Summary
The AMP-activated protein kinase (AMPK) is a sensor of cellular energy and nutrient status, expressed almost universally in eukaryotes as heterotrimeric complexes comprising catalytic (α) and regulatory (β and γ) subunits. Along with the mechanistic target-of-rapamycin complex-1 (mTORC1), AMPK may have been one of the earliest signaling pathways to have arisen during eukaryotic evolution. Recent crystal structures have provided insights into the mechanisms by which AMPK is regulated by phosphorylation and allosteric activators. Another recent development has been the realization that activation of AMPK by the upstream kinase LKB1 may primarily occur not in the cytoplasm but at the surface of the lysosome, where AMPK and mTORC1 are regulated in a reciprocal manner by the availability of nutrients. It is also becoming clear that there is a substantial amount of crosstalk between the AMPK pathway and other signaling pathways that promote cell growth and proliferation, and this will be discussed.
The AMP-activated protein kinase (AMPK) is a ubiquitous sensor of cellular energy and nutrient status in eukaryotic cells. Because of the adenylate kinase reaction (2ADP ↔ ATP +AMP), which appears to be maintained close to equilibrium in essentially all cells, increases in the cellular ADP:ATP ratio (signalling a deficit in cellular energy), are always accompanied by even larger increases in the AMP:ATP ratio (Gowans et al., 2013). AMPK monitors the ratio of AMP:ATP (and, to a lesser extent, ADP:ATP) and, if energy deficit is detected, acts to restore energy homeostasis by switching on alternate catabolic pathways that produce ATP, while switching off biosynthetic pathways and other non-essential processes consuming ATP. The pathway appears to have arisen very early during eukaryotic evolution, because AMPK orthologs are found in essentially all eukaryotes, including protists, fungi, plants and animals. One interesting exception that “proves the rule” is the microsporidian parasite Encephalitozoon cuniculi, a eukaryote that lives as an obligate parasite within mammalian cells. It has an extremely small genome encoding only 29 conventional protein kinases, which lacks any genes encoding AMPK subunits (Miranda-Saavedra et al., 2007). Its genome does encode a glycolytic pathway, but it lacks functional mitochondria and appears to “steal” ATP from its host cell by means of ATP:ADP translocases located in its plasma membrane (Tsaousis et al., 2008). Rather than being an ancestral eukaryote that never contained AMPK, it therefore seems more likely that E. cuniculi, which has been able to strip its genome down to the bare minimum due to its existence as an obligate parasite, has dispensed with AMPK because its host cell contains AMPK and can maintain energy homeostasis on its behalf.
A major event within the last year was the publication of the first complete crystal structure for a human AMPK complex (Xiao et al., 2013), and I discuss below how this and related structural studies have illuminated the mechanism by which the AMPK system senses cellular energy. While AMPK was viewed at one time as a system that operated rather independently of other signalling pathways, as the field matured it was perhaps inevitable that an increasing amount of crosstalk with pathways regulating cell growth and proliferation would be found. The past year or so has also seen major developments in this area, particularly in the interaction with the mechanistic target-of-rapamycin (mTORC1) pathway. This will form another major theme of this review. Readers interested in other aspects of AMPK function, such as the identification of downstream targets, its anti-cancer or anti-inflammatory effects, or its role in the regulation of whole body energy balance, should consult other recent reviews (Hardie, 2014; Hardie and Ashford, 2014; Hardie et al., 2012; O'Neill and Hardie, 2013; Steinberg et al., 2013).
Canonical regulation by phosphorylation and by adenine nucleotides
AMPK appears to exist in all eukaryotes as heterotrimeric complexes comprising catalytic α subunits and regulatory β and γ subunits. In mammals these occur as multiple subunit isoforms (α1/α2, β1/β2, γ1/γ2/γ3) encoded by distinct genes, which can form at least 12 distinct heterotrimeric combinations. The kinase activity of the α subunit increases >100-fold following phosphorylation at a conserved threonine residue within the activation loop of the kinase domain (usually called Thr172 due to its position in rat α1/α2 (Hawley et al., 1996)). The major upstream kinase phosphorylating this site was identified as a heterotrimeric complex containing the tumour suppressor kinase LKB1, the pseudokinase STRAD and the scaffold protein MO25 (Hawley et al., 2003; Shaw et al., 2004; Woods et al., 2003). Subsequently, an alternate upstream pathway was discovered in which Thr172 is phosphorylated by calmodulin-dependent kinase kinases (CaMKKs), especially CaMKKβ (Hawley et al., 2005; Hurley et al., 2005; Woods et al., 2005). The Ca2+-dependent and AMP-dependent pathways occur independently, although because AMP acts in part by inhibiting Thr172 dephosphorylation (see below) the two can act synergistically if both Ca2+ and AMP are elevated (Fogarty et al., 2010).
Phosphorylation by the LKB1 complex is also required for the activity of at least twelve AMP-related kinases, which have kinase domains related to AMPK but do not have the other domains or subunits associated with it (Lizcano et al., 2004). The activities of LKB1 or these AMPK-related kinases do not appear to change under conditions of metabolic stress where AMPK is activated in an LKB1-dependent manner, such as during muscle contraction (Sakamoto et al., 2004; Sakamoto et al., 2005). This implies that regulation of AMPK under these circumstances is primarily due to signals acting on the downstream kinase (AMPK) rather than on the upstream kinase (LKB1). As in other situations of metabolic stress, muscle contraction is accompanied by increases in the AMP:ATP and ADP:ATP ratios (Sakamoto et al., 2005), and numerous studies in cell-free systems have revealed that binding of AMP and/or ADP to AMPK activates the kinase by three complementary mechanisms, all of which are antagonized by binding of ATP:
-
1)
AMP binding promotes Th172 phosphorylation by the LKB1 complex. It has been suggested that binding of ADP also promotes phosphorylation by CaMKKβ (Oakhill et al., 2010; Oakhill et al., 2011), but the author’s group found, using both nucleotides at 300 µM, that this effect was specific for AMP, and only operated with LKB1 and not CaMKKβ (Gowans et al., 2013). Major new insights into the mechanism by which LKB1 activates AMPK in intact cells have been obtained within the last year, and will be discussed later.
-
2)
AMP binding causes a conformational change that inhibits dephosphorylation of Thr172 by protein phosphatases (Davies et al., 1995). It was subsequently reported that binding of ADP, as well as AMP, can produce this effect (Xiao et al., 2011). The author’s group finds that the effect of ADP requires about ten-fold higher concentrations than those of AMP, although this is compensated by the fact that cellular concentrations of ADP are usually also about ten-fold higher than AMP. Both nucleotides may therefore contribute to the effect in intact cells, although we have argued that AMP may be the more important physiological activator because the changes in AMP in cells undergoing stress are always much larger than those in ADP (Gowans et al., 2013).
-
3)
Binding of AMP, but not ADP, causes allosteric activation of the kinase. Under appropriate conditions the degree of allosteric activation by AMP can be substantial (>10-fold), even in the presence of physiological ATP concentrations (5 mM) (Gowans et al., 2013).
Activation by natural products and synthetic activators
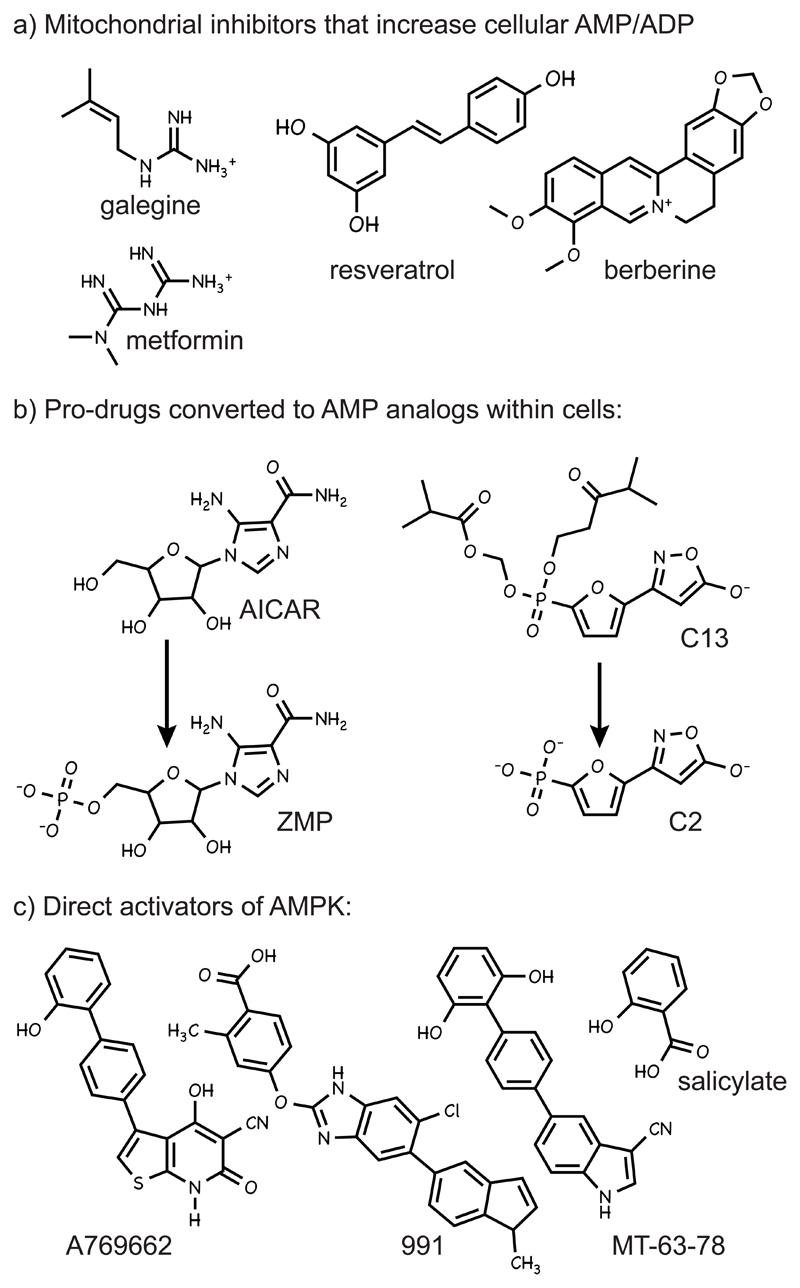
Given the great potential of AMPK activators as therapeutic agents (Hardie, 2013), many high-throughput screens have been conducted and numerous activators found; the structures of some of these are shown in Fig. 1. They can be divided into three major classes according to their mechanism of action:
-
1)
Indirect activators that inhibit cellular ATP production and thus increase cellular AMP:ATP and ADP:ATP ratios (Fig. 1a). Most of these compounds reduce ATP production by inhibiting the respiratory chain, although resveratrol, quercetin and oligomycin inhibit the mitochondrial ATP synthase (F1F0-ATPase) (Gledhill et al., 2007), while 2-deoxyglucose inhibits glycolysis. This mechanism is most clearly demonstrated using cells expressing a mutation in the AMPK-γ subunit (e.g. R531G in γ2) that prevents binding of AMP at the critical activating site; this class of compound fails to activate the mutant kinase. This approach has been used to show that the anti-diabetic drug metformin, the glycolytic inhibitor 2-deoxyglucose, and the plant products, resveratrol, berberine and galegine (metformin was derived from the latter), all act via this mechanism (Hawley et al., 2010). In the last two or three years a bewildering variety of natural plant products, many of which (like berberine) have been used in traditional Asian medicine, have been shown to activate AMPK. The author recently compiled a list of over 50 such compounds (Hardie, 2014). While their mechanism of action has not been established in most cases, the author suspects that many of them are mitochondrial poisons produced by plants as a defensive mechanism. Following the aphorism of Paracelsus that “the dose makes the poison”, at high doses these compounds may act as mitochondrial poisons that deter grazing of the plant by animals or insects, or infection by plant pathogens, while at lower doses they may have useful therapeutic properties by causing a mild inhibition of ATP synthesis that activates AMPK.
-
2)
Compounds that are pro-drugs converted into AMP analogs inside cells. The founder member of this class is 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR), a nucleoside that is taken up into cells by adenosine transporters (Gadalla et al., 2004) and then phosphorylated to the equivalent nucleotide ZMP (Fig. 1b). ZMP is an AMP analog that mimics all three effects of AMP on the AMPK system (Corton et al., 1995) and binds to the same site(s) as AMP, so the effects of AICAR are also abolished by the R531G mutation (Hawley et al., 2010). For many years, AICAR was the only member of this class, but recently a new AMP analog, 5-(5-hydroxyl-isoxazol-3-yl)-furan-2-phosphonic acid or C2, was reported to be 100-fold more potent than AMP, and over 1000-fold more potent than ZMP, as an AMPK activator in cell-free assays. Since it carries a negatively charged phosphonate group, C2 is not cell-permeable, but it can be administered to cells in the form of the pro-drug C13 (Fig. 1b) in which the phosphonate group is esterified; C13 is converted into C2 by intracellular esterases. Unlike ZMP, C2 has the advantage that it does not affect other AMP-sensitive metabolic enzymes, such as glycogen phosphorylase or fructose-1,6-bisphosphatase (Gomez-Galeno et al., 2010). Like AMP and ZMP, C2 causes both allosteric activation and protection against Thr172 dephosphorylation, and its effects are abolished by the R531G mutation, but unlike AMP and ZMP it is a selective activator of complexes containing AMPK-α1 rather than -α2 (Hunter et al., 2014).
-
3)
Direct AMPK activators whose binding involves the β subunit carbohydrate-binding module. The recently recognized binding site for this class of AMPK activator is described in more detail in the section on AMPK structure below. The first activator in this class was the thienopyridone A769662 (Cool et al., 2006), which directly activates AMPK, mimicking the effects of AMP to cause allosteric activation and protect against Thr172 dephosphorylation (Goransson et al., 2007; Sanders et al., 2007). A769662 acts in an AMP-independent manner, since it activates AMPK in cells expressing the AMP-insensitive R531G mutant (Hawley et al., 2010). Even before the exact binding site had been clarified, it was clear that it involved the β subunit, because activation was more effective with β1 than β2 complexes (Scott et al., 2008) and was abolished by a mutation within β1 (S108A) that prevents autophosphorylation of that serine residue (Sanders et al., 2007). Recently, two new activators, “991” and “MT-63-78”, have been described (Fig. 1c). Crystallography showed that 991 binds in the same site as A769662 (Xiao et al., 2013), while MT-63-78 also shows selectivity for β1 complexes (Zadra et al., 2014) and is likely therefore to bind the same site. The presence of a well-defined binding site for these synthetic activators raises the intriguing question as to whether there are natural ligands that bind this site. Salicylate (derived originally from the bark of the white willow, Salix alba) has been used as a medicine for thousands of years, and is the natural product from which aspirin was derived. Aspirin is an acetyl ester that is rapidly broken down to salicylate following its adsorption from the gut. Its acetyl group (not present in salicylate) irreversibly inactivates the cyclo-oxygenases (COX1 and COX2) that catalyse the key step in prostanoid biosynthesis, which are generally regarded as the main targets of aspirin. However, the very short half-life of aspirin in the circulation (a few minutes) compared with salicylate (several hours) raises doubts as to whether all of the therapeutic effects of aspirin are mediated by this mechanism (Steinberg et al., 2013). Salicylate, but not aspirin, activates AMPK in intact cells at concentrations that occur in plasma of humans taking high doses of aspirin (Hawley et al., 2012). Like A769662 or 991, activation by salicylate is selective for β1 complexes and is abolished by an S108A mutation in β1, so these compounds all appear to bind at the same site. In mice, treatment with either salicylate or A769662 caused a more rapid switch from carbohydrate to fat oxidation at the onset of fasting, but these metabolic effects were absent in β1 knockout mice, providing strong evidence that they were mediated by AMPK activation (Hawley et al., 2012). While salicylate is a natural ligand, it is a plant product and this leaves open the question as to whether there are naturally occurring ligands binding at the A769662 site that are produced by mammals.
Figure 1. Structures of a selection of three different classes of AMPK-activating compounds.
Allosteric activation in the absence of Thr172 phosphorylation
Since phosphorylation of AMPK at Thr172 increases its activity by >100-fold, the dephosphorylated enzyme has generally been regarded as inactive, and many researchers have used the degree of Thr172 phosphorylation (assessed using phosphospecific antibodies) as a surrogate for AMPK activation. However, recent results suggest that this may not always be valid (Scott et al., 2014). A769662 was found to cause a very large allosteric activation of either wild type AMPK not phosphorylated on Thr172, or of a non-phosphorylatable T172A mutant (65- or 100-fold respectively). However, maximal activation required either prior autophosphorylation of Ser108, or a phosphomimetic mutation (S108E) at that residue. Remarkably, a T172A/S108E double mutant was activated ≈900-fold by A797662, and in the presence of AMP (which caused 20-fold activation on it own) this increased to ≈2,500-fold, reaching a final activity comparable with that obtained with the maximally phosphorylated wild type kinase. Thus, combined allosteric activation by A-769662 and AMP can achieve the same final activity with or without Thr172 phosphorylation, although this does require Ser108 to be phosphorylated. In the same study, it was shown that Ser108 was modified by cis autophosphorylation and that, following a pulse of CaMKKβ activation in COS7 cells, dephosphorylation of Ser108 was slower than that of Thr172. This implies that, following an event that caused a transient Thr172 phosphorylation (which would trigger Ser108 autophosphorylation), the binding of activators at both the A769662 and AMP sites might then cause a hyper-activated state that could continue even after Thr172 was dephosphorylated, as long as Ser108 remained phosphorylated. Another interesting possibility is that there might be a distinct upstream kinase for Ser108. While these findings are intriguing, their true physiological significance may only become clear when a natural ligand that binds the A769662 site has been identified. However, they do show that relying on Thr172 phosphorylation alone as a marker for AMPK activation is inadvisable, and that the phosphorylation of a downstream target such as acetyl-CoA carboxylase should also be monitored. While unlikely to be relevant to the mechanism described by Scott et al (2014), others have found situations in vivo where changes in AMPK activity measured in immunoprecipitates (and therefore not due to allosteric effects) did not appear to be associated with changes in Thr172 phosphorylation (Dagon et al., 2012; Pulinilkunnil et al., 2011).
AMPK – structure and regulation
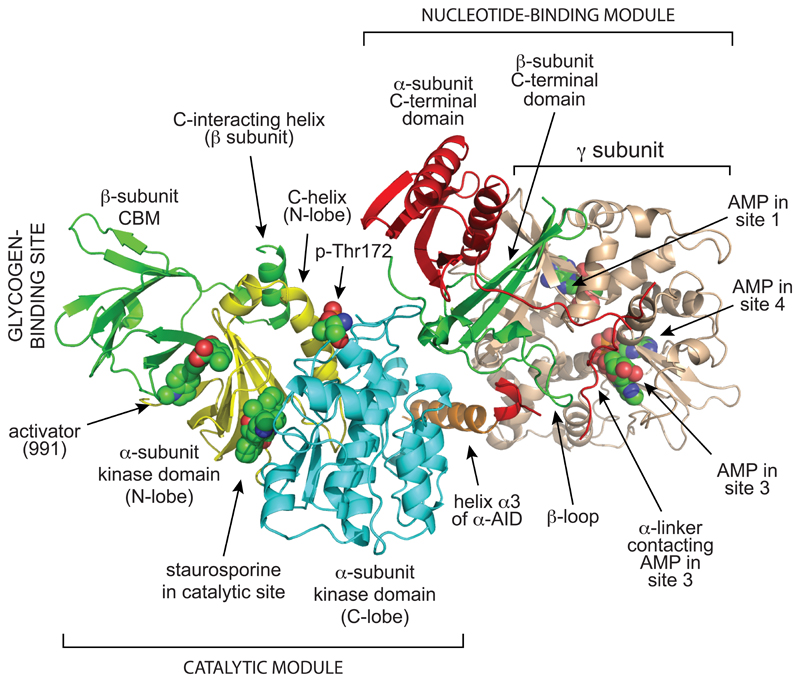
I will now discuss how the recent crystal structure of a complete human α2β1γ1 complex (Xiao et al., 2013), together with analysis of previous partial structures, has illuminated the regulatory mechanisms described above. The structure of the heterotrimer can be divided into two major sections, i.e. the “catalytic module” (Fig. 2, bottom left) and the “nucleotide-binding module” (Fig. 2, top right). The critical phosphorylation site, Thr172, lies in the cleft between these two modules, where access to upstream kinases and phosphatases may be particularly sensitive to conformational changes. The structures of the three subunits within the heterotrimer will now be discussed in turn.
Figure 2. Structure of complete α2β1γ1 heterotrimer of AMPK.
The model was created with MacPyMol using PDB file 4CFE. The α, β and γ subunits are shown in “cartoon view” with different domains color coded, while the activator 991, the kinase inhibitor staurosporine, the three bound molecules of AMP, and phospho-Thr172 are shown in “sphere view” with C atoms green, O red, N blue and P orange. Various features labeled are discussed in the text.
The α subunit
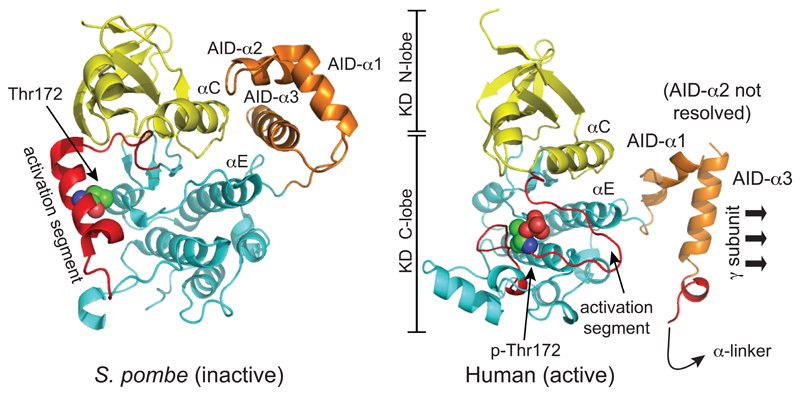
The kinase domain at the N-terminus of the α subunit has the small N-lobe and larger C-lobe of a typical serine/threonine kinase domain, with the active site that binds MgATP (which in this structure contained the kinase inhibitor staurosporine) located in the cleft between them. Despite this bound inhibitor the kinase domain was in an active conformation, because Thr172 had been phosphorylated prior to crystallization, and because the construct was also crystallized in the presence of two activating ligands, i.e. AMP and 991. In addition, the four hydrophobic residues that form the “regulatory spine” (Leu68 and Leu79 from the N-lobe, and His137 and Phe158 from the C-lobe) were aligned as in other active kinases (Taylor and Kornev, 2011). Within the α subunit sequence the kinase domain (KD) is immediately followed by the auto-inhibitory domain (AID), so-called because KD-AID constructs are around ten-fold less active than constructs containing the KD alone, even after phosphorylation on Thr172 (McBride et al., 2009; Pang et al., 2007). Structures for a KD-AID construct from the fission yeast Schizosaccharomyces pombe (Chen et al., 2009), and for two isolated rat and human AIDs (Chen et al., 2013), show that AIDs form a compact bundle of three α-helices. In the structure of the S. pombe KD-AID (Chen et al., 2009), where the kinase is in an inactive conformation with the “regulatory spine” out of alignment, helix α3 of the AID interacts with the “rear” of the kinase domain (i.e. the opposite side to the active site), forming interactions with residues in the C-helix of the N-lobe and the E-helix of the C-lobe that appear to maintain the kinase domain in an inactive conformation (Fig. 3, left). Although the AID was not fully resolved in the human structure (Xiao et al., 2013), it was clear by comparison with the S. pombe KD-AID structure that the AID had undergone a major rotation, so that helix α3 within it now interacts with the γ subunit, rather than with the N- and C-lobes of the kinase domain (Fig. 3, right). The first part of the linker between the AID and the C-terminal domain of the α subunit (the “α-linker”) forms the “hinge” between the “catalytic module” and the “nucleotide-binding module”, and the binding of this linker to the AMP-bound γ subunit appears to cause this reorientation of the AID. This is discussed in more detail below, in the section describing the γ subunit.
Figure 3. Comparison of structure of the inactive KD:AID fragment from S. pombe, and the active KD:AID fragment from human α2, showing the dramatic change in the position of the AID.
The models were created with McPymol using PDB entries 3H4J and 4CFE (although the latter was a complete heterotrimer, only the KD and AID are shown). The kinase domains are viewed from approximately the same angle, so that the change in the orientation of the AID is more obvious. The N- and C-lobes of the kinase domain are in yellow and cyan respectively, except that the activation segments within the C-lobe (which have markedly different conformations in the two structures) are in red. The AIDs are in orange, with their three α-helices labeled AID-α1 to -α3 (AID-α2 was poorly resolved in the human structure). In the inactive S. pombe structure, AID-α3 interacts with αC (C-helix) from the N-lobe and αE (E-helix) from the C-lobe, whereas in the active human structure it interacts mainly with the γ subunit (not shown) instead.
The globular C-terminal domain of the α subunit terminates with an α-helix that carries a well-defined nuclear export sequence. A similar sequence is found in both the α1 and α2 isoforms, although it has only been shown to be functional as a nuclear export sequence in the case of α2 (Kazgan et al., 2010). Just prior to this α-helix there is a long serine/threonine-rich loop that has not been resolved in any of the crystal structures. We refer to this as the “ST loop”, and its function will be discussed later.
The β subunit
Within the β1 subunit, only the two conserved regions found in β subunits from all species, i.e. the carbohydrate-binding module (CBM) and the C-terminal domain, were fully resolved in the structure (Xiao et al., 2013). The CBM is related to non-catalytic domains found in enzymes that bind starch and glycogen, and has been shown to be responsible for the binding of a proportion of AMPK to the surface of glycogen particles (Hudson et al., 2003; McBride and Hardie, 2009; Polekhina et al., 2003; Polekhina et al., 2005). The physiological role of glycogen binding is not completely clear, although it would co-localize AMPK with glycogen synthase, whose phosphorylation by AMPK causes inactivation of both the muscle and liver isoforms and thus inhibits glycogen synthesis (Bultot et al., 2012; Carling and Hardie, 1989; Jorgensen et al., 2004). One of the most intriguing features of the new heterotrimer structure is that the CBM sits next to the N-lobe of the kinase domain, with the cleft between them providing the binding site for the synthetic AMPK activators 991 and A769662 (Fig. 2). Ser108 within the CBM, which must be autophosphorylated to allow full activation by A769662 and salicylate, is not directly involved in binding of these ligands, but when phosphorylated it forms electrostatic interactions between the CBM and the N-lobe of the kinase domain that stabilize the binding cleft. The CBM is followed in the β1 subunit sequence by the “C-interacting helix”, which interacts with the C-helix of the N-lobe of the kinase domain. It seems likely that this interaction transmits the effects of binding of A769662-like activators to the active site, because a change in the position of the N-terminal end of the C-helix is one of the critical steps that must be achieved for activation of many kinases (Taylor and Kornev, 2011).
Turning to the C-terminal domain of the β subunit, the first portion has an extended conformation that interacts with the C-terminal domain of the α subunit. It then takes an excursion termed the β-loop, which contacts one quadrant of the γ subunit (see below), before terminating with two strands of a three-stranded β-sheet, the third strand being contributed by the γ subunit. This method of linking the three subunits is conserved in AMPK orthologs from humans, fission yeast and budding yeast (Amodeo et al., 2007; Townley and Shapiro, 2007; Xiao et al., 2007). The N-terminal region of the β subunit (containing the myristoylation site), and the 30 residues connecting the C-interacting helix and the C-terminal domain, have not been resolved in any structure to date.
The γ subunit
It has been known for many years that the γ subunit contains the regulatory binding sites for adenine nucleotides (Cheung et al., 2000; Scott et al., 2004). Following N-terminal regions that vary greatly in length and sequence between different isoforms, and a short segment providing the β-strand that interacts with the C-terminal domain of the β-subunit, γ subunits from all species contain four tandem cystathione β-synthase (CBS) repeats. CBS repeats occur in a small number of other human proteins (including cystathione β-synthase itself) (Bateman, 1997), usually as just two repeats that form a structure known as a Bateman domain, with a cleft between the repeats that often binds ligands containing adenosine (Scott et al., 2004). The two Bateman domains in the AMPK-γ subunits assemble in a pseudo-symmetrical head-to-head manner, forming a flattened disk with one CBS repeat in each quadrant. This arrangement provides four symmetrical clefts near the center of the disk where the regulatory nucleotides could potentially bind; by convention (Kemp et al., 2007) these are numbered 1-4 according to the number of the CBS repeat providing a conserved aspartate side chain that interacts with the ribose ring of the bound nucleotide. However, in mammalian AMPK only sites 1, 3 and 4 are utilized, with site 2 always unoccupied (interestingly, CBS2 has an arginine in place of the conserved aspartate). The phosphate groups of adenine nucleotides bound at sites 1, 3 and 4 lie close together in the center of the disk. Some amino acid side chains interact with nucleotides at more than one site, or even with nucleotides in different sites depending on which nucleotide is bound (Chen et al., 2012; Xiao et al., 2007; Xiao et al., 2011). This arrangement suggests that there would be complex interactions between the three sites; agreeing with this, mutations of any of the three sites reduces the effects of AMP on both Thr172 phosphorylation and allosteric activation (Oakhill et al., 2010). Sites 1 and 3 are able to bind AMP, ADP and ATP in competition, but there is some disagreement about site 4. It was originally suggested that site 4 only binds AMP in a “non-exchangeable” manner, with sites 1 and 3 being the sites where ADP or ATP exchanged with AMP when they were soaked into crystals made using AMP (Xiao et al., 2007; Xiao et al., 2011). However, it was subsequently reported that if the core of the heterotrimeric complex was crystallized in the presence of ATP rather than AMP, ATP was found at sites 1 and 4, rather than 1 and 3 (Chen et al., 2012). Binding of ATP rather than AMP was also observed to cause a conformational change in which the two Bateman domains (formed by CBS1/CBS2 and CBS3/CBS4) rotated towards each other by 5°, which had not been observed in the previous crystal soaking experiments. Since AMPK complexes purified from bacteria in the absence of added nucleotides predominantly have AMP rather than ATP bound at site 4, Chen et al (2012) argued that this site must have a higher affinity for AMP than ATP. As discussed in the next paragraph, site 3 appears to be the site where AMP binding causes activation, but analysis of the crystals obtained in the presence of ATP also suggested that binding of ATP at site 4 would preclude binding of any other nucleotide at site 3 (Chen et al., 2012). Thus, to obtain binding of AMP at the critical site 3, AMP may have to be already bound at site 4, suggesting a co-operative interaction between the two sites.
Intriguingly, the linker peptide between the AID and the C-terminal domain of the α subunit wraps around the face of the γ subunit that contains sites 2 and 3 (Xiao et al., 2013; Xiao et al., 2011). One well-conserved sequence motif within this linker (α-regulatory subunit interacting motif-1, α-RIM1) contacts the unused site 2, while another (α-RIM2) contacts AMP bound in site 3 (Xin et al., 2013). These authors proposed that the binding of α-RIM1 and α-RIM2 to the γ subunit when AMP is bound in site 3 would force the AID to dissociate from the kinase domain, thus relieving its inhibitory effects. Since this linker also forms the hinge between the “catalytic” and the “nucleotide-binding” modules, this change may also pull these modules together, reducing the accessibility of Thr172 to protein phosphatases and protecting it against dephosphorylation. Although this model is quite compelling, more structures of mammalian heterotrimers in inactive states will be required to confirm it. The role of nucleotide binding at site 1, and the reason why ADP mimics only one of the three effects of AMP, remain unclear. Also unclear is the mechanism by which AMP binding promotes phosphorylation of Thr172 by LKB1, although the next section describes some intriguing new twists in this story.
LKB1 may activate AMPK at the lysosomal surface in a reciprocal manner with mTORC1
Using a highly purified, reconstituted cell-free system, LKB1 can phosphorylate Thr172 and activate AMPK in a reaction stimulated by AMP, without requiring any additional components (Gowans et al., 2013). However, recent remarkable results from the group of Shengcai Lin suggest that in intact cells this interaction may be facilitated by the assembly of these complexes, with the help of various adapter proteins, at a two dimensional surface, i.e. the cytoplasmic surface of the lysosomal membrane (Zhang et al., 2014; Zhang et al., 2013). These studies also suggest that there are close reciprocal interactions between the regulation of the AMPK system, which is activated by lack of energy or nutrients, and that of mTORC1, which is activated under the opposite circumstance, i.e. by availability of nutrients.
Initial clues came from studies of axin, a scaffold protein originally identified as a negative regulator of Wnt signaling (Zeng et al., 1997). Unexpectedly, knockdown of axin in mouse liver using siRNAs caused accumulation of triglycerides that was accompanied by reduced AMPK activation (Zhang et al., 2013). In cell lines in which axin was knocked down, the activation of AMPK by treatments such as glucose deprivation, 2-deoxyglucose or AICAR was also defective. Moreover, co-precipitation studies suggested that axin enhanced Thr172 phosphorylation by assembling in a signaling complex with AMPK and LKB1, whose formation was enhanced by prior glucose deprivation of the cells. In cell lysates, co-precipitation of axin and AMPK (but not of axin and LKB1) was promoted by AMP and inhibited by ATP, while ADP had no effect. These results suggested a model in which LKB1 and axin form a constitutive complex, while AMPK is recruited to this complex when it binds AMP, thus enhancing the phosphorylation of Thr172 and causing AMPK activation.
Following up this study, the same group performed a two-hybrid screen to detect axin-interacting proteins and detected LAMTOR1 (Zhang et al., 2014), a protein that is tethered to the lysosomal membrane by N-terminal myristoylation and palmitoylation, and which is one of the components of the Ragulator complex. The latter is a pentameric complex, which under nutrient-rich conditions acts as a guanine nucleotide exchange factor (GEF) for the small G proteins RagA or RagB (Bar-Peled and Sabatini, 2014). In their GTP-bound state RagA or B (together with their heterodimer partners RagC or RagD) recruit mTORC1 to the lysosomal membrane. The Ragulator also interacts with the vacuolar ATPase (v-ATPase), the protein that acidifies the lumen of the lysosome by pumping protons into it. This complex is required to sense nutrients (particularly amino acids) within the lysosomal lumen, because v-ATPase inhibitors block mTORC1 activation by nutrients, although the molecular details of the sensing mechanism remain unclear (Bar-Peled and Sabatini, 2014).
Lin and colleagues (Zhang et al., 2014) also generated mice with floxed alleles of the LAMTOR1 gene, and used these to knock out LAMTOR1 conditionally in various tissues and cells. Remarkably, AMPK activation was defective in response to starvation in LAMTOR1-deficient liver, in response to exercise in LAMTOR1-deficient muscle, and in response to glucose deprivation in LAMTOR1-deficient mouse embryo fibroblasts (MEFs). In lysates derived from MEFs, anti-axin antibodies pulled down LAMTOR1, LKB1 and AMPK, and this co-precipitation was enhanced by prior glucose deprivation of the cells. The interaction between LAMTOR1 and axin required the N-terminal region of axin (1-400) whereas only the C-terminal region (507-731) was required for the interaction with LKB1 and AMPK. Thus, axin appears to be an adaptor protein that recruits LKB1 and AMPK to the Ragulator, and hence the lysosome, under nutrient-poor conditions. Interestingly, LAMTOR1 deficiency did not affect activation of AMPK by the Ca2+ ionophore A23817, and CaMKKβ did not co-precipitate with axin or LAMTOR1, suggesting that LAMTOR1 and axin are not involved in the alternate, Ca2+-mediated pathway for AMPK activation. By sub-cellular fractionation, the LAMTOR1:axin:LKB1 complex was found in “late endosomal” and “detergent-resistant/lipid raft” membrane fractions following glucose deprivation. Interestingly, AMPK was detected both in these membrane fractions and in cytosolic fractions in control incubations, and the amount recovered in the membrane fractions did not increase upon glucose deprivation, although its phosphorylation on Thr172 did. By fluorescence microscopy, both LKB1 and axin also co-localized with the lysosomal membrane marker LAMP2 following glucose deprivation, and the translocation of LKB1 to the lysosomal membrane following glucose deprivation required axin, but not AMPK.
Interestingly, AMP promoted co-precipitation of axin and AMPK in detergent lysates of the “late endosomal” fraction from MEFs, but this occurred at much lower AMP concentrations if the membrane fractions had been prepared from glucose-deprived cells. These results suggest that the Ragulator complex somehow senses glucose deprivation and, while this acts independently of the canonical activation of AMPK by LKB1 in the presence of AMP, the two regulatory systems can interact with and reinforce each other. The Ragulator-dependent mechanism seems to require the v-ATPase, because it was affected by siRNA knockdown of subunits of the v-ATPase, while the v-ATPase inhibitor concanamycin A promoted translocation of axin and LKB1 to the lysosome, mimicking the effects of glucose deprivation.
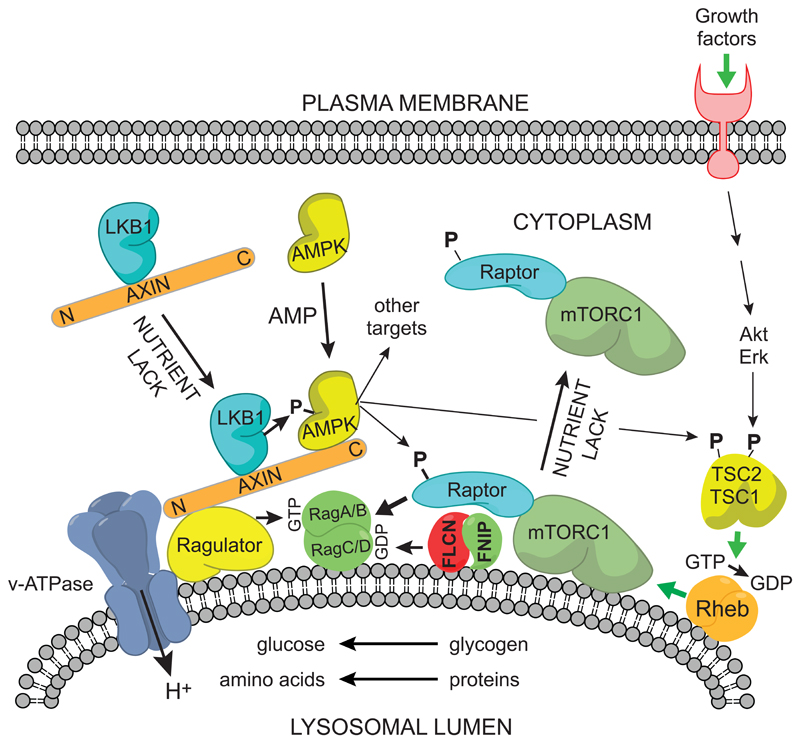
Taken together, these results suggest that the Ragulator complex, which is known to be important for recruiting mTORC1 to the lysosome under nutrient-rich conditions, is also involved in the reciprocal recruitment of the axin-LKB1 complex to the lysosome under nutrient-poor or low energy conditions, leading to phosphorylation and activation of AMPK (Fig. 4). Axin deficiency was also found to reduce the dissociation of mTORC1 from the lysosome following a switch from nutrient-rich to nutrient-poor conditions. It was already known that activation of AMPK causes inactivation of mTORC1 by phosphorylating Raptor (Gwinn et al., 2008) and TSC2 (Inoki et al., 2003). Raptor is a key component of the mTORC1 complex that interacts with RagA/BGTP:RagC/DGDP, thus recruiting mTORC1 to the lysosome (Sancak et al., 2008). Phosphorylation of Raptor by AMPK may therefore be part of the mechanism by which mTORC1 dissociates from the lysosome under nutrient-poor conditions.
Figure 4. Proposed mechanism for reciprocal regulation of mTORC1 and AMPK at the cytoplasmic surface of the lysosome.
The model is based on (Bar-Peled and Sabatini, 2014) and (Zhang et al., 2014). (1) Activation of mTORC1: nutrient availability (amino acids and/or glucose) within the lysosome are sensed by a mechanism requiring the v-ATPase proton pump, and this effect is transmitted to the Ragulator, which converts RagA/BGDP to RagA/BGTP, while the folliculin:FNIP complex converts RagC/DGTP to RagC/DGDP. The RagAGTP:RagCGDP complex binds the Raptor component of mTORC1, whose activation also requires Rheb:GTP. Formation of the latter complex is promoted by growth factors via phosphorylation and dissociation from the lysosome of TSC1:TSC2, a Rheb-GAP (Menon et al., 2014). (2) Activation of AMPK: nutrient lack causes the Ragulator to recruit the axin:LKB1 complex, and elevated AMP also recruits AMPK to this complex. LKB1 phosphorylates and activates AMPK, which then presumably dissociates from the complex to phosphorylate downstream targets. Two of these targets are Raptor and the TSC1:TSC2 complex. Phosphorylation of Raptor by AMPK, coupled with conversion of RagAGTP:RagCGDP to RagAGDP:RagCGTP, may promote the dissociation of mTORC1 from the lysosome. Phosphorylation of the TSC1:TSC2 complex appears to increase its Rheb-GAP activity, thus converting Rheb:GTP to the inactive Rheb:GDP form, although the exact mechanism remains unclear.
At first sight, these findings of Lin and colleagues (Zhang et al., 2014; Zhang et al., 2013) seemed quite surprising, because neither axin nor LAMTOR1 have been found to interact with LKB1 or AMPK in protein interaction screens by other laboratories, and because their proposed new mechanism (Fig. 4) also represents a radical departure from the existing view of AMPK activation by LKB1. However, there are other recent findings that might be consistent with their hypothesis:
-
1)
Another component of mTORC1 regulation is the folliculin complex, containing folliculin and folliculin-interacting protein-1 or -2 (FNIP1/2). Loss-of-function mutations in the folliculin gene (FLCN) in humans cause Birt-Hogg-Dubé syndrome, characterized by the formation of frequent benign tumors of hair follicles (hence the name), as well as increased risk of development of lung and kidney cysts and renal cancer; folliculin is therefore a tumor suppressor (Tee and Pause, 2013). The folliculin complex acts as a GTPase activator protein (GAP) for RagC/D, the heterodimer partners of RagA/B (Tsun et al., 2013). Since recruitment of mTORC1 to the lysosome requires RagA/B to be in the GTP-bound form and (unusually) RagC/D to be in the GDP-bound form, the Ragulator and folliculin complexes are both positive regulators of mTORC1, even though one is a GEF and the other a GAP. Interestingly, it was shown in 2006 that AMPK interacts with FNIP1, and it was suggested that folliculin was phosphorylated by AMPK, although direct phosphorylation site(s) have not been identified (Baba et al., 2006; Wang et al., 2010). Very recently it has been reported that AMPK is constitutively activated in folliculin-deficient MEFs, and that this is associated with various metabolic changes, including increased mitochondrial biogenesis. Similar changes in gene expression were observed by immunohistochemistry in renal cell carcinomas from Birt-Hogg-Dubé patients (Yan et al., 2014). Thus, folliculin appears to be a positive regulator of mTORC1 but a negative regulator of AMPK.
-
2)
The major splice variant of LKB1 expressed in most cell types (Towler et al., 2008) contains a C-terminal CKQQ sequence where the cysteine residue is modified by farnesylation. Recently, a knock-in mouse with a C→S mutation that prevents farnesylation was generated (Houde et al., 2014). The lack of farnesylation did not affect the intrinsic activity of LKB1 measured using a peptide substrate, but the activation of AMPK and the phosphorylation of downstream targets in isolated hepatocytes in response to AICAR, and during in situ contraction or AICAR treatment in skeletal muscle in vivo, was impaired. There was also a significantly smaller proportion of the farnesylation-deficient mutant associated with membrane fractions in mouse liver or MEFs. These authors proposed that the farnesylation of LKB1 and the β-subunit myristoylation of AMPK might cause them to associate together at a membrane surface, thus facilitating their interaction (Houde et al., 2014).
-
3)
On glucose deprivation of COS7 cells expressing wild type GFP-tagged AMPK, GFP fluorescence changed from a diffuse cytoplasmic signal that was lost when the plasma membrane was permeabilized with digitonin, to a speckled perinuclear pattern that was digitonin-resistant. By contrast, in cells expressing a non-myristoylatable G2A mutant the cytoplasmic fluorescence was unaffected by glucose deprivation, and the speckled perinuclear pattern was not seen (Oakhill et al., 2010). Based on the recent findings of Lin and colleagues, it is tempting to speculate that the speckled perinuclear location to which AMPK migrates on glucose deprivation is the lysosome, and that the farnesylation of LKB1 might also help to co-localize it on the lysosomal surface with axin.
Taking all of these observations together, the lysosomal surface may represent a key locus where nutrients are sensed in a reciprocal manner by both the mTORC1 and the AMPK signaling pathways. mTORC1 is activated by the availability of nutrients, and promotes cell growth and proliferation and the associated metabolic changes (e.g. expression of HIF-1α, leading to increased glucose uptake and aerobic glycolysis (Shackelford et al., 2009)), whereas AMPK is activated by lack of nutrients, inhibits cell growth and proliferation, and promotes the more energy-efficient oxidative metabolism exhibited by most quiescent cells. By phosphorylating Raptor, AMPK may also be involved in promoting dissociation of mTORC1 from the lysosomal membrane under nutrient-poor conditions.
Why might AMPK and mTORC1 sense nutrients at the lysosome?
One can make several evolutionary arguments at to why nutrient sensing might have developed at the lysosome. Many heterotrophic protists, such as flagellates, ciliates and amoebas, feed primarily by phagocytosis, which delivers macromolecules directly to lysosomes or vacuoles without passage through the cytoplasm. Lysosomes or vacuoles in unicellular organisms can therefore be regarded as the equivalent of the gut in a multicellular organism. All eukaryotic cells also carry out autophagy, the process by which organelles and macromolecular complexes, such as protein aggregates or glycogen particles, are engulfed by autophagosomes and delivered to lysosomes for digestion. Indeed, AMPK plays a fundamental role in switching on autophagy during starvation by phosphorylating ULK1, the protein kinase that initiates the process (Egan et al., 2011). Plant and fungal cells contain vacuoles, which are essentially very large lysosomes that can be used for long-term storage of nutrients and other purposes. Interestingly, in the budding yeast Saccharomyces cerevisiae there appear to be two pools of glycogen, a cytoplasmic pool that can be rapidly mobilized by phosphorylase (encoded by GPH1) during early stationary phase, and a vacuolar pool mobilized by α-glucosidase (encoded by SGA1) during late stationary phase (Wilson et al., 2010). These two pools can be distinguished by mutations in GPH1 or SGA1 (which both cause increased glycogen accumulation, but at different stages during stationary phase), and because the vacuolar pool does not form in strains with mutations in genes required for autophagy. Intriguingly, deletion of any one of twelve genes required for the structure or assembly of the v-ATPase also leads to glycogen accumulation, which may occur because acidification of the vacuole fails to occur, and this is necessary for the activity of the vacuolar α-glucosidase encoded by SGA1 (Wilson et al., 2002). Glycogen breakdown can also occur in lysosomes in mammals, particularly during the neonatal period. Thus, human cells also express a lysosomal α-glucosidase, whose genetic loss is fatal during the first year of life. Interestingly, following the birth of rats numerous glycogen-containing autophagosomes rapidly appear in the liver and muscle and then disappear within a few hours (Schiaffino et al., 2008). In addition, mice with knockouts in the autophagy genes Atg5 and Atg7 are born at the expected frequency, but die within one day of birth (Komatsu et al., 2005; Kuma et al., 2004). Mice with a knock-in mutation in RagA that inhibits its GTPase activity and locks it in its GTP-bound state, thus activating mTORC1 irrespective of nutrient supply, also die within a few hours of birth. This appears to occur because they are unable to respond to the profound hypoglycemia that follows the loss of nutrient supply from the placenta, by switching off mTORC1 and inducing autophagy (Efeyan et al., 2013). It appears that lysosomal glycogen breakdown may be particularly important in newborn mammals, which must rapidly switch from the placenta to the gut for their source of nutrients. All of these observations suggest that the lysosome or vacuole can be an important site where sensing of glucose has to occur.
Crosstalk between AMPK and other pathways
Phosphorylation of the ST loop of AMPK by Akt, PKA and GSK3
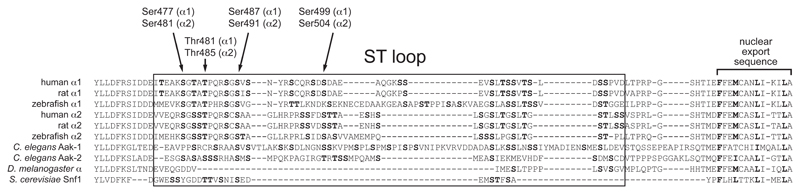
When the sequences of AMPK-α subunits from different species are aligned, α1 and α2 sequences from vertebrates and Caenorhabditis elegans can be seen to contain a serine/threonine-rich insert of about 55 amino acids, just preceding the final α-helix with its nuclear export sequence. This “ST loop” is absent or truncated in sequences from other invertebrates such as Drosophila melanogaster or Saccharomyces cerevisiae (Fig. 5), and is not resolved in any existing crystal structures, suggesting that it is mobile within the crystals. I will now discuss the evidence that phosphorylation at one or more sites within this loop down-regulates AMPK signaling.
Figure 5. Alignment of sequences at the C-terminal end of the α subunits, showing the location of the ST loop.
Sequences were aligned using CLC Main Workbench 6 using a “gap open cost” of 10 and a “gap extension cost” of 1. Serine and threonine residues within the ST loop are highlighted in bold type, with residues where there is evidence for phosphorylation shown with arrows (human residue numbering). Also shown are the nuclear export sequences at the extreme C-termini, with critical hydrophobic residues highlighted in bold.
The phosphatidylinositol (PI) 3-kinase → Akt pathway is activated by binding of insulin or insulin-growth factor-1 (IGF1) to their cognate receptors, with PI 3-kinase generating the Akt-activating second messenger, PI-3,4,5-trisphosphate (PIP3). These hormones are signals emanating from elsewhere in the body indicating either that nutrients are available (insulin), or that conditions are suitable for cell growth (IGF1). In general, insulin promotes the uptake of nutrients (glucose, fatty acids and amino acids) and their anabolic conversion to macromolecular storage forms, i.e. glycogen, triglycerides and proteins. Although AMPK can also stimulate uptake of glucose and fatty acids, it does so for a different purpose, i.e. their catabolic breakdown to generate ATP. The insulin/IGF1 and AMPK pathways are therefore essentially antagonistic, and it is perhaps not surprising that there should be mechanisms by which Akt down-regulates AMPK. In 2006, it was reported that Akt phosphorylated Ser485 of rat α1, which lies within the ST loop. Evidence was presented that this phosphorylation inhibited the phosphorylation of Thr172 by LKB1 and, consistent with this, prior treatment of perfused rat hearts with insulin caused phosphorylation of Ser485, and this was associated with reduced Thr172 phosphorylation during subsequent ischemia (Horman et al., 2006). Our group has recently confirmed and extended these observations, and provided evidence for a molecular mechanism for the effect (Hawley et al., 2014). We were able to confirm that Ser487 on human α1 (corresponding to Ser485 in rat α1; to avoid confusion I will use human residue numbering in the following text, and in Fig. 5) was a good substrate for Akt. However, the equivalent residue on human α2, Ser491, was an extremely poor substrate for Akt although subject to rapid autophosphorylation. Consistent with this, Ser487 on α1 became phosphorylated when cells were incubated with IGF1 to activate Akt, but not when incubated with berberine to activate AMPK, whereas the converse was true for Ser491 on α2. Pre-treatment of cells expressing wild type α1 with IGF1 greatly reduced subsequent Thr172 phosphorylation in response to A769662, but this effect was completely abolished in cells expressing an S487A mutant. To place these observations in the context of cancer, in three different tumor cell lines in which Akt was hyper-activated due to loss of PTEN (the phosphatase that breaks down PIP3), there was only a modest Thr172 phosphorylation and AMPK activation in response to A769662, but full effects could be restored either by adding a selective Akt inhibitor, or be re-expressing PTEN. These results show that a previously unrecognized effect of Akt hyper-activation in tumor cells (which can also occur due to mutations in PI 3-kinase, or upstream receptors), is that AMPK activation is down-regulated, removing the restraining influences that AMPK might otherwise have on cell growth and proliferation.
We also established the likely mechanism for these effects of Ser487 phosphorylation (Hawley et al., 2014). Although the ST loop is not resolved in the crystal structures, the residues at the ends of the “missing” loop lie quite close to Thr172. This suggested that when the ST loop was phosphorylated it could interact with the kinase domain, blocking access of upstream kinases to Thr172. We noticed a series of basic residues within the C-helix of the N-lobe of the kinase domain (Arg64, Lys 71 and Arg74) that are conserved in all vertebrate AMPK-α subunits, but not in most other kinases. Our hypothesis is that phosphorylated side chains on the ST loop interact with these basic residues, anchoring the loop to the N-lobe of the kinase domain and thus blocking access to Thr172. We tested this using two approaches: (i) a synthetic peptide corresponding to the ST loop inhibited Thr172 phosphorylation and activation of a human α1β2γ1 complex, but only when phosphorylated on the residue equivalent to Ser487; (ii) the inhibitory effect of Ser487 phosphorylation was completely abolished by a triple mutation of the C-helix (R64A/K71A/R74A), although kinase activity and Ser487 phosphorylation were unaffected. Prior Ser487 phosphorylation also inhibited Thr172 phosphorylation by CaMKKβ as well as LKB1, supporting a mechanism involving physical exclusion of the upstream kinases from Thr172. How Ser487 phosphorylation might affect dephosphorylation of Thr172 has not yet been addressed.
Other protein kinases may also phosphorylate the ST loop. Hurley et al (2006) reported that Ser487/491 on AMPK-α1 or -α2 (the isoform was not specified) was phosphorylated in response to the cyclic AMP-elevating agents in the pancreatic β cell line, INS-1. A recombinant AMPK-α1 peptide could also be phosphorylated at Ser487 by cyclic AMP-dependent protein kinase (PKA) in cell-free assays. Complicating this story, however, CaMKKβ appeared to be inactivated by cyclic AMP-elevating agents in INS-1 cells, and their effects on AMPK were abolished in CaMKKβ-null MEFs, suggesting that they were mediated by modulation of CaMKKβ, rather than AMPK (Hurley et al., 2006). To add further complications, using a bacterially expressed AMPK complex (α1β1γ1), PKA was found to phosphorylate not only Ser487 but also Ser499 (also within the ST loop), as well as Ser175 (within the activation loop and adjacent to Thr174, the residue equivalent to Thr172 in human α1). Various mutations at these sites were generated, and their analysis suggested that it was phosphorylation at Ser175, rather than Ser487 or Ser499, that blocked subsequent Thr174 phosphorylation and AMPK activation. These authors also provided evidence that this mechanism of down-regulation of AMPK by PKA limited the ability of AMPK to inhibit lipolysis in white adipocytes (Djouder et al., 2010). A puzzling feature of this paper is that the authors did not observe any effect on subsequent Thr172 phosphorylation when Ser487 was phosphorylated by PKA, even though this has been observed by other laboratories when the same residue was phosphorylated by Akt.
In addition to Ser487 and Ser499, the ST loop may be regulated by phosphorylation at other sites. Suzuki et al (2013) reported that the protein kinase GSK3β phosphorylated Thr481 and Ser477 (within the ST loop just N-terminal to Ser487), that this was “primed” by phosphorylation of Ser487, and that this further inhibited AMPK by promoting Thr172 dephosphorylation. While this is an interesting proposal, the physiological rationale underlying inhibition of AMPK by GSK3 is difficult to grasp, because both GSK3 isoforms (α and β) are inactivated by phosphorylation by Akt, and because GSK3 generally acts, like AMPK and in contrast to Akt, to inhibit rather than promote anabolic pathways. For example, GSK3 and AMPK both inactivate glycogen synthase and hence glycogen synthesis (Carling and Hardie, 1989; Jorgensen et al., 2004; McManus et al., 2005).
As discussed earlier, Ser487 on α1 is a good target for Akt, whereas the equivalent site on α2, Ser491, is a very poor substrate for Akt and appears to be modified by autophosphorylation instead (Hawley et al., 2014). It will therefore be important to examine the role of ST loop phosphorylation using the individual α subunit isoforms of AMPK, rather than simply assuming that results obtained with one isoform will be applicable to the other. For example, it has been suggested that p70 S6 kinase directly phosphorylates Ser491 in a recombinant α2β1γ1 complex, and that this explains how leptin inhibits AMPK in the hypothalamus (Dagon et al., 2012). While this is an attractive hypothesis, the evidence that S6 kinase directly phosphorylates Ser491 was based mainly on the acquisition of signal using an anti-pS491 phosphospecific antibody, which gives no information about the real extent of phosphorylation. Given our own findings (Hawley et al., 2014), it is also puzzling as to why these authors did not observe autophosphorylation when the α2β1γ1 complex was incubated without S6 kinase. In addition, there is no direct evidence that phosphorylation of Ser491 on α2 down-regulates Thr172 phosphorylation, which has currently only been demonstrated for Ser487 on α1.
Crosstalk between the AMPK and the Ras-Raf-MEK-ERK pathways
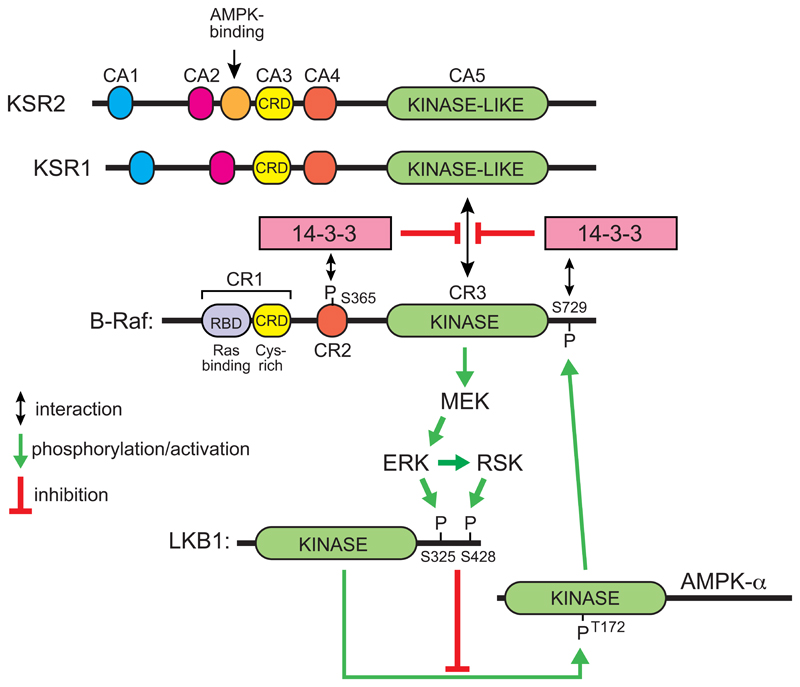
A-Raf, B-Raf and c-Raf are paralogous protein kinases involved in the Ras-Raf-MEK-ERK pathway, by which the effects of many growth factor receptors on cell growth and proliferation are transduced. The role of B-Raf in cancer was highlighted by findings that a V600E mutation in its activation loop, which causes constitutive activation, is present in around 50% of all cases of malignant melanomas, and in smaller proportions of other cancers (Davies et al., 2002). It was shown many years ago that AMPK phosphorylates a site near the C-terminus of c-Raf, Ser621, in cell-free assays (Sprenkle et al., 1997), and recently this has been re-investigated. Despite the fact that the sequence around Ser621 is a good fit for the consensus recognition motif for AMPK (Gwinn et al., 2008; Scott et al., 2002), treatment of MEFs with the AMPK activator A769662 did not appear to cause increased phosphorylation of c-Raf at Ser621. However, it did cause increased phosphorylation of B-Raf at the equivalent site, Ser729, as well as attenuation of ERK activation, and both effects were lost in AMPK knockout MEFs (Shen et al., 2013). When B-Raf was re-expressed in B-Raf-deficient MEFs, the ability of AMPK activators to inhibit ERK activation was regained using wild type B-Raf but not an S729A mutant; a keratinocyte cell line expressing the S729A mutant B-Raf also proliferated more rapidly than cells expressing the wild type. Phosphorylation of Ser729 on B-Raf does not directly inhibit its kinase activity, but instead promotes 14-3-3 binding and thus prevents association of B-Raf with KSR1, a scaffold protein with a kinase-like domain related to the Raf kinases, on which the latter normally assemble with their downstream kinases MEK and ERK to trigger downstream signaling (Shen et al., 2013) (Fig. 6).
Figure 6. Proposed reciprocal feedback loops operating between the Raf-MEK-ERK-RSK and LKB1-AMPK signaling pathways.
In the inactive state of B-Raf, phosphorylation of Ser365 and Ser729 [the latter by AMPK (Shen et al., 2013)] causes binding of 14-3-3 proteins that prevent association of B-Raf with the plasma membrane and with the Raf-like scaffold proteins KSR1 or KSR2. When Ras is activated it binds to the Ras-binding region of the B-Raf CR1 domain, relieving the inhibitory effect of the latter, and if 14-3-3 proteins are not bound KSR1 or KSR2 then forms a complex with B-Raf and MEK, causing phosphorylation and activation of the latter by B-Raf. This in turn triggers a cascade of phosphorylation and activation of the downstream kinases, ERK and RSK. The latter phosphorylate LKB1 at Ser325 and Ser428 respectively, reducing its ability to activate AMPK via phosphorylation of Thr172 (Zheng et al., 2009). Activated AMPK then phosphorylates B-Raf at Ser729, causing its interaction with 14-3-3 proteins and down-regulation once again. For more details of the conserved domains on B-Raf, KSR1 and KSR2, see Udell et al (2011).
Inhibitors of oncogenic B-RafV600E, such as vemurafenib and dabrafenib, are initially very effective in treating melanomas carrying the mutation, but unfortunately resistance to these drugs develops rapidly and disease returns. However, following their findings suggesting that AMPK down-regulated B-Raf signaling, the same group showed that treatment of melanoma cells with a combination of PLX4720 (a relative of vemurafenib) and phenformin (a more cell-permeable relative of metformin) enhanced apoptosis and reduced cell viability in a synergistic manner, delayed the emergence of PLX4720 resistance in a cell culture model, and induced tumor regression in xenograft and genetically engineered (B-RafV600E/PTEN-/-) mouse models of melanoma (Yuan et al., 2013). These results suggest that the AMPK activators metformin or phenformin might be useful adjuncts for therapy of human melanoma patients with B-Raf inhibitors.
Interestingly, the AMPK signaling pathway was also reported to be down-regulated in cells expressing the B-Raf V600E mutation; this was associated with phosphorylation by ERK and RSK of two sites on LKB1 that were proposed to negatively regulate phosphorylation of AMPK (Zheng et al., 2009). Thus, there appear to be two opposing negative feedback loops operating between B-Raf and AMPK (Fig. 6).
Although the molecular details are less clear, there are other possible connections between the Ras-Raf-MEK-ERK and AMPK pathways. KSR2 is a paralog of KSR1; protein interaction screens identified several AMPK subunits (α1, α2, β1, γ1) as binding partners that had a preference for KSR2 over KSR1 (Costanzo-Garvey et al., 2009). Although only KSR1 is detectable in MEFs, neither ksr1-/- nor ksr2-/- MEFs were able to oxidize exogenous fatty acids, but this was restored by over-expression of either KSR1 or KSR2; this required the region of these proteins (between domains CA2 and CA3) that interact with AMPK. Interestingly, KSR2 knockout mice become obese, glucose-intolerant and insulin-resistant as adults, which appeared to be due to reduced energy expenditure rather than increased food intake. The phosphorylation of AMPK in response to AICAR in explants of white adipose tissue from wild type mice was lost in the KSR2-/- mice, and genes involved in oxidative metabolism were also down-regulated in white adipose tissue. Interestingly, this function of KSR2 appears to translate into humans (Pearce et al., 2013), because in a screen of around 2000 obese humans, a significantly higher proportion had heterozygous mutations in the KSR2 gene compared with lean controls. Heterozygous carriers of human KSR2 mutations had lower basal metabolic rates and higher respiratory quotients, the latter indicating a relative deficit in fat oxidation. Finally, fatty acid oxidation was enhanced in myotubes that had been differentiated from C2C12 myoblasts transfected with DNA encoding wild type KSR2, but this was partially or completely abolished using most of the human KSR2 mutants, although the defect could be restored in every case by treating the cells with metformin (Pearce et al., 2013). Thus, KSR2 appears to be required for the ability of AMPK to promote fatty acid oxidation. However, the molecular mechanisms underlying this remain obscure, particularly because by Western blotting KSR2 is only detectable in mouse brain; very low levels of mRNA are detectable in liver and muscle, and essentially none in adipose tissue (Costanzo-Garvey et al., 2009). It is therefore not clear that the effect of KSR2 loss on AMPK activation in vivo is a cell-autonomous process.
Conclusions and future challenges
The “canonical’ regulation of AMPK by adenine nucleotides and by phosphorylation of Thr172 is now well understood, and the recent crystal structure of a complete human AMPK heterotrimer (Xiao et al., 2013) has provided many insights into how this regulation operates at the molecular level. However, elucidation of some details of the activation mechanism, such as the exact roles of the three individual adenine nucleotide-binding sites, how they distinguish between AMP, ADP and ATP, and the mechanism by which combinations of allosteric activators (AMP and A769662) can mimic the effects of phosphorylation, remain for the future. Another remaining challenge is the identification of natural ligands (other than salicylate) that bind at the same site as the synthetic activator A769662.
Particularly exciting developments in the past year have concerned the mechanisms by which the AMPK pathway talks to other signaling pathways. It appears that that LKB1 and AMPK do not simply wait to collide within the three dimensions of the cytoplasm, but are brought together on the two-dimensional surface of the lysosome by the adapter proteins axin and LAMTOR1, where the reciprocal regulation of mTORC1 and AMPK may be intimately linked (Zhang et al., 2014). Although both are absent in the obligate parasite E. cuniculi (Miranda-Saavedra et al., 2007), genes encoding the mTORC1 and AMPK pathways are present in the genome of Giardia lamblia, which has the smallest kinome of any free-living eukaryote and appears to have branched very early from the main eukaryotic lineage (Manning et al., 2011). The mTORC1 and AMPK pathways were therefore probably among the first signaling pathways to have developed during evolution of eukaryotes, where their functions may have been to sense to the availability, or lack of availability, of nutrients that were delivered to the lysosome or vacuole by phagocytosis or autophagy, and to regulate cell growth and metabolism accordingly. However, the complex cross-talk between AMPK and other signaling pathways that arose later during the development of multicellular animals, such as the insulin/IGF1 and Ras-Raf-MEK-ERK pathways, is also now being unraveled.
Acknowledgements
DGH is funded by a Senior Investigator Award (097726) from the Wellcome Trust and a Programme Grant (C37030/A15101) from Cancer Research UK. I would like to thank all of the current members of my lab (Romana Auciello, Alex Gray, Fiona Ross, Fiona Russell, Graeme Gowans, Simon Hawley and Diana Vara-Ciruelos) for discussions that have helped in putting this review together.
References
- Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449:492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, 3rd, Hartley JL, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.03.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- Bultot L, Guigas B, Von Wilamowitz-Moellendorff A, Maisin L, Vertommen D, Hussain N, Beullens M, Guinovart JJ, Foretz M, Viollet B, et al. AMP-activated protein kinase phosphorylates and inactivates liver glycogen synthase. Biochem J. 2012;443:193–203. doi: 10.1042/BJ20112026. [DOI] [PubMed] [Google Scholar]
- Carling D, Hardie DG. The substrate and sequence specificity of the AMP-activated protein kinase. Phosphorylation of glycogen synthase and phosphorylase kinase. Biochim Biophys Acta. 1989;1012:81–86. doi: 10.1016/0167-4889(89)90014-1. [DOI] [PubMed] [Google Scholar]
- Chen L, Jiao ZH, Zheng LS, Zhang YY, Xie ST, Wang ZX, Wu JW. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang J, Zhang YY, Yan SF, Neumann D, Schlattner U, Wang ZX, Wu JW. AMP-activated protein kinase undergoes nucleotide-dependent conformational changes. Nat Struct Mol Biol. 2012;19:716–718. doi: 10.1038/nsmb.2319. [DOI] [PubMed] [Google Scholar]
- Chen L, Xin FJ, Wang J, Hu J, Zhang YY, Wan S, Cao LS, Lu C, Li P, Yan SF, et al. Conserved regulatory elements in AMPK. Nature. 2013;498:E8–E10. doi: 10.1038/nature12189. [DOI] [PubMed] [Google Scholar]
- Cheung PCF, Salt IP, Davies SP, Hardie DG, Carling D. Characterization of AMP-activated protein kinase g subunit isoforms and their role in AMP binding. Biochem J. 2000;346:659–669. [PMC free article] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-Aminoimidazole-4-carboxamide ribonucleoside: a specific method for activating AMP-activated protein kinase in intact cells? Eur J Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- Costanzo-Garvey DL, Pfluger PT, Dougherty MK, Stock JL, Boehm M, Chaika O, Fernandez MR, Fisher K, Kortum RL, Hong EG, et al. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 2009;10:366–378. doi: 10.1016/j.cmet.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagon Y, Hur E, Zheng B, Wellenstein K, Cantley LC, Kahn BB. p70S6 Kinase phosphorylates AMPK on serine 491 to mediate leptin's effect on food intake. Cell Metab. 2012;16:104–112. doi: 10.1016/j.cmet.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Davies SP, Helps NR, Cohen PTW, Hardie DG. 5'-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2Ca and native bovine protein phosphatase-2Ac. FEBS Lett. 1995;377:421–425. doi: 10.1016/0014-5793(95)01368-7. [DOI] [PubMed] [Google Scholar]
- Djouder N, Tuerk RD, Suter M, Salvioni P, Thali RF, Scholz R, Vaahtomeri K, Auchli Y, Rechsteiner H, Brunisholz RA, et al. PKA phosphorylates and inactivates AMPKalpha to promote efficient lipolysis. EMBO J. 2010;29:469–481. doi: 10.1038/emboj.2009.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty S, Hawley SA, Green KA, Saner N, Mustard KJ, Hardie DG. Calmodulin-dependent protein kinase kinase-beta activates AMPK without forming a stable complex - synergistic effects of Ca2+ and AMP. Biochem J. 2010;426:109–118. doi: 10.1042/BJ20091372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadalla AE, Pearson T, Currie AJ, Dale N, Hawley SA, Randall AD, Hardie DG, Frenguelli BG. Distinct mechanisms underlie the activation of rat brain AMP-activated protein kinase and the inhibition of excitatory synaptic transmission by AICA riboside (Acadesine) in area CA1 of rat hippocampus. J Neurochem. 2004;88:1272–1282. doi: 10.1046/j.1471-4159.2003.02253.x. [DOI] [PubMed] [Google Scholar]
- Gledhill JR, Montgomery MG, Leslie AG, Walker JE. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc Natl Acad Sci USA. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Galeno JE, Dang Q, Nguyen TH, Boyer SH, Grote MP, Sun Z, Chen M, Craigo WA, van Poelje PD, MacKenna DA, et al. A potent and selective AMPK activator that inhibits de novo lipogenesis. ACS Med Chem Lett. 2010;1:478–482. doi: 10.1021/ml100143q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goransson O, McBride A, Hawley SA, Ross FA, Shpiro N, Foretz M, Viollet B, Hardie DG, Sakamoto K. Mechanism of action of A-769662, a valuable tool for activation of AMP-activated protein kinase. J Biol Chem. 2007;282:32549–32560. doi: 10.1074/jbc.M706536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 2013;18:556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMPK: a target for drugs and natural products with effects on both diabetes and cancer. Diabetes. 2013;62:2164–2172. doi: 10.2337/db13-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: maintaining energy homeostasis at the cellular and whole body levels. Ann Rev Nutrition. 2014;34 doi: 10.1146/annurev-nutr-071812-161148. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ashford ML. AMPK: regulating energy balance at the cellular and whole body levels. Physiology. 2014;29:99–107. doi: 10.1152/physiol.00050.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Rev Mol Cell Biol. 2012;13:251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRADa/b and MO25a/b are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. Characterization of the AMP-activated protein kinase kinase from rat liver, and identification of threonine-172 as the major site at which it phosphorylates and activates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Fullerton MD, Ross FA, Schertzer JD, Chevtzoff C, Walker KJ, Peggie MW, Zibrova D, Green KA, Mustard KJ, et al. The ancient drug salicylate directly activates AMP-activated protein kinase. Science. 2012;336:918–922. doi: 10.1126/science.1215327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Chevtzoff C, Green KA, Evans A, Fogarty S, Towler MC, Brown LJ, Ogunbayo OA, Evans AM, et al. Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 2010;11:554–565. doi: 10.1016/j.cmet.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley SA, Ross FA, Gowans GJ, Tibarewal P, Leslie NR, Hardie DG. Phosphorylation by Akt within the ST loop of AMPK-α1 down-regulates its activation in tumour cells. Biochem J. 2014;459:275–287. doi: 10.1042/BJ20131344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman S, Vertommen D, Heath R, Neumann D, Mouton V, Woods A, Schlattner U, Wallimann T, Carling D, Hue L, et al. Insulin antagonizes ischemia-induced Thr172 phosphorylation of AMP-activated protein kinase alpha-subunits in heart via hierarchical phosphorylation of Ser485/491. J Biol Chem. 2006;281:5335–5340. doi: 10.1074/jbc.M506850200. [DOI] [PubMed] [Google Scholar]
- Houde VP, Ritorto MS, Gourlay R, Varghese J, Davies P, Shpiro N, Sakamoto K, Alessi DR. Investigation of LKB1 Ser431 phosphorylation and Cys433 farnesylation using mouse knockin analysis reveals an unexpected role of prenylation in regulating AMPK activity. Biochem J. 2014;458:41–56. doi: 10.1042/BJ20131324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ER, Pan DA, James J, Lucocq JM, Hawley SA, Green KA, Baba O, Terashima T, Hardie DG. A novel domain in AMP-activated protein kinase causes glycogen storage bodies similar to those seen in hereditary cardiac arrhythmias. Current Biol. 2003;13:861–866. doi: 10.1016/s0960-9822(03)00249-5. [DOI] [PubMed] [Google Scholar]
- Hunter RW, Foretz M, Bultot L, Fullerton MD, Deak M, Ross FA, Hawley SA, Shpiro N, Viollet B, Barron D, et al. Mechanism of action of Compound-13: an α1-selective small molecule activator of AMPK. Chem Biol. 2014 doi: 10.1016/j.chembiol.2014.05.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmoldulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem. 2005;280:29060–29066. doi: 10.1074/jbc.M503824200. [DOI] [PubMed] [Google Scholar]
- Hurley RL, Barre LK, Wood SD, Anderson KA, Kemp BE, Means AR, Witters LA. Regulation of AMP-activated protein kinase by multisite phosphorylation in response to agents that elevate cellular cAMP. J Biol Chem. 2006;281:36662–36672. doi: 10.1074/jbc.M606676200. [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- Jorgensen SB, Nielsen JN, Birk JB, Olsen GS, Viollet B, Andreelli F, Schjerling P, Vaulont S, Hardie DG, Hansen BF, et al. The a2-5'AMP-activated protein kinase is a site 2 glycogen synthase kinase in skeletal muscle and is responsive to glucose loading. Diabetes. 2004;53:3074–3081. doi: 10.2337/diabetes.53.12.3074. [DOI] [PubMed] [Google Scholar]
- Kazgan N, Williams T, Forsberg LJ, Brenman JE. Identification of a nuclear export signal in the catalytic subunit of AMP-activated protein kinase. Mol Biol Cell. 2010;21:3433–3442. doi: 10.1091/mbc.E10-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp BE, Oakhill JS, Scott JW. AMPK structure and regulation from three angles. Structure. 2007;15:1161–1163. doi: 10.1016/j.str.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Lizcano JM, Göransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Mäkelä TP, Hardie DG, et al. LKB1 is a master kinase that activates 13 protein kinases of the AMPK subfamily, including the MARK/PAR-1 kinases. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Reiner DS, Lauwaet T, Dacre M, Smith A, Zhai Y, Svard S, Gillin FD. The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol. 2011;12:R66. doi: 10.1186/gb-2011-12-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A, Ghilagaber S, Nikolaev A, Hardie DG. The glycogen-binding domain on AMP-activated protein kinase is a regulatory domain that allows the kinase to act as a sensor of glycogen structure. Cell Metab. 2009;9:23–34. doi: 10.1016/j.cmet.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A, Hardie DG. AMP-activated protein kinase--a sensor of glycogen as well as AMP and ATP? Acta Physiol (Oxf) 2009;196:99–113. doi: 10.1111/j.1748-1716.2009.01975.x. [DOI] [PubMed] [Google Scholar]
- McManus EJ, Sakamoto K, Armit LJ, Ronaldson L, Shpiro N, Marquez R, Alessi DR. Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. EMBO J. 2005;24:1571–1583. doi: 10.1038/sj.emboj.7600633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Saavedra D, Stark MJ, Packer JC, Vivares CP, Doerig C, Barton GJ. The complement of protein kinases of the microsporidium Encephalitozoon cuniculi in relation to those of Saccharomyces cerevisiae and Schizosaccharomyces pombe. BMC Genomics. 2007;8:309. doi: 10.1186/1471-2164-8-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- Oakhill JS, Chen ZP, Scott JW, Steel R, Castelli LA, Ling N, Macaulay SL, Kemp BE. beta-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK) Proc Natl Acad Sci USA. 2010;107:19237–19241. doi: 10.1073/pnas.1009705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332:1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- Pang T, Xiong B, Li JY, Qiu BY, Jin GZ, Shen JK, Li J. Conserved alpha-helix acts as autoinhibitory sequence in AMP-activated protein kinase alpha subunits. J Biol Chem. 2007;282:495–506. doi: 10.1074/jbc.M605790200. [DOI] [PubMed] [Google Scholar]
- Pearce LR, Atanassova N, Banton MC, Bottomley B, van der Klaauw AA, Revelli JP, Hendricks A, Keogh JM, Henning E, Doree D, et al. KSR2 mutations are associated with obesity, insulin resistance, and impaired cellular fuel oxidation. Cell. 2013;155:765–777. doi: 10.1016/j.cell.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polekhina G, Gupta A, Michell BJ, van Denderen B, Murthy S, Feil SC, Jennings IG, Campbell DJ, Witters LA, Parker MW, et al. AMPK b-Subunit targets metabolic stress-sensing to glycogen. Current Biol. 2003;13:867–871. doi: 10.1016/s0960-9822(03)00292-6. [DOI] [PubMed] [Google Scholar]
- Polekhina G, Gupta A, van Denderen BJ, Feil SC, Kemp BE, Stapleton D, Parker MW. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure (Camb) 2005;13:1453–1462. doi: 10.1016/j.str.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Pulinilkunnil T, He H, Kong D, Asakura K, Peroni OD, Lee A, Kahn BB. Adrenergic regulation of AMP-activated protein kinase in brown adipose tissue in vivo. J Biol Chem. 2011;286:8798–8809. doi: 10.1074/jbc.M111.218719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Goransson O, Hardie DG, Alessi DR. Activity of LKB1 and AMPK-related kinases in skeletal muscle: effects of contraction, phenformin, and AICAR. Am J Physiol Endocrinol Metab. 2004;287:E310–E317. doi: 10.1152/ajpendo.00074.2004. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, McCarthy A, Smith D, Green KA, Hardie DG, Ashworth A, Alessi DR. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 2005;24:1810–1820. doi: 10.1038/sj.emboj.7600667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MJ, Ali ZS, Hegarty BD, Heath R, Snowden MA, Carling D. Defining the mechanism of activation of AMP-activated protein kinase by the small molecule A-769662, a member of the thienopyridone family. J Biol Chem. 2007;282:32539–32548. doi: 10.1074/jbc.M706543200. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Mammucari C, Sandri M. The role of autophagy in neonatal tissues: just a response to amino acid starvation? Autophagy. 2008;4:727–730. doi: 10.4161/auto.6143. [DOI] [PubMed] [Google Scholar]
- Scott JW, Hawley SA, Green KA, Anis M, Stewart G, Scullion GA, Norman DG, Hardie DG. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Ling N, Issa SM, Dite TA, O′Brien MT, Chen ZP, Galic S, Langendorf CG, Steinberg GR, Kemp BE, et al. Small molecule drug A-769662 and AMP synergistically activate naive AMPK independent of upstream kinase signaling. Chem Biol. 2014 doi: 10.1016/j.chembiol.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol. 2002;317:309–323. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- Scott JW, van Denderen BJ, Jorgensen SB, Honeyman JE, Steinberg GR, Oakhill JS, Iseli TJ, Koay A, Gooley PR, Stapleton D, et al. Thienopyridone drugs are selective activators of AMP-activated protein kinase beta1-containing complexes. Chem Biol. 2008;15:1220–1230. doi: 10.1016/j.chembiol.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, Wu CL, Vera DR, Shaw RJ. mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci USA. 2009;106:11137–11142. doi: 10.1073/pnas.0900465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CH, Yuan P, Perez-Lorenzo R, Zhang Y, Lee SX, Ou Y, Asara JM, Cantley LC, Zheng B. Phosphorylation of BRAF by AMPK impairs BRAF-KSR1 association and cell proliferation. Mol Cell. 2013;52:161–172. doi: 10.1016/j.molcel.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenkle AB, Davies SP, Carling D, Hardie DG, Sturgill TW. Identification of Raf-1 Ser621 kinase activity from NIH 3T3 cells as AMP-activated protein kinase. FEBS Lett. 1997;403:254–258. doi: 10.1016/s0014-5793(97)00062-8. [DOI] [PubMed] [Google Scholar]
- Steinberg GR, Dandapani M, Hardie DG. AMPK: mediating the metabolic effects of salicylate-based drugs? Trends Endocrinol Metab. 2013;24:481–487. doi: 10.1016/j.tem.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Bridges D, Nakada D, Skiniotis G, Morrison SJ, Lin JD, Saltiel AR, Inoki K. Inhibition of AMPK catabolic action by GSK3. Mol Cell. 2013;50:407–419. doi: 10.1016/j.molcel.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Pause A. Birt-Hogg-Dube: tumour suppressor function and signalling dynamics central to folliculin. Fam Cancer. 2013;12:367–372. doi: 10.1007/s10689-012-9576-9. [DOI] [PubMed] [Google Scholar]
- Towler MC, Fogarty S, Hawley SA, Pan DA, Martin D, Morrice NA, McCarthy A, Galardo MN, Meroni SB, Cigorraga SB, et al. A novel short splice variant of the tumour suppressor LKB1 is required for spermiogenesis. Biochem J. 2008;416:1–14. doi: 10.1042/BJ20081447. [DOI] [PubMed] [Google Scholar]
- Townley R, Shapiro L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science. 2007;315:1726–1729. doi: 10.1126/science.1137503. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, Kunji ER, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udell CM, Rajakulendran T, Sicheri F, Therrien M. Mechanistic principles of RAF kinase signaling. Cell Mol Life Sci. 2011;68:553–565. doi: 10.1007/s00018-010-0520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kobayashi T, Piao X, Shiono M, Takagi Y, Mineki R, Taka H, Zhang D, Abe M, Sun G, et al. Serine 62 is a phosphorylation site in folliculin, the Birt-Hogg-Dube gene product. FEBS Lett. 2010;584:39–43. doi: 10.1016/j.febslet.2009.11.033. [DOI] [PubMed] [Google Scholar]
- Wilson WA, Roach PJ, Montero M, Baroja-Fernandez E, Munoz FJ, Eydallin G, Viale AM, Pozueta-Romero J. Regulation of glycogen metabolism in yeast and bacteria. FEMS Microbiol Rev. 2010;34:952–985. doi: 10.1111/j.1574-6976.2010.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WA, Wang Z, Roach PJ. Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol Cell Proteomics. 2002;1:232–242. doi: 10.1074/mcp.m100024-mcp200. [DOI] [PubMed] [Google Scholar]
- Woods A, Dickerson K, Heath R, Hong SP, Momcilovic M, Johnstone SR, Carlson M, Carling D. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, et al. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Carmena D, Bright NJ, Haire LF, Underwood E, Patel BR, Heath RB, Walker PA, Hallen S, et al. Structural basis of AMPK regulation by small molecule activators. Nature Commun. 2013;4:3017. doi: 10.1038/ncomms4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin FJ, Wang J, Zhao RQ, Wang ZX, Wu JW. Coordinated regulation of AMPK activity by multiple elements in the alpha-subunit. Cell Res. 2013;23:1237–1240. doi: 10.1038/cr.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Gingras MC, Dunlop EA, Nouet Y, Dupuy F, Jalali Z, Possik E, Coull BJ, Kharitidi D, Dydensborg AB, et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J Clin Invest. 2014 doi: 10.1172/JCI71749. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Ito K, Perez-Lorenzo R, Del Guzzo C, Lee JH, Shen CH, Bosenberg MW, McMahon M, Cantley LC, Zheng B. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc Natl Acad Sci U S A. 2013;110:18226–18231. doi: 10.1073/pnas.1317577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadra G, Photopoulos C, Tyekucheva S, Heidari P, Weng QP, Fedele G, Liu H, Scaglia N, Priolo C, Sicinska E, et al. A novel direct activator of AMPK inhibits prostate cancer growth by blocking lipogenesis. EMBO Mol Med. 2014 doi: 10.1002/emmm.201302734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL, 3rd, Lee JJ, Tilghman SM, Gumbiner BM, Costantini F. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1 that constitutes a switch between catabolism and anabolism. Cell Metab. 2014 doi: 10.1016/j.cmet.2014.06.014. in press. [DOI] [PubMed] [Google Scholar]
- Zhang YL, Guo H, Zhang CS, Lin SY, Yin Z, Peng Y, Luo H, Shi Y, Lian G, Zhang C, et al. AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metab. 2013;18:546–555. doi: 10.1016/j.cmet.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, Cantley LC. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell. 2009;33:237–247. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]