Abstract
DDX3X is a conserved DEAD-box RNA helicase involved in translation initiation and other processes of RNA metabolism. Mutations in human DDX3X and deregulation of its expression are linked to tumorigenesis and intellectual disability. The protein is also targeted by diverse viruses. Previous studies demonstrated helicase and NTPase activities for DDX3X, but important biochemical features of the enzyme remain unclear. Here, we systematically characterize enzymatic activities of human DDX3X and compare these to its closely related S. cerevisiae ortholog Ded1p. We show that DDX3X, like Ded1p, exclusively utilizes adenosine triphosphates to unwind helices, oligomerizes to function as efficient RNA helicase, and does not unwind DNA duplexes. The ATPase activity of DDX3X is markedly stimulated by RNA and weaker by DNA, although DNA binds to the enzyme. For RNA unwinding, DDX3X shows a greater preference than Ded1p for substrates with unpaired regions 3′ to the duplex over those with 5′ unpaired regions. DDX3X separates longer RNA duplexes faster than Ded1p and is less potent than Ded1p in facilitating strand annealing. Our results reveal that the biochemical activities of human DDX3X are typical for DEAD-box RNA helicases, but diverge quantitatively from its highly similar S. cerevisiae ortholog Ded1p.
Graphical abstract
INTRODUCTION
Human DDX3X belongs to the DEAD-box RNA helicase family of enzymes, which bind and remodel RNA in an ATP-dependent fashion (1, 2). Like all DEAD-box RNA helicases, DDX3X contains a helicase core with two similar RecA-like domains that harbor 13 characteristic helicase sequence motifs, including the eponymous motif II, which reads D–E-A–D in single letter code (3, 4). The helicase core of DDX3X is flanked by an N-terminus with a nuclear export signal and regions of low sequence complexity and a C-terminus that also contains regions of low sequence complexity (Suppl. Fig.S1) (2, 4).
Virtually all sequenced eukaryotes encode one or more highly similar DDX3X orthologs (2, 5). DDX3X shows 51% sequence identity to its S. cerevisiae ortholog Ded1p (2, 5) (Suppl. Fig.S1). All DDX3X orthologs have N- and C-terminal regions of low sequence complexity. In these regions, the sequence varies more than in the helicase core (2, 5). Crystal structures of the helicase core of DDX3X have been reported (3, 4, 6). A recent structure revealed defined interactions between the two RecA-like domains of DDX3X that are specific to the DDX3X/Ded1p subfamily (4). DDX3X and several of its orthologs have been shown to function in translation initiation, mRNA export and in other aspects of RNA metabolism (7–13). DDX3X has also been implicated in signal transduction and is part of cytoplasmic ribonucleoprotein-granules (14–16).
DDX3X is encoded on the X-chromosome (2). Mutations in DDX3X are linked to developmental disorders and intellectual disability (17, 18). Deregulation of DDX3X expression and mutations in the helicase core of DDX3X are associated with tumors, including medulloblastoma, T-cell lymphoma, lung, colorectal, breast, oral, liver and prostate cancer (19–25), and DDX3X is a potential therapeutic target for cancer treatment (26). DDX3X is also directly targeted by multiple pathogenic viruses including HIV, HCV, other flaviviridae, poxviridae, and HBV (27). The enzyme has been suggested as candidate for the development of broad spectrum antiviral drugs (28).
The central role of DDX3X in RNA biology and its link to various diseases emphasize the need to clearly define the biochemical activities of the enzyme. Previous work demonstrated ATPase and helicase functions for DDX3X (3, 4). It was also reported that DDX3X displays high levels of RNA-independent ATPase, DNA-stimulated ATPase, and DNA unwinding activities (29, 30), features that would markedly differ from other DEAD-box helicases. However, no systematic investigation of the biochemical characteristics of DDX3X has yet been reported.
Here, we systematically characterize the basic biochemical activities of DDX3X and compare obtained parameters to those of the S. cerevisiae ortholog Ded1p. We find extensive similarities between DDX3X and Ded1p, including the exclusive utilization of adenosine triphosphates to unwind RNA-RNA and RNA-DNA helices, inability to separate DNA-DNA duplexes, and the requirement of oligomerization for efficient duplex unwinding. In addition, we show that the ATPase activity of DDX3X depends on nucleic acids. RNA stimulates ATPase activity of DDX3X markedly more than DNA, although DNA readily binds the enzyme. However, DDX3X separates shorter duplexes (≥ 16 bp) slower than Ded1p, and longer duplexes faster. Compared to Ded1p, DDX3X shows a more pronounced preference for substrates with unpaired regions 3′ to the duplex than to those with 5′ unpaired regions. Moreover, DDX3X is less potent than Ded1p in facilitating strand annealing. Our data reveal biochemical features of DDX3X that are typical for DEAD-box RNA helicases, but also show differences between DDX3X and Ded1p, highlighting that even highly similar orthologs vary in their activities.
RESULTS
DDX3X harbors weaker strand annealing activity than Ded1p
To characterize the basic biochemical activities of human DDX3X, we first tested unwinding of a 3′ tailed RNA substrate under pre-steady state conditions (Fig.1). Unwinding was seen with, but not without ATP (Fig.1). At identical concentrations and reaction conditions, Ded1p unwound the duplex at a similar rate as DDX3X (Fig.1). The data demonstrate clear RNA unwinding activity of DDX3X.
Figure 1. RNA unwinding by DDX3X and Ded1p.
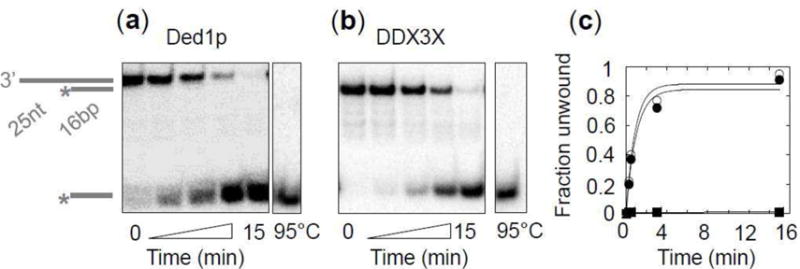
(a) Representative PAGE for unwinding reactions with Ded1p and (b) DDX3X (ATP: 2 mM, RNA: 0.5 nM, Ded1p, DDX3X: 75 nM). Cartoons mark duplex RNA substrate and unwound product, asterisks show the radiolabel.
(c) Representative unwinding time courses for Ded1p (open circles) and DDX3X (filled circles) for the RNA substrate and the reaction conditions shown in panels (a,b). The squares show a representative reaction without ATP for DDX3X. Lines mark fits to the integrated first order rate equation (DDX3X: kunw = 0.83 ± 0.37 min−1, Ded1p: kunw = 0.95 ± 0.39 min−1).
Ded1p promotes strand annealing, in addition to duplex unwinding (31). We therefore examined whether DDX3X also catalyzes strand annealing (Fig.2). Without ATP, DDX3X increased the bimolecular rate constant of duplex formation by roughly a factor of two over the basal level (Fig.2a,b). In contrast, Ded1p increased the bimolecular rate constant for duplex formation by approximately three orders of magnitude (Fig.2a,b), consistent with previous observations (31). These data suggest that Ded1p is a markedly more potent strand annealer than DDX3X, in the absence of ATP.
Figure 2. Strand annealing by DDX3X and Ded1p.
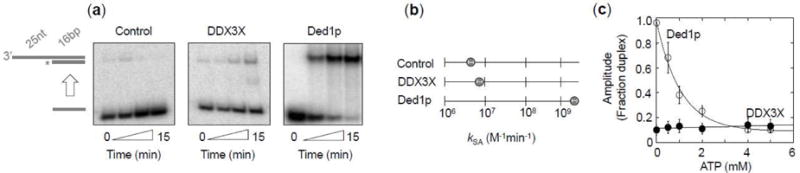
(a) Representative PAGE for strand annealing reactions (control: no protein, DDX3X and Ded1p: 150 nM). Cartoons on the left show unpaired and duplex RNA substrate, asterisks mark the radiolabel.
(b) Second order rate constants for strand annealing for reactions without proteins (kSA = 4.1 ± 0.1·106 M−1min−1), with DDX3X (150 nM, kSA = 8.9 ± 0.2·106 M−1min−1) and with Ded1p (150 nM, kSA = 5.4 ± 0.6·109 M−1min−1). The rate constants refer to the formation of the duplex from the individual RNA strands. Data points represent an average of three independent experiments, error bars mark one standard deviation.
(c) Dependence of the reaction amplitude of annealing reactions on the ATP concentration for Ded1p (150 nM, open circles) and DDX3X (150 nM, filled circles). Amplitudes were determined from the time courses at each ATP concentration after 15 minutes. Data points represent an average of three independent experiments, error bars mark one standard deviation. The lines show the trend.
To assess strand annealing activity with ATP, we measured amplitudes for strand annealing reactions with increasing ATP concentrations (Fig.2c). For Ded1p, the amplitude decreased with increasing concentrations of ATP (Fig.2c), consistent with previous reports (31). This amplitude decrease reflects a modest decrease in the rate constant of strand annealing and a large enhancement of the rate constant for duplex unwinding with increasing ATP concentration (31). For DDX3X, the amplitude for strand annealing reactions was largely insensitive to the ATP concentration, suggesting comparably little impact of ATP on the ratio between strand annealing and unwinding (Fig.2c). Annealing activity of DDX3X did not significantly increase between concentrations of 150 and 600 nM, the highest experimentally accessible concentration (data not shown). Collectively, the data show that DDX3X harbors significantly weaker strand annealing activity than Ded1p.
DDX3X preferentially unwinds substrates with 3′ single-stranded overhangs
We next examined unwinding activity by DDX3X in more detail. We measured unwinding rate constants for two substrates with identical 16 bp duplexes and 3′ and 5′ unpaired tails (25 nt, Fig.3). For the 3′ tailed substrate, unwinding rate constants scaled in a sigmoidal fashion with the DDX3X concentration (Fig.3a). A sigmoidal scaling was also seen for Ded1p (Fig.3b), consistent with previous observations (31, 32). Of note, unwinding rate constants at Ded1p concentrations above 300 nM, which likely exceed kunw > 6 min−1 at the reaction conditions, could not be reliably measured with the manual experimental approach used here. For this reason, unwinding rate constants at enzyme saturation could not be determined, and functional binding isotherms for Ded1p could not be quantitatively evaluated.
Figure 3. Impact of tail polarity on unwinding by DDX3X and Ded1p.
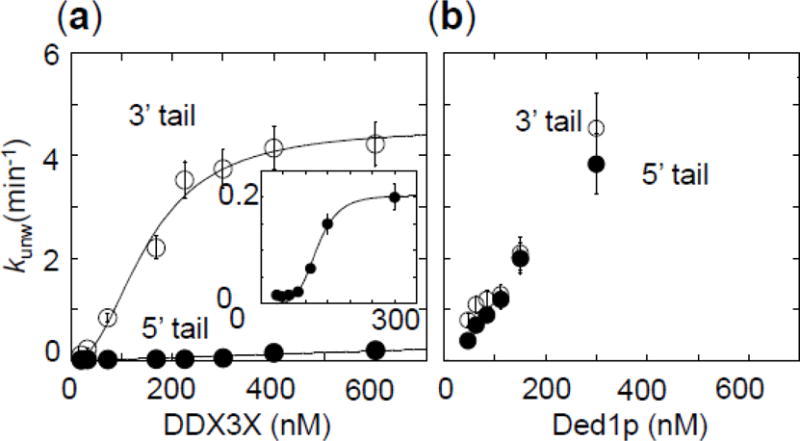
(a) Functional binding isotherms for DDX3X for 16 bp substrates with 25 nt unpaired tails 3′ (open circles) or 5′ (closed circles) to the duplex. The inset shows the isotherm for the 5′ tailed substrate with magnified y-axis. Data points represent averages of at least three independently measured unwinding rate constants, error bars mark one standard deviation. Lines show best fits to the Hill equation (3′ tailed substrate: K1/2 = 112 ± 6 nM, kunwmax = 4.2 ± 0.2 min−1, H = 3.1 ± 0.6 ; 5′ tailed substrate: K1/2 = 127 ± 20 nM, kunwmax = 0.20 ± 0.02 min−1, H = 3.2 ± 1.2.
(b) Functional binding isotherms for Ded1p for 16 bp substrates with 25 nt unpaired tails 3′ (open circles) or 5′ (closed circles) to the duplex. Data points represent averages of at least three independently measured unwinding rate constants, error bars mark one standard deviation.
The data suggest that DDX3X unwinds 3′-tailed RNA duplexes as an oligomer containing at least three protomers (H = 3.1 ± 0.6, Fig.3a). This observation mirrors previous data for Ded1p (32). The maximal unwinding rate constant at enzyme saturation for the 5′ tailed substrate was lower by a factor of roughly 25, compared to the 3′ tailed substrate (Fig.3a). However, functional affinities of DDX3X were similar for 5′ and 3′ tailed substrates, and the sigmoidal functional binding isotherm suggests that DDX3X unwinds the 5′ tailed substrate also as an oligomer. The marked reduction in the maximal unwinding rate constant for the 5′ tailed substrate contrasts with Ded1p, where unwinding of 5′ and 3′ tailed substrates does not differ as markedly as with DDX3X. (Fig.3b).
Strand separation rate constants scale with duplex stability
Unwinding rate constants for virtually all examined DEAD-box helicases decrease with duplex length (1, 33, 34). However, the degree by which unwinding rate constants scale with duplex lengths varies for different enzymes (35, 36). To probe how unwinding rate constants scaled with duplex length for DDX3X, we measured unwinding rate constants for a series of substrates with duplex regions from 13 to 19 bp with 25 nt 3′ or 5′ tails (Fig.4a,b).
Figure 4. Impact of duplex length and stability on unwinding by DDX3X.
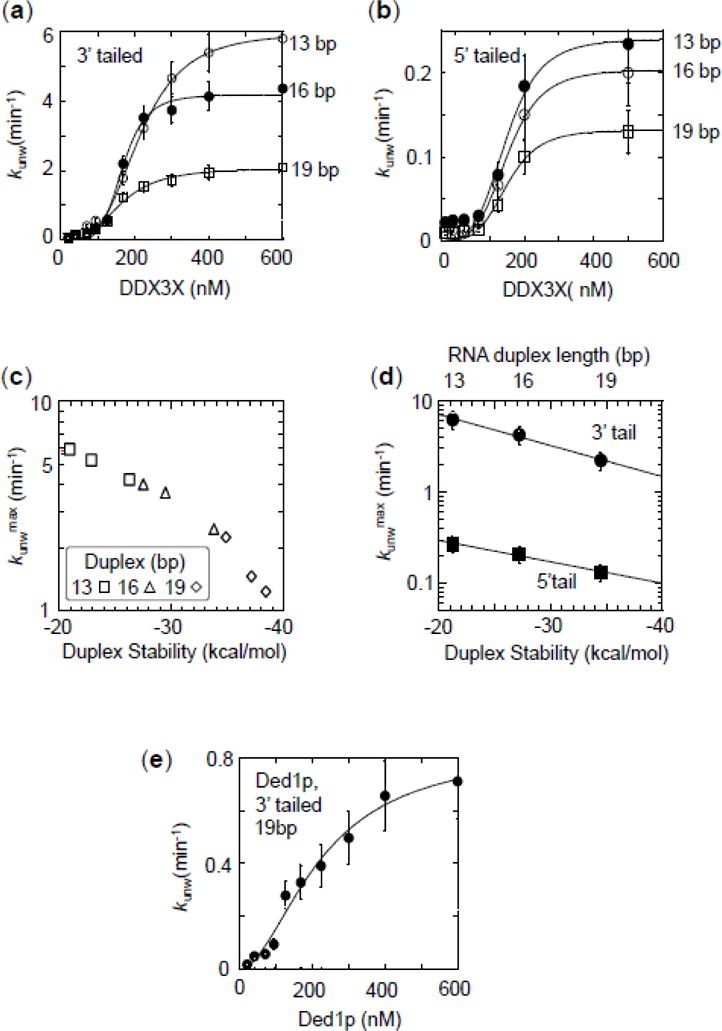
(a) Functional binding isotherms for DDX3X for substrates with 25 nt unpaired tails 3′ to duplexes with 13 (open circles), 16 (closed circles) and 19 bp (open squares). Data points represent averages of at least three independently measured unwinding rate constants, error bars mark one standard deviation. Lines show best fits to the Hill equation (K1/213bp = 165 ± 29 nM, H13bp = 3.1 ± 0.3, kunwmax(13bp) = 6.2 ± 0.2 min−1; K1/216bp = 118 ± 18 nM, H16 bp = 3.0 ± 0.2, kunwmax(16bp) = 4.2 ± 0.3 min−1; K1/219bp = 136 ± 30 nM, H19bp = 3.1 ± 0.2, kunwmax(19bp)= 2.1 ± 0.1 min−1).
(b) Functional binding isotherms for DDX3X for substrates with 25 nt unpaired tails 5′ to duplexes with 13 (open circles), 16 (closed circles) and 19 bp (open squares). Data points represent averages of at least three independently measured unwinding rate constants, error bars mark one standard deviation. Lines show best fits to the Hill equation (K1/213bp = 139 ± 21 nM, H13bp = 3.0 ± 0.2; kunwmax(13bp) = 0.29 ± 0.02 min−1; K1/216bp = 119 ± 35 nM, H16bp = 3.1 ± 0.2, kunwmax(16bp) = 0.21 ± 0.04 min−1; K1/219bp = 166 ± 18 nM, H19bp = 3.1 ± 0.3, kunwmax(19bp) = 0.13 ± 0.02 min−1).
(c) Correlation of maximal unwinding rate constants for 25 nt, 3′ tailed substrates with calculated duplex stabilities (x-axis) and duplex length (inset). For sequences of the substrates see Table 1, Materials and Methods. Data points represent averages of at least three independently measured maximal unwinding rate constants (kunwmax) at enzyme saturation, error bars one standard deviation. RNA stability was calculated according to Mathews et al. (52). Note the logarithmic scale of the y-axis. Data points represent kunwmax and RNA stability (ΔG) for individual substrates, as indicated in the inset.
(d) Dependence of maximal unwinding rate constants for DDX3X (circles: 25 nt 3′ tails, squares 25 nt 5′ tails). Data points represent rate constants reported in panels (a) and (b). Error bars mark the standard error for the respective calculated maximal rate constant. Note the logarithmic scale of the y-axis. Lines mark linear trends.
(e) Functional binding isotherms for Ded1p for the substrates with 19 bp (25 nt, 3′ tail). Data points represent averages of at least three independently measured unwinding rate constants, error bars mark one standard deviation. The line shows the best fit to the Hill equation (K1/2 = 175 ± 12 nM, kunwmax = 0.7 ± 0.1 min−1, H = 3.6 ± 0.3)
Unwinding rate constants for all substrates scaled in a sigmoidal fashion with the DDX3X concentration (Fig.4a,b). Functional affinities and Hill coefficients were similar for all substrates (Fig.4a,b). The maximal unwinding rate constant decreased with increasing duplex length (Fig.4a,b). The maximal unwinding rate constants were significantly lower for all 5′ tailed substrates, compared to equivalent 3′-tailed substrates (Fig.4b). Together, the observations indicate scaling of the maximal unwinding rate constants with duplex length, but insensitivity of functional affinities of DDX3X to duplex length. These characteristics are typical for DEAD-box helicases (1, 33, 34).
The maximal unwinding rate constant reflects the strand separation step (32). The observed scaling of maximal unwinding rate constants with duplex length is thought to result from an increase in duplex stability, which increases with duplex length (37). However, a connection between duplex stability and unwinding rate constant has been shown explicitly only for few DEAD-box helicases (38). We therefore set out to directly examine the link between duplex length, stability and maximal unwinding rate constant for DDX3X. We measured maximal unwinding rate constants for additional 3′-tailed substrates with different sequences in the duplex region. Substrates were designed with differing duplex stabilities for identical length. We observed clear scaling of the maximal unwinding rate constants with duplex stability (Fig.4c). Duplex length per se did not markedly impact the maximal unwinding rate constant (Fig.4c). The data support the notion that the decrease of the maximal unwinding rate constant with duplex length is due to increased stability of longer duplexes.
A logarithmic plot of maximal unwinding rate constants vs. duplex stability revealed similar slopes for 5′- and 3′-tailed substrates, although the maximal rate constants for 5′-tailed substrates were substantially lower than for 3′-tailed duplexes (Fig.4d). This observation suggests that duplex stability impacts 3-′ and 5′-tailed substrates similarly, and raises the possibility that the different maximal rate constants for 3-′ and 5′-tailed substrates are due to differences in the positioning of the unwinding protomer on 5′- and 3′-tailed substrates.
We noted that Ded1p unwound the 19 bp substrate with a lower maximal rate constant (kunwmax) than DDX3X (Fig.4e, compare to Fig4.a), even though Ded1p unwound the 16 bp substrate with a higher maximal rate constant than DDX3X (Fig.3a,b). These results indicate that the maximal unwinding rate constants scale differently with duplex stability for Ded1p and DDX3X. Since the scaling of the maximal unwinding rate constant with duplex stability correlates with the dwell time of the locally opened duplex and the number of basepairs that are actively separated by the helicase (32), these observations suggest that DDX3X actively opens more basepairs than Ded1p, keeps opened basepairs separated longer, or both.
Unpaired substrate regions impact the functional affinity of DDX3X
We next examined how the length of the single stranded tail affected the unwinding activity of DDX3X. To this end, we measured unwinding rate constants for a series of substrates with sequentially extended unpaired 3′ tails (5 – 35 nt, 16 bp duplex) (Fig.5). Substrates with 25 and 35 nt tails showed similar dependencies on the DDX3X concentration (Fig.5). The substrate with the 15 nt tail was unwound with a maximal unwinding rate constant similar to the substrates with 25 and 35 nt tails, but with a lower functional affinity than for the substrates with the longer tails (Fig.5).The substrate with the 5 nt tail could not be saturated with DDX3X at experimentally accessible concentrations (Fig.5). The observations reveal decreasing functional affinity for tails with less than 25 nt. The maximal unwinding rate constant remains largely unaffected by the tail length for the substrates that could be saturated with DDX3X. These characteristics mirror previous observations for Ded1p (34). The data support the notion that the unpaired tail facilitates assembly of the unwinding oligomer for both, Ded1p and DDX3X (32).
Figure 5. Impact of tail-length on unwinding by DDX3X.
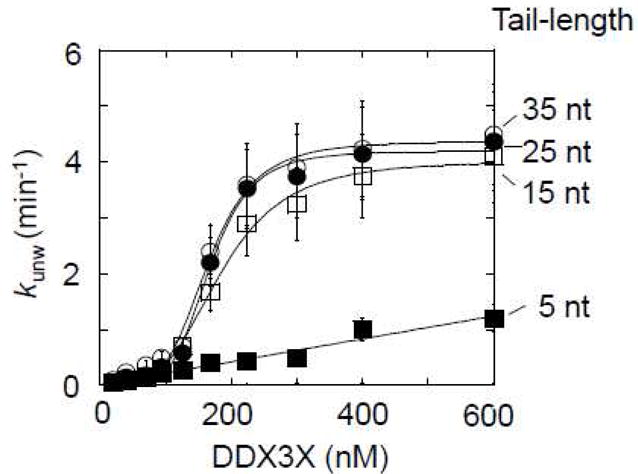
Functional binding isotherms for DDX3X for substrates with 16 bp and 3′ tails of increasing length, as indicated. Data points represent averages of at least three independently measured unwinding rate constants, error bars mark one standard deviation. Lines show best fits to the Hill equation for the substrates with 35 -15 nt tails [35 nt tail (open circles): kunwmax = 4.2 ± 0.1 min−1, H = 3.2 ± 0.2, K1/2 = 126 ± 4 nM; 25 nt tail (filled circles): kunwmax = 4.2 ± 0.1 min−1, H = 3.2 ± 0.4, K1/2 = 118 ± 5 nM; 15 nt tail (open squares): kunwmax = 3.8 ± 0.1 min−1. H = 3.2 ± 0.7, K1/2 = 298 ± 4 nM]. For the substrate with the 5 nt tail, the line marks a trend.
DDX3X interacts with DNA
Having detected marked similarities in the basic unwinding features of Ded1p and DDX3X, we tested how DDX3X interacted with DNA. A previous report suggested that DDX3X unwinds DNA-DNA duplexes and that unpaired DNA stimulates the ATPase activity of DDX3X to a level seen with RNA (30). These characteristics would be unusual for DEAD-box helicases and would differ markedly from Ded1p, which does not unwind DNA-DNA duplexes (39), and whose ATPase activity is not equally stimulated by DNA and RNA (40).
We first measured unwinding rate constants for tailed DNA-DNA and RNA-DNA hybrid substrates with identical sequences (Fig.6). DDX3X readily unwound the RNA-DNA substrates, including one with an unpaired DNA tail. However, DDX3X did not separate the tested DNA-DNA complexes at an appreciable rate (Fig.6). Identical unwinding patterns had been seen for Ded1p (31, 34). Our findings thus contrast with the prior report regarding DNA helicase activity of DDX3X (30). Nevertheless, the efficient unwinding of the substrate with the DNA tail suggests that DDX3X, like Ded1p, can bind unpaired DNA, and that DNA facilitates assembly of an unwinding-competent DDX3X oligomer.
Figure 6. Unwinding of DNA-containing substrates by DDX3X.
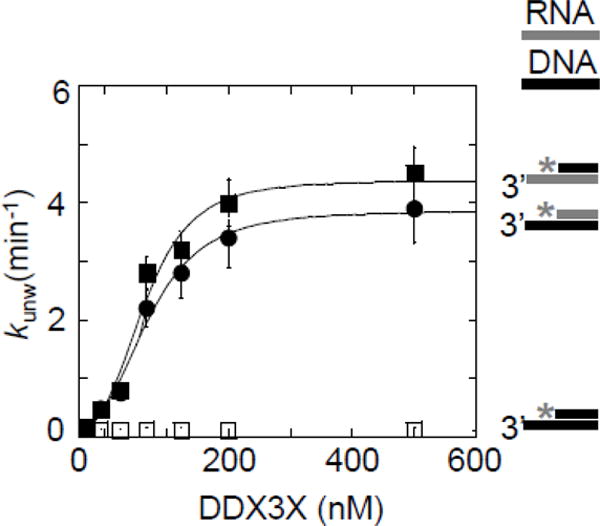
Functional binding isotherms for DDX3X for DNA/RNA and DNA/DNA substrates with 16 bp and 25 nt 3′ tails. Data points represent averages of at least three independently measured unwinding rate constants, error bars mark one standard deviation. Lines show best fits to the Hill equation. (Filled squares: DNA top, RNA bottom strand: kunwmax = 4.3 ± 0.1 min−1, K1/2 = 129 ± 5 nM, H = 3.0 ± 0.7, filled circles: RNA top, DNA bottom strand: kunwmax = 3.9 ± 0.1 min−1, K1/2 = 134 ± 4 nM, H = 3.5 ± 0.3. Open squares: DNA top and bottom strand: no significant strand separation.
To confirm and extend this observation, we examined DDX3X binding to single stranded DNA. We performed a competition assay, measuring unwinding of an RNA substrate in the presence of increasing concentrations of an unpaired DNA oligonucleotide (Fig.7a). We observed inhibition of the apparent unwinding rate constant that scaled with the DNA concentration, consistent with binding of the DNA to DDX3X. However, an RNA oligonucleotide with the same sequence bound with higher apparent affinity compared to the DNA (Fig.7b). Under identical conditions, Ded1p also bound the DNA. The equivalent RNA was bound with higher apparent affinity than the DNA (Fig.7c). The data show that both orthologs bind single stranded DNA, but bind equivalent RNA significantly tighter.
Figure 7. Functional binding of unpaired DNA and RNA to DDX3X and Ded1p.
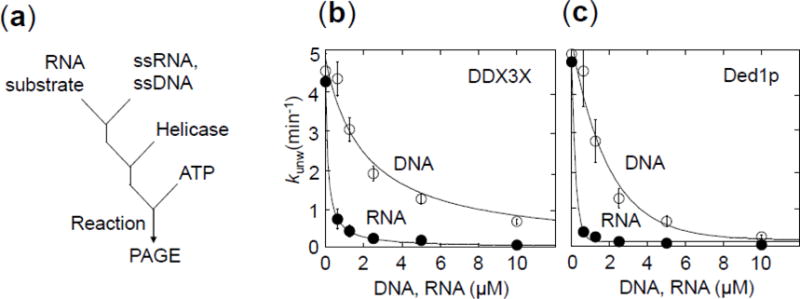
(a) Reaction scheme of the competition reaction to measure functional binding of unpaired DNA and RNA to DDX3X and Ded1p.
(b) Unwinding reactions with DDX3X (150 nM, ATP: 2 mM) RNA substrate: 16 bp duplex RNA substrate with 25 nt 3′ tail, 0.5 nM) with increasing concentrations of a 41 nt DNA (open circles) and a 41 nt RNA (closed circles). Data points represent averages of at least three independent measurements, error bars mark one standard deviation. Functional affinities Ki for unpaired DNA and RNA were calculated as in panel (b) (KiDNA = 3,560 ± 100 nM, KiRNA = 71 ± 5 nM).
(c) Unwinding reactions with Ded1p (150 nM, ATP: 2 mM). RNA substrate: 16 bp duplex RNA substrate with 25 nt 3′ tail, 0.5 nM) with increasing concentrations of a 41 nt DNA (open circles) and a 41 nt RNA (closed circles). Data points represent averages of at least three independent measurements, error bars mark one standard deviation. The lines mark a trend. Functional affinities could not be calculated because the unwinding rate constant at enzyme saturation without inhibitor could not be determined (Fig.3b).
Unpaired DNA stimulates ATP hydrolysis by DDX3X markedly less than unpaired RNA
Having shown that DDX3X and Ded1p bind DNA, but do not unwind DNA duplexes tested, we measured to which extent DNA stimulates the ATPase activity of DDX3X (Fig.8). For both DDX3X and Ded1p, no significant ATPase activity was seen in the absence of nucleic acid (Vmax < 10−6 mM∙min−1, Fig.8a). This observation is consistent with previous data for Ded1p (32), but contrasts with reports of marked ATPase activity of DDX3X in the absence of nucleic acid (29). A tailed duplex DNA substrate stimulated the ATPase activity of DDX3X markedly less than an equivalent RNA at identical concentration (Fig.8a).
Figure 8. ATP hydrolysis by DDX3X with RNA and DNA.
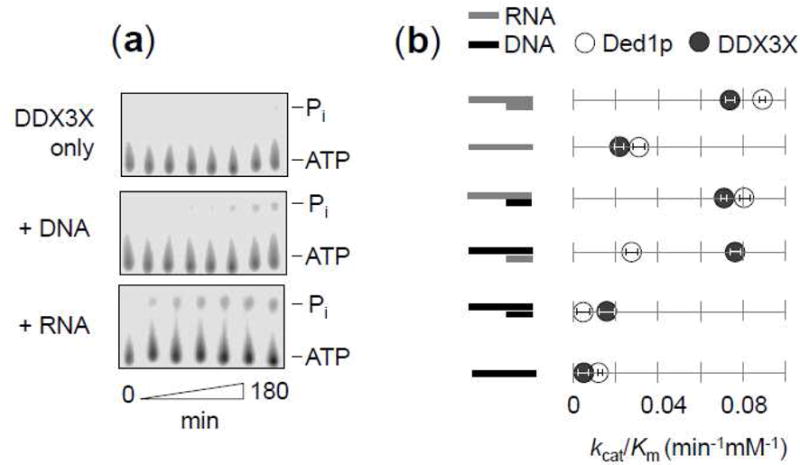
(a) Representative ATPase reactions of DDX3X (450nM, ATP 2mM) without nucleic acid (upper panel), DNA (middle panel, 16 bp substrate with 25 nt 3′ tailed substrate, 2.5 μM), or RNA (lower panel, 16 bp substrate with 25 nt 3′ tailed substrate, 2.5 μM).
(b) ATPase activity of DDX3X (450 nM, filled circles) and Ded1p (450 nM, open circles) for RNA/RNA, DNA/RNA, and DNA/DNA duplex substrates (16 bp, 25 nt 3′ tail). Data points represent independently measured kcat and Km values (Suppl. Table 1). Error bars mark one standard deviation of the kcat/Km value obtained from fits of ATP titrations to the Michaelis-Menten equation (Suppl. Table 1).
We next measured steady state Michaelis-Menten parameters for ATP hydrolysis by DDX3X and Ded1p for unpaired DNA and RNA using a series of tailed DNA-DNA, RNA-RNA and DNA-RNA duplexes (Fig.8b, Suppl. Table 1). Unpaired RNA and tailed RNA duplexes stimulated the ATPase activity for both DDX3X and Ded1p, with Ded1p showing slightly higher activity than DDX3X (Fig.8b). A DNA-RNA hybrid substrate with unpaired RNA tail stimulated the ATPase for both proteins to a similar degree as the complete RNA duplex (Fig.8b). These observations for DDX3X mirror previous data for Ded1p (32).
A notable difference between DDX3X and Ded1p was seen for the DNA-RNA hybrid substrate with unpaired DNA tail (Fig.8b). This substrate stimulated the ATPase activity of DDX3X almost to the level seen with the complete RNA substrate, whereas the stimulation for Ded1p was markedly lower (Fig.8b). For Ded1p, the data indicate that ATP hydrolysis mostly occurs at the unpaired tail, as previously noted (32). For DDX3X, this observation suggests that RNA in the duplex segment either (i) stimulates the ATPase activity of DDX3X more than for Ded1p, (ii) induces ATP hydrolysis of the DDX3X protomers bound to the DNA tail, or both. Despite the different levels of ATPase stimulation seen for Ded1p and DDX3X with this substrate, this DNA-tailed DNA-RNA hybrid duplex is unwound by both, Ded1p (32), and DDX3X (Fig.6).
A complete DNA-DNA substrate and a single stranded DNA stimulated the ATPase activity of both Ded1p and DDX3X to a markedly lower level than substrates containing RNA (Fig.8b). The level of stimulation was more than an order of magnitude lower, compared to substrates containing RNA, but ATPase activity was detectable. Since no appreciable unwinding of the DNA substrate was seen (Fig.6), the data indicate that nucleic-acid-stimulated ATPase activity does not necessarily lead to duplex unwinding. For DDX3X, the markedly lower ATPase stimulation by unpaired DNA, compared to RNA, contrasts with a previous report of similar levels of ATPase stimulation by DNA and RNA (29).
Unwinding activity of DDX3X depends on adenosine nucleoside triphosphates
DDX3X has been reported to display relaxed nucleoside specificity (29). We therefore examined the selectivity of DDX3X for adenosine nucleoside triphosphates for its unwinding activity. We first tested which of the four basic NTPs promoted unwinding (Fig.9a). Appreciable unwinding was seen only with ATP, but not with GTP, CTP and UTP (Fig.9a). Specificity for adenosine nucleoside triphosphates in unwinding reactions is also observed with Ded1p and other DEAD-box helicases (1).
Figure 9. Unwinding with different nucleoside triphosphates.
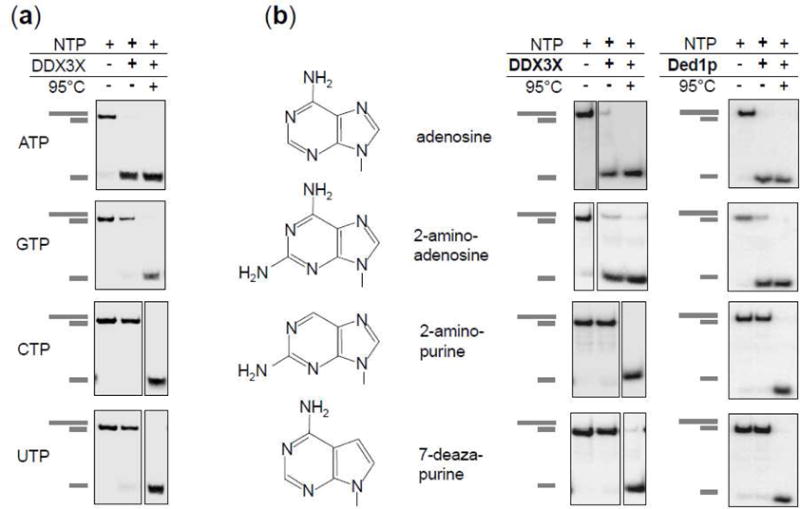
(a) Representative unwinding reactions by DDX3X (150nM) with ATP, GTP, CTP and UTP (2 mM each). (RNA: 16 bp, 25 nt 3′ tail, 0.5 nM reaction time: 15 min). Cartoons on the left mark duplex RNA substrate and unwound product.
(b) Unwinding with ATP, 2-amino adenosine triphosphate, 2-amino purine triphosphate, and 7-deaza purine triphosphate. Chemical structures of the nucleobases are depicted. Gel panels show representative unwinding reactions by DDX3X (left column) (150 nM) and Ded1p (right column, 150nM). (RNA: 16 bp, 25 nt 3′ tail, 0.5 nM, reaction time: 15 min). Cartoons on the left mark duplex RNA substrate and unwound product.
We then probed the role of the N7 and the N6 amino group of the adenine base (Fig.9b). These two positions have been implied in specific contacts between DEAD-box helicases and the adenosine base (41). Substitution of the N7 nitrogen with carbon abrogated unwinding activity in both, DDX3X and Ded1p (Fig.9b). Removal of the N6 amino group also eliminated unwinding activity in DDX3X and Ded1p (Fig.9b). These observations indicate that the nucleobase selectivity of DDX3X in the unwinding reaction does not markedly differ from Ded1p and other DEAD-box helicases, consistent with crystal structures of numerous helicase cores, including that of DDX3X (1, 4).
DISCUSSION
We have characterized biochemical features of human DDX3X focusing on its unwinding activity. Our data provide a basic, systematic definition of the biochemical properties of full length human DDX3X, an RNA helicase with critical roles in RNA metabolism that has also been implicated in numerous diseases (2). The findings designate DDX3X as bona-fide DEAD-box helicase, but also reveal biochemical differences between the similar orthologs DDX3X and Ded1p.
DDX3X behaves like a typical DEAD-box helicase with respect to basic unwinding mechanism and nucleotide preference (42–46). Unwinding rate constants decrease with increasing duplex stability. RNA substrate affinities increase with the length of the unpaired tail. Marked unwinding activity is seen only with adenosine triphosphates. The nitrogen at position 7 and the amino group at position 6 of the adenine base are critical for unwinding, consistent with numerous crystal structures of DEAD-box helicases (1). Like other DEAD-box helicases, DDX3X does not unwind DNA duplexes. Yet, we identify biochemical features of DDX3X that are not widely shared among DEAD-box helicases, including a pronounced preference for unwinding 3′-tailed duplex substrates, and ATPase activity stimulated by an DNA tailed RNA-DNA hybrid substrate. A further notable difference between DDX3X and Ded1p is the absence of pronounced strand annealing activity by DDX3X. This observation parallels a recent report for the DEAD-box helicase orthologs Dbp2p from S.cerevisae and DDX5X from human, where the yeast ortholog also shows stronger strand annealing activity than the human protein (47).
Both, Ded1p and DDX3X unwind RNA duplexes most efficiently as oligomers. A Hill coefficent of roughly H ≈ 3 suggests that the DDX3X oligomer, like Ded1p, contains at least three protomers. For both enzymes, functional affinity depends in a similar manner on the length of the unpaired RNA tail. Unpaired DNA also promotes oligomerization. These similarities suggest an evolutionary conservation of oligomerization in the DDX3/Ded1p subfamily and thus a biologically conserved function for the helicase oligomers.
However, Ded1p and DDX3X oligomers interact with 5′-tailed substrates differently. Although a 5′ tail facilitates oligomerization for both proteins, the unwinding protomers of Ded1p and DDX3X appear to interact with the duplex differently. As a result, rate constants for strand separation for DDX3X are markedly lower for 5′-tailed substrates than for those with a 3′ tail. For Ded1p, rate constants for strand separation are only slightly higher for 3′-tailed substrates, compared to those with a 5′ tail (34). Since functional affinities of DDX3X for 3′ and 5′-tailed substrates are similar, we speculate that unwinding of 5′-tailed substrates by DDX3X is associated with an energy penalty at the strand separation step, perhaps caused by an unfavorable orientation of the unwinding protomer. The biological relevance for DDX3X’s preference for 3′-tailed duplexes is not clear.
Despite different discrimination against 5′-tailed substrates, the actual process of strand separation appears to be identical for Ded1p and DDX3X.The rate constant for strand separation decreases for both proteins with duplex stability. However, the extent to which strand separation rate constants scale with duplex stability differs between Ded1p and DDX3X (Fig.4). The data are consistent with a scenario where DDX3X either opens more basepairs than Ded1p, keeps these basepairs separated longer, or both. Opening of different numbers of basepairs could be caused by slight differences in the RNA binding sites, by differences in the interaction with RNA by the less conserved C and N-termini, or by a combination of these factors. Further examination of links between slight structural variations in the RNA helicase core and functional differences might illuminate principles according to which structural elements dictate biochemical functions of DEAD-box helicases.
In contrast to potential differences in RNA binding between Ded1p and DDX3X, both proteins appear to interact with ATP similarly. Both proteins unwind RNA only with adenosine nucleoside triphosphates. The N6 amino group and the N7 position are critical for unwinding (Fig.9b), as expected from structural data for many DEAD-box helicases (1). Our data do not rule out the ability of either or both helicases to interact and perhaps even hydrolyze non-adenosine nucleotides, as has been shown for Mss116p (48). However, a previously reported relaxed nucleoside specificity of DDX3X (29) does not impact its unwinding activity. Strand separation firmly requires adenosine nucleoside triphosphate.
While our ATPase data show similarity between Ded1p and DDX3X, some of our results differ from two previously published observations. First, in contrast to a prior report (29), we did not observe pronounced ATPase activity for DDX3X without nucleic acid. In this respect, DDX3X behaves in our hands like Ded1p and other DEAD-box helicases (2, 35). Second, we observed clear stimulation of ATPase activity by unpaired RNA (Fig.8b). This result contrasts with another recent report, where, however, a truncated DDX3X variant was used and tested RNAs contained only 10 nt (3). We speculate that DDX3X, like Ded1p, requires a minimal RNA length to interact with the nucleic acid in a manner that promotes ATP hydrolysis, possibly requiring oligomerization on the RNA (32).
A final, notable difference to Ded1p is the ability of DDX3X to hydrolyze ATP in the presence of a substrate with an unpaired DNA tail connected to an RNA-DNA hybrid duplex (Fig.8b).These data support the notion that both orthologs vary slightly in their nucleic acid binding characteristics. Both, DDX3X and Ded1p hydrolyze ATP in the presence of DNA, although this activity is markedly lower than with RNA. Stimulation of ATPase activity by DNA has not been widely reported for DEAD-box helicases, and it is thus possible that a link between DNA and ATP hydrolysis by DDX3X and Ded1p serves a specific, yet unknown biological purpose.
In sum, the reported data provide a basic characterization of the biochemical features of full-length, human DDX3X. Despite biochemical differences between DDX3X and Ded1p, DDX3X can at least partially complement a Ded1p defect in yeast (49). This observation suggests that the biological function of the helicase in yeast tolerates variation in biochemical parameters under the conditions examined in the complementation experiments. Our results suggest that differences between DDX3X and Ded1p likely arise from perhaps only slight variations in the way both helicases interact with nucleic acids. In contrast, the interaction with ATP appears largely identical between the orthologs. These insights have potential implications for the development of specific inhibitors for DDX3X and perhaps other DEAD-box helicases, suggesting that the ATP binding site may be more conserved between DEAD-box helicases than the RNA binding sites. Instead of targeting the highly conserved ATP binding site with inhibitors, it might be more promising to target nucleic acid binding sites, or regions that couple ATP and RNA binding.
MATERIALS AND METHODS
Protein Expression and Purification
Ded1p was expressed, purified and characterized as previously described (31). Full-length DDX3X with an N-terminal TEV-cleavable His6-tag was cloned into a pET-22b vector and expressed in E.coli BL21 (37°C). Cells were processed as previously described for Ded1p (31). Lysates were passed through pre-equilibrated nickel-agarose beads and washed with solutions of increasing imidazole concentrations (5 – 60 mM). DDX3X was eluted with 250 mM imidazole. The His6-tag was cleaved using TEV protease in 50 mM Tris-Cl (pH 8.0), 0.5 mM EDTA, 1 mM DTT. DDX3X was further purified by adsorption to phosphocellulose resin (P11, Whatman) and elution with NaCl, as described for Ded1p (31). Eluted fractions were analyzed by SDS-PAGE and Western blotting with the anti-His antibody to confirm removal of the His6-tag. DDX3X concentration was determined using the Bradford assay. DDX3X fractions were supplemented to 40% (v/v) glycerol, flash-frozen in liquid nitrogen, and stored at −80 °C.
RNA Substrate Preparation
RNA oligonucleotides were purchased from DHARMACON, and duplex substrates were prepared as described (31). Top strands of duplexes were radiolabeled with γ32PATP, and RNA was quantified by scintillation counting. Substrate sequences are listed in Table 1.
Table 1.
Sequences of oligonucleotides used in this study. R: RNA, D: DNA.
| Oligonucleotide | Sequence (5′-3′) | |
|---|---|---|
| 1 | R13-3′a | AGC ACC GUA AAG C |
| 2 | R13-5′a | CGA AAU GCC ACG A |
| 3 | R13-3′b | U GCA UGA UUG UCG |
| 4 | R13-3′c | U CGU GGC AUU UCU |
| 5 | R16-3′a | G GCA GAA AUG CCA CGA |
| 6 | R16-3′b | U CGU GGC AUU UCU GCG |
| 7 | R16-3′c | C CGU CUU UAC GGU GCU |
| 8 | R16-5′a | AGC ACC GUA AAG ACG C |
| 9 | R19-3′a | C GAC GCA GAA AUG CCA CGA |
| 10 | R19-3′b | G CAG UCA CCU GAC AUA CGC |
| 11 | R19-3′c | A CGA GGG AGA CGA GGA GAC |
| 12 | R19-5′c | CAG AGG AGC AGA GGG AGC A |
| 13 | R38-3′a | GCU UUA CGG UGC UUA AAA CAA AAC AAA ACA AAA CAA AA |
| 14 | R38-5′a | AAA ACA AAA CAA AAC AAA ACA AAA UUC GUG GCA UUU CG |
| 15 | R38-3′c | AGA AAU GCC ACG AU GCG CCG GGC CGG GCC ACA ACC AAA |
| 16 | R38-3′b | CGA CAA UCA UGC AAU UUU GUU UUG UUU UGU UUU GUU UU |
| 17 | R41-3′b | CGC AGA AAU GCC ACG AAU UUU GUU UUG UUU UGU UUU GUU UU |
| 18 | R41-3′c | AGC ACC GUA AAG ACG GUA AAA CAA AAC AAA ACA AAA CAA AA |
| 19 | R41-3′a | UCG UGG CAU UUC UGC C AU CCC GUU UGG UUU UGU UUU GUU UU |
| 20 | R44-3′b | GCG UAU GUC AGG UGA CUG CUA AAA CAA AAC AAA ACA AAA CAA AA |
| 21 | R44-3′c | GUC UCC UCG UCU CCC UCG UCA GCA UCA AUG ACA UCA GCA UCA AA |
| 22 | R44-5′c | AA ACU ACG ACU ACA GUA ACU ACG ACU GCU CCC UCU GCU CCU CUG |
| 23 | R44-3′a | UCG UGG CAU UUC UGC GUC GUU CUU UUC UUU UCU UUU CUU UUA CG |
| 24 | R41-5′a | AAA ACA AAA CAA AAC AAA ACA AAA UGC GUC UUU ACG GUG CU |
| 25 | D41-3′a | CG CAG AAA TGC CAC GAT AAA ACA AAA CAA AAC AAA ACA AAA |
| 26 | D16-3′b | AGC ACC GTA AAG ACG C |
| 27 | D16-3′a | T CGT GGC ATT TCT GCG |
| 28 | D41-3′b | GCG TCT TTA CGG TGC TTA AAA CAA AAC AAA ACA AAA CAA AA |
Sequences are listed 5′ to 3′. The first number in the oligonucleotide name indicates length (nt), the number after the dash indicates the tail orientation in the duplex. The letter marks the duplex sequence. For example, R38-3′a is the 38 nt bottom strand that base pairs with R13a to form a 13 bp duplex with a 25 nt 3′ tail. Duplex regions in the bottom strands are underlined. Substrates with different length overhangs used in Fig.5 were generated by appending or deleting (A4C) repeats to R41-3′c.
Unwinding Reactions
Reaction mixtures (30 μL), containing 40 mM Tris-HCl (pH 8.0), 50 mM NaCl, 0.5 mM MgCl2, 2 mM DTT, 1 unit/mL RNasin, 0.01% (v/v) IGEPAL, and 0.5 nM radiolabeled substrate were incubated for 5 min with the indicated concentrations of Ded1p or DDX3X. Unwinding reactions were initiated by adding an equimolar mixture of MgCl2 and ATP (2 mM final concentration, unless otherwise stated). Reactions for both, DDX3X and Ded1p were performed in a temperature-controlled aluminum block at 30°C. At the times indicated, aliquots were removed, and the reaction was stopped with a solution containing 0.1% (w/v) SDS, 50 mM EDTA, 0.01% (w/v) xylenecyanol, 0.01% (w/v) bromophenol blue, and 20% (v/v) glycerol. Aliquots were applied to non-denaturing PAGE. Duplex and single stranded RNA was separated at room temperature at 20 V/cm. Gels were dried, and the radiolabeled RNAs were visualized and quantified with a PhosphorImager by using ImageQuant 5.2 software (Molecular Dynamics).Data were fit using the Kaleidagraph software (Synergy). Unwinding reactions were tested for ATP-independent strand separation and for strand separation in the presence of the non hydrolyzable ATP analog AMPPNP and in the presence of ADP. No significant strand separation was observed under these conditions in any case. Unwinding rate constants were calculated from unwinding reaction timecourses as described (31). Functional binding parameters were calculated from functional binding isotherms using the Hill-equation as described (32).
Apparent inhibitor binding constants (Ki) for functional DNA binding reactions (Fig.7b) were calculated according to a competitive inhibition model:
kunw is the observed unwinding rate constant in the presence of increasing concentrations of unpaired DNA or RNA, kmax is the unwinding rate constant in the absence of unpaired DNA or RNA at enzyme saturation, K1/2 is the apparent affinity of Ded1p or DDX3X for the RNA substrate in the unwinding reaction, [S] is the concentration of DDX3X or Ded1p, [I] is the concentration of unpaired DNA or RNA, and Ki is the apparent inhibition constant, which corresponds to the functional affinity of the unpaired RNA or DNA to Ded1p or DDX3X in the presence of ATP.
Strand Annealing Reactions
Duplex RNA substrates were denatured at 95 °C for 2 min to generate single-stranded RNAs. Reaction mixtures (30 μL) containing buffer identical to that used in unwinding reactions were incubated with 600 nM Ded1p or DDX3X and, where indicated, with equimolar Mg-ATP concentrations for 5 min. Reactions were performed in a temperature-controlled aluminum block at 30°C. Annealing reactions were initiated by addition of 0.5 nM of the denatured substrate strands. Aliquots (3 μL) were removed at the times indicated, and reactions were stopped with the buffer used to quench unwinding reactions. Single-stranded and duplex RNAs were separated and quantified as described for the unwinding assays. Strand annealing rate constants and second order annealing rate constants were calculated from reaction timecourses as described (31).
ATPase Measurements
ATPase measurements were performed in buffer identical to that used in unwinding reactions at 30°C. Hydrolysis of γ32PATPwas monitored by TLC, as described (50, 51). Initial observed reaction rates were calculated from the linear part of the plot of product versus time, Michaelis-Menten parameters by plotting initial observed reaction rates at constant Ded1p or DDX3X over varying ATP concentration, as previously described (51).
Supplementary Material
Highlights.
Systematic characterization of helicase activity of full-length, human DDX3X.
DDX3X behaves like a typical DEAD-box helicase and does not unwind DNA duplexes
DDX3X, like its yeast ortholog Ded1p, unwinds RNA as oligomer
DDX3X preferentially unwinds 3′-tailed RNA substrates and does not promote strand annealing
Acknowledgments
We thank the members of our group for discussions, Akshay Tambe for initial experiments on expression and purification of full-length DDX3X, and Fei Liu for initial experiments testing RNA unwinding by Ded1p with ATP analogs. This work was supported by the NIH (GM118088 to E.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 2.Sharma D, Jankowsky E. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol. 2014;49:343–360. doi: 10.3109/10409238.2014.931339. [DOI] [PubMed] [Google Scholar]
- 3.Epling LB, Grace CR, Lowe BR, Partridge JF, Enemark EJ. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J Mol Biol. 2015;427:1779–1796. doi: 10.1016/j.jmb.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floor SN, Condon KJ, Sharma D, Jankowsky E, Doudna JA. Autoinhibitory Interdomain Interactions and Subfamily-specific Extensions Redefine the Catalytic Core of the Human DEAD-box Protein DDX3. J Biol Chem. 2016;291:2412–2421. doi: 10.1074/jbc.M115.700625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarn WY, Chang TH. The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol. 2009;6:17–20. doi: 10.4161/rna.6.1.7440. [DOI] [PubMed] [Google Scholar]
- 6.Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, Flores A, Karlsson Hedestam GB, Schiavone LH. Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol. 2007;372:150–159. doi: 10.1016/j.jmb.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 7.Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 8.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai MC, Lee YH, Tarn WY. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res. 2015;25:1196–1205. doi: 10.1101/gr.191601.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh S, Flynn RA, Floor SN, Purzner J, Martin L, Do BT, Schubert S, Vaka D, Morrissy S, Li Y, Kool M, Hovestadt V, Jones DT, Northcott PA, Risch T, Warnatz HJ, Yaspo ML, Adams CM, Leib RD, Breese M, Marra MA, Malkin D, Lichter P, Doudna JA, Pfister SM, Taylor MD, Chang HY, Cho YJ. Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget. 2016;7:28169–28182. doi: 10.18632/oncotarget.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruciat CM, Dolde C, de Groot RE, Ohkawara B, Reinhard C, Korswagen HC, Niehrs C. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-beta-catenin signaling. Science. 2013;339:1436–1441. doi: 10.1126/science.1231499. [DOI] [PubMed] [Google Scholar]
- 15.Gu L, Fullam A, Brennan R, Schroder M. Human DEAD box helicase 3 couples IkappaB kinase epsilon to interferon regulatory factor 3 activation. Mol Cell Biol. 2013;33:2004–2015. doi: 10.1128/MCB.01603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih JW, Wang WT, Tsai TY, Kuo CY, Li HK, Wu Lee YH. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J. 2012;441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 17.McRae JF, et al. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MR, Venselaar H, Helsmoortel C, Cho MT, Hoischen A, Vissers LE, Koemans TS, Wissink-Lindhout W, Eichler EE, Romano C, Van Esch H, Stumpel C, Vreeburg M, Smeets E, Oberndorff K, van Bon BW, Shaw M, Gecz J, Haan E, Bienek M, Jensen C, Loeys BL, Van Dijck A, Innes AM, Racher H, Vermeer S, Di Donato N, Rump A, Tatton-Brown K, Parker MJ, Henderson A, Lynch SA, Fryer A, Ross A, Vasudevan P, Kini U, Newbury-Ecob R, Chandler K, Male A, Dijkstra S, Schieving J, Giltay J, van Gassen KL, Schuurs-Hoeijmakers J, Tan PL, Pediaditakis I, Haas SA, Retterer K, Reed P, Monaghan KG, Haverfield E, Natowicz M, Myers A, Kruer MC, Stein Q, Strauss KA, Brigatti KW, Keating K, Burton BK, Kim KH, Charrow J, Norman J, Foster-Barber A, Kline AD, Kimball A, Zackai E, Harr M, Fox J, McLaughlin J, Lindstrom K, Haude KM, van Roozendaal K, Brunner H, Chung WK, Kooy RF, Pfundt R, Kalscheuer V, Mehta SG, Katsanis N, Kleefstra T. Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am J Hum Genet. 2015;97:343–352. doi: 10.1016/j.ajhg.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heerma van Voss MR, Schrijver WA, Ter Hoeve ND, Hoefnagel LD, Manson QF, van der Wall E, Raman V, van Diest PJ. The prognostic effect of DDX3 upregulation in distant breast cancer metastases. Clin Exp Metastasis. 2017;34:85–92. doi: 10.1007/s10585-016-9832-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG, Pan CM, Hu Y, Cai CP, Dong Y, Huang JY, Wang L, Shen Y, Meng G, Zhou JF, Hu JD, Wang JF, Liu YH, Yang LH, Zhang F, Wang JM, Wang Z, Peng ZG, Chen FY, Sun ZM, Ding H, Shi JM, Hou J, Yan JS, Shi JY, Xu L, Li Y, Lu J, Zheng Z, Xue W, Zhao WL, Chen Z, Chen SJ. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47:1061–1066. doi: 10.1038/ng.3358. [DOI] [PubMed] [Google Scholar]
- 21.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL, Greulich H, Lawrence MS, Lennon NJ, McKenna A, Meldrim J, Ramos AH, Ross MG, Russ C, Shefler E, Sivachenko A, Sogoloff B, Stojanov P, Tamayo P, Mesirov JP, Amani V, Teider N, Sengupta S, Francois JP, Northcott PA, Taylor MD, Yu F, Crabtree GR, Kautzman AG, Gabriel SB, Getz G, Jager N, Jones DT, Lichter P, Pfister SM, Roberts TM, Meyerson M, Pomeroy SL, Cho YJ. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su CY, Lin TC, Lin YF, Chen MH, Lee CH, Wang HY, Lee YC, Liu YP, Chen CL, Hsiao M. DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectal cancer. Oncotarget. 2015;6:18602–18612. doi: 10.18632/oncotarget.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee CH, Lin SH, Yang SF, Yang SM, Chen MK, Lee H, Ko JL, Chen CJ, Yeh KT. Low/negative expression of DDX3 might predict poor prognosis in non-smoker patients with oral cancer. Oral Dis. 2014;20:76–83. doi: 10.1111/odi.12076. [DOI] [PubMed] [Google Scholar]
- 24.Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC, Wu Lee YH. DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene. 2006;25:1991–2003. doi: 10.1038/sj.onc.1209239. [DOI] [PubMed] [Google Scholar]
- 25.Xie M, Vesuna F, Tantravedi S, Bol GM, Heerma van Voss MR, Nugent K, Malek R, Gabrielson K, van Diest PJ, Tran PT, Raman V. RK-33 Radiosensitizes Prostate Cancer Cells by Blocking the RNA Helicase DDX3. Cancer Res. 2016;76:6340–6350. doi: 10.1158/0008-5472.CAN-16-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bol GM, Xie M, Raman V. DDX3, a potential target for cancer treatment. Mol Cancer. 2015;14:188. doi: 10.1186/s12943-015-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valiente-Echeverria F, Hermoso MA, Soto-Rifo R. RNA helicase DDX3: at the crossroad of viral replication and antiviral immunity. Rev Med Virol. 2015;25:286–299. doi: 10.1002/rmv.1845. [DOI] [PubMed] [Google Scholar]
- 28.Brai A, Fazi R, Tintori C, Zamperini C, Bugli F, Sanguinetti M, Stigliano E, Este J, Badia R, Franco S, Martinez MA, Martinez JP, Meyerhans A, Saladini F, Zazzi M, Garbelli A, Maga G, Botta M. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc Natl Acad Sci U S A. 2016;113:5388–5393. doi: 10.1073/pnas.1522987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franca R, Belfiore A, Spadari S, Maga G. Human DEAD-box ATPase DDX3 shows a relaxed nucleoside substrate specificity. Proteins. 2007;67:1128–1137. doi: 10.1002/prot.21433. [DOI] [PubMed] [Google Scholar]
- 30.Garbelli A, Beermann S, Di Cicco G, Dietrich U, Maga G. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS One. 2011;6:e19810. doi: 10.1371/journal.pone.0019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 32.Putnam AA, Gao Z, Liu F, Jia H, Yang Q, Jankowsky E. Division of Labor in an Oligomer of the DEAD-Box RNA Helicase Ded1p. Mol Cell. 2015;59:541–552. doi: 10.1016/j.molcel.2015.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jankowsky E, Putnam A. Duplex unwinding with DEAD-box proteins. Methods Mol Biol. 2010;587:245–264. doi: 10.1007/978-1-60327-355-8_18. [DOI] [PubMed] [Google Scholar]
- 34.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Putnam AA, Jankowsky E. DEAD-box helicases as integrators of RNA, nucleotide and protein binding. Biochim Biophys Acta. 2013;1829:884–893. doi: 10.1016/j.bbagrm.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell R, Jarmoskaite I, Lambowitz AM. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol. 2013;10:44–55. doi: 10.4161/rna.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 39.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 40.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 41.Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreou AZ, Klostermeier D. The DEAD-box helicase eIF4A: paradigm or the odd one out? RNA Biol. 2013;10:19–32. doi: 10.4161/rna.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarmoskaite I, Russell R. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem. 2014;83:697–725. doi: 10.1146/annurev-biochem-060713-035546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma WK, Cloutier SC, Tran EJ. The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J Mol Biol. 2013;425:3824–3838. doi: 10.1016/j.jmb.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma WK, Paudel BP, Xing Z, Sabath IG, Rueda D, Tran EJ. Recruitment, Duplex Unwinding and Protein-Mediated Inhibition of the Dead-Box RNA Helicase Dbp2 at Actively Transcribed Chromatin. J Mol Biol. 2016;428:1091–1106. doi: 10.1016/j.jmb.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samatanga B, Klostermeier D. DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res. 2014;42:10644–10654. doi: 10.1093/nar/gku747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing Z, Wang S, Tran EJ. Characterization of the mammalian DEAD-box protein DDX5 reveals functional conservation with Scerevisiae ortholog Dbp2 in transcriptional control and glucose metabolism. RNA. 2017;23:1125–1138. doi: 10.1261/rna.060335.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mallam AL, Sidote DJ, Lambowitz AM. Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. Elife. 2014;3:e04630. doi: 10.7554/eLife.04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senissar M, Le Saux A, Belgareh-Touze N, Adam C, Banroques J, Tanner NK. The DEAD-box helicase Ded1 from yeast is an mRNP cap-associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res. 2014;42:10005–10022. doi: 10.1093/nar/gku584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Putnam AA, Jankowsky E. AMP sensing by DEAD-box RNA helicases. J Mol Biol. 2013;425:3839–3845. doi: 10.1016/j.jmb.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Z, Putnam AA, Bowers HA, Guenther UP, Ye X, Kindsfather A, Hilliker AK, Jankowsky E. Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. Elife. 2016;5 doi: 10.7554/eLife.16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded Sequence Dependence of Thermodynamic Parameters Improves Prediction of RNA Secondary Structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


