Abstract
Genotoxic stresses activate intracellular signaling molecules, which lead to growth arrest, DNA repair, and/or apoptosis. Among these molecules are the growth arrest and DNA damage protein 34 (GADD34) and the Src-related protein tyrosine kinase Lyn. Here, we report that these two proteins physically and functionally interact to regulate DNA damage-induced apoptosis. Multiple isolates of GADD34 and the related murine protein MyD116 were identified as binding partners of Lyn in a yeast two-hybrid screen. The specific interaction was confirmed by in vitro association of GADD34 with glutathione S-transferase fusion proteins containing the Src Homology 3 (SH3) domain of Lyn, as well as coimmunoprecipitation of GADD34 and Lyn from mammalian cells. GADD34 was tyrosine-phosphorylated in vivo in a Lyn-dependent manner. Lyn efficiently phosphorylated affinity-purified GADD34 in vitro. Lyn negatively regulated the proapoptotic function of GADD34 in a kinase-dependent manner. Expression of wild-type, but not kinase-inactive, Lyn weakened promotion of apoptosis by GADD34 following treatment with methyl-methanesulfonate or ionizing radiation in HEK293 and HeLa cells. In contrast, pretreatment of cells with the Src-specific tyrosine kinase inhibitor PP1 strengthened promotion of apoptosis by GADD34. We propose that Lyn regulates the proapoptotic function of GADD34 by binding and phosphorylating it.
Genotoxic agents such as alkylating drugs or ionizing radiation injure cells primarily by damaging their DNA. The two outcomes of genotoxic injury, survival or apoptosis, are determined by DNA damage response pathways. Several key mediators in these pathways, such as DNA-dependent protein kinase DNA-PK, phosphatidylinositol 3-kinase family member ATM, protein tyrosine kinase c-Abl, global transcriptional regulator of stress responses p53, and members of the Bcl-2 family have been studied in considerable detail (1). The roles of other DNA damage-related molecules are not as well understood. GADD34, a member of the protein family whose expression is up-regulated by Growth Arrest and DNA Damage (2) is one of such molecules. GADD34 transcript levels increase in response to medium depletion or DNA damage (3), heat shock (4), and several other treatments that induce apoptosis (5). Transcriptional induction of GADD34 strongly correlates with apoptosis (3, 5). Overexpression of GADD34 enhances the apoptotic response to DNA damage (6). The kinetics of GADD34 induction by DNA-damaging agents is similar to that of the proapoptotic protein Bax; however, unlike Bax, GADD34 is induced via a p53-independent pathway (3). GADD34 could therefore play a major role in the alternative, p53-independent mechanism of genotoxic apoptosis in drug-resistant cancers associated with the loss of p53. MyD116, the GADD34-related mouse gene, is transcriptionally activated during interleukin-6-induced terminal myeloid differentiation of the M1 myeloblasts, a process involving growth arrest and culminating in apoptosis (7). The C termini of GADD34/MyD116 proteins are highly homologous to the herpes simplex virus γ(1)34.5 protein whose role is to prevent apoptosis in host cells during viral infection. Moreover, the C terminus of MyD116 can functionally substitute for the corresponding domain of the γ(1)34.5 in the context of the viral genome (8). Association with proliferating cell nuclear antigen suggests that GADD34 might inhibit proliferation (9). HRX leukemic fusion oncogenes, but not wild-type HRX protein, the human homologue of Drosophila trithorax (trx) gene, bind GADD34 to negatively regulate the apoptotic response (6). These data suggest that GADD34 is a positive regulator of several physiologically distinct apoptotic processes whose activity could be modulated by interaction with other proteins. The mechanism of action and regulation of GADD34, however, remain unknown.
The Src-related protein tyrosine kinase Lyn has also been implicated in response to DNA damage. Lyn, but not other Src kinases, is activated after genotoxic treatments (10). This activation has been associated with cell cycle arrest (11) and apoptosis (12). Studies in Lyn-deficient cells demonstrate that Lyn is required for apoptosis induced by topoisomerase II inhibitors but not ionizing radiation (13). In addition to a role in DNA damage responses, Lyn plays a role in other processes such as glutamate receptor signaling in neurons (14), cytoskeletal rearrangement and motility in mast cells (15), cytokine responses in hematopoietic cells, and B lymphocyte receptor signaling. In hematopoietic cells, cytokine-mediated activation of Lyn stimulates proliferation (16–18) and inhibits apoptosis (19–22). In B lymphocytes, Lyn negatively regulates B cell receptor signaling and promotes growth arrest, but not apoptosis (23, 24). Thus, Lyn's effects on survival, proliferation, or apoptosis may depend on the cell type and the nature of the activating stimulus.
In this report, we demonstrate that GADD34 associates with Lyn and can be phosphorylated in a Lyn-dependent manner. Whereas separate expression of either GADD34 or Lyn stimulates genotoxic apoptosis, their joint expression has an inhibitory effect. We propose that Lyn acts as a negative regulator of GADD34 in DNA damage-induced cell death.
Materials and Methods
Yeast Two-Hybrid Screen.
The HybriZap yeast two-hybrid system (Stratagene) was used as directed by the manufacturer. We screened the cDNA library of granulocyte–macrophage colony-stimulating factor-treated 32Dcl3 custom-made by Stratagene, and the Matchmaker human bone marrow library (CLONTECH) with the pGBT9-Lyn, made by inserting codons 1–226 of human Lyn (25) into the EcoRI–BamHI sites of pGBT9 (CLONTECH). Following transformation of YRG-2 yeast with the bait and library DNA, positive clones were selected in the absence of leucine, tryptophan, and histidine. Library plasmids were isolated from positive clones and retransformed into YRG-2 together with either the pGBT9-Lyn or the negative control bait p53. cDNA inserts of library plasmids conferring histidine-independent growth with pGBT9-Lyn, but not with p53, were sequenced. Matches of the identified sequences to the GenBank database were found by using the BLAST search algorithm.
Cells, Plasmids, Transfection, and Immunoblots.
Daudi, HEK293, BaF-3, and HeLa cell lines were obtained from American Type Culture Collection. Chicken B cell line DT40 and its lyn−/− derivative D33 were obtained from Dr. T. Kurosaki (Kansai University, Japan). HEK293 and HeLa cells were transfected by using the calcium phosphate system or Lipofectin-Plus (GIBCO) and processed 40 h and 60 h posttransfection, respectively. DT40 and D33 cells were transfected by electroporation at 960 μF, 300 V in 4-mm gap cuvettes. To construct pFLAG-CMV2-GADD34, the BamHI–SalI fragment of the library isolate containing codons 30–672 of the human GADD34 (3) was inserted into BamHI–SmaI sites of pFLAG-CMV2 (Sigma). The resulting plasmid was cut with HindIII–XbaI, and the HindIII–XbaI cut product of Daudi mRNA reverse transcription (RT)–PCR amplification with primers CCCAAGCTTATGGCCCCAGGCCAAGCACC and CTGCTCCTTTTACTGCTTCCAC was inserted. pcDNA3-Lyn and its K275R mutant have been described (17). Glutathione S-transferase (GST)-Lyn bacterial expression constructs are derivatives of pGEX-2T (Amersham Pharmacia) carrying Lyn cDNA fragments (26). All DNA constructs were verified by sequencing. FLAG-tagged GADD34 and Lyn were detected by immunoblotting with the anti-FLAG monoclonal M2 (Sigma), anti-GADD34 polyclonal S-20 and anti-Lyn monoclonal H6 (Santa Cruz Biotechnology), or anti-Lyn polyclonal (Upstate Biotechnology, Lake Placid, NY) antibodies and tyrosine phosphorylation with the 4G10 antibody (Upstate Biotechnology). Following incubation with primary antibodies and with appropriate secondary horseradish peroxidase-conjugated antibodies, blots were developed by using ECL reagent and film imaging.
In Vitro Binding Assay.
GST and GST-Lyn fusion proteins were expressed in Escherichia coli and purified by adsorption to glutathione-agarose. HEK293 cells transiently expressing FLAG-GADD34 were lysed with 20 mM Tris, pH 7.6/150 mM NaCl/0.1% Nonidet P-40/1 mM phenylmethylsulfonyl fluoride/1 μg/ml leupeptin/1 μg/ml aprotinin. Lysates cleared by centrifugation for 10 min at 10,000 × g were incubated 2 h at 4°C with glutathione agarose beads containing 5 μg of either GST or GST-Lyn fusion proteins. Beads were collected and washed with the lysis buffer, and bound proteins were analyzed by Western blotting with the anti-FLAG antibody.
Immunoprecipitation.
Immunoprecipitations were carried out for 2 h at 4°C with antibodies added to 1 μg/ml followed by 1 h with protein A Sepharose beads. To examine GADD34 phosphorylation in vivo, cells were lysed with RIPA buffer (150 mM NaCl/50 mM Tris, pH 8.0/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS) in the presence of protease inhibitors and the tyrosine phosphatase inhibitors pervanadate (200 μM) and phenylarsine oxide (1 mM). Following adjustment of NaCl concentration to 300 mM to reduce nonspecific binding, the lysates were incubated for 2 h at 4°C with anti-FLAG agarose beads (Sigma). The beads were washed with RIPA buffer, and bound proteins were analyzed by Western blotting.
Affinity Purification of FLAG-GADD34.
FLAG-GADD34 expressed in HEK293 cells was adsorbed on anti-FLAG agarose beads, washed with 20 mM Tris, pH 7.6/150 mM NaCl/1 mM EDTA/10% glycerol, and eluted with the wash buffer containing 100 μg/ml FLAG peptide (Sigma). Fractions containing FLAG-GADD34 were combined and concentrated to 50 μg/ml by dialysis against wash buffer containing 50% (wt/wt) polyethylene glycol 8000. Aliquots of the purified protein were stored at −70°C.
Immune Complex Kinase Assay.
Anti-Lyn immunoprecipitates were incubated for 20 min at 24°C with exogenous substrates in protein kinase buffer (50 mM Hepes, pH 7.4/10 mM MgCl2/10 mM MnCl2/50 μM ATP/250 μCi/ml [γ-32P]ATP; 4,500 Ci/mmol). Before addition to reactions, FLAG-GADD34 was passed through Sephadex G-50 spin column equilibrated with protein kinase buffer without ATP. Reaction products were analyzed by SDS/PAGE and autoradiography.
Apoptosis Assays.
For internucleosomal DNA fragmentation assay, cells were harvested 12 h following treatment with 100 μg/ml methyl-methanesulfonate (MMS) in growth medium or irradiation at 20 Gy on Gammacell 1000 Elite (Nordion, Kanata, Ontario, Canada). Cells were lysed 10 min on ice with 20 mM Tris, pH 8.0/10 mM EDTA/100 mM NaCl/0.5% Triton X-100. Lysates were incubated 3–12 h at 37°C with 50 μg/ml proteinase K and 1% SDS, extracted with phenol-chloroform, and DNA was precipitated with ethanol. Following incubation with 20 μg/ml RNaseA for 30 min, samples were run on 2% agarose-ethidium bromide gels, and gels were photographed under UV illumination. For nuclear condensation assays, cell samples were collected 2 h posttreatment and stained with 0.1 μg/ml Hoechst 33342; cells with compact fluorescent nuclei were counted on an Axiovert fluorescent microscope (Zeiss). For sub-G1 population analysis, cells were collected 12 h posttreatment, fixed with 70% ethanol, and stained with 50 μg/ml propidium iodide/20 μg/ml RNaseA in PBS. Cells were analyzed by flow cytometry on the FACSCalibur cell analyzer (Becton Dickinson).
Results
Association of GADD34 with Lyn.
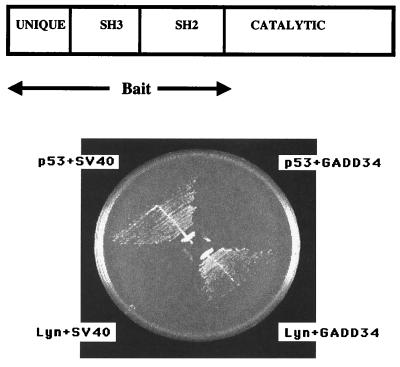
To investigate Lyn's role in apoptosis and proliferation, we sought to identify Lyn-binding proteins as potential signaling partners by using the yeast two-hybrid technique. As a typical Src kinase, Lyn has a C-terminal catalytic domain and the N-terminal regulatory domain consisting of unique, Src homology 3 (SH3) and Src homology 2 (SH2) domains (Fig. 1Upper). SH3 and SH2 domains bind proline-rich and phosphotyrosine-containing sequences, respectively, thus facilitating association with other proteins. We used the unique SH3–SH2 region of human Lyn as bait to screen two yeast two-hybrid libraries, our custom-made mouse myeloid 32Dcl3 library, and the human bone marrow library. About 200 primary isolates were tested for specific interaction as recommended by manufacturers. Briefly, a library plasmid isolate was considered as encoding a protein specifically interacting with Lyn if after retransformation in yeast it showed interaction with Lyn, but did not show interaction with the unrelated p53 bait. Specifically interacting clones included known Lyn binding partners such as ribonucleoprotein hnRNP K and adaptor protein c-Cbl. There were three isolates of MyD116 cDNA from the 32Dcl3 library and one isolate of GADD34 cDNA from the human bone marrow library. Specific interaction between Lyn and GADD34 is shown in Fig. 1Lower. Comparison of sequences at 5′ breakpoints of MyD116/GADD34 clones with database sequences demonstrated that all four clones were derived from independent, near full-size cDNAs (data not shown).
Figure 1.
Yeast two-hybrid interaction between GADD34 and Lyn. (Upper) Block diagram of Lyn's domains. Unique, SH3, SH2, and catalytic domains as well as the portion of Lyn used as bait are shown. (Lower) YRG-2 yeast transformants harboring pGBT9-Lyn, the human bone marrow library GADD34 plasmid, and control plasmids p53 and pSV40 as indicated were streaked on a plate lacking leucine, tryptophan, and histidine. The plate was incubated at 30°C for 3 days. Histidine-independent growth indicates the two-hybrid interaction between plasmid-encoded proteins.
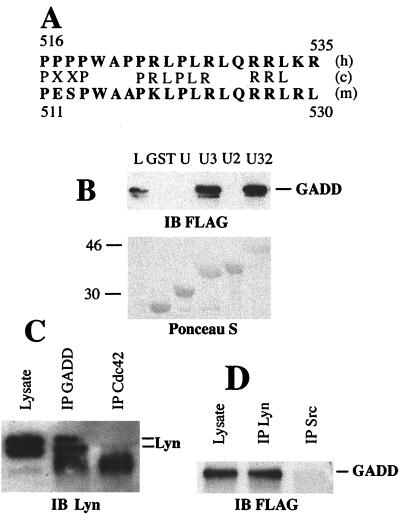
The most likely interaction in a yeast two-hybrid screen with noncatalytic Lyn bait is between the Lyn's SH3 domain and proline-rich clusters of library proteins. Indeed, most of the cDNAs isolated in our screens encode proteins with proline-rich clusters (data not shown). The analysis of amino acid sequences of human GADD34 and mouse MyD116 reveals a conserved proline-rich cluster (residues 516–535 and 511–530 of human GADD34 and mouse MyD116, respectively) upstream of the γ(1)34.5 homology region. This site has two PXXP motifs and is rich in leucine and arginine residues found in typical Lyn SH3 binding sites (refs. 27 and 28; Fig. 2A). The role of the SH3 domain of Lyn in binding GADD34 is underscored by the lack of interaction with the Lyn unique SH2 bait in the yeast two-hybrid assay (data not shown). GADD34 did not show yeast two-hybrid interaction with noncatalytic Src (data not shown).
Figure 2.
GADD34–Lyn interaction in vitro and in mammalian cells. (A) Alignment of human GADD34 (h) and mouse MyD116 (m) proline-rich clusters with Lyn SH3 binding site consensus sequence (c). Single-letter amino acid code is used. Residue numbers are shown. (B) GST pull-down assay with GST or GST fusions with unique (U), unique SH3 (U3), unique SH2 (U2), and unique SH3-SH2 (U32) domains of Lyn, and lysates of HEK293 cells transiently transfected with pFLAG-CMV2-GADD34. Proteins from lysate bound to GST fusion beads were analyzed by using immunoblot (IB). (Upper) Immunoblot probed with the anti-FLAG antibody. (Lower) Blot stained with Ponceau S to show equal load of GST fusions. L, total cell lysate lane shown to indicate position of FLAG-GADD34. Size markers on the left are in kilodaltons. The minor band migrating slightly faster than FLAG-GADD34 is a result of partial degradation. (C) Immunoblot of Daudi cell lysate, anti-GADD34 (IP GADD), and control (IP Cdc42) immunoprecipitates probed with the monoclonal anti-Lyn antibody H-6. The diffuse bands at 46 kDa in immunoprecipitate lanes are because of the presence of crossreacting Ig heavy chain. Position of Lyn doublet is indicated. (D) Immunoblot of the lysate of 293 cells transfected with pFlag-CMV2-GADD34, anti-Lyn, and anti-Src immunoprecipitates probed with the anti-GADD34 antibody. Position of GADD34 is indicated.
To confirm the interaction and examine the role of Lyn's SH3 domain, we performed in vitro binding assays with GST fusion proteins containing various domains of the regulatory region of Lyn. For this purpose, GST fusions with unique, unique SH2, unique SH3, and unique SH3–SH2 domains of Lyn were expressed in E. coli and purified by adsorption to glutathione agarose. Beads with adsorbed fusion proteins were incubated with lysates of HEK293 cells transiently expressing the FLAG epitope-tagged GADD34. Beads were recovered and binding of GADD34 examined by SDS/PAGE and Western blotting with the anti-FLAG antibody. GADD34 bound only to GST fusion proteins containing the SH3 domain of Lyn (Fig. 2B). Therefore, GADD34 associates with Lyn in vitro, and this association, like the yeast two-hybrid interaction, requires the SH3 domain of Lyn.
To examine whether GADD34 and Lyn associate in mammalian cells, we performed coimmunoprecipitation from Daudi or HEK293 lysates. The Daudi cell line was chosen because of its high levels of endogenous GADD34 and Lyn (data not shown). Daudi cells were lysed with nonionic detergent buffer, and resulting lysates were immunoprecipitated with either rabbit polyclonal anti-GADD34 antibodies or an irrelevant rabbit polyclonal antibody. Immune complexes collected on protein A Sepharose were analyzed by Western blotting with anti-Lyn monoclonal antibodies. Lyn was coimmunoprecipitated by anti-GADD34, but not control antibodies (Fig. 2C). In a reciprocal experiment with HEK293 cells transiently expressing FLAG-GADD34, this protein coimmunoprecipitated with Lyn but not Src (Fig. 2D). HEK 293 cells express comparable levels of endogenous Lyn and Src (data not shown). Thus, we have demonstrated specific association of GADD34 and Lyn in mammalian cells.
Lyn-Dependent Phosphorylation of GADD34.
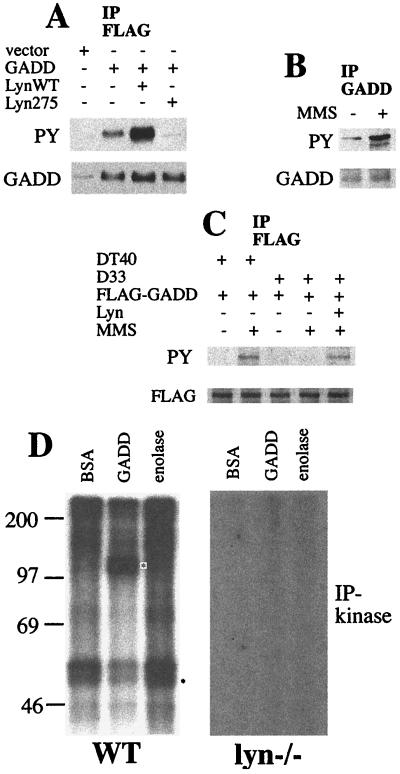
Association of GADD34 and Lyn may indicate that GADD34 might serve as a phosphorylation target of Lyn. To test whether Lyn affects phosphorylation of GADD34 in vivo, HEK293 cells were transiently transfected to express FLAG-GADD34 and either wild-type Lyn or the kinase-inactive K275R mutant of Lyn. Lysates of transfected cells were incubated with anti-FLAG antibodies immobilized on beads, and the resulting immunoprecipitates were subjected to Western blotting with either antiphosphotyrosine antibodies or with anti-GADD34 antibodies. Expression of wild-type, but not kinase-inactive, Lyn increased the level of tyrosine phosphorylation of GADD34 (Fig. 3A). Because Lyn is activated by DNA damage, we determined whether the tyrosine phosphorylation GADD34 is increased by genotoxic stress. In BaF-3 cells, tyrosine phosphorylation of endogenous GADD34 increased in cells treated with the DNA-damaging agent MMS (Fig. 3B). A similar increase was observed in DT40 cells transiently expressing FLAG-GADD34. Stimulus-induced phosphorylation of GADD34 was not detected in the lyn-deficient line D33. Ectopic expression of Lyn in D33 cells restored the MMS-induced phosphorylation (Fig. 3C). Thus, GADD34 is phosphorylated in vivo in a DNA damage- and Lyn-dependent manner. To examine the possibility of direct phosphorylation, we performed anti-Lyn immune complex-kinase assays with GADD34 as an exogenous substrate. FLAG-GADD34 protein expressed in HEK293 cells was purified by anti-FLAG affinity chromatography to near homogeneity (data not shown). Lysates of DT40 cells or lyn-deficient D33 cells were immunoprecipitated with anti-Lyn antibody. The resulting immunoprecipitates were incubated with [γ-32P]ATP and affinity-purified FLAG-GADD34, or acid-denatured enolase, a routinely used in vitro substrate of Src kinases, or a nonsubstrate BSA. Reaction products were analyzed by SDS/PAGE and autoradiography. Autophosphorylation of Lyn and phosphorylation of the exogenous substrates was observed in the immunoprecipitates from wild type but not from lyn-deficient cells, ruling out the contribution of protein kinases other than Lyn. GADD34, but not BSA, was as efficiently phosphorylated by Lyn as acid-denatured enolase, a known in vitro Src substrate (Fig. 3B). Thus, GADD34 is a substrate of Lyn.
Figure 3.
Lyn-mediated phosphorylation of GADD34. (A) Antiphosphotyrosine (PY) and anti-GADD34 (GADD) immunoblots of anti-FLAG immunoprecipitates of HEK293 cells transfected with empty vectors, pFLAG-CMV2-GADD34 (5 μg), and pcDNA3-Lyn constructs (5 μg) as indicated. Vector DNA was added to make 10 μg total whenever required. Equal amounts of GADD34 protein were recovered in the immunoprecipitates of GADD34-transfected cells. (B) Antiphosphotyrosine (PY) and anti-GADD34 (GADD) immunoblots of anti-GADD34 immunoprecipitates of BaF-3 cells untreated or treated with 50 μg/ml MMS for 1 h. (C) Antiphosphotyrosine (PY) and anti-FLAG (FLAG) immunoblots of anti-FLAG immunoprecipitates of DT40 and D33 cells transfected with pFLAG-CMV2-GADD34 (5 μg) and pcDNA3-Lyn (5 μg) and treated with 50 μg/ml MMS as indicated. (D) Anti-Lyn immune kinase assays with the exogenous substrates BSA, FLAG-GADD34 (GADD), and acid-denatured enolase. Lysates were prepared from DT40 wild-type or D33 Lyn-deficient cells and immunoprecipitated with the polyclonal anti-Lyn antibody. Equal amounts of protein from immunoprecipitates were used. Asterisks indicate positions of phosphorylated substrates. Lyn is a prominent doublet of 53 and 56 kDa. BSA is expected to appear as a 68-kDa band. Size markers on the left are in kilodaltons.
Lyn Modulates Promotion of Apoptosis by GADD34.
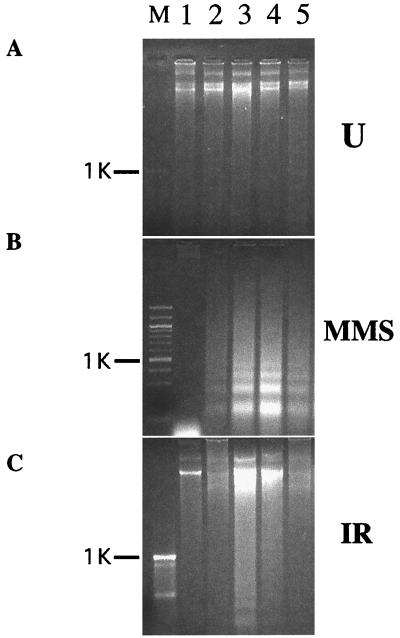
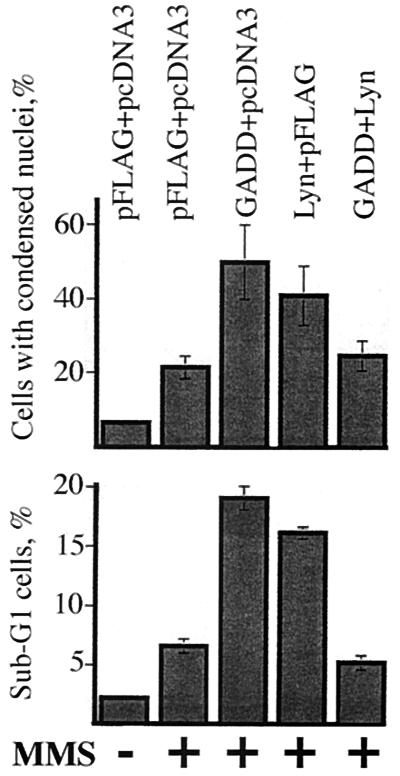
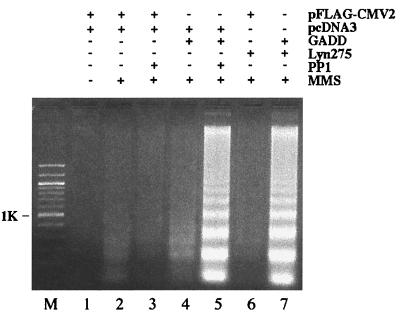
High levels of GADD34 have been reported to promote the DNA damage-induced apoptosis (5, 6). We took advantage of this observation to examine whether levels or activity of Lyn affect function of GADD34. To address the effects of high levels of Lyn on promotion of apoptosis by GADD34, we transiently expressed both proteins in HEK293 cells or HeLa cells and examined induction of apoptosis in the resulting transfectants by two DNA-damaging agents, MMS and ionizing radiation (IR). As judged by the absence of apoptosis-specific internucleosomal DNA fragmentation, expression of either GADD34 or Lyn, or both, did not appreciably induce apoptosis in the absence of DNA damage (Fig. 4A). As expected, expression of GADD34 augmented the apoptotic internucleosomal DNA fragmentation, compared with vector-transfected controls, in HEK293 (Fig. 4 B and C, lanes 2 and 3) and HeLa cells (data not shown) following treatment with MMS or IR. Expression of Lyn also augmented the genotoxic apoptosis (Fig. 4 B and C, lane 4). Coexpression of GADD34 and Lyn resulted in a significantly weaker response compared with cells expressing either of the two proteins alone (Fig. 4 B and C, lane 5, data for HeLa cells not shown). To corroborate these findings, we assayed genotoxic apoptosis in cells transiently expressing GADD34 or Lyn, or both, by using nuclear condensation and appearance of cells with sub-G1 DNA content as independent manifestations of the apoptotic process. These experiments confirmed results of the internucleosomal DNA fragmentation assay (Fig. 5). Thus, three different assays demonstrate that expression of either GADD34 or Lyn promotes, but their joint expression inhibits, genotoxic-induced apoptosis. This paradoxical result suggests that when GADD34 and Lyn are expressed together, they may negatively regulate each other. A possible mechanism for this effect could be steric obstruction of GADD34 and Lyn because of their association, rendering both proteins nonfunctional. Data in Fig. 6, however, argue against this mechanism because they demonstrate the importance of Lyn kinase activity rather than Lyn protein levels. First, expression of the kinase-deficient Lyn mutant failed to down-regulate promotion of MMS-induced apoptosis by GADD34; instead, it augmented the apoptotic response (Fig. 6, lane 7). Second, treatment with the Src family specific inhibitor PP1 markedly increased apoptosis in cells transfected with GADD34 (Fig. 6, lanes 4 and 5), but not with an empty vector (Fig. 6, lanes 2 and 3). Altogether, our data argue that low levels of Lyn activity strengthen, and high levels weaken, promotion of DNA damage-induced apoptosis by GADD34, thus implicating Lyn kinase as a negative regulator of GADD34.
Figure 4.
DNA fragmentation apoptosis assay following DNA damage in cells expressing GADD34 and Lyn. Agarose gel electropherograms of DNA from HEK293 cells transfected with 5 μg each of pcDNA3.1 and pFLAG-CMV2 (lanes 1 and 2), pFLAG-CMV2-GADD34 + pcDNA3.1 (lane 3), pFLAG-CMV2 + pcDNA3-Lyn (lane 4), pFLAG-CMV2-GADD34 + pcDNA3-Lyn (lane 5), and either left untreated (A) or treated with MMS (B) or IR (C). FLAG-GADD34 and Lyn expression levels were equal in double and single transfectants, as judged by Western blots (data not shown). DNA was extracted 12 h following treatment because at this time point, DNA laddering is most pronounced (data not shown). M, molecular size marker lane; position of 1-kb band is indicated. Two other repeats of the same experiment produced similar results (data not shown).
Figure 5.
Nuclear condensation and DNA content assays for genotoxic apoptosis in cells expressing GADD34 and Lyn. (Upper) HEK293 cells transfected with 5 μg each of pFLAG-CMV2, pcDNA3, pFLAG-CMV2-GADD34, and pcDNA3-Lyn as indicated were treated with MMS for 2 h and stained with Hoechst 33342. Cells with bright compact nuclei and total cells were counted by using epifluorescence and differential interference contrast illumination, respectively. (Lower) HeLa cells transfected as above were treated with MMS for 12 h, fixed, and stained with propidium iodide. Percentage of cells with sub-G1 DNA content was determined by flow cytometry. The data shown are average ± standard error of three independent experiments.
Figure 6.
Inhibition of Lyn augments apoptosis in cells expressing GADD34. HEK293 cells were transfected with 5 μg each of constructs indicated. Western blots showed equal levels of FLAG-GADD34 in transfectants (data not shown). Cells were left untreated or treated with MMS as indicated for 12 h. Where indicated,10 μM PP1 was added to cells 10 min before MMS. DNA was extracted and analyzed by electrophoresis through agarose gel. Results are representative of three independent experiments.
Discussion
We isolated GADD34/MyD116 as an interacting partner of Lyn by screening two different yeast two-hybrid libraries with the noncatalytic region of Lyn as bait. Multiple independent isolates that we describe provide strong evidence for GADD34–Lyn interaction in yeast. Other proteins isolated in our screens include known signaling partners of Lyn, such as the ribonucleoprotein hnRNP K and the adaptor protein c-Cbl, suggesting that GADD34–Lyn interaction in yeast may also reflect physiological interaction in mammalian cells. We demonstrated association between GADD34 and Lyn by three methods: yeast two-hybrid interaction, in vitro association in the GST pull-down assay, and in vivo association in mammalian cells in the coimmunoprecipitation assay. Results obtained in Daudi cells are of special value because they demonstrate association between endogenous GADD34 and Lyn under physiological conditions without overexpressing either of the proteins, suggesting physiologically relevant interaction.
Lyn's SH3 domain is essential for the interaction. GADD34–Lyn interaction in yeast as well as association in vitro in the GST pull-down assay were not observed in the absence of the SH3 domain of Lyn. As these assays do not measure the relative strength of association, we cannot exclude contribution of sequences outside the SH3 domain to efficient binding. The 516–535 proline-rich cluster of human GADD34 and the homologous 511–530 cluster of MyD116 are structurally the most likely sites responsible for the interaction with the SH3 domain of Lyn because they conform the consensus Lyn SH3 binding site (26, 27). Interestingly, a deletion overlapping the proline-rich cluster of GADD34 has been reported to abrogate binding of HRX leukemic fusion proteins (6), implicating this region of GADD34 as a site of multiple protein interactions.
GADD34 expressed in HEK293 cells is tyrosine-phosphorylated, and levels of this phosphorylation increase when cells also express wild-type, but not kinase-inactive, Lyn. GADD34 phosphorylation is increased by DNA damage, the condition known to activate Lyn. Moreover, MMS-induced phosphorylation of GADD34 does not occur in lyn-deficient D33 cells. These data implicate Lyn as the kinase that phosphorylates GADD34 in vivo, although they do not rule out phosphorylation by other Src kinases. Lyn efficiently phosphorylates exogenous FLAG-GADD34 in vitro. GADD34 acts as a specific substrate because other tyrosine-containing proteins, BSA and immunoglobulins, are not detectably phosphorylated by Lyn. Although in vivo experiments do not prove direct phosphorylation by Lyn and in vitro assays do not prove GADD34 being a physiological substrate of Lyn in vivo, the combination of in vivo and in vitro data strongly argues for direct and physiological phosphorylation of GADD34 by Lyn in living cells.
Expression of GADD34 in the colorectal cancer cell line SW480 has been reported to enhance the IR-induced apoptosis (6). We further extend this observation by demonstrating similar effect in two other cell lines, HEK293 and HeLa, and in cells treated with a different DNA-damaging agent, MMS. These results provide additional support to the idea that GADD34 protein is a DNA damage-inducible positive regulator of apoptosis. Here, we report that levels and/or activity of Lyn modulate promotion of genotoxic apoptosis by GADD34. Expression of wild-type Lyn inhibits the proapoptotic effect of GADD34, whereas lower levels of Lyn activity resulting from expression of the kinase-inactive Lyn or inhibition by PP1 augment the proapoptotic effect of GADD34. This, together with Lyn-dependent phosphorylation of GADD34 in vivo and in vitro, implicates phosphorylation by Lyn as a likely mechanism of negative regulation of GADD34.
Inhibition of apoptosis in cells coexpressing GADD34 and Lyn was unexpected because each of these proteins promotes apoptosis when expressed alone. We propose that activation of Lyn by DNA damage may serve, depending on the cell type and the dose of genotoxic treatment, to activate survival functions and promote recovery. Several observations support this idea. First, activation of Lyn occurs at lower doses of radiation and with faster kinetics than activation of GADD34 (3, 10), consistent with roles of Lyn and GADD34 in sustainable and lethal damage, respectively. Second, activation of Lyn by DNA-damaging agents may promote cell cycle arrest (11), which can be regarded as checkpoint activation, a pro-survival response. Third, receptor-mediated activation of Lyn has pro-proliferative and antiapoptotic effects in several cell types (16–22). Similar effects could occur following activation of Lyn by DNA damage. Promotion of apoptosis by Lyn following lethal DNA damage that we and others observed could be a consequence of inappropriate activation of survival functions. Inhibition of GADD34 by Lyn may therefore play a role in preventing apoptosis following sublethal DNA damage. Similar interaction has been reported between the proapoptotic protein kinase JNK and heat shock proteins of the HSP70 family. HSP70 proteins act as molecular chaperones to facilitate recovery from protein-denaturing stresses. Binding of HSP70 proteins to JNK prevents activation of the latter, thus inhibiting the apoptotic development following sublethal stresses (29–31). As in the case of JNK and HSP70, GADD34–Lyn interaction may play a role in determining the cell fate following stress-inflicted damage.
Acknowledgments
We thank Vera Roginskaya for excellent technical assistance and Drs. Dan Johnson and Baskaran Rajasekaran for critically reading the manuscript. This work was supported by the Hirtzel Foundation (to A.V.G.), a Bear Necessities Pediatric Cancer Foundation grant (to A.V.G.), an American Cancer Society grant (to S.J.C.), and by a National Institutes of Health R29 Grant (to S.J.C.). A.V.G. is Fellow of Cure for Lymphoma Foundation. S.J.C. is a National Institutes of Health Independent Scientist.
Abbreviations
- GST
glutathione S-transferase
- IR
ionizing radiation
- MMS
methyl-methanesulfonate
- GADD
growth arrest and DNA damage
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rich T, Allen R L, Wyllie A H. Nature (London) 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 2.Zhan Q, Lord K A, Alamo I, Jr, Hollander M C, Carrier F, Ron D, Kohn K W, Hoffman B, Liebermann D A, Fornace A J., Jr Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollander M C, Zhan Q, Bae I, Fornace A J., Jr J Biol Chem. 1997;272:13731–13737. doi: 10.1074/jbc.272.21.13731. [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa T, Xiao H, Hamajima F, Isobe K. Biochem J. 2000;352:795–800. [PMC free article] [PubMed] [Google Scholar]
- 5.Hollander M C, Saeed Sheikh M, Yu K, Zhan Q, Iglesias M, Woodworth C, Fornace A J., Jr Int J Cancer. 2001;96:22–31. doi: 10.1002/1097-0215(20010220)96:1<22::aid-ijc3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 6.Adler H T, Chinery R, Wu D Y, Kussick S J, Payne J M, Fornace A J, Jr, Tkachuk D C. Mol Cell Biol. 1999;19:7050–7060. doi: 10.1128/mcb.19.10.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lord K A, Hoffman-Liebermann B, Liebermann D A. Nucleic Acids Res. 1990;18:2823. doi: 10.1093/nar/18.9.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He B, Chou J, Liebermann D A, Hoffman B, Roizman B. J Virol. 1996;70:84–90. doi: 10.1128/jvi.70.1.84-90.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown S M, MacLean A R, McKie E A, Harland J. J Virol. 1997;71:9442–9449. doi: 10.1128/jvi.71.12.9442-9449.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kharbanda S, Yuan Z M, Rubin E, Weichselbaum R, Kufe D. J Biol Chem. 1994;269:20739–20743. [PubMed] [Google Scholar]
- 11.Yuan Z M, Huang Y, Kraeft S K, Chen L B, Kharbanda S, Kufe D. Oncogene. 1996;13:939–946. [PubMed] [Google Scholar]
- 12.Yoshida K, Weichselbaum R, Kharbanda S, Kufe D. Mol Cell Biol. 2000;20:5370–5380. doi: 10.1128/mcb.20.15.5370-5380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maruo A, Oishi I, Sada K, Nomi M, Kurosaki T, Minami Y, Yamamura H. Int Immunol. 1999;11:1371–1380. doi: 10.1093/intimm/11.9.1371. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, Umemori H, Mishina M, Yamamoto T. Nature (London) 1999;397:72–76. doi: 10.1038/16269. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Shoji S, Yamamoto K, Nada S, Okada M, Yamamoto T, Honda Z. J Immunol. 1998;161:3694–3701. [PubMed] [Google Scholar]
- 16.Linnekin D, DeBerry C S, Mou S. J Biol Chem. 1997;272:27450–27455. doi: 10.1074/jbc.272.43.27450. [DOI] [PubMed] [Google Scholar]
- 17.Corey S J, Dombrosky-Ferlan P M, Zuo S, Krohn E, Donnenberg A D, Zorich P, Romero G, Takata M, Kurosaki T. J Biol Chem. 1998;273:3230–3235. doi: 10.1074/jbc.273.6.3230. [DOI] [PubMed] [Google Scholar]
- 18.Dahl M E, Arai K I, Watanabe S. Genes Cells. 2000;5:143–153. doi: 10.1046/j.1365-2443.2000.00312.x. [DOI] [PubMed] [Google Scholar]
- 19.Yosefi S, Hoessli D C, Blaser K, Mills G B, Simon H U. J Exp Med. 1996;183:1407–1414. doi: 10.1084/jem.183.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katagiri K, Yokoyama K K, Yamamoto T, Omura S, Irie S, Katagiri T. J Biol Chem. 1996;271:11557–11562. doi: 10.1074/jbc.271.19.11557. [DOI] [PubMed] [Google Scholar]
- 21.Wei S, Liu J H, Epling-Burnette P K, Gamero A M, Ussery D, Pearson E W, Elkabani M E, Diaz J I, Djeu J Y. J Immunol. 1996;157:5155–5162. [PubMed] [Google Scholar]
- 22.Pazdrak K, Olszewska-Pazdrak B, Stafford S, Garofalo R P, Alam R. J Exp Med. 1998;188:421–429. doi: 10.1084/jem.188.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsueh R C, Scheuermann R H. Adv Immunol. 2000;75:283–316. doi: 10.1016/s0065-2776(00)75007-3. [DOI] [PubMed] [Google Scholar]
- 24.Chan V W, Meng F, Soriano P, DeFranco A L, Lowell C A. Immunity. 1997;7:69–81. doi: 10.1016/s1074-7613(00)80511-7. [DOI] [PubMed] [Google Scholar]
- 25.Corey S J, Shapiro D N. Leukemia. 1994;8:1914–1917. [PubMed] [Google Scholar]
- 26.Dombrosky-Ferlan P M, Corey S J. Oncogene. 1997;14:2019–2024. doi: 10.1038/sj.onc.1201031. [DOI] [PubMed] [Google Scholar]
- 27.Alexandropoulous K, Cheng G, Baltimore D. Proc Natl Acad Sci USA. 1995;92:3110–3114. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rickles R J, Botfield M C, Weng Z, Taylor J A, Green O M, Brugge J S, Zoller M J. EMBO J. 1994;13:5598–5604. doi: 10.1002/j.1460-2075.1994.tb06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosser D D, Caron A W, Bourget L, Denis-Larose C, Massie B. Mol Cell Biol. 1997;17:5317–5327. doi: 10.1128/mcb.17.9.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buzzard K A, Giaccia A J, Killender M, Anderson R L. J Biol Chem. 1998;273:17147–17153. doi: 10.1074/jbc.273.27.17147. [DOI] [PubMed] [Google Scholar]
- 31.Mosser D D, Caron A W, Bourget L, Merlin A B, Sherman M Y, Morimoto R I, Massie B. Mol Cell Biol. 2000;20:7146–7159. doi: 10.1128/mcb.20.19.7146-7159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]