5):1805–1809. doi: 10.1128/aem.61.5.1805-1809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle RJ, Stannett PJ, Cunliffe RN. Legionella pneumophila: comparison of isolation from water specimens by centrifugation and filtration. Epidemiology and Infection. 1987;99(2):241–247. doi: 10.1017/s0950268800067704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder S, Trebesius K, Heesemann J. Evaluation of detection of Legionella spp. in water samples by fluorescence in situ hybridization, PCR amplification and bacterial culture. International Journal of Medical Microbiology. 2002;292(3–4):241–245. doi: 10.1078/1438-4221-00213. [DOI] [PubMed] [Google Scholar]
- Buse HY, Schoen ME, Ashbolt NJ. Legionellae in engineered systems and use of quantitative microbial risk assessment to predict exposure. Water Research. 2012;46(4):921–933. doi: 10.1016/j.watres.2011.12.022. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Guidelines for environmental infection control in health-care facilities. Georgia, Altanta, USA: 2003. [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Procedures for the recovery of Legionella from the environment. 2005 http://www.cdc.gov/legionella/health-depts/inv-tools-cluster/lab-inv-tools/procedures-manual.pdf. Accessed in Dec, 2016.
- Centers for Disease Control Prevention (CDC) Legionellosis — United States, 2000–2009. Morbidity and Mortality Weekly Report. 2011;60(11):1083–1086. [PubMed] [Google Scholar]
- Centers for Disease Control Prevention (CDC) Surveillance for waterborne disease outbreaks associated with drinking water and other nonrecreational water - United States, 2009–2010. Morbidity and Mortality Weekly Report. 2013;62(35):714–720. [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Sampling procedure and potential sampling sites. Protocol for collecting environmental samples for Legionella culture during a cluster or outbreak investigation or when cases of disease may ba associated with a facility. 2015 https://www.cdc.gov/legionella/downloads/cdc-sampling-procedure.pdf Accessed on Dec, 2016.
- Chang CW, Lu LW, Kuo CL, Hung NT. Density of environmental Acanthamoeba and their responses to superheating disinfection. Parasitology Research. 2013;112(11):3687–3696. doi: 10.1007/s00436-013-3556-3. [DOI] [PubMed] [Google Scholar]
- Chang CW, Wu YC, Ming KW. Evaluation of real-time PCR methods for quantification of Acanthamoeba in anthropogenic water and biofilms. Journal of Applied Microbiology. 2010;109(3):799–807. doi: 10.1111/j.1365-2672.2010.04708.x. [DOI] [PubMed] [Google Scholar]
- Chang SL. Resistance of pathogenic Naegleria to some common physical and chemical agents. Applied and Environmental Microbiology. 1978;35(2):368–375. doi: 10.1128/aem.35.2.368-375.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao YQ, Mao YP, Wang ZP, Zhang T. Diversity and functions of bacterial community in drinking water biofilms revealed by high-throughput sequencing. Scientific Reports. 2015;5 doi: 10.1038/srep03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron D, Bedard E, Lalancette C, Laferriere C, Prevost M. Impact of electronic faucets and water quality on the cccurrence of Pseudomonas aeruginosa in water: A multi-hospital study. Infection Control and Hospital Epidemiology. 2015;36(3):311–319. doi: 10.1017/ice.2014.46. [DOI] [PubMed] [Google Scholar]
- Chen NT, Chang CW. Rapid quantification of viable legionellae in water and biofilm using ethidium monoazide coupled with real-time quantitative PCR. Journal of Applied Microbiology. 2010;109(2):623–634. doi: 10.1111/j.1365-2672.2010.04678.x. [DOI] [PubMed] [Google Scholar]
- Chern EC, King D, Haugland R, Pfaller S. Evaluation of quantitative polymerase chain reaction assays targeting Mycobacterium avium, M. intracellulare, and M. avium subspecies paratuberculosis in drinking water biofilms. Journal of Water and Health. 2015;13(1):131–139. doi: 10.2166/wh.2014.060. [DOI] [PubMed] [Google Scholar]
- Cheunoy W, Prammananan T, Chaiprasert A, Foongladda S. Comparative evaluation of polymerase chain reaction and restriction enzyme analysis: Two amplified targets, hsp65 and rpoB, for identification of cultured mycobacteria. Diagnostic Microbiology and Infectious Disease. 2005;51(3):165–171. doi: 10.1016/j.diagmicrobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Chimara E, Ferrazoli L, Ueky SYM, Martins MC, Durham AM, Arbeit RD, Leao SC. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in a reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiology. 2008;8:48. doi: 10.1186/1471-2180-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Kim MH, Cho MS, Kim BK, Kim JY, Kim C, Park DS. Improved PCR for identification of Pseudomonas aeruginosa. Applied Microbiology and Biotechnology. 2013;97(8):3643–3651. doi: 10.1007/s00253-013-4709-0. [DOI] [PubMed] [Google Scholar]
- Contreras PJ, Urrutia H, Sossa K, Nocker A. Effect of PCR amplicon length on suppressing signals from membrane-compromised cells by propidium monoazide treatment. Journal of Microbiological Methods. 2011;87(1):89–95. doi: 10.1016/j.mimet.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Conza L, Casati S, Gaia V. Detection limits of Legionella pneumophila in environmental samples after co-culture with Acanthamoeba polyphaga. BMC Microbiology. 2013;13:49. doi: 10.1186/1471-2180-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope JR, Ratard RC, Hill VR, Sokol T, Causey JJ, Yoder JS, Mirani G, Mull B, Mukerjee KA, Narayanan J, Doucet M, Qvarnstrom Y, Poole CN, Akingbola OA, Ritter JM, Xiong Z, da Silva AJ, Roellig D, Van Dyke RB, Stern H, Xiao L, Beach MJ. The first association of a primary amebic meningoencephalitis death with culturable Naegleria fowleri in tap water from a US treated public drinking water system. Clinical Infectious Diseases. 2015;60(8):e36–42. doi: 10.1093/cid/civ017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert TC, Rodgers MR, Reyes AL, Stelma GN. Occurrence of nontuberculous mycobacteria in environmental samples. Applied and Environmental Microbiology. 1999;65(6):2492–2496. doi: 10.1128/aem.65.6.2492-2496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craun GF, Brunkard JM, Yoder JS, Roberts VA, Carpenter J, Wade T, Calderon RL, Roberts JM, Beach MJ, Roy SL. Causes of outbreaks associated with drinking water in the United States from 1971 to 2006. Clinical Microbiology Reviews. 2010;23(3):507–528. doi: 10.1128/CMR.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca G, Stampi S, Lezzi L, Zanetti F. Effect of heat and acid decontamination treatments on the recovery of Legionella pneumophila from drinking water using two selective media. The New Microbiologica. 1999;22(3):203–208. [PubMed] [Google Scholar]
- Declerck P, Behets J, Margineanu A, van Hoef V, De Keersmaecker B, Ollevier F. Replication of Legionella pneumophila in biofilms of water distribution pipes. Microbiol Research. 2009;164(6):593–603. doi: 10.1016/j.micres.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Delafont V, Mougari F, Cambau E, Joyeux M, Bouchon D, Hechard Y, Moulin L. First evidence of amoebae-mycobacteria association in drinking water network. Environmental Science & Technology. 2014;48(20):11872–11882. doi: 10.1021/es5036255. [DOI] [PubMed] [Google Scholar]
- Delgado-Viscogliosi P, Simonart T, Parent V, Marchand G, Dobbelaere M, Pierlot E, Pierzo V, Menard-Szczebara F, Gaudard-Ferveur E, Delabre K, Delattre JM. Rapid method for enumeration of viable Legionella pneumophila and other Legionella spp. in water. Applied and Environmental Microbiology. 2005;71(7):4086–4096. doi: 10.1128/AEM.71.7.4086-4096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Viscogliosi P, Solignac L, Delattre JM. Viability PCR, a culture-independent method for rapid and selective quantification of viable Legionella pneumophila cells in environmental water samples. Applied and Environmental Microbiology. 2009;75(11):3502–3512. doi: 10.1128/AEM.02878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditommaso S, Giacomuzzi M, Gentile M, Moiraghi AR, Zotti CM. Effective environmental sampling strategies for monitoring Legionella spp contamination in hot water systems. American Journal of Infection Control. 2010;38(5):344–349. doi: 10.1016/j.ajic.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Ditommaso S, Giacomuzzi M, Ricciardi E, Zotti CM. Viability-qPCR for detecting Legionella: Comparison of two assays based on different amplicon lengths. Molecular and Cellular Probes. 2015;29(4):237–243. doi: 10.1016/j.mcp.2015.05.011. [DOI] [PubMed] [Google Scholar]
- Donohue MJ, O'Connell K, Vesper SJ, Mistry JH, King D, Kostich M, Pfaller S. Widespread molecular detection of Legionella pneumophila Serogroup 1 in cold water taps across the United States. Environmental Science & Technology. 2014;48(6):3145–3152. doi: 10.1021/es4055115. [DOI] [PubMed] [Google Scholar]
- Eichler S, Christen R, Höltje C, Westphal P, Bötel J, Brettar I, Mehling A, Höfle MG. Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-Based 16S rRNA Gene Fingerprinting. Applied and Environmental Microbiology. 2006;72(3):1858–1872. doi: 10.1128/AEM.72.3.1858-1872.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enrico DL, Manera MG, Montagna G, Cimaglia F, Chiesa M, Poltronieri P, Santino A, Rella R. SPR based immunosensor for detection of Legionella pneumophila in water samples. Optics Communications. 2013;294:420–426. [Google Scholar]
- Environmental Protection Agency (EPA) Fact Sheet: Final third drinking water contaminant candidate list (CCL3) 2009 EPA 815F09001, Office of Water(4607M) [Google Scholar]
- Falkinham JO., III Hospital water filters as a source of Mycobacterium avium complex. Journal of Medical Microbiology. 2010;59(10):1198–1202. doi: 10.1099/jmm.0.022376-0. [DOI] [PubMed] [Google Scholar]
- Falkinham JO., III Nontuberculous mycobacteria from household plumbing of patients with nontuberculous mycobacteria disease. Emerging Infectious Diseases. 2011;17(3):419–424. doi: 10.3201/eid1703.101510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham JO, III, Iseman MD, de Haas P, van Soolingen D. Mycobacterium avium in a shower linked to pulmonary disease. J Water Health. 2008;6(2):209–213. doi: 10.2166/wh.2008.032. [DOI] [PubMed] [Google Scholar]
- Falkinham JO., III Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. Journal of Applied Microbiology. 2009;107(2):356–367. doi: 10.1111/j.1365-2672.2009.04161.x. [DOI] [PubMed] [Google Scholar]
- Falkinham JO, III, Norton CD, LeChevallier MW. Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other mycobacteria in drinking water distribution systems. Applied and Environmental Microbiology. 2001;67(3):1225–1231. doi: 10.1128/AEM.67.3.1225-1231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. Opportunistic pathogens enriched in showerhead biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(38):16393–16398. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferroni A, Nguyen L, Pron B, Quesne G, Brusset MC, Berche P. Outbreak of nosocomial urinary tract infections due to Pseudomonas aeruginosa in a paediatric surgical unit associated with tap-water contamination. Journal of Hospital Infection. 1998;39(4):301–307. doi: 10.1016/s0195-6701(98)90295-x. [DOI] [PubMed] [Google Scholar]
- Fittipaldi M, Codony F, Adrados B, Camper AK, Morato J. Viable real-time PCR in environmental samples: Can all data be interpreted directly? Microbial Ecology. 2011;61(1):7–12. doi: 10.1007/s00248-010-9719-1. [DOI] [PubMed] [Google Scholar]
- Flanders WD, Kirkland KH, Shelton BG. Effects of holding time and measurement error on culturing Legionella in environmental water samples. Water Research. 2014;62:293–301. doi: 10.1016/j.watres.2014.05.025. [DOI] [PubMed] [Google Scholar]
- Flannery B, Gelling LB, Vugia DJ, Weintraub JM, Salerno JJ, Conroy MJ, Stevens VA, Rose CE, Moore MR, Fields BS, Besser RE. Reducing Legionelia colonization of water systems with monochloramine. Emerging Infectious Diseases. 2006;12(4):588–596. doi: 10.3201/eid1204.051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French Republic and Ministry of Health and Solidarity (FRMHS) Water in Health Care Facilities (Technical Guide) Department of Hospitalization and Organization of Care, Directorate General of Health; Paris, France: p. 2005. [Google Scholar]
- French Republic and Ministry of Labor (FRML) Order of 1 February 2010 relating to the surveillance of legionella in installations for the production, storage and distribution of domestic hot water (JORF No 0033 of 9 February 2010) 2010 [Google Scholar]
- Fuechslin HP, Koetzsch S, Keserue HA, Egli T. Rapid and quantitative detection of Legionella pneumophila applying immunomagnetic separation and flow cytometry. Cytometry Part A. 2010;77A(3):264–274. doi: 10.1002/cyto.a.20858. [DOI] [PubMed] [Google Scholar]
- Garcia A, Goni P, Cieloszyk J, Teresa Fernandez M, Calvo-Begueria L, Rubio E, Francisca Fillat M, Luisa Peleato M, Clavel A. Identification of free-living amoebae and amoeba-associated bacteria from reservoirs and water treatment plants by molecular techniques. Environmental Science & Technology. 2013;47(7):3132–3140. doi: 10.1021/es400160k. [DOI] [PubMed] [Google Scholar]
- Garcia MT, Jones S, Pelaz C, Millar RD, Abu Kwaik Y. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environmental Microbiology. 2007;9(5):1267–1277. doi: 10.1111/j.1462-2920.2007.01245.x. [DOI] [PubMed] [Google Scholar]
- Gensberger ET, Polt M, Konrad-Koeszler M, Kinner P, Sessitsch A, Kostic T. Evaluation of quantitative PCR combined with PMA treatment for molecular assessment of microbial. Water Research. 2014;67:367–376. doi: 10.1016/j.watres.2014.09.022. [DOI] [PubMed] [Google Scholar]
- Gensberger ET, Sessitsch A, Kostic T. Propidium monoazide-quantitative polymerase chain reaction for viable Escherichia coli and Pseudomonas aeruginosa detection from abundant background microflora. Analytical Biochemistry. 2013;441(1):69–72. doi: 10.1016/j.ab.2013.05.033. [DOI] [PubMed] [Google Scholar]
- Girones R, Ferrús MA, Alonso JL, Rodriguez-Manzano J, Calgua B, de Abreu Corrêa A, Hundesa A, Carratala A, Bofill-Mas S. Molecular detection of pathogens in water – The pros and cons of molecular techniques. Water Research. 2010;44(15):4325–4339. doi: 10.1016/j.watres.2010.06.030. [DOI] [PubMed] [Google Scholar]
- Goldschmidt P, Degorge S, Saint-Jean C, Year H, Zekhnini F, Batellier L, Laroche L, Chaumeil C. Resistance of Acanthamoeba to classic DNA extraction methods used for the diagnosis of corneal infections. British Journal of Ophthalmology. 2008;92(1):112–115. doi: 10.1136/bjo.2007.125898. [DOI] [PubMed] [Google Scholar]
- Gomez-Smith CK, LaPara TM, Hozalski RM. Sulfate reducing bacteria and mycobacteria dominate the biofilm communities in a chloraminated drinking water distribution system. Environmental Science & Technology. 2015;49(14):8432–8440. doi: 10.1021/acs.est.5b00555. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Rolando M, Neou M, Dervins-Ravault D, Demirtas J, Rouy Z, Moore RJ, Chen HL, Petty NK, Jarraud S, Etienne J, Steinert M, Heuner K, Gribaldo S, Medigue C, Glockner G, Hartland EL, Buchrieser C. Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease. Genome Biology. 2014;15(11):505. doi: 10.1186/s13059-014-0505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clinical Microbiology Reviews. 2004;17(2):413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemet TA, Levesque B, Gauvin D, Brousseau N, Giroux JP, Cantin P. Assessment of real-time PCR for quantification of Legionella spp. in spa water. Letters in Applied Microbiology. 2010;51(6):639–644. doi: 10.1111/j.1472-765X.2010.02947.x. [DOI] [PubMed] [Google Scholar]
- Halabi M, Wiesholzer-Pittl M, Schoberl J, Mittermayer H. Non-touch fittings in hospitals: a possible source of Pseudomonas aeruginosa and Legionella spp. Journal of Hospital Infection. 2001;49(2):117–121. doi: 10.1053/jhin.2001.1060. [DOI] [PubMed] [Google Scholar]
- Hata A, Katayama H, Kitajima M, Visvanathan C, Nol C, Furumai H. Validation of internal controls for extraction and amplification of nucleic acids from enteric viruses in water samples. Applied and Environmental Microbiology. 2011;77(13):4336–4343. doi: 10.1128/AEM.00077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Health Facilities Scotland. Part C: TVC Testing Protocol. NHS National Services Scotland; 2011. Scottish Health Technical Memorandum 04-01: The control of Legionella, hygiene, ‘safe’ hot water, cold water and drinking water systems; pp. 1–15. [Google Scholar]
- Helbig JH, Luck PC, Kunz B, Bubert A. Evaluation of the Duopath Legionella lateral flow assay for identification of Legionella pneumophila and Legionella species culture isolates. Applied and Environmental Microbiology. 2006;72(6):4489–4491. doi: 10.1128/AEM.00346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holinger EP, Ross KA, Robertson CE, Stevens MJ, Harris JK, Pace NR. Molecular analysis of point-of-use municipal drinking water microbiology. Water Research. 2014;49:225–235. doi: 10.1016/j.watres.2013.11.027. [DOI] [PubMed] [Google Scholar]
- Hong PY, Hwang CC, Ling FQ, Andersen GL, LeChevallier MW, Liu WT. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Applied and Environmental Microbiology. 2010;76(16):5631–5635. doi: 10.1128/AEM.00281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SW, Hsu BM. Survey of Naegleria from Taiwan recreational waters using culture enrichment combined with PCR. Acta Tropica. 2011;119(2–3):114–118. doi: 10.1016/j.actatropica.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Hubbs S. Addressing Legionella: Public health enemy #1 in US water systems. 2014 http://www.waterandhealth.org/addressing-legionella-public-health-enemy-1-water-systems/, Accessed Dec, 2013.
- Hussein Z, Landt O, Wirths B, Wellinghausen N. Detection of non-tuberculous mycobacteria in hospital water by culture and molecular methods. International Journal of Medical Microbiology. 2009;299(4):281–290. doi: 10.1016/j.ijmm.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Huws SA, McBain AJ, Gilbert P. Protozoan grazing and its impact upon population dynamics in biofilm communities. Journal of Applied Microbiology. 2005;98(1):238–244. doi: 10.1111/j.1365-2672.2004.02449.x. [DOI] [PubMed] [Google Scholar]
- Hwang C, Ling F, Andersen GL, LeChevallier MW, Liu WT. Evaluation of methods for the extraction of DNA from drinking water distribution system biofilms. Microbes and Environments. 2012;27(1):9–18. doi: 10.1264/jsme2.ME11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijo T, Izumi Y, Nakamoto S, Yamaguchi N, Nasu M. Distribution and Respiratory activity of mycobacteria in household water system of healthy volunteers in Japan. PLoS ONE. 2014;9(10):e110554. doi: 10.1371/journal.pone.0110554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IDEXX. IDEXX Pseudalert® Approved for Pseudomonas aeruginosa by UBA. 2015 http://www.rapidmicrobiology.com/news/idexx-pseudalert-approved-pseudomonas-aeruginosa/ Accessed on Dec, 2016.
- Inkinen J, Kaunisto T, Pursiainen A, Miettinen IT, Kusnetsou J, Riihinen K, Keinanen-Toivola MM. Drinking water quality and formation of biofilms in an office building during its first year of operation, a full scale study. Water Research. 2014;49:83–91. doi: 10.1016/j.watres.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Inoue H, Takama T, Yoshizaki M, Agata K. Detection of Legionella Species in environmental water by the quantitative PCR method in combination with ethidium monoazide rreatment. Biocontrol Science. 2015;20(1):71–74. doi: 10.4265/bio.20.71. [DOI] [PubMed] [Google Scholar]
- International Standards Organization (ISO) ISO 11731-2:2004 Water quality – Detection and enumeration of Legionella – Part 2: Direct membrane filtration method for waters with low bacterial counts. Geneva; Switzerland: 2004. [Google Scholar]
- International Standards Organization (ISO) ISO/TS 12869:2012 Water quality – Detection and quantification of Legionella spp. and/or Legionella pneumophila by concentration and genic amplification by quantitative polymerase chain reaction (qPCR) Geneva; Switzerland: 2012. [Google Scholar]
- Ithoi I, Ahmad AF, Nissapatorn V, Lau YL, Mahmud R, Mak JW. Detection of Naegleria species in environmental samples from Peninsular Malaysia. PLoS ONE. 2011;6(9):e24327. doi: 10.1371/journal.pone.0024327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Parks J, Edwards MA, Pruden A. Impact of Water Chemistry, Pipe Material and Stagnation on the Building Plumbing Microbiome. PLoS ONE. 2015;10(10):e0141087. doi: 10.1371/journal.pone.0141087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlisch L, Henne K, Draheim J, Brettar I, Hofle MG. High-resolution in situ genotyping of Legionella pneumophila populations in drinking water by multiple-locus variable-number tandem-repeat analysis using environmental DNA. Applied and Environmental Microbiology. 2010;76(18):6186–6195. doi: 10.1128/AEM.00416-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keserue HA, Baumgartner A, Felleisen R, Egli T. Rapid detection of total and viable Legionella pneumophila in tap water by immunomagnetic separation, double fluorescent staining and flow cytometry. Microbial Biotechnology. 2012;5(6):753–763. doi: 10.1111/j.1751-7915.2012.00366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilvington S, Gray T, Dart J, Morlet N, Beeching JR, Frazer DG, Matheson M. Acanthamoeba keratitis: The role of domestic tap water contamination in the United Kingdom. Investigative Ophthalmology & Visual Science. 2004;45(1):165–169. doi: 10.1167/iovs.03-0559. [DOI] [PubMed] [Google Scholar]
- Kyle DE, Noblet GP. Vertical distribution of potentially pathogenic free-living amoebae in freshwater lakes. The Journal of protozoology. 1985;32(1):99–105. doi: 10.1111/j.1550-7408.1985.tb03022.x. [DOI] [PubMed] [Google Scholar]
- Lautenschlager K, Boon N, Wang Y, Egli T, Hammes F. Overnight stagnation of drinking water in household taps induces microbial growth and changes in community composition. Water Research. 2010;44(17):4868–4877. doi: 10.1016/j.watres.2010.07.032. [DOI] [PubMed] [Google Scholar]
- Lavenir R, Sanroma M, Gibert S, Crouzet O, Laurent F, Kravtsoff J, Mazoyer MA, Cournoyer B. Spatio-temporal analysis of infra-specific genetic variations among a Pseudomonas aeruginosa water network hospital population: invasion and selection of clonal complexes. Journal of Applied Microbiology. 2008;105(5):1491–1501. doi: 10.1111/j.1365-2672.2008.03907.x. [DOI] [PubMed] [Google Scholar]
- Lee CS, Wetzel K, Buckley T, Wozniak D, Lee J. Rapid and sensitive detection of Pseudomonas aeruginosa in chlorinated water and aerosols targeting gyrB gene using real-time PCR. Journal of Applied Microbiology. 2011a;111(4):893–903. doi: 10.1111/j.1365-2672.2011.05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ES, Lee MH, Kim BS. Evaluation of propidium monoazide-quantitative PCR to detect viable Mycobacterium fortuitum after chlorine, ozone, and ultraviolet disinfection. International Journal of Food Microbiology. 2015;210:143–148. doi: 10.1016/j.ijfoodmicro.2015.06.019. [DOI] [PubMed] [Google Scholar]
- Lee JV, Lai S, Exner M, Lenz J, Gaia V, Casati S, Hartemann P, Lueck C, Pangon B, Ricci ML, Scaturro M, Fontana S, Sabria M, Sanchez I, Assaf S, Surman-Lee S. An international trial of quantitative PCR for monitoring Legionella in artificial water systems. Journal of Applied Microbiology. 2011b;110(4):1032–1044. doi: 10.1111/j.1365-2672.2011.04957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtola MJ, Torvinen E, Kusnetsov J, Pitkanen T, Maunula L, von Bonsdorff CH, Martikainen PJ, Wilks SA, Keevil CW, Miettinen IT. Survival of Mycobacterium avium, Legionella pneumophila, Escherichia coli, and caliciviruses in drinking water-associated biofilms grown under high-shear turbulent flow. Applied and Environmental Microbiology. 2007;73(9):2854–2859. doi: 10.1128/AEM.02916-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni E, Legnani PP. Comparison of selective procedures for isolation and enumeration of Legionella species from hot water systems. Journal of Applied Microbiology. 2001;90(1):27–33. doi: 10.1046/j.1365-2672.2001.01178.x. [DOI] [PubMed] [Google Scholar]
- Leoni E, Legnani PP, Sabattini MAB, Righi F. Prevalence of Legionella spp. in swimming pool environment. Water Research. 2001;35(15):3749–3753. doi: 10.1016/s0043-1354(01)00075-6. [DOI] [PubMed] [Google Scholar]
- Lesnik B. Immunoassay techniques in environmental analyses. In: Meyers RA, editor. Encyclopedia of analytical chemistry: Applications, Theory and Instrumentation, Environment: Water and Waste. Vol. 3. John Wiley and Sons; New York: 2000. 2000. p. 2653. [Google Scholar]
- Levi K, Smedley J, Towner KJ. Evaluation of a real-time PCR hybridization assay for rapid detection of Legionella pneumophila in hospital and environmental water samples. Clinical Microbiology and Infection. 2003;9(7):754–758. doi: 10.1046/j.1469-0691.2003.00666.x. [DOI] [PubMed] [Google Scholar]
- Liu RY, Yu ZS, Guo HG, Liu MM, Zhang HX, Yang M. Pyrosequencing analysis of eukaryotic and bacterial communities in faucet biofilms. Science of the Total Environment. 2012;435:124–131. doi: 10.1016/j.scitotenv.2012.07.022. [DOI] [PubMed] [Google Scholar]
- Lucas CE, Taylor TH, Jr, Fields BS. Accuracy and precision of Legionella isolation by US laboratories in the ELITE program pilot study. Water Research. 2011;45(15):4428–4436. doi: 10.1016/j.watres.2011.05.030. [DOI] [PubMed] [Google Scholar]
- Madarova L, Trnkova K, Feikova S, Klement C, Obernauerova M. A real-time PCR diagnostic method for detection of Naegleria fowleri. Experimental Parasitology. 2010;126(1):37–41. doi: 10.1016/j.exppara.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Mansi A, Amori I, Marchesi I, Marcelloni AM, Proietto AR, Ferranti G, Magini V, Valeriani F, Borella P. Legionella spp. survival after different disinfection procedures: Comparison between conventional culture, qPCR and EMA-qPCR. Microchemical Journal. 2014;112:65–69. [Google Scholar]
- Marciano-Cabral F, Jamerson M, Kaneshiro ES. Free-living amoebae, Legionella and Mycobacterium in tap water supplied by a municipal drinking water utility in the USA. Journal of Water and Health. 2010;8(1):71–82. doi: 10.2166/wh.2009.129. [DOI] [PubMed] [Google Scholar]
- Martinelli F, Caruso A, Moschini L, Turano A, Scarcella C, Speziani F. A comparison of Legionella pneumophila occurrence in hot water tanks and instantaneous devices in domestic, nosocomial, and community environments. Current Microbiology. 2000;41(5):374–376. doi: 10.1007/s002840010152. [DOI] [PubMed] [Google Scholar]
- Mathys W, Stanke J, Harmuth M, Junge-Mathys E. Occurrence of Legionella in hot water systems of single-family residences in suburbs of two German cities with special reference to solar and district heating. International Journal of Hygiene and Environmental Health. 2008;211(1–2):179–185. doi: 10.1016/j.ijheh.2007.02.004. [DOI] [PubMed] [Google Scholar]
- McCarty SC, Atlas RM. Effect of amplicon size on PCR detection of bacteria exposed to chlorine. PCR Methods and Applications. 1993;3(3):181–185. doi: 10.1101/gr.3.3.181. [DOI] [PubMed] [Google Scholar]
- McCoy WF, Downes EL, Leonidas LF, Cain MF, Sherman DL, Chen K, Devender S, Neuille MJ. Inaccuracy in Legionella tests of building water systems due to sample holding time. Water Research. 2012;46(11):3497–3506. doi: 10.1016/j.watres.2012.03.062. [DOI] [PubMed] [Google Scholar]
- Moore MR, Pryor M, Fields B, Lucas C, Phelan M, Besser RE. Introduction of monochloramine into a municipal water system: Impact on colonization of buildings by Legionella spp. Applied and Environmental Microbiology. 2006;72(1):378–383. doi: 10.1128/AEM.72.1.378-383.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morio F, Corvec S, Caroff N, Le Gallou F, Drugeon H, Reynaud A. Real-time PCR assay for the detection and quantification of Legionella pneumophila in environmental water samples: Utility for daily practice. International Journal of Hygiene and Environmental Health. 2008;211(3–4):403–411. doi: 10.1016/j.ijheh.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Mull BJ, Narayanan J, Hill VR. Improved method for the detection and quantification of Naegleria fowleri in water and sediment using immunomagnetic separation and real-time PCR. Journal of parasitology research. 2013;2013:608367. doi: 10.1155/2013/608367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Iatta R, Fasano F, Marsico T, Montagna MT. Variable bacterial load of Legionella spp. in a hospital water system. Science of the Total Environment. 2009;408(2):242–244. doi: 10.1016/j.scitotenv.2009.09.039. [DOI] [PubMed] [Google Scholar]
- National Research Coucil (NRC) Drinking water distribution systems: Assessing and reducing risks. The national academies press; Washington DC: 2006. [Google Scholar]
- Nazarian EJ, Bopp DJ, Saylors A, Limberger RJ, Musser KA. Design and implementation of a protocol for the detection of Legionella in clinical and environmental samples. Diagnostic Microbiology and Infectious Disease. 2008;62(2):125–132. doi: 10.1016/j.diagmicrobio.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Neumann M, SchulzeRobbecke R, Hagenau C, Behringer K. Comparison of methods for isolation of mycobacteria from water. Applied and Environmental Microbiology. 1997;63(2):547–552. doi: 10.1128/aem.63.2.547-552.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocker A, Burr M, Camper AK. Synthesis document on molecular techniques for the drinking water industry. American Water Works Association Research Foundation. 2009 http://www.waterrf.org/Pages/Projects.aspx?PID=3110.
- Nocker A, Sossa KE, Camper AK. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. Journal of Microbiological Methods. 2007;70(2):252–260. doi: 10.1016/j.mimet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Paris T, Skali-Lami S, Block JC. Effect of wall shear rate on biofilm deposition and grazing in drinking water flow chambers. Biotechnology and Bioengineering. 2007;97(6):1550–1561. doi: 10.1002/bit.21321. [DOI] [PubMed] [Google Scholar]
- Parthuisot N, Binet M, Touron-Bodilis A, Pougnard C, Lebaron P, Baudart J. Total and viable Legionella pneumophila cells in hot and natural waters as measured by immunofluorescence-based assays and solid-phase cytometry. Applied and Environmental Microbiology. 2011;77(17):6225–6232. doi: 10.1128/AEM.00393-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival SL, Knapp JS, Wales DS, Edyvean RGJ. The effect of turbulent flow and surface roughness on biofilm formation in drinking water. Journal of Industrial Microbiology & Biotechnology. 1999;22(3):152–159. [Google Scholar]
- Pernin P, Pélandakis M, Rouby Y, Faure A, Siclet F. Comparative recoveries of Naegleria fowleri amoebae from seeded river water by filtration and centrifugation. Applied and Environmental Microbiology. 1998;64(3):955–959. doi: 10.1128/aem.64.3.955-959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor CR, Gachter M, Kotzsch S, Rolli F, Sigrist R, Walser JC, Hammes F. Biofilms in shower hoses - choice of pipe material influences bacterial growth and communities. Environmental Science-Water Research & Technology. 2016;2(4):670–682. [Google Scholar]
- Pruden A, Edwards M, Falkinham JO., III Research needs for opportunisitc pathogens in premise plumbing WRF # 4379 Final Report. 2013 http://www.waterrf.org/PublicReportLibrary/4379.pdf.
- Pussard M, Pons R. Morphology of the cystic wall and taxonomy of the genus Acanthamoeba (Protozoa, Amoebida) Protistologica. 1977;13(4):557–598. [Google Scholar]
- Puzon GJ, Lancaster JA, Wylie JT, Plumb JJ. Rapid detection of Naegleria fowleri in water distribution pipeline biofilms and drinking water samples. Environmental Science & Technology. 2009;43(17):6691–6696. doi: 10.1021/es900432m. [DOI] [PubMed] [Google Scholar]
- Qin X, Emerson J, Stapp J, Stapp L, Abe P, Burns JL. Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli from patients with cystic fibrosis. Journal of Clinical Microbiology. 2003;41(9):4312–4317. doi: 10.1128/JCM.41.9.4312-4317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnstrom Y, Visvesvara GS, Sriram R, da Silva AJ. Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. Journal of Clinical Microbiology. 2006;44(10):3589–3595. doi: 10.1128/JCM.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomski N, Lucas FS, Moilleron R, Cambau E, Haenn S, Moulin L. Development of a real-time qPCR method for detection and enumeration of Mycobacterium spp. in surface water. Applied and Environmental Microbiology. 2010;76(21):7348–7351. doi: 10.1128/AEM.00942-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomski N, Roguet A, Lucas FS, Veyrier FJ, Cambau E, Accrombessi H, Moilleron R, Behr MA, Moulin L. atpE gene as a new useful specific molecular target to quantify Mycobacterium in environmental samples. BMC Microbiology. 2013;13:277. doi: 10.1186/1471-2180-13-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regierung D. Regulation on the quality of water intended for human consumption (Drinking water regulation - TrinkwV 2001) German Drinking Water Ordinance 2001 [Google Scholar]
- Reinthaler FF, Sattler J, Schaffler-Dullnig K, Weinmayr B, Marth E. Comparative study of procedures for isolation and cultivation of Legionella pneumophila from tap water in hospitals. Journal of Clinical Microbiology. 1993;31(5):1213–1216. doi: 10.1128/jcm.31.5.1213-1216.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveiller FL, Varenne MP, Pougnard C, Cabanes PA, Pringuez E, Pourima B, Legastelois S, Pernin P. An enzyme-linked immunosorbent assay (ELISA) for the identification of Naegleria fowleri in environmental water samples. Journal of Eukaryotic Microbiology. 2003;50(2):109–113. doi: 10.1111/j.1550-7408.2003.tb00244.x. [DOI] [PubMed] [Google Scholar]
- Riviere D, Szczebara FM, Berjeaud JM, Frere J, Hechard Y. Development of a real-time PCR assay for quantification of Acanthamoeba trophozoites and cysts. Journal of Microbiological Methods. 2006;64(1):78–83. doi: 10.1016/j.mimet.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Robinson BS, Monis PT, Dobson PJ. Rapid, sensitive, and discriminating identification of Naegleria spp. by real-time PCR and melting-curve analysis. Applied and Environmental Microbiology. 2006;72(9):5857–5863. doi: 10.1128/AEM.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lazaro D, D’Agostino M, Herrewegh A, Pla M, Cook N, Ikonomopoulos J. Real-time PCR-based methods for detection of Mycobacterium avium subsp paratuberculosis in water and milk. International Journal of Food Microbiology. 2005;101(1):93–104. doi: 10.1016/j.ijfoodmicro.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Rogues AM, Boulestreau H, Lasheras A, Boyer A, Gruson D, Merle C, Castaing Y, Bebear CM, Gachie JP. Contribution of tap water to patient colonisation with Pseudomonas aeruginosa in a medical intensive care unit. Journal of Hospital Infection. 2007;67(1):72–78. doi: 10.1016/j.jhin.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Sartory DP, Pauly D, Garrec N, Bonadonna L, Semproni M, Schell C, Reimann A, Firth SJ, Thom C, Hartemann P, Exner M, Baldauf H, Lee S, Lee JV. Evaluation of an MPN test for the rapid enumeration of Pseudomonas aeruginosa in hospital waters. Journal of Water and Health. 2015;13(2):427–436. doi: 10.2166/wh.2014.187. [DOI] [PubMed] [Google Scholar]
- Schoen ME, Ashbolt NJ. An in-premise model for Legionella exposure during showering events. Water Research. 2011;45(18):5826–5836. doi: 10.1016/j.watres.2011.08.031. [DOI] [PubMed] [Google Scholar]
- Schwake DO, Garner E, Strom OR, Pruden A, Edwards MA. Legionella DNA markers in tap water coincident with a spike in Legionnaires' Disease in Flint, MI. Environmental Science & Technology Letters. 2016;3(9):311–315. [Google Scholar]
- Schwartz T, Volkmann H, Kirchen S, Kohnen W, Schon-Holz K, Jansen B, Obst U. Real-time PCR detection of Pseudomonas aeruginosa in clinical and municipal wastewater and genotyping of the ciprofloxacin-resistant isolates. FEMS Microbiology Ecology. 2006;57(1):158–167. doi: 10.1111/j.1574-6941.2006.00100.x. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nature Biotechnology. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Slimani S, Robyns A, Jarraud S, Molmeret M, Dusserre E, Mazure C, Facon JP, Lina G, Etienne J, Ginevra C. Evaluation of propidium monoazide (PMA) treatment directly on membrane filter for the enumeration of viable but non cultivable Legionella by qPCR. Journal of Microbiological Methods. 2012;88(2):319–321. doi: 10.1016/j.mimet.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Sobral D, Le Cann P, Gerard A, Jarraud S, Lebeau B, Loisy-Hamon F, Vergnaud G, Pourcel C. High-throughput typing method to identify a non-outbreak-involved Legionella pneumophila strain colonizing the entire water supply system in the Town of Rennes, France. Applied and Environmental Microbiology. 2011;77(19):6899–6907. doi: 10.1128/AEM.05556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Harrington GW, Xagoraraki I, Goel R. Factors affecting bulk to total bacteria ratio in drinking water distribution systems. Water Research. 2008;42(13):3393–3404. doi: 10.1016/j.watres.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Stockman LJ, Wright CJ, Visvesvara GS, Fields BS, Beach MJ. Prevalence of Acanthamoeba spp. and other free-living amoebae in household water, Ohio, USA-1990-1992. Parasitology Research. 2011;108(3):621–627. doi: 10.1007/s00436-010-2120-7. [DOI] [PubMed] [Google Scholar]
- Stout JE, Yu VL, Yee YC, Vaccarello S, Diven W, Lee TC. Legionella pneumophila in residential water supplies: environmental surveillance with clinical assessment for Legionnaires’ disease. Epidemiology and Infection. 1992;109(1):49–57. [PMC free article] [PubMed] [Google Scholar]
- Streby A, Mull BJ, Levy K, Hill VR. Comparison of real-time PCR methods for the detection of Naegleria fowleri in surface water and sediment. Parasitology Research. 2015;114(5):1739–1746. doi: 10.1007/s00436-015-4359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora JL, Keleti G, Martinez AJ. Occurrence and pathogenicity of Naegleria fowleri in artificially heated waters. Applied and Environmental Microbiology. 1983;45(3):974–979. doi: 10.1128/aem.45.3.974-979.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta AC, Stout JE, Yu VL, Wagener MM. Comparison of culture methods for monitoring Legionella species in hosptial potable water systems and recommendations for standardization of such methods. Journal of Clinical Microbiology. 1995;33(8):2118–2123. doi: 10.1128/jcm.33.8.2118-2123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Ross K, Bentham R. Legionella, protozoa, and biofilms: Interactions within complex microbial systems. Microbial Ecology. 2009;58(3):538–547. doi: 10.1007/s00248-009-9514-z. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Bentham RH, Ross KE. Limitations of using propidium monoazide with qPCR to discriminate between live and dead Legionella in biofilm samples. Microbiology Insights. 2014;7:15–24. doi: 10.4137/MBI.S17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Association Française de Normalisation (AFNOR) NF T90-471 Water quality - Detection and quantification of Legionella and / or Legionella pneumophila by concentration and gene amplification by real-time polymerase chain reaction (qPCR) 2015 Jun; 2015. [Google Scholar]
- Thomas JM, Ashbolt NJ. Do free-living amoebae in treated drinking water systems present an emerging health risk? Environmental Science & Technology. 2011;45(3):860–869. doi: 10.1021/es102876y. [DOI] [PubMed] [Google Scholar]
- Thomas JM, Thomas T, Stuetz RM, Ashbolt NJ. Your garden hose: A potential health risk due to Legionella spp. Growth facilitated by free-living amoebae. Environmental Science & Technology. 2014;48(17):10456–10464. doi: 10.1021/es502652n. [DOI] [PubMed] [Google Scholar]
- Thomas V, Herrera-Rimann K, Blanc DS, Greub G. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Applied and Environmental Microbiology. 2006;72(4):2428–2438. doi: 10.1128/AEM.72.4.2428-2438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas V, Loret JF, Jousset M, Greub G. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environmental Microbiology. 2008;10(10):2728–2745. doi: 10.1111/j.1462-2920.2008.01693.x. [DOI] [PubMed] [Google Scholar]
- Thomson R, Carter R, Gilpin C, Coulter C, Hargreaves M. Comparison of methods for processing drinking water samples for the isolation of Mycobacterium avium and Mycobacterium intracellulare. Applied and Environmental Microbiology. 2008;74(10):3094–3098. doi: 10.1128/AEM.02009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler NE, Pfunder M, Herzog K, Frey JE, Altwegg M. Rapid detection and species identification of Mycobacterium spp. using real-time PCR and DNA-Microarray. Journal of Microbiological Methods. 2006;66(1):116–124. doi: 10.1016/j.mimet.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Toriyama K, Suzuki T, Inoue T, Eguchi H, Hoshi S, Inoue Y, Aizawa H, Miyoshi K, Ohkubo M, Hiwatashi E, Tachibana H, Ohashi Y. Development of an immunochromatographic assay kit using fluorescent silica nanoparticles for rapid diagnosis of Acanthamoeba Keratitis. Journal of Clinical Microbiology. 2015;53(1):273–277. doi: 10.1128/JCM.02595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torvinen E, Suomalainen S, Lehtola MJ, Miettinen IT, Zacheus O, Paulin L, Katila ML, Martikainen PJ. Mycobacteria in water and loose deposits of drinking water distribution systems in Finland. Applied and Environmental Microbiology. 2004;70(4):1973–1981. doi: 10.1128/AEM.70.4.1973-1981.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann M, Michalsky T, Wiedeck H, Radosavljevic V, Ruhnke M. Tap water colonization with Pseudomonas aeruginosa in a surgical intensive care unit (ICU) and relation to Pseudomonas infections of ICU patients. Infection Control and Hospital Epidemiology. 2001;22(1):49–52. doi: 10.1086/501828. [DOI] [PubMed] [Google Scholar]
- Tyndall RL, Hand RE, Jr, Mann RC, Evans C, Jernigan R. Application of flow cytometry to detection and characterization of Legionella spp. Applied and Environmental Microbiology. 1985;49(4):852–857. doi: 10.1128/aem.49.4.852-857.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndall RL, Ironside KS, Metler PL, Tan EL, Hazen TC, Fliermans CB. Effect of thermal additions on the density and distribution of thermophilic amoebae and pathogenic Naegleria fowleri in a newly created cooling lake. Applied and Environmental Microbiology. 1989;55(3):722–732. doi: 10.1128/aem.55.3.722-732.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.K. Department of Health. Part B: operational management. London: The stationery office; 2006. Water systems: Health technical memorandum 04-01: The control of Legionella, hygiene, “safe” hot water, cold water and drinking water systems. http://www.whtlimited.com/doc/lib/98/htm-04-01-part-b-20061009113435.pdf Accessed in Dec, 2016. [Google Scholar]
- U.K. Department of Health. Water systems health technical memorandum 04-01: Addendum. Pseudomonas aeruginosa - advice of augmented care units. 2013 https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/140105/Health_Technical_Memorandum_04-01_Addendum.pdf Accessed in Dec, 2016.
- U.K. Environmental Agency. The determination of Legionella bacteria in waters and other environmental samples (2005)- Part 1 - Rationale of surveying and sampling - Methods for the examination of waters and associated materials. 2005 https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/316814/book_200_1028650.pdf Accessed in Dec, 2016.
- van der Mee-Marquet N, Bloc D, Briand L, Besnier JM, Quentin R. Non-touch fittings in hospitals: a procedure to eradicate Pseudomonas aeruginosa contamination. Journal of Hospital Infection. 2005;60(3):235–239. doi: 10.1016/j.jhin.2004.11.023. [DOI] [PubMed] [Google Scholar]
- van der Wielen PWJJ, van der Kooij D. Nontuberculous mycobacteria, fungi, and opportunistic pathogens in unchlorinated drinking water in the Netherlands. Applied and Environmental Microbiology. 2013;79(3):825–834. doi: 10.1128/AEM.02748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veterans Health Administration (VHA) Prevention of healthcare-associated Legionella disease and scald injury from potable water distributioon systems. VHA Directive. 2014;1061 [Google Scholar]
- Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunology and Medical Microbiology. 2007;50(1):1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- Visvesvara GS, Peralta MJ, Brandt FH, Wilson M, Aloisio C, Franko E. Production of monoclonal antibodies to Naegleria fowleri, agent of primary amebic meningoencephalitis. Journal of Clinical Microbiology. 1987;25(9):1629–1634. doi: 10.1128/jcm.25.9.1629-1634.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker S, Schreiber C, Kistemann T. Drinking water quality in household supply infrastructure-A survey of the current situation in Germany. International Journal of Hygiene and Environmental Health. 2010;213(3):204–209. doi: 10.1016/j.ijheh.2010.04.005. [DOI] [PubMed] [Google Scholar]
- von Baum H, Bommer M, Forke A, Holz J, Frenz P, Wellinghausen N. Is domestic tap water a risk for infections in neutropenic patients? Infection. 2010;38(3):181–186. doi: 10.1007/s15010-010-0005-4. [DOI] [PubMed] [Google Scholar]
- Wang H, Edwards M, Falkinham JO, Pruden A. Molecular survey of occurrence of Legionella spp., Mycobacterium spp., Pseudomonas aeruginosa and amoeba hosts in two chloraminated drinking water distribution systems. Applied and Environmental Microbiology. 2012;78(17):6285–6294. doi: 10.1128/AEM.01492-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Proctor CR, Edwards MA, Pryor M, Domingo JWS, Ryu H, Camper AK, Olson A, Pruden A. Microbial community response to chlorine conversion in a chloraminated drinking water distribution system. Environmental Science & Technology. 2014;48(18):10624–10633. doi: 10.1021/es502646d. [DOI] [PubMed] [Google Scholar]
- Wang H, Pryor M, Edwards M, Falkinham JOI, Pruden A. Effect of GAC pre-treatment and disinfectant on microbial community structure and opportunistic pathogen occurrence. Water Research. 2013;47(15):5760–5772. doi: 10.1016/j.watres.2013.06.052. [DOI] [PubMed] [Google Scholar]
- Wellinghausen N, Frost C, Marre R. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Applied and Environmental Microbiology. 2001;67(9):3985–3993. doi: 10.1128/AEM.67.9.3985-3993.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley H, Keegan A, Fallowfield H, Bentham R. The presence of opportunistic pathogens, Legionella spp., L.pneumophila and Mycobacterium avium complex, in South Australian reuse water distribution pipelines. Journal of Water and Health. 2015;13(2):553–561. doi: 10.2166/wh.2014.317. [DOI] [PubMed] [Google Scholar]
- Winck MAT, Caumo K, Rott MB. Prevalence of Acanthamoeba from tap water in Rio Grande do Sul, Brazil. Current Microbiology. 2011;63(5):464–469. doi: 10.1007/s00284-011-0003-5. [DOI] [PubMed] [Google Scholar]
- Winthrop KL, McNelley E, Kendall B, Marshall-Olson A, Morris C, Cassidy M, Saulson A, Hedberg K. Pulmonary nontuberculous mycobacterial disease prevalence and clinical features an emerging public health disease. American Journal of Respiratory and Critical Care Medicine. 2010;182(7):977–982. doi: 10.1164/rccm.201003-0503OC. [DOI] [PubMed] [Google Scholar]
- Wullings BA, Bakker G, van der Kooij D. Concentration and diversity of uncultured Legionella spp. in two unchlorinated drinking water supplies with different concentrations of natural organic matter. Applied and Environmental Microbiology. 2011;77(2):634–641. doi: 10.1128/AEM.01215-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullings BA, van der Kooij D. Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15 degrees C. Applied and Environmental Microbiology. 2006;72(1):157–166. doi: 10.1128/AEM.72.1.157-166.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez MA, Nocker A, Soria-Soria E, Murtula R, Martinez L, Catalan V. Quantification of viable Legionella pneumophila cells using propidium monoazide combined with quantitative PCR. Journal of Microbiological Methods. 2011;85(2):124–130. doi: 10.1016/j.mimet.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Yaradou DF, Hallier-Soulier S, Moreau S, Poty F, Hillion Y, Reyrolle M, Andre J, Festoc G, Delabre K, Vandenesch F, Etienne J, Jarraud S. Integrated real-time PCR for detection and monitoring of Legionella pneumophila in water systems. Applied and Environmental Microbiology. 2007;73(5):1452–1456. doi: 10.1128/AEM.02399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JS, Straif-Bourgeois S, Roy SL, Moore TA, Visvesvara GS, Ratard RC, Hill VR, Wilson JD, Linscott AJ, Crager R, Kozak NA, Sriram R, Narayanan J, Mull B, Kahler AM, Schneeberger C, da Silva AJ, Poudel M, Baumgarten KL, Xiao LH, Beach MJ. Primary amebic meningoencephalitis deaths associated with sinus irrigation using contaminated tap water. Clinical Infectious Diseases. 2012;55(9):E79–E85. doi: 10.1093/cid/cis626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, McCann C, Hanrahan J, Jencson A, Joyce D, Fyffe S, Piesczynski S, Hawks R, Stout JE, Yu VL, Vidic RD. Legionella control by chlorine dioxide in hospital water systems. Journal American Water Works Association. 2009;101(5):117–127. [Google Scholar]


