Abstract
Endometriosis is a heritable, complex chronic inflammatory disease, for which much of the causal pathogenic mechanism remain unknown. Genome-wide association studies (GWAS) to date have identified 12 single nucleotide polymorphisms or SNPs at 10 independent genetic loci associated with endometriosis. Most of these were more strongly associated with rAFS stage III/IV, rather than I/II. The loci are almost all located in inter-genic regions that are known to play a role in the regulation of expression of target genes yet to be identified. To identify the target genes and pathways perturbed by the implicated variants, studies are required involving functional genomic annotation of the surrounding chromosomal regions, in terms of transcriptor factor binding, epigenetic modification (e.g. DNA methylation and histone modification) sites, as well as their correlation with RNA transcription. These studies need to be conducted in tissue types relevant to endometriosis – in particular endometrium. In addition, to allow biologically and clinically relevant interpretation of molecular profiling data, they need to be combined and correlated with detailed, systematically collected phenotypic information (surgical and clinical). The WERF Endometriosis Phenome and Biobanking Harmonization project (EPHect) is a global standardisation initiative that has produced consensus data and sample collection protocols for endometriosis research. These now pave the way for collaborative studies integrating phenomic with genomic data, to identify informative subtypes of endometriosis that will enhance understanding of the pathogenic mechanisms of the disease and discovery of novel, targeted treatments.
Keywords: endometriosis, genes, genome-wide association study (GWAS), epigenetic, phenotype, EPHect
Introduction
Endometriosis is a chronic inflammatory condition affecting an estimated 176M women worldwide in their reproductive years1, associated with pelvic pain (dysmenorrhea, dyspareunia and chronic pelvic pain) as well as reduced fertility2. It is characterised by the presence of tissue resembling endometrium outside the uterine cavity. This is present mainly on pelvic surfaces such as the ovaries, uterosacral ligaments, peritoneal surface, and in deeper tissue such as the rectovaginal septum or the bladder. Definitive diagnosis can only be established through surgical (laparoscopic) visualisation of the lesions ideally with histological verification3, resulting in reported average diagnostic delays worldwide of 7-10 years from onset of symptoms4. The disease is most commonly staged using the revised American Fertility Society (AFS)/American Society for Reproductive Medicine (ASRM) stage I-IV classification5 6, based on lesion number and size, presence of adhesions and ovarian vs. peritoneal involvement. Higher stages are assigned with the occurrence of ovarian cysts and adhesions, but do not correlate well with pain severity7. Current treatment options are limited to surgical removal (often repeated) of lesions, and/or hormone medication with considerable side-effects2.
The exact prevalence of endometriosis in the population is unknown, because of the need for a surgical diagnosis, but it is estimated to affect approximately 5-10% of women during the reproductive years8; the prevalence increases to 35-50% in women having a laparoscopy for endometriosis-associated symptoms of pelvic pain and subfertility9. The condition has substantial long-term impact on the quality of life of sufferers and their families, affecting general wellbeing, daily life, and work productivity4. It also carries a substantial economic burden, with direct and indirect costs combined estimated at €9500/woman/year, and treatment costs similar to those of type II diabetes and rheumatoid arthritis10. Therefore, endometriosis is an area of major unmet personal, clinical and societal need, for which novel treatments and methods of non-invasive diagnosis urgently need to be identified.
The path to providing new therapeutic options that benefit patients lies in improved understanding of the fundamental pathogenic processes underlying endometriosis. Although advances have been made in the past decade shedding more light on mechanisms that are likely to promote the maintenance and growth of lesions, the actual causes remain largely unclear. The most widely accepted origin of peritoneal lesions is through retrograde menstruation, in which menstrual debris containing viable endometrial cells enter the pelvic cavity via the Fallopian tubes11. This hypothesis is supported by prospective epidemiological studies showing that increased ‘exposure to menstruation’ (e.g. early age at menarche, increased menstrual flow, shorter cycle length)12 increases disease risk. As retrograde menstruation is common13, research has focused on providing explanations for the selective adherence of endometrial cells and progressive growth into lesions, including the potential role of endometrial aberrations in women with endometriosis, altered peritoneal environment, impaired immune surveillance and increased angiogenic capacity2 14. Certainly, inflammatory and immune responses, angiogenesis, and apoptosis are altered in affected women favouring the survival and maintenance of endometriotic tissue15. In particular, lesions produce estradiol, in a feedback cycle that favours the overexpression of P450 aromatase and decreases the expression of 17β-HSD2, while failing to respond to progesterone (‘progesterone resistance’) required to counteract estradiol effects15. This mechanism is considered one key process through which lesion maintenance and growth may be promoted in endometriosis but, crucially, it is not known whether these processes are a necessary cause, or rather an effect of the disease process.
One method to improve understanding of complex disease pathogenesis is through the identification of genetic factors (DNA sequence variants) underlying disease risk. As germ-line (inherited) genetic variants in DNA are not subject to environmental influences, or a result of the disease process, a significant association between a variant and disease implies a role in its pathophysiology – provided the study is well-designed and there are no systematic biases in genetic profiles between cases and controls16. The involvement of genetic factors in the development of endometriosis is supported by numerous studies17–20, and its heritability (the proportion of variability in disease risk in the population due to genetic factors) is estimated at 47%21 - 52%18.
The first types of study design to unravel genetic factors underlying disease were so-called ‘linkage’ studies in families containing multiple affected members. Linkage studies considered the statistical evidence of sharing of chromosomal regions between multiple cases within a family, and between families. They assume that most of the disease risk in a family is attributable to the same genetic variant(s) shared by cases, and that other affected families have disease-causing variants in the same gene (‘monogenic disease’). Linkage studies were extremely successful in the 1980/90s to uncover single rare variants responsible for monogenic diseases (e.g. cystic fibrosis22, Huntington’s disease23), however, they proved to be problematic in elucidating the genetic basis for complex diseases, because of their multifactorial origin (multiple cases in the same and between families may not have the same underlying aetiology of disease). Although significant linkage to chromosomes 10 and 7 was observed for endometriosis, the (likely rare) variants responsible for these linkages remain to be identified24 25. Subsequently, candidate gene association studies based on biological hypotheses became common-place to identify specific DNA variants associated with disease. Indeed, in endometriosis hundreds such studies have been conducted but due to inherent issues in study design (e.g. validity of the biological hypothesis; few variants tested per gene; varying case definitions; small sample sizes) very few of their results have been replicated19 20.
Given the disappointments with linkage and candidate gene association studies in complex disease, enthusiasm then grew for genome-wide association studies (GWAS). GWAS are hypothesis-free and test genetic variants across the entire genome for association with a disease or trait. From the year 2000 onwards, several major parallel developments paved the way for GWAS to become feasible: 1) The sequencing of the human genome (The Human Genome Project26); 2) the discovery of common single base-pair variants that are highly abundant across the genome (single nucleotide polymorphisms or SNPs)27 and that can be used for genome-wide association testing; and 3) the development of technology to genotype these SNPs at large-scale, and ever decreasing cost. Subsequent developments included the International HapMap project28 and more recently The 1000 Genomes Project29 that provided data on the correlation between SNPs in close proximity to each other (linkage disequilibrium) in different ethnic populations, thus allowing the statistical inference (‘imputation’) of most of the known common SNPs in the human genome based on genotyping of only a few 100K SNPs. In addition, statistical methods and guidelines were developed for the appropriate analysis of GWAS – which arise from a large multiple-testing issue that could lead to many false positive results. After much debate, a general genome-wide significance threshold per SNP of p < 5 x 10-8 was generally accepted by journals to indicate a statistically robust association. Early GWAS findings were from analyses performed in individual and/or relatively small datasets (e.g., ~2000 cases versus 2000 controls)30, and were therefore required to be supplemented by evidence of replication in independent datasets.
However, the high statistical significance threshold coupled with the current practice of meta-analysis of multiple large independent GWAS datasets makes the possibility of a false positive association signal being produced by random sampling variation extremely low. Furthermore, studies have shown that joint analysis of GWA studies is more powerful than 2-stage (discovery and replication) GWA study designs31
These developments led to wide-scale adoption of GWAS for the investigation of common genetic variants underlying complex disease. Indeed, since the first study in 2005, GWAS have been successful in identifying common DNA sequence variants associated with human diseases and phenotypes, with thousands of risk loci identified across hundreds of traits32. Given the considerable uncertainty about the causes of endometriosis, and its substantial heritability, GWAS also have provided a promising approach in improving the understanding of the genetic contribution to the disease.
GWAS in endometriosis
Overview of studies to date
Five GWAS analyses have been published for endometriosis, comprising four independent datasets – one on women of Japanese ancestry, and three on women of European ancestry33. The first, smallest, GWAS study published in 2010 including 696 cases and 825 controls yielded no genome-wide significant associations34. This was quickly followed in the same year by Uno et al.35, including 1,907 Japanese cases and 5,292 controls from Biobank Japan (BBJ). They reported one genome-wide significant association in CDKN2B-AS1 (P=5.57x10-12; OR=1.44 95% CI: 1.30 – 1.59) and two suggestive associations (rs13271465 in an inter-genic region on chromosome 8, P=9.84x10-6; and rs16826658 16.6 kb away from WNT4, P=1.66x10-6).
In 2011, the International Endogene Consortium (IEC) published the first GWAS in women of European ancestry (3,194 cases; 7,060 controls, with replication in 2,392 cases and 2,271 controls)36. They observed one genome-wide significant locus, in an intergenic region on chromosome 7 (rs12700667; P=1.4x10-9, OR=1.20 95% CI: 1.13 – 1.32), and meta-analysing their results for WNT4 with those reported by Uno et al.35, reported genome-wide significant association also for this locus (rs7521902, P=4.2x10-8 OR=1.19 95% CI: 1.12 – 1.27). A subsequent collaborative genome-wide meta-analysis37 of the IEC and BBJ datasets in 2012 identified 7 genome-wide significant loci, confirming 7p15.2, WNT4, and CDKN2B-AS1 while finding 4 new loci in GREB1 at 2p25.1 (rs13394619 P=6.1x10-8, OR=1.15 95% CI: 1.09 – 1.20), near VEZT at 12q22 (rs10859871 P=5.5x10-9, OR=1.20 95% CI: 1.14 – 1.26), near ID4 (rs7739264 P=3.6x10-10, OR=1.17 95% CI: 1.11 – 1.23), and rs4141819 in an intergenic region on 2p14 (P=8.5x10-8, OR=1.15 95% CI: 1.09 – 1.21). A recent further analysis of these datasets using 1000 Genomes imputation of genotypes38 additionally identified IL1A (OR: 1.21; 95% confidence interval = 1.13 – 1.29; P=3.43 × 10-8). The most recent GWAS in women of European ancestry (2,019 cases; 14,471 controls) was published in 2013, confirming WNT4 and reporting a new intergenic locus on 2q23.3 (P=4.7x10-8, OR: 1.20 95% CI: 1.13 – 1.29)39.
Meta-analysis33 of the reported loci across all four independent GWAS and four replication datasets, totalling >11,000 cases and >30,000 controls, confirmed consistency of effects across datasets and populations for six of the reported loci (rs12700667 on 7p15.2, rs7521902 near WNT4, rs10859871 near VEZT, rs1537377 near CDKN2B-AS1, rs7739264 near ID4, rs13394619 in GREB1), with an additional two (rs1250248 in FN1 and rs4141819 on 2p14) showing evidence close to the genome-wide significance threshold of p < 5 x 10-8. Details of all the loci and evidence of association are provided in Table 1.
Table 1.
Overview of all 10 genetic loci (12 SNPs) associated with endometriosis in genome-wide association studies to date, ranked by meta-analysis association p-value including all endometriosis cases.
| SNP ID | Location (chr: bp) a | Genomic annotationc | Nearest gene (kb distance) | Meta-analysis p-value all endo (stage III/IV) d | All Endometriosis | Stage III/IV Endometriosis | ||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | Sample size (cases:controls) | OR (95% CI) | Sample size (cases:controls) | |||||
| rs7521902 | 1:22490474 | Intergenic |
WNT4 (21Kb) |
1.8x10-15 (1.8x10-10) |
1.18 (1.13-1.23) | 9,114:30,407 | 1.25 (1.16-1.33) | 2,432 : 24,241 |
| rs10859871 | 12:95711626 | Intergenic; Regulatory region variant |
VEZT (17Kb) |
4.8x10-15 (6.8x10-7) |
1.18 (1.13-1.22) | 7,680:26,866 | 1.19 (1.11-1.27) | 2,212 : 21,531 |
| rs7739264 | 6:19785338 | Intergenic; Non-coding transcript variant |
ID4 (52Kb) |
6.2x10-10 (1.2x10-8) |
1.11 (1.08-1.15) | 10,072:29,137 | 1.20 (1.13-1.28) | 2,212 : 21,531 |
| rs12700667 | 7:25901389 | Intergenic |
NFE2L3 (290/331Kb) |
1.6x10-9 (3.6x10-8) |
1.13 (1.08-1.17) | 11,506:32,678 | 1.22 (1.14-1.31) | 2,432 : 24,241 |
| rs7798431 | 7:25860562 | Intergenic | 5.4x10-9 (9.7x10-8) |
1.13 (1.09-1.18) | 8,734:24,633 | 1.24 (1.14-1.33) | 2,212 : 21,531 | |
| rs1537377 | 9:22169450 | Intergenic |
CDKN2BAS1 (48/4Kb) |
1.0x10-8 (2.3x10-7) |
1.12 (1.08-1.17) | 7,680:26,866 | 1.18 (1.11-1.26) | 2,212 : 21,531 |
| rs1333049 | 9:22125253 | Intergenic | 0.25 (0.60) |
1.04 (0.98-1.10) | 6,681:21,531 | 1.03 (0.94-1.12) | 1,364 : 7,060 | |
| rs13394619 | 2:11727257 | Intronic splice variant |
GREB1 (0Kb) |
2.9x10-8 (2.1x10-3) |
1.13 (1.07-1.20) | 8,376:27,691 | 1.18 (1.11-1.24) | 2,212 : 21,531 |
| rs6542095 | 2:113529183 | Downstream gene variant |
IL1A (2.3Kb) |
3.6x10-7 (3.4×10-8) |
1.15 (1.09-1.22) | 5,300:10,218 | 1.21 (1.13-1.29) | 1,357 : 10,218 |
| rs4141819 | 2:67864425 | Intergenic; Non-coding transcript variant |
ETAA1 (227Kb) | 8.8x10-6 (9.2x10-8) |
1.08 (1.04-1.12) | 7,680:26,866 | 1.16 (1.09-1.24) | 2,212 : 21,531 |
| rs1250248 | 2:216286843 | Intronic |
FN1 (0Kb) |
1.1x10-4 (8.0x10-8) |
1.11 (1.04-1.18) | 8,443:14,190 | 1.26 (1.16-1.38) | 1,584 : 9,770 |
| rs6734792 | 2:151624632 | Intergenic | RND3 (281Kb) | 2.2x10-6 (2.7x10-5) |
1.10 (1.06-1.16) | 6,636:22,849 | 1.10 (1.05-1.15) | 2,212 : 21,531 |
Human Genome Build 38
Meta-analysis p-value is from Rahmioglu et al. 2014 (ref). For rs4141819, rs1250248, rs6542095 the meta-analysis p-values including only stage III/IV cases is given in parenthesis, which reach borderline significance.
Annotation according to Ensembl genome browser: www.ensembl.org and UCSC genome browser: https://genome.ucsc.edu/.
Insight into disease origin heterogeneity
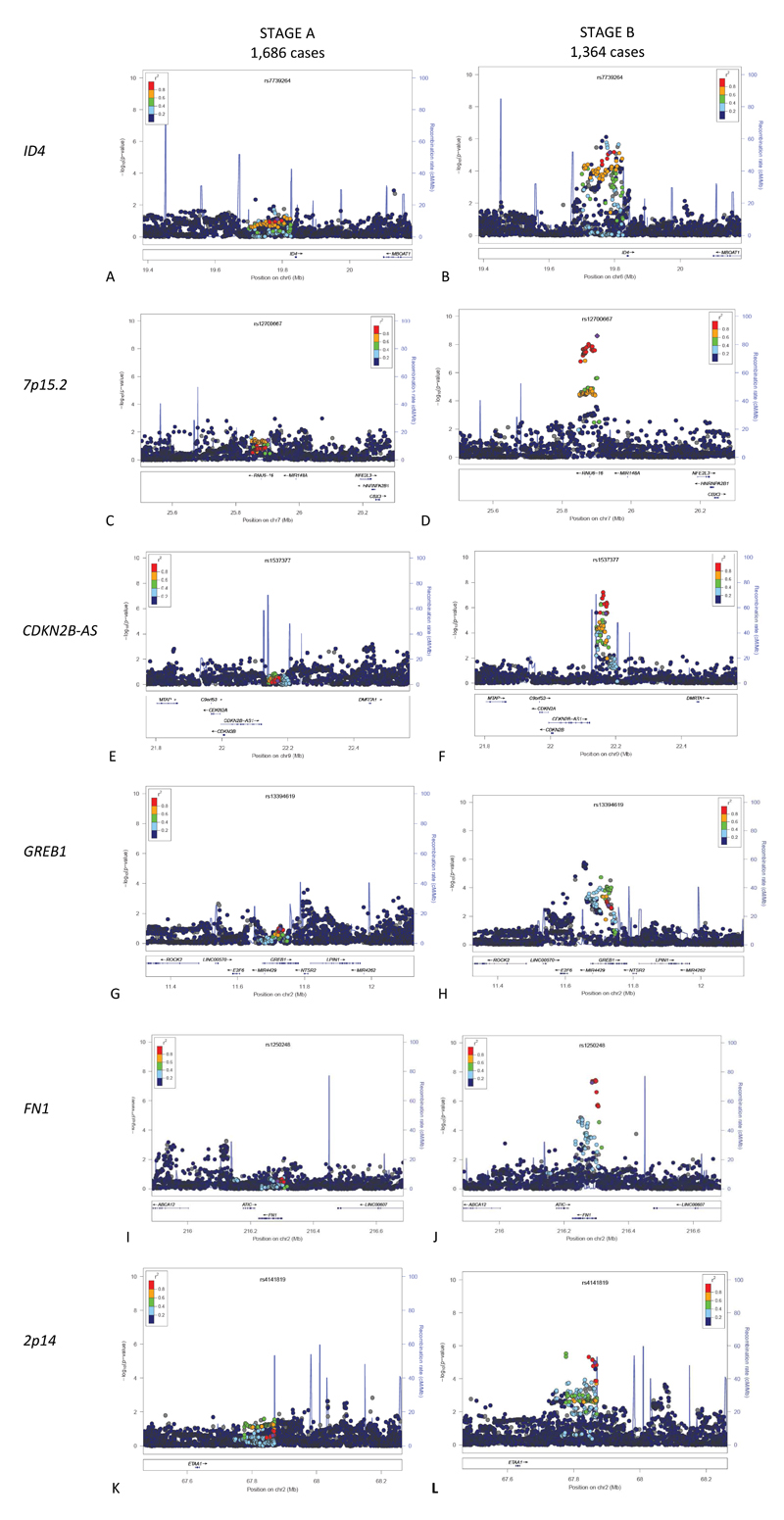
GWAS have to be large in sample size (typically > 2,000 cases) to detect, with genome-wide significance, the modest effect sizes that common genetic variants exert in complex disease40. Most case datasets are therefore collected retrospectively, and often lack detailed phenotypic information. Of the endometriosis GWAS conducted to date, two studies36 39 involving three datasets included surgically confirmed cases (IEC QIMR; IEC Oxford; Utah). In the IEC datasets, retrospective evaluation of records only allowed broad classification into stage B (rAFS stage III/IV defined by large lesion volume and/or large ovarian disease with many adhesions), and stage A (stage I/II defined by small lesion volume and allowing for only small ovarian disease with a few adhesions). The IEC used various statistical methods to show that stage B disease is more ‘genetically driven’ than stage A: the relative contributions of common genetic variation assayed in the GWAS to stage B vs. stage A disease were 31% vs. 15%, respectively36, results that were supported by subsequent analyses41. The genetic distinction between origins of stage A vs. stage B became further apparent when considering the association at individual genetic loci (Figure 1).
Figure 1.
Regional ‘locuszoom’ chromosomal plots showing contrasting evidence of association for individual SNPs with stage A (left column; 1,686 cases) vs. stage B (right column; 1,364 cases) endometriosis vs. 7,060 controls. Data were from the International Endogene Consortium GWAS dataset36 after imputation to the 1000 Genomes panel version 3. Each data point signifies a variant (SNP), and SNPs are colour coded according to their correlation (r2) with the top associated genotyped SNP (purple diamond) from meta-analysis (Table 1).The x-axis shows the genomic location, relative to gene locations. The y-axis shows the significance of association (-log10 of p-value).
Unfortunately, no other phenotypic information was collected in the individual case datasets, hampering further dissection of genetic heterogeneity between the surgically-defined stages. Whether these differences can be attributed to the pathogenesis of ovarian disease or adhesion development cannot be determined. Furthermore, this rAFS stage-based dichotomization does not provide insight into genetic heterogeneity by different surgically or symptomatically defined case definitions.
Insights into biology
Of the 12 ‘index’ SNPs (defined as the SNP with the lowest p-value at a GWAS locus) that have been associated with endometriosis, only two are located within genes, in introns, while the remaining are intergenic at varying distances from known genes (Table 1). Because of this, it has been difficult to highlight specific causal molecular mechanisms through which endometriosis-associated variants impact on disease, and the genes through which their effects are mediated. For a more detailed review of the biology implicated by these loci, we refer to Rahmioglu et al.42.
Genetic variants in the identified loci are likely to have roles in regulation of expression of near or distantly located genes. Genomic annotation provided by the Encyclopedia of DNA Elements (ENCODE) project43 on a wide variety of cell types and tissues is useful to investigate the putative regulatory function of these variants. ENCODE is an international collaboration of research groups funded by the National Human Genome Research Institute (USA) to build a comprehensive list of functional elements in the human genome, including elements that act at the protein and RNA levels, and regulatory elements that control cells and circumstances in which a gene is active (https://www.encodeproject.org/). Regulatory elements are typically investigated through DNA hypersensitivity assays, assays of DNA methylation, and immunoprecipitation (IP) of proteins that interact with DNA and RNA, i.e., modified histones, transcription factors, chromatin regulators, and RNA-binding proteins, followed by sequencing. Though information on endometrial tissue and cells is very limited, a major limitation for endometriosis research, many of the genomic annotation features are shared across different cell and tissue types.
The following genomic loci have been associated with endometriosis in GWAS:
Rs13394619 is an intronic splice variant (affecting the formation of different transcripts) in GREB1 (growth regulation by estrogen in breast cancer 1). GREB1 encodes for an early response gene involved in estrogen induced growth of breast cancer cells44. Its role in estrogen induced endometrium proliferation remains to be uncovered.
Rs1250248 is located in an intron of FN1 (Fibronectin 1), which is a transcription factor binding site (ENCODE). FN1 mediates various cellular interactions with the extracellular matrix and has important roles in cell adhesion, migration, growth and differentiation45. It has been shown that SOX2, a transcription factor that targets FN1, is involved in mediating cell migration in ovarian cancer46.
Rs7521902 is an intergenic variant that is located 21Kb downstream of WNT4. WNT4 (Wingless-type MMTV integration site family member 4) encodes for a protein imperative for development of the female reproductive tract47. Moreover, WNT4 is shown to be expressed in normal peritoneum, suggesting that endometriosis can arise through metaplasia using developmental pathways involved in development of the female reproductive tract48.
Rs7739264 is an intergenic variant located 52Kb downstream of ID4 (Inhibitor of DNA binding 4). ID4 is an oncogene that is over-expressed in most primary ovarian cancers but not in normal ovary and fallopian tubes. Moreover, it’s been shown to be overexpressed in most ovarian, endometrial and breast cancer cell lines49. ID4 can potentially regulate HOXA9 and CDKN1A genes, which are mediators of cell proliferation and differentiation. More specifically, HOXA genes are involved in differentiation of fallopian tubes, uterus, cervix and vagina50.
Rrs1537377 and rs1333049 are intergenic variants located 48Kb and 4Kb upstream of CDKN2B-AS1. CDKN2B-AS1 (Cyclin-dependent kinase inhibitor 2B antisense RNA) has been shown to mediate tumour suppressor genes, namely CDKN2B, CDKN2A and ARF expression51–53. Inactivation of CDKN2A has been previously reported in endometriosis, adenomyosis, endometrial cancer through loss of heterozygosity or hypermethylation of its promoter54–56.
Rs10859871 is an intergenic variant, located in a regulatory region (DNAse hypersensitivity cluster present in 71/125 cell types, ENCODE), 17Kb upstream of VEZT. VEZT (Vezatin) encodes an adherens junction transmembrane protein. It is also a putative tumour suppressor gene57 that targets cell migration, cell adhesion genes and TCF19, a cell cycle progression gene which has been associated with lymphocyte count, mean cell haemoglobin, white blood cell count, haematocrit count and easinophil count58. The collective evidence suggests VEZT can have a role in maintaining immunological balance through regulation of TCF19.
Rs6542095 is a downstream region variant located 2.3kb of IL1A. IL1A (Interleukin 1A) encodes for a member of the interleukin I cytokine family that is involved in pro-inflammatory immune processes and haematopoiesis. This protein may have a role in the inflammatory component of endometriosis that has been suggested by co-occurrence of autoimmune disease59 60 and endometriosis as well as success of anti-inflammatory drugs on relieving endometriosis related pelvic pain61.
The remaining four variants are in three independent intergenic regions that have no genes within 200Kb:
Rs12700667 is an intergenic variant in chromosome 7p15.2, and located in histone modification (acetylation and methylation) marks H3K27ac and H3K4me1 (ENCODE). The region contains multiple potentially relevant genes including (1) a transcription factor called NFE2L3 (Nuclear factor erythroid derived 2-like 3) that regulates cell differentiation, inflammation and carcinogenesis62, (2) a microRNA, miRNA_148a which is involved in regulation of Wnt/beta-catenin signalling pathway 63 that has a documented role in communication of epithelial and stromal cells of the endometrium 64, in infertility associated with endometriosis through sex hormone regulation 65 66, fibrogenesis67 and adipogenesis63, (3) two members of a transcription factors family called homeobox A, HOXA10 and HOXA11, that have a role in uterine development68 69.
The intergenic signal on 2p14, is 227Kb away from ETAA1 (Ewing’s tumor-associated antigen 1) gene, which encodes for a Ewing family tumor (EFT) specific cell surface antigen70. Likely more relevant is that this variant sits in a 188Kb long non-coding RNA called, AC007422.1, the function of which remains unknown.
The intergenic signal on 2q23.3 is located 280Kb upstream of RND3 (Rho Family GTPase 3) gene, encoding a member of Rho family of small GTP-binding proteins that is involved in regulating actin cytoskeleton and stress fibre formation. It is also a regulator of cell-cycle progression, transformation and cell migration 71 72.
We considered the magnitude of the p-value testing association for each of these loci with stage A vs. stage B endometriosis in the IEC GWAS36, imputed to the latest 1000 Genomes panel (version 3). Of the 10 loci, six showed a strong signal for stage B disease, but no statistically significant association at all with stage A (Figure 1): ID4, GREB1, 7p15.2, CDKN2B-AS1, FN1, and 2p14. WNT4 showed weaker association for stage A, much reduced compared to stage B (not shown). VEZT showed a similar strength of association for stage A and B, as did IL1A albeit with different SNP sets (not shown). Lastly, the SNPs reported on 2q23.3 showed limited evidence of association in the IEC dataset (not shown), nor did it show consistent evidence in the meta-analysis by Rahmioglu et al33. Thus, the majority of GWAS loci identified to date are implicated most strongly in the origins of stage B (rASRM stage III/IV) disease. The discrepancy in association signals with stage I/II may be driven by inclusion of stage I cases, as our previous work showed stage II and III to be similar in terms of genetic burden41.
Two of the signals, WNT4 and 7p15.2 point at WNT signalling as a key pathway involved in endometriosis pathogenesis. Interestingly, the 7p15.2 locus was also found to be genome-wide significantly associated with fat distribution (waist-to-hip ratio adjusted for body mass index; WHRadjBMI) in an entirely independent GWAS73. This led to a study leveraging the two GWAS datasets showing, for the first time, evidence of shared genetic origins and pathways between endometriosis and fat distribution, mostly through the WNT/β catenin signalling pathway74. In a similar type of analysis, shared genetic origins between endometriosis and ovarian cancer were explored, showing strong genetic correlations between endometriosis and clear-cell, endometrioid, and low-grade serous ovarian cancer75.
The GWAS SNPs associated with endometriosis to date explain less than 2% of disease risk variability. In contrast, half of the estimated 50% heritability (26%) is estimated to be due to common variants that can be captured through GWAS76. Successful examples from other complex diseases have shown that by increasing sample sizes, many more common genetic loci (with population frequencies > 1%) can be discovered. For instance, the most recent GWAS analyses in breast cancer involving 62,000 cases identified more than 90 GWAS loci, explaining 16% of disease risk77. For Crohn’s disease, 140 common genetic loci have been found, explaining 14% of disease risk, in meta-analyses involving 20,000 cases. Extended endometriosis consortium efforts are currently ongoing to conduct further meta-GWAS studies including datasets with endometriosis diagnostic information worldwide, increasing sample size 4-fold to around 20,000, as well as the first “exome-wide” analyses focusing on variants that directly impact gene protein products and allow more direct biological interpretation.
Utility of endometriosis GWAS results: What are The Big Questions?
Broadly speaking, there are three areas of potential utility for GWAS results for a disease/trait: 1) Improve understanding of pathogenesis; 2) Dissection of phenotypic heterogeneity (discovery of ‘subtypes’); and 3) Risk prediction. We will discuss the endometriosis GWAS results for each of these areas below.
Understanding of pathogenesis
Making headway with inferring causality and functional mechanisms for pathogenesis from any GWAS results is hampered by two issues. Firstly, the most significantly associated SNP with a disease may not be the causal variant, but may be correlated (in linkage disequilibrium - LD) with the causal variant40. Due to common ancestry of individuals in a population, SNPs located near each other are typically correlated (‘in LD’). LD can stretch over many thousands of DNA base pairs and include 10-1000s of SNPs. Although there are various ways to narrow down the region of association, inclusing sequencing and fine-mapping as well as statistical methods (e.g. through trans-ethnic mapping78 or calculation of so-called ‘credible sets’ of SNPs79 that have the highest probability to include the causal variant) these methods need to be supplemented by other functional evidence that associate a specific variant to actual transcription variation. Secondly, as mentioned before, most variants at GWAS loci are now known to be located in distal intergenic or intronic (regulatory) regions80, with unknown effects on transcription of protein-coding genes and pathways. This feature, characteristic of GWAS variants, represents a major challenge we face in translating our endometriosis GWAS signals into clinically meaningful results. Understanding the effects of genetic variants on perturbation of protein-coding ‘effector’ transcripts and pathways requires integration with genomic annotation data, including gene expression (RNA) and DNA modification profiles, in tissues/cells relevant to the disease.
Recently, there have been major advances in understanding the regulatory architecture of the human genome, and, in particular, in the development of techniques for assessing the relationships between regulatory elements and the genes they control. Studies integrating gene expression data from a diverse range of tissues with genetic data have provided important insights into DNA variants that regulate gene expression (‘expression quantitative trait loci SNPs (eQTL SNPs)’), showing that many eQTLs are tissue-specific and that this tissue-specificity is an important feature of disease susceptibility variants81–83. Studies have similarly shown that genetic variants can also affect transcription through altering DNA methylation profiles (metQTLs), and – of particular significance – can affect how DNA is ‘packaged’ by altering chromatin signatures that can create or destroy transcription factor binding sites and thereby alter the chromatin signature with long-range (100kbs) effects on gene expression transcription factor binding84. Many GWAS associations map to ‘DNAse I hypersensitive’ sites (DHS) that mark chromatin accessibility for transcription factor binding, which in turn have been associated with changes in RNA expression80 85.
The importance of identifying which variant controls which gene was recently highlighted by analyses of regulatory control at the FTO locus (influencing body mass index and obesity). The causal variants map intronic to FTO and it had been widely-assumed that they regulate expression of this gene. However, it was subsequently shown that these FTO intronic SNPs lie within a region that regulates expression of a gene 500kb away (IRX3)86, demonstrating that enhancers (DNA sequences affecting levels of transcription) of one gene can lie in the introns of another.
The crucial need for genomic annotation data to advance our knowledge of the mode of action of GWAS variants is now fully recognised. Large-scale initiatives such as ENCODE87, the Genotype-Tissue Expression (GTEx) project88 and the NIH Roadmap Epigenomics Project89 are underway and aimed at building public databases providing tissue- and cell-specific genomic profiling landscapes (e.g. RNA transcription, DNA methylation, DHS) to enable investigations of the impact of genetic variation on these. However, none of these sources include endometrium and its constituent cell types – the target tissue for endometriosis – preventing systematic progress on the translation of endometriosis GWAS signals. Also, none of the genome-wide expression studies of endometrium or endometrial cell types published to date have included genomic data that would allow the investigation of eQTLs.
In order to make headway with understanding the causality and functionality of endometriosis loci, genomic profiling (including eQTLs) of endometrium and endometrial cell types from 100s of women with and without the disease needs to be conducted. Tissue-based profiling has the advantage of assessment of profiles reflecting inter-cellular communications in their ‘natural biological state’, however cellular heterogeneity means that for this type of study in particular, sufficiently large sample sizes are required to detect signals. Such sample sizes can more easily be achieved in collaborative efforts between research groups, however, differences in case definitions and phenotypic data collected, as well as protocols for tissue sampling, have made combination and comparison of the data difficult. The WERF Endometriosis Phenome and Biobanking Harmonisation Project (EPHect) (see below), which has harmonised and standardised data and sample collection protocols for endometriosis research, now enables these large-scale collaborative studies to be conducted.
Dissection of phenotypic heterogeneity
The endometriosis GWAS to date have provided the first biological evidence that rAFS stage III/IV disease may arise from distinct pathophysiology compared to stage I/II. However the limited information available from retrospectively assessed surgical records did not allow analysis of the disease element driving the staging for these cases nor other surgical/clinical phenotypes (e.g. deep infiltrating vs. peritoneal disease), as these data were not systematically collected. Moreover, the limited detail of surgical/clinical data also begged the question as to what the genetically dissected sub-types rAFS ‘I/II’ vs. ‘III/IV’ signified. For example, since ovarian endometriosis (endometrioma) typically falls into the rAFS III/IV category, did the GWAS results associate predominantly to this disease entity; or to other disease aspects captured by this categorisation? To allow this question to be answered, systematic collection of surgical and clinical data needs to accompany genetic profiling of samples.
Identification of functionally distinct disease sub-types based on molecular profiles will allow much-needed patient stratification for focused endometriosis drug target and biomarker discovery programmes. In cancer, there are many successful examples of this approach, e.g. the gene expression based identification of basal, ER+/-, HER2+/-, and ‘Normal’ breast cancer subtypes90, which have different risk-factor/survival rate profiles91 and targeted drug regimes (e.g. Tamoxifen for ER+; Trastuzumab for HER2 tumours). The Cancer Genome Atlas (TCGA)92 93 has embarked on comprehensive, large-scale molecular profiling (DNA genomic, DNA methylation, exome, mRNA/miRNA sequencing) to uncover further cancer sub-types and targeted treatments. To date, only one such study has been conducted in endometriosis - limited to using expression profiling of eutopic endometrium to distinguish cases from controls, and rAFS stage III/IV from I/II94. There is therefore clear scope for similar detailed molecular dissection studies in endometriosis, and to assess to what extent detailed surgical and clinical data are correlated with - or can capture - such molecularly defined subtypes. Again, to allow such studies to be conducted we need detailed and standardised data collection on women with biological samples collected through standardised protocols.
Endometriosis risk prediction
With the successes in GWAS, disease risk prediction based on genetic profiles (‘genetic risk scores’) in complex disease has been a rapidly emerging field of interest as the proportion of disease risk explained has been increasing due to larger studies performed95–98. At present, the genetic loci discovered for endometriosis do not have utility in terms of disease risk prediction or screening. This is because, together, they explain less than 2% of all disease risk variance. As the sample size for future planned endometriosis GWAS grows, the proportion of variance explained by new loci will increase. Whether these will have utility in screening tools will depend on the proportion of disease risk they explain at a population level. More likely, screening tools will require combinations of genetic variants with other molecular profile changes between cases and controls (e.g. distinct epigenomic or transcriptomic patterns in eutopic endometrium), though whether such markers will be detectable in readily accessible samples such as blood is also unknown. In addition, inclusion of standardized collection of phenotypic profiles (e.g. clinical, symptomatic, metabolic) in screening tools that correlate with distinct molecular profiles between endometriosis cases and controls may be of benefit.
Towards the integration of Genomics with Phenomics: WERF EPHect
As we highlighted above, the translation of endometriosis GWAS results in all three major areas of potential utility – elucidation of causal mechanisms, identification of subtypes of disease, and even risk prediction – will require integration with other molecular profiling as well as detailed phenotypic data. To allow the investigation of the full spectrum of potential disease-related effects, such phenotypic data can (and should) include surgical, clinical (symptomatic) and image-based manifestations of disease, as well as disease correlates such as comorbidities, covariate characteristics (e.g. body mass index and fat distribution, cigarette smoking), and demographic factors. Houle et al. 99 referred to such deep phenotyping (the acquisition of high-dimensional phenotypic data on an organism-wide scale) as Phenomics. Key to the valid acquisition and cross-study comparability of phenomic data is standardisation of tools and measurements of data collection. Similarly, robust molecular phenotyping that allows comparisons among studies and centres requires standardisation of the protocols with which samples are obtained. The WERF Endometriosis Phenotyping and Biobanking Harmonisation Project (EPHect; endometriosisfoundation.org/ephect) was designed for exactly these objectives: to standardise and harmonise data collection instruments and sample collection protocols for endometriosis research, to reduce data variability, facilitate comparability, and encourage large-scale clinical and basic research collaborations100–103.
WERF EPHect was established in 2013, through a global collaboration between 34 academic endometriosis research centres and 3 industry partners. Through two workshops and several consultation rounds, the Working Group reached a consensus on freely available instruments for surgical 100 and clinical data collection101, as well as standard operating protocols (SOPs) for the collection of 10 biological sample types (tissue103 and fluids102) in endometriosis research. All instruments have standard, as well as minimum versions.
The WERF EPHect Surgical Standard Form (SSF)100 was developed with the aim to collect all currently deemed relevant and important information describing the visual endometriosis phenotype and surgical treatment that would allow clinically and scientifically meaningful studies104. It includes detailed information about clinical covariates: current menstrual cycle, current hormone treatment, and history of previous endometriosis surgery, as well as any imaging findings before the procedure. The second part concentrates on intraoperative findings including the type and duration of the procedure; and the extent, exact location, and colour of endometriotic lesions, with a particular focus on size of endometrioma and endometriotic nodules. It allows for an exact description of tissue biopsies, including location and appearance, and surgical treatment of lesions. The WERF EPHect Endometriosis Phenotype Questionnaire Standard (EPQ-S)101 was developed to capture standardised clinical patient information, and includes validated instruments to assess pelvic pain, subfertility and reproductive history, menstrual history and hormone use, medical and surgical history, medication use, and personal information. For both the SSF and EPQ-S, minimum versions (MSF and EPQ-M) were developed for centres without research support, however the standard tools are recommended as it allows a much broader phenotypic exploration.
The WERF EPHect standardised biological sample collection protocols were developed by systematic comparison and review of evidence using 1) information provided by the Working Group centres; 2) publicly available SOPs from general large-scale biobanking efforts (e.g., UK Biobank); large biorepositories (International Society for Biological and Environmental Biorepositories; the NCI Biorepositories and Biospecimen Research Branch; and the Australian Biospecimen Network); and 3) systematic literature searches in PubMed, of reference lists of retrieved articles, and of online material from biobanks and biorepositories. Based on review of the evidence, consensus SOPs were developed for the collection, processing, and storage of six biofluid types: blood and its derivatives (serum, plasma, and red/white blood cells), urine, saliva, peritoneal fluid, endometrial fluid, and menstrual effluent102. Similarly, consensus on SOPs for collection, processing and storage of five tissue types was reached (ectopic and eutopic endometrium, myometrium, and peritoneum)103. For most steps in the SOPs, two tiers were agreed upon: standard and minimum required. While following standard protocol steps was recommended whenever possible, the minimum required SOP steps were offered to provide the fundamentals for standardization that need to be adhered to as an absolute minimum requirement given unavoidable logistical and budgetary circumstances.
Feedback on the WERF EPHect instruments is strongly encouraged through its website; the tools will be reviewed and updated, first after one year and then every three years. In addition, to facilitate collaboration, WERF EPHect has implemented a voluntary registry for centres using the tools after having obtained local IRB approval (endometriosisfoundation.org/ephect).
Conclusions
The path to providing new therapeutic options for endometriosis, targeted to subtypes of disease, lies in improved understanding of the fundamental pathogenic processes underlying the disease. Although GWAS SNP risk loci typically have small effects, they can reveal novel insights into disease pathogenesis and do not imply that their implicated targets will have low therapeutic value105. GWAS results for endometriosis have delivered novel identification of potential pathophysiologic pathways involved (e.g. WNT signalling). However, as the study sample sizes have been relatively modest, the number of GWAS loci robustly identified to date has been limited to seven; further loci will no doubt be discovered in ongoing meta-GWAS analyses that will increase sample size four-fold.
As is typical for GWAS signals – most of the endometriosis loci reside in intergenic or intronic regions that are likely to regulate gene expression elsewhere. These regulatory pathways, and the causal driver variants at each locus, remain to be uncovered; for example through integrated gene-expression (eQTL) studies in endometrium. The loci robustly associated with endometriosis in GWAS studies to date have also suggested that the pathogenesis of rAFS stage III/IV disease is likely to be distinct from stage I/II. To understand this observation, further phenotypic dissection requires the collection of much more detailed, standardised surgical and clinical data, integrated with genomic and other molecular profiling of endometrium and other relevant samples from the same women. The WERF EPHect standardised data collection instruments and sample collection protocols now allow such data to be collected and compared across different endometriosis research centres, paving the way for studies focused on the translation of GWAS results into results that are meaningful for patients and practitioners: novel treatments that target subtypes of disease.
Funding statements
N.R. was supported by an MRC grant (MR/K011480/1). A.P.M. is a Wellcome Trust Senior Fellow in Basic Biomedical Science (WT098017). G.W.M. is supported by the NHMRC Fellowships Scheme (339446, 619667).
References
- 1.Adamson GD, Kennedy SH, Hummelshoj L. Creating solutions in endometriosis: global collaboration through the World Endometriosis Research Foundation. J Endometriosis. 2010;2(1):3–6. [Google Scholar]
- 2.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364(9447):1789–99. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 3.Dunselman GA, Vermeulen N, Becker C, et al. ESHRE guideline: management of women with endometriosis. Hum Reprod. 2014;29(3):400–12. doi: 10.1093/humrep/det457. [DOI] [PubMed] [Google Scholar]
- 4.Nnoaham KE, Hummelshoj L, Webster P, et al. Impact of endometriosis on quality of life and work productivity: a multicenter study across ten countries. Fertil Steril. 2011;96(2):366–73. doi: 10.1016/j.fertnstert.2011.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Society AF. Revised American fertility society classification of endometriosis: 1985. Fertil Steril. 1985;43(3):351–52. doi: 10.1016/s0015-0282(16)48430-x. [DOI] [PubMed] [Google Scholar]
- 6.Medicine ASfR. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67(5):817–21. doi: 10.1016/s0015-0282(97)81391-x. [DOI] [PubMed] [Google Scholar]
- 7.Vercellini P, Trespidi L, De Giorgi O, Cortesi I, Parazzini F, Crosignani PG. Endometriosis and pelvic pain: relation to disease stage and localization. Fertil Steril. 1996;65(2):299–304. [PubMed] [Google Scholar]
- 8.Missmer SA, Hankinson SE, Spiegelman D, Barbieri RL, Marshall LM, Hunter DJ. Incidence of laparoscopically confirmed endometriosis by demographic, anthropometric, and lifestyle factors. Am J Epidemiol. 2004;160(8):784–96. doi: 10.1093/aje/kwh275. [DOI] [PubMed] [Google Scholar]
- 9.Eskenazi B, Warner ML. Epidemiology of endometriosis. Obstet Gynecol Clin North Am. 1997;24:235–58. doi: 10.1016/s0889-8545(05)70302-8. [DOI] [PubMed] [Google Scholar]
- 10.Simoens S, Dunselman G, Dirksen C, et al. The burden of endometriosis: costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. 2012;27(5):1292–99. doi: 10.1093/humrep/des073. [DOI] [PubMed] [Google Scholar]
- 11.Sampson JA. Peritoneal endometriosis due to the menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol. 1927;14:422–69. [Google Scholar]
- 12.Missmer SA, Hankinson SE, Spiegelman D, et al. Reproductive history and endometriosis among premenopausal women. Obstet Gynecol. 2004;104(5 Pt 1):965–74. doi: 10.1097/01.AOG.0000142714.54857.f8. [DOI] [PubMed] [Google Scholar]
- 13.Halme J, Hammond MG, Hulka JF, Raj SG, Talbert LM. Retrograde menstruation in healthy women and in patients with endometriosis. Obstet Gynecol. 1984;64:151–54. [PubMed] [Google Scholar]
- 14.Missmer SA, Cramer DW. The epidemiology of endometriosis. Obstet Gynecol Clin North Am. 2003;30(1):1–19. doi: 10.1016/s0889-8545(02)00050-5. vii. [DOI] [PubMed] [Google Scholar]
- 15.Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–79. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- 16.Zondervan KT, Cardon LR, Kennedy SH. What makes a good case-control study? Design issues for complex traits such as endometriosis. Hum Reprod. 2002;17:1415–23. doi: 10.1093/humrep/17.6.1415. [DOI] [PubMed] [Google Scholar]
- 17.Simpson JL, Elias S, Malinak LR, Buttram VCJ. Heritable aspects of endometriosis. I. Genetic studies. Am J Obstet Gynecol. 1980;137:327–31. doi: 10.1016/0002-9378(80)90917-5. [DOI] [PubMed] [Google Scholar]
- 18.Treloar SA, O'Connor DT, O'Connor VM, Martin NG. Genetic influences of endometriosis in an Australian twin sample. Fertil Steril. 1999;71(4):701–10. doi: 10.1016/s0015-0282(98)00540-8. [DOI] [PubMed] [Google Scholar]
- 19.Rahmioglu N, Missmer SA, Montgomery GW, Zondervan KT. Insights into Assessing the Genetics of Endometriosis. Curr Obstet Gynecol Rep. 2012;1(3):124–37. doi: 10.1007/s13669-012-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rahmioglu N, Montgomery GW, Zondervan KT. Genetics of endometriosis. Womens Health (Lond Engl) 2015;11(5):577–86. doi: 10.2217/whe.15.41. [DOI] [PubMed] [Google Scholar]
- 21.Saha R, Pettersson HJ, Svedberg P, et al. Heritability of endometriosis. Fertil Steril. 2015;104(4):947–52. doi: 10.1016/j.fertnstert.2015.06.035. [DOI] [PubMed] [Google Scholar]
- 22.Kerem B, Rommens JM, Buchanan JA, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245(4922):1073–80. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 23.Gilliam TC, Tanzi RE, Haines JL, et al. Localization of the Huntington's disease gene to a small segment of chromosome 4 flanked by D4S10 and the telomere. Cell. 1987;50(4):565–71. doi: 10.1016/0092-8674(87)90029-8. [DOI] [PubMed] [Google Scholar]
- 24.Treloar SA, Wicks J, Nyholt DR, et al. Genomewide linkage study in 1,176 affected sister pairs identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am J Hum Genet. 2005;77(3):365–76. doi: 10.1086/432960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zondervan KT, Treloar SA, Lin J, et al. Significant evidence of one or more susceptibility loci for endometriosis with near-Mendelian inheritance on chromosome 7p13-15. Hum Reprod. 2007;22(3):717–28. doi: 10.1093/humrep/del446. [DOI] [PubMed] [Google Scholar]
- 26.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 27.Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409(6822):928–33. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 28.The International HapMap C. A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 32.Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahmioglu N, Nyholt DR, Morris AP, Missmer SA, Montgomery GW, Zondervan KT. Genetic variants underlying risk of endometriosis: insights from meta-analysis of eight genome-wide association and replication datasets. Hum Reprod Update. 2014;20(5):702–16. doi: 10.1093/humupd/dmu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adachi S, Tajima A, Quan J, et al. Meta-analysis of genome-wide association scans for genetic susceptibility to endometriosis in Japanese population. J Hum Genet. 2010;55(12):816–21. doi: 10.1038/jhg.2010.118. [DOI] [PubMed] [Google Scholar]
- 35.Uno S, Zembutsu H, Hirasawa A, et al. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42:707–10. doi: 10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- 36.Painter JN, Anderson CA, Nyholt DR, et al. Genome-wide association study identifies a locus at 7p15.2 associated with the development of endometriosis. Nat Genet. 2011;43:51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyholt DR, Low SK, Anderson CA, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat Genet. 2012;44(12):1355–9. doi: 10.1038/ng.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sapkota Y, Low SK, Attia J, et al. Association between endometriosis and the interleukin 1A (IL1A) locus. Hum Reprod. 2015;30(1):239–48. doi: 10.1093/humrep/deu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albertsen HM, Chettier R, Farrington P, Ward K. Genome-wide association study link novel loci to endometriosis. PLoS One. 2013;8(3):e58257. doi: 10.1371/journal.pone.0058257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5(2):89–100. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]
- 41.Sapkota Y, Attia J, Gordon SD, et al. Genetic burden associated with varying degrees of disease severity in endometriosis. Mol Hum Reprod. 2015;21(7):594–602. doi: 10.1093/molehr/gav021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahmioglu N, Nyholt DR, Morris AP, Missmer SA, Montgomery GW, Zondervan KT. Genetic variants underlying risk of endometriosis: insights from meta-analysis of eight genome-wide association and replication datasets. Hum Reprod Update. 2014;20(5):702–16. doi: 10.1093/humupd/dmu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92(2):141–9. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 45.Pankov R, Yamada KM. Fibronectin at a glance. Journal of cell science. 2002;115(Pt 20):3861–3. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 46.Lou X, Han X, Jin C, et al. SOX2 targets fibronectin 1 to promote cell migration and invasion in ovarian cancer : new molecular leads for therapeutic intervention. Omics : a journal of integrative biology. 2013;17(10):510–8. doi: 10.1089/omi.2013.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397(6718):405–9. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 48.Gaetje R, Holtrich U, Engels K, et al. Endometriosis may be generated by mimicking the ontogenetic development of the female genital tract. Fertil Steril. 2007;87(3):651–6. doi: 10.1016/j.fertnstert.2006.07.1533. [DOI] [PubMed] [Google Scholar]
- 49.Ren Y, Cheung HW, von Maltzhan G, et al. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Science translational medicine. 2012;4(147):147ra12. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kobayashi A, Behringer RR. Developmental genetics of the female reproductive tract in mammals. Nat Rev Genet. 2003;4(12):969–80. doi: 10.1038/nrg1225. [DOI] [PubMed] [Google Scholar]
- 51.Jarinova O, Stewart AF, Roberts R, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arteriosclerosis, thrombosis, and vascular biology. 2009;29(10):1671–7. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Sanoff HK, Cho H, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4(4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer research. 2007;67(8):3963–9. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 54.Goumenou AG, Arvanitis DA, Matalliotakis IM, Koumantakis EE, Spandidos DA. Loss of heterozygosity in adenomyosis on hMSH2, hMLH1, p16Ink4 and GALT loci. International journal of molecular medicine. 2000;6(6):667–71. doi: 10.3892/ijmm.6.6.667. [DOI] [PubMed] [Google Scholar]
- 55.Guida M, Sanguedolce F, Bufo P, et al. Aberrant DNA hypermethylation of hMLH-1 and CDKN2A/p16 genes in benign, premalignant and malignant endometrial lesions. European journal of gynaecological oncology. 2009;30(3):267–70. [PubMed] [Google Scholar]
- 56.Martini M, Ciccarone M, Garganese G, et al. Possible involvement of hMLH1, p16(INK4a) and PTEN in the malignant transformation of endometriosis. International journal of cancer. Journal international du cancer. 2002;102(4):398–406. doi: 10.1002/ijc.10715. [DOI] [PubMed] [Google Scholar]
- 57.Miao R, Guo X, Zhi Q, et al. VEZT, a novel putative tumor suppressor, suppresses the growth and tumorigenicity of gastric cancer. PLoS One. 2013;8(9):e74409. doi: 10.1371/journal.pone.0074409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferreira MA, Hottenga JJ, Warrington NM, et al. Sequence variants in three loci influence monocyte counts and erythrocyte volume. Am J Hum Genet. 2009;85(5):745–9. doi: 10.1016/j.ajhg.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrier BF. Immunology of endometriosis. Clinical obstetrics and gynecology. 2010;53(2):397–402. doi: 10.1097/GRF.0b013e3181db7c33. [DOI] [PubMed] [Google Scholar]
- 60.Mathur SP. Autoimmunity in endometriosis: relevance to infertility. American journal of reproductive immunology. 2000;44(2):89–95. doi: 10.1111/j.8755-8920.2000.440204.x. [DOI] [PubMed] [Google Scholar]
- 61.Vercellini P, Crosignani P, Somigliana E, Vigano P, Frattaruolo MP, Fedele L. 'Waiting for Godot': a commonsense approach to the medical treatment of endometriosis. Hum Reprod. 2011;26(1):3–13. doi: 10.1093/humrep/deq302. [DOI] [PubMed] [Google Scholar]
- 62.Chevillard G, Blank V. NFE2L3 (NRF3): the Cinderella of the Cap'n'Collar transcription factors. Cell Mol Life Sci. 2011;68(20):3337–48. doi: 10.1007/s00018-011-0747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin L, Chen Y, Niu Y, et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC genomics. 2010;11:320. doi: 10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tulac S, Nayak NR, Kao LC, et al. Identification, characterization, and regulation of the canonical Wnt signaling pathway in human endometrium. J Clin Endocrinol Metab. 2003;88(8):3860–6. doi: 10.1210/jc.2003-030494. [DOI] [PubMed] [Google Scholar]
- 65.Matsuzaki S, Darcha C, Maleysson E, Canis M, Mage G. Impaired down-regulation of E-cadherin and beta-catenin protein expression in endometrial epithelial cells in the mid-secretory endometrium of infertile patients with endometriosis. J Clin Endocrinol Metab. 2010;95(7):3437–45. doi: 10.1210/jc.2009-2713. [DOI] [PubMed] [Google Scholar]
- 66.Wang Y, van der Zee M, Fodde R, Blok LJ. Wnt/Beta-catenin and sex hormone signaling in endometrial homeostasis and cancer. Oncotarget. 2010;1(7):674–84. doi: 10.18632/oncotarget.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuzaki S, Darcha C. In vitro effects of a small-molecule antagonist of the Tcf/ss-catenin complex on endometrial and endometriotic cells of patients with endometriosis. PLoS One. 2013;8(4):e61690. doi: 10.1371/journal.pone.0061690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor HS, Bagot C, Kardana A, Olive D, Arici A. HOX gene expression is altered in the endometrium of women with endometriosis. Hum Reprod. 1999;14(5):1328–31. doi: 10.1093/humrep/14.5.1328. [DOI] [PubMed] [Google Scholar]
- 69.Wu Y, Halverson G, Basir Z, Strawn E, Yan P, Guo SW. Aberrant methylation at HOXA10 may be responsible for its aberrant expression in the endometrium of patients with endometriosis. American journal of obstetrics and gynecology. 2005;193(2):371–80. doi: 10.1016/j.ajog.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 70.Borowski A, Dirksen U, Lixin L, Shi RL, Gobel U, Schneider EM. Structure and function of ETAA16: a novel cell surface antigen in Ewing's tumours. Cancer immunology, immunotherapy : CII. 2006;55(4):363–74. doi: 10.1007/s00262-005-0017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chardin P. Function and regulation of Rnd proteins. Nature reviews Molecular cell biology. 2006;7(1):54–62. doi: 10.1038/nrm1788. [DOI] [PubMed] [Google Scholar]
- 72.Guasch RM, Scambler P, Jones GE, Ridley AJ. RhoE regulates actin cytoskeleton organization and cell migration. Molecular and cellular biology. 1998;18(8):4761–71. doi: 10.1128/mcb.18.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 novel loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rahmioglu N, Macgregor S, Drong AW, et al. Genome-wide enrichment analysis between endometriosis and obesity-related traits reveals novel susceptibility loci. Hum Mol Genet. 2015;24(4):1185–99. doi: 10.1093/hmg/ddu516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu Y, Cuellar-Partida G, Painter JN, et al. Shared genetics underlying epidemiological association between endometriosis and ovarian cancer. Hum Mol Genet. 2015;24(20):5955–64. doi: 10.1093/hmg/ddv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee SH, Harold D, Nyholt DR, et al. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer's disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22(4):832–41. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Michailidou K, Beesley J, Lindstrom S, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–80. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35(8):809–22. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maller JB, McVean G, Byrnes J, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44(12):1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grundberg E, Small KS, Hedman AK, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44(10):1084–9. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grundberg E, Meduri E, Sandling JK, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93(5):876–90. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Min JL, Nicholson G, Halgrimsdottir I, et al. Coexpression network analysis in abdominal and gluteal adipose tissue reveals regulatory genetic loci for metabolic syndrome and related phenotypes. PLoS Genet. 2012;8(2):e1002505. doi: 10.1371/journal.pgen.1002505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lower KM, De Gobbi M, Hughes JR, et al. Analysis of sequence variation underlying tissue-specific transcription factor binding and gene expression. Hum Mutat. 2013;34(8):1140–8. doi: 10.1002/humu.22343. [DOI] [PubMed] [Google Scholar]
- 85.Degner JF, Pai AA, Pique-Regi R, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482(7385):390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smemo S, Tena JJ, Kim KH, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Consortium G. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4(3):317–24. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamimi RM, Colditz GA, Hazra A, et al. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;131(1):159–67. doi: 10.1007/s10549-011-1702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Network TCGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tamaresis JS, Irwin JC, Goldfien GA, et al. Molecular classification of endometriosis and disease stage using high-dimensional genomic data. Endocrinology. 2014;155(12):4986–99. doi: 10.1210/en.2014-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jostins L, Barrett JC. Genetic risk prediction in complex disease. Hum Mol Genet. 2011;20(R2):R182–8. doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yarwood A, Han B, Raychaudhuri S, et al. A weighted genetic risk score using all known susceptibility variants to estimate rheumatoid arthritis risk. Ann Rheum Dis. 2015;74(1):170–6. doi: 10.1136/annrheumdis-2013-204133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ibrahim-Verbaas CA, Fornage M, Bis JC, et al. Predicting stroke through genetic risk functions: the CHARGE Risk Score Project. Stroke. 2014;45(2):403–12. doi: 10.1161/STROKEAHA.113.003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mavaddat N, Pharoah PD, Michailidou K, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst. 2015;107(5) doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11(12):855–66. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 100.Becker CM, Laufer MR, Stratton P, et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: I. Surgical phenotype data collection in endometriosis research. Fertil Steril. 2014;102(5):1213–22. doi: 10.1016/j.fertnstert.2014.07.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vitonis AF, Vincent K, Rahmioglu N, et al. World Endometriosis Research Foundation Endometriosis Phenome and biobanking harmonization project: II. Clinical and covariate phenotype data collection in endometriosis research. Fertil Steril. 2014;102(5):1223–32. doi: 10.1016/j.fertnstert.2014.07.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rahmioglu N, Fassbender A, Vitonis AF, et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: III. Fluid biospecimen collection, processing, and storage in endometriosis research. Fertil Steril. 2014;102(5):1233–43. doi: 10.1016/j.fertnstert.2014.07.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fassbender A, Rahmioglu N, Vitonis AF, et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonisation Project: IV. Tissue collection, processing, and storage in endometriosis research. Fertil Steril. 2014;102(5):1244–53. doi: 10.1016/j.fertnstert.2014.07.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rogers PA, D'Hooghe TM, Fazleabas A, et al. Defining future directions for endometriosis research: workshop report from the 2011 World Congress of Endometriosis In Montpellier, France. Reprod Sci. 2013;20(5):483–99. doi: 10.1177/1933719113477495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Barrett JC, Dunham I, Birney E. Using human genetics to make new medicines. Nat Rev Genet. 2015;16(10):561–2. doi: 10.1038/nrg3998. [DOI] [PubMed] [Google Scholar]