This study illuminates the locations of water channels within Photosystem II, a large membrane protein complex.
Abstract
Photosystem II (PSII), a unique membrane-bound oxidoreductase, catalyzes light-driven oxidation of water to molecular oxygen. Although high-resolution structures of PSII are known, the exact path of the substrate water molecules to the catalytic Mn4CaO5 center within the PSII complex remains poorly understood. PSII produces reactive oxygen species (ROS), responsible for the frequent damage and turnover of this megacomplex that occur under physiological conditions. Such ROS are known to specifically modify PSII proteins. Using high-resolution tandem mass spectrometry, we identified oxidative modifications on 36 amino acid residues on the lumenal side of PSII, in the core PSII proteins D1, D2, and CP43 of the cyanobacterium Synechocystis sp. PCC 6803. Remarkably, these oxidized residues clustered into three nearly continuous formations, tracking the pathways of ROS diffusion from the manganese center all the way out to the surface of PSII. We suggest that these profiles of oxidized residues reveal the locations of water channels within PSII. Our results provide the most comprehensive experimental evidence to date of physiologically relevant oxidized residues in PSII and illuminate three possible channels for water between the catalytic Mn cluster in the PSII complex and the bulk medium around it.
INTRODUCTION
Photosystem II (PSII) is an oxidoreductase found in the thylakoid membrane of organisms that perform oxygenic photosynthesis. The fully assembled PSII complex consists of approximately 20 protein subunits and multiple redox-active cofactors that allow PSII to function as a catalyst for the light-driven oxidation of water and concomitant reduction of plastoquinone (1–4). This conversion of sunlight to chemical energy initiates the photosynthetic electron transport chain and sustains nearly all life on Earth.
The active site for water oxidation, a Mn4CaO5 cluster (henceforth Mn cluster), is buried within the PSII complex near the lumen-membrane interface (5). This buried position limits water access to the active site, a key feature that promotes the reaction. When PSII is modified to allow an unrestricted flow of water to the active site, incomplete water oxidation occurs, forming hydrogen peroxide (H2O2) that is reduced to the harmful hydroxyl radical (HO•) (6–9). The buried active site minimizes, but does not completely prevent, such a side reaction pathway, allowing some reactive oxygen species (ROS) to be produced under physiological conditions (10, 11). ROS-induced damage to PSII is the major mechanism believed to be responsible for the frequent PSII turnover that occurs in cells (2, 12), and oxidative modifications of PSII residues near the Mn cluster have been detected (11, 13–17).
Although PSII is the only known enzyme capable of complete water oxidation, it is not the only enzyme for which water is more than just a solvent. Aquaporins transport water across membranes; cytochrome c oxidases produce water as a by-product while pumping protons; and the haloalkane dehalogenases use specific interactions with carefully placed internal water molecules to achieve catalysis. For these and many other enzymes, it is becoming increasingly clear that water dynamics is exquisitely tuned by the protein scaffold to fit the enzyme’s function. For example, specific channels regulate the accessibility, geometrical positioning, and flow rate of water molecules within the abovementioned enzymes and in many others (18–21).
Water transport within PSII is of prime interest, given water’s unprecedented role as substrate and the problems associated with unrestricted water access to the active site. A number of computational studies have searched for water channels by examining cavities within the static PSII crystal structure (22–24) or by performing molecular dynamics (MD) simulations (25–30). Pathways for removal of the reaction products, dioxygen and protons, have also been considered (22–24, 27, 28, 30) because the extended presence of dioxygen can lead to protein damage (from conversion to singlet dioxygen) (31), and of protons, to inhibition of catalysis (due to improper redox leveling) (32, 33). These computational studies have identified several putative channel systems within PSII.
Here, we took an experimental approach to identify oxidized residues on the lumenal side of PSII from the cyanobacterium Synechocystis sp. PCC 6803 (henceforth Synechocystis) by using high-resolution tandem mass spectrometry (MS). We reasoned that after generation near the Mn cluster, ROS would diffuse through putative channels that lead away from the cluster and modify most readily the residues that line the walls of these channels. The approach is related to a typical hydroxyl radical footprinting experiment, in which hydroxyl radicals are generated from solvent water molecules by an x-ray or laser pulse (34–37) or via a metal-catalyzed reaction (36, 38–40) and then modify nearby residues. The modified residues, detected by MS, leave a trail of oxidative damage that, when identified, can illuminate structural aspects of the system being studied. Hence, the ROS produced at the Mn cluster serve as a built-in footprinting reagent. Using this approach, we identified three nearly continuous formations of oxidized residues that are centered at the Mn cluster and radiate outward all the way to the bulk solvent. Hydrogen peroxide and the hydroxyl radical are the two major ROS known to be produced at the Mn cluster (6, 11). The hydroxyl radical, by far the more reactive species, is short-lived but can diffuse several tens of angstroms after generation at a protein site (38, 41, 42). Given the similar size and hydrophilicity of HO• and water (43), an HO• diffusion pathway is likely to be favorable for water as well. We conclude that the three ROS channels identified in our study represent three possible water channels in PSII. We discuss the implications of our results for the delivery of water to the site of its oxidation in PSII.
RESULTS
To track the oxidative damage trail, we used bottom-up proteomics on a high-performance mass spectrometer. Briefly, this method involves digestion of the protein sample into peptides, chromatographic separation of the peptides, and online injection into a mass spectrometer. Automated analysis of the MS data, combined with manual verification for rigor, enables confident identification of the peptide sequences in the sample. During the analysis of our MS data, we focused on the D1, D2, and CP43 subunits of PSII because these proteins are most closely associated with the Mn cluster (5), with additional targeted analysis of residues in the CP47 subunit, as described below. We obtained 35, 37, and 46% sequence coverage of the D1, D2, and CP43, respectively. Our coverage is appreciably higher than was typically reported for these proteins (44–47). In an earlier study of oxidative modifications in higher plant PSII, Frankel et al. (48) found 24, 27, and 26% coverage for D1, D2, and CP43, respectively. Our higher coverage can be attributed to the increased sensitivity and speed of the Q-Exactive (QE) Plus mass spectrometer we used, compared to the LTQ-FTICR, earlier Orbitrap, and MALDI-TOF instruments used previously. The high mass accuracy (~0.02 Da) of the product ion (MS/MS) spectra recorded on the QE enabled highly confident peptide identification and residue-level localization of posttranslational modifications to an extent not possible on earlier instruments lacking high mass accuracy for product ions formed in MS/MS. We were able to locate unambiguously all oxidative modifications shown below at the resolution of a single amino acid residue. Example product ion (MS/MS) spectra (shown in Fig. 1 and figs. S1 to S3) testify to the quality of the data obtained in this study (see table S1 for a list of the types of oxidative modifications included in the searches).
Fig. 1. Example of product-ion (MS/MS) spectra that identify oxidative modifications of PSII residues.
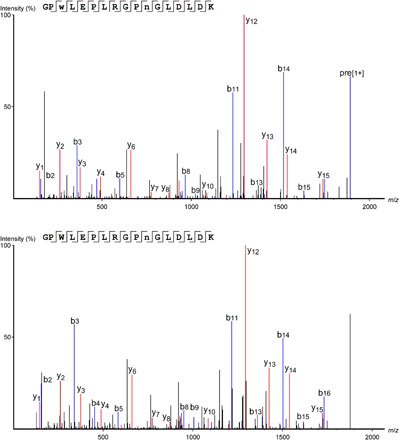
Product-ion (MS/MS) spectra of the oxidized CP43 peptide 362GPwLEPLRGPNGLDLD378K (top) and the unmodified form of the peptide (bottom). The oxidation (+15.9949 Da) was localized to 364W. Note that 372N contains the common deamidation modification (+0.9840 Da) according to both spectra. The fragment maps and corresponding labeled b- and y-ions show the convincing fragmentation series obtained at high mass accuracy (~0.02 Da), allowing highly confident peptide identification and unambiguous residue-level localization of the oxidative modification. Lowercase lettering in the amino acid sequence indicates the site of oxidation (see figs. S1 to S3 for additional example spectra). m/z, mass/charge ratio.
We obtained MS coverage of 472 residues on D1, D2, and CP43 in the MS analyses and identified oxidative modifications on 50 residues (11%) (tables S2 and S3; residue numbering follows the Synechocystis 6803 sequence unless otherwise noted). Of these 50 modified residues, 19 were detected only in the light-incubated samples (whereas only 7 were detected in the dark-incubated samples but not in the light-incubated samples). This finding strengthens the concept that PSII photochemistry leads to oxidative damage of its protein components. Of all the PSII proteins, D1 is believed to be the primary site of photodamage (1) and, as a result, is turned over the fastest (49). Vermaas and co-workers (49) found that in Synechocystis, D1, D2, and CP43 have increasing half-lives of <1, 3.3 ± 1, and 6.5 ± 1.5 hours, respectively, with a longer half-life presumably reflecting a lower damage rate. Matching this trend, we found that D1 showed the most marked response to light incubation, with a 3-fold increase in the number of oxidized residues detected after light incubation, followed by D2 (2.5-fold increase), and, finally, CP43 (1.2-fold increase) (table S2). We note that, owing to the varying ionization efficiency of modified and unmodified forms of a peptide, our data did not allow us to quantify the fraction of a particular peptide that is modified (50).
Of the 50 oxidized residues, 36 are on the lumenal side of PSII and 14 are on the cytosolic side. We focused our analysis on the 36 lumen-side residues because they are proximate to the Mn cluster. We mapped the oxidized lumen-side residues onto the 1.9 Å crystal structure of PSII from Thermosynechococcus vulcanus [Protein Data Bank (PDB): 3WU2] (5) (Fig. 2). This comparison is justified by the extremely high degree of conservation between PSII protein sequences of Synechocystis and T. vulcanus (and across all oxygenic photosynthetic organisms). The D1, D2, and CP43 proteins of these two organisms share 86, 90, and 87% sequence identity and 94, 99, and 94% sequence similarity, respectively.
Fig. 2. Residues with oxidative modifications detected in this study.
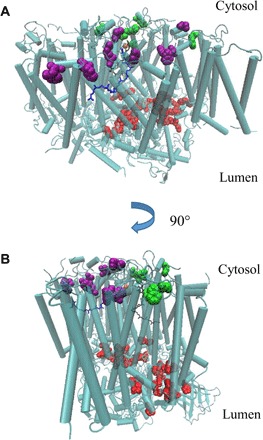
The modified residues on D1, D2, and CP43 were mapped onto the 1.9 Å crystal structure of PSII from T. vulcanus (PDB: 3WU2). Red, modified residues on the lumenal side of PSII; green, modified residues on the cytosolic side of PSII, in the vicinity of QA; purple, modified residues on the cytosolic side of PSII, in the vicinity of QB.
To be oxidized, a residue must have been exposed to ROS. As has been pointed out by Frankel et al. (13, 48), it is reasonable that surface-exposed residues could be oxidized by ROS in the bulk solvent, but buried oxidized residues should reflect the location of ROS production within the complex. Because the Mn cluster can produce ROS, we expected that the oxidized lumen-side residues would cluster particularly around this metal center, as has been observed in several studies (13–17). Figure 3A locates the 36 lumen-side oxidized residues. Examination of these data shows that 13 of these residues are in the vicinity (within 15 Å) of the Mn cluster, but the remaining 23 are not (the full range of distances is 2 to 30 Å from the cluster). However, most of the oxidized residues lie in three nearly continuous formations (“arms”) that are centered on the Mn cluster and radiate outward in different directions (although we note that the arm 2 group shows less continuity than arms 1 and 3) (Fig. 3, A and B). Arm 1 consists of D1 and D2 residues (and also several CP47 residues identified afterward, as described below); arm 2 consists of D1, D2, and CP43 residues; and arm 3 consists only of CP43 residues (Fig. 3B and table S2). To ensure that this distinct pattern is not skewed by limited MS coverage of lumen-side residues, we mapped all 255 lumen-side residues (unmodified and modified) identified in our study onto the crystal structure (Fig. 3C). This depiction clearly showed that a broad coverage of lumen-side residues was achieved, and the observed geometrical arrangement of the oxidized residues is not simply a quirk of limited MS coverage.
Fig. 3. Lumen-side oxidized residues detected in this study.
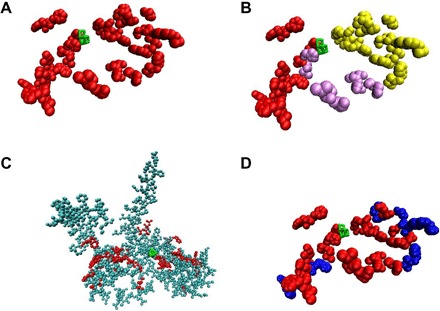
(A) Depiction of the 42 lumen-side oxidized residues detected in this study (red). (B) Same depiction as in (A), but arm 1 is shown in red, arm 2 in purple, and arm 3 in yellow. (C) All the 255 lumen-side residues that were covered by MS in this study, plus the covered and oxidized CP47 residues discussed in the text (see table S3). Some residues are obscured by others and are not visible in this view. Red, oxidized residues; cyan, nonoxidized residues. (D) Same depiction as in (A), except that in this view, the surface-exposed residues are colored blue, and buried residues are colored red. Visual Molecular Dynamics (VMD) (66) was used to determine whether a residue is surface-exposed or buried. Throughout the figure, the Mn cluster is shown in green.
An explanation for the accessibility of ROS to these residues is that they line the walls of channels within PSII through which ROS are able to diffuse after their generation at the Mn cluster. The outermost oxidized residues of these arms are surface-exposed and in contact with bulk solvent (Fig. 3D); thus, these outer residues indicate near-continuous trajectories for water travel from the surface of PSII all the way to the Mn cluster (with the exception that the arm 2 residues extend to one amino acid length away from the surface). As mentioned above, water channels have long been believed to be important for efficient water oxidation in PSII, given the buried active site and the need to regulate water access to it, although experimental evidence for their locations has remained sparse. As mentioned above, HO• is the major ROS known to be produced at the Mn cluster (6, 11), and because of their similar size and hydrophilicity (43), diffusion pathways of HO• are likely to be favorable for water as well. We conclude that the three ROS channels identified in our study represent three possible water channels in PSII.
DISCUSSION
Comparison with channels identified in computational studies
We compared each of the channel systems that were identified in computational studies with the oxidized residue arms that we observed to identify any overlap. Some of the calculated channels are strikingly consistent with our results, whereas others are not (Fig. 4, tables 1 and S4, and movie S1). It should be kept in mind that the computational studies using static-structure calculations do not account for protein dynamics that could alter channel topology and that even the MD studies do not account for conformational changes known to occur during the Mn cluster’s S-state transitions (51). We refer mainly to the channel names in Ho et al. (23) and Vassiliev et al. (26, 27) in our analysis below, but see Table 1 for nomenclature correspondence with the other computational studies.
Fig. 4. Superposition of channels identified in previous MD studies onto the lumen-side oxidized residues.
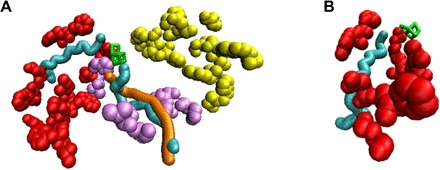
(A) A segment of the oxidized arm 1 residues partially surrounds water channel 3 (cyan; left) identified in MD studies (26, 27). The oxidized arm 2 residues partially surround water channel 4a (cyan; right) and oxygen channel 4ai (orange) identified by Vassiliev et al. (26, 27). The oxidized CP47 residues mentioned in the discussion are included in both parts of this figure (see movie S1 for a full rotation of this image about the y axis). (B) A closer view of the correspondence between oxidized arm 1 residues and channel 3 (26, 27). The 1.9 Å PSII crystal structure PDB: 3ARC was used for correspondence with channels in the study by Vassiliev et al. (26, 27). It should be noted that the depictions of channels 3, 4a, and 4ai in Figs. 4 and 5 are fixed-diameter representations. Because of the natural protein dynamics, the size and exact local shape of the actual accessible pathway within the protein complex may vary slightly from the depiction. (A) and (B): Red, arm 1 residues that correspond closely to channel 3; light purple, arm 2 residues; yellow, arm 3 residues; green, Mn cluster.
Table 1. Correspondence between channels identified in computational studies and this work.
Channel names are listed followed by the species they were assigned to conduct in the original study. Channels in the same column are nearly identical unless otherwise noted.
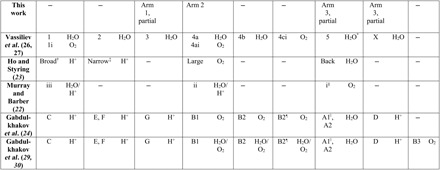
*The authors calculated a high (~22 kcal/mol) activation energy barrier for water travel through this channel, rendering it “virtually impermeable” to water (see main text for discussion).
†Encompasses approximately two-thirds of channel 1 length.
‡Encompasses approximately three-fourths of channel 2 length.
§Encompasses approximately one-half of channel 5 length.
¶Encompasses approximately three-fourths of channel 4ci length.
||Encompasses approximately two-thirds of channel 5 length.
We found significant correspondence between the arm 1 residues and channel “3,” a water channel identified by Vassiliev and co-workers (26, 27) through MD simulations (Table 1 and table S4). Channel 3 travels alongside arm 1 residues for around 20 Å, starting from the Mn cluster (Fig. 4B). The last ~15 Å interval of channel 3 angles away from arm 1 (although see below for additional correspondence from oxidized CP47 residues), whereas arm 1 continues to extend an additional 13 Å past the branching point. Channel 3 corresponds closely to channel “G” identified by another group using calculations on the static PSII crystal structure (24) (Table 1).
We also found significant correspondence between the arm 2 residues and channels “4a” and “4ai,” which are nearly identical water and dioxygen channels, respectively, identified by Vassiliev and co-workers (26, 27) in their MD simulations (Table 1 and table S4). The arm 2 residues we identified travel alongside channels 4a and 4ai for their initial 7 Å path from the Mn cluster and surround them for their final 18 Å stretch to the surface of PSII, with a residue gap in their middle 8 Å portion (Fig. 4A). Channels 4a and 4ai correspond closely to the “large channel” (23), channel “ii” (22), and channel “B1” (24, 29, 30) identified by other groups in static-structure calculations and MD simulations (Table 1).
The arm 3 residues do not overlap significantly with channels from computational studies. The arm 3 pathway(s) may become accessible to ROS diffusion as a result of conformational dynamics, including those related to the S-state cycle transitions (51), which are not captured in the computational studies.
The “back,” “broad,” and “narrow” channels (23), which correspond to channels 5, 1/1i, and 2 in the studies by Vassiliev et al. (26, 27), respectively, do not overlap strongly with our oxidized residue formations, although we obtained MS peptide coverage in those areas. Using MD simulations, the back channel has been calculated to be essentially impermeable to water transport because of a high energetic barrier of ~22 kcal/mol (27), and we would not expect diffusion of HO• to be favored either. The broad channel is believed to function as a proton exit pathway (52–54), consistent with our finding that it is not conducive to ROS diffusion. The narrow channel has been proposed to transport water to the active site (55). We obtained coverage of 11 of 17 of the narrow channel residues and found three that are oxidized (D1-E333, CP43-L336, and -M355). However, D1-E333 is a ligand to the Mn cluster, and thus, its proximity to this site does not assist significantly in describing the unique path of the narrow channel. Our results therefore do not strongly support water transport through the narrow channel, but also do not strongly reject this possibility, given the low level of oxidation that was observed and the fact that 6 of 17 channel residues remained uncovered by MS.
Comparison with earlier experimental studies of oxidized PSII residues
In a study that appeared during the preparation of this manuscript, Kale et al. (11) identified oxidative modifications on D1 and D2 from spinach PSII before and after light exposure, allowing us to compare our results with theirs. This comparison should be valid because there is extremely high sequence conservation of the core PSII proteins between these species: 84 and 85% sequence identity and 95 and 97% sequence similarity for D1 and D2, respectively. The 19 lumen-side residues they found to be oxidized occur in a pattern nearly identical to arm 1 and include 7 residues that we also found oxidized (D1-T316, W317, M331, H332, and E333 and D2-W328 and M329) (Fig. 5A). We note that D1-H332, an inner-sphere ligand of Mn-1, is oxidized upon illumination in both their study and ours. The authors hypothesized that Mn-1 is responsible for the reduction of H2O2 to HO•, leading to the oxidation of D1-H332 followed by other residues (11).
Fig. 5. Comparison of lumen-side oxidized residues detected by Bricker and co-workers (11, 13) and in the current study.
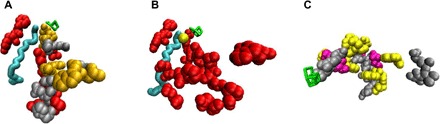
(A) Lumen-side oxidized residues detected on the D1 and D2 proteins in the study by Kale et al. (11) and in arm 1 in this study and their correspondence to channel 3 (26). (B) D1/D2 arm 1 residues in this study and the additional oxidized CP47 residues identified in a targeted analysis based on the results of Frankel et al. (13) and their correspondence to channel 3 (26, 27). (C) Oxidized residues in the arm 3 region detected by Frankel et al. (13) and in the current study. Note: Some additional lumen-side residues detected by Frankel et al. (13) that are not relevant to this discussion are not shown in this figure. Gray, residues only found oxidized by Bricker and co-workers (11, 13); orange (A) and purple (C), residues found oxidized in the studies by Kale et al. (11) or Frankel et al. (13), respectively, and the current study; red, D1/D2 arm 1 residues; yellow, Cl− (B) and arm 3 residues (C); cyan, channel 3 (26); green, Mn cluster.
In an earlier study from the same group using spinach PSII, Frankel et al. (13) identified 10 oxidized CP47 residues in the vicinity of arm 1. Because the positioning of these residues and their distance from the Mn cluster do not clearly indicate a pathway leading away from it, the authors did not propose one (13). However, our arm 1 residues connect their oxidized CP47 residues to the Mn cluster, showing that their results are part of a near-continuous path from the Mn cluster to the PSII surface. We performed a targeted analysis of CP47 residues in this region and found that two of these residues (CP47-M359 and E364) are also oxidized in our results. Three additional residues in this immediate vicinity are also oxidized (CP47-F362, M370, and T371). These five residues extend the correspondence between the arm 1 residues and channel 3 (26) and have been included in Fig. 4. The resulting arm 1 pattern shows a formation that originates at the Mn cluster, radiates outward along the channel 3 path for ~20 Å, and then diverges in two directions: The first continues an additional ~8 Å along the final segment of channel 3, and the second extends ~13 Å along a previously unidentified pathway (Fig. 5B). This formation seems to describe two different water intake pathways that merge and lead to a common destination.
Frankel et al. (13) also detected four oxidized CP43 residues within 15 Å of the Mn cluster and proposed that these residues constitute a part of an ROS exit channel leading from the cluster to the surface of PSII. However, they did not find additional oxidized residues that could complete this channel and trace a path all the way to the surface of the complex. In contrast, the oxidized CP43 residues we identified on arm 3 do lead in a nearly continuous fashion all the way from the Mn cluster to the surface of PSII, supporting and elaborating on the original proposal by Frankel et al. (13). The arm 3 residues include two (T354 and M355) of the four identified in the earlier study. Superimposing the two studies’ results shows that our arm 3 residues “fill in the gaps” between the oxidized CP43 residues in the study by Frankel et al. (13) and the Mn cluster (Fig. 5C). Their results strengthen the geometrical formation of the arm 3 residues we observed, showing a path of oxidized residues that originates at the Mn cluster and widens somewhat as it radiates outward toward the surface.
Bricker and co-workers (48) studied their system further by performing synchrotron radiolysis of water and exposing their PSII sample to the resulting HO• species for 0, 4, 8, or 16 s. They reasoned that, for a residue to be oxidized in this experiment, it must be in contact with water, from which HO• are generated. Mapping buried oxidized residues on the PSII crystal structure could therefore indicate the location of water channels within PSII. Only 5 (14%) of the 36 oxidized residues that we detected were found to be oxidized in their study before the radiolysis experiment (0-s irradiation). After 4, 8, or 16 s of irradiation, however, they found oxidation at a total of 23 (64%) of the 36 oxidized residues that we detected. Of these, 14 are buried, indicating that at least 14 of the 23 buried oxidized residues that we detected are in contact with internal water molecules. As has been pointed out, water molecules are dispersed widely throughout the lumenal side of PSII and most likely do not all comprise substrate channels, but this finding nonetheless supports our above conclusions that our oxidized residue formations represent water-filled pathways within PSII. This experiment uses HO• generated from solvent water that could travel inward toward the Mn cluster, whereas our experiment uses HO• generated in situ near the Mn cluster and that traveled outward. The correspondence between the results supports the assertion that the oxidized residue formations represent water channels connecting the Mn cluster with the bulk solvent.
We note that Wang (56) has recently reported that careful analysis of the PSII crystal structure 3ARC (5) reveals that around 10% of the residues are oxidized. Some of these residues overlap with the ones we have experimentally determined to be oxidized, and it is likely that at least some reflect oxidative damage incurred by PSII. However, as suggested in this report, many of these oxidized residues might have alternatively originated from the energetic x-rays used during crystallographic analysis and so do not necessarily reflect natural pathways of ROS diffusion away from the Mn cluster.
The PSII crystal structures reveal that there are four water ligands to the Mn cluster, and these are crucial for optimizing its hydrogen-bonding network to enable water oxidation (55, 57–59). All of these water molecules are positioned at the openings of arms 1 and 2, suggesting that these are the pathways that deliver these water molecules. The possibility of two distinct water delivery channels is also indicated by kinetic isotope exchange experiments (52, 60). The most current theoretical models of the water oxidation mechanism present two options for the key substrate water binding step during the S2→S3 transition (55). One option involves the substrate water forming an initial hydrogen bond with O4 and the other option with water-3 (W3). Both of these options are consistent with water delivery through arms 1 and 2 because arm 1 would deliver a water molecule to O4 and arm 2 to W3. The arm 2 residues we detected correspond to the large channel system (23), which has been proposed to deliver the water molecule to W3 according to the latter option. Our results provide experimental evidence for the accessibility of this channel system. According to the former option, the narrow channel (23) has been proposed to deliver the substrate water molecule to O4. As mentioned above, we did not find strong evidence from oxidation patterns for the narrow channel, but we also cannot rule it out. However, our arm 1 pathway would deliver water molecules to the same end location at the Mn cluster as the narrow channel but via a different trajectory. We propose that water delivery to the O4 site could occur through arm 1 [which is highly similar to channel 3 in the study by Vassiliev et al. (26)] instead of the narrow channel. The Cl− ion bound ~6 Å from the Mn cluster is found within the arm 1 pathway and could partially restrict water access (Fig. 5B). Removal of Cl− is known to increase water accessibility to the active site, leading to incomplete oxidation and ROS formation (11, 61). Substrate water delivery occurring through the arm 1 pathway would thus provide a plausible explanation for how Cl− restricts water access to the active site to promote efficient water oxidation.
In conclusion, we have found that the total number of oxidized residues in the PSII proteins D1, D2, and CP43 increases by around 60% following light incubation. The D1 protein showed the most pronounced increase (threefold), consistent with the fact that it undergoes photodamage the fastest (1) and has the shortest half-life of all PSII proteins (30). The lumen-side oxidized residues we detected are arranged in three nearly continuous arms originating at the Mn cluster and radiating outward all the way to the surface of PSII. Because the major oxidizing species is likely HO•, with transport properties highly similar to those of water, our oxidized residue formations indicate accessible water channels within PSII. This study represents the most comprehensive experimental characterization to date of oxidized residues in PSII, providing new information and also synthesizing results from multiple earlier studies in a single experiment. Our arm 1 pathway matches a formation found recently in spinach PSII (11), our arm 3 pathway extends the description of a previously proposed formation in a different study in spinach PSII (13), and our arm 2 pathway has not been proposed previously through experiment.
Channel 3 and channels 4a/4ai (26, 27) [large channel in the study by Ho and Styring (23)], which are putative water channels identified in computational studies, are highly similar to our arm 1 and arm 2 pathways, respectively (Table 1). Both are well-positioned to deliver ligated water molecules to their sites around the Mn cluster, and we propose that both serve this function. Previous proposals that the large channel plays this role are consistent with our results. Our results did not support a water delivery function for the back or broad channels, which is consistent with previous analyses using other methods (27, 52–54). The narrow channel has also been proposed to deliver water (55), and we cannot rule out this possibility, but we propose that our arm 1 pathway may serve this role instead by delivering water to the same end location at the Mn cluster. Water delivery through arm 1 provides a molecular basis for the increased water accessibility to the Mn cluster caused by Cl− removal, as Cl− is found within arm 1. Most of the arm 3 pathway does not overlap strongly with channels from computational studies, although Frankel and co-workers (13, 48) previously found overlapping oxidized residues. The functional significance of this pathway remains to be investigated.
MATERIALS AND METHODS
Cyanobacterial culture and PSII purification
The HT3 (His47) strain of Synechocystis 6803 was a gift from T. Bricker (Louisiana State University, Baton Rouge, LA) (62). The cultures were grown in BG11 medium at 30°C under 30-μmol photons m−2 s−1. Histidine-tagged PSII complexes were purified as described previously (63) with minor modifications and stored in 25% glycerol, 10 mM MgCl2, 5 mM CaCl2, 50 mM MES buffer (pH 6.0) (RB buffer).
Sample preparation and proteolytic digestion
Two His47 PSII samples (two biological replicates) consisting of 1 μg of chlorophyll a were precipitated using the 2D cleanup kit (GE Healthcare) to remove salt and detergent. For the light-incubated condition, the PSII samples were incubated at 30°C and 55-μmol photons m−2 s−1 for 6 hours before precipitation. The dark-incubated samples were treated identically to the light-incubated samples, but they were wrapped in aluminum foil during the incubation. After precipitation, the protein pellets were resuspended in 15 μl of 8 M urea. Disulfide reduction was performed by incubation in a final concentration of 2.5 mM tris(2-carboxyethyl)phosphine for 30 min at 37°C. Alkylation of cysteines was then performed by the addition of iodoacetamide to a final concentration of 5 mM, with incubation at room temperature in the dark for 30 min. Digestion was performed in two stages: First, lysyl endopeptidase was added at around 1 μg /20 μg protein, followed by incubation for 2 hours at 37°C. After 2 hours, samples were diluted 1:8 in 100 mM tris (pH 8.5) and 10 mM CaCl2 trypsin buffer, with a final ratio of around 1-μg trypsin/20-μg protein, and incubated at 37°C overnight. The digestion was stopped by acidifying the sample to a final concentration of 1% formic acid. Samples were then analyzed by LC-MS/MS.
Liquid chromatography–tandem mass spectrometry
Samples were analyzed on a QE Plus mass spectrometer (Thermo Fisher), as described by Weisz et al. (64), with the following adjustments. For LC, a linear 82-min gradient from 2 to 43% solvent B (80% acetonitrile, 20% water, and 0.1% formic acid) was used, followed by a linear 30-min gradient from 43 to 98% solvent B, then a 5-min hold at 95% solvent B. Automatic gain control for MS/MS was set at 2.5 × 105 ions and a maximal injection time of 100 ms. Charge states other than 2 to 7 were excluded, and each sample was analyzed in triplicate.
Data analysis
The raw LC-MS/MS files were searched for Synechocystis PSII proteins using PEAKS (version 7.0, Bioinformatics Solutions Inc.), with the built-in decoy fusion database used for false discovery rate calculation (65). The oxidative modifications listed in table S1 were included in the search, as well as carbamidomethylation (C, D, H, K, and E, peptide N terminus), +57.0215 Da; deamidation (N and Q), +0.9840 Da; acetylation of protein N terminus, +42.0106 Da; and carbamylation (K, peptide N terminus), +43.0058 Da. Additional search parameters are as follows: precursor-ion mass tolerance, 10.0 parts per million (ppm); fragment-ion mass tolerance, 0.02 Da; maximum three variable modifications per peptide; maximum three missed cleavages; maximum one nonspecific cleavage. Peptides were identified with a 0.1% false discovery rate (−10 log P ≥ 40). For every candidate oxidized residue that met the 0.1% false discovery threshold, product-ion (MS/MS) spectra were inspected manually to confirm data quality, and spectra were rejected if the oxidation could not be localized to a single residue, if the precursor-ion mass error was greater than 5.0 ppm, or if the peptide was a product of a nonspecific cleavage. MS data are available through Chorus (http://chorusproject.org), project #1388. Protein visualization was performed using VMD (66).
Supplementary Material
Acknowledgments
We thank Dr. Hao Zhang for the helpful discussions. Funding: This work was supported by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, United States Department of Energy (DOE) (grant DE-FG02-99ER20350 to H.B.P.); the Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the U.S. DOE, Office of Basic Energy Sciences (grant DE-SC 0001035 to H.B.P. and M.L.G.); and the NIH (grant 2P41GM103422 to M.L.G.). Author contributions: D.A.W., M.L.G., and H.B.P. conceived and designed the experiments. D.A.W. performed sample collection and preparation. D.A.W. performed data acquisition. D.A.W. performed data analysis. D.A.W., M.L.G., and H.B.P. wrote and edited the manuscript. H.B.P. supervised the project. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The proteomic data have been deposited and are publicly available online at http://chorusproject.org, project #1388, under the title, “Oxidative modifications of Photosystem II in Synechocystis 6803.” All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/eaao3013/DC1
fig. S1. Example 1 of product-ion (MS/MS) spectra detected that identify oxidative modifications of PSII residues.
fig. S2. Example 2 of product-ion (MS/MS) spectra detected that identify oxidative modifications of PSII residues.
fig. S3. Example 3 of product-ion (MS/MS) spectra detected that identify oxidative modifications of PSII residues.
table S1. Oxidative modifications included as variable modifications in the MS database searches.
table S2. Oxidative modifications of PSII detected in this study.
table S3. List of oxidized peptides detected in this study.
table S4. Oxidized residues that correspond to channels determined in previous MD studies.
movie S1. Rotation of Fig. 4A about the y axis.
Reference (67)
REFERENCES AND NOTES
- 1.Nickelsen J., Rengstl B., Photosystem II assembly: From cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Weisz D. A., Gross M. L., Pakrasi H. B., The use of advanced mass spectrometry to dissect the life-cycle of Photosystem II. Front. Plant Sci. 7, 617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinz S., Liauw P., Nickelsen J., Nowaczyk M., Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857, 274–287 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Järvi S., Suorsa M., Aro E.-M., Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847, 900–909 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Umena Y., Kawakami K., Shen J.-R., Kamiya N., Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Pospíšil P., Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 7, 1950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yadav D. K., Pospíšil P., Role of chloride ion in hydroxyl radical production in photosystem II under heat stress: Electron paramagnetic resonance spin-trapping study. J. Bioenerg. Biomembr. 44, 365–372 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Wydrzynski T., Hillier W., Messinger J., On the functional significance of substrate accessibility in the photosynthetic water oxidation mechanism. Physiol. Plant. 96, 342–350 (1996). [Google Scholar]
- 9.Thompson L. K., Blaylock R., Sturtevant J. M., Brudvig G. W., Molecular basis of the heat denaturation of photosystem II. Biochemistry 28, 6686–6695 (1989). [DOI] [PubMed] [Google Scholar]
- 10.Pospíšil P., Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta 1787, 1151–1160 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Kale R., Hebert A. E., Frankel L. K., Sallans L., Bricker T. M., Pospíšil P., Amino acid oxidation of the D1 and D2 proteins by oxygen radicals during photoinhibition of Photosystem II. Proc. Natl. Acad. Sci. U.S.A. 114, 2988–2993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pospíšil P., The role of metals in production and scavenging of reactive oxygen species in Photosystem II. Plant Cell Physiol. 55, 1224–1232 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Frankel L. K., Sallans L., Limbach P. A., Bricker T. M., Identification of oxidized amino acid residues in the vicinity of the Mn4CaO5 cluster of Photosystem II: Implications for the identification of oxygen channels within the photosystem. Biochemistry 51, 6371–6377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasson T. M. D., Rexroth S., Barry B. A., Light-induced oxidative stress, N-formylkynurenine, and oxygenic photosynthesis. PLOS ONE 7, 42220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreaden T. M., Chen J., Rexroth S., Barry B. A., N-formylkynurenine as a marker of high light stress in photosynthesis. J. Biol. Chem. 286, 22632–22641 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson L. B., Maderia M., Ouellete A. J. A., Putnam-Evans C., Higgins L., Krick T., MacCoss M. J., Lim H., Yates J. R. III, Barry B. A., Posttranslational modifications in the CP43 subunit of photosystem II. Proc. Natl. Acad. Sci. U.S.A. 99, 14676–14681 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma J., Panico M., Shipton C. A., Nilsson F., Morris H. R., Barber J., Primary structure characterization of the photosystem II D1 and D2 subunits. J. Biol. Chem. 272, 33158–33166 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Tani K., Fujiyoshi Y., Water channel structures analysed by electron crystallography. Biochim. Biophys. Acta 1840, 1605–1613 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Yang L., Skjevik Å. A., Du W.-G. H., Noodleman L., Walker R. C., Gӧtz A. W., Water exit pathways and proton pumping mechanism in B-type cytochrome c oxidase from molecular dynamics simulations. Biochim. Biophys. Acta 1857, 1594–1606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugitani R., Stuchebrukhov A. A., Molecular dynamics simulation of water in cytochrome c oxidase reveals two water exit pathways and the mechanism of transport. Biochim. Biophys. Acta 1787, 1140–1150 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klvana M., Pavlova M., Koudelakova T., Chaloupkova R., Dvorak P., Prokop Z., Stsiapanava A., Kuty M., Kuta-Smatanova I., Dohnalek J., Kulhanek P., Wade R. C., Damborsky J., Pathways and mechanisms for product release in the engineered haloalkane dehalogenases explored using classical and random acceleration molecular dynamics simulations. J. Mol. Biol. 392, 1339–1356 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Murray J. W., Barber J., Structural characteristics of channels and pathways in photosystem II including the identification of an oxygen channel. J. Struct. Biol. 159, 228–237 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Ho F. M., Styring S., Access channels and methanol binding site to the CaMn4 cluster in photosystem II based on solvent accessibility simulations, with implications for substrate water access. Biochim. Biophys. Acta 1777, 140–153 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Gabdulkhakov A., Guskov A., Broser M., Kern J., Müh F., Saenger W., Zouni A., Probing the accessibility of the Mn4Ca cluster in photosystem II: Channels calculation, noble gas derivatization, and cocrystallization with DMSO. Structure 17, 1223–1234 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Vassiliev S., Comte P., Mahboob A., Bruce D., Tracking the flow of water through photosystem II using molecular dynamics and streamline tracing. Biochemistry 49, 1873–1881 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Vassiliev S., Zaraiskaya T., Bruce D., Exploring the energetics of water permeation in photosystem II by multiple steered molecular dynamics simulations. Biochim. Biophys. Acta 1817, 1671–1678 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Vassiliev S., Zaraiskaya T., Bruce D., Molecular dynamics simulations reveal highly permeable oxygen exit channels shared with water uptake channels in photosystem II. Biochim. Biophys. Acta 1827, 1148–1155 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Zaraiskaya T., Vassiliev S., Bruce D., Discovering oxygen channel topology in photosystem II using implicit ligand sampling and wavefront propagation. J. Comput. Sci. 5, 549–555 (2014). [Google Scholar]
- 29.Gabdulkhakov A. G., Kljashtorny V. G., Dontsova M. V., Molecular dynamics studies of pathways of water movement in cyanobacterial photosystem II. Crystallogr. Rep. 60, 83–89 (2015). [Google Scholar]
- 30.Gabdulkhakov A. G., Kljashtorny V. G., Dontsova M. V., Analysis of molecular oxygen exit pathways in cyanobacterial photosystem II: Molecular dynamics studies. Crystallogr. Rep. 60, 884–888 (2015). [Google Scholar]
- 31.Cardona T., Sedoud A., Cox N., Rutherford A. W., Charge separation in photosystem II: A comparative and evolutionary overview. Biochim. Biophys. Acta 1817, 26–43 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Linke K., Ho F. M., Water in Photosystem II: Structural, functional, and mechanistic considerations. Biochim. Biophys. Acta 1837, 14–32 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Klauss A., Haumann M., Dau H., Alternating electron and proton transfer steps in photosynthetic water oxidation. Proc. Natl. Acad. Sci. U.S.A. 109, 16035–16040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liuni P., Zhu S., Wilson D. J., Oxidative protein labeling with analysis by mass spectrometry for the study of structure, folding, and dynamics. Antioxid. Redox Signal. 21, 497–510 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Wang L., Chance M. R., Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting. Anal. Chem. 83, 7234–7241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu G., Chance M. R., Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Hambly D. M., Gross M. L., Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J. Am. Soc. Mass Spectrom. 16, 2057–2063 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Cheal S. M., Ng M., Barrios B., Miao Z., Kalani A. K., Meares C. F., Mapping protein–protein interactions by localized oxidation: Consequences of the reach of hydroxyl radical. Biochemistry 48, 4577–4586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee S., Young N. L., Whetstone P. A., Cheal S. M., Benner W. H., Lebrilla C. B., Meares C. F., Method to site-specifically identify and quantitate carbonyl end products of protein oxidation using oxidation-dependent element coded affinity tags (O-ECAT) and nanoliquid chromatography Fourier transform mass spectrometry. J. Proteome Res. 5, 539–547 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Bridgewater J. D., Lim J., Vachet R. W., Using metal-catalyzed oxidation reactions and mass spectrometry to identify amino acid residues within 10 Å of the metal in Cu-binding proteins. J. Am. Soc. Mass Spectrom. 17, 1552–1559 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Mukherjee S., Sousa R., Use of site-specifically tethered chemical nucleases to study macromolecular reactions. Biol. Proced. Online 5, 78–89 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winterbourn C. C., Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Takamoto K., Chance M. R., Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Nakamura T., Dohmae N., Takio K., Characterization of a digested protein complex with quantitative aspects: An approach based on accurate mass chromatographic analysis with Fourier transform-ion cyclotron resonance mass spectrometry. Proteomics 4, 2558–2566 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Aro E.-M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N., Rintamäki E., Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56, 347–356 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Liu H., Chen J., Huang R. Y.-C., Weisz D., Gross M. L., Pakrasi H. B., Mass spectrometry-based footprinting reveals structural dynamics of Loop E of the chlorophyll-binding protein CP43 during photosystem II assembly in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 288, 14212–14220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H., Zhang H., Niedzwiedzki D., Prado M., He G., Gross M. L., Blankenship R. E., Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 342, 1104–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frankel L. K., Sallans L., Bellamy H., Goettert J. S., Limbach P. A., Bricker T. M., Radiolytic mapping of solvent-contact surfaces in Photosystem II of higher plants: Experimental identification of putative water channels within the photosystem. J. Biol. Chem. 288, 23565–23572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao D. C. I., Brune D. C., Vermaas W. F. J., Lifetimes of photosystem I and II proteins in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 586, 169–173 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Verrastro I., Pasha S., Jensen K. T., Pitt A. R., Spickett C. M., Mass spectrometry-based methods for identifying oxidized proteins in disease: Advances and challenges. Biomolecules 5, 378–411 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kupitz C., Basu S., Grotjohann I., Fromme R., Zatsepin N. A., Rendek K. N., Hunter M. S., Shoeman R. L., White T. A., Wang D., James D., Yang J.-H., Cobb D. E., Reeder B., Sierra R. G., Liu H., Barty A., Aquila A. L., Deponte D., Kirian R. A., Bari S., Bergkamp J. J., Beyerlein K. R., Bogan M. J., Caleman C., Chao T.-C., Conrad C. E., Davis K. M., Fleckenstein H., Galli L., Hau-Riege S. P., Kassemeyer S., Laksmono H., Liang M., Lomb L., Marchesini S., Martin A. V., Messerschmidt M., Milathianaki D., Nass K., Ros A., Roy-Chowdhury S., Schmidt K., Seibert M., Steinbrener J., Stellato F., Yan L., Yoon C., Moore T. A., Moore A. L., Pushkar Y., Williams G. J., Boutet S., Doak R. B., Weierstall U., Frank M., Chapman H. N., Spence J. C. H., Fromme P., Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 513, 261–265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogt L., Vinyard D. J., Khan S., Brudvig G. W., Oxygen-evolving complex of Photosystem II: An analysis of second-shell residues and hydrogen-bonding networks. Curr. Opin. Chem. Biol. 25, 152–158 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Ishikita H., Saenger W., Loll B., Biesiadka J., Knapp E.-W., Energetics of a possible proton exit pathway for water oxidation in photosystem II. Biochemistry 45, 2063–2071 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Bondar A.-N., Dau H., Extended protein/water H-bond networks in photosynthetic water oxidation. Biochim. Biophys. Acta 1817, 1177–1190 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Askerka M., Brudvig G. W., Batista V. S., The O2-evolving complex of Photosystem II: Recent insights from quantum mechanics/molecular dynamics (QM/MM), extended X-ray absorption fine structure (EXAFS), and femtosecond X-ray crystallography data. Acc. Chem. Res. 50, 41–48 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Wang J., X-ray radiation-induced addition of oxygen atoms to protein residues. Protein Sci. 25, 1407–1419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young I. D., Ibrahim M., Chatterjee R., Gul S., Fuller F., Koroidov S., Brewster A. S., Tran R., Alonso-Mori R., Kroll T., Michels-Clark T., Laksmono H., Sierra R. G., Stan C. A., Hussein R., Zhang M., Douthit L., Kubin M., de Lichtenberg C., Long Vo P., Nilsson H., Cheah M. H., Shevela D., Saracini C., Bean M. A., Seuffert I., Sokaras D., Weng T.-C., Pastor E., Weninger C., Fransson T., Lassalle L., Bräuer P., Aller P., Docker P. T., Andi B., Orville A. M., Glownia J. M., Nelson S., Sikorski M., Zhu D., Hunter M. S., Lane T. J., Aquila A., Koglin J. E., Robinson J., Liang M., Boutet S., Lyubimov A. Y., Uervirojnangkoorn M., Moriarty N. W., Liebschner D., Afonine P. V., Waterman D. G., Evans G., Wernet P., Dobbek H., Weis W. I., Brunger A. T., Zwart P. H., Adams P. D., Zouni A., Messinger J., Bergmann U., Sauter N. K., Kern J., Yachandra V. K., Yano J., Structure of photosystem II and substrate binding at room temperature. Nature 540, 453–457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X., Siegbahn P. E. M., Alternative mechanisms for O2 release and O–O bond formation in the oxygen evolving complex of photosystem II. Phys. Chem. Chem. Phys. 17, 12168–12174 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Siegbahn P. E. M., Water oxidation mechanism in photosystem II, including oxidations, proton release pathways, O—O bond formation, and O2 release. Biochim. Biophys. Acta 1827, 1003–1019 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Hillier W., Wrydrzynski T., Substrate water interactions within the photosystem II oxygen evolving complex. Phys. Chem. Chem. Phys. 6, 4882–4889 (2004). [Google Scholar]
- 61.Arató A., Bondarava N., Krieger-Liszkay A., Production of reactive oxygen species in chloride- and calcium-depleted photosystem II and their involvement in photoinhibition. Biochim. Biophys. Acta 1608, 171–180 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Bricker T. M., Morvant J., Masri N., Sutton H. M., Frankel L. K., Isolation of a highly active Photosystem II preparation from Synechocystis 6803 using a histidine-tagged mutant of CP 47. Biochim. Biophys. Acta 1409, 50–57 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., Pakrasi H. B., Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Weisz D. A., Liu H., Zhang H., Thangapandian S., Tajkhorshid E., Gross M. L., Pakrasi H. B., Mass spectrometry-based cross-linking study shows that the Psb28 protein binds to cytochrome b559 in Photosystem II. Proc. Natl. Acad. Sci. U.S.A. 114, 2224–2229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G. A., Ma B., PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 11, M111.010587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Humphrey W., Dalke A., Schulten K., VMD: Visual Molecular Dynamics. J. Mol. Graphics 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 67.Renzone G., Salzano A. M., Arena S., D’Ambrosio C., Scaloni A., Mass spectrometry-based approaches for structural studies on protein complexes at low-resolution. Curr. Proteomics 4, 1–16 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/11/eaao3013/DC1
fig. S1. Example 1 of product-ion (MS/MS) spectra detected that identify oxidative modifications of PSII residues.
fig. S2. Example 2 of product-ion (MS/MS) spectra detected that identify oxidative modifications of PSII residues.
fig. S3. Example 3 of product-ion (MS/MS) spectra detected that identify oxidative modifications of PSII residues.
table S1. Oxidative modifications included as variable modifications in the MS database searches.
table S2. Oxidative modifications of PSII detected in this study.
table S3. List of oxidized peptides detected in this study.
table S4. Oxidized residues that correspond to channels determined in previous MD studies.
movie S1. Rotation of Fig. 4A about the y axis.
Reference (67)


