Abstract
Background
Diagnosis and monitoring of localized prostate cancer requires discovery and validation of noninvasive biomarkers. Nuclear magnetic resonance (NMR)-based metabolomics of seminal plasma reportedly improves diagnostic accuracy, but requires validation in a high-risk clinical cohort.
Materials and methods
Seminal plasma samples of 151 men being investigated for prostate cancer were analyzed with 1H-NMR spectroscopy. After adjustment for buffer (add-to-subtract) and endogenous enzyme influence on metabolites, metabolite profiling was performed with multivariate statistical analysis (principal components analysis, partial least squares) and targeted quantitation.
Results
Seminal plasma metabolites best predicted low- and intermediate-risk prostate cancer with differences observed between these groups and benign samples. Lipids/lipoproteins dominated spectra of high grade samples with less metabolite contributions. Overall prostate cancer prediction using previously described metabolites was not validated.
Conclusion
Metabolomics of seminal plasma in vitro may assist urologists with diagnosis and monitoring of either low or intermediate grade prostate cancer. Less clinical benefit may be observed for high-risk patients. Further investigation in active surveillance cohorts, and/or in combination with in vivo magnetic resonance spectroscopic imaging may further optimize localized prostate cancer outcomes.
Keywords: Biomarker, Metabolomics, Nuclear Magnetic Resonance (NMR), Prostate Cancer, Seminal Fluid
1. Introduction
Accurate prostate cancer (CaP) diagnosis to prolong life with minimal morbidity is a daily challenge for urologists. Although early treatment of localized clinically significant CaP (csCaP) with curative intent reduces mortality and metastases,1 harms associated with overdetection and treatment of indolent CaP driven by injudicious use of serum prostate specific antigen (PSA) and prostate biopsy have reduced overall CaP detection.2 Limitations of serum PSA have driven advancements in multiparametric magnetic resonance imaging and biomarkers in serum (e.g., Prostate Health Index) and urine [prostate cancer antigen 3 (PCA3), TMPRSS2:ERG fusion gene].3, 4, 5, 6 However, due to cost-effectiveness concerns, these are used as adjunctive tests rather than as standalone detection tests despite their improved diagnostic accuracy.5, 6
Prostatic fluid, produced as seminal plasma (SP) after physiological prostatic smooth muscle contraction, contains the clinical biomarkers PSA and prostatic acid phosphatase (PAP).7, 8 Malignant prostatic cells in ejaculates of men with CaP have been shown to express genes (PCA3, Hepsin) and microRNAs that improve detection compared with serum PSA.9, 10, 11 Metabolomics is a modern biomarker approach that quantifies small metabolites, most commonly using nuclear magnetic resonance (NMR) spectroscopy or mass spectrometry.12, 13 NMR-based metabolomics is highly sensitive and reproducible with affordable sample-to-sample costs.12 SP metabolite profiles improve PSA-based diagnosis,14, 15 but require clinical validation.
This study investigates the feasibility of SP analysis using NMR-based metabolomics for the prediction of csCaP in a high-risk clinical cohort and compares metabolite profile CaP diagnosis against prostate biopsy and radical prostatectomy (RP) histology.
2. Materials and methods
Ethical approval was obtained from the University of Queensland Medical Research Ethics Committee (Project no. 2006000262) and the Royal Brisbane and Women's Hospital Human Research Ethics Committee (HREC/09/QRBW/320, HREC/09/QRBW/305 and 1995/088B).
2.1. Patients and clinical data
Male patients (n = 154) attending either the Royal Brisbane and Women's Hospital Urology outpatient department or local private consulting rooms for investigation of elevated PSA and/or abnormal digital rectal examination between January 2007 and February 2013 were enrolled in this prospective cohort study. Following informed consent, patients provided ejaculate specimens on site or at home prior to or at least 1 month after prostate biopsy, prior to commencement of any treatment. No specifications to time of day, relation to voiding, urethral meatus sterilization, or other parameters were provided to patients to simplify the sample collection process. Patients denied surgical treatment for benign prostatic hyperplasia and subsequent retrograde ejaculation prohibiting sample collection.
Patient data collected included age, serum PSA and detailed prostate biopsy, and radical prostatectomy histology records. Biopsy and RP specimens were reported by uropathologists according to the 2005 International Society of Urological Pathology classification.16 Patients were monitored for biopsy progression, such as CaP detection following initial false negative biopsy or upgraded Gleason score with further biopsy or RP (n = 60).
Risk stratification (low, intermediate, high risk) was performed according to the D’Amico criteria recommended in the American Urological Association Guidelines17 and used to determine csCaP presence (intermediate, high risk requiring treatment; Table 1). Given established disparity between biopsy and RP histopathology, risk classification accuracy was optimized using whichever histopathology best described tumor characteristics.
Table 1.
Demographic information for patients based on biopsy and radical prostatectomy (RP) histology.
| Age (y) | Serum PSA (ng/mL) | Pathological stage, n |
||||
|---|---|---|---|---|---|---|
| pT2 | pT3a | pT3b | ||||
| Biopsy | n = 151 | |||||
| Overall | 61 (55–66) | 6.5 (4.3–9.2) | ||||
| CaP status | Positive (n = 98) | 60.5 (55–65) | 6.4 (4.5–11) | |||
| Negative (n = 53) | 62 (55.75–68.25)NS | 6.5 (3.6–7.9)NS | ||||
| csCaP status | Present (n = 82) | 61 (55–66) | 6.75 (4.5–11.9) | |||
| Absent (n = 69) | 61 (55–67)NS | 6.0 (3.6–8.1)* | ||||
| RP | n = 60 | |||||
| Overall | 57 (54–64) | 6.15 (4.1–9.1) | ||||
| ISUP group | 1 (n = 2) | 56 (54–57) | 6.5 (4–9 ) | 2 | ||
| 2 (n = 30) | 57.5 (53–64) | 5.5 (4–7) | 26 | 3 | 1 | |
| 3 (n = 20) | 57 (55–61) | 7.3 (5–12) | 15 | 5 | ||
| 4 (n = 1) | 55 | 19 | 1 | |||
| 5 (n = 7) | 60 (55–68) | 10 (6–12) | 3 | 1 | 3 | |
| Primary/ tertiary pattern | ≥ 4 (n = 33) | 57 (55–64) | 7.3 (5–12) | 23 | 7 | 3 |
| 3 (n = 27) | 57 (51–63)NS | 5.4 (4–7)** | 25 | 2 | 1 | |
Median and interquartile range are shown for age and serum PSA. All comparisons were made using the Mann–Whitney U test (two-tailed).
* P < 0.05, ** P < 0.01.
CaP, prostate cancer; csCaP, clinically significant prostate cancer; ISUP, International Society of Urological Pathology; NS, not significant; PSA, prostate specific antigen.
2.2. Specimen processing
Ejaculate specimens were deposited directly into sterile microurine jars containing 20 mL Hanks Balanced Salt Solution (HBSS; Gibco, Life Technologies Australia, Mulgrave, Australia) for the first 117 patients used initially for cytology and RNA analyses, which was thereafter replaced by phosphate buffered saline (PBS) (in-house preparation) because glucose in HBSS interfered with preliminary metabolomics analysis. All specimens were provided to the laboratory without cooling as soon as logistically possible by the patients and were processed in the laboratory within 2 hours of production. Specimens were combined with 20 mL HBSS or PBS, layered over 10 mL isotonic Percoll (GE Healthcare-Pharmacia, Rydalmere, Australia) and centrifuged at 974g for 30–60 minutes at 4°C. Isolated supernatants, referred to as SP, were snap-frozen on dry ice in 1-mL aliquots and stored at –80°C.
2.3. Sample preparation
SP samples were thawed on wet ice and distributed in 100-μL aliquots. Eighty μL of PBS solution were added along with 20 μL D2O as lock substance that contained 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) as internal chemical shift standard and 1,1-difluoro-1-trimethylsilanyl methylphosphonic acid as internal pH indicator leading to final concentrations of 100μM, resulting in 200 μL total sample volume. Samples were transferred to sterile 3-mm NMR tubes (Bruker Biospin, Rheinstetten, Germany).
2.4. NMR spectroscopy
NMR spectra of SP samples were measured on a Bruker Avance 900 spectrometer operating at a 1H frequency of 900.13 MHz (Bruker Biospin), equipped with a 5-mm self-shielded z-gradient triple resonance cryoprobe and SampleJet sample changer. One-dimensional (1D) nuclear Overhauser effect spectroscopy (NOESY) spectra were acquired at 298 K with the “noesypr1d” pulse sequence, accumulating 200 transients (following 8 dummy scans) at 32,738 data points with a spectral width of 14 ppm.18 The transmitter frequency was set to the water resonance, which was suppressed by continuous wave irradiation during the NOESY mixing time of 0.1 seconds and relaxation delay of 3.0 seconds. Tuning/matching, shimming, and data acquisition were performed automatically with the IconNMR interface for high-throughput automation. Samples were measured in one batch per sample collection buffer and ordered randomly within these batches.
2.5. Spectral processing
NMR spectra were processed in TopSpin 3.2 (Bruker Biospin). The free induction decays were baseline corrected by a Gaussian function (0.1 ppm filter width) for postacquisition water deconvolution,19 followed by multiplication with an exponential window function (0.1 Hz line broadening), and Fourier transformation to 65,536 points. Subsequently, the spectra were manually phased, manually baseline corrected with a cubic spline curve, and referenced to DSS at 0.0 ppm. For all further data manipulation, the spectra were truncated to δ = 10.0–0.25 ppm, exported into MATLAB 2015b (The Mathworks Inc., Natick, Massachusetts, USA), and scaled according to the Bruker NC_proc parameter.
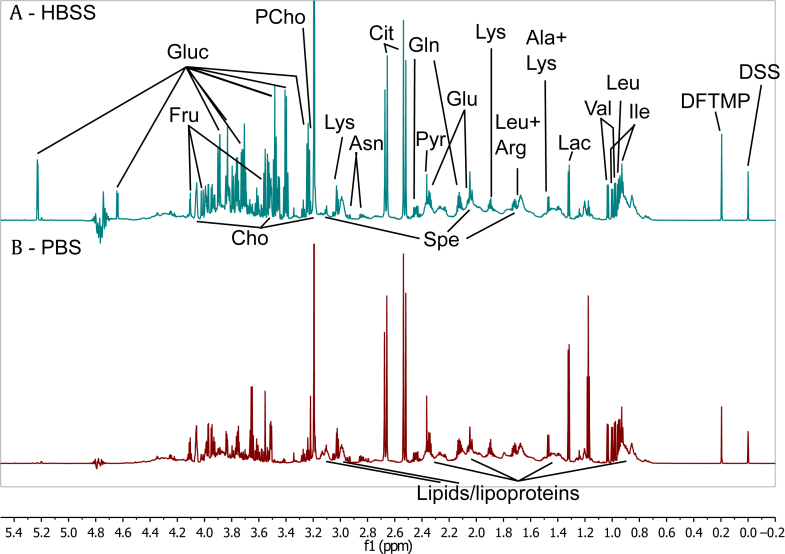
2.6. “Add-to-subtract” glucose exclusion
Preliminary analysis revealed glucose at sometimes dominant levels in most samples (Fig. 1). As HBSS contains 1 g/L D-glucose and ejaculate volumes were varied, the exogenous glucose concentration and its influence on subsequent multivariate statistical analysis (MVSA) was unpredictable. Thus, we used the “add-to-subtract” method20 to exclude glucose signals from the NMR spectra: (1) we added 1 μL of 1M D-glucose in PBS to each sample and repeated NMR measurement with identical experimental parameters, leading to a total of 302 spectra for 151 patients (151 original, 151 with additional glucose); (2) using Topspin’s multiple display, we determined the corresponding scaling factor between Spectrum 2 and Spectrum 1 for each sample that ensures elimination of the glucose signal upon subtraction; (3) then the exported Spectra 1 and 2 for each sample were aligned using “icoshift”21 on the glucose peaks at 3.37–3.44 ppm and then along 10 equal segments; and (4) for each sample Spectrum 2 was scaled with the scaling factor recorded in Topspin and subtracted from Spectrum 1. The resulting difference spectra were stored in a separate matrix.
Fig. 1.
Sections of one-dimensional (1D) nuclear Overhauser effect spectroscopy (NOESY) spectra from seminal plasma (SP) measured at 900 MHz. (A) 1D NOESY spectrum of SP collected in HBSS, with additional peaks due to exogenous glucose present. (B) 1D NOESY spectrum of SP collected in PBS. Ala, alanine; Arg, arginine; Asn, asparagine; bPBS, Panel B – PBS (panel labelling); Cho, choline; Cit, citrate; DFTMP, 1,1-difluoro-1-trimethylsilanyl methylphosphonic acid; DSS, 4,4-dimethyl-4-silapentane-1-sulfonic acid; f1, Chemical shift; Fru, fructose; Gln, glutamine; Gluc, glucose; Glu, glutamate; Ile, isoleucine; Lac, lactate; Leu, leucine; Lys, lysine; PCho, phosphocholine; Pyr, pyruvate; SP, seminal plasma; Spe, spermine; Val, valine.
2.7. Spectral alignment and data reduction
The peaks of all difference spectra were aligned at full resolution using “icoshift”, initially on the lactate doublet at 1.32 ppm and subsequently on manually defined segments. No shifting artefacts were identified. Using an in-house MATLAB script, the aligned difference spectra were data reduced to buckets of 0.01 ppm width over the range 10.0–5.08 ppm and 4.52–0.25 ppm, excluding the water signal region.
2.8. Multivariate statistical analysis
Metabolite data (X) matrices containing original and difference (add-to-subtract) data were quantile normalized with the “affy” package22 in R version 3.2.223 and imported into SIMCA P+ 12.0 (Umetrics, Umeå, Sweden) for MVSA together with clinical data variables (Y-matrix).
X-matrices were Pareto-scaled before unsupervised principal components analysis (PCA).24 To determine which metabolite signatures were associated with clinical data (cancer/risk status; Y-matrix), supervised partial least squares (PLS) was performed.24 Multivariate model quality was judged by the R2 (“goodness of fit”) and Q2 (“goodness of prediction”) figures of merit (Table S1). PLS models were validated by 200-fold response permutation.
Traditional statistical analysis, including the nonparametric Mann–Whitney U test, logistic regression, and receiver operating characteristic analysis were performed in MedCalc 12.7 for Windows (MedCalc Software, Ostend, Belgium).
2.9. Targeted metabolite profiling
SP metabolites were quantified using NMR suite 8.1 (Chenomx Inc., Edmonton, Canada)25 using DSS as internal concentration standard. Logistic regression on the SP metabolite concentrations was performed in MedCalc, similar to that previously described.15
3. Results
3.1. Clinical cohort demographics
From 151 patients who provided SP samples, 80 were initially diagnosed with CaP and an additional 18 patients diagnosed during the follow-up period. Within these 98 patients, 82 met csCaP criteria. Sixty patients underwent RP for localized CaP in which 59 were determined to be high risk per the D’Amico criteria, with six upgraded from low risk. The Gleason grade subgroups according to International Society of Urological Pathology (ISUP) category with corresponding stage based on RP histology are presented in Table 1. Primary Gleason Pattern 4 or higher or tertiary Pattern 5 was present in 34 patients based on RP histology. The demographic information for each group (Table 1) demonstrated that serum PSA was higher in those with high D’Amico risk or who were ineligible for active surveillance. The remaining 38 patients received radiation based therapy (n = 17), androgen deprivation therapy for metastatic disease (n = 2) or embarking on conservative management (active surveillance, watchful waiting; n = 12), although seven were lost to follow up.
3.2. Unsupervised multivariate statistical analysis
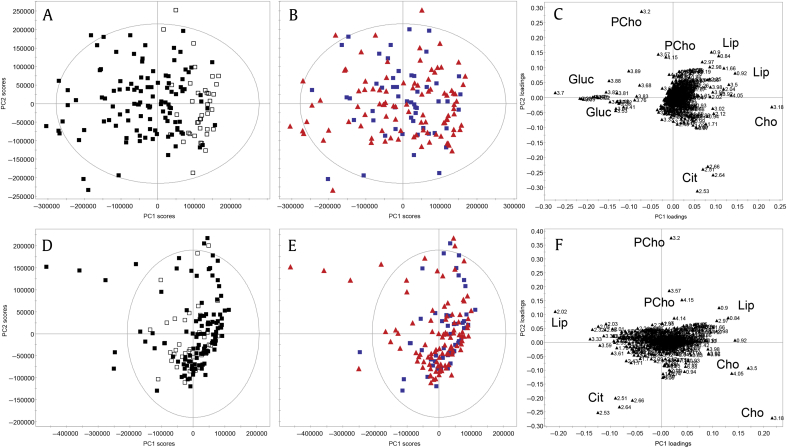
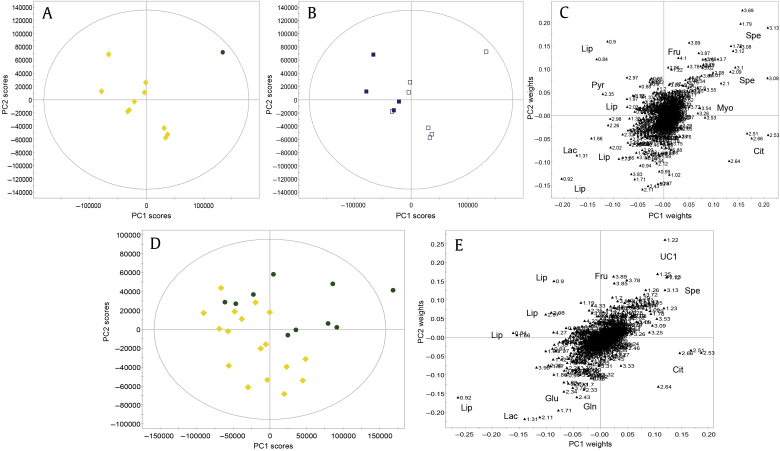
The SP samples were analyzed with 1H NMR spectroscopy. One-dimensional NOESY spectra were measured, aligned, and data reduced to 0.01 buckets. For initial PCA, buckets corresponding to ethanol, resulting from sample preparation, were excluded, as were spectra that were outliers due to broad resonances (n = 2). PCA yielded a model (Table S1 M1) with six principal components (PCs), in which samples clustered per the buffer solution used (PC1/PC2; Figs. 2A–2C), with higher glucose levels in samples prepared in HBSS. In higher PCs, sample variation was observed due to intersample differences of lipids/lipoproteins, phosphocholine, choline, and citrate, as well as spermine (data not shown), which were unrelated to CaP in this analysis.
Fig. 2.
Principal components analysis of seminal plasma NMR spectra from men being investigated for prostate cancer (n = 151), prepared with different buffer solutions (Hanks Balanced Salt Solution: filled square, phosphate buffered saline: empty square). (A–C) Initial sample clustering is observed due to the difference in buffer solutions and resulting sample glucose content (principal component 1) and intersample differences in metabolite (citrate, choline, lipids/lipoproteins and phosphocholine) variation (principal component 2). (D–F) After add-to-subtract elimination of glucose, the previously observed effects of different buffer solutions are no longer apparent (D). No clustering was present according to CaP status (blue squares = benign; red triangles = CaP). (A, B, D, E) Scores plots. (C, F) Loadings plots. CaP, prostate cancer; Cho, choline; Cit, citrate; Gluc, glucose; Lip, lipids/lipoproteins ; NMR, nuclear magnetic resonance; PCho, phosphorylcholine; PC, principal component.
The “add-to-subtract” method20 was used to remove glucose signals from NMR spectra. Following measurement of a “baseline spectrum” (Spectrum 1), glucose was added in high concentration to the sample in the same NMR tube and a second spectrum was measured (Spectrum 2). Spectrum 2 was subtracted from Spectrum 1 with an appropriate scaling factor to remove glucose signals but preserve signals of all other compounds in the resulting difference spectrum. The method assumes that introduction of the compound of interest does not change sample conditions, preserving sample matrix, line shapes, and signal frequencies.
PCA of the difference spectra (Figs. 2D–2F, Table S1 M2) showed no sample grouping due to differences in buffer used (Fig. 2D). The predominant drivers of sample variation were lipids/lipoproteins (PC1), an inverse relationship between choline and phosphocholine as well as citrate. An association with csCaP was suggested by the presence of lipids/lipoproteins, although separation between clinical groups was not observed in any PC.
Given that the inverse relationship observed between phosphocholine and choline is due to PAP-mediated hydrolysis, a reaction which was not inhibited in these samples,26 choline-based metabolites (choline, phosphocholine, and glycerophosphocholine) were excluded to remove their effect of unbalanced regulation on the MVSA. However, subsequent PCA (Fig. 3, Table S1 M3) showed no obvious clustering, with most variation due to lipids/lipoproteins, citrate, and serine (Figs. 3A, 3B). Fructose and spermine were other significant sources of variation in PC3/PC4 (Figs. 3C, 3D).
Fig. 3.
Principal components analysis after exclusion of choline containing metabolites demonstrated that lipids/lipoproteins, citrate, and serine were influential metabolites (A, B) as well as fructose and spermine (C, D). No clustering was present according to CaP status (blue squares = benign; red triangles = CaP). (A, C) Scores plots. (B, D) Loadings plots. CaP, prostate cancer; Cit, citrate; Fru, fructose; Lip, lipids/lipoproteins; PC, principal component; Ser, serine; Spe, spermine.
3.3. Supervised multivariate statistical analysis
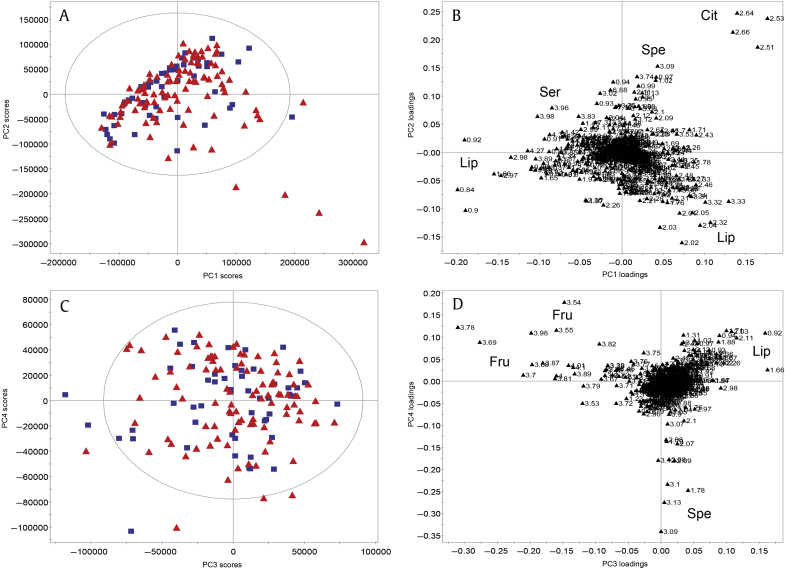
In the unsupervised PCAs, which determine sources of variation potentially independent of underlying biology, no sample clustering into clinical groups was seen, prompting the need for supervised MVSA. First, the presence of csCaP according to the D’Amico criteria based on biopsy was used as the predictive variable in PLS analysis (Fig. 4, Table S1 M4) and demonstrated lipids/lipoproteins to be associated with variation for csCaP, which were mostly limited high-risk patients. Furthermore, there was potential subgrouping among the D’Amico risk groups (Fig. 4B).
Fig. 4.
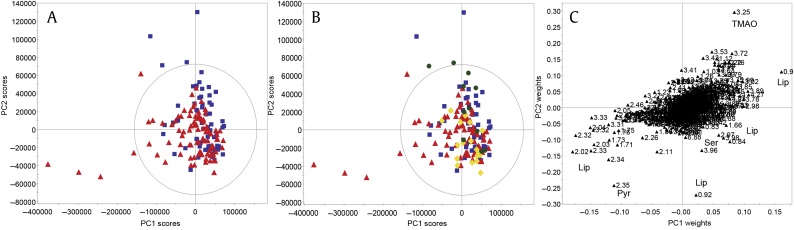
Supervised, partial least squares analysis of seminal plasma NMR spectra in predicting csCaP (according to D’Amico criteria) following add-to-subtract. Minimal separation is seen according to csCaP (blue squares = benign, red triangles = csCaP; A). When coloured according to risk subgroups (blue squares = benign; green dots = low risk; yellow diamonds = intermediate risk; red triangles = high risk/cancer present; panel B), potential intragroup clustering was seen due to pyruvate, serine and lipids/lipoproteins (high/intermediate risk), and TMAO (low risk). (A, B) Scores plots. (C) Loadings plot. csCaP, clinically significant prostate cancer; Lip, lipids/lipoproteins; NMR, nuclear magnetic resonance; Pyr, pyruvate; Ser, serine; TMAO, trimethylamine N-oxide; PC, principal component.
Based on these results and reports that maximal metabolite disturbances are observed in low- and intermediate-risk tumors, we analyzed with PLS a subgroup of 11 samples correlating to these grades confirmed by RP histology only (Figs. 5A–5C, Table S1 M5). The single low-risk sample was separated from the intermediate-risk samples due to reduced lactate, pyruvate and lipids/lipoproteins and increased citrate, myo-inositol, spermine and fructose (Figs. 5A, 5C). Within these low/intermediate-risk samples, separation was seen in accordance with primary Gleason Pattern 4, associated with higher levels of lipids/lipoproteins, lactate, and pyruvate as well as lower levels of citrate, spermine, and myo-inositol (Figs. 5B, 5C). These relationships were observed when classifiers based on all low/intermediate-risk patients, determined by biopsy or RP, were performed (Figs. 5D, 5E; Table S1 M6). When benign samples were considered with risk group combinations and Primary Gleason Pattern 4 presence, minimal separation was observed and models were weak/nonpredictive (Fig. S1, Table S1 M7–M10).
Fig. 5.
Supervised, partial least squares analysis of seminal plasma NMR spectra in predicting CaP risk (according to D’Amico criteria) following add-to-subtract. Separation between low- (green dots) and intermediate- (yellow diamonds) risk patients based on RP histology due to elevated lactate, lipids/lipoproteins and pyruvate and reduced citrate, fructose, myo-inositol, and spermine in intermediate-risk samples (A,C). These relationships were observed when expanded to all low and intermediate risk patients (D, E). Discrimination within the low/intermediate group was observed due to primary Gleason Pattern 4 (filled square = present; empty square = absent) and higher lipids/lipoproteins, lactate, and pyruvate as well as reduced citrate, myo-inositol and spermine (B,C). (A, B, D) Scores plots. (C, E) Loadings plots. CaP, prostate cancer; Cit, citrate; Fru, fructose; Gln, glutamine; Glu, glutamate, Lac, lactate; Lip, lipids/lipoproteins; Myo, myo-inositol; Pyr, pyruvate; RP, radical prostatectomy; Spe, spermine; UC1, unknown compound 1.; PC, principal component.
Analysis of only the samples collected in PBS, unaffected by any external glucose (Fig. S2, Table S1 M11–17), showed similar relationships to those seen for the full cohort. Specifically, valid models were obtained for separation between low- and intermediate-risk samples (M12, Figs. S2C, S2D; limited by sample size) and low-risk and benign samples (M13, Figs. S2E, S2F). Findings were confirmed with PCA (M15–17, Fig. S3) and driven by lactate levels (Fig. S4). The presence of the TMPRSS2:ERG fusion gene, detected in the epithelial cell fraction of SP, used as Y variable was weakly but nonpredictively associated with lipid/macromolecule resonances (Fig. S5, Table S1 M18).
3.4. Targeted metabolite profiling
SP metabolite quantification with subsequent logistic regression showed that citrate or myo-inositol were not significant predictors of CaP status (Table 2). Significant metabolites for CaP status (choline, leucine) and csCaP (leucine, valine) did not significantly improve diagnosis compared with serum PSA metabolite predictability.
Table 2.
Logistic regression weightings following targeted metabolite quantification using Chenomx, similar to that reported by Li et al.18
| Metabolite | Mean (± SE) (mM) | Logistic regression (log base 10) |
ROC analysis |
||||
|---|---|---|---|---|---|---|---|
| P | Coefficient | SE | AUC | Std. Error | |||
| Prostate cancer status | Alanine | 0.1734 (0.0208) | 0.7998 | 0.5211 | 2.0552 | 0.555 | 0.0498 |
| Choline | 1.3326 (0.1392) | 0.0291 | 2.0211 | 0.9263 | 0.556 | 0.0495 | |
| Citrate | 2.9243 (0.2643) | 0.1490 | −1.2433 | 0.8616 | 0.542 | 0.0490 | |
| Creatine | 0.1156 (0.0114) | 0.8786 | −0.1850 | 1.2113 | 0.565 | 0.0492 | |
| Fructose | 1.0591 (0.1004) | 0.9631 | −0.0413 | 0.8938 | 0.603 | 0.0510 | |
| Glucose | 3.2694 (0.2118) | 0.9144 | −0.0262 | 0.2439 | 0.629 | 0.0487 | |
| Glutamine | 0.5802 (0.0604) | 0.8559 | −0.3076 | 1.6943 | 0.541 | 0.0509 | |
| Glycerophosphocholine | 0.2259 (0.0236) | 0.0951 | −1.0546 | 0.6318 | 0.603 | 0.0485 | |
| Lactate | 0.8520 (0.0645) | 0.5879 | −0.5962 | 1.1003 | 0.579 | 0.0484 | |
| Leucine | 0.4067 (0.0416) | 0.0008 | −12.3505 | 3.6772 | 0.572 | 0.0499 | |
| Myo-inositol | 0.3251 (0.0238) | 0.9287 | −0.1095 | 1.2238 | 0.592 | 0.0472 | |
| Phosphocholine | 0.1810 (0.0459) | 0.1042 | 0.6905 | 0.4250 | 0.543 | 0.0499 | |
| Serum PSA | 8.0867 (0.6075)* | 0.0601 | 1.5605 | 0.8299 | 0.593 | 0.0472 | |
| Pyruvate | 0.3709 (0.0373) | 0.9736 | −0.0286 | 0.8672 | 0.546 | 0.0493 | |
| Uridine | 0.1793 (0.0167) | 0.7146 | 0.1528 | 0.4180 | 0.568 | 0.0495 | |
| Valine | 0.3206 (0.0392) | 0.0013 | 1.4923 | 1.6868 | 0.534 | 0.0503 | |
| Clinically significant prostate cancer | Alanine | 0.1734 (0.0208) | 0.1629 | −2.2913 | 1.6421 | 0.592 | 0.0466 |
| Choline | 1.3326 (0.1392) | 0.1595 | 1.1536 | 0.8200 | 0.584 | 0.0467 | |
| Citrate | 2.9243 (0.2643) | 0.1147 | −1.4430 | 0.9147 | 0.580 | 0.0466 | |
| Creatine | 0.1156 (0.0114) | 0.8808 | −0.1703 | 1.1364 | 0.603 | 0.0465 | |
| Fructose | 1.0591 (0.1004) | 0.5227 | −0.5368 | 0.8397 | 0.611 | 0.0470 | |
| Glucose | 3.2694 (0.2118) | 0.3728 | 0.1970 | 0.2211 | 0.584 | 0.0475 | |
| Glutamine | 0.5802 (0.0604) | 0.4141 | 1.1168 | 1.3674 | 0.572 | 0.0473 | |
| Glycerophosphocholine | 0.2259 (0.0236) | 0.4243 | −0.4494 | 0.5624 | 0.599 | 0.0463 | |
| Lactate | 0.8520 (0.0645) | 0.8719 | −0.1501 | 0.9313 | 0.582 | 0.0467 | |
| Leucine | 0.4067 (0.0416) | 0.0025 | −9.2502 | 3.0562 | 0.597 | 0.0466 | |
| Myo-inositol | 0.3251 (0.0238) | 0.4242 | 0.9335 | 1.1681 | 0.609 | 0.0459 | |
| Phosphocholine | 0.1810 (0.0459) | 0.2600 | 0.4343 | 0.3856 | 0.506 | 0.0477 | |
| Serum PSA | 8.0867 (0.6075)a) | 0.0228 | 1.7942 | 0.7880 | 0.617 | 0.0456 | |
| Pyruvate | 0.3709 (0.0373) | 0.6820 | −0.3337 | 0.8146 | 0.569 | 0.0472 | |
| Uridine | 0.1793 (0.0167) | 0.2795 | 0.4318 | 0.3993 | 0.568 | 0.0473 | |
| Valine | 0.3206 (0.0392) | 0.0030 | 9.7952 | 3.2968 | 0.571 | 0.0471 | |
Among 151 patients, CaP status (positive 98, negative 53) and D'Amico risk (high = 82, low = 69) were used as dependent variables for logistic regression analysis (P to enter 0.05, P to exclude >0.1).
AUC, area under the curve; CaP, prostate cancer; PSA, prostate specific antigen; ROC, receiver operating characteristic; SE, standard error.
Bold indicate P < 0.05.
Serum PSA determined using immunoassay, units ng/mL.
4. Discussion
In this paper, we present the largest validation study of SP-based metabolite prediction of CaP using high resolution NMR spectroscopy, having analyzed SP metabolite profiles from 151 men being investigated for CaP. Undue influence of exogenous glucose contained in the HBSS buffer used for RNA analyses was successfully excluded by applying add-to-subtract and revealed inherent variation due to enzyme-dependent changes in choline-based metabolites. SP metabolites best predicted low- and intermediate-risk CaP with differences observed between these groups and benign and high-risk samples. Metabolites previously reported to determine CaP, such as citrate, spermine, and myo-inositol, showed minimal predictive ability in this clinically applicable cohort. These findings were confirmed with targeted metabolite quantification.
Well described prostatic metabolite changes due to CaP, specifically reduced citrate and polyamines (e.g., myo-inositol, spermine) and increased intracellular lactate, choline, and creatine,12 were not predictive in this study due to the following underlying clinical and biological factors. Clinically, the study population presented here contains patients suspected of harboring CaP, encountered in daily urological practice (Table 1). In earlier reports where SP metabolites significantly improved CaP detection, CaP-positive samples were compared with healthy controls or men unlikely to have CaP, suggested by marked discrepancies in serum PSA between groups. Our population contained heterogeneous disease states, inclusive of all tumor grades with predominance toward high-risk CaP. Although group separation was observed between CaP risk groups (Fig. 5, Figs. S2, S3), we could not truly exclude CaP in patients with a negative biopsy due to limitations in biopsy-based CaP detection and known metabolic changes in early tumorigenesis, which may lead to confounding overlap between groups and invalid statistical models. To exclude uncertainty among the control group, a subanalysis of the presence or absence of Gleason Pattern 4 on RP histology showed overlap of groups (M10, Figs. S1G, S1H), likely owing to reduced metabolite influence in poorly differentiated tumors. Similarly, limitations of biopsy-based risk stratification given known upstaging at RP in up to 40% of patients may confound the accuracy of risk subgroup analyses.27 When analyses based only on RP-based diagnosis were expanded to include biopsy-based diagnosis to increase sample size, sample grouping was less obvious despite similar metabolite patterns being observed in the loadings plot (Fig. 5). Thus, a larger low-/intermediate-risk RP cohort would be expected to accurately “upclassify” (upstage) low-risk samples with metabolite patterns similar to intermediate-/high-risk samples, as shown elsewhere.28
Biologically, gene expression and metabolite alterations occur early in tumorigenesis and are more pronounced in lower grade (Gleason ≤ 7) compared with higher grade (Gleason ≥ 8) tumors,28 supported by our analysis of low- and intermediate-risk patients (Fig. 5, Figs. S2, S3). In addition to direct metabolic changes, the inverse relationship between lactate and fructose resulting in group separation between low- and intermediate-risk and benign samples may indicate disturbed SP homeostasis of anions (zinc) or enzymes (PSA, PAP) known to improve sperm function,29 resulting in impaired sperm glycolysis. Indeed, poor discrimination of metabolite profiles from high-risk tumors was demonstrated here and in other studies, likely due to accumulated genetic alterations with disease progression.28, 30 Thus, patients with lower grade tumors may be amenable to SP-mediated in vitro or magnetic resonance spectroscopic imaging (MRSI)-mediated in vivo assessment or monitoring as a potential substitute for repeat biopsy in active surveillance.28, 31
Altered metabolite homeostasis correlates with increased fatty acid synthesis, due to or in association with TMPRSS2:ERG fusion gene translocation associated with aggressive CaP, may account for the overwhelming influence of lipids/lipoproteins in high-risk patients in this study, similar to that reported by others.32 Higher grade tumors overexpress the oncogene MYC, which is associated with dysregulated lipid metabolism33 and display altered cholesterol metabolism to increase energy storage.30 Upregulated lipid subclasses have been described between normal, localized, and metastatic prostatic cells, with choline kinase α implicated in de novo lipogenesis in aggressive metastatic cells.34 Systemically, lipid and energy metabolites in serum have been strongly associated with aggressive CaP35 and may improve CaP detection.
This study was an opportunistic analysis of SP samples collected initially for cytology and subsequently epithelial cell RNA analyses. The exogenous glucose contained in the HBSS required significant correction using add-to-subtract, which did not introduce further influence into the MVSA. Subsequently, the uninhibited changes in choline-based metabolites showed significant influence in the preliminary MVSA. These metabolite peaks were excluded because PAP-catalyzed hydrolysis of phosphocholine to choline is a rapid, endogenous reaction to enhance spermatozoal function and protection.26 Variations in time from sample production to processing, despite most being done within 2 hours, are likely to cause significant variation among these metabolites independent of underlying CaP due to the unknown degree of reaction completion. Given the postulated role of choline in tumor progression, as indicated by elevated in vivo levels, reliable quantification of choline-based metabolites in SP is desired. Thus, a sample collection/storage protocol should be implemented that limits the PAP reaction to 2–3% progression, such as our recommendation that ejaculate samples be collected in a sterile urine jar containing 5mM tartrate in 20 mL PBS solution cooled to 4°C.26 Although malignant prostatic metabolite contribution to SP, considering concurrent contributions from multiple organs and resulting proteolysis, may intuitively be minimal or diluted, our findings are similar to those seen in tissue extracts and in vivo,28, 30 likely to be enhanced by spectral acquisition at 900 MHz. Although prostatitis is known to reduce prostatic citrate and zinc content12 and potentially affect MVSA, this influence in the current study would be minimal due to the focus on RP-based and malignant pathology, as well as only being confirmed histologically for two patients.
In conclusion, metabolomics of seminal plasma in vitro may assist diagnosis and monitoring of either low or intermediate grade prostate cancer. Lipids/lipoproteins dominated spectra of high grade samples with fewer contributions from other metabolites. As a validation study, we were unable to replicate previous performance of SP-based metabolite prediction of CaP in 151 men being investigated for CaP. Dedicated metabolomics protocols ideally in serial collections may maximize information recovery. The value of metabolomics analysis of SP for CaP currently appears to be in active surveillance of low- or intermediate-grade tumors suspicious of understaging, in which in vivo correlation with MRSI and monitoring in vitro with SP or in vivo with MRSI may further clinical practice.
Conflicts of interest
All authors have no conflicts of interest to declare.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.prnil.2017.03.005.
Contributor Information
Horst Joachim Schirra, Email: h.schirra@uq.edu.au.
Robert A. Gardiner, Email: f.gardiner@uq.edu.au.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Schröder F.H., Hugosson J., Roobol M.J., Tammela T.L.J., Zappa M., Nelen V. Screening and prostate cancer mortality: results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet. 2014;384:2027–2035. doi: 10.1016/S0140-6736(14)60525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhindi B., Mamdani M., Kulkarni G.S., Finelli A., Hamilton R.J., Trachtenberg J. Impact of the U.S. Preventive Services Task Force recommendations against prostate specific antigen screening on prostate biopsy and cancer detection rates. J Urol. 2015;193:1519–1524. doi: 10.1016/j.juro.2014.11.096. [DOI] [PubMed] [Google Scholar]
- 3.Pokorny M.R., de Rooij M., Duncan E., Schroder F.H., Parkinson R., Barentsz J.O. Prospective study of diagnostic accuracy comparing prostate cancer detection by transrectal ultrasound-guided biopsy versus magnetic resonance (MR) imaging with subsequent MR-guided biopsy in men without previous prostate biopsies. Eur Urol. 2014;66:22–29. doi: 10.1016/j.eururo.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Roberts M.J., Richards R.S., Chow C.W., Doi S.A., Schirra H.J., Buck M. Prostate-based biofluids for the detection of prostate cancer: a comparative study of the diagnostic performance of cell-sourced RNA biomarkers. Prostate Int. 2016;4:97–102. doi: 10.1016/j.prnil.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephan C., Jung K., Semjonow A., Schulze-Forster K., Cammann H., Hu X. Comparative assessment of urinary prostate cancer antigen 3 and TMPRSS2:ERG gene fusion with the serum [-2]proprostate-specific antigen-based prostate health index for detection of prostate cancer. Clin Chem. 2013;59:280–288. doi: 10.1373/clinchem.2012.195560. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson A., Mahon J., Boland A., Beale S., Dwan K., Fleeman N. The clinical effectiveness and cost-effectiveness of the PROGENSA(R) prostate cancer antigen 3 assay and the Prostate Health Index in the diagnosis of prostate cancer: a systematic review and economic evaluation. Health Technol Assess. 2015;19 doi: 10.3310/hta19870. i–xxxi, 1–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong H.Y., Byun J. Emerging roles of human prostatic acid phosphatase. Biomol Ther (Seoul) 2013;21:10–20. doi: 10.4062/biomolther.2012.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts M.J., Richards R.S., Gardiner R.A., Selth L.A. Seminal fluid: a useful source of prostate cancer biomarkers? Biomark Med. 2015;9:77–80. doi: 10.2217/bmm.14.110. [DOI] [PubMed] [Google Scholar]
- 9.Gardiner R.A., Samaratunga M.L., Gwynne R.A., Clague A., Seymour G.J., Lavin M.F. Abnormal prostatic cells in ejaculates from men with prostatic cancer—a preliminary report. Brit J Urol. 1996;78:414–418. doi: 10.1046/j.1464-410x.1996.00089.x. [DOI] [PubMed] [Google Scholar]
- 10.Roberts M.J., Chow C.W., Schirra H.J., Richards R., Buck M., Selth L.A. Diagnostic performance of expression of PCA3, hepsin and miR biomarkers inejaculate in combination with serum PSA for the detection of prostate cancer. Prostate. 2015;75:539–549. doi: 10.1002/pros.22942. [DOI] [PubMed] [Google Scholar]
- 11.Selth L.A., Roberts M.J., Chow C.W., Marshall V.R., Doi S.A., Vincent A.D. Human seminal fluid as a source of prostate cancer-specific microRNA biomarkers. Endocr Relat Cancer. 2014;21:L17–L21. doi: 10.1530/ERC-14-0234. [DOI] [PubMed] [Google Scholar]
- 12.Roberts M.J., Schirra H.J., Lavin M.F., Gardiner R.A. Metabolomics: a novel approach to early and noninvasive prostate cancer detection. Korean J Urol. 2011;52:79–89. doi: 10.4111/kju.2011.52.2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke R.A., Schirra H.J., Catto J.W., Lavin M.F., Gardiner R.A. Markers for detection of prostate cancer. Cancers (Basel) 2010;2:1125–1154. doi: 10.3390/cancers2021125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kline E.E., Treat E.G., Averna T.A., Davis M.S., Smith A.Y., Sillerud L.O. Citrate concentrations in human seminal fluid and expressed prostatic fluid determined via 1H nuclear magnetic resonance spectroscopy outperform prostate specific antigen in prostate cancer detection. J Urol. 2006;176:2274–2279. doi: 10.1016/j.juro.2006.07.054. [DOI] [PubMed] [Google Scholar]
- 15.Serkova N.J., Gamito E.J., Jones R.H., O'Donnell C., Brown J.L., Green S. The metabolites citrate, myo-inositol, and spermine are potential age-independent markers of prostate cancer in human expressed prostatic secretions. Prostate. 2008;68:620–628. doi: 10.1002/pros.20727. [DOI] [PubMed] [Google Scholar]
- 16.Epstein J.I., Allsbrook W.C., Jr., Amin M.B., Egevad L.L. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 17.D'Amico A.V., Whittington R., Malkowicz S., Schultz D., Blank K., Broderick G.A. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280:969–974. doi: 10.1001/jama.280.11.969. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Wijffels G., Yu Y., Nielsen L.K., Niemeyer D.O., Fisher A.D. Altered fatty acid metabolism in long duration road transport: an NMR-based metabonomics study in sheep. J Proteome Res. 2010;10:1073–1087. doi: 10.1021/pr100862t. [DOI] [PubMed] [Google Scholar]
- 19.Marion D., Ikura M., Bax A. Improved solvent suppression in one- and two-dimensional NMR spectra by convolution of time-domain data. J Magn Reson (1969) 1989;84:425–430. [Google Scholar]
- 20.Ye T., Zheng C., Zhang S., Gowda G.A.N., Vitek O., Raftery D. “Add to subtract”: a simple method to remove complex background signals from the 1H nuclear magnetic resonance spectra of mixtures. Anal Chem. 2011;84:994–1002. doi: 10.1021/ac202548n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savorani F., Tomasi G., Engelsen S.B. Icoshift: a versatile tool for the rapid alignment of 1D NMR spectra. J Magn Res. 2010;202:190–202. doi: 10.1016/j.jmr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 23.Kohl S., Klein M., Hochrein J., Oefner P., Spang R., Gronwald W. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics. 2012;8:146–160. doi: 10.1007/s11306-011-0350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trygg J., Holmes E., Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 25.Weljie A.M., Newton J., Mercier P., Carlson E., Slupsky C.M. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78:4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 26.Roberts M.J., Hattwell J.P., Chow C.W., Lavin M., Pierens G.K., Gardiner R.A. Tartrate inhibition of prostatic acid phosphatase improves seminal fluid metabolite stability. Metabolomics. 2016;12 [Google Scholar]
- 27.Epstein J.I., Feng Z., Trock B.J., Pierorazio P.M. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol. 2012;61:1019–1024. doi: 10.1016/j.eururo.2012.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen A.F., Sandsmark E., Rye M.B., Wright A.J., Bertilsson H., Richardsen E. Presence of TMPRSS2-ERG is associated with alterations of the metabolic profile in human prostate cancer. Oncotarget. 2016;7:42071–42085. doi: 10.18632/oncotarget.9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milardi D., Grande G., Vincenzoni F., Castagnola M., Marana R. Proteomics of human seminal plasma: identification of biomarker candidates for fertility and infertility and the evolution of technology. Mol Reprod Dev. 2013;80:350–357. doi: 10.1002/mrd.22178. [DOI] [PubMed] [Google Scholar]
- 30.Meller S., Meyer H.A., Bethan B., Dietrich D., Maldonado S.G., Lein M. Integration of tissue metabolomics, transcriptomics and immunohistochemistry reveals ERG- and gleason score-specific metabolomic alterations in prostate cancer. Oncotarget. 2016;7:1421–1438. doi: 10.18632/oncotarget.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts M.J., Yaxley J.W., Coughlin G.D., Gianduzzo T.R., Esler R.C., Dunglison N.T. Can atorvastatin with metformin change the natural history of prostate cancer as characterized by molecular, metabolomic, imaging and pathological variables? A randomized controlled trial protocol. Contemp Clin Trials. 2016;50:16–20. doi: 10.1016/j.cct.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 32.Madhu B., Shaw G.L., Warren A.Y., Neal D.E., Griffiths J.R. Response of Degarelix treatment in human prostate cancer monitored by HR-MAS 1H NMR spectroscopy. Metabolomics. 2016;12 doi: 10.1007/s11306-016-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priolo C., Loda M. Untargeted metabolomics for profiling oncogene-specific metabolic signatures of prostate cancer. Mol Cell Oncol. 2015;2 doi: 10.1080/23723556.2014.1001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burch T.C., Isaac G., Booher C.L., Rhim J.S., Rainville P., Langridge J. Comparative metabolomic and lipidomic analysis of phenotype stratified prostate cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mondul A.M., Moore S.C., Weinstein S.J., Karoly E.D., Sampson J.N., Albanes D. Metabolomic analysis of prostate cancer risk in a prospective cohort: the alpha-tocolpherol, beta-carotene cancer prevention (ATBC) study. Int J Cancer. 2015;137:2124–2132. doi: 10.1002/ijc.29576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.